The histone binding WD40-repeat protein MSI1 forms a complex with a histone deacetylase that represses abscisic acid receptor genes, thus affecting sensitivity to this phytohormone in Arabidopsis.
Abstract
MSI1 belongs to a family of histone binding WD40-repeat proteins. Arabidopsis thaliana contains five genes encoding MSI1-like proteins, but their functions in diverse chromatin-associated complexes are poorly understood. Here, we show that MSI1 is part of a histone deacetylase complex. We copurified HISTONE DEACETYLASE19 (HDA19) with MSI1 and transcriptional regulatory SIN3-like proteins and provide evidence that MSI1 and HDA19 associate into the same complex in vivo. These data suggest that MSI1, HDA19, and HISTONE DEACETYLATION COMPLEX1 protein form a core complex that can integrate various SIN3-like proteins. We found that reduction of MSI1 or HDA19 causes upregulation of abscisic acid (ABA) receptor genes and hypersensitivity of ABA-responsive genes. The MSI1-HDA19 complex fine-tunes ABA signaling by binding to the chromatin of ABA receptor genes and by maintaining low levels of acetylation of histone H3 at lysine 9, thereby affecting the expression levels of ABA receptor genes. Reduced MSI1 or HDA19 levels led to increased tolerance to salt stress corresponding to the increased ABA sensitivity of gene expression. Together, our results reveal the presence of an MSI1-HDA19 complex that fine-tunes ABA signaling in Arabidopsis.
INTRODUCTION
Proteins related to budding yeast (Saccharomyces cerevisiae) MSI1 form a family of WD40 domain proteins found in all eukaryotes (Ruggieri et al., 1989; Hennig et al., 2005). MSI1-like proteins bind histones and can tether various protein complexes that act on histones or chromatin to their substrates (reviewed in Hennig et al., 2005). In Arabidopsis thaliana, this protein family contains the five members MSI1 to MSI5 (Ach et al., 1997; Kenzior and Folk, 1998; Hennig et al., 2003). Arabidopsis MSI4 and MSI5 associate with the histone deacetylases HDA5 and HDA6 and are involved in deacetylation and silencing of FLOWERING LOCUS C and endogenous small interfering RNA-directed DNA methylation target loci (Ausín et al., 2004; Kim et al., 2004; Gu et al., 2011; Luo et al., 2015). Arabidopsis MSI1 is a subunit of chromatin assembly factor CAF-1 and of various Polycomb-repressive complexes (Kaya et al., 2001; Köhler et al., 2003; De Lucia et al., 2008; Derkacheva et al., 2013). Recently, MSI1 was shown to also copurify with HISTONE DEACETYLASE19 (HDA19) (Derkacheva et al., 2013), but it is not known whether MSI1 functions in histone deacetylation.
HDA19 and HDA6 belong to the RPD3/HDA1 superfamily containing homologs of REDUCED POTASSIUM DEPENDENCY3 (RPD3) from budding yeast (reviewed in Hollender and Liu, 2008). RPD3 is a core subunit of the SWI-INDEPENDENT3 (SIN3) complex that is conserved in yeast and animals (Carrozza et al., 2005; Keogh et al., 2005; Grzenda et al., 2009). In animals, the core SIN3 complex includes SIN3, an RPD3 homolog, the SIN3-associated proteins SAP18 and SAP30, and an MSI-like protein (Hassig et al., 1997; Zhang et al., 1997). Arabidopsis contains six homologs of SIN3: the SIN3-like proteins SNL1 to SNL6 (Bowen et al., 2010). HDA19 was shown to interact with SNL3 (Song et al., 2005) in yeast two-hybrid assays and with SNL1 in bimolecular fluorescence complementation assays (Wang et al., 2013). In yeast two-hybrid assays, Arabidopsis HDA6 and HDA19 both also interacted with a homolog of the animal SIN3 complex subunit SAP18 (Song and Galbraith, 2006; Hill et al., 2008). The yeast SIN3 complex also contains the subunit RXT3 (Carrozza et al., 2005), and the Arabidopsis RXT3-domain protein HISTONE DEACETYLATION COMPLEX1 (HDC1) was shown to interact with HDA19 and HDA6 in bimolecular fluorescence complementation assays (Perrella et al., 2013). These findings suggest that SIN3-related complexes are present in plants, but additional biochemical and molecular evidence is needed.
Histone deacetylases can be recruited to target genes by DNA binding transcriptional regulators and are often involved in a wide range of processes. However, the six SIN3-like, five MSI1-like, and 12 RPD3-like proteins in Arabidopsis may form a large number of SIN3-like complexes with potentially specialized functions, and more work is needed to assign physiological functions to distinct histone deacetylase complexes. Both HDA6 and HDA19 have been implicated in abscisic acid (ABA)-mediated responses to drought and high salinity (Chen et al., 2010; Chen and Wu, 2010; Luo et al., 2012b). Similarly, HDC1 was shown to affect responses to abiotic stress and ABA (Perrella et al., 2013). Part of HDA19’s function is mediated by its association with ERF transcription repressors to regulate gene expression in response to abiotic stresses (reviewed in Luo et al., 2012a). However, it is likely that HDA19 complexes can also be recruited by other DNA binding proteins. Interestingly, a role in abiotic stress responses has also been reported for MSI1, which appears to be independent of the well studied MSI1-containing CAF-1 and Polycomb-repressive complexes (Alexandre et al., 2009). In particular, reduction in MSI1 levels led to increased expression of many genes with ABA-responsive elements (ABREs) in their promoters (Alexandre et al., 2009). Because MSI1 copurified with HDA19 and SIN3-like proteins (Derkacheva et al., 2013) and both MSI1 and HDA19 have been implicated in abiotic stress responses, we hypothesized that MSI1 affects ABA responsive genes via functioning in a SIN3 histone deacetylase complex.
Here, we tested this hypothesis and provide several lines of experimental evidence in its support. First, biochemical evidence for a MSI1-HDA19 SIN3-like complex is shown: MSI1 and HDA19 can interact in yeast and exist in complex(es) of ∼600 kD in plants. HDA19 interacting partners were purified in vivo, and MSI1 as well as a common set of proteins that interact with both MSI1 and HDA19 were identified. Reduction of MSI1 or HDA19 levels caused hypersensitivity of ABA-regulated genes. MSI1 and HDA19 bind to the chromatin of ABA receptor genes PYL4, PYL5, and PYL6 where they reduce histone acetylation and transcription, suggesting a model in which the effects of MSI1 and HDA19 on ABA sensitivity of gene expression are caused by increased expression of ABA receptor genes. Finally, genetic double mutant analysis confirmed that MSI1 and HDA19 function together in ABA receptor gene expression and ABA sensitivity of gene expression.
RESULTS
MSI1 Copurifies with HDA19 and SNLs
HDA19 is a candidate interaction partner of MSI1 (Derkacheva et al., 2013). To characterize HDA19 complexes in vivo, we used an Arabidopsis line that expressed GFP-tagged HDA19 (Fong et al., 2006) and immunoaffinity purified an HDA19:GFP complex (see Methods for details). An Arabidopsis line expressing GFP (Derkacheva et al., 2013) was used as control. The purified fractions of four independent experiments were analyzed by high-resolution tandem mass spectrometry (MS/MS). Only proteins identified with at least two peptides in all four replicates but not in control samples were taken into account. HDA19 was found to copurify with all six SNL proteins (SNL1 to SNL6; Table 1), suggesting that HDA19 is a part of SIN3 complexes in plants. Biochemical identification of SNL1, SNL3, and HDC1 as interaction partners of HDA19 is in agreement with earlier reports based on yeast two-hybrid and bimolecular fluorescence complementation assays (Song et al., 2005; Perrella et al., 2013; Wang et al., 2013). In addition, the chaperonin ATTCP-1 (AT3G20050) was consistently found together with HDA19. This is consistent with a report that the mammalian TCP-1 Ring complex binds to the RPD3-like HDAC3 (Guenther et al., 2002). Finally, MSI1 but not MSI2-5 was found to copurify with HDA19 in vivo (Table 1). This finding, together with our previous results (Derkacheva et al., 2013), firmly establish that MSI1 acts together with HDA19 in Arabidopsis.
Table 1. HDA19 Copurifies with Homologs of Subunits of the SIN3 Complex.
| Gene ID | Name | No. of Unique Peptides IP1-IP2-IP3-IP4 | Sequence Coverage (%) IP1-IP2-IP3-IP4 | Sequest Score IP1-IP2-IP3-IP4 |
|---|---|---|---|---|
| AT4G38130 | HDA19 | 19-7-15-5 | 52-15.8-54.3-12.8 | 213-181-190-46 |
| AT5G58230 | MSI1 | 5-3-5-4 | 22.6-7.8-17.2-14.4 | 16.7-18.3-22.7-34 |
| AT3G01320 | SNL1 | 5-4-3-2 | 5.4-4.1-4.3-3 | 23.6-20.9-27.9-21 |
| AT5G15020 | SNL2 | 20-10-14-8 | 19.6-9.3-14-8.6 | 97.8-54.7-121-67 |
| AT1G24190 | SNL3 | 15-5-6-2 | 13.8-5.3-4.9-1.9 | 49-28.5-32.8-6.3 |
| AT1G70060 | SNL4 | 4-2-0-1 | 4.8-2.6-0-1 | 20-19.6-0-2.6 |
| AT1G59890 | SNL5 | 24-9-13-8 | 25.8-9.7-14.7-9.2 | 84.9-46.9-120-96 |
| AT1G10450 | SNL6 | 11-14-8-5 | 11.9-13-9.2-5.2 | 42.5-101-105-29.3 |
| AT5G08450 | HDC1 | 14-14-14-0 | 18.3-16.3-23.2-0 | 44.3-60-89-0 |
| AT3G20050 | ATTCP-1 | 10-5-3-4 | 22-9.5-5.7-7.5 | 30.7-19.7-10.3-12.8 |
HDA19 binding proteins were identified by immunoaffinity purification of HDA19:GFP from Pro35S:HDA19:GFP plants and mass spectrometry. The experiment was performed with four biological replicates. Shown are all proteins identified in the Pro35S:HDA19:GFP but not in a Pro35S:GFP line.
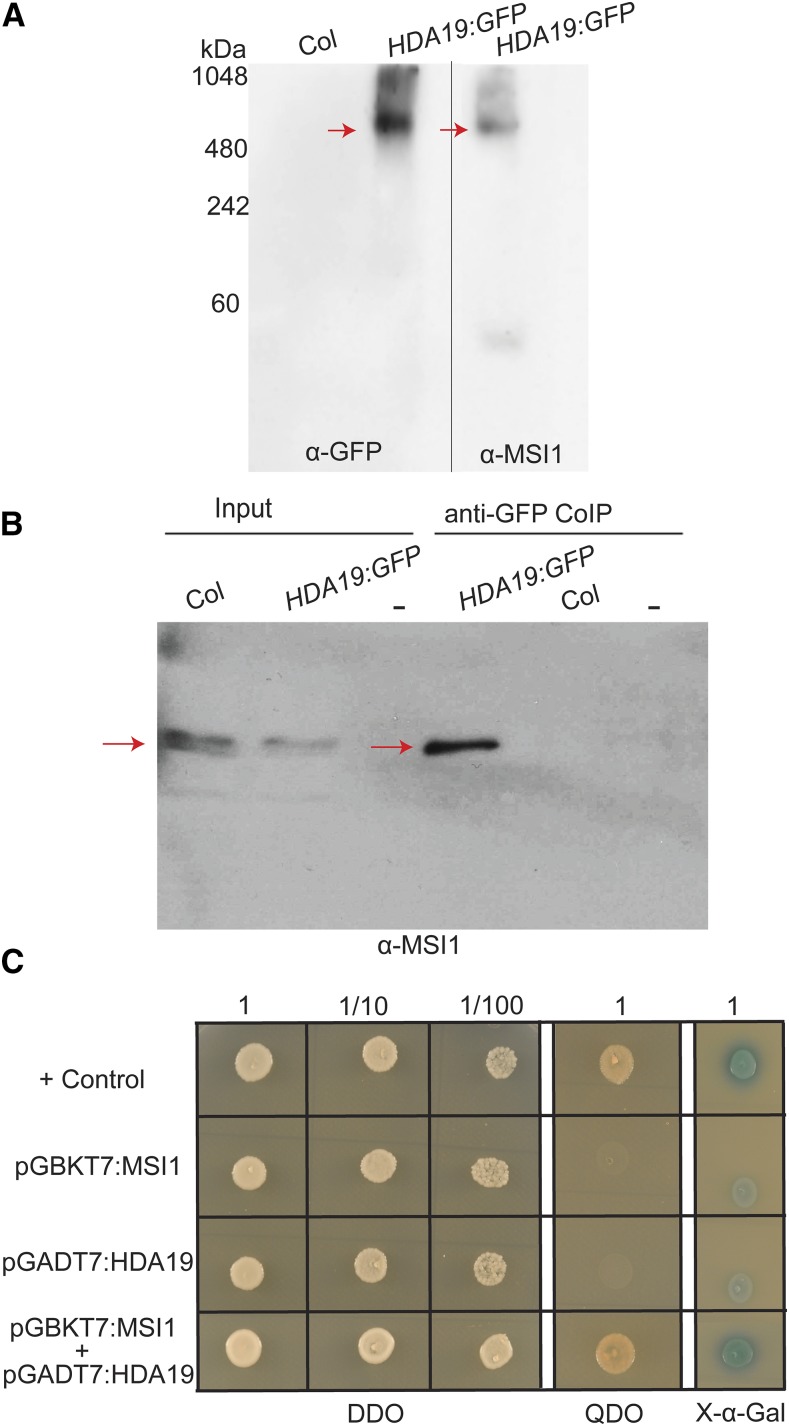
To further support that MSI1 and HDA19 are part of a common protein complex, we analyzed MSI1- and HDA19-containing complexes by native PAGE. Indeed, both HDA19 and MSI1 migrate as part of complexes of similar molecular weight (∼600 kD; Figure 1A). Whereas HDA19 was mostly present in the high molecular weight fraction, for MSI1, signals of monomers were clearly detected as well. We also performed a coimmunoprecipitation assay using the HDA19:GFP line. Immunoblot analysis showed that MSI1 coprecipitated with HDA19, demonstrating that these proteins associate into the same complexes in vivo (Figure 1B). In addition, MSI1 and HDA19 also interacted in yeast two-hybrid assays, suggesting that the interaction is direct (Figure 1C).
Figure 1.
HDA19 Associates into the Same Complex with MSI1 in Vivo.
(A) MSI1 and HDA19 are components of the same histone deacetylase complex. HDA19:GFP and MSI1 were found to be part of 500- to 600-kD complexes (red arrows) detected by anti-GFP and anti-MSI1 using native PAGE. No anti-GFP signal was detected in wild-type Col. Specificity of the anti-MSI1 antibody was shown before (Hennig et al., 2003).
(B) MSI1 copurifies with HDA19. HDA19:GFP (red arrows) was precipitated from inflorescences of wild-type and Pro35S:HDA19:GFP plants. Precipitates were analyzed by immunoblotting using anti-MSI1 antibodies.
(C) MSI1 interacts with HDA19 in a yeast two-hybrid assay. Only yeast cotransformed with bait (pGBKT7:MSI1) and prey (pGADT7:HDA19) plasmids grow on selective medium (GDO, quadruple dropout, SD/–Ade/–His/–Leu/–Trp) and give a positive X-Gal signal (DDO, double dropout, SD/–Leu/–Trp without or with X-α-Gal).
Next, we tested whether MSI1 and HDA19 are coexpressed, which is required for a joint function. Using a compendium of transcript data for 79 developmental stages, organs, and tissues (Schmid et al., 2005), we observed a strong positive Pearson correlation of 0.95 for MSI1 and HDA19 expression (Supplemental Figure 1). In fact, HDA19 was one of the genes most strongly correlated in expression with MSI1, and only 11 of the 21,500 probed genes had stronger correlation. This result demonstrates that MSI1 and HDA19 expression allows MSI1-HDA19 complex formation.
Common interaction partners of both MSI1 and HDA19 suggest that MSI1, HDA19, SNL2, SNL3, SNL4, and HDC1 could function in the same complex(es) in vivo. MSI1, HDA19, and HDC1 could form a core SIN3-like complex in vivo, while diverse associating SNLs could provide targeting specificity via binding to different transcription factors.
Reduction of MSI1 and HDA19 Levels Causes Upregulation of ABA-Responsive Genes
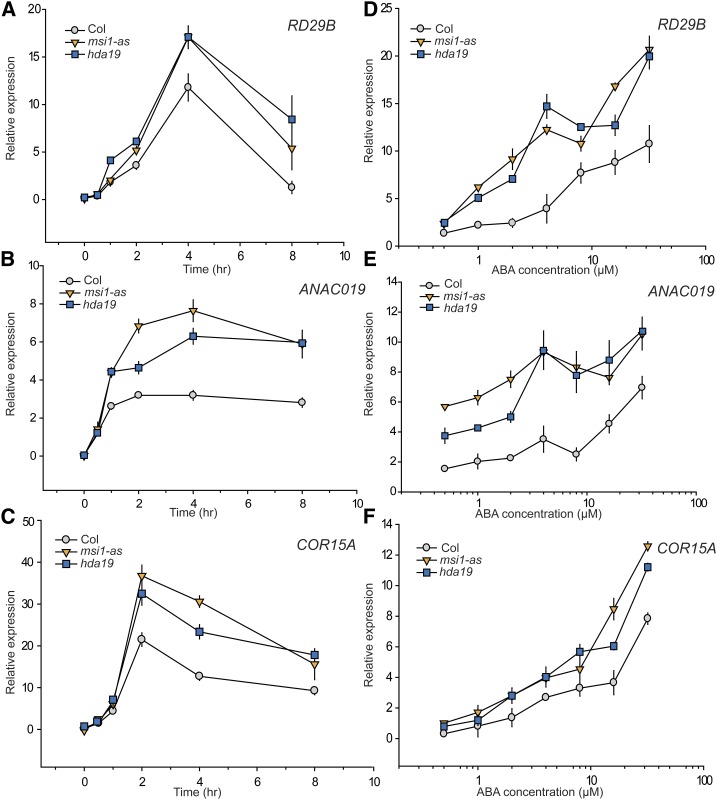
Various components of histone deacetylase complexes have been implicated in plant responses to biotic and abiotic stress (Chen and Wu, 2010; Perrella et al., 2013; Wang et al., 2013), and reduction of MSI1 levels can affect the expression of genes with ABREs in their promoters (Alexandre et al., 2009). Because ABA is a major mediator of responses to abiotic stress, we tested a potential role of MSI1 and HDA19 in activation of ABA-responsive genes using MSI1 antisense (msi1-as) and HDA19 RNAi (hda19) Arabidopsis lines. Ten-day-old seedlings of the wild type, msi1-as, and hda19 were treated with 5 μM ABA hydroponically for 8 h, and aliquots were harvested for RNA extraction every 2 h. Expression of three ABA-responsive genes was tested: RD29B (RESPONSIVE TO DESSICATION29B) (Msanne et al., 2011), ANACO19 (NAC DOMAIN CONTAINING PROTEIN19) (Jensen et al., 2010), and COR15A (COLD REGULATED15A) (Baker et al., 1994, Steponkus et al., 1998), which all have ABREs in their promoter regions (Choi et al., 2000). After exposure to ABA, transcript levels of all three genes were considerably higher in msi1-as and hda19 lines compared with the wild type at all time points (Figures 2A to 2C). Notably, the activation kinetics were not grossly changed but remained similar to that in wild-type plants.
Figure 2.
Both msi1-as and hda19 Mutants Show Upregulation of ABA-Responsive Genes over Time in an ABA Dose-Dependent Manner.
(A) to (C) Expression kinetics of RD29B, ANACO19, and COR15A. Relative change in expression over 8 h of RD29B, ANAC019, and COR15A in the wild type (gray circles), msi1-as (orange triangles), and hda19 (blue squares), respectively. Differences from the wild type are significant (two-sided t test, P ≤ 0.05) for all genes for time points at 3 h and longer and for ANAC019 also at 1 h.
(D) to (F) ABA dose dependency of RD29B, ANACO19, and COR15A expression. Relative change in expression of RD29B, ANAC019, and COR15A in the wild type (gray circles), msi1-as (orange triangles), and hda19 (blue squares), respectively, treated with different concentrations of ABA for 4 h. Differences from the wild type are significant (P ≤ 0.05) for RD29B above 0.5 µM ABA, for ANAC019 at all concentrations, and for COR15A above 10 µM ABA. Expression levels are relative to PP2A. Graphs show the mean ± se of three biological replicates.
To test whether the higher expression of stress-responsive genes in the msi1-as and hda19 lines reflected increased sensitivity of gene expression to ABA, we treated 10-d-old seedlings of the wild type, msi1-as, and hda19 with concentration between 0.5 and 32 μM of ABA hydroponically for 4 h before harvesting material for RNA extraction. Consistent with the time course expression results, at most ABA concentrations, we observed higher expression of RD29B, ANAC019, and COR15A in msi1-as and hda19 compared with the wild type (Figures 2D to 2F).
Together, these results demonstrate that several ABA-responsive genes respond much more strongly to ABA if MSI1 or HDA19 levels are decreased.
msi1-as and hda19 Plants Have Increased Transcript Levels of ABA Receptor Genes
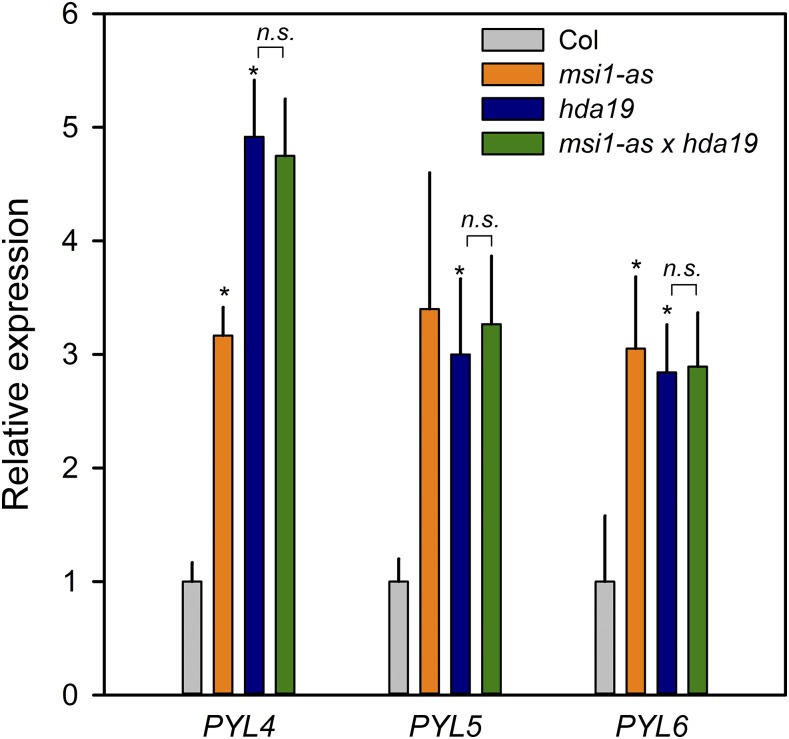
A family of PYR/PYL receptor proteins mediates ABA signaling in plants (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). ABA binding to PYR/PYLs increases their ability to bind and inhibit type 2C protein phosphatases (PP2Cs). In the absence of ABA, PP2Cs bind and inactivate SNF1-related kinases (SnRK2 kinases) (Umezawa et al., 2009). Inactivation of PP2Cs by PYR/PYLs leads to activation of SnRK2, which in turn causes activation of basic leucine-zipper transcription factors called ABFs/AREBs that bind ABREs and switch on stress response genes (Kobayashi et al., 2005; Furihata et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007). PYL4 interacts with PP2CA in an ABA-dependent manner (Lackman et al., 2011; Pizzio et al., 2013) and shows high expression levels in many tissues, and its inactivation is needed to generate ABA-insensitive mutants (Gonzalez-Guzman et al., 2012). This indicates that PYL4 is one of the key players in the ABA receptor family. Interestingly, PYL4 acetylation and expression were previously reported to be affected by altered HDC1 levels (Perrella et al., 2013). We tested a potential role of the MSI1-HDA19 complex in ABA signaling at the level of receptor gene expression by measuring transcripts of PYL4 and its two close homologs, PYL5 and PYL6, in msi1-as, hda19, and the wild type. Indeed, PYL4, PYL5, and PYL6 transcript levels were increased in the lines with reduced levels of MSI1 and HDA19 (Figure 3). PYL4, PYL5, and PYL6 are also examples of genes that are downregulated by ABA (Goda et al., 2008; Perrella et al., 2013). Notably, this repression by ABA was considerably stronger in msi1-as and hda19 than in the wild type, demonstrating that not only ABA-induced but also ABA-repressed genes react more strongly to ABA in plants with reduced levels of MSI1 and HDA19 (Supplemental Figure 2). These results show that overexpression of PYL4, PYL5, and PYL6 in msi1-as and hda19 is alleviated by ABA and argue that although MSI1 and HDA19 affect the expression of PYL4, PYL5, and PYL6, they are not required for their ABA-dependent repression.
Figure 3.
Increased Transcript Levels of ABA Receptors in msi1-as and hda19.
Relative expression of PYL4, PYL5, and PYL6 in the wild type (gray), msi1-as (orange), hda19 (blue), and msi1-as hda19 (green). Expression levels are relative to PP2A and further normalized to the wild type. Graphs show the mean ± se of three biological replicates. Asterisks indicate values that are significantly larger (P ≤ 0.05) than in the wild type. n.s. indicates no significant difference between msi1-as hda19 and hda19.
Increased expression of genes for ABA receptors is consistent with the increased ABA sensitivity of ABA-activated and ABA-repressed genes in plants with reduced MSI1 and HDA19 levels. To test whether MSI1 and HDA19 act in the same genetic pathway to affect PYL4, PYL5, and PYL6 expression, we crossed the msi1-as and hda19 lines and measured PYL4, PYL5, and PYL6 expression (Figure 3). There was no further increase in PYL gene expression in the double mutant, suggesting that MSI1 and HDA19 function together in the same genetic pathway to affect PYL gene transcription, consistent with the protein-protein interaction data. Because ABA responses are sensitive to the dosage of PYL genes (Gonzalez-Guzman et al., 2012) and because overexpression of PYL4 or PYL5 increases ABA sensitivity (Santiago et al., 2009; Pizzio et al., 2013), increased expression of PYL4, PYL5, and PYL6 can explain the increased ABA sensitivity of gene expression in msi1-as and hda19 plants.
MSI1 Binds to Chromatin at the PYL4, PYL5, and PYL6 Genes and Maintains Low H3K9 Acetylation
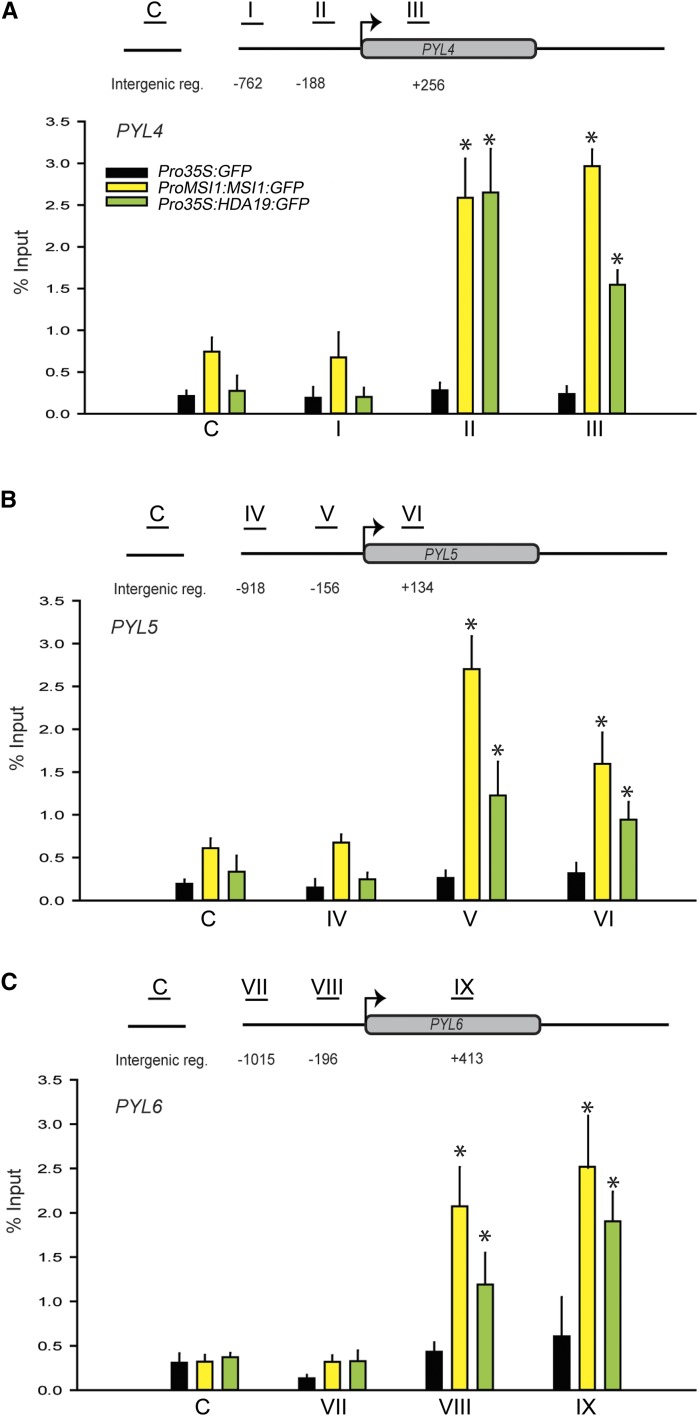
Next, we tested whether regulation of ABA receptor gene expression by MSI1 and HDA19 was direct. We performed a chromatin immunoprecipitation (ChIP) assay using a line expressing GFP-tagged MSI1 in an msi1 mutant background (Alexandre et al., 2009) and the HDA19:GFP line. High MSI1 and HDA19 recovery was observed around the PYL4, PYL5, and PYL6 transcriptional start sites (Figure 4). In contrast, only background signals were observed in regions more distant to the transcriptional start sites in a GFP control line and in IgG control reactions (Figure 4; Supplemental Figure 3). Thus, the effects of MSI1 and HDA19 on PYL4, PYL5, and PYL6 expression involve direct binding to chromatin at the ABA receptor genes.
Figure 4.
MSI1 and HDA19 Bind to Chromatin at ABA Receptor Genes.
ChIP was performed using 14-d-old Pro35S:GFP, ProMSI1:MSI1:GFP, and Pro35S:HDA19:GFP plants and anti-GFP antibodies. The graphs show MSI1 (yellow) and HDA19 (green) recovery at PYL4, PYL5, and PYL6 compared with a Pro35S:GFP control (black). Schemes above the diagrams represent the recovery at PYL4 (A), PYL5 (B), and PYL6 (C) loci. Black lines with Roman numerals represent the regions probed by PCR. Results of IgG control ChIP assays were plotted separately (Supplemental Figure 2). Values are recovery as percent of input; shown are mean ± se of three biological replicates. Asterisks indicate significant (P ≤ 0.001) difference from Pro35S:GFP controls.
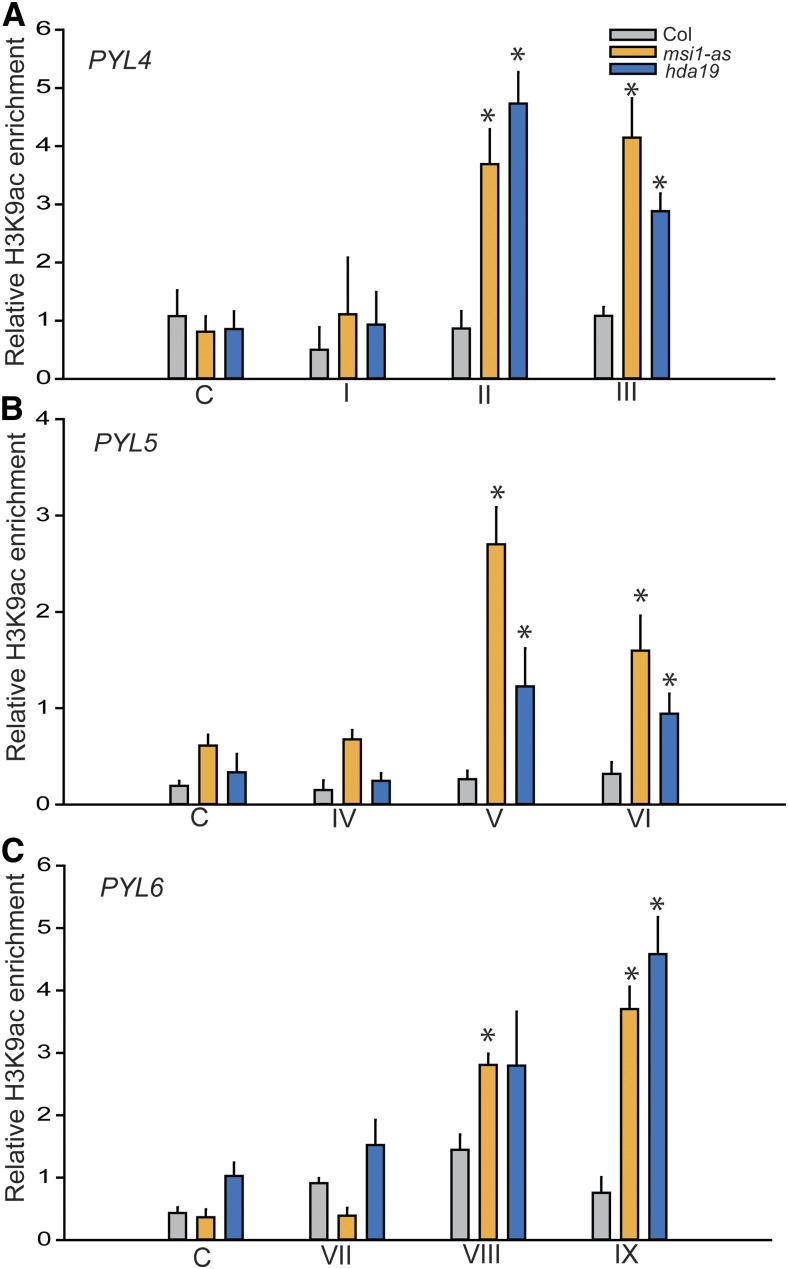
MSI1 and HDA19 function together in a complex in Arabidopsis. It is possible that altered expression of PYL4, PYL5, and PYL6 in msi1-as and hda19 is associated with increased histone acetylation. To test this hypothesis, we performed ChIP with antibodies against histone H3 acetylated at lysine 9 (H3K9ac) in wild-type, msi1-as, and hda19 plants. In the wild type, levels of H3K9ac at PYL4, PYL5, and PYL6 were not considerable higher than the level in an intergenic control region or IgG control values (Figure 5; Supplemental Figure 4). In contrast, in both msi1-as and hda19, H3K9ac levels were considerably increased around the transcriptional start site (Figure 5). Thus, MSI1 and HDA19 are both needed to maintain low histone acetylation at PYL4, PYL5, and PYL6.
Figure 5.
MSI1 and HDA19 Affect the H3K9 Acetylation Status at ABA Receptor Genes.
The graphs show increased H3K9ac levels in msi1-as (orange) and hda19 (blue) compared with the wild type (gray) at the PYL4, PYL5, and PYL6 locus, respectively. Fragment labels below the graphs are as in Figure 4. Anti-H3K9ac ChIP-qPCR was performed using 14-d-old plants. Values represent recovery expressed as relative enrichment of H3K9ac compared with H3 levels. Results of IgG control ChIP assays were plotted separately (Supplemental Figure 3). Shown are means ± se of three biological replicates. Asterisks indicate significant (P ≤ 0.01) difference from the wild type.
The notion that the MSI1-HDA19 complex is recruited to chromatin at PYL4, PYL5, and PYL6 genes and represses their transcription implies that HDAC inhibitors should mirror the effects of reduced MSI1 or HDA19 dosage. To test this hypothesis, we measured the effect of the HDAC inhibitor trichostatin A (TSA) on PYL4, PYL5, and PYL6 expression in wild-type, msi1-as, and hda19 plants (Supplemental Figure 5). In the wild type, TSA indeed caused an increase in expression of the three genes similar to that observed in hda19 plants. In hda19 plants, however, TSA did not lead to a considerable further increase in PYL4, PYL5, and PYL6 transcript levels, suggesting that HDA19 is the major HDAC involved in regulating PYL4, PYL5, and PYL6 expression. The msi1-as plants were still responsive to TSA in this assay, probably because of the remaining MSI1 levels (∼30% of wild-type levels) in this line (Exner et al., 2006). In contrast, TSA strongly increased ANAC019, RD29B, and COR15A transcript levels in unstressed wild-type, msi1-as, and hda19 plants (Supplemental Figure 6), showing that TSA treatment has effects on msi1-as and hda19 plants and suggesting that HDACs other than HDA19 maintain low ANAC019, RD29B, and COR15A expression in unstressed plants. Together, these results are consistent with the notion that HDAC activity mediates the effects of MSI1 and HDA19 on PYL gene expression.
Loss of MSI1 or HDA19 Leads to Increased Tolerance to Salt Stress
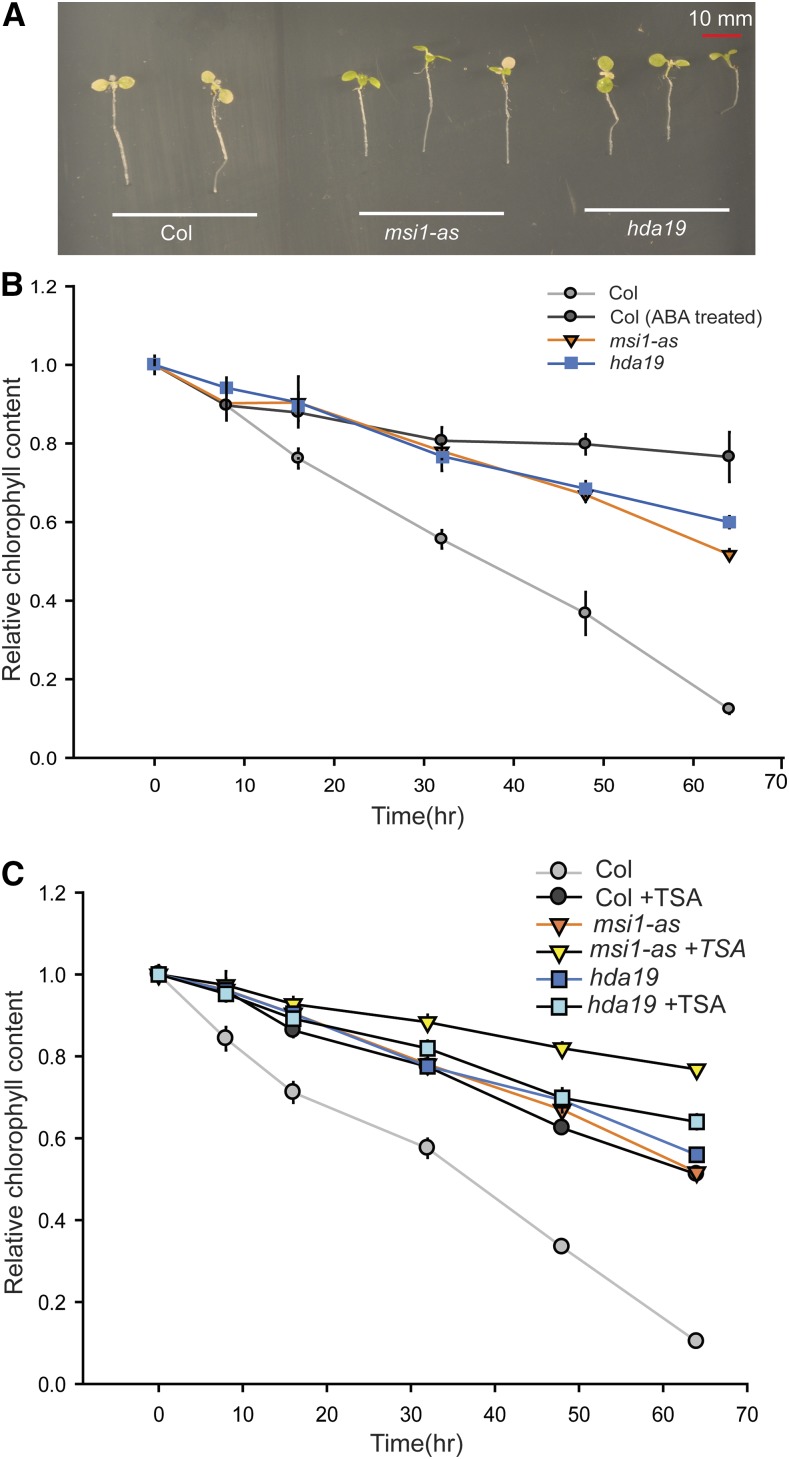
Because MSI1 and HDA19 dampen ABA-responsive gene expression in seedlings, it was possible that the gene expression response to abiotic stress was also altered in msi1-as and hda19 plants. To test this hypothesis, we exposed wild-type, msi1-as, and hda19 plants to salt and tested the expression of the salt-responsive gene RD29B. Consistent with the increased ABA sensitivity of gene expression, we detected increased salt sensitivity of gene expression as well (Supplemental Figure 7). Next, we tested whether the physiological response to salt was also altered in msi1-as and hda19 plants. Seven-day-old seedlings were transferred onto medium containing 150 mM NaCl. This severe stress causes chlorophyll loss, which can be considerably delayed by ABA (Figure 6). Similarly, msi1-as and hda19 plants tolerated 150 mM salt much longer than the wild type, as evident from the delayed chlorophyll loss (Figure 6). TSA pretreatment for 48 h delayed chlorophyll loss in the wild type considerably but had only a minor effect on hda19, consistent with the notion that HDAC activity, mainly contributed by HDA19, is required for normal chlorophyll loss upon salt exposure (Figure 6C). Notably, TSA affected chlorophyll loss in msi1-as, which is similar to its effects on PYL4, PYL5, and PYL6 expression in this background. Together, the results indicate that the increased ABA sensitivity of gene expression in msi1-as and hda19 plants was accompanied by increased tolerance to salt stress.
Figure 6.
msi1-as and hda19 Have Increased Tolerance to Salt Stress.
(A) Performance of plants grown on NaCl. msi1-as and hda19 plants remain green longer on 150 mM NaCl compared with the wild type.
(B) Change in chlorophyll content over time on 150 mM NaCl. msi1-as (orange), hda19 (blue), and ABA-treated wild type (dark gray) have substantially more chlorophyll after 64 h than the wild type (gray). Differences from the wild type are significant (two-sided t test, P ≤ 0.05) for time points at 16 h and longer.
(C) Effect of pretreatment with TSA for 48 h on changes in chlorophyll content during exposure to salt (150 mM). Wild type treated with TSA (dark gray) show delayed loss of chlorophyll compared with the untreated wild type (gray). Differences from the wild type are significant (P ≤ 0.05) for time points at 8 h and longer. Shown are means ± se for three biological replicates.
DISCUSSION
HDAC protein complexes play key roles in the regulation of gene expression in diverse eukaryotes, such as budding yeast, Drosophila melanogaster, and mammals (Yang and Seto, 2008). In Arabidopsis, HDA6 and HDA19 are the most widely studied HDACs, and both are involved in a wide range of developmental processes and environmental responses (Tian and Chen, 2001; Tian et al., 2003; Zhou et al., 2005; Long et al., 2006; Chen and Wu, 2010; Jang et al., 2011). Given that HDA6 and HDA19 are closely related, it is not surprising that they have partly overlapping functions. Both enzymes are involved in pathogen and jasmonic acid responses (Zhou et al., 2005; Wu et al., 2008) as well as in suppression of embryonic programs after germination (Tanaka et al., 2008), abiotic stress responses (Chen and Wu, 2010), promotion of flowering, and senescence (Wu et al., 2008; Yu et al., 2011). However, HDA6 also has specific functions in silencing of transposons, transgenes, and rRNA genes (Aufsatz et al., 2007; To et al., 2011; Liu et al., 2012). HDA6 forms complexes with MSI4 and its homolog MSI5 (Gu et al., 2011). These complexes are recruited to target genes where they lead to histone deacetylation and transcriptional gene silencing (Gu et al., 2011). In contrast to HDA6, HDA19 is presumably localized to euchromatin and is excluded from the nucleolus (Fong et al., 2006). We demonstrate that HDA19 and MSI1 form a complex and share common biological functions. Our data did not reveal any association of MSI1 with HDA6 or of MSI4/5 with HDA19, suggesting that these proteins form separate RPD3-like HDAC complexes in plants.
Arabidopsis MSI1 has been reported to be part of CAF-1 and several Polycomb-Repressive Complex 2 (PRC2)-like complexes such as the FERTILIZATION INDEPENDENT SEED, VERNALIZATION, and EMBRYONIC FLOWER complexes (Kaya et al., 2001; Köhler et al., 2003; De Lucia et al., 2008; Derkacheva et al., 2013). Because we found that MSI1 forms a complex with HDA19, it is possible that MSI1 links HDA19 to PRC2-like complexes or CAF-1. Notably, earlier work had suggested an association of RPD3 with PRC2 subunits in Drosophila (van der Vlag and Otte, 1999; Tie et al., 2001, 2003). In contrast, our work did not reveal any association of HDA19 with Arabidopsis PRC2 components. Similarly, a purification of the Arabidopsis VERNALIZATION PRC2 complex did not reveal an association with HDACs (De Lucia et al., 2008). This suggests that a potential association of the MSI1-HDA19 core complex with Arabidopsis PRC2 is not prevalent. Also, no other CAF-1 subunits or other known chromatin-related proteins were found to associate with HDA19, suggesting that the functions of the MSI1-HDA19 complex do not involve prevalent stable interactions with other chromatin-related protein complexes. Extending what has been known about MSI1 in PRC2 and CAF-1, we provide four key lines of evidence to support a role of MSI1 in a histone deacetylation complex to repress transcription. (1) MSI1 and HDA19 form a complex in vivo. (2) Reduction of MSI1 and HDA19 levels leads to increase transcript levels of the ABA receptor genes PYL4, PYL5, and PYL6. (3) Reduction of MSI1 and HDA19 levels leads to increased H3K9 acetylation levels at PYL4, PYL5, and PYL6. (4) MSI1 and HDA19 bind to the chromatin at PYL4, PYL5, and PYL6.
Loss of MSI1 is lethal and/or strong downregulation leads to severe developmental defects, making such lines inappropriate for the assays used here (Hennig et al., 2003; Köhler et al., 2003; Guitton et al., 2004). The report that MSI1 binds to the chromatin of the ABA-responsive gene RD20, which is upregulated even without exogenous ABA in adult plants of an Arabidopsis line with greatly reduced MSI1 levels and strongly altered development (Alexandre et al., 2009), is consistent with a role of MSI1 in gene expression that goes beyond the effects of an MSI1-HDA19 complex on PYL4, PYL5, and PYL6, which was the focus of this study. To limit confounding effects of altered development, an antisense line of MSI1 was used here, in which MSI1 levels were reduced to ∼30% of wild-type levels and which has only mild developmental alterations (Exner et al., 2006, 2008). However, the residual MSI1 levels in msi1-as may allow the formation of residual MSI1-HDA19 complexes that may still be sufficient to perform some functions. This could explain why msi1-as plants were responsive to TSA in some assays.
This study revealed a robust upregulation of PYL4, PYL5, and PYL6 in plants with reduced MSI1 or HDA19 levels. An earlier transcriptome study had not identified these genes as upregulated in plants with reduced MSI1 levels, possibly due to the limited power of microarrays to robustly detect weakly expressed genes (Alexandre et al., 2009). Because inhibition of HDAC activity by TSA did not lead to activation of PYL4, PYL5, and PYL6 beyond that observed in hda19 plants, PYL4, PYL5, and PYL6 expression appears to be mainly regulated by HDA19. In contrast, TSA treatment strongly activated RD29B, COR15A, and ANAC019, even in an hda19 background. This is consistent with the notion that another HDAC complex can regulate these ABA-responsive genes. As MSI4 represses COR15A expression, binds to COR15A chromatin, and associates with HDA6 (Ausín et al., 2004; Kim et al., 2004; Gu et al., 2011; Jeon and Kim, 2011), it is possible that an MSI4-HDA6 complex directly regulates COR15A, while the MSI1-HDA19 complex indirectly regulates COR15A via affecting PYL4, PYL5, and PYL6 expression.
In Arabidopsis, several components of potential RPD3-like HDAC complexes have been implicated to affect development. For example, SNL1 affects seed dormancy via changing the expression of ABA- and ethylene-responsive genes (Wang et al., 2013). Similarly, HDC1 affects plant growth via modulation of ABA-sensitive genes (Perrella et al., 2013). HDA19 and HSL1 may act together to repress the expression of seed maturation genes during germination (Zhou et al., 2013). Notably, many of the developmental roles that were described for HDA19 or other complex subunits may be explained by altered ABA signaling. We argue that at least some of these effects are mediated by repression of ABA receptor genes by an MSI1-HDA19 SIN3-like complex. First, our protein purification data establish that MSI1, HDA19, HDC1, and SNL1-6 can form a complex in vivo. Second, we found that MSI1 and HDA19 affect ABA sensitivity of gene expression via modulation of ABA receptor gene expression. These results are also consistent with earlier reports of the effects of MSI1 on the expression of ABA-responsive genes (Alexandre et al., 2009) and of ABA hypersensitivity of plants with reduced SNL3 levels (Song et al., 2005). Plants accumulate ABA in response to diverse abiotic stresses such as drought, cold, and salt; ABA then regulates the expression of many stress-responsive genes, promoting stress tolerance (reviewed in Nakashima and Yamaguchi-Shinozaki, 2013). Consistently, increased ABA sensitivity of gene expression in plants with reduced MSI1 or HDA19 levels was associated with increased tolerance to salt stress.
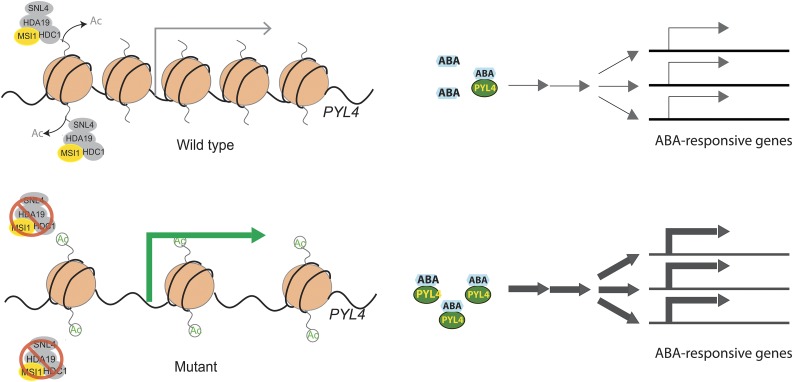
Together, we propose a model of how MSI1 functions as part of a histone deacetylase complex to fine-tune ABA-responsive gene expression (Figure 7). In wild-type plants, MSI1 together with HDA19 directly represses the expression of ABA receptor genes such as PYL4, which causes dampening of ABA signaling. However, in the absence of MSI1 or HDA19, increased expression of ABA receptor genes leads to increased sensitivity of expression of ABA-responsive genes. However, ABA levels are high enough to efficiently activate ABA receptor-dependent activation of gene expression only upon stress or ABA treatment, while under control conditions, ABA-responsive genes have only small changes in basal expression. Because ABA represses PYL4, PYL5, and PYL6, their overexpression is alleviated by extended exposure to ABA, suggesting that the MSI1-HDA19 complex most strongly affects a subset of the initial responses to ABA in unstressed plants. Future work will focus on establishing how specificity in complex formation and target gene selection is established for the MSI1-HDA19 versus the MSI4/5-HDA6 complex.
Figure 7.
Proposed Model of MSI1 Function in ABA Signaling.
In the wild type, a MSI1-HDA19 SIN3-like complex deacetylates chromatin at ABA receptor genes such as PYL4 and thus maintains low transcription of the receptor genes (gray arrow). Increased expression of ABA receptor genes (green arrow) in msi1 or hda19 mutant plants causes increased expression of ABA-responsive genes in the presence of ABA (blue hexagons). Reduction of MSI1 or HDA19 levels has no strong effect on the expression of ABA-responsive genes in the absence of ABA.
METHODS
Plant Material
Seeds of Arabidopsis thaliana (accession Columbia) wild-type control and mutants were sterilized, stratified, and germinated on half-strength Murashige and Skoog medium and 0.8% agar at pH 5.7 in controlled growth rooms with 20 to 22°C and long-day conditions (16 h light/8 h dark; 110 µmol m−2 s−1 PAR full-spectrum white light supplied by Osram Lumilux cool white fluorescent tubes), unless stated otherwise. Ten-day-old seedlings were used for RNA extraction and ChIP experiments. For ABA and NaCl dose–response experiments, 10-d-old seedlings (five to eight) were transferred onto a 16-well hydroponic plate supplemented with either ABA, NaCl, or TSA at the concentration mentioned in the figures. Chlorophyll was assayed as described (Moran and Porath, 1980). The Pro35S:GFP, Pro35S:HDA19:GFP, ProMSI1:MSI1:GFP, HDA19 RNAi (hda19), and MSI1 antisense (msi1-as) plant lines were described before (Hennig et al., 2003; Zhou et al., 2005; Exner et al., 2006; Fong et al., 2006; Alexandre et al., 2009; Derkacheva et al., 2013). In the HDA19 RNAi lines, HDA19 transcript levels are reduced to ∼10% of the value in the wild type, and there is no considerable off-target effect on the HDA19 homolog HDA6 (Zhou et al., 2005; Supplemental Figure 8). Similarly, there are no considerable off-target effects on MSI1 homologs in msi1-as lines (Hennig et al., 2003).
Immunoprecipitation and Protein Immunoblot Analyses
For immunoprecipitation (IP) followed by mass spectrometry, 10 g of plant material was ground in a mortar with liquid nitrogen; for coimmunoprecipitation, 2 g of plant material was used. Soluble proteins were extracted in two volumes of extraction buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% IGEPAL, 1% Triton X-100, and protease inhibitors [Roche]) at 4°C for 30 min with gentle rocking. The centrifuged (4500g) supernatant was precleared with 30 μL of prewashed protein A Sepharose beads (GE Healthcare) at 4°C for 20 min with gentle rocking. An input aliquot was taken from the precleared centrifuged (2000g) supernatant before the rest of the supernatant was subjected to IP with 50 μL of bead-coupled antibodies at 4°C for 2 h with gentle rocking. The precipitate was washed six times with extraction buffer and eluted in 2× Laemmli buffer. Anti-GFP Trap-A beads (gta-10; Chromotek) were used for IP. For protein immunoblots, proteins were separated by 12% SDS-PAGE and transferred to polyvinylidene fluoride membrane (Roth) by semidry blotting in 25 mM Tris-HCl, pH 8.3, 150 mM glycine, and 10% methanol for 1 h at 15 V. Enhanced chemiluminescence detection was performed as recommended by the manufacturer (GE Healthcare). Mouse anti-MSI1 (Hennig et al., 2003) antibodies, 1:1000, were used for immunoblotting.
MS/MS Analyses
After IP, the proteins were separated by 12% SDS-PAGE, lanes were cut into three sections, and in-gel digestion was performed (Shevchenko et al., 1996). The peptides from each tryptic digest were dissolved in 10 μL of 0.1% formic acid (FA). Five microliters of each sample was injected on an EASY-nanoLC-system (Thermo Scientific). The enzymatic peptides were separated in reversed phase on a 10-cm-long C18-A2 column (i.d. 75 µm; Thermo Scientific) using mobile phase A = 0.1% FA and B = 0.1% FA, 99.9% acetonitrile, and were then electrosprayed on-line to the mass spectrometer. Mass spectrometry measurements were performed on an LTQ Orbitrap Velos ETD mass spectrometer (Thermo Finnigan). MS/MS was performed, applying collision-induced dissociation in the LTQ mass spectrometer. MS/MS spectra were searched with Sequest (Eng et al., 1994) against the UNIPROT protein datABAse, using Arabidopsis as a target organism. Carbamidomethylation of cysteine was set as fixed modification, and oxidation of methionine was set as a variable modification. The data were analyzed by Proteome Discoverer (Proteome Software). The cutoff for data analyses was set to a medium and high confidence for peptide identification. Proteins identified with at least two unique peptides in at least two replicates but never in control samples were taken into account.
Native PAGE of Nuclear Proteins
Native protein was extracted from 1 g of 10-d-old wild-type and HDA19:GFP seedlings in 10 mL MEB extraction buffer (0.775 M hexylene glycol, 20 mM PIPES-KOH, pH 7.6, 10 mM MgCl2, 0.1 mM EGTA, 60 mM KCl, 1× Protease inhibitor, 0.5% Triton, and 5 mM β-mercaptoethanol) using a MACS dissociator (Miltenyi Biotech). The extract was filtered through two layers of Miracloth, and the filtrate was centrifuged at 1500g at 4°C for 10 min. The extracted nuclei were suspended in 500 mL of protein extraction buffer (65 mM Tris-HCl, pH 7.4, 10% glycerol, and 0.05% bromophenol blue). Proteins were precipitated with 4 volumes of ice-cold acetone and redissolved in 100 μL extraction buffer. After centrifugation at 14,000g, 20 μL of each sample was loaded onto precast 4 to 15% Mini-Protean TGX gels (Bio-Rad) and separated using Tris-glycine buffer (25 mM Tris and 190 mM glycine) at 70 V for 2 h. Transfer to polyvinylidene fluoride membranes was done in 25 mM Tris, 190 mM glycine, and 10% methanol at 30 V overnight in a wet-blot transfer cell. Proteins were detected using mouse anti-MSI1 (Hennig et al., 2003) (1:3000) and anti-GFP (Clontech; 1:3000) antibodies.
ChIP
For ChIP, 100 mg of 10-d-old seedlings was collected and cross-linked. Nuclei were extracted as described by Shu et al. (2013). For anti-MSI1:GFP ChIP, extracted nuclei were resuspended in 1 mL of PBS, pH 8.0, with 10 mM dimethyladipimidate dihydrochloride (Sigma-Aldrich) at 4°C for 2 h. To stop cross-linking, 50 mM of Tris-HCl, pH 7.5, was added followed by incubation with gentle rotation at 4°C for 20 min. Cross-linked nuclei were collected by centrifugation at 1500g for 10 min. Extracted nuclei were washed once with ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, and 1× Complete EDTA-free protease inhibitor [Roche]) and resuspended in 100 μL of lysis buffer (50 mM Tris-HCl, pH 8.0, 1% SDS, and 1× Complete EDTA-free protease inhibitor). Chromatin was sheared by seven cycles of sonication of 30 s On/30 s Off using a Bioruptor (Diagenode) and diluted 10-fold with ChIP dilution buffer, followed by clearing by centrifugation (4500g, 5 min, 4°C). Twenty-five microliters of chromatin was used as input control and 250 μL of chromatin was used in one immunoprecipitation reaction with 1 µg of antibody and incubated overnight. Immunoprecipitated complexes were collected using 20 μL of Dynabeads Protein A (Life Technologies) per reaction after incubation for 1.5 h at 4°C. Beads were washed two times for 5 min with 300 μL of low salt wash buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 1 × Complete EDTA-free protease inhibitor) and two times for 5 min with 300 μL of high salt wash buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 1× Complete EDTA-free protease inhibitor). Bound complexes were eluted using an IPure kit (Diagenode). Antibodies used were anti-GFP (Molecular Probes Invitrogen), IgG (Sigma-Aldrich), antihistone H3 (Millipore), and anti-H3K9ac (Millipore). DNA recovery after ChIP was quantified as percentage of input or relative to anti-H3 signal (relative abundance). qPCR with gene-specific primers (Supplemental Table 1) was performed using a CFX Connect system (Bio-Rad) and SsoAdvance Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions.
Yeast Two-Hybrid Assay
Constructs for yeast two-hybrid analysis were generated using the Matchmaker Gold Yeast Two-Hybrid System (Clontech) vectors pGBKT7 and pGADT7, which express protein fusions to the GAL4 DNA binding domain or transcriptional-activation domain, respectively. cDNA inserts encoding MSI1 and HDA19 were introduced in pGBKT7 and pGADT7 using the Gateway cloning system and Matchmaker Gold Yeast Two-Hybrid System as described in the user manuals. The analysis was performed in strain AH109 carrying HIS3 and MEL1 reporters for reconstituted GAL4 activity.
RNA Isolation and RT-qPCR
RNA extraction and RT-qPCR were performed using an RNeasy Plant Mini Kit (Qiagen), followed by cDNA synthesis using a RevertAid first-strand cDNA synthesis kit (Thermo Scientific) on biological triplicates. qPCR with gene-specific primers (Supplemental Table 2) was performed using a CFX Connect system and SsoAdvance Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Data were analyzed using a method described by Simon (2003).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MSI1 (AT5G58230), HDA19 (AT4G38130), HDC1 (AT5G08450), SNL1 (AT3G01320), SNL2 (AT5G15020), SNL3 (AT1G24190), SNL4 (AT1G70060), SNL5 (AT1G59890), SNL6 (AT1G10450), ATTCP-1 (AT3G20050), PYL4 (AT2G38310), PYL5 (AT5G05440), PYL6 (AT2G40330), RD29B (AT5G52300), COR15A (AT2G42540), and ANAC019 (AT1G52890).
Supplemental Data
Supplemental Figure 1. Expression of MSI1 and HDA19 is strongly correlated.
Supplemental Figure 2. Effect of MSI1 and HDA19 on the expression of the ABA-repressed genes PYL4, PYL5, and PYL6.
Supplemental Figure 3. IgG controls of anti-GFP ChIP.
Supplemental Figure 4. IgG controls of anti-H3K9ac ChIP.
Supplemental Figure 5. Effect of TSA on the expression of ABA receptor genes.
Supplemental Figure 6. Effect of TSA on the expression of ABA-responsive genes.
Supplemental Figure 7. Effect of salt on RD29B expression.
Supplemental Figure 8. Absence of off-target effect on HDA6 in HDA19 RNAi lines.
Supplemental Table 1. Sequences of primers used for ChIP-qPCR.
Supplemental Table 2. Sequences of gene-specific primers used for RT-qPCR.
Supplementary Material
Acknowledgments
We thank Keqiang Wu (National Taiwan University) for providing seeds of the Arabidopsis Pro35S:HDA19:GFP and hda19 RNAi lines. This work was supported by grants from ETH Zurich, the Swedish Research Council VR (2011-5010 and 621-2011-4423), The Science for Life Laboratory, MS-Based Proteomics Platform Uppsala, and the Knut and Alice Wallenberg Foundation.
AUTHOR CONTRIBUTIONS
M.D., S.M., and L.H. designed the research. M.D., S.M., M.R., and L.K. performed research. M.D., M.R., J.B., and L.H. analyzed data. S.M., M.D., J.B., and L.H. wrote the article.
Glossary
- ABA
abscisic acid
- ABRE
ABA-responsive element
- PP2C
type 2C protein phosphatase
- ChIP
chromatin immunoprecipitation
- TSA
trichostatin A
- IP
immunoprecipitation
- MS/MS
tandem mass spectrometry
- FA
formic acid
References
- Ach R.A., Taranto P., Gruissem W. (1997). A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell 9: 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre C., Möller-Steinbach Y., Schönrock N., Gruissem W., Hennig L. (2009). Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol. Plant 2: 675–687. [DOI] [PubMed] [Google Scholar]
- Aufsatz W., Stoiber T., Rakic B., Naumann K. (2007). Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene 26: 5477–5488. [DOI] [PubMed] [Google Scholar]
- Ausín I., Alonso-Blanco C., Jarillo J.A., Ruiz-García L., Martínez-Zapater J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36: 162–166. [DOI] [PubMed] [Google Scholar]
- Baker S.S., Wilhelm K.S., Thomashow M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24: 701–713. [DOI] [PubMed] [Google Scholar]
- Bowen A.J., Gonzalez D., Mullins J.G., Bhatt A.M., Martinez A., Conlan R.S. (2010). PAH-domain-specific interactions of the Arabidopsis transcription coregulator SIN3-LIKE1 (SNL1) with telomere-binding protein 1 and ALWAYS EARLY2 Myb-DNA binding factors. J. Mol. Biol. 395: 937–949. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Workman J.L. (2005). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Chen L.T., Luo M., Wang Y.Y., Wu K. (2010a). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61: 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.T., Wu K. (2010b). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5: 1318–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275: 1723–1730. [DOI] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M., Greb T., Dean C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 105: 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M., Steinbach Y., Wildhaber T., Mozgová I., Mahrez W., Nanni P., Bischof S., Gruissem W., Hennig L. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32: 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Exner V., Gruissem W., Hennig L. (2008). Control of trichome branching by chromatin assembly factor-1. BMC Plant Biol. 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Taranto P., Schönrock N., Gruissem W., Hennig L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133: 4163–4172. [DOI] [PubMed] [Google Scholar]
- Fong P.M., Tian L., Chen Z.J. (2006). Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro. Cell Res. 16: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernández M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., Rodriguez P.L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzenda A., Lomberk G., Zhang J.S., Urrutia R. (2009). Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., Jiang, D., Yang, W., Jacob, Y., Michaels, S.D., and He, Y. (2011). Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 7: e1002366. [DOI] [PMC free article] [PubMed]
- Guenther M.G., Yu J., Kao G.D., Yen T.J., Lazar M.A. (2002). Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 16: 3130–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton A.E., Page D.R., Chambrier P., Lionnet C., Faure J.E., Grossniklaus U., Berger F. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981. [DOI] [PubMed] [Google Scholar]
- Hassig C.A., Fleischer T.C., Billin A.N., Schreiber S.L., Ayer D.E. (1997). Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89: 341–347. [DOI] [PubMed] [Google Scholar]
- Hennig L., Bouveret R., Gruissem W. (2005). MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15: 295–302. [DOI] [PubMed] [Google Scholar]
- Hennig L., Taranto P., Walser M., Schönrock N., Gruissem W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565. [DOI] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185. [DOI] [PubMed] [Google Scholar]
- Hollender C., Liu Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50: 875–885. [DOI] [PubMed] [Google Scholar]
- Jang I.C., Chung P.J., Hemmes H., Jung C., Chua N.H. (2011). Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196. [DOI] [PubMed] [Google Scholar]
- Jeon J., Kim J. (2011). FVE, an Arabidopsis homologue of the retinoblastoma-associated protein that regulates flowering time and cold response, binds to chromatin as a large multiprotein complex. Mol. Cells 32: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22: 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142. [DOI] [PubMed] [Google Scholar]
- Kenzior A.L., Folk W.R. (1998). AtMSI4 and RbAp48 WD-40 repeat proteins bind metal ions. FEBS Lett. 440: 425–429. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., et al. (2005). Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Hyun Y., Park J.Y., Park M.J., Park M.K., Kim M.D., Kim H.J., Lee M.H., Moon J., Lee I., Kim J. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36: 167–171. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44: 939–949. [DOI] [PubMed] [Google Scholar]
- Lackman P., et al. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. USA 108: 5891–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu C.W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Luo M., Liu X., Singh P., Cui Y., Zimmerli L., Wu K. (2012a). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 129–136. [DOI] [PubMed] [Google Scholar]
- Luo M., Tai R., Yu C.W., Yang S., Chen C.Y., Lin W.D., Schmidt W., Wu K. (2015). Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J. 82: 925–936. [DOI] [PubMed] [Google Scholar]
- Luo M., Wang Y.Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. (2012b). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63: 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Moran R., Porath D. (1980). Chlorophyll determination in intact tissues using n,n-dimethylformamide. Plant Physiol. 65: 478–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msanne J., Lin J., Stone J.M., Awada T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234: 97–107. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32: 959–970. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Yoshida T., Kitahata N., Asami T., Shinozaki K., Hirayama T. (2007). ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50: 935–949. [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G., Lopez-Vernaza M.A., Carr C., Sani E., Gosselé V., Verduyn C., Kellermeier F., Hannah M.A., Amtmann A. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25: 3491–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio G.A., Rodriguez L., Antoni R., Gonzalez-Guzman M., Yunta C., Merilo E., Kollist H., Albert A., Rodriguez P.L. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri R., Tanaka K., Nakafuku M., Kaziro Y., Toh-e A., Matsumoto K. (1989). MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86: 8778–8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S.Y., Márquez J.A., Cutler S.R., Rodriguez P.L. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60: 575–588. [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Shu H., Gruissem W., Hennig L. (2013). Measuring Arabidopsis chromatin accessibility using DNase I-polymerase chain reaction and DNase I-chip assays. Plant Physiol. 162: 1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P. (2003). Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19: 1439–1440. [DOI] [PubMed] [Google Scholar]
- Song C.P., Agarwal M., Ohta M., Guo Y., Halfter U., Wang P., Zhu J.K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.P., Galbraith D.W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60: 241–257. [DOI] [PubMed] [Google Scholar]
- Steponkus P.L., Uemura M., Joseph R.A., Gilmour S.J., Thomashow M.F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95: 14570–14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146: 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Chen Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Wang J., Fong M.P., Chen M., Cao H., Gelvin S.B., Chen Z.J. (2003). Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F., Furuyama T., Prasad-Sinha J., Jane E., Harte P.J. (2001). The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275–286. [DOI] [PubMed] [Google Scholar]
- Tie F., Prasad-Sinha J., Birve A., Rasmuson-Lestander A., Harte P.J. (2003). A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol. Cell. Biol. 23: 3352–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, T.K., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7: e1002055. [DOI] [PMC free article] [PubMed]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J., Otte A.P. (1999). Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23: 474–478. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cao H., Sun Y., Li X., Chen F., Carles A., Li Y., Ding M., Zhang C., Deng X., Soppe W.J., Liu Y.X. (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59: 225–234. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Seto E. (2008). The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9: 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Nishimura N., Kitahata N., Kuromori T., Ito T., Asami T., Shinozaki K., Hirayama T. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. (1997). Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89: 357–364. [DOI] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. (2013). HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.