The authors wish to alert readers of the following:
1. The original legend for Figure 1, panel C could potentially be misleading and is therefore corrected as follows (modifications are indicated in bold):
(C) RNA blot analyses of high- and low-molecular-weight RNAs using DNA probes against GFP (upper panels) or the PVX-specific coat protein (CP) (lower panels) sequences. RNA extracted from GFP transgenic plants was used as a positive control in lane 1 only for the two high-molecular-weight RNA blots. RNA extracted from the AMP line was used as a negative control in lane 2 for the high- and low-molecular-weight RNA blots. The same RNA samples were used for the four RNA blots, hence the use of a unique rRNA equal loading control panel. (rRNA) Ethidium bromide staining of ribosomal RNAs; (gRNA) PVX genomic RNA; (sgRNA) subgenomic RNA; (CP) PVX coat protein.
2. A comparison of the published figures and the original raw data has revealed several mounting mistakes, which have been shared with the editor-in-chief and systematically corrected based on the original files provided. The authors wish to issue the following corrections:
Figure 2, panel D:
FIGURE 2.
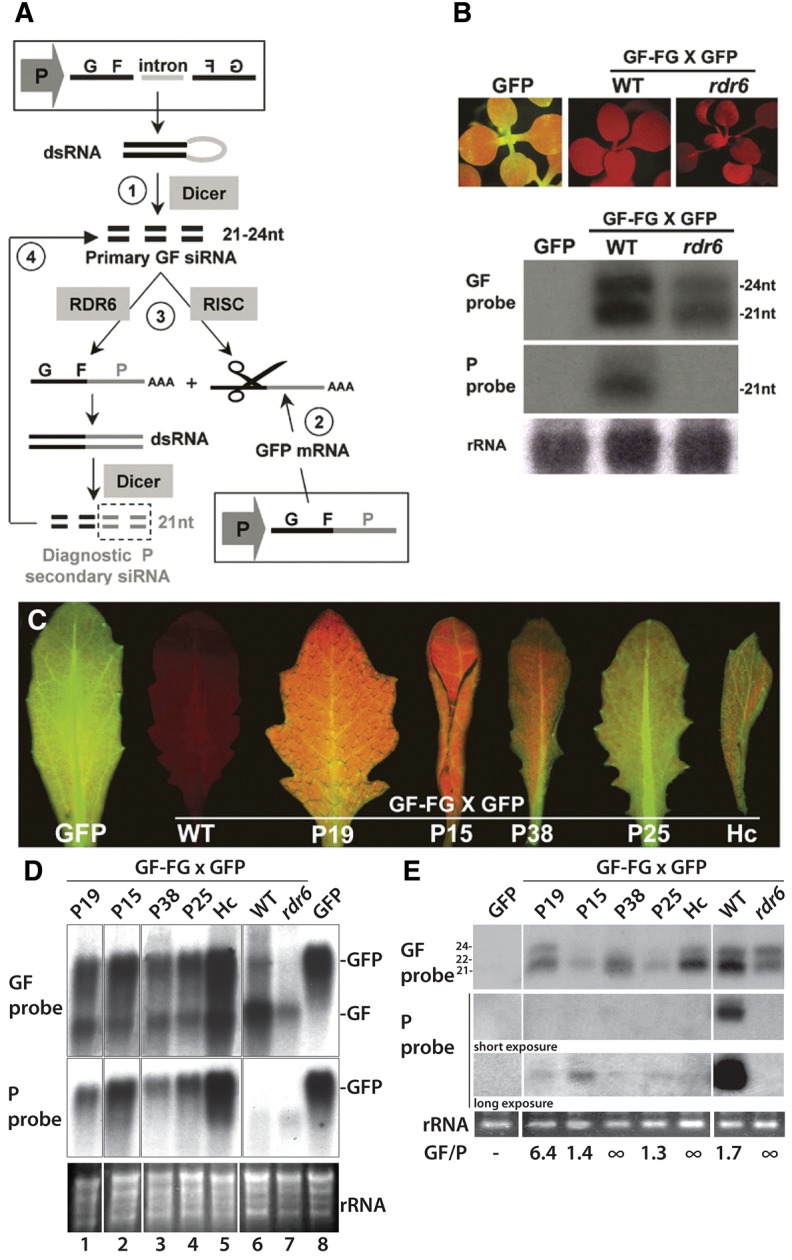
The blot used to mount this panel has been spliced to remove unnecessary data but this operation was not disclosed. We have retrieved the original blot and remounted the figure with white vertical bands to specify the spliced regions. Also, the rRNA loading control panel has been corrected, and the contrast of the “GF” and “P” panels has now been decreased for background signals to be visible. These corrections do not affect the original conclusions drawn from this panel.
Figure 2, panel E:
The GFP control for this figure comes from a separate blot, which was not specified in the published figure, creating an offset in the rRNA loading control. This has now been corrected with the original blot. Moreover, to be consistent with panel D of Figure 2, only the same WT sample is now shown in the amended panel E. These corrections do not affect the original conclusions drawn from this panel.
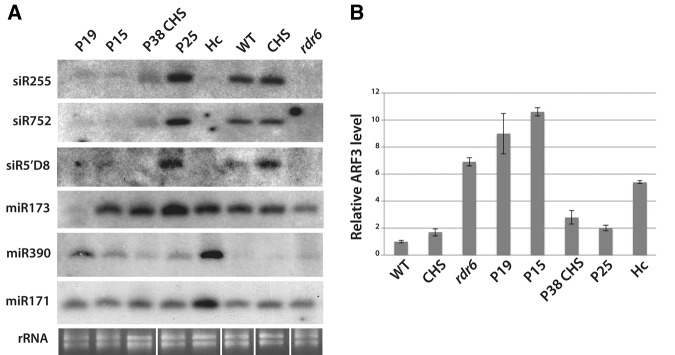
FIGURE 3.
This figure was originally believed to depict the same blot as in Figure 2E, stripped and rehybridized for various endogenous small RNAs. This led to the use of the same rRNA loading control as in Figure 2E. The confusion is confirmed in reading the original text: “We further tested the accumulation of endogenous trans-acting siRNAs in the various suppressor-transgenic lines used in the above study.” However, inspection of the original data revealed that none of the VSRs used in the experiments depicted in Figure 3 were in the GFFG–GFP background used in Figure 2E. Instead, the VSRs were all in the WT background, except P38, which was expressed in the context of a second transgene that causes RNAi of the chalcone synthase (CHS) mRNA (not stipulated in the original figure). As a specific negative control for the P38 plants, we thus used the cognate CHS RNAi background without P38 (also not stipulated in the original figure). We have now retrieved hybridizations corresponding to each of the segments composing the blot in Figure 3, as well as the original, cognate rRNA loading control to produce an amended figure that does not contradict any of the original claims made with this figure. Please note a mistake concerning the original siR5′D7 section of the blot, which, in fact, corresponds to a hybridization against siR5′D8. This has now been corrected in the panel labeling.
We therefore would like to rephrase the legend of Figure 3 as follows (again, modifications are indicated in bold):
Figure 3. Effects of silencing suppressors on accumulation of endogenous, RDR6-dependent trans-acting siRNAs and RDR6-independent miRNAs. (A) Low-molecular-weight RNA analyses in transgenic plants expressing the five silencing suppressors, using DNA oligonucleotide probes specific for endogenous small RNAs. Enhanced contrast has been applied onto the siR5′D8 blot. (rRNA) Same as in Figure 1. (B) Quantitative-RT PCR analyses of ARF3 (At2g33860) accumulation in suppressor-expressing plants relative to WT. The chalcone synthase RNAi (CHS) line is used as a control for the P38 CHS line. All the other silencing suppressors are in a WT background. The rdr6 line is used as a control. The primers flanked the second complementary sequence of the TAS3 tasiRNA59D7, as found in the ARF3 mRNA.
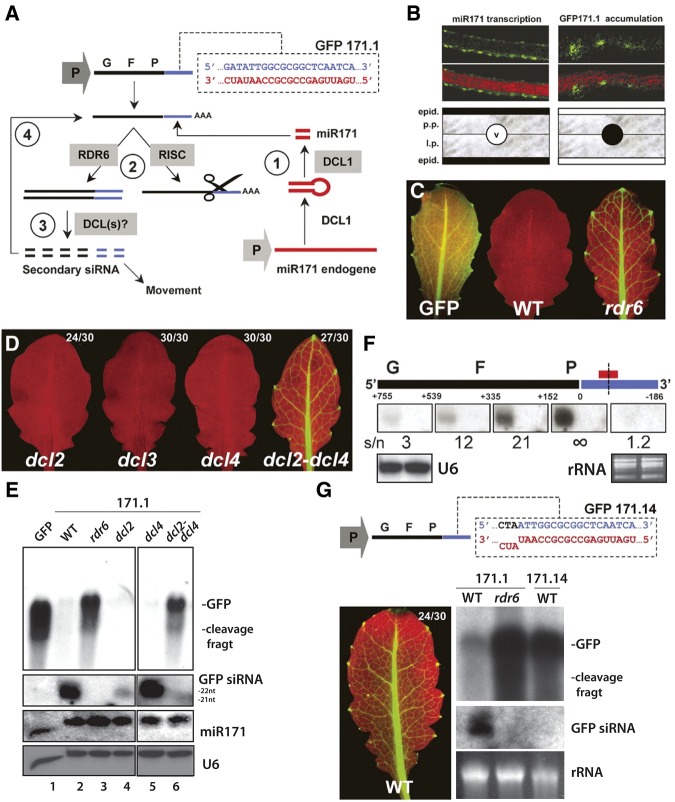
Figure 4, panel E:
FIGURE 4.
The blot used to mount this panel has been spliced to remove unnecessary data but this operation was not disclosed. We have retrieved the originals and remounted the figure using white vertical bands to specify blot splicing. While inspecting the corresponding GFP siRNA blot, we realized that the dcl3 track was mistakenly kept in place of the adjacent dcl2 track. This mistake likely originates from the fact that the 21-nt siRNA signal in the dcl3 track is significantly stronger than in the dcl2 track. We now have retrieved a longer exposure of the same blot, which clearly shows the cognate 21-nt siRNA signal in the dcl2 track, and have amended the figure accordingly. Finally, we have detected a problem in the miR171 lane, in which the aberrant migration pattern of the leftmost signal was not scored as a bona fide signal, leading to an offset of the lane and of the corresponding U6 loading control. This has been corrected in the new version of Figure 4E. These corrections do not affect the original conclusions drawn from this panel.
Figure 4, panel F:
Ethidium bromide staining of ribosomal RNAs (rRNAs) is depicted to show the equal loading between the GFP171.1/WT (left signal) and GFP171.1/rdr6 (right signal) tracks on the rightmost blot (GFP-3′), which derived from a membrane different than the one used for the four other hybridizations depicted. These used U6 hybridization as an equal loading signal. The corrections do not affect the original conclusions drawn from this panel.
Figure 4, panel G:
The originally published high- and low-molecular-weight RNA blots showing GFP mRNA and siRNA accumulation could not be retrieved. However, we have found different original blots depicting the same result. Panel G is now corrected using the cognate ethidium bromide staining of ribosomal RNAs (rRNAs) as equal loading control.
All of the authors have approved these corrections and apologize for any resulting inconvenience.
doi: 10.1261/rna.055012.115