Abstract
The Drosophila melanogaster sex-peptide (melSP) is a seminal fluid component that induces postmating responses (PMR) of females via the sex-peptide receptor (SPR) . Although SP orthologs are found in many Drosophila species, their functions remain poorly characterized. It is unknown whether SP functions are conserved across species or rather specific to each species. Here, we developed a GFP-tagged melSP (G-SP) and used it to visualize cross-species binding activity to the female reproductive system of various species. First we demonstrated that ectopically expressed G-SP induced PMR in D. melanogaster females and bound to the female reproductive system, most notably to the common oviduct. No binding occurred in the females lacking SPR, indicating that G-SP binding was dependent on SPR. Next we tested whether G-SP binds to the common oviducts from 11 Drosophila species using dissected reproductive tracts. The binding was observed in six species belonging to the D. melanogaster species group, but not to those outside the group. Injection of melSP reduced the receptivity of females belonging to the D. melanogaster species group, but not of those outside the group, being consistent with the ability to bind G-SP. Thus the SP-mediated PMR appears to be limited to this species group. SPR was expressed in the oviducts at high levels in this group; therefore, we speculate that an enhanced expression of SPR in the oviduct was critical to establish the SP-mediated PMR during evolution.
Keywords: sex-peptide, SP receptor, GFP-tag, postmating response
THE mating behavior and physiology of Drosophila melanogaster females are dramatically modified after copulation: they reject courting males by extruding their ovipositor, a behavior not seen in virgin females and start to lay many eggs (Kubli 1992). Sex-peptide (SP), a seminal fluid peptide, has been shown to play a major role in eliciting postmating response (PMR). Injection of SP into the abdominal cavity of virgin females reduces receptivity and stimulates oviposition (Chen et al. 1988) and these phenotypes can be induced by ectopic expression of SP in virgin females (Aigaki et al. 1991). Furthermore, experiments involving an SP null mutant generated by gene targeting and dsRNAi-mediated gene knockdown unambiguously demonstrated that SP is a major component in inducing changes in receptivity, ovulation, and oviposition in mated females (Liu and Kubli 2003; Chapman et al. 2003). In addition, SP stimulates food intake (Hanin et al. 2011) and activates immune response genes (Peng et al. 2005b).
An extensive transgenic RNAi screen identified a receptor for sex-peptide (SPR), a G-protein-coupled receptor broadly expressed in the female reproductive tracts and in some neural tissues (Yapici et al. 2008). Mutant females deleted for SPR do not respond to mating, accept repeated mating, and maintain a low level of oviposition (Yapici et al. 2008). It has been demonstrated that SPR expression in a limited number of pickpocket-expressing neurons in the female common oviduct was sufficient to rescue the mutant phenotype, suggesting that these neurons are targeted by SP (Yang et al. 2009; Hanin et al. 2012). Interestingly, neuronally expressed SP reduced receptivity and increased oviposition in the absence of SPR, suggesting an additional pathway capable of inducing PMR (Haussmann et al. 2013).
Several studies have shown that myoinhibitory peptides (MIPs) are also potent ligands for SPR (Kim et al. 2010; Poels et al. 2010; Yamanaka et al. 2010). Recently, it has been demonstrated that MIP and SPR are essential for sleep stabilization in both male and female flies (Oh et al. 2014). Although MIP and SPR exist in most of the sequenced insect genomes, SP occurs in the family Drosophilidae only. Therefore, it has been thought that SP arose in an ancestral species of the Drosophilidae and evolved as a PMR regulator by hijacking SPR-mediated signaling (Kim et al. 2010). The interesting feature of SP function is that it is beneficial to males, but on the other hand, it reduces females’ fitness (Wigby and Chapman 2005). Thus SP and SPR are likely to mediate sexual conflict, which facilitate rapid evolutionary changes in the relevant genes. Of 12 Drosophila species whose genomes have been sequenced, D. mojavensis and D. grimshawi do not seem to contain SP orthologs in their genomes, suggesting that the SP-dependent system was abandoned in these species (Kim et al. 2010). On the other hand, species belonging to D. melanogaster species subgroup contain Dup99B, an SP paralog, and D. ananassae contain three copies of SP orthologs. It is interesting to note that the frequency of female’s remating is variable depending on the species of Drosophila: D. subobscura, D. acanthoptera, and D. silvestris females mate only once in their lifetimes, whereas D. hydei or D. nigrospiracula remate up to four times in a given morning (Markow and O’Grady 2005). These suggest that the role of SP could be variable among the species. However, the extent of the functional divergence between different species remains unknown.
Here, we developed a GFP-tagged SP (G-SP) derived from D. melanogaster and used it to visualize cross-species binding activity to female reproductive system of 11 species. G-SP binds to the oviducts of six species belonging to the D. melanogaster species group, but not to those outside the group. We also assessed the biological activity of SPs from different species by injecting them into D. melanogaster or conspecific females. Based on the results of these experiments, we concluded that the SP-mediated PMR was established in the lineage of D. melanogaster species group.
Materials and Methods
GFP fusion constructs
GFP cDNA was PCR amplified using pQESP::GFPS65T (Villella et al. 2006) as a template with a set of primers gfpc1 (5′-ccatctagatccacctcctttgtatagttcatccatgcc-3′) and the T3 primer and subcloned into pQE vector (Qiagen, Hilden, Germany). An amino-acid substitution, V63A, was accidentally incorporated during PCR. Since it was brighter than GFPS65T alone, we used GFPS65T, V63A cDNA for the fusion constructs and refer to this as GFP in this study.
To construct SPn::GFP::SPc (G-SP), GFP was inserted between the N-terminal 10 amino-acid residues (SPn) and the C-terminal 26 amino acids (SPc). The fragment borders were determined based on previous studies on functional dissection with a synthetic peptide (Schmidt et al. 1993) and a potential trypsin cleavage site (Peng et al. 2005a). N- and C-terminal fragments were fused to GFP to construct SPn::GFP (G-SPn) and GFP::SPc (G-SPc), respectively. GFP alone was used as a control. All constructs were fused to the signal sequence of SP (SPs) to be secreted from cells. Five- to seven-amino-acid-long spacers were placed between GFP and SP fragments. The following primer sets were used for PCR to generate peptide fragments using pBluescript SK+ plasmid containing SP cDNA (Chen et al. 1988) as a template: sps1 (5′-ctgaattctgggactggaccaagccgagtac) and T3 (5′-aatacgactcactatag) for SPs; spn1 (5′-ccgtctagatctcaaaatgaaaactctagccctattc) and spn2 (5′-gaggaattcgatcctgtaggcttcctattccacgg) for SPs + SPn; and spc1 (5′-cgcactagtggaagtaagtttccaattccaagcccc) and T3 for SPc. To construct SPs + SPn::GFP, a 99-bp-long SPs + SPn cDNA flanked by XbaI/EcoRI was subcloned into pBluescript SK+. To construct SPs::GFP::SPc and SPs+SPn::GFP::SPc, a 114-bp SPc cDNA flanked by SpeI/BamHI was subcloned into the EcoRI/BamHI sites of pUC18 together with a 730-bp EcoRI–XbaI fragment of GFP, whose stop codon was eliminated by PCR mutagenesis using the primer gfpc1 (ccatctagatccacctcctttgtatagttcatccatgcc) and T3. An 865-bp GFP::SPc fragment was subcloned into BamHI/SalI sites of pQE31, and an 860-bp GFP::SPc fragment excised from the resulting plasmid was then subcloned into EcoRI/SalI-digested SPs::GFP and SPs + SPn::GFP plasmids to generate SPs::GFP::SPc and SPs + SPn::GFP::SPc, respectively. To construct SPs::GFP (sGFP), a 66-bp-long SPs cDNA flanked by XbaI/EcoRI was subcloned into pBluescript SK+. The resulting plasmid vector was digested with EcoRI/SalI and ligated with a 737-bp EcoRI–SalI fragment of GFP cDNA. All constructs were digested with BglII and SalI and subcloned into the BglII/XhoI sites of the pUAST vector (Brand et al. 1994).
DNA constructs were purified using QIA-tip 20 columns (Qiagen). The P-element-mediated germline transformation was essentially done according to the standard technique (Rubin and Spradling 1982) using the y1 Df(1)w67C23 strain as a recipient. Multiple independent transgenic lines were established for each construct, and at least three lines per construct were used for biological activity assays and for microscopic observation to localize GFP fusion proteins.
Fly stocks
paired–GAL4 (Xue and Noll 2002), sca–GAL4 (Klaes et al. 1994), GMR–GAL4 (Freeman 1996), ptc–GAL4 (Staehling-Hampton et al. 1994), UAS–SP (Nakayama et al. 1997), and YSX YL, In(1)EN, y stock were obtained from the Kyoto Stock Center. YSX YL, In(1)EN, y was used to generate X/O males carrying the SP::GFP transgene. All Df(1)Exel6234 stocks were used as SPR-deficient stock (Yapici et al. 2008). UAS–SPR flies (Yapici et al. 2008) were from Dickson. Unless otherwise stated, D. melanogaster y1 Df(1)w67C23 strain was used for all experiments. D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, and D. virilis stocks were obtained from Kyorin-fly, Kyorin University. Flies were reared on a standard cornmeal-glucose medium at 25°.
Receptivity assay
For D. melanogaster, D. simulans, and D. sechellia, five females (5-day-old virgins) were placed together with seven males (4- or 5-day-old) in an empty vial, and the number of copulations achieved within 60 min was counted. Receptivity was expressed as a percentage of the copulated females (Chen et al. 1988). At least 30 females were used for each assay. For the remaining species, 14-day-old flies were used for all assays unless otherwise stated. z-test was used to assess statistical difference between two groups in assays for receptivity, ovulation, and immune response.
Ovulation assay
Five-day-old virgin females were examined for ovulation by gently squeezing the tip of the abdomen with forceps, so that females bearing an egg in their uterus would eject it through their ovipositor (Chen et al. 1988). The ovulation level was expressed as the percentage of ovulating females. At least 30 females were used for each assay.
Immune response assay
mtk expression levels were determined using real-time polymerase chain reaction. Total RNA was extracted from 5-day-old adult flies using TRIzol reagent (Invitrogen). After DNase treatment, cDNA was synthesized with a SuperScript VILO cDNA synthesis Kit (Invitrogen). Quantitative PCR was performed using SyBER premix EX Taq (Takara, Shiga, Japan) and a Chromo4 Detector (Bio-Rad) with the following primers: mtk (5′-TCTTGGAGCGATTTTTCTGG-3′) and (5′-GGTTAGGATTGAAGGGCGAC-3′) and rp49 (5′-GCTAAGCTGTCGCACAAATG-3′) and (5′-TGTGCACCAGGAACTTCTTG-3′). Data were normalized with the level rp49 transcript in each sample.
Observation of GFP signal
Mating of transgenic flies expressing GFP fusion genes with females was observed under a Leica MZFLIII fluorescent dissecting microscope. Reproductive organs and other tissues were dissected in Ringer’s solution and observed under either an epifluorescent microscope (BX60-FLA, Olympus, Tokyo, Japan) or a laser scanning confocal microscope (C1s, Nikon, Kawasaki, Japan). Reproductive organs dissected from various Drosophilidae species were incubated with a head homogenate of GMR > G-SP flies in phosphate-buffered saline containing 0.03% Triton X-100 (PBST) for 10 min at room temperature and observed under an epifluorescent microscope.
Immunohistochemistry
Reproductive organs were dissected in phosphate-buffered saline (PBS) and fixed in 4% formaldehyde/PBS for 2 hr at room temperature. After three washes in PBST, the tissues were blocked in 10% goat serum/PBST for 2 hr at room temperature and incubated with an anti-SPR antibody (Yapici et al. 2008) at a dilution of 1:500 in PBST at 4° overnight. After three washes in PBST for 10 min, the tissues were incubated with an Alexa 568-conjugated anti-rabbit IgG (Molecular Probes) at a dilution of 1:1000 in PBST for 2 hr at room temperature. The samples were washed in PBST three times and mounted with 80% glycerol/PBS. The samples were observed by confocal microscopy.
Identification of SP orthologs
The NCBI TBLSTN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search a genome sequence contig with a homology to D. melanogaster SP against the whole-genome shotgun assembly from each of 11 other sequenced Drosophila species (http://www.ncbi.nlm.nih.gov/Traces/wgs/). Assuming that the exon/intron structure is conserved among SP orthologs, a putative intron was defined based on the location, the length, and the consensus sequences (5′-GTAAGT-3′ for donors and 5′-AG-3′ for acceptors). A start codon “ATG” was defined based on the distance from the 5′ end of the intron and the presence of signal peptide coding sequence. Protein coding sequences in two exons were merged to generate a full-length SP ortholog. Putative signal sequences for secretion were predicted by using SignalP 4.0 Server (Petersen et al. 2011).
Expression levels of SPR orthologs
Total RNA was extracted from the female reproductive tracts without ovaries using Trizol reagent (Invitrogen). Real-time PCR was performed as described above. The following primers were used to determine the SPR ortholog expression levels in the D. melanogaster group species, D. pseudoobscura, and D. persimilis: SPR (5′-AGCAAGAAGAGCATGGCCA-3′ and 5′-ACGGAGATGGTGTGGCACA-3′), rp49 (5′-AAGAAGCGCACCAAGCACT-3′ and 5′-TAACCGATGTTGGGCATCA-3′). SPR ortholog expression levels in D. willistoni, D. mojavensis, and D. virilis were measured with the following primers and compared to that in D. pseudoobscura; wil and pse SPR (5′-TCCATGTGTCTGGCCTA-3′ and 5′-TACTCGTGCACCCACAT-3′), moj and pse SPR (5′-AGCTCGCTGATCATCGA-3′ and 5′-TCTCCTTGAAGGTCTCG-3′), wil, moj, and pse rp49 (5′-AAGAAGCGCACCAAGCACT-3′ and 5′-TTGAAGCCAGTGGGCAGCAT-3′), vir and pse SPR (5′-ATGTGCCACACCATCTC-3′ and 5′-ATGTTGAGCGTCACCAG-3′), and vir and pse rp49 (5′-AAGAAGCGCACCAAGCACT-3′ and 5′-TAACCGATGTTGGGCATCA-3′). Data were normalized with the level rp49 transcript in each sample.
Injection of synthetic SPs
The following SP orthologs were synthesized (Sigma): D. mel SP (WEWPWNRKPTKFPIPSPNPRDKWCRLNLGPAWGGRC), D. pse SP (WGRMTSRRPTPKQSQAQFQKWCRLNFGPAWGGRGC), D. wil SP (NPNPERGGDKGKWCRLNLGPAYGGRC), and D. vir SP (EYKTTKWPRYPNKWCRLNYGPYLGGRC). Synthesized SPs were purified with a reversed-phase HPLC after incubating in 0.01 M ammonium bicarbonate (pH 8) containing 3% DMSO for 36 hr (Yapici et al. 2008). Virgin females (5 days old for D. melanogaster, D. simulans, and D. sechellia, and 10 or 11 days old for the remaining species) were anesthetized on a chilling block, and a synthetic SP was injected into their abdominal cavity with a fine needle (5 pmol/fly, or 50 pmol/fly, if the response was unclear). The flies were transferred into vials with fresh food and were subjected to bioassays 3–4 hr after injection.
Results and Discussion
Construction of the GFP-tagged SP
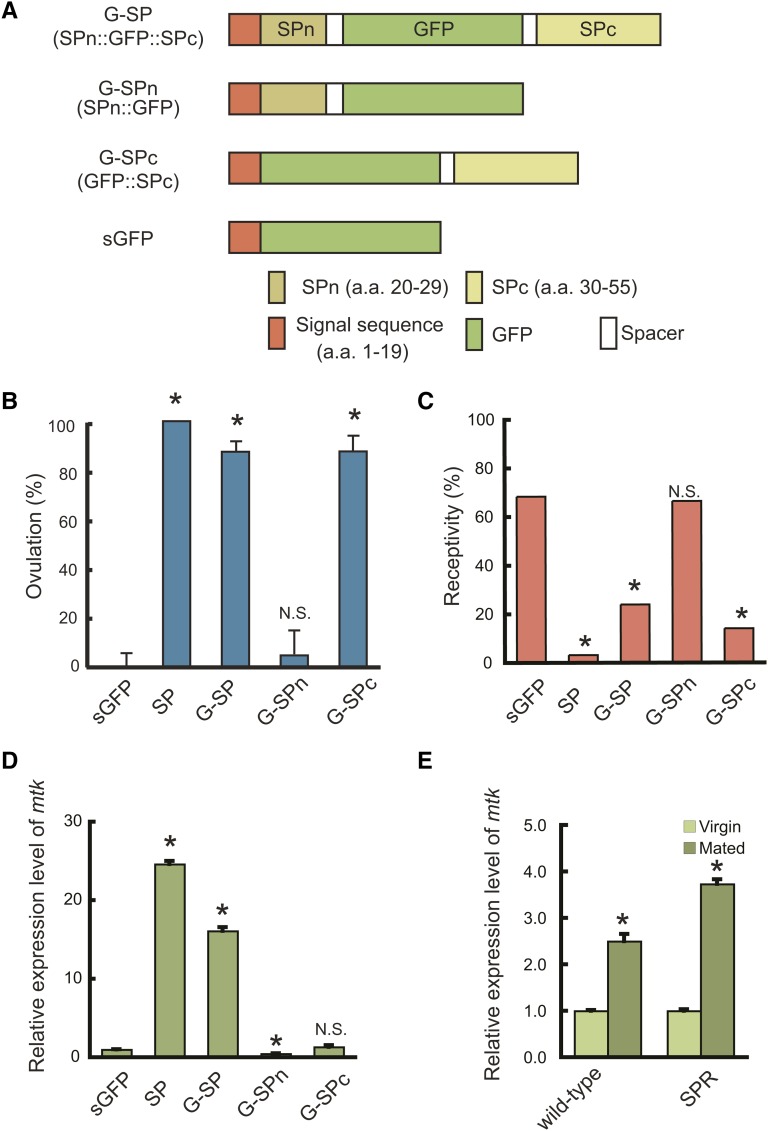
The SP and SPR are important components of the PMR mechanism in D. melanogaster. A GFP-tagged SP is useful in visualizing the interaction between the two molecules and exploring the evolutionary aspects of SP/SPR-mediated PMR. In a previous study, we generated transgenic males expressing the SP–GFP fusion gene (SP promoter-SP::GFP) and used them to visualize the transfer of seminal fluid from males to females (Villella et al. 2006). However, the males expressing SP–GFP in an SP null background did not stimulate oviposition (Domanitskaya et al. 2007), raising the possibility that the C-terminally fused GFP impairs the biological activity of SP. To produce a biologically active and fluorescent SP, we inserted GFP between threonin and lysine at positions 29 and 30 of SP; thus, both the N and C termini remain free. The fusion protein contains the SP signal sequence (SP1–19), SPn (SP20–29), GFP, and SPc (SP30–55) and is referred to as G-SP (Figure 1A). Immunoblot analysis with anti-GFP antibody revealed that all of the fusion proteins and GFP alone were expressed as products with expected sizes (Supporting Information, Figure S1). To assess the biological activity of the fusion protein, the construct was expressed in virgin females using the GAL4–UAS system. We used a sca–GAL4 driver to induce pan-neuronal overexpression of transgenes (Mlodzik et al. 1990). Transgenic females expressing G-SP exhibited a high level of ovulation and reduced receptivity as observed in those expressing the wild-type SP (sca>SP). Ovulation levels and receptivity did not change in the control females expressing the SP signal sequence-GFP fusion protein (sGFP), indicating that G-SP is biologically active (Figure 1, B and C).
Figure 1.
Structure and biological activities of GFP-tagged SP (G-SP). (A) Schematic representation of G-SP fusion proteins. All fusion proteins contain the SP signal sequence (SP1–19) at their N termini and were expressed using the GAL4–UAS system. SPn and SPc indicate SP20–29 and SP30–55 of mature peptides, respectively. (B) Effects of fusion proteins on ovulation and (C) sexual receptivity, and (D) expression of an immune response gene, mtk. sca–GAL4 was used to induce ectopic expression of the fusion constructs. Wild-type SP was used as a positive control. mtk expression levels were determined by quantitative RT–PCR. G-SP and G-SPc stimulate ovulation and reduce receptivity, whereas no change was observed with G-SPn. G-SP also induces mtk expression, but no response was observed with G-SPn or G-SPc. (E) Mating-induced immune response in wild-type and SPR mutant females. SPR is not required for the mating-induced immune response. Values are the mean ±SEM of three experiments (*, P < 0.01; N.S., not significant).
Functional dissection of SP
To dissect the functions of the N- and C-terminal regions of SP, we constructed SPn::GFP and GFP::SPc, which were referred to as G-SPn and G-SPc, respectively (Figure 1A). We found that G-SPc reduced receptivity and increased ovulation levels, while G-SPn did not affect either aspect (Figure 1, B and C). These results indicate that the C-terminal part of SP is essential for the induction of PMR (reduction of receptivity and stimulation of ovulation) and are consistent with those of previous experiments involving the injection of synthetic SP and fragments thereof (Schmidt et al. 1993; Kim et al. 2010). The C-terminal part likely interacts with SPR to elicit PMR (Schmidt et al. 1993; Kim et al. 2010).
In addition to the above-mentioned PMR, SP can stimulate the innate immune system via the Toll and Imd pathways inducing antibacterial peptides, such as metchnikowin (mtk) (Peng et al. 2005b). We examined whether the N- or C-terminal part is specifically involved in this functional aspect. Ectopic expression of G-SP in virgin females induces mtk expression (Figure 1D). However, no increase in mtk expression levels was observed with either G-SPn or G-SPc (Figure 1D), suggesting that the lack of either region impairs the ability to induce an immune response. It has been shown that SP hydroxyproline residues are responsible for eliciting antimicrobial peptide synthesis (Domanitskaya et al. 2007). SPn contains the most N-terminally located hydroxyproline residue, while SPc contains the remaining four residues. It is possible that an SP-induced immune response requires all five hydroxyproline residues. In addition, we found that SPR mutant females exhibit mtk expression after mating (Figure 1E), indicating that SPR is not required for the SP-induced immune response.
Sexual transfer of G-SP into the sperm storage organs
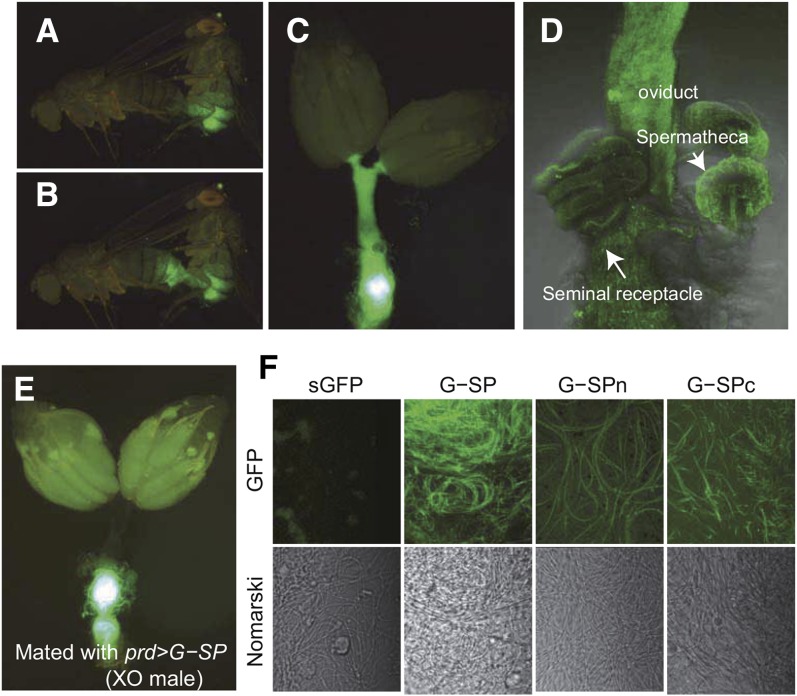
We first utilized G-SP to visualize its sexual transfer from males to females during mating. G-SP was expressed in the male accessory glands using paired-GAL4 as a driver (Xue and Noll 2002). When these males were mated with virgin females, a gradual transfer of fluorescent protein into the females was clearly visible under a fluorescent stereo microscope (Figure 2, B and C, and File S1). Initially, a high level of GFP signal was detected in the uterus and the oviduct (Figure 2C) and then in the sperm storage organs, seminal receptacles, and the spermathecae (Figure 2D).
Figure 2.
Visualization of transfer and localization of G-SP. Transgenic males expressing G-SP were mated with wild-type females. (A) Mated pair soon after the beginning of copulation and (B) 1 min later. (C) Dissected female reproductive tracts soon after the end of copulation. (D) Confocal image of reproductive organs 2 hr after the end of copulation. The GFP signal was detected in the sperm storage organs (spermathecae and seminal receptacles). (E) Distribution of G-SP in the females mated with X/O males. Strong GFP signal is from the uterus, while no or little signal was detected in seminal receptacle, spermatheca, and oviduct. (F) Binding of G-SP to the sperm. Sperm from the females mated with transgenic males expressing indicated fusion protein. Confocal (top) and Nomarski images (bottom). Both N- and C-terminal sequences contribute to G-SP binding to sperm.
Next, we examined whether sperm has any role in the transfer of G-SP into the female reproductive tracts using X/O males that do not produce motile sperm. These males can transfer G-SP into mated females, but its distribution was clearly distinct from that in females mated with wild-type males: no fluorescent signal was detected in the seminal receptacle or spermathecae, although the uterus was fluorescent (Figure 2E). This suggests that motile sperm are necessary for the transfer of SP into the sperm storage organs.
Sperm-binding properties of G-SP
The postmating responses of naturally mated females lasts about 1 week (Kubli 1992), whereas the effects of injection or ectopic expression of SP last 1 or 2 days (Chen et al. 1988; Aigaki et al. 1991). It has been shown that SP binds to sperm via its N-terminal sequence and this binding is important to prolong the period of post mating responses (Peng et al. 2005a). We examined whether G-SP could bind to sperm. We observed the sperm collected from the reproductive organs of females that mated with transgenic males expressing G-SP or sGFP as a control. The sperm collected from the former had strong fluorescence, whereas those from the latter had little fluorescence, indicating that the SP sequence is responsible for the binding of G-SP to sperm (Figure 2F). We also found that both G-SPn and G-SPc could bind to sperm, but the fluorescence signal was weaker than that of G-SP (Figure 2F). These results suggest that both the N- and C-terminal sequences of SP contribute to sperm binding.
Receptor-mediated binding of G-SP
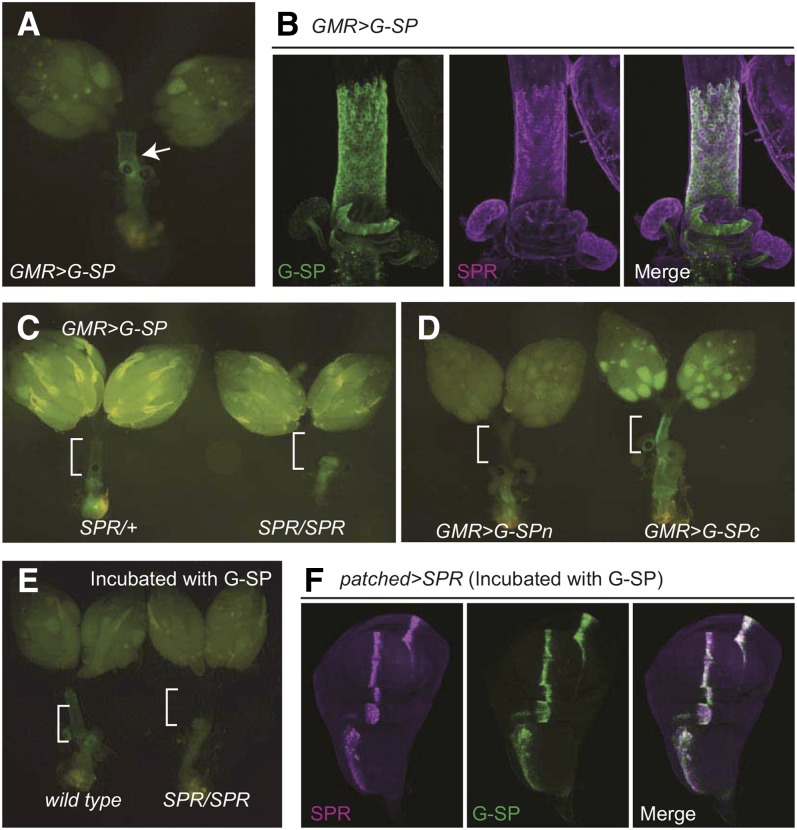
SPR is involved in the inhibition of mating and stimulation of ovulation and oviposition (Yapici et al. 2008). G-SP might be useful in visualizing the binding to SPR expressed in the female reproductive system. We investigated whether G-SP secreted into the hemolymph could localize to the reproductive tracts where SPR is expressed. Forced expression of G-SP was induced in eye imaginal discs using GMR-GAL4 as a driver. The G-SP fluorescent signal was clearly detected in the oviduct and the spermathecae (Figure 3A), which is consistent with those stained with anti-SPR antibody (Figure 3B). No such localization was observed in the SPR mutant females (Figure 3C), indicating that this localization was mediated by SPR. In support of this hypothesis, SPR-mediated localization was observed with ectopically expressed G-SPc but not G-SPn or sGFP (Figure 3D), suggesting that the G-SP-binding property of oviducts and spermathecae is associated with the biological activity that induces PMR. Although SPR is expressed in broad regions of brain, such as ventral regions of the suboesophgeal ganglions, the cervical connective, and many nerve roots (Yapici et al. 2008), there was no obvious G-SP fluorescent signal in those regions.
Figure 3.
SPR-mediated localization of G-SP. (A) G-SP localizes to the oviduct and spermathecae in the females expressing G-SP in the eye discs using GMR-GAL4 (GMR > G-SP). (B) Confocal images of female reproductive tracts. Left: GFP. Middle: anti-SPR antibody. Right: merged. (C) Distribution of G-SP in an SPR mutant background. Left: SPR/+. Right: SPR/SPR. SPR is required for the localization of G-SP to the oviduct and spermathecae. (D) Distribution of G-SPn and G-SPc in the female reproductive system. A GFP signal was detected in the oviduct and spermathecae of GMR > G-SPc, but not of GMR > G-SPn females. (E) Co-incubation of dissected reproductive organs with G-SP. Dissected female reproductive organs from the wild-type (left) and SPR mutants (right) were co-incubated with a tissue homogenate containing G-SP in the same chamber. G-SP localizes to the oviduct and spermathecae in wild type, but not in SPR mutants. (F) SPR-dependent localization of G-SP in wing imaginal discs. Wing imaginal discs expressing SPR (patched > SPR) were co-incubated with a tissue homogenate containing G-SP. The fluorescent signal was detected in a patched expression pattern.
To exclude the possibility that GMR–GAL4 is expressed in the reproductive tracts, we developed an in vitro incubation assay. We prepared a homogenate from the head of GMR > G-SP flies, hereafter referred to as G-SP homogenate. The G-SP homogenate was incubated with dissected reproductive organs of wild-type and SPR mutant females. We found that the fluorescent protein was specifically localized to the oviduct and spermathecae of wild type, but not of the SPR mutant females (Figure 3E). These results demonstrate that G-SP binds to the target tissues in an SPR-dependent manner.
We next examined whether SPR expression is sufficient to localize G-SP in nonreproductive organs. The G-SP homogenate was incubated with wing imaginal discs in which SPR was ectopically expressed using patched–GAL4 (Figure 3F). The fluorescent signal was detected in the pattern of ectopically expressed SPR. Therefore, ectopic expression of SPR is sufficient to localize G-SP in wing imaginal disc cells that are irrelevant to PMR.
Evolutionary aspects of SP and SPR
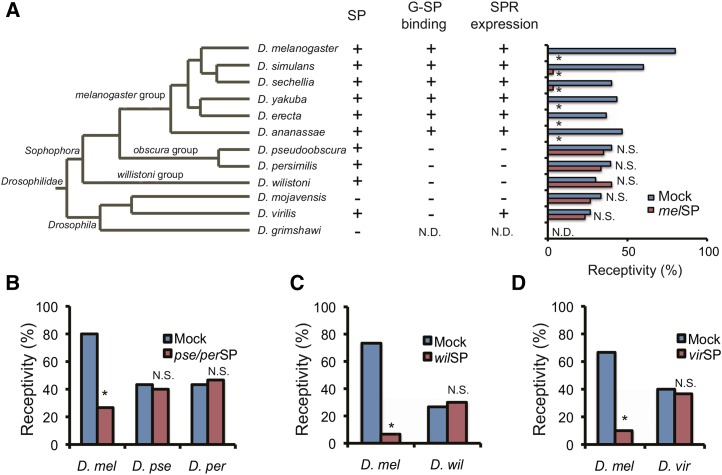
Recently, MIPs were identified as a second family of SPR ligands (Kim et al. 2010; Poels et al. 2010; Yamanaka et al. 2010). MIPs and SPRs were present in all sequenced insect genomes, with two exceptions: the honeybee Apis mellifera and the parasitoid wasp Nasonia vitripennis (Kim et al. 2010). Unlike MIP or SPR, SP only occurs in the family Drosophilidae. We identified putative orthologs in nine Drosophila species. All of them met the following criteria: (1) it has a conserved C-terminal sequence known to be important for SP functions in D. melanogaster; (2) it has a conserved exon/intron structure; and (3) it has a signal sequence for secretion (Figure S2, A and B). Thus, it is likely that SP has arisen in the ancestor of the Drosophilidae species and evolved as a regulator of PMR by utilizing SPR-mediated signaling. However, the apparent loss of SP orthologs in D. mojavensis and D. grimshawi suggests that functional importance of SP varies depending on the species (Figure 4A). Although SP has been shown to interact with SPRs from a wide range of species in vitro using cultured CHO cells (Yapici et al. 2008), whether or not the interaction occurs in vivo and to what extent the function is conserved across species is unknown.
Figure 4.
Evolution of SP-dependent PMR in Drosophila. (A) The phylogenetic tree of 12 sequenced Drosophila species was adapted from http://flybase.org/blast/. Presence or absence of an SP-like sequence in each species is indicated by + or −, respectively (SP). Whether or not the D. melanogaster G-SP binding occurs in the common oviduct of each species is indicated by + or −, respectively (G-SP binding). Expression level of SPR orthologs in female reproductive tracts of each species is indicated by + (strong) or − (weak), respectively. Effects of D. melanogaster SP on sexual receptivity of virgin females from various species. The peptide was injected into virgin females of each species. At least 30 females were used for one assay. N.D. indicates not determined. Effects of SP orthologs from (B) D. pseudoobscura and D. persimilis, (C) D. willistoni, and (D) D. virilis on the receptivity of D. melanogaster and their conspecific females. None of the SP orthologs reduced receptivity in conspecific females, while they all reduced receptivity in D. melanogaster females. The amino acid sequences of SP orthologs are identical for D. pseudoobscura and D. persimilis. Therefore, the same peptide was used for both species (*; P < 0.01; N.S., not significant) .
G-SP binding properties of female reproductive tracts
With an in vitro incubation assay, we examined whether G-SP, whose sequence is derived from D. melanogaster, could bind to the female reproductive systems from other Drosophila species (Figure 4A and Figure S3). After incubation, the fluorescent signal was clearly detected in the oviducts and spermathecae in all species examined from the D. melanogaster species group, D. simulans, D. sechellia, D. yakuba, D. erecta, and D. ananassae, whereas no signal was detected in D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, or D. virilis. These results suggest that the SP-binding ability of the female reproductive system is evolutionarily conserved at least within the D. melanogaster species group. On the other hand, the SP/SPR system may not be conserved in the other species. Considering that SPR expression was critical in localizing G-SP, it is possible that SPR orthologs are not expressed in the oviduct of those species. We performed a series of quantitative PCR analyses using dissected oviducts from various species and found that SPR orthologs are highly expressed in the D. melanogaster species group but not in the distant species except for D. virilis (Figure 4A and Figure S4). The results are unlikely due to technical errors of PCR, since expression of SPR orthologs were detected in whole-body extracts from both sexes in all tested species (Figure S5). Interestingly, SP orthologs were expressed in the male accessory glands of all species, with exception of D. mojavensis whose genome does not contain any SP ortholog. Therefore, although SP orthologs are likely to be involved in the male reproductive function, they may exert their function via SPR that is expressed in tissues other than oviduct or independently from SPR.
Evolutionary aspects of SP/SPR-mediated PMR
To assess the functional conservation of SP/SPR-mediated PMR, we injected a synthetic D. melanogaster SP (melSP) into virgin females of various species. We measured the receptivity as a reliable PMR, since stimulated ovulation is not consistently observed in other species (Aigaki and Ohba 1984). We did not measure SP-dependent activation of innate immunity, because it is difficult to discriminate from the effects of the injection itself (Wigby et al. 2008). Of the 11 species examined, receptivity was significantly reduced in six species of the D. melanogaster species group, whereas the remaining five species did not respond even at concentrations 10 times higher (Figure 4A).
The lack of response to D. melanogaster SP suggested that there might be a high degree of species specificity of SP and SPR systems in the species not belonging to the D. melanogaster species group. Thus, to test the role of SP in the regulation of receptivity, we synthesized orthologs of D. pseudoobscura, D. persimilis, D. willistoni, and D. virilis SP and tested whether they affected the receptivity of D. melanogaster or conspecifics (Figure 4, B–D). Injection of these SP orthologs clearly reduced the receptivity of D. melanogaster females, indicating that SP orthologs from these species are potent ligands for D. melanogaster SPR in vivo. Indeed, all of these peptides had C-terminal sequences that are highly orthologous to that of D melanogaster SP, an essential sequence for PMR-inducing activity and for interaction with SPR (Figure S2A). We then tested whether SP orthologs could reduce the receptivity in their conspecifics. However, none of these species showed an obvious reduction in receptivity when injected with their conspecific SPs (Figure 4, B–D). These results suggest that the SP/SPR-mediated PMR is distinctly evolved within the D. melanogaster species group. However, it has been shown that injection of D. melanogaster SP can elicit some aspects of PMR in virgin females of Heliconverpa armigera, such as inhibition of pheromone production, suppression of calling behavior, and a reduction of PBAN-receptor gene expression (Hanin et al. 2012). The melSP-induced PMR in the moth appeared to be mediated by HaeSP-R, the moth ortholog of SPR (Hanin et al. 2012). There is an SP-like peptide in the male accessory glands (Hanin et al. 2011). Identification of endogenous ligand in the moth would be crucial to understand the evolutionary aspects of the SP-SPR system as a regulator of PMR in insects.
The ability to inhibit remating in females is thought to be advantageous for males in terms of sperm competition. SP likely plays a central role in sexual conflict (Wigby and Chapman 2005). SP transfer is of obvious benefit for males as it increases paternity. It also decreases the fitness and lifespan of females. Sexual conflict may lead to sexually antagonistic coevolution with relatively rapid evolutionary changes within species. The SP-response system seems to be highly variable: although all Drosophila species contain SPR, at least two species have lost SP; females of species outside the D. melanogaster group do not respond to conspecific SP; and those belonging to the D. melanogaster group respond to all SP orthologs examined. The variability of the SP/SPR system among the Drosophilidae species supports the hypothesis that the SP-dependent mechanisms evolved rapidly.
What could be the function of SP orthologs in the species whose females do not show obvious response to conspecific peptides? Considering that all of these SP orthologs contain a conserved C-terminal sequence essential for D. melanogaster PMR (Figure S2A), and were indeed capable of inducing PMR in D. melanogaster, they might also be involved in PMR, but the peptide alone may not be sufficient to induce PMR. Expression levels of SPR orthologs in female reproductive tracts seem to be positively correlated with responsiveness to SP (Figure S4). Therefore, the decreased SPR expression levels may be the cause of reduced responsiveness to SPs. However, D. virilis express SP at a level comparable to some of the SP-responding species. It is unclear why D. virilis respond poorly to SP orthologs. The SP orthologs of the poor-responding species may require additional factors that promote interaction between SP and SPR in vivo. In fact, several seminal proteins are known to play crucial roles in reproduction by regulating the release, stability, and localization of SP within the female reproductive organs (Ram and Wolfner 2009; Wolfner 2009).
Alternatively, SP orthologs in the species outside the D. melanogaster group may be contributing to functions independent of SPR, such as eliciting the innate immune response. Recently, SP has been shown to affect female reproductive behavior in an SPR independent manner, proposing that SP has additional receptors (Haussmann et al. 2013). Conservation of SP within Drosophila species suggests that SP might have other roles in complex reproductive processes through these receptors.
In contrast to the conserved C-terminal region of SP orthologs, the N-terminal region is highly variable among the species (Figure S2A). In D. melanogaster, the N-terminal region has been shown to stimulate juvenile hormone synthesis and is dispensable for PMR induction (Moshitzky et al. 1996). The sequence divergence among the orthologs suggests that the N-terminal part of SP might be involved in the species-specific aspects of the reproductive process.
Supplementary Material
Acknowledgments
We thank the Kyorin-fly, Tokyo, the Drosophila Genetic Resource Center, Kyoto, Japan, and the Bloomington Stock Center for providing the fly stocks. This work was supported by a special grant from the Tokyo Metropolitan Government and KAKENHI (a Grant-in-Aid for Scientific Research) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to T. Aigaki.
Footnotes
Communicating editor: M. F. Wolfner
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177550/-/DC1.
Literature Cited
- Aigaki T., Ohba S., 1984. Individual analysis of age-associated changes in reproductive activity and lifespan of Drosophila virilis. Exp. Gerontol. 19: 13–23. [DOI] [PubMed] [Google Scholar]
- Aigaki T., Fleischmann I., Chen P. S., Kubli E., 1991. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 7: 557–563. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Manoukian A. S., Perrimon N., 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44: 635–654. [DOI] [PubMed] [Google Scholar]
- Chapman T., Bangham J., Vinti G., Seifried B., Lung O., et al. , 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100: 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. S., Stumm-Zollinger E., Aigaki T., Balmer J., Bienz M., et al. , 1988. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54: 291–298. [DOI] [PubMed] [Google Scholar]
- Domanitskaya E. V., Liu H., Chen S., Kubli E., 2007. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 274: 5659–5668. [DOI] [PubMed] [Google Scholar]
- Freeman M., 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87: 651–660. [DOI] [PubMed] [Google Scholar]
- Hanin O., Azrielli A., Zakin V., Applebaum S., Rafaeli A., 2011. Identification and differential expression of a sex-peptide receptor in Helicoverpa armigera. Insect Biochem. Mol. Biol. 41: 537–544. [DOI] [PubMed] [Google Scholar]
- Hanin O., Azrielli A., Applebaum S. W., Rafaeli A., 2012. Functional impact of silencing the Helicoverpa armigera sex-peptide receptor on female reproductive behaviour. Insect Mol. Biol. 21: 161–167. [DOI] [PubMed] [Google Scholar]
- Haussmann I. U., Hemani Y., Wijesekera T., Dauwalder B., Soller M., 2013. Multiple pathways mediate the sex-peptide-regulated switch in female Drosophila reproductive behaviours. Proc. Biol. Sci. 280: 20131938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Bartalska K., Audsley N., Yamanaka N., Yapici N., et al. , 2010. MIPs are ancestral ligands for the sex peptide receptor. Proc. Natl. Acad. Sci. USA 107: 6520–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaes A., Menne T., Stollewerk A., Scholz H., Klambt C., 1994. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell 78: 149–160. [DOI] [PubMed] [Google Scholar]
- Kubli E., 1992. The sex-peptide. BioEssays 14: 779–784. [DOI] [PubMed] [Google Scholar]
- Liu H., Kubli E., 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A., O’Grady P. M., 2005. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu. Rev. Genet. 39: 263–291. [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Baker N. E., Rubin G. M., 1990. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 4: 1848–1861. [DOI] [PubMed] [Google Scholar]
- Moshitzky P., Fleischmann I., Chaimov N., Saudan P., Klauser S., et al. , 1996. Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum. Arch. Insect Biochem. Physiol. 32: 363–374. [DOI] [PubMed] [Google Scholar]
- Nakayama S., Kaiser K., Aigaki T., 1997. Ectopic expression of sex-peptide in a variety of tissues in Drosophila females using the P[GAL4] enhancer-trap system. Mol. Gen. Genet. 254: 449–455. [DOI] [PubMed] [Google Scholar]
- Oh Y., Yoon S.-E., Zhang Q., Chae H.-S., Daubnerová I., et al. , 2014. A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS Biol. 12: e1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Chen S., Busser S., Liu H., Honegger T., et al. , 2005a Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Curr. Biol. 15: 207–213. [DOI] [PubMed] [Google Scholar]
- Peng J., Zipperlen P., Kubli E., 2005b Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 15: 1690–1694. [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H., 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Poels J., Van Loy T., Vandersmissen H. P., Van Hiel B., Van Soest S., et al. , 2010. Myoinhibiting peptides are the ancestral ligands of the promiscuous Drosophila sex peptide receptor. Cell. Mol. Life Sci. 67: 3511–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram K. R., Wolfner M. F., 2009. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proc. Natl. Acad. Sci. USA 106: 15384–15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Schmidt T., Choffat Y., Klauser S., Kubli E., 1993. The Drosophila melanogaster sex-peptide: a molecular analysis of structure-function relationship. J. Insect Physiol. 39: 361–368. [Google Scholar]
- Staehling-Hampton K., Jackson P. D., Clark M. J., Brand A. H., Hoffmann F. M., 1994. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 5: 585–593. [PubMed] [Google Scholar]
- Villella A., Peyre J. B., Aigaki T., Hall J. C., 2006. Defective transfer of seminal-fluid materials during matings of semi-fertile fruitless mutants in Drosophila. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 192: 1253–1269. [DOI] [PubMed] [Google Scholar]
- Wigby S., Chapman T., 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15: 316–321. [DOI] [PubMed] [Google Scholar]
- Wigby S., Domanitskaya E. V., Choffat Y., Kubli E., Chapman T., 2008. The effect of mating on immunity can be masked by experimental piercing in female Drosophila melanogaster. J. Insect Physiol. 54: 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M. F., 2009. Battle and ballet: molecular interactions between the sexes in Drosophila. J. Hered. 100: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Noll M., 2002. Dual role of the Pax gene paired in accessory gland development of Drosophila. Development 129: 339–346. [DOI] [PubMed] [Google Scholar]
- Yamanaka N., Hua Y. J., Roller L., Spalovska-Valachova I., Mizoguchi A., et al. , 2010. Bombyx prothoracicostatic peptides activate the sex peptide receptor to regulate ecdysteroid biosynthesis. Proc. Natl. Acad. Sci. USA 107: 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Rumpf S., Xiang Y., Gordon M. D., Song W., et al. , 2009. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici N., Kim Y. J., Ribeiro C., Dickson B. J., 2008. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451: 33–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.