Abstract
Premature translation-termination codons (PTCs) elicit rapid degradation of the mRNA by a process called nonsense-mediated mRNA decay (NMD). NMD appears to be significantly more efficient for mRNAs of genes belonging to the immunoglobulin superfamily, which frequently acquire PTCs during VDJ rearrangment, than for mRNAs of other genes. To identify determinants for efficient NMD, we developed a minigene system derived from a mouse immunoglobulin μ gene (Ig-μ) and measured the effect of PTCs at different positions on the mRNA level. This revealed that PTCs located downstream of the V–D junction in the VDJ exon of Ig-μ minigenes and of endogenous Ig-μ genes elicit very strong mRNA downregulation, whereas NMD efficiency decreases gradually further upstream in the V segment where a PTC was inserted. Interestingly, two PTCs are in positions where they usually do not trigger NMD (<50 nt from the 3′-most 5′ splice site) still resulted in reduced mRNA levels. Using a set of hybrid constructs comprised of Ig-μ and an inefficient substrate for NMD, we identified a 177 nt long element in the V segment that is necessary for efficient downregulation of PTC-containing hybrid transcripts. Moreover, deletion of this NMD-promoting element from the Ig-μ minigene results in loss of strong NMD.
INTRODUCTION
Quality control mechanisms at different steps of gene expression are important to prevent accumulation of malfunctioning, deleterious proteins in a cell. On the post-transcriptional level, eukaryotic cells possess a translation-dependent quality control system referred to as nonsense-mediated mRNA decay (NMD) or mRNA surveillance that recognizes aberrant mRNAs with premature translation-termination codons (PTCs) and selectively degrades these nonsense mRNAs (1–5). By reducing the steady-state levels of PTC-containing mRNA (hereafter called as PTC+ mRNA), NMD prevents accumulation of C-terminally truncated proteins, which are toxic for cells when they act as dominant-negative inhibitors of the wild-type (wt) protein. Therefore, NMD plays a vital role in improving the fidelity of gene expression to the level required for complex organisms to function properly.
But what are the rules for deciding whether a translation-termination codon is premature (i.e. a PTC) or whether it is the correct physiological stop codon? Analysis of mRNA levels from triose phosphate isomerase (TPI), mouse major urinary protein (MMUP), glutathione peroxidase 1 (GPx1) and β-globin genes with PTCs at many different positions revealed that only stop codons located more than 50–55 nt upstream of the 3′-most exon-exon junction mediate a reduction in mRNA abundance (6–10). This ‘50 nucleotides boundary rule’ for NMD is corroborated further by the finding that nearly all physiological stop codons in the mRNAs of a variety of organisms reside either in the last exon or within the 3′-most 50 nt of the second-last exon (11). It was subsequently discovered that the exon junction complex (EJC), a protein complex that is deposited on the mRNA during splicing ∼22 nt upstream of the exon-exon junction (12,13), provides a binding platform for NMD factors (14). Based on these findings, the current mechanistic models for NMD in mammals (15–18) propose that the ribosome displaces or modifies all EJCs upstream of the stop codon during the first round of translation. It is postulated further that, if there remains a (unmodified) EJC on the mRNA downstream of the stop codon, an interaction between the terminating ribosome and this EJC triggers rapid degradation of the mRNA by an hitherto not known mechanism.
Among the relatively small number of genes for which the effects of PTCs has been systematically investigated, transcripts encoded by genes of the immunoglobulin superfamily differ remarkably from transcripts of other genes in several aspects. For example, steady-state levels of PTC+ T-cell receptor β (TCR-β) transcripts, as well as of PTC+ transcripts encoding immunoglobulin heavy and light chains, are downregulated several fold more efficiently than for example PTC+ β-globin or TPI mRNAs (19). Because PTCs arise very frequently in TCR and immunoglobulin genes as a consequence of programmed V(D)J rearrangements during lymphocyte maturation, whereas somatic mutation leading to PTCs in other genes is relatively a rare event, it is conceivable that specific signals might have evolved in genes of the immunoglobulin superfamily that trigger a particularly efficient mode of NMD to avoid production of truncated TCR and immunoglobulin polypeptide chains. In support of this hypothesis, the VDJ exon together with immediately flanking intron sequences of two differently rearranged TCR-β genes have recently been shown to elicit strong downregulation when inserted into a PTC+ TPI gene (19).
TCR-β transcripts also differ from other mammalian mRNAs in that they violate the ‘50 nucleotides boundary rule’. TCR-β mRNAs with PTCs closer than 50 nt to the 3′-most exon–exon junction are still downregulated, although less efficient than TCR-β mRNAs with PTCs upstream of the boundary (20,21). This indicates that there may exist different modes by which PTCs can decrease steady-state mRNA levels.
To investigate further these extraordinary effects of PTCs on transcripts of the immunoglobulin superfamily, we have developed an Ig-μ minigene system and analyzed the effects of PTCs at many different positions on steady-state mRNA levels. We find that only PTCs located downstream of the V-D junction in the VDJ exon can cause a strong downregulation in the mRNA levels, whereas the extent of mRNA downregulation gradually decreases further upstream where the PTC is located in the V segment. Remarkably and like TCR-β, Ig-μ transcripts do not comply with the ‘50 nucleotides boundary rule’. Furthermore, a 177 nt long sequence element comprising the 5′-half of the VDJ exon could be identified that is necessary for strong downregulation of PTC+ mRNA.
MATERIALS AND METHODS
Cell lines and plasmids
Mouse Ig-μ hybridoma cell lines used in this study are described in (22). The Ig-μ minigene is derived from the plasmid pR-Sp6 described in (23). The intron between VDJ and C1 exons was shortened by fusing the 5′ part of the Ig-μ gene (comprising the VL and VDJ exon, from the translation start codon to 32 nt downstream of the VDJ 5′ splice site) to the 3′ part of the Ig-μ gene (comprising the exons C1–C4, starting from 281 nt upstream of the exon C1 3′ splice site). Exact sequences of all constructs are available on request. Nonsense and frameshift mutations in the Ig-μ minigene were generated by PCR-mediated site-directed mutagenesis using ‘QuikChange® XL Site-Directed Mutagenesis Kit’ (Stratagene). Oligonucleotide sequences used are available on request. PTCs at amino acid positions 32, 57, 73, 310, 440, 452 and 459 were generated by mutations of AAG to TAG (construct Ter32), AAG to TAG (construct Ter57), GGA to TGA (construct Ter73), GGA to TGA (construct Ter310), GAA to TAA (construct Ter440), TCA to TGA (construct Ter452) and TCA to TGA (construct Ter459), respectively. Construct Ter108 was obtained by insertion of an adenosine between codon 104 and 105, generating a −1 frameshift. The Sxl minigene comprising exons 2–4 is based on the TE2343′sspCA sequence described in (24), which contains two PTCs at amino acid positions 43 and 47 in exon3. The two PTCs were changed into Trp and Gln by site-directed mutagenesis using ‘QuikChange® Site-Directed Mutagenesis Kit’ (Stratagene). To keep the open reading frame (ORF) in the hybrid constructs intact, the third nucleotide (guanosine) upstream of the Sxl exon 2 5′ splice site was deleted, and a guanosine, one nucleotide after the Sxl exon 3 3′ splice site was inserted. The various deletions of parts of the Ig-μ VDJ exon were generated by fusion PCR. The 177 bp sequence comprising the NPE was amplified using primers 5′-GACTCCCGGGACTGCCAGGTCCAGCTGCAG-3′ and 5′-GACTCCCGGGTATTAACATTTCCAGGATAAATCCATC-3′ (Ava1 site in italics), or primers 5′-CTGCAGAACCATCCCAATGGCTGCCAGGTCCAGCTGCAG-3′ and 5′-CTGCAGAACCATTGGGATGGGTATTAACATTTCCAGGATAAATCCATC-3′ (BstX1 site in italics), and inserted into the Ava1 or BstX1 site of miniμΔNPE to generate miniμΔNPE/NPEC1 and miniμΔNPE/NPEC3, respectively. The coding parts and any other relevant parts of all constructs used in this study were verified by sequencing.
Cell culture and transfection
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen), supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin (Amimed).
HeLa cells were grown in 6-well plates and transfected at a confluency of ∼80% with 100 ng plasmid DNA, using 3 μl LipofectAmine (Invitrogen). In transient transfection experiments, cells were harvested 48 h post transfection. Polyclonal populations of stably transfected HeLa cells were obtained by selection with 500 μg/ml G418 (Roche) for 2–3 weeks. The results from stably transfected polyclonal cell pools have been reproduced for each construct with at least two independently transfected cell pools.
Real-time RT–PCR
Total cellular RNA (1 μg) isolated with ‘Absolutely RNA™ RT–PCR Miniprep Kit’ (Stratagene) was reverse transcribed in 50 μl Stratascript first strand buffer in the presence of 0.4 mM dNTPs, 300 ng random hexamers, 40 U RNasIn (Promega) and 50 U Stratascript (Stratagene) according to the manufacturer's protocol. For real-time PCR, reverse transcribed material corresponding to 40 ng RNA was amplified in 30 μl Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems) and with the primers and TaqMan probes described below. Ig-μ mRNA and mRNA of hyb3 was measured over the exonC2/exonC3 junction with 800 nM 5′-GTCTCACCTTCTTGAAGAACGTGTC-3′, 800 nM 5′-GGGATGGTGAAGGTTAGGATGTC-3′ and 200 nM 5′-FAM-CACATGTG-CTGCCAGTCCCTCCAC-TAMRA-3′. Sxl mRNA and mRNA of hyb4, hyb6 and hyb8-14 was measured over the Sxl exon 3/exon 4 junction using 400 nM 5′-TTTGAATCGAGGACACCTCCA-3′, 400 nM 5′-TGGCAGAGAATGGGACATCC-3′ and 200 nM 5′-FAM-AGCCCTTGGTGGGCGTGGATTT-TAMRA-3′. Neo mRNA was measured using 800 nM 5′-TGTGACATAATTGGACAAACTACCTACA-3′, 800 nM 5′-CATTCCACCACTGCTC-CCA-3′ and 200 nM 5′-FAM-AGATTTAAAGCTCTAAG-ATTCCAACCTATGGAACTGATGATAMRA-3′. GAPDH mRNA and 18S rRNA were measured using pre-developed assay reagents from Applied Biosystems. For 18S rRNA measurements, 400 pg cDNA instead of 40 ng was used. Real time PCR was run on the GeneAmp 5700 Sequence Detection System (Applied Biosystems) using the standard thermal profile.
Northern blot analysis
Total cellular RNA (15 μg) was separated on a 1.2% agarose gel containing 1× MOPS and 1% formaldehyde. RNA was transferred to positively charged nylon membranes (Roche) in 20× SSC by standard capillary blotting method. Following UV crosslinking of the RNA to the nylon filter, pre-hybridization and hybridization were carried out in 6× SSC, 5× Denhardt's reagent and 0.5% SDS at 60°C. Pre-hybridization was with 50 μg/ml denatured herring sperm DNA and 100 μg/ml denatured calf thymus DNA. For hybridization, 100 ng hyb4+ or miniμ ter310 DNA was labeled with [α-32P]dCTP using the Ready-To-Go™ DNA labeling Kit (Amersham). After overnight hybridization, membranes were washed twice with 2× SSC/0.2% SDS and twice with 0.2× SSC/0.1% SDS at 60°C before exposure to a PhosphorImager screen.
RT–PCR
Total cellular RNA was isolated and reverse transcribed as for real-time RT–PCR, except that 6 ng/μl of oligo 5′-(dT)30VN-3′ or 244 nM of reverse primer 5′-CAGTTGCTCACGAGCTGGTGGC-3′ was used instead of random hexamers. Reverse transcribed material corresponding to 100 ng RNA was amplified with ‘PCR Core Kit’ (Qiagen) in 50 μl PCR buffer containing 200 nM dNTPs, 5 U Taq polymerase and 244 nM forward (5′-GGCTAGTGTTTGTGTGGAAGACTGGAATAAC-3′) and reverse primer to analyze the C3/C4 region. RT–PCR (10 μl) reaction was separated on a 2% agrose gel, and the rest was used to clone into the TOPO vector pCRII (Invitrogen) for subsequent sequencing. For TOPO cloning of the Vleader/VDJ/C1 region in miniμwt and miniμΔNPE mRNA, cDNA was amplified using 300 nM of primers 5′-ACTGCATTGTCGACCTCACCATGGGATGGAG-3′ and 5′-AGCGCCTAGGGAGGTGGCTAGGTACTTGCCCCCTG-3′.
Western blot analysis
Whole cell lysates were electrophoresed on a 10% SDS-PAGE. Protein was transferred to Optitran BA-S 85 reinforced nitrocellulose (Schleicher & Schuell) and probed with a 1:500 dilution of affinity-purified goat anti-mouse IgM (μ chain-specific, Jackson ImmunoResearch Laboratories). The primary antibody was detected with a 1:4000 diluted horseradish peroxidase-conjugated donkey anti-goat IgG antibody (Promega) using ECL+ western blotting detection system (Amersham Pharmacia Biotech).
RESULTS
Efficiency of NMD increases with 5′ to 3′ polarity
For studying in more detail effects of premature termination codons (PTCs) on stability (NMD) and on splicing (NAS) of Ig-μ transcripts, a minigene construct was derived from the productively rearranged mouse Ig-μ gene expressed in the hybridoma cell line Sp6 (23), which produces IgM against the hapten 2,4,6-trinitrophenyl (25). The Ig-μ minigene comprises the leader exon, the VDJ exon and the four exons of the constant region (C1–C4), but lacks the two final exons (M1 and M2) that encode the membrane-binding domain of the heavy chain (Figure 1A). The 5.7 kb long intron between the VDJ and the C1 exon of the endogenous gene was shortened to 307 bp, thereby deleting the μ enhancer and flanking matrix attachment regions. In the expression vector, the Ig-μ minigene is transcribed from the human β-actin promoter. To investigate the efficiency of NMD elicited by PTCs at different positions, Ig-μ minigene constructs with a PTC at amino acid positions 32, 57, 73, 108 and 310 were generated by site-directed mutagenesis (Figure 1A). Relative mRNA levels of these constructs expressed in HeLa cells after transient (Figure 1B and C) or stable transfection (Figure 1D) were measured by real-time PCR using TaqMan chemistry and analyzed by northern blotting (Figure 1C and data not shown). Irrespectively whether relative Ig-μ mRNA levels were determined from transient transfections by normalizing to plasmid-encoded neomycin mRNA (Figure 1B) or from stably transfected polyclonal cell pools by normalizing to endogenous GAPDH mRNA (Figure 1D), the result was very similar, demonstrating that the stably transfected cell pools represent true polyclonal ‘average populations’. The northern blots confirmed that all constructs produced one major mRNA species of the expected size. Over all, the pattern of mRNA reduction in response to a PTC is very similar in the minigene system (Figure 1B–D) and in the hybridoma cells that express from the endogenous promoter the full-length Ig-μ gene with PTCs at different positions (Figure 1E).
Figure 1.
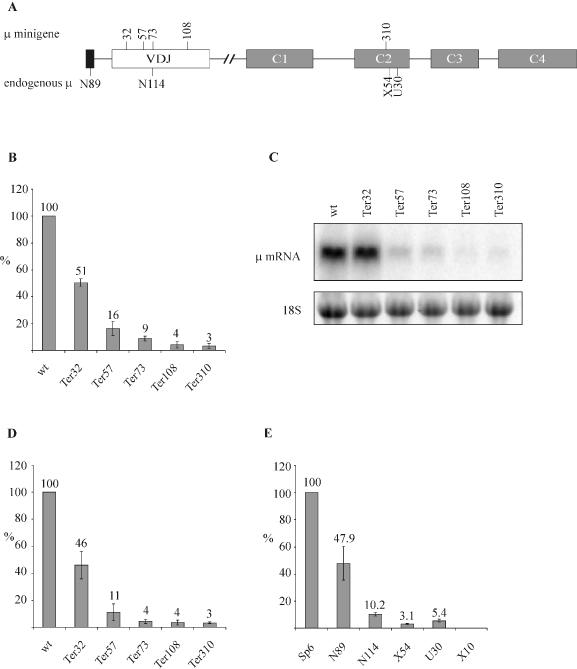
Efficiency of NMD increases with 5′ to 3′ polarity. (A) Schematic representation of the Ig-μ gene, with numbers above depicting the amino acid positions where PTCs were inserted in the μ-minigene. The position of the PTC in the endogenous Ig-μ gene in the different hybridoma cell lines is shown below the diagram. The original clone names of the hybridoma cell lines are used throughout this paper (22). The PTC in cell line N89, N114, X54 and U30 is located at amino acid positions 3, 73, 325 and 335, respectively. (B) Relative Ig-μ mRNA levels of HeLa cells transiently transfected with the indicated Ig-μ minigenes under control of the human β-actin promotor were analyzed 48 h post transfection by real-time RT–PCR. The indicated relative Ig-μ mRNA levels were normalized to neomycin mRNA encoded on the transfected plasmid. ‘wt’ denotes the PTC-free Ig-μ minigene, the various PTC+ constructs are named ‘Ter’ (for termination) followed by a number indicating the amino acid position of the PTC. (C) Analysis of the same RNA used in (B) by northern blotting using a 32P-labeled probe for Ig-μ mRNA. As a loading control, the 18S rRNA band from the ethidium bromide-stained gel before blotting is shown in the lower panel. (D) HeLa cells were transfected with the same Ig-μ minigene constructs as in (B) and stably transfected polyclonal cell pools were generated by selection with Geneticin. Relative Ig-μ mRNA levels were determined by real-time RT–PCR as in (B) and normalized to relative endogenous GAPDH mRNA levels. (E) RNA of the parental hybridoma cell line Sp6, encoding a productively rearranged full-length Ig-μ mRNA, and of five Sp6-derived, mutated cell clones was analyzed. X10 has deleted the entire Ig-μ gene and serves as a negative control. Relative Ig-μ mRNA levels were determined by real-time RT–PCR as in (B) and normalized to relative 18S ribosomal RNA levels. In (B), (D) and (E), average values of three real-time PCR runs with cDNAs of one representative experiment are shown. Error bars indicate standard deviations.
In comparison to other well-studied NMD substrates, PTC+ transcripts of TCR-β, Ig-κ and Ig-μ genes are in general downregulated much more efficiently (19). Here, we find that the mRNAs of Ig-μ minigenes expressed in HeLa cells under the control of the β-actin promoter are as efficiently downregulated as the endogenous Ig-μ mRNAs in the hybridoma cells with a PTC at a corresponding position, suggesting that all signals for efficient downregulation reside in the mRNA and that neither the promoter nor the cell type significantly influences the efficiency of Ig-μ NMD. Most interestingly, the efficiency of NMD depends on the position of the PTC and increases as the PTC is moved further downstream in the Ig-μ gene, as reflected by the decreasing PTC+ mRNA levels. This 5′ to 3′ polarity of NMD efficiency was apparent irrespectively of whether the constructs were transiently transfected (Figure 1B) or whether mRNA of polyclonal populations of stably transfected cells was analyzed (Figure 1D). The same 5′ to 3′ polarity of NMD efficiency was also observed in Sp6-derived hybridoma cell lines expressing Ig-μ genes with PTCs at different positions (22,26). In these cells, the mRNA level of a Ig-μ gene with a PTC at amino acid position 3 (cell line N89) is only 2-fold lower compared with the wt Ig-μ mRNA (cell line Sp6), whereas a PTC at amino acid position 73 (cell line N114) results in a 10-fold reduction of mRNA. PTCs further downstream in the Ig-μ gene at amino acid positions 325 and 335 (cell lines X54 and U30, respectively) reduced the mRNA level even further to 3.1 and 5.4% of wt Ig-μ mRNA, respectively. Cell line X10 has the Ig-μ gene deleted and serves as a negative control. Collectively, our results suggest that only PTCs located downstream from the middle of the VDJ exon have the capacity to elicit maximal Ig-μ mRNA downregulation (see also below).
Testing the ‘50 nucleotides boundary rule’ for PTC+ Ig-μ transcripts
To examine whether PTC+ Ig-μ mRNA also conforms to the ‘50 nucleotides boundary rule’ (11), we determined mRNA levels of stably or transiently transfected Ig-μ minigene constructs with a PTC 67 (Ter440), 31 (Ter452) or 10 nt (Ter459) upstream of the 3′-most exon–exon junction (Figure 2 and data not shown). Ter440 mRNA (Figure 2B) was reduced to a similar extent as Ter73, Ter108 and Ter310 mRNA (Figure 1D), indicating that NMD is still fully functioning just upstream of the ‘50 nucleotides boundary’. When the PTC is located closer than 50 nt to the 3′-most exon–exon junction (Ter452 and Ter459), the mRNA is still downregulated about 3-fold compared with wt Ig-μ mRNA. Thus, Ig-μ shows an intermediate phenotype with regard to the ‘50 nucleotides boundary’: on the one hand, the 3-fold reduced mRNA levels of Ter452 and Ter459 represent a clear violation of the boundary rule. For comparison, in β-globin, TPI, MMUP or GPx1, PTCs downstream of the boundary do not reduce mRNA levels at all (6,7,10,27). On the other hand, Ter452 and Ter459 mRNA levels are downregulated about 4- to 5-fold less than Ter440 mRNA, indicating that the ‘50 nucleotides boundary’ nevertheless has an effect on Ig-μ mRNAs (see Discussion). Northern blot analysis confirmed that the point mutations introduced to generate the PTCs did not cause large alterations of the splicing pattern of the transcripts (Figure 2C). To exclude that the mutations close to the 5′ splice site activated a cryptic 5′ splice site nearby, we examined this region of the transcripts also by RT–PCR. As shown in Figure 2D, the three nonsense mRNAs and the wt Ig-μ mRNA all gave an amplification product corresponding in length to the 163 bp predicted for the correct removal of the C3/C4 intron. Correct joining of exons C3 and C4 during splicing was further confirmed by cloning and sequencing of the RT–PCR products (data not shown).
Figure 2.
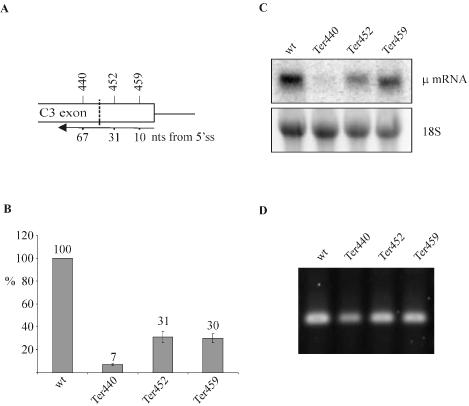
Testing the ‘50 nucleotides boundary rule’ for NMD of Ig-μ minigenes. (A) PTCs were introduced into exon C3 of the Ig-μ minigene at the amino acid positions indicated above the diagram. The numbers below depict the distance of these PTCs from the 3′-most 5′ splice site. The ‘50 nucleotides boundary’ is marked by the dashed line. (B) RNA of polyclonal HeLa cell pools stably transfected with the indicated Ig-μ minigenes under control of the human β-actin promotor were analyzed by real-time RT–PCR. The indicated relative Ig-μ mRNA levels were normalized to endogenous GAPDH mRNA. Average values of three real-time PCR runs of a typical experiment are shown. Error bars indicate standard deviations. (C) Northern blot analysis of RNA harvested 48 h post transfection with a 32P-labeled probe for Ig-μ mRNA. As a loading control, the 18S rRNA band from the ethidium bromide-stained gel before blotting is shown in the lower panel. (D) RT–PCR analysis of the RNA used in (B) to check for potential cryptic splicing between exon C3 and exon C4. Correct joining of exon C3 to exon C4 is predicted to give a PCR product of 163 bp.
The 5′ half of the Ig-μ VDJ exon is essential for strong mRNA downregulation of PTC+ hybrid constructs
As mentioned above, TCR-β and immunoglobulin transcripts, which commonly harbor PTCs as a result of programmed DNA rearrangement during lymphocyte maturation, are downregulated much more effectively in response to PTCs than other known mammalian gene transcripts. This efficient mRNA downregulation appears to depend on signals encoded within the RNA (19). To identify sequence elements that influence the efficiency of PTC+ mRNA downregulation, hybrid genes were generated between sequences of the Ig-μ minigene and of a Drosophila Sex-lethal (Sxl) minigene. The Sxl minigene used here comprises exons 2–4 of the Sxl gene and is derived from TE234 3′ss pCA (24), which contains a mutation in the proximal 3′ splice site of exon 3 and therefore exclusively uses the distal 3′ splice site of exon 3 (24). Notably, this Sxl minigene lacks the sequences that code for the RNA binding domain present in the female-specific functional Sxl protein. Thus, neither the PTC+ nor the PTC-version can interfere with its own pre-mRNA splicing. In HeLa cells, because of the absence of functional Sxl protein, the PTC-containing male-specific exon 3 is always included in the Sxl mRNA. Compared with a construct where the two in-frame stop codons in exon 3 (ter43 and ter47) were removed, the PTC+ Sxl mRNA was downregulated only about 3-fold. In contrast, PTC+ Ig-μ transcripts were downregulated more than 20-fold (Figures 1 and 3A).
Figure 3.
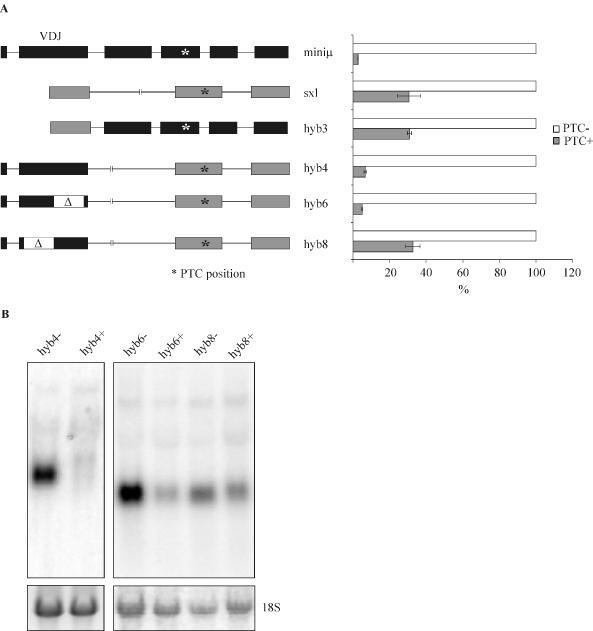
The 5′ half of the Ig-μ minigene VDJ exon is necessary for efficient NMD of Ig-μ/Sxl hybrid transcripts. (A) Relative hybrid mRNA levels of HeLa cells stably expressing the constructs schematically represented on the left were determined by real-time RT–PCR and are shown in the right panel. For each pair of hybrid constructs, the PTC− mRNA level (white bars) was set as 100% and the PTC+ mRNA level (gray bars) was calculated relative to it. The hybrid mRNA values were normalized to endogenous GAPDH mRNA. Average values of three real-time PCR runs of a typical cell pool are shown, and error bars indicate standard deviations. In the left panel, exons originating from Ig-μ are depicted in black and Sxl exons are depicted in gray. The position of the PTC is indicated by an asterisk and corresponds to ter310 in miniμ or ter43 in Sxl, respectively. The white regions labeled with Δ mark deletions. (B) Northern blot analysis of 15 μg of total cellular RNA isolated from HeLa cells stably transfected with the indicated hybrid constructs. As a loading control, the 18S rRNA band from the ethidium bromide-stained gel before blotting is shown in the lower panel.
For the first hybrid constructs, we fused the 5′ half of the Ig-μ minigene (VL and VDJ exons) with the 3′ half of Sxl (exons 3 and 4), resulting in the construct hyb4. Vice versa, the 5′ half of Sxl (exon 2) was fused to the 3′ half of Ig-μ (exons C1–C4), giving the construct hyb3 (Figure 3A). HeLa cells were stably transfected with PTC− or PTC+ versions of these fconstructs, and mRNA levels were determined from polyclonal cell populations. When compared with the corresponding PTC− mRNA, PTC+ mRNA levels of hyb4 were nearly as strongly downregulated as PTC+ mRNA of the parental Ig-μ minigene. In contrast, hyb3 mRNA was only modestly downregulated (∼3-fold) in response to a PTC, similar to the Sxl minigene. This indicates that sequences in the 5′ half of the Ig-μ minigene, comprising VL and VDJ exons, are necessary and sufficient to enhance downregulation of PTC+ mRNA ∼6-fold.
To more precisely map sequences responsible for efficient NMD, we next deleted either the 5′ half or the 3′ half of the VDJ exon in hyb4, giving rise to hyb8 and hyb6, respectively. Because we wanted to avoid alteration of splicing, these deletions did not include nucleotides close to the splice sites. As shown in Figure 3A, PTC+ hyb6 mRNA was downregulated to the same extent as PTC+ mRNA of the parental Ig-μ minigene or hyb4, suggesting that the NMD-promoting activity resides in the 5′ half of the VDJ exon. Consistent with this notion, deletion of the 5′ half of the VDJ exon (hyb8) resulted in loss of strong NMD. In fact, PTC+ hyb8 mRNA was downregulated to almost exactly the same extent as hyb3 and the Sxl minigene, which both lack the Ig-μ VDJ exon. Northern blot analysis revealed one predominant band of the expected size for all hybrid constructs (Figure 3B), suggesting that none of the hybrid constructs induced any unpredicted major alterations in splice site usage. Collectively, these results demonstrate that a sequence of 177 bp, between amino acid positions 19 and 76 of the VDJ exon, possesses an NMD-enhancing activity that is necessary for the efficient downregulation of hyb4.
Smaller deletions within the 5′ half of the Ig-μ VDJ exon did not affect its NMD-promoting activity
Remarkably, the VDJ exons together with some immediately flanking intronic sequences of two differently rearranged TCR-β genes have recently been shown to possess a similar downregulatory-promoting activity (19). Since genes of both gene families more often than not acquire PTCs by the programmed DNA rearrangements during normal lymphocyte development, it seemed reasonable to hypothesize that a common mechanism might have evolved to efficiently downregulate PTC+ TCR-β and immunoglobulin mRNA. A common putative NMD-promoting factor could interact with the target RNA either through a conserved sequence motif or through a conserved secondary structure.
To search for conserved sequence motifs, we aligned the rearranged VDJ exon sequences of several different TCR-β and Ig-μ genes. As expected for sequences encoding the variable regions of their respective proteins, these sequences were rather different and no highly conserved sequence motif was detectable. Yet, we noticed some short stretches of higher conservation in a region corresponding to amino acid positions 52–70 in our Ig-μ gene (Figure 4A, underlined), which maps to the 3′ end of the 177 bp long NMD-promoting element (NPE). However, deletion of these 57 bp from hyb4 and hyb6, giving hyb9 and hyb10, respectively, did not affect the downregulatory activity of the VDJ sequences in the context of the Ig-μ/Sxl hybrid genes (Figure 4B, compare hyb4 with hyb9, and hyb6 with hyb10).
Figure 4.
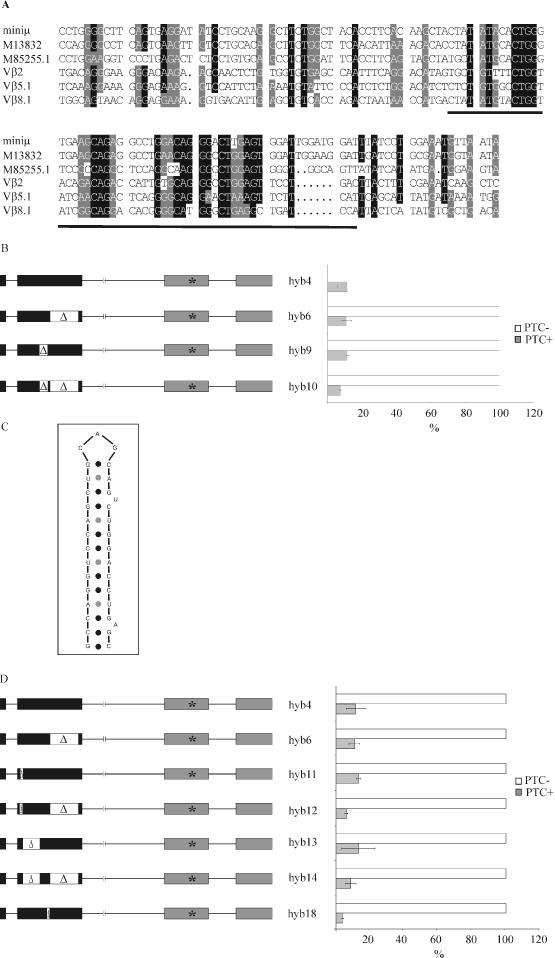
Smaller deletions within the 5′ half of the Ig-μ VDJ exon did not affect its NMD-promoting activity. (A) Alignment of VDJ exon sequences of several different human and mouse Ig-μ and TCR-β genes. ‘Miniμ’ is from the mouse Ig-μ of the hybridoma cell line Sp6 (23). Accession numbers M13832 and M85255.1 are rearranged VDJ regions of mouse and human Ig-μ, respectively. Vβ2 (accession no. X04331), Vβ5.1 (19) and Vβ8.1 (19) are mouse TCR-β sequences. The underlined sequence corresponding to amino acid positions 52–70 in our Ig-μ minigene contains conserved motifs and is deleted in hyb9 and hyb10. (B) Total cellular RNA of HeLa cells transiently transfected with the indicated Ig-μ/Sxl hybrid genes were analyzed 48 h post transfection by real-time RT–PCR. Relative Ig-μ mRNA levels were normalized to neomycin mRNA encoded on the transfected plasmid. Average values of three real-time PCR runs of a typical experiment are shown. Error bars indicate standard deviations. (C) Predicted stem–loop structure located between amino acid positions 19 and 30 of Ig-μ. (D) The 33 bp comprising the predicted stem–loop structure depicted in (C) were deleted from hyb4 and hyb6, giving hyb11 and hyb12, respectively. The 63 bp between the predicted stem–loop and the underlined motif in (A) were deleted from hyb4 and hyb6, giving hyb13 and hyb14, respectively. The 18 bp comprising amino acid positions 71–76 were deleted from hyb4, giving hyb18. Transfection and real-time RT–PCR were as described in (B).
Next, we applied the MFold program (28) to identify predicted secondary structures in the 5′ half of the Ig-μ VDJ exon. A predicted stem–loop structure consisting of a 14 bp stem with two single nucleotide bulges in the descending strand, a CAG loop, and a ΔG of −20.6 kcal mole−1 caught our attention (Figure 4C). This putative secondary structure is located at the 5′ end of the VDJ exon between amino acid 19 and 30, again within the 177 bp element that harbors the NMD-promoting activity. However, no similar secondary structure was predicted by MFold in the TCR-β VDJ exons studied by Gudikote and Wilkinson (data not shown). Nevertheless, we deleted the 33 bp comprising the predicted stem–loop in hyb4 and hyb6, giving hyb11 and hyb12, respectively, and determined the difference in steady-state mRNA from a PTC+ and a PTC− version of each hybrid construct transiently transfected into HeLa cells. As shown in Figure 4D, deletion of the predicted stem-loop sequence did not interfere with the NMD-promoting activity of the VDJ sequences in the context of the Ig-μ/Sxl hybrid genes. Compared with the corresponding PTC− mRNA, PTC+ hyb11 mRNA and PTC+ hyb12 mRNA were downregulated to the same extent than PTC+ hyb4 mRNA and PTC+ hyb6 mRNA, respectively.
Since neither deletion of the 5′-most 33 bp nor deletion of 57 bp in the 3′ end of the NPE interfered with the activity of this element, we next deleted the remaining 63 bp in the middle of the NPE (amino acid positions 31–51) in hyb4 and in hyb6, giving hyb13 and hyb14, respectively. Surprisingly, also this deletion did not interfere with the NMD-promoting activity, and neither did the deletion of the 18 bp comprising amino acid positions 71–76 (Figure 4D). Thus, we conclude that the NMD-promoting activity exerted by the 5′ half of the Ig-μ VDJ exon must consist of at least two redundant determinants.
The 5′ half of the Ig-μ VDJ exon is required for strong downregulation of PTC+ Ig-μ mRNA
The Ig-μ/Sxl hybrid constructs allowed us to map an NMD-promoting activity to the 5′ half of the Ig-μ VDJ exon. Next, we asked whether this sequence element was also necessary for efficient downregulation of mRNA expressed from a PTC+ Ig-μ minigene. To this end, the 177 bp long NPE was deleted from the PTC-free and the ter310 version of the Ig-μ minigene, giving miniμΔNPE− and miniμΔNPE+, respectively. Figure 5A shows the relative Ig-μ mRNA levels in total RNA from polyclonal pools of HeLa cells stably expressing the PTC− or the PTC+ version of miniμwt and miniμΔNPE, respectively. As for the Ig-μ/Sxl hybrid transcripts (Figure 3), deletion of the NPE from miniμ results in the loss of strong NMD: miniμΔNPE+ mRNA was only ∼3-fold reduced compared with miniμΔNPE−, whereas the PTC+ mRNA of the full-length miniμ is downregulated more than 20-fold.
Figure 5.
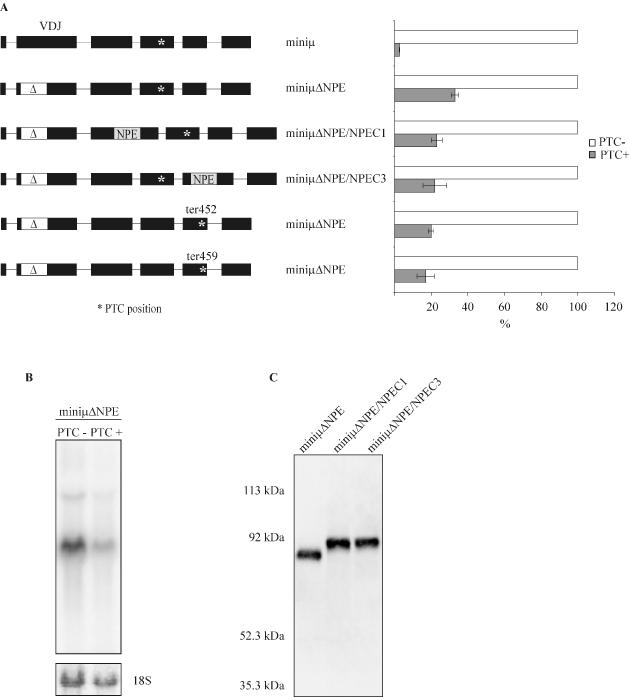
The NMD-promoting element is necessary for efficient NMD of Ig-μ minigene mRNA and functions in a position-dependent manner. (A) Relative Ig-μ mRNA levels of HeLa cells stably expressing the constructs schematically represented on the left were determined by real-time RT–PCR and are shown in the right panel. For all constructs, the PTC− mRNA level (white bars) was set as 100% and the PTC+ (ter310) mRNA level (gray bars) was calculated relative to it. The Ig-μ values were normalized to endogenous GAPDH mRNA. Average values of three real-time PCR runs of one representative cell pool are shown, and error bars indicate standard deviations. The deletion in all miniμΔNPE constructs, indicated by the white region marked with Δ, is identical to the deletion in hyb8 (Figure 3) and comprises 177 bp from amino acid positions 19–76. MiniμΔNPE/NPEC1 and miniμΔNPE/NPEC3 were generated by insertion of the NPE into exon C1 or into exon C3 of the PTC− and the PTC+ version of miniμΔNPE, respectively. (B) Northern blot analysis of 15 μg RNA from the PTC− and PTC+ miniμΔNPE expressing cells analyzed in (A). As a loading control, the 18S rRNA band from the ethidium bromide-stained gel before blotting is shown in the lower panel. (C) Detection of the Ig-μ polypeptide in lysate of cells expressing the PTC− version of the indicated constructs by western blotting confirms the intactness of the respective ORFs in these mRNAs. MiniμΔNPE/NPEC1 and miniμΔNPE/NPEC3 encode for a 593 amino acids long polypeptide, the polypeptide encoded by miniμΔNPE is 59 amino acids shorter.
Next, we re-inserted into miniμΔNPE the NPE into the C1 or the C3 exon in order to test if the NPE can also function from a different position within the mRNA. Although the PTC+ Ig-μ mRNA levels appeared to be reduced slightly more in these two constructs compared with miniμΔNPE, downregulation of these PTC+ mRNAs was still much less efficient than when the NPE is located in its correct place in the 5′-half of the VDJ exon (Figure 5A). We conclude that NPE function is position-dependent and that the NPE is inactive when placed in the constant region of the Ig-μ gene.
Finally, we wanted to know whether the NPE was required for the reduced Ig-μ mRNA levels observed in constructs with a PTC downstream of the 50 nt boundary (Figure 2). Therefore, we inserted into miniμΔNPE a PTC 31 (Ter452) or 10 nucleotides (Ter459) upstream of the 3′-most 5′ splice site and measured the relative mRNA levels of these constructs stably expressed in polyclonal HeLa cell pools. As shown in Figure 5A, even in the absence of the NPE, Ter452 and Ter459 still led to ∼5-fold reduced mRNA levels compared with the PTC− miniμΔNPE. Thus, violation of the 50 nt boundary rule by PTC+ Ig-μ transcripts does not require the NPE.
To confirm that deletion of the NPE did not alter splicing of the miniμΔNPE transcripts, we analyzed the PTC− and the PTC+ miniμΔNPE mRNA by northern blotting and sequenced the cDNA region comprising Vleader, VDJ and C1 exons of miniμ and miniμΔNPE. Northern blotting gave a single major band of the expected size for miniμ ΔNPE mRNA (Figure 5B), and cDNA sequencing confirmed correct splicing of the VDJ exon to its neighbouring exons (data not shown). In addition, intactness of the ORF in the PTC− versions of miniμΔNPE, miniμΔNPE/NPEC1 and miniμΔNPE/NPEC3 mRNA was confirmed by detecting the Ig-μ polypeptide by western blotting (Figure 5C). In accordance with miniμΔNPE lacking 59 amino acids compared with miniμΔNPE/NPEC1 and miniμΔNPE/NPEC3, the polypeptide encoded by miniμΔNPE migrates slightly faster than the two other polypeptides.
In summary, we conclude that the 177 bp long sequence in the 5′ half of the VDJ exon is a bona fide NMD-promoting element that is required for efficient downregulation of PTC+ mRNA both in the heterologous context of the hybrid constructs as well as in the Ig-μ minigene. Furthermore, we showed that the NPE functions in a position-dependent manner and that it is not required for NMD elicited by PTCs downstream of the 50 nt boundary.
DISCUSSION
The Ig-μ minigene system described and analyzed here behaves very similar to the parental, endogenous Ig-μ gene with respect to the relative abundance of mRNAs bearing PTCs at different positions. Because the minigene was expressed in HeLa cells under the control of the human β-actin promoter, this result demonstrates that the major determinants for the efficiency of PTC-mediated mRNA downregulation reside within the mRNA sequence, and that the position of the PTC within the mRNA is an important determinant for the efficiency of NMD. Although similar observations were made previously for TCR-β minigenes (19), NMD of β-globin was abrogated when the gene was expressed from the thymidine kinase promoter of herpes simplex virus type 1 (29). Since NMD of neither Ig-μ nor TCR-β genes expressed from this viral promoter has been analyzed, we cannot exclude that certain promoters could affect NMD efficiency of Ig-μ or TCR-β.
When we analyzed the extent of mRNA reduction caused by PTCs at different positions in Ig-μ, we found that downregulation of the mRNA levels of both the endogenous gene and the minigene gradually increased with increasing distance of the PTC from the 5′ end of the transcript until to the middle of the VDJ exon (22,26,30). PTCs located downstream of this position gave maximal downregulation (>20-fold). Interestingly, the boundary for maximal downregulation appears to be between amino acid positions 73 and 108, which overlaps with the position where the V-segment was joined to the D-segment during B-cell maturation (amino acid position 86/87). Since out-of-frame VDJ-rearrangements are the major source for PTCs in immunoglobulins, it would make sense that signals to ensure efficient downregulation of such non-productively rearranged transcripts would have evolved to act on PTCs downstream of the V–D junction. Possible mechanisms by which the NMD-promoting sequence element identified in this study might function to specifically enhance downregulation of PTCs downstream of the V–D junction are discussed below.
Alternatively, inefficient NMD of Ig-μ mRNAs with PTCs close to the 5′ end could also result from translation re-initiation at an in-frame start codon downstream of the PTC, as was reported for TPI transcripts (8). For Ig-μ expressed in hybridoma cells, translation re-initiation has also been reported to occur at codon 100 (Met100) with an efficiency of ∼4% the normal rate when a PTC is located at amino acid position 3 (30). However, translation re-initiation at Met100 cannot readily explain why upstream PTCs at different positions cause a gradual 5′-to-3′ increase in mRNA downregulation, ranging from 2-fold (Ter32) to 25-fold (Ter73) for Ig-μ minigenes, and from 2-fold (N89, ter3) to 10-fold (N114, ter73) for endogenous Ig-μ genes (Figure 1). Furthermore, we were not able to detect in Ter32, Ter57 or Ter73-expressing cells an Ig-μ polypeptide corresponding in size to re-initiation at Met100, whereas we could detect this polypeptide as reported previously (30) in N89, which we used as our positive control (data not shown). Therefore, we favor the hypothesis that the NPE identified in this study is fully effective only when the PTC is located downstream of it, and that its effect gradually decreases further upstream where a PTC is located within the NPE.
According to the current models, NMD is triggered by an interaction between a ribosome at the translation–termination codon and components of the EJC, of which the 3′-most is located 22 nt upstream of the 3′-most exon–exon junction (12). Taking into account the sizes of the ribosome and the EJC, one would predict that translation-termination codons <20–40 nt upstream of the 3′-most EJC should not trigger NMD. This prediction is also supported by the empirically determined ‘50 nucleotides boundary rule’ (11). In partial agreement with this prediction, PTC+ Ig-μ mRNA levels increased ∼4- to 5-fold when the PTC was located downstream of this boundary. But it was unexpected that the PTCs located only 34 and 10 nt upstream of the 3′-most exon-exon junction still led to 3- to 5-fold reduced mRNA levels (Figure 2). This suggests that either some PTC+ Ig-μ mRNA downregulation occurs independently of the EJC, or that additional EJC components bind the Ig-μ mRNA in the final exon. Importantly, PTCs in the endogenous Ig-μ gene behave very similar, based on Ig-μ mRNA quantifications in a large set of mutated Sp6 hybridoma cell lines (31). In that study, Baumann and colleagues measured 3- to 10-fold reduced Ig-μ mRNA levels in hybridoma cells expressing Ig-μ genes with PTCs located 10 nt upstream of or across the 3′-most exon-exon junction. As mentioned earlier, in addition to Ig-μ, TCR-β mRNAs with PTCs closer than 50 nt to the 3′-most exon–exon junction are also still downregulated 2- to 5-fold compared with PTC-TCR-β mRNA (20,21), suggesting that this ‘boundary-independent’ effect of PTCs might be part of the very efficient PTC+ mRNA downregulation response typical for genes belonging to the immunoglobulin superfamily (see below). However, in contrast to the gradual decrease in NMD efficiency with decreasing distance to the 3′-most exon-exon junction reported for TCR-β transcripts (21), NMD efficiency of Ig-μ transcripts appears to sharply decrease when the PTC is located beyond the 50 nt boundary (Figure 2). This difference between TCR-β and Ig-μ might reflect a difference in the molecular mechanisms responsible for ‘Super-NMD’ of transcripts of these two genes, but more work is required to clarify this point.
To identify cis-acting signals in Ig-μ that are responsible for the very strong NMD response, we have generated and analyzed a large set of hybrid PTC+ and PTC− constructs comprising parts of the efficient NMD-substrate Ig-μ and the poor NMD-substrate Sxl (Figures 3 and 4). This approach led to the identification of a 177 bp long sequence of Ig-μ (amino acid positions 19–76), essentially corresponding to the 5′ half of the VDJ exon, which is necessary (hyb8 and miniμ ΔNPE) and sufficient (hyb6) to increase the NMD efficiency by a factor of 4- to 7-fold. Further support for the requirement of the NPE for efficient NMD of Ig-μ mRNAs comes from our analysis of an alternatively spliced Ig-μ transcript that is produced at very low level (M.Bühler and O.Mühlemann, unpublished observation). This Ig-μ alt-mRNA, resulting from joining the 5′ splice site of the leader exon to an alternative 3′ splice site at amino acid position 96 in the VDJ exon, lacks the NPE and generates a frameshift that leads to a PTC further downstream in the mRNA (data not shown). Interestingly in the context of the study here, introduction of a frameshift mutation into the Ig-μ minigene (construct Ter108) that removes the PTC in alt-mRNA results only in a moderate increase of alt-mRNA between 1.6- and 3-fold (data not shown), which is similar to the 3-fold reduction in mRNA caused by the PTC in hyb8 and miniμ ΔNPE, but much less than the over 20-fold downregulation observed for NPE-containing mRNAs. We noticed that all tested PTCs that elicit strong NMD are located downstream of the NPE, which may indicate that the NPE can only promote NMD of PTCs located downstream of it (see above). The fact that all tested partial deletions within the NPE did not affect NMD efficiency suggests that this 177 bp long sequence contains at least two separate, functionally redundant motifs with NMD-promoting activity.
Since two different TCR-β VDJ exons (including immediately flanking intronic sequences) were also recently found to promote PTC+ mRNA downregulation in a heterologous context (19), it is tempting to speculate that a common mechanism would function through conserved cis-acting signals in Ig-μ and TCR-β transcripts. However, except for a short sequence with some conserved motifs (Figure 4A) that is dispensable for NMD-promoting activity (Figure 4B), we could not detect any conserved sequence motifs or predicted secondary structures. Thus, we postulate that different RNA-binding factors with different sequence-specificity can promote NMD by interacting through a conserved domain with a common NMD factor. Alternatively, interaction of a NMD-promoting trans-acting factor with the transcript might be position-specific rather than sequence-specific. Both explanations would be consistent with the evidence for the existence of multiple different NMD-promoting signals in the 5′-half of the Ig-μ VDJ exon, but position-dependence alone cannot explain why such NMD-promoting activities are only found in Ig-μ and TCR-β mRNA, but not in other transcripts investigated so far.
In the case of TCR-β, the downregulation-promoting VDJ exon also functioned when placed between the C2.1 and the C2.2 exon, provided that the PTC was located downstream of it (19). In contrast, when we shifted the NPE of Ig-μ and placed it upstream (exon C1) or downstream (exon C3) of the PTC (ter310 in exon C2), NPE activity was almost completely lost. Thus, unlike TCR-β, the NPE identified in our Ig-μ minigene appears to function in a position-dependent manner.
Apart from the differences discussed above, obvious similarities between TCR-β (19) and Ig-μ (this study) concerning the requirements for efficient NMD should also be pointed out. In both genes, the NMD-promoting element maps to the VDJ exon and appears to promote downregulation of mRNAs only when the PTC is located downstream of the element. Furthermore, TCR-β and Ig-μ transcripts are to the best of our knowledge the only known examples where PTCs downstream of the ‘50 nucleotides boundary’ still cause a reduction in steady-state mRNA levels (20,21), suggesting a mode of PTC-mediated mRNA downregulation that is EJC-independent. However, for both TCR-β and for Ig-μ, this downregulation of mRNAs with PTCs downstream of the ‘50 nucleotides boundary’ is significantly less efficient than when the PTC is located upstream of the boundary. Based on these observations, we propose that the highly efficient downregulation of PTC+ TCR-β and Ig-μ mRNAs is the net result of (i) ordinary EJC-dependent NMD plus (ii) an additional PTC-specific process that recognizes PTCs independently of their relative position to EJC. We provide here evidence that the NPE of Ig-μ enhances the efficiency of ordinary NMD, but does not seem to influence the postulated additional, EJC-independent process (see Figures 2B and 5A). The challenge with this hypothesis of two distinct downregulatory processes will be to explain how a PTC can be distinguished from the physiological termination codon in the EJC-independent process. In our future studies, we will also try to identify factors that interact with the NMD-promoting element in order to gain insight into the molecular mechanism by which this sequence element contributes to the highly efficient downregulation of PTC+ Ig-μ mRNA levels.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Marc Shulman and Juan Valcarcel for providing cell lines and plasmids, respectively. This work was financed by the Swiss National Research Foundation (grant 3100-61720.00 and 3100A0-102159), the Kanton Bern, the Novartis Foundation for Biomedical Research and the Helmut Horten Foundation.
REFERENCES
- 1.Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- 3.Maquat L.E. (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev. Mol. Cell Biol., 5, 89–99. [DOI] [PubMed] [Google Scholar]
- 4.Mendell J.T. and Dietz,H.C. (2001) When the message goes awry. Disease-producing mutations that influence mRNA content and performance. Cell, 107, 411–414. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz C.J., Wang,W. and Peltz,S.W. (2001) Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev., 15, 2781–2785. [DOI] [PubMed] [Google Scholar]
- 6.Belgrader P. and Maquat,L.E. (1994) Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol. Cell. Biol., 14, 6326–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J., Belgrader,P., Zhou,X. and Maquat,L.E. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J. and Maquat,L.E. (1997) Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J., 16, 826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998) Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- 12.Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Hir H., Moore,M.J. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 14.Le Hir H., Gatfield,D., Izaurralde,E. and Moore,M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lykke-Andersen J. (2001) mRNA quality control: marking the message for life or death. Curr. Biol., 11, R88–R91. [DOI] [PubMed] [Google Scholar]
- 16.Wilusz C.J., Wormington,M. and Peltz,S.W. (2001) The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell Biol., 2, 237–246. [DOI] [PubMed] [Google Scholar]
- 17.Schell T., Kulozik,A.E. and Hentze,M.W. (2002) Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol., 3, REVIEWS1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreyfuss G., Kim,V.N. and Kataoka,N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol., 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 19.Gudikote J.P. and Wilkinson,M.F. (2002) T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J., 21, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter M.S., Li,S. and Wilkinson,M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Gudikote,J.P., Olivas,O.R. and Wilkinson,M.F. (2002) Boundary-independent polar nonsense-mediated decay. EMBO Rep., 15, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connor A., Wiersma,E. and Shulman,M.J. (1994) On the linkage between RNA processing and RNA translatability. J. Biol. Chem., 269, 25178–25184. [PubMed] [Google Scholar]
- 23.Ochi A., Hawley,R.G., Hawley,T., Shulman,M.J., Traunecker,A., Kohler,G. and Hozumi,N. (1983) Functional immunoglobulin M production after transfection of cloned immunoglobulin heavy and light chain genes into lymphoid cells. Proc. Natl Acad. Sci. USA, 80, 6351–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penalva L.O., Lallena,M.J. and Valcarcel,J. (2001) Switch in 3′ splice site recognition between exon definition and splicing catalysis is important for sex-lethal autoregulation. Mol. Cell. Biol., 21, 1986–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler G. and Milstein,C. (1976) Derivation of specific antibody-producing tissue culture and tumor lines by cell fusion. Eur. J. Immunol., 6, 511–519. [DOI] [PubMed] [Google Scholar]
- 26.Muhlemann O., Mock-Casagrande,C.S., Wang,J., Li,S., Custodio,N., Carmo-Fonseca,M., Wilkinson,M.F. and Moore,M.J. (2001) Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell, 8, 33–43. [DOI] [PubMed] [Google Scholar]
- 27.Sun X., Moriarty,P.M. and Maquat,L.E. (2000) Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J., 19, 4734–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enssle J., Kugler,W., Hentze,M.W. and Kulozik,A.E. (1993) Determination of mRNA fate by different RNA polymerase II promoters. Proc. Natl Acad. Sci. USA, 90, 10091–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buzina A. and Shulman,M.J. (1999) Infrequent translation of a nonsense codon is sufficient to decrease mRNA level. Mol. Biol. Cell, 10, 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumann B., Potash,M.J. and Kohler,G. (1985) Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J., 4, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]


