Summary
Discrimination among pathogenic and beneficial microbes is essential for host organism immunity and homeostasis. Here, we show that chemosensory detection of two secondary metabolites produced by Pseudomonas aeruginosa modulates a neuroendocrine signaling pathway that promotes avoidance behavior in the simple animal host Caenorhabditis elegans. Secondary metabolites phenazine-1-carboxamide and pyochelin activate a G protein-signaling pathway in the ASJ chemosensory neuron pair that induces expression of the neuromodulator DAF-7/TGF-β. DAF-7, in turn, activates a canonical TGF-β signaling pathway in adjacent interneurons to modulate aerotaxis behavior and promote avoidance of pathogenic P. aeruginosa. Our data provide a chemical, genetic, and neuronal basis for how the behavior and physiology of a simple animal host can be modified by the microbial environment, and suggest that secondary metabolites produced by microbes may provide environmental cues that contribute to pathogen recognition and host survival.
Keywords: host-microbe interactions, Caenorhabditis elegans, Pseudomonas aeruginosa, TGF-β signaling, neuropeptides, pathogen avoidance behavior, bacterial secondary metabolites
Introduction
The recognition of microbial pathogens and the corresponding danger they represent is essential for the survival of host organisms. Innate immune systems have evolved to respond to foreign structures derived from microbes, which help the host distinguish microbes from self. However, such molecular patterns do not necessarily help the host discriminate a microbe that is pathogenic from one that is commensal. Microbial molecular patterns such as lipopolysaccharide are found on pathogen and commensal alike. In view of the diversity of olfactory receptors present even in relatively simple host organisms, chemosensory responses offer the potential to detect a far greater set of relevant microbial molecules. Candidate receptors and specific bacterial cues that modulate host physiology and avoidance behavior have begun to be explored (Pradel et al., 2007; Rivière et al., 2009; Stensmyr et al., 2012).
The soil dwelling nematode Caenorhabditis elegans is a simple host organism that forages on decomposing organic matter for bacterial food (Félix and Duveau, 2012). The presence of bacterial food affects diverse behaviors of C. elegans such as feeding, locomotion, thermotaxis, and aerotaxis (Avery and Horvitz, 1990; Chang et al., 2006; Hedgecock and Russell, 1975; Sawin et al., 2000), and differences in the species composition of the food supply can alter aspects of their physiology and behavior (Gusarov et al., 2013; MacNeil et al., 2013; Shtonda and Avery, 2006; Watson et al., 2014). Pathogenic bacteria kill C. elegans and induce an aversive learning response (Zhang et al., 2005) that promotes protective behavioral avoidance (Chang et al., 2011; Pradel et al., 2007; Pujol et al., 2001; Reddy et al., 2009). Bacterial molecules including Serratia marcescens serrawettin and Pseudomonas aeruginosa quorum-sensing regulators have been implicated in the behavioral avoidance of bacterial lawns (Beale et al., 2006; Pradel et al., 2007). As such, C. elegans is emerging as a useful model for dissecting the genetic and biochemical mechanisms underlying microbial discrimination in animal hosts.
In this study we focus on how the DAF-7/TGF-β pathway functions in chemosensory neurons of C. elegans to regulate behavior in response to changes in the microbial environment. DAF-7 functions in the neuroendocrine regulation of diverse aspects of organismal development and physiology, including the dauer developmental decision, foraging and aggregation behaviors, quiescence, metabolism, and longevity (de Bono et al., 2002; Gallagher et al., 2013; Greer et al., 2008; Milward et al., 2011; Shaw et al., 2007; Swanson and Riddle, 1981). In addition, another C. elegans TGF-β-family ligand, DBL-1, has been shown to regulate olfactory aversive learning and antifungal defenses (Zhang and Zhang, 2012; Zugasti and Ewbank, 2009). Expression of daf-7 is limited to the ASI pair of chemosensory neurons and has been shown to respond to the availability of bacterial food (Ren et al., 1996; Schackwitz et al., 1996). Here, we show that chemosensory recognition of the pathogenic bacterium P. aeruginosa dramatically alters the neuronal expression pattern of DAF-7 to promote avoidance behavior. Through a forward genetic screen we identify conserved components of G protein signaling that act cell-autonomously in the ASJ neurons to activate transcription of daf-7 in response to P. aeruginosa. Finally, we show that the ASJ neurons respond to the secondary metabolites phenazine-1-carboxamide and pyochelin secreted by P. aeruginosa during stationary phase. Our findings demonstrate how specific bacteria can exert effects on host behavior and physiology, and point to how secondary metabolites may serve as environmental cues that contribute to pathogen discrimination and avoidance.
Results
DAF-7/TGF-β is required for behavioral avoidance of Pseudomonas aeruginosa
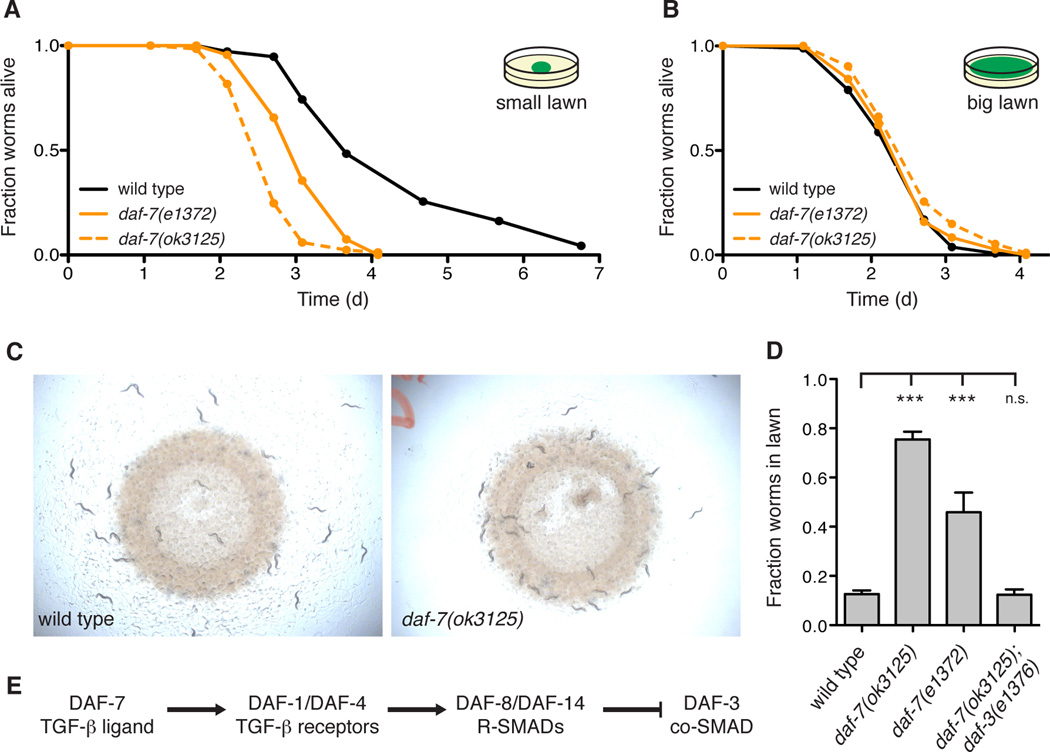
We previously observed that mutants defective in the DAF-7/TGF-β pathway display enhanced susceptibility to infection by P. aeruginosa (Reddy et al., 2009). In the standard pathogenesis assay, daf-7 mutants die faster than the wild-type strain N2 and exhibit a failure to avoid the bacterial lawn (Figures 1A and 1C). Initially, animals begin inside the lawn, but by 15 h wild-type N2 animals avoid the lawn of pathogenic bacteria, while daf-7 mutants remain inside the lawn of P. aeruginosa (Figures 1C and 1D). In a modified pathogenesis assay in which C. elegans are unable to avoid the lawn of pathogenic bacteria, daf-7 mutants display the same susceptibility phenotype as wild-type animals, demonstrating that the failure to avoid P. aeruginosa is the principal determinant in the susceptibility to infection of daf-7 animals (Figure 1B). We also confirmed that daf-7 animals were indeed being exposed to a higher dose of pathogenic bacteria by repeating the experiment with a P. aeruginosa lawn containing red fluorescent beads that serve as markers of the bacterial load in the intestine (Figure S1A).
Figure 1. DAF-7/TGF-β is required for the protective behavioral avoidance response to P. aeruginosa.
(A) Fraction animals alive after being transferred to plates seeded with P. aeruginosa strain PA14. (B) Fraction animals alive after being transferred to a “Big Lawn” of P. aeruginosa in which the bacteria has been spread to the edges of the plate. All data points represent the average of at least three independent replicates. (C) Photographs of wild-type and daf-7 animals after 15 h exposure to P. aeruginosa. (D) Lawn occupancy of animals on P. aeruginosa after 15 h. (E) The canonical DAF-7/TGF-β signaling pathway in C. elegans. *** P < 0.001 as determined by one-way ANOVA followed by Dunnett’s Multiple Comparison Test. n.s. = not significant. Values represent means of at least three independent experiments. Error bars indicate standard error. See also Figure S1.
Once secreted, DAF-7 binds to the TGF-β type I receptor DAF-1 and the TGF-β type II receptor DAF-4, which then act to antagonize the co-SMAD DAF-3 (Figure 1E) (Estevez et al., 1993; Georgi et al., 1990; Patterson et al., 1997). Other mutants in the DAF-7/TGF-β-signaling pathway such as daf-1 and the R-SMAD daf-8 also display P. aeruginosa avoidance defects (Figure S1B), and we observed complete suppression of the daf-7(ok3125) avoidance defect by a mutation in daf-3 (Figure 1D). These results indicate that the DAF-7 pathway is specifically required for avoidance of P. aeruginosa, functioning through the canonical DAF-3 signaling pathway to promote survival by limiting host exposure to pathogenic bacteria.
P. aeruginosa induces daf- 7 expression in the ASJ neuron pair and promotes avoidance behavior
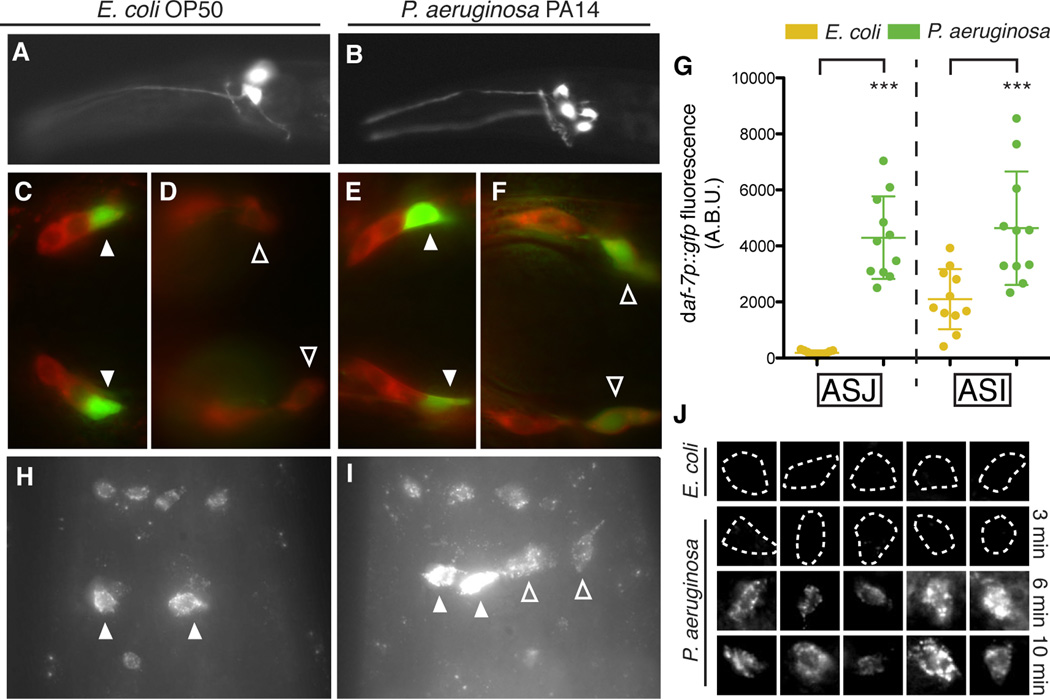
The TGF-β ligand daf-7 was previously shown to be expressed exclusively in the ASI pair of ciliated chemosensory neurons, with expression levels responsive to changes in the availability of standard E. coli food and C. elegans pheromones (Ren et al., 1996; Schackwitz et al., 1996). To monitor daf-7 expression in the presence of P. aeruginosa we used a transgenic strain carrying a daf-7p∷gfp transcriptional reporter. Unexpectedly, we observed that exposure to P. aeruginosa induced fluorescence in four cells—the ASI neuron pair as well as a second bilaterally symmetric pair of ciliated chemosensory neurons (Figures 2A–2B). Through co-localization experiments with the lipophilic dye DiI, we identified the additional cells as the ASJ chemosensory neurons (Figures 2C–2F). We quantified the daf-7p∷gfp fluorescence increase in the ASJ neurons and found that the reporter was induced at least 1000-fold on P. aeruginosa relative to E. coli (Figure 2G). We also observed that the daf-7p∷gfp fluorescence in the ASI neurons increased 2-fold on P. aeruginosa (Figure 2G).
Figure 2. daf-7 expression is induced in the ASJ neurons upon exposure to P. aeruginosa.
(A–B) daf-7p∷gfp expression pattern in C. elegans on E. coli (A) and P. aeruginosa (B). (C–F) Co-localization of daf-7p∷gfp expression and DiI staining (red) in animals on E. coli, dorsal view (C) and ventral view (D), and in animals on P. aeruginosa, dorsal view (E) and ventral view (F). Filled triangles indicate ASI neurons; empty triangles indicate ASJ neurons. (G) Maximum fluorescence values of daf-7p∷gfp in the ASJ or ASI neurons after 16 h exposure to indicated bacteria. *** P < 0.001 as determined by one-way ANOVA followed by Tukey’s Multiple Comparison Test. Error bars indicate standard deviation. (H–I) daf-7 FISH in wild-type N2 animals on E. coli (H) and P. aeruginosa (I). Filled triangles indicate ASI neurons; empty triangles indicate ASJ neurons. Additional cells expressing daf-7 mRNA are the OLQ neurons (top) and the ADE neurons (bottom). (J) daf-7 FISH in the ASJ neurons (co-localized with trx-1p∷gfp) on E. coli and P. aeruginosa after various exposure times. Dashed lines indicate cell boundaries. Each image represents an individual animal. See also Figure S2.
We used fluorescent in situ hybridization of mRNA molecules to corroborate our observations made with the daf-7p∷gfp transcriptional reporter. We did not detect fluorescence in the ASJ neuron pair when the laboratory wild-type strain N2 was propagated on E. coli OP50 (Figures 2H and 2J). In contrast, upon exposure of C. elegans to P. aeruginosa, we observed daf-7 mRNA expression in two additional cells corresponding to the ASJ neuron pair (Figures 2I and 2J). Confirming the specificity of the fluorescence for daf-7 mRNA, we did not detect daf-7 mRNA in a daf-7(ok3125) mutant strain carrying a 664 bp deletion, using probes designed to hybridize exclusively to the sequence within the deletion (Figure S2A). Of note, we observed that when the wild-type strain was propagated on E. coli, in addition to previously described expression in the ASI neuron pair, daf-7 mRNA was also present in six other sensory neurons, which we identified using reporter co-localization experiments as the ADE neuron pair and the OLQ neurons (Figures 2H–2I and S2B–S2G). ADE and OLQ are mechanosensory neurons that have been implicated in bacterial food sensing (Hart et al., 1995; Sawin et al., 2000), and likely represent additional previously unknown sites of DAF-7/TGF- β expression.
To follow the kinetics of the endogenous daf-7 transcriptional response to P. aeruginosa in the ASJ neurons, we fixed animals following exposures to P. aeruginosa ranging in duration from 3 min to 24 h and probed for daf-7 mRNA. On E. coli or following a 3 min exposure to P. aeruginosa we did not detect daf-7 mRNA in the ASJ neurons (Figure 2J). However after a 6 min exposure to P. aeruginosa daf-7 mRNA was present in the ASJ neurons and did not appear to increase further over time (Figure 2J). The rapid kinetics of this transcriptional response, far faster than the kinetics of intestinal infection or aversive learning behavior (Tan et al., 1999; Zhang et al., 2005), led us to hypothesize that the ASJ chemosensory neurons may be responding directly to specific P. aeruginosa cues.
We determined that transgenic overexpression of daf-7 under the native daf-7 promoter or the ASJ-specific trx-1 promoter, as well as under the ASI-specific str-3 promoter, were each sufficient to restore wild-type pathogen avoidance and susceptibility in the daf-7(ok3125) mutant background (Figures 3A and 3C), consistent with the activity of DAF-7 as a secreted ligand. We also observed that the genetic ablation of the ASJ neuron pair conferred a partial deficit in P. aeruginosa avoidance and susceptibility (Figures 3B and 3D), demonstrating that the ASJ neurons are necessary for the complete protective response to P. aeruginosa. To determine if the ASJ-ablation avoidance phenotype was due to a loss of DAF-7 secretion from those neurons, we introduced mutations in the downstream co-SMAD daf-3 that is epistatic to daf-7 (Figure 1E). Mutant daf-3 alleles were able to partially suppress the avoidance defect of both ASJ-ablation lines, consistent with the ASJ-ablation phenotype being due in part to a loss of P. aeruginosa-induced DAF-7 secretion (Figure 3D). These data establish a functional role for the induction of daf-7 in the ASJ neuron pair upon exposure to P. aeruginosa.
Figure 3. DAF-7 from the ASJ neuron pair signals to the RIM/RIC interneurons to alter aerotaxis behavior and promote avoidance of P. aeruginosa.
(A–B) Fraction animals alive after being transferred to plates seeded with P. aeruginosa. All data points represent the average of at least three independent replicates. (C–F) Lawn occupancy of animals on P. aeruginosa after 15 h. *** P < 0.001, ** P < 0.01, * P < 0.05 as determined by one-way ANOVA followed by Dunnett’s Multiple Comparison Test. n.s. = not significant. Error bars indicate standard deviation. See also Figure S3.
DAF-7 signals to the RIM/RIC interneurons adjacent to the ASJ neurons to promote pathogen avoidance
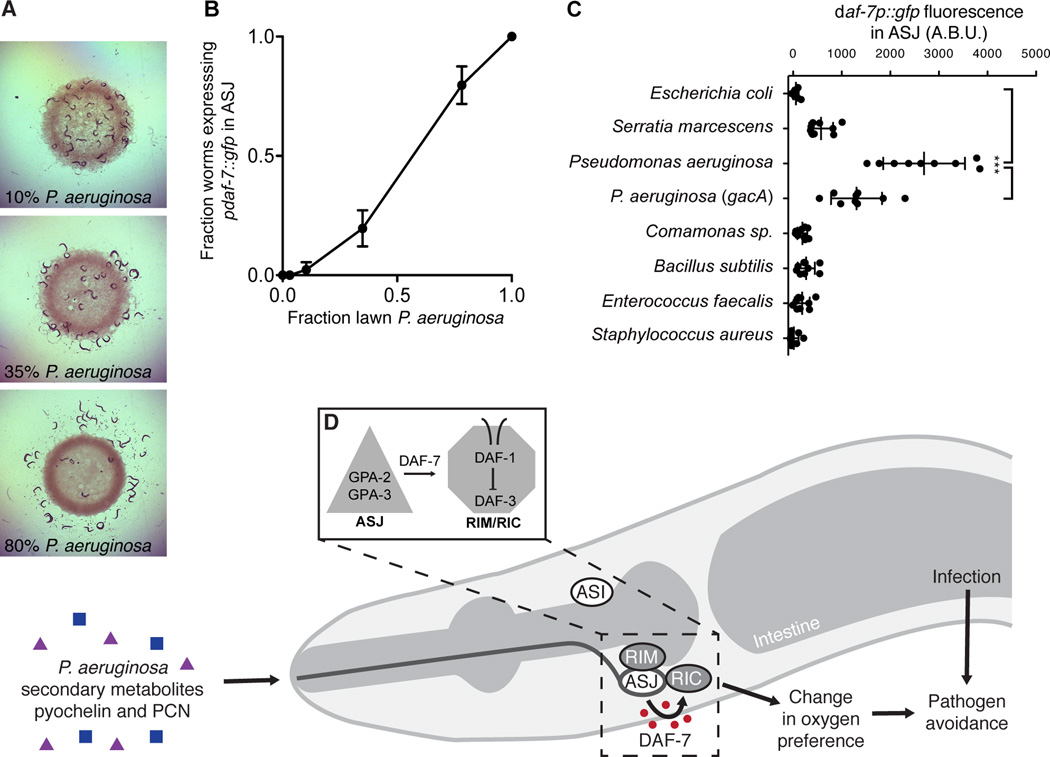
To understand how expression of daf-7 from the ASJ neurons promotes pathogen avoidance, we proceeded to examine the involvement of downstream signaling components of the canonical DAF-7/TGF-β pathway in the behavioral avoidance of P. aeruginosa. The daf-1 gene encodes the TGF-β Type I receptor that binds DAF-7 and is expressed in over 80 neurons including ciliated sensory neurons, pharyngeal neurons, and interneurons (Gunther et al., 2000). We found that daf-1 mutants, like daf-7 mutants, are deficient in avoidance of P. aeruginosa (Figure 3E). Using daf-1 mutant strains in which transgenic expression of daf-1 is directed in subsets of neurons, we observed that daf-1 expression under its native promoter or the panneuronal egl-3 promoter rescued P. aeruginosa avoidance to wild-type levels, but no rescue was observed when daf-1 was expressed in ~60 ciliated sensory neurons (including ASI and ASJ) using either the osm-6 or bbs-1 promoters (Figure 3E). Expression of daf-1 in only the RIM/RIC interneurons under the tdc-1 promoter was sufficient to restore pathogen avoidance behavior in the daf-1 mutant to wild-type levels (Figure 3E). Previous work showed that DAF-1 also acts in the RIM/RIC interneurons to mediate wild-type development, feeding rate, and quiescence (Gallagher et al., 2013; Greer et al., 2008), suggesting that despite the broad expression of daf-1 the RIM/RIC neurons may be the primary site of DAF-1 activity with regard to regulation of C. elegans behavior and physiology. The close proximity of the left and right ASJ neurons to the corresponding RIM/RIC interneurons on the ventral side of the amphid are in striking contrast to the location of the ASI neuron pair on the far dorsal side (White et al., 1986), and suggest an anatomical basis for how P. aeruginosa promotes enhanced DAF-7/TGF-β-dependent signaling (Figure 7D).
Figure 7. Microbial discrimination of P. aeruginosa activates daf-7 transcription in the ASJ neurons and promotes avoidance behavior.
(A) C. elegans after 16 h exposure to bacterial lawns consisting of E. coli OP50 and P. aeruginosa PA14. Fraction of bacteria that were P. aeruginosa when daf-7p∷gfp was assayed is indicated. (B) Fraction of animals expressing daf-7p∷gfp in the ASJ neurons after 16 h exposure to E. coli, P. aeruginosa, or mixtures of E. coli and P. aeruginosa. Error bars indicate standard deviation. (C) Maximum fluorescence values of daf-7p∷gfp in ASJ neurons after 16 h exposure to indicated bacteria. *** P < 0.001 as determined by one-way ANOVA followed by Tukey’s Multiple Comparison Test. Error bars indicate standard deviation. (D) In response to P. aeruginosa exposure, or P. aeruginosa metabolites phenazine-1-carboxamide (PCN) and pyochelin, daf-7 expression is activated via G-protein alpha subunits GPA-3 and GPA-2 in the ASJ neurons. Secreted DAF-7 signals to the TGF-β receptor DAF-1 the adjacent RIM/RIC interneurons. DAF-7/TGF-β signaling acts to alter aerotaxis behavior and promote avoidance of pathogenic bacteria.
DAF-7 promotes pathogen avoidance through the modulation of aerotaxis behavior
DAF-7-dependent regulation of dauer entry, feeding rate, and fat storage similarly rely on repression of the co-SMAD DAF-3 in the RIM/RIC interneurons, but downstream regulation of each of these physiological processes diverges genetically and utilizes distinct sets of neurons (Greer et al., 2008). Mutations in the steroid hormone receptor daf-12, for example, are able to suppress the constitutive dauer entry phenotype of daf-7 but not the increased fat storage or decreased feeding rate phenotypes. Similarly, mutations in the tyramine and octopamine synthesizing enzymes tdc-1 and tbh-1 suppress only the feeding rate phenotype of daf-7, while mutations in the Goα signaling molecules goa-1 and dgk-1 suppress only the fat storage phenotype of daf-7 (Greer et al., 2008). We determined that mutations in daf-12, tdc-1, tbh-1, goa-1, and dgk-1 were all unable to suppress the avoidance phenotypes of daf-7 mutants, indicating the existence of an additional signaling output by the RIM/RIC interneurons (Figure 3F).
In the presence of bacterial food, the laboratory wild-type strain of C. elegans N2 does not exhibit a distinct oxygen preference, and in particular does not avoid high atmospheric oxygen concentrations (i.e. 20% O2). In contrast, daf-7 mutants have altered aerotaxis behavior preferring oxygen concentrations near 8% and avoiding higher atmospheric oxygen levels (Chang et al., 2006). We have previously shown that the P. aeruginosa lawn is hypoxic, and that altered oxygen preference can affect bacterial lawn avoidance (Reddy et al., 2009; 2011). To determine whether a similar mechanism underlies the pathogen avoidance behavior promoted by daf-7, we introduced a mutation in the soluble guanylate cyclase gcy-35, which is responsible for responding to increases in oxygen concentration (Zimmer et al., 2009), and observed full suppression of the daf-7 avoidance defect on P. aeruginosa (Figure 3F). In addition, we performed the pathogen avoidance assay in an oxygen chamber at low oxygen concentrations, such that the surrounding environment would fall within the preferred oxygen concentration range of the animals. In this context daf-7 mutants show no avoidance defect relative to wild-type animals, supporting our conclusion that daf-7 mutants fail to avoid P. aeruginosa due to aberrant aerotaxis behavior (Figure S3A). We also conducted further experiments to demonstrate that the observed effects of DAF-7 on aerotaxis behavior and pathogen avoidance are not limited to the laboratory-adapted N2 strain (Figure S3B). Our model suggests that by activating daf-7 expression in the ASJ neurons in response to P. aeruginosa, inhibition of DAF-3 activity in the adjacent RIM/RIC interneurons is increased, modifying the response of C. elegans to oxygen levels and promoting exit from the lawn of P. aeruginosa.
G protein-dependent signaling activates daf-7 expression in the ASJ neuron pair in response to P. aeruginosa
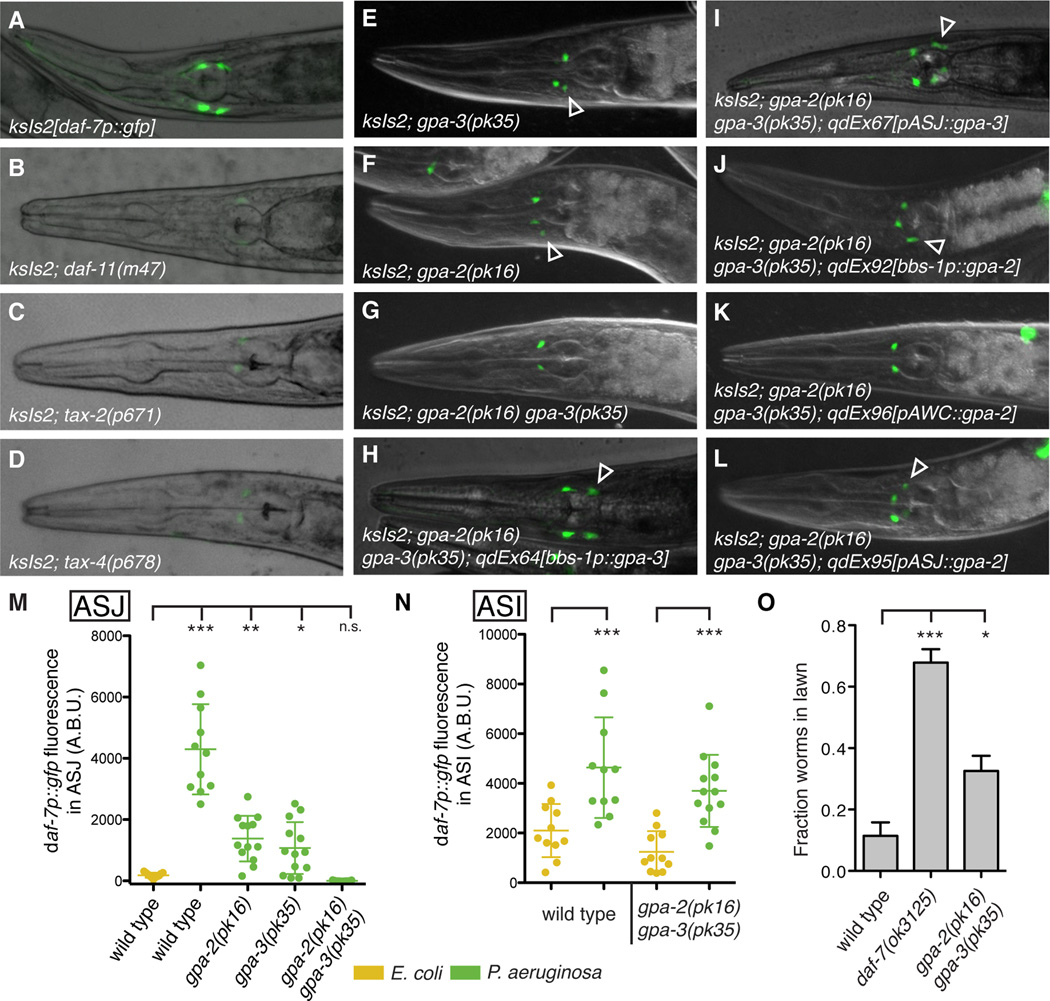
The rapid kinetics and functional consequence of P. aeruginosa-induced daf-7 expression in the ASJ neuron pair motivated use of the daf-7p∷gfp reporter to identify signaling mechanisms coupling P. aeruginosa exposure to this robust transcriptional response. We began by analyzing mutants known to have reduced daf-7 expression levels in the ASI neuron pair, such as the guanylate cyclase DAF-11 and the cyclic nucleotide-gated channel encoded by tax-2/tax-4 (Chang et al., 2006; Murakami et al., 2001), which are downstream of G protein signaling in C. elegans chemosensory neurons (Bargmann, 2006). No daf-7 expression was observed in either the ASI or ASJ neurons on P. aeruginosa in these mutant backgrounds, consistent with daf-11, tax-2, and tax-4 acting upstream of daf-7 expression in all tissues (Figures 4A–4D and S4A–S4C). These results suggest that the ASJ response to P. aeruginosa is downstream of G protein signaling and further motivate the identification of genes that specifically regulate daf-7 expression in the ASJ neurons.
Figure 4. GPA-3 and GPA-2 function cell-autonomously in the ASJ neurons to activate daf-7 expression in response to P. aeruginosa.
(A–L) daf-7p∷gfp expression on P. aeruginosa in various genetic backgrounds. Empty triangles indicate ASJ neurons. (M–N) Maximum fluorescence values of daf-7p∷gfp in the ASJ (M) or ASI (N) neurons after 16 h exposure to indicated bacteria. (O) Lawn occupancy of animals on P. aeruginosa after 15 h. *** P < 0.001, ** P < 0.01, * P < 0.05 as determined by one-way ANOVA followed by Dunnett’s Multiple Comparison Test. Error bars indicate standard deviation. See also Figure S4.
We performed a forward genetic screen to identify mutants in which the daf-7p∷gfp reporter failed to be expressed in the ASJ neurons on P. aeruginosa but GFP fluorescence remained unchanged in the ASI neurons. One such mutant, qd262 (Figures S4D–S4E), mapped to a region of chromosome V containing the G protein alpha subunit gpa-3. Sequencing revealed that the qd262 mutant carries a missense mutation in gpa-3 that converts a glycine residue (conserved from yeast to humans) into aspartic acid. Confirming the identity of the qd262 mutant as an allele of gpa-3, we crossed the gpa-3 deletion allele pk35 into the daf-7p∷gfp reporter and observed reduced fluorescence in the ASJ neurons on P. aeruginosa (Figures 4E and 4M). Mutants carrying a deletion in a second highly homologous G protein alpha subunit, gpa-2, also displayed reduced daf-7p∷gfp fluorescence in ASJ (Figures 4F and 4M), and the gpa-2 gpa-3 double mutant was completely deficient in the ASJ-response to P. aeruginosa (Figures 4G and 4M). Interestingly, the gpa-2(pk16) gpa-3(pk35) double mutant was still able to upregulate expression of daf-7p∷gfp in the ASI neuron on P. aeruginosa, indicating that the ASJ and ASI responses to P. aeruginosa are genetically distinct (Figure 4N). The C. elegans nervous system is equipped with approximately 1,300 G protein-coupled receptors that act as chemoreceptors (Thomas and Robertson, 2008), and we hypothesize that GPA-3 and GPA-2 may be acting downstream of one or more GPCRs responsible for sensing P. aeruginosa.
gpa-3 is expressed in at least ten pairs of amphid sensory neurons (including ASJ), and its known functions include mediating responses to C. elegans pheromone in ASK, odorant attraction in AWA and AWC, and odorant avoidance in ASH (Hilliard et al., 2004; Jansen et al., 1999; Kim et al., 2009; Lans et al., 2004; Zwaal et al., 1997). To determine if GPA-3 acts cell-autonomously in the ASJ neurons to mediate the response to P. aeruginosa, we expressed wild-type copies of gpa-3 cDNA under heterologous promoters in the gpa-2(pk16) gpa-3(pk35) background. Under control of either the pan-ciliated neuronal promoter bbs-1, or the ASJ-specific promoter trx-1, gpa-3 cDNA was able to rescue the daf-7 ASJ expression defect, indicating that GPA-3 acts cell-autonomously in ASJ to mediate the response to P. aeruginosa (Figures 4H–4I and S4F–S4I).
gpa-2 is reported to be expressed in only one pair of chemosensory neurons (AWC) and functions in the processes of olfaction and dauer induction (Lans et al., 2004; Zwaal et al., 1997). To determine the site of action for gpa-2 in the response to P. aeruginosa, we expressed wild-type copies of gpa-2 cDNA under heterologous promoters in the gpa-2(pk16) gpa-3(pk35) background. To our surprise, expression of gpa-2 driven by either the pan-ciliated neuronal promoter bbs-1 or the ASJ-specific promoter trx-1 was able to rescue the daf-7p∷gfp phenotype, but expression of gpa-2 in the AWC neurons under the ceh-36 promoter was not able to rescue the gpa-2 gpa-3 mutant phenotype (Figures 4J–4L and S4J–S4K). This indicates that GPA-2, like GPA-3, also acts cell-autonomously in ASJ to induce daf-7 expression in response to P. aeruginosa. To confirm that gpa-2 was indeed expressed in the ASJ neurons we used single molecule FISH to probe for gpa-2 mRNA. Using probes specific for gpa-2, we observed 5–10 mRNA molecules in each ASJ neuron (data not shown). This novel expression supports our neuron-specific rescue data and indicates that GPA-2 and GPA-3 act together in the ASJ neurons to activate daf-7 expression in response to P. aeruginosa. Finally, we tested the gpa-2 gpa-3 double mutant for a P. aeruginosa avoidance defect, and observed a partial deficit in lawn avoidance that we hypothesize is due to a loss of DAF-7 secretion from the ASJ neurons (Figure 4O). These experiments identify a conserved G protein signaling pathway that acts cell-autonomously in the ASJ neurons to induce daf-7 expression in response to P. aeruginosa.
Chemosensory recognition of the P. aeruginosa secondary metabolites phenazine-1-carboxamide and pyochelin by the ASJ neuron pair of C. elegans
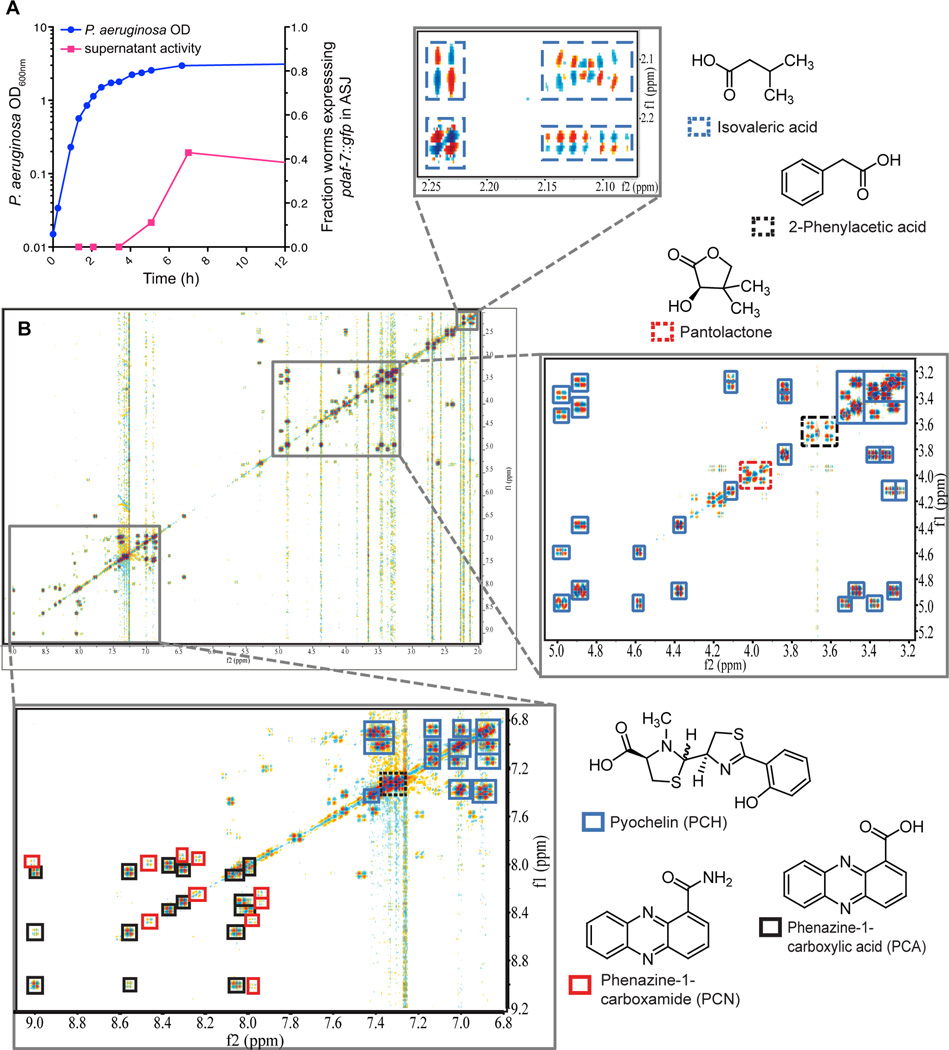
We next sought to identify the specific bacterial cues inducing expression of daf-7 in the ASJ neuron pair of C. elegans. We observed that exposure of C. elegans to filtered P. aeruginosa supernatant was sufficient to induce daf-7 expression (Figures 5A and 6A–6B). Furthermore, by testing supernatants from liquid cultures at various growth stages we determined that P. aeruginosa in stationary phase induced daf-7 expression in ASJ neurons most dramatically (Figure 5A), leading us to hypothesize that specific secondary metabolites produced under high cell density were being sensed by C. elegans. Metabolite identification was accomplished via activity-guided fractionation followed by 2D NMR spectroscopic profiling. Supernatant from large volumes of stationary phase P. aeruginosa was fractionated using an automated chromatography system, and individual fractions were tested for activity in the daf-7p∷gfp assay. 1D and 2D NMR (dqfCOSY, HSQC, HMBC) spectroscopic data of the active fractions were then analyzed. Such NMR spectroscopic analysis of metabolome fractions of lowered complexity shifts focus onto testing individual identified components in bioassays, thereby reducing the need for further time-intensive purification (Taggi et al., 2004).
Figure 5. Supernatant from P. aeruginosa in stationary phase activates daf-7 expression and contains secondary metabolites.
(A) Growth curve of P. aeruginosa as measured by OD600nm (blue circles) and activity of bacterial supernatant in inducing daf-7p∷gfp expression in the ASJ neurons (magenta squares). (B) DQF-COSY spectrum of active metabolome fraction of P. aeruginosa media extract. Enlarged sections show cross-peaks of the most abundant metabolites present in this fraction. See also Figure S5.
Figure 6. The ASJ neurons respond to P. aeruginosa secondary metabolites phenazine-1-carboxamide and pyochelin.
(A–B) daf-7p∷gfp expression in the ASJ or ASI neurons after 16 h exposure to E. coli, P. aeruginosa, or E. coli supplemented with indicated material. Data represent (A) the fraction of animals expressing daf-7p∷gfp in ASJ above background or (B) the maximum fluorescence values of daf-7p∷gfp in ASJ and ASI. All compounds were dissolved in DMSO. *** P < 0.001, ** P < 0.01, * P < 0.05 as determined by one-way ANOVA followed by Dunnett’s Multiple Comparison Test. n.s. = not significant. Error bars indicate standard deviation. (C–D) GCaMP5 expression in the ASJ neurons immediately prior to (C) or following (D) addition of PCN. Filled triangle indicates the ASJ cell body and open triangle indicates the ASJ sensory projection. (E) GCaMP5 fluorescence changes in individual animals following the addition of PCN or DMSO at t = 0 seconds. (F) Average changes in GCaMP fluorescence in the 2–6 s following addition of either PCN or DMSO. *** P < 0.001 as determined by unpaired t-Test. Error bars indicate standard deviation. See also Figure S6.
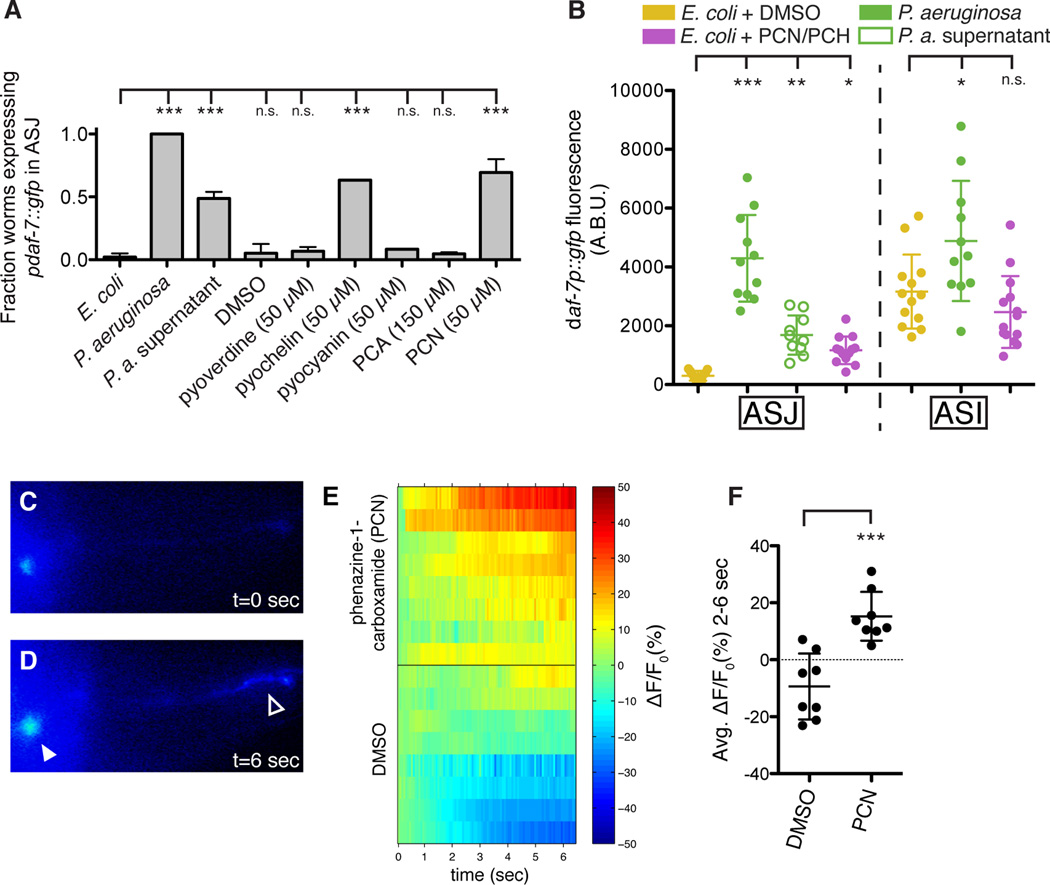
2D NMR spectra of the active fractions revealed the presence of pantolactone (Nakata et al., 2013), isovaleric acid (Niu et al., 2008), phenazine-1-carboxylic acid (Mehnaz et al., 2013), phenazine-1-carboxamide (PCN, see Table S1), the siderophore pyochelin (see Table S2) and phenyl acetic acid (Korsager et al., 2013) as major components (Figures 5B, S5A, and S5B), in addition to small quantities of monoacyl glycerides and β-hydroxy fatty acids. When synthetic and HPLC-purified samples of these compounds were tested, PCN and pyochelin displayed concentration-dependent activity in the daf-7p∷gfp assay, regardless of solvent (Figures 6A and S6), whereas the other identified metabolites were not active in this assay. Furthermore, we found that other P. aeruginosa-produced phenazines (e.g. pyocyanin and phenazine-1-carboxylic acid) and pyoverdine, another unrelated P. aeruginosa siderophore, do not induce daf-7 expression in the ASJ neurons (Figure 6A), indicating that general properties of these compounds are likely not the cause of their activity. We quantified the daf-7p∷gfp fluorescence increase in the ASJ neurons following addition of pyochelin and PCN and found the response was similar to that induced by P. aeruginosa supernatant (Figure 6B). Interestingly we found that these compounds had no effect on daf-7p∷gfp fluorescence in the ASI neurons, further decoupling the responses of the ASJ and ASI neurons to P. aeruginosa (Figure 6B).
Given the rapid transcriptional response of the ASJ neurons to the presence of P. aeruginosa secondary metabolites, we tested the ability of phenazine-1-carboxamide (PCN) to activate the ASJ neurons. We constructed a transgenic strain in which the genetically encoded calcium indicator GCaMP5 (Akerboom et al., 2012) is expressed exclusively in the ASJ neuron pair. Upon administration of PCN, but not the carrier control DMSO, we observed increased GCaMP5 fluorescence in both the ASJ cell body and ASJ ciliated projection that is exposed to the environment (Figures 6C–6D). We quantified the change in fluorescence in the ASJ cell body and observed a significant difference between PCN and the carrier control (Figures 6E–6F). These data suggest that ASJ may be sensing the P. aeruginosa metabolite PCN directly and, in turn, activating daf-7 transcription.
Microbial discrimination and specificity in the chemosensory response to P. aeruginosa
In the natural environment C. elegans are unlikely to be presented with homogeneous bacterial lawns consisting of a single bacterial species, and so we wondered how sensitive the daf-7p∷gfp response is to the concentration of P. aeruginosa. We created heterogeneous bacterial lawns consisting of E. coli and P. aeruginosa and assayed daf-7p∷gfp fluorescence. We observed that the induction of daf-7 expression does not function as a binary switch but rather can respond in a more subtle manner that is proportional to the fraction of P. aeruginosa present in the mixed E. coli/P. aeruginosa lawn (Figures 7A–7B). Interestingly, the P. aeruginosa concentration range over which daf-7 is activated corresponds to the range sufficient to induce the C. elegans behavioral avoidance response (Figures 7A–7B).
Finally, we investigated how specific the induction of daf-7 transcription in the ASJ neuron pair was to P. aeruginosa as compared to other environmental microbes. We exposed daf-7p∷gfp animals to a wide array of bacterial species and strains, covering pathogenic and non-pathogenic alpha-, beta-, and gammaproteobacteria as well as gram-positive bacterial species. We observed that a number of non-E. coli species can induce daf-7 expression in the ASJ neuron pair on the order of 10-fold, but that the response is 1–2 orders of magnitude greater upon exposure to P. aeruginosa PA14 (Figure 7C). This result indicates that the identity of the microbial species is the principal determinant in inducing daf-7 expression in the ASJ neuron pair, likely due to the relatively species-specific production of P. aeruginosa metabolites PCN and pyochelin (see Discussion). The presence of low-level activity in bacterial strains other than P. aeruginosa is consistent with the existence of additional, unidentified bacterial determinants that also contribute to the induction of daf-7 in the ASJ neuron pair.
PCN and pyochelin are secondary metabolites produced by P. aeruginosa at high cell density that promote biofilm formation in soil as well as chronic infections in human lungs (Cornelis and Dingemans, 2013; Price-Whelan et al., 2006). In P. aeruginosa production of phenazines and pyochelin are both positively regulated by GacA, a global activator of cell-density-dependent gene expression and virulence (Reimmann et al., 1997; Wei et al., 2013). We observed that the P. aeruginosa gacA mutant was deficient in inducing daf-7p∷gfp expression in the ASJ neurons relative to wild-type bacteria (Figure 7C). As such, these molecules may serve as bacterial growth-stage-specific cues for C. elegans, alerting the host to the presence of bacteria in a “pathogenic state,” and inducing a correspondingly beneficial behavioral avoidance response.
Discussion
Our data suggest that C. elegans responds to secondary metabolites produced by bacterial pathogens using G protein signaling pathways in their chemosensory neurons. C. elegans lack the antigen-specific responses of vertebrate immunity, but the utilization of chemosensory neurons and the repertoire of an estimated 1,300 GPCRs may provide a means by which a simple host organism can detect microbial pathogens. Bacterial secondary metabolism can generate a wide range of molecules that are often largely specific with regard to producer organism and regulated by environmental and growth conditions. Redox-active phenazine-1-carboxamide and the siderophore pyochelin are two such secondary metabolites of P. aeruginosa, produced under conditions of high cell density and low oxygen. Interestingly, while a number of other bacterial species produce phenazines, such as Burkholderia, Brevibacterium, and Streptomyces, the modifying enzyme responsible for producing phenazine-1-carboxamide, phzH, is not part of the canonical phenazine operon, but rather found elsewhere in the genome and limited to P. aeruginosa and P. chlororaphis (Chin-A-Woeng et al., 2001; Mavrodi et al., 2001). Similarly, pyochelin production is restricted to Pseudomonas and Burkholderia species (Gross and Loper, 2009). As such, these two metabolites may act as pathogen-specific cues for C. elegans navigation, providing a molecular readout not only of the presence of P. aeruginosa itself, but the presence of P. aeruginosa in a “pathogenic state” that is more strongly associated with virulence.
Our data demonstrate that DAF-7 neuroendocrine signaling is necessary for the behavioral avoidance response to P. aeruginosa. The decision to occupy or avoid a lawn of P. aeruginosa integrates multiple sensory inputs including chemosensation of bacterial compounds, chemosensation of oxygen, and mechanosensation (Chang et al., 2011; Ha et al., 2010; Pradel et al., 2007; Reddy et al., 2009). Once C. elegans have learned to avoid a particular food source, a process that requires the association of bacterial infection with specific odors and occurs many hours after exposure to pathogenic bacteria (Zhang et al., 2005), they are confronted with conflicting environmental stimuli: the odor of pathogenic bacteria drives animals out of the lawn, while the relatively low oxygen concentration of the bacterial environment keeps animals inside the lawn. The DAF-7 pathway suppresses this latter tendency, and thus increasing the activity of DAF-7 in response to pathogenic bacteria promotes subsequent avoidance behavior. For C. elegans, which must balance attraction to bacterial food with avoidance of microbial pathogens, suppressing behaviors such as aerotaxis that keep the animal inside the bacterial lawn may be just as essential as activating behaviors that drive it away.
Whereas defining the connectivity of the C. elegans nervous system has provided the anatomical foundation for functional studies of neuronal circuitry (White et al., 1986), genetic studies of behavior have revealed pivotal roles for neuromodulators, such as neurotransmitters and neuropeptides, which modify the activity of synaptic circuits in response to environmental cues (Bargmann, 2012). Prior studies have illustrated how a change in the neuronal expression pattern of a single component of neuromodulator signaling can have dramatic effects on complex behavior (Lim et al., 2004; Pocock and Hobert, 2010). Our study establishes that patterns of neuromodulator expression and activity may be subject to dramatic modification by the microbial environment. There has been an emerging appreciation of the profound influence that the animal microbiota can have on host organisms (Clemente et al., 2012; Lyte, 2013), and our study provides a genetic, neuronal, chemical basis for how microbes may influence host neuroendocrine physiology and behavior.
Experimental Procedures
C. elegans Strains
C. elegans was maintained on E. coli OP50 as previously described (Brenner, 1974). Constitutive dauer strains were grown at the permissive temperature of 16°C until they had passed the dauer developmental decision. For assays in which a synchronized population of animals was required, strains were egg-prepped in bleach and arrested overnight in M9 buffer at the L1 larval stage. For a complete list of strains used in this study see Table S3.
daf-7p∷gfp Induction Assays and Bacterial Strains
For testing the dynamics of daf-7 expression on P. aeruginosa PA14, an overnight culture of PA14 was grown in 2–3 mL LB at 37°C, and the following morning 7 µL of culture was seeded onto 3.5 cm SKA plates as previously described (Tan et al., 1999). PA14 plates were grown overnight at 37°C and then grown for an additional day at room temperature. Animals at the L4 larval stage were then picked onto the center of the bacterial lawn, incubated at 25°C, and scored for changes in GFP expression 15–20 h later. Cell identification was performed using the lipophilic dye DiI from Molecular Probes. Images were acquired with an Axioimager Z1 microscope using animals anaesthetized in 50 mM sodium azide. To quantify GFP fluorescence animals were imaged at 40× magnification and the maximum intensity value within the ASJ neuron was determined using FIJI software. The same procedure (with the indicated modifications) was followed for: Escherichia coli OP50, Serratia marcescens DB10, Bacillus subtilis PY79, Comamonas sp. DA1877, Enterococcus faecalis OG1RF (grown overnight in Brain Heart Infusion Broth), and Staphylococcus aureus NCTC8325 (grown overnight in Tryptic Soy Broth). To test the activity of bacterial supernatant, fractionated supernatant, and purified compounds, 3.5 cm SKA plates were seeded with 5 µL E. coli OP50. Prior to adding C. elegans, 25 µL of test material was added to the bacterial lawn and allowed to dry. All compounds were dissolved in DMSO. To generate bacterial lawns that contained mixtures of E. coli and P. aeruginosa, plates were seeded with E. coli and allowed to grow for two days as above, at which time 7 µL of liquid P. aeruginosa culture (diluted to varying degrees) was added to the E. coli lawn and allowed to dry. After the experiment had concluded the fraction of the lawn corresponding to each bacterial species was measured by counting CFUs of E. coli and P. aeruginosa (distinguished by colony morphology).
P. aeruginosa Avoidance and Killing Assays
Plates for P. aeruginosa avoidance assays were prepared as above. 30 animals at the L4 larval stage were transferred to the center of PA14 lawns, incubated at 25°C, and scored for avoidance 15 h later. For avoidance experiments in hypoxia, plates were placed in a Coy Laboratory Products Inc. Hypoxic In Vitro Cabinet and incubated at 1%–4% O2 at room temperature. Plates for measuring accumulation of red fluorescent beads were prepared as above with addition of Fluoresbrites 0.2 µm microspheres (Polyscience, Inc.) to the PA14 culture prior to seeding at a ratio of 50:1 (bacteria:beads). Plates for P. aeruginosa slow killing assays were prepared as above with the addition of 50 µg/mL 5-fluorodeoxyuridine (FUdR). 30 animals at the L4 larval stage were transferred to the center of PA14 lawns, incubated at 25°C, and scored for killing over the course of 5 days. For “Big Lawn” slow killing assays the following modifications were made: 15 µL of PA14 culture was spread to the edges of the SKA plate and 75 animals were added to each plate. Statistical analysis was performed using GraphPad Prism Software.
Single Molecule Fluorescent In Situ Hybridization
smFISH was performed as previously described (Raj et al., 2008). Briefly, C. elegans were fixed in 4% formaldehyde for 45 min at room temperature. After washing with PBS, larvae were resuspended in 70% EtOH and incubated overnight at 4°C. The following day fixed larvae were transferred into hybridization solution with the smFISH probe and incubated overnight at 30°C. The daf-7 and gpa-2 probes were constructed by pooling 25 unique DNA oligos that tile the coding regions deleted in the daf-7(ok3125) and gpa-2(pk16) mutants and coupling them to Cy5 dye. For a complete list of oligos see Table S4. Images were acquired with a Nikon Eclipse Ti Inverted Microscope outfitted with a Princeton Instruments PIXIS 1024 camera. Data were analyzed using FIJI software; images presented in Figures 2H, 2I, and S2A are maximum intensity z-projections of 30 stacked exposures.
Generation of Transgenic Animals
The daf-7 promoter (3.1 kb), trx-1 promoter (1.1 kb) (Fierro González et al., 2011), str-3 promoter (2.8 kb), and ceh-36 promoter (<1 kb) (Kim et al., 2010) were amplified by PCR from genomic DNA. The bbs-1 promoter (1.9 kb) was amplified by PCR from plasmid pKA40 (laboratory of K. Ashrafi). daf-7 cDNA was amplified by PCR from an ORFeome RNAi clone. gpa-3 and gpa-2 cDNAs were amplified by PCR from cDNA generated with an Ambion RETROscript Kit. The GCaMP5G gene was amplified from Plasmid 31788: pCMV-GCaMP5G (Addgene). The unc-54 3-prime UTR was amplified by PCR from Fire Vector pPD95.75. DNA constructs (promoter∷cDNA∷unc-54 3’UTR) were synthesized using PCR fusion as previously described (Hobert, 2002). PCR fusion products were cloned into pGEM-T Easy plasmids, sequenced to confirm identity, and injected into animals at a concentration of 50 ng/µL along with a plasmid carrying either ges-1p∷gfp or ofm-1p∷gfp (50 ng/µL). At least three independent transgenic lines were analyzed for each rescue construct. For a complete list of primers used in this study see Table S4.
Biochemical Identification of P. aeruginosa Metabolites
To generate P. aeruginosa supernatant, overnight bacterial cultures were grown in 500 mL LB at 37°C shaking at 200 rpm. Once the cultures had reached OD600 = 3.0 they were passed through a 0.22 µm PES filter system. The filtrate was lyophilized and the residue was extracted twice with 50 mL of 4:1 dichloromethane:methanol mixture over 12 h. The resulting suspension was centrifuged at 4,750 rpm at 4°C for 15 min and the supernatant liquid was collected. The solvent was evaporated in vacuo at room temperature to produce the P. aeruginosa supernatant extract used for chromatographic separations and analysis. For details, see Supplemental Experimental Procedures. NMR spectra were recorded on a Varian Inova 600 MHz NMR spectrometer equipped with an HCN indirect detection probe. Spectra were baseline corrected, phased and calibrated to the solvent peak (CHCl3 singlet at 7.26 ppm) using Varian VNMR and MestreLab’s Mnova software packages. Non-gradient phase-cycled double-quantum filtered correlation spectroscopy (DQF-COSY) spectra were acquired using the following parameters: 0.6 s acquisition time, 512 complex increments, 16 scans per increment. DQF-COSY spectra were zero filled to 8,192 × 4,096, and 90°-shifted sine bell window functions were applied in both dimensions before Fourier transformation. Heteronuclear single quantum coherence spectroscopy (HSQC) and heteronuclear multiple bond correlation spectroscopy (HMBC) spectra were acquired using the following parameters: 0.25 s acquisition time, 400–600 complex increments, 16 scans per increment. HSQC and HMBC spectra were zero filled to 2,048 × 2,048, and 90°-shifted sine bell window functions were applied in both dimensions. Unit mass-resolution HPLC-MS was performed using an Agilent 1100 Series HPLC system equipped with a diode array detector and connected to a Quattro II mass spectrometer (Micromass/Waters). MassLynx software was used for MS data acquisition and processing. UV-Vis spectra were recorded on Agilent Technologies 8453 UV-Vis spectrophotometer. For testing the activity of candidate compounds, phenazine-1-carboxamide was purchased from Princeton Biomolecular Research (PBMR030086), phenazine-1-carboxylic acid was purchased from Apollo Scientific (OR01490), pyochelin was purchased from Cfm Oskar Tropitzsch (164104-31-8/164104-32-9), and pyocyanin (P0046) and pyoverdine (P8374) were purchased from Sigma-Aldrich.
Calcium Imaging
To measure the activity of the ASJ neurons GCaMP5G was expressed exclusively in the ASJ neurons under the trx-1 promoter. Animals were immobilized on NGM agar pads using Surgi-lock 2oc instant tissue adhesive (Meridian) and imaged at 40X using a Zeiss AxioVert S100 inverted microscope outfitted with an Andor iXon EMCCD camera. Ten seconds after imaging had begun approximately 3 µL of stimulant (either PCN 10 mg/mL or DMSO) was added to the agar pad. In Figures 6C–6F time = 0 refers to the addition of stimulant. Data were analyzed using custom MATLAB software written by Nikhil Bhatla.
Supplementary Material
Highlights.
C. elegans chemosensory neurons respond to specific bacterial secondary metabolites
Host neuromodulator gene expression pattern is altered by the microbial environment
Detection of P. aeruginosa metabolites promotes protective behavioral avoidance
Acknowledgements
We thank J. Alcedo, K. Ashrafi, F. Ausubel, H. R. Horvitz, the C. elegans Knockout Consortium, and the Caenorhabditis Genetics Center (which is supported by the NIH, Office of the Director) for providing strains and reagents. We thank N. Bhatla, C. Engert, C. Pender, and S. Sando for technical assistance. J.D.M. was supported by a Graduate Research Fellowship from the National Science Foundation. This study was supported by the NIH (GM084477 to D.H.K. and GM088290 to F.C.S.) and an Ellison New Scholar Award (to D.H.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.D.M. performed C. elegans and P. aeruginosa experiments. O.P. and P.M. performed fractionation and NMR analysis. J.D.M., O.P., F.C.S., and D.H.K, analyzed the data and interpreted results. J.D.M. and D.H.K. wrote the paper with input from O.P. and F.C.S.
References
- Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. Journal of Neuroscience. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J. Exp. Zool. 1990;253:263–270. doi: 10.1002/jez.1402530305. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Chemosensation in C. elegans. WormBook. 2006:1–29. doi: 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Beyond the connectome: how neuromodulators shape neural circuits. Bioessays. 2012;34:458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- Beale E, Li G, Tan MW, Rumbaugh KP. Caenorhabditis elegans Senses Bacterial Autoinducers. Applied and Environmental Microbiology. 2006;72:5135–5137. doi: 10.1128/AEM.00611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A Distributed Chemosensory Circuit for Oxygen Preference in C. elegans. Plos Biol. 2006;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Paek J, Kim DH. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 2011;480:525–529. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-A-Woeng TF, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant Microbe Interact. 2001;14:1006–1015. doi: 10.1094/MPMI.2001.14.8.1006. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. 2002;419:899–903. doi: 10.1038/nature01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M, Attisano L, Wrana JL, Albert PS, Massagué J, Riddle DL. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature. 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- Félix M-A, Duveau F. Population dynamics and habitat sharing ofnatural populations of Caenorhabditis elegans andC. briggsae. BMC Biol. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro González JC, Cornils A, Alcedo J, Miranda-Vizuete A, Swoboda P. The Thioredoxin TRX-1 Modulates the Function of the Insulin-Like Neuropeptide DAF-28 during Dauer Formation in Caenorhabditis elegans. PLoS ONE. 2011;6:e16561. doi: 10.1371/journal.pone.0016561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T, Kim J, Oldenbroek M, Kerr R, You Y-J. ASI Regulates Satiety Quiescence in C. elegans. Journal of Neuroscience. 2013;33:9716–9724. doi: 10.1523/JNEUROSCI.4493-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi LL, Albert PS, Riddle DL. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- Greer ER, Pérez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and Molecular Dissection of a C. elegans Sensory Circuit that Regulates Fat and Feeding. Cell Metabolism. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- Gunther CV, Georgi LL, Riddle DL. A Caenorhabditis elegans type I TGF beta receptor can function in the absence of type II kinase to promote larval development. Development. 2000;127:3337–3347. doi: 10.1242/dev.127.15.3337. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Ha H-I, Hendricks M, Shen Y, Gabel CV, Fang-Yen C, Qin Y, Colón-Ramos D, Shen K, Samuel ADT, Zhang Y. Functional Organization of a Neural Network for Aversive Olfactory Learning in Caenorhabditis elegans. Neuron. 2010;68:1173–1186. doi: 10.1016/j.neuron.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Russell RL. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.a. 1975;72:4061–4065. doi: 10.1073/pnas.72.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bergamasco C, Arbucci S, Plasterk RHA, Bazzicalupo P. Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. Embo J. 2004;23:1101–1111. doi: 10.1038/sj.emboj.7600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat. Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim R, Sengupta P. The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C. elegans. Development. 2010;137:963–974. doi: 10.1242/dev.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, Clardy J, Touhara K, Sengupta P. Two Chemoreceptors Mediate Developmental Effects of Dauer Pheromone in C. elegans. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsager S, Taaning RH, Skrydstrup T. Effective palladium-catalyzed hydroxycarbonylation of aryl halides with substoichiometric carbon monoxide. J. Am. Chem. Soc. 2013;135:2891–2894. doi: 10.1021/ja3114032. [DOI] [PubMed] [Google Scholar]
- Lans H, Rademakers S, Jansen G. A network of stimulatory and inhibitory Galpha-subunits regulates olfaction in Caenorhabditis elegans. Genetics. 2004;167:1677–1687. doi: 10.1534/genetics.103.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MM, Wang Z, Olazábal DE, Ren X, Terwilliger EF, Young LJ. Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature. 2004;429:754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- Lyte M. Microbial Endocrinology in the Microbiome-Gut-Brain Axis: How Bacterial Production and Utilization of Neurochemicals Influence Behavior. PLoS Pathog. 2013;9:e1003726. doi: 10.1371/journal.ppat.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJM. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153:240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnaz S, Saleem RSZ, Yameen B, Pianet I, Schnakenburg G, Pietraszkiewicz H, Valeriote F, Josten M, Sahl H-G, Franzblau SG, et al. Lahorenoic acids A–C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013;76:135–141. doi: 10.1021/np3005166. [DOI] [PubMed] [Google Scholar]
- Milward K, Busch KE, Murphy RJ, de Bono M, Olofsson B. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2011;108:20672–20677. doi: 10.1073/pnas.1106134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Koga M, Ohshima Y. DAF-7/TGF-beta expression required for the normal larval development in C. elegans is controlled by a presumed guanylyl cyclase DAF-11. Mech. Dev. 2001;109:27–35. doi: 10.1016/s0925-4773(01)00507-x. [DOI] [PubMed] [Google Scholar]
- Nakata K, Gotoh K, Ono K, Futami K, Shiina I. Kinetic resolution of racemic 2-hydroxy-γ-butyrolactones by asymmetric esterification using diphenylacetic acid with pivalic anhydride and a chiral acyl-transfer catalyst. Org. Lett. 2013;15:1170–1173. doi: 10.1021/ol303453j. [DOI] [PubMed] [Google Scholar]
- Niu DF, Xiao LP, Zhang AJ, Zhang GR, Tan QY, Lu JX. Electrocatalytic carboxylation of aliphatic halides at silver cathode in acetonitrile. Tetrahedron. 2008 [Google Scholar]
- Patterson GI, Koweek A, Wong A, Liu Y, Ruvkun G. The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes & Development. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock R, Hobert O. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nature Publishing Group. 2010;13:610–614. doi: 10.1038/nn.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.a. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LEP, Newman DK. Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Meth. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Hunter RC, Bhatla N, Newman DK, Kim DH. Caenorhabditis elegans NPR-1-mediated behaviors are suppressed in the presence of mucoid bacteria. Proceedings of the National Academy of Sciences. 2011;108:12887–12892. doi: 10.1073/pnas.1108265108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-β Dauer Pathway Regulates Longevity via Insulin Signaling. Current Biology. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- Swanson MM, Riddle DL. Critical periods in the development of the Caenorhabditis elegans dauer larva. Dev. Biol. 1981;84:27–40. doi: 10.1016/0012-1606(81)90367-5. [DOI] [PubMed] [Google Scholar]
- Taggi AE, Meinwald J, Schroeder FC. A new approach to natural products discovery exemplified by the identification of sulfated nucleosides in spider venom. J. Am. Chem. Soc. 2004;126:10364–10369. doi: 10.1021/ja047416n. [DOI] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.a. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Robertson HM. The Caenorhabditis chemoreceptor gene families. BMC Biol. 2008;6:42. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJM. Interspecies Systems Biology Uncovers Metabolites Affecting C. elegans GeneExpression and Life History Traits. Cell. 2014;156:759–770. doi: 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Huang X, Tang L, Wu D, Xu Y. Global Control of GacA in Secondary Metabolism, Primary Metabolism, Secretion Systems, and Motility in the Rhizobacterium Pseudomonas aeruginosa M18. J. Bacteriol. 2013;195:3387–3400. doi: 10.1128/JB.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang Y. DBL-1, a TGF-β, is essential for Caenorhabditis elegans aversive olfactory learning. Proceedings of the National Academy of Sciences. 2012;109:17081–17086. doi: 10.1073/pnas.1205982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron. 2009;61:865–879. doi: 10.1016/j.neuron.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-β signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol. 2009;10:249–256. doi: 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Mendel JE, Sternberg PW, Plasterk RH. Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans Dauer-inducing pheromone. Genetics. 1997;145:715–727. doi: 10.1093/genetics/145.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.