Abstract
In Arabidopsis thaliana, acyl-CoA-binding proteins (ACBPs) are encoded by six genes, and they display varying affinities for acyl-CoA esters. Recombinant ACBP4 and ACBP5 have been shown to bind oleoyl-CoA esters in vitro. In this study, the subcellular localizations of ACBP4 and ACBP5 were determined by biochemical fractionation followed by western blot analyses using anti-ACBP4 and anti-ACBP5 antibodies and immuno-electron microscopy. Confocal microscopy of autofluorescence-tagged ACBP4 and ACBP5, expressed transiently in onion epidermal cells and in transgenic Arabidopsis, confirmed their expression in the cytosol. Taken together, ACBP4 and ACBP5 are available in the cytosol to bind and transfer cytosolic oleoyl-CoA esters. Lipid profile analysis further revealed that an acbp4 knockout mutant showed decreases in membrane lipids (digalactosyldiacylglycerol, monogalactosyldiacylglycerol, phosphatidylcholine, phosphatidylethanolamine and phosphatidylinositol) while acbp4-complemented lines attained levels similar to wild type, suggesting that ACBP4 plays a role in the biosynthesis of membrane lipids including galactolipids and phospholipids.
Keywords: acyl-CoA-binding protein, autofluorescence-tagged protein, cytosol, galactolipids, lipid metabolism, phospholipids
Introduction
Unlike animals and fungi in which fatty acid biosynthesis occurs in the cytosol, de novo fatty acid biosynthesis in higher plants is found exclusively in the chloroplasts (Ohlrogge and Browse 1995). Plastidial fatty acids are subsequently exported as acyl-CoAs, palmitoyl-CoA (16:0-CoA) and oleoyl-CoA (18:1-CoA), to the endoplasmic reticulum (ER) for the biosynthesis of non-plastidial membrane lipids and triacylglycerol (Ohlrogge and Browse, 1995). Subsequently, the diacylglycerol moieties from the ER pathway are sent to the plastids for the synthesis of glycerolipids in the thylakoid membranes (Browse et al. 1986). Hence, extensive transfer of acyl-CoA derivatives occurs between the outer chloroplast envelope and the ER membrane via the cytosol (Ohlrogge and Browse, 1995). A highly-conserved 10-kDa acyl-CoA-binding protein (ACBP) has been proposed to facilitate transport and maintenance of long-chain acyl-CoA esters in the cytosol, and in the protection of cytosolic acyl-CoAs from hydrolysis by cellular acyl-CoA hydrolases (Engeseth et al. 1996). This 10-kDa ACBP, prevalent in eukaryotes, has also been reported in protists and in some pathogenic bacteria (Burton et al. 2005).
In Arabidopsis thaliana, besides the 10-kDa ACBP (Engeseth et al. 1996) which we have designated as ACBP6 (Xiao et al. 2008), there are five larger forms (designated ACBP1 to ACBP5, ranging in size from 37.5 to 73.1 kDa) (Chye 1998; Chye et al. 2000; Leung et al. 2004, 2006). All these ACBPs are conserved at the acyl-CoA-binding domain (Leung et al. 2004). The presence of additional structural domains such as ankyrin repeats in ACBP1 and ACBP2, and kelch motifs in ACBP4 and ACBP5, suggests that these ACBPs interact with protein partners (Leung et al. 2004; Li and Chye 2004). ACBP1 (Chye 1998; Chye et al. 1999) and ACBP2 (Li and Chye 2003) are membrane-associated proteins while ACBP3 is an extracellularly-targeted protein (Leung et al. 2006). Western blot analysis of Arabidopsis subcellular fractions and immuno-electron microscopy using anti-ACBP1 antibodies, as well as confocal laser scanning microscopy of autoflurescence-tagged ACBP1 transiently expressed in onion epidermal cells, localized ACBP1 to the ER and the plasma membrane (Chye 1998; Chye et al. 1999; Li and Chye 2003). Western blot analysis of Arabidopsis subcellular fractions and confocal microscopy of autofluorescence-tagged ACBP2 located ACBP2 to the ER and the plasma membrane (Chye et al. 2000; Li and Chye 2003). The extracellular localization of ACBP3 was experimentally verified by confocal laser scanning microscopy using autofluorescence-tagged ACBP3, transiently expressed in tobacco BY-2 cells and onion epidermal cells (Leung et al. 2006).
We have demonstrated by in vitro site-directed mutagenesis that the acyl-CoA-binding domain in each of ACBP1 to ACBP5 functions in binding long-chain acyl-CoA esters, implying that these ACBPs can participate in the subcellular transportation of acyl-CoA esters in the plant cell (Chye 1998; Chye et al. 2000; Leung et al. 2004, 2006). Their preferential affinities for various acyl-CoA esters suggest that they have various cellular roles (Chye 1998; Chye et al. 2000; Leung et al. 2004, 2006). By using in vitro binding assays, recombinant (His)6-ACBP4 and (His)6-ACBP5 expressed in Escherichia coli were observed to bind oleoyl-CoA esters well; thus ACBP4 and ACBP5 are likely candidates that can transfer oleoyl-CoA esters from the chloroplasts to the ER (Leung et al. 2004). To substantiate their biological functions in the cytosol related to the transfer of oleoyl-CoA esters in plant lipid metabolism, the subcellular localizations of ACBP4 and ACBP5 were addressed in this study.
Materials and Methods
Plant materials and growth conditions
Onions (Allium cepa L.) were obtained from a local supermarket for particle gun bombardment. Unless otherwise stated, Arabidopsis thaliana ecotype Columbia (Col-0) was grown under 16 h light (23 °C)/8 h dark (21 °C) cycles.
Western blot analysis
Protein extracts were prepared by homogenizing Arabidopsis tissues in ice-cold extraction buffer (0.1 M TES, pH 7.8, 0.2 M NaCl, 1 mM EDTA, 2% β-mercaptoethanol and 1 mM PMSF). Total proteins were separated on SDS-PAGE and transferred onto Hybond-C membranes (Amersham). The blots were blocked in TTBS (TBS plus 0.05% Tween 20) containing 5% nonfat milk for 2 h and incubated for an additional 2 h with anti-ACBP4 or anti-ACBP5 primary antibodies. The blots were washed three times with TTBS and then incubated with secondary antibody for 1 h. Either the Amplified Alkaline Phosphatase Goat Anti-rabbit Immuno-blot Assay Kit (BioRad) or the ECL Western Blotting Detection Kit (Amersham) was used following the manufacturer’s instructions to detect cross-reacting bands. To generate ACBP4- and ACBP5-specific antibodies, synthetic peptides (RMQTLQLRQELGEAE corresponding to amino acids 566 to 580 of ACBP4, and KEELAEIDTRNTE corresponding to amino acids 554 to 566 of ACBP5) were used for rabbit immunization.
Subcellular fractionation of Arabidopsis proteins by differential centrifugation
Subcellular fractionation of Arabidopsis proteins was carried out following the protocols as described (Smith et al. 1988; Zhang et al. 2007) with minor modifications. Three-week-old wild-type (Col-0) Arabidopsis rosettes (2–3 g) were ground to fine powder in liquid nitrogen using a mortar with a pestle. The powder was transferred into 10 ml grinding buffer (0.3 M sucrose, 40 mM Tris-HCl (pH 7.8), 5 mM MgCl2, 1 mM PMSF) and swelled on ice for 5 min. Homogenization was performed for two 30-second pulses at high-speed setting. The homogenate was filtered through two layers of Miracloth (Tetko, Elmsford, N.Y., USA) and was subsequently separated by centrifugation at 350 g for 10 min at 4 °C. The pellet (crude nuclear) was further layered onto 1 ml of 2.3 M sucrose, 50 mM Tris-HCl (pH 8.8), 5 mM MgCl2 in an Eppendorf tube for centrifugation at 15,000 g for 10 min at 4 °C, to obtain the nuclear fraction in the derived pellet. Supernatants from the first low-speed centrifugation (350 g) were centrifuged at 12,000 g for 20 min at 4 °C. The pellet contained large particles including mitochondria, chloroplasts and peroxisomes. The supernatant was further centrifuged at 100,000 g for 1 h at 4 °C to yield the soluble cytosol fraction in the resulting supernatant. The pellet representing the membrane fraction was resuspended in 0.1 ml grinding buffer. Protein concentration in the extract was determined following the method of Bradford (1976) using the Bio-Rad Protein Assay Kit I.
Immuno-electron microscopy
Arabidopsis roots and leaves were fixed in a solution of 4% (v/v) paraformaldehyde and 0.5% (v/v) glutaradehyde in 0.1 M phosphate buffer (pH 7.2) for 20 min under vacuum and then a further 3 h at room temperature. The specimens were then dehydrated in a graded ethanol series, infiltrated in stepwise increments of LR white resin (London Resin, Theal, Berkshire, UK) and polymerized at 45 °C for 24 h. Materials for immuno-gold labeling were prepared according to the procedure of Varagona and Raikhel (1994) with modification as described. Specimens (90 nm) were sectioned using a Leica Reichert Ultracut S microtome and mounted on formvar-coated slotted grids. Grids were incubated in a blocking solution of TTBS containing 1% (w/v) fish skin gelatin and 1% (w/v) BSA for 30 min. Anti-ACBP4 and anti-ACBP5 antibodies diluted 1:50 in blocking solution were added and incubated at room temperature for 2 h. The grids were then rinsed three times, each for 5 min, in TTBS and then incubated for 2 h in 10 nm gold-conjugated goat anti-rabbit IgG secondary antibody (Sigma), diluted 1:20 with blocking solution. Grids were rinsed three times, each for 5 min in TTBS, following by three 5-min rinses in distilled water. After staining in 2% (w/v) uranyl acetate for 6 min followed by 2% (w/v) lead citrate for 6 min, the sections were visualized and photographed using Philips EM208s electron microscope operating at 80 kV. Control sections were prepared by replacing the primary antibody with blocking solution.
Construction of autofluorescence-tagged plasmids
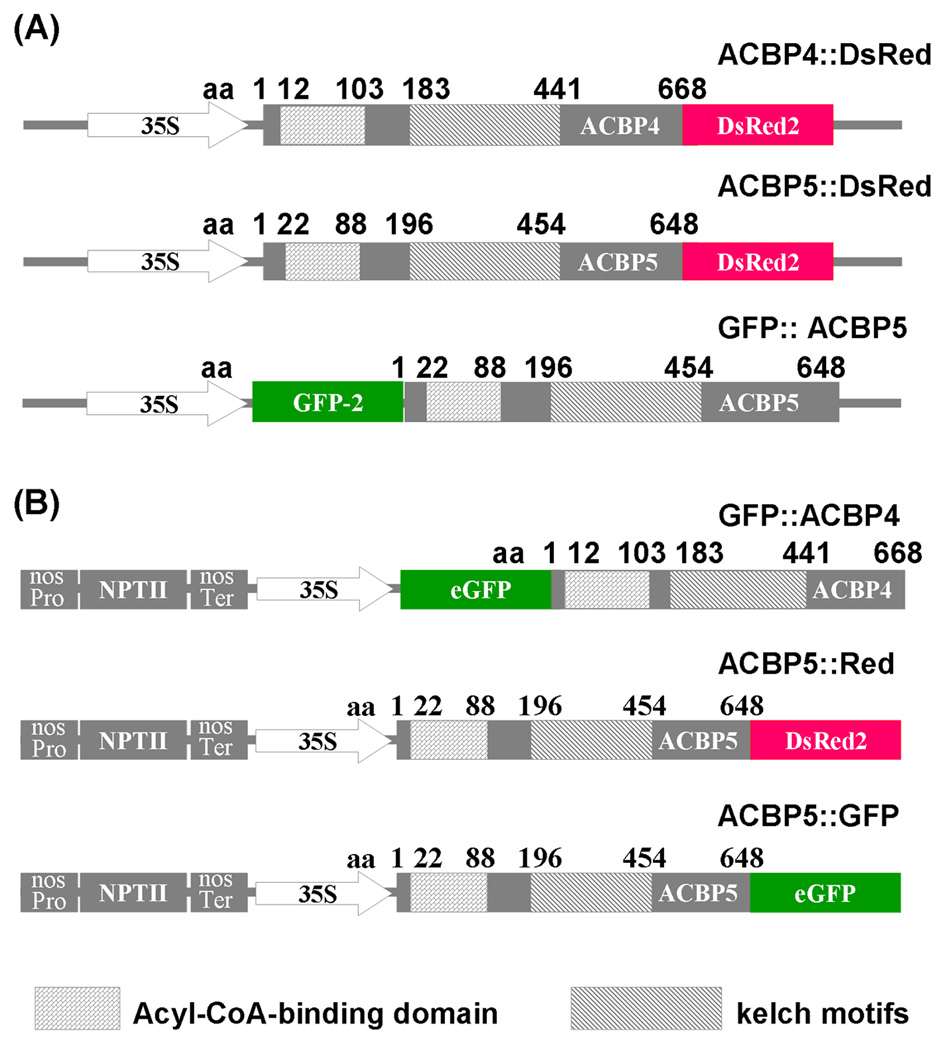
All binary vectors used in this study were derived from plasmids pRGD, pGDG and pBI-eGFP which contain genes encoding the autofluorescent proteins DsRed2, GFP and eGFP, respectively (Goodin et al. 2002; Leung et al. 2006; Shi et al. 2005). For the pACBP4∷DsRed construct, a 2-kbp XhoI-BamHI fragment encoding the complete ACBP4 coding sequence was generated by PCR using plasmid pAT181 (Leung et al. 2004) as template. For the pACBP5∷DsRed construct, a 1.9-kbp XhoI-BamHI fragment encoding the complete ACBP5 coding sequence was generated by PCR using plasmid pAT182 (Leung et al. 2004) as template. The primers used in PCR were as follows: for ACBP4, ML350 (5’-CCTCGAGAATGGCTATGCCTAGGGC-3’, XhoI site underlined) and ML682 (5’-GGATCCACAAGGCGAATCATCATCT-3’, BamHI site underlined) and for ACBP5, ML683 (5’- CTCGAGATGGCTCACATGGTGAGAGCG −3’, XhoI site underlined) and ML684 (5’- GGATCCACATGTTTTAGGCGGAGGAG −3’, BamHI site underlined). The fragments were subsequently cloned in pGEM-T Easy vector to generate plasmids pAT280 and pAT281, respectively. The 2-kbp XhoI-BamHI ACBP4 fragment derived from plasmid pAT280 was cloned into the XhoI and BamHI sites of pRGD to produce pACBP4∷DsRed. The 1.9-kbp XhoI-BamHI ACBP5 fragment derived from plasmid pAT281 was cloned into corresponding restriction endonuclease sites on pRGD to generate pACBP5∷DsRed. To generate the pGFP∷ACBP5 construct, the 1.9-kbp XhoI-BamHI fragment released from pAT189 (a pBluescript II SK (−) derivative containing a 1.9-kbp ACBP5 full-length cDNA) was cloned into similar sites of pGDG. In the above plasmids, DsRed2 was fused to the C-terminus of ACBP4 or ACBP5, and GFP was fused to the N-terminus of ACBP5.
The pGFP∷ACBP4 fusion was constructed by generating a 2-kbp ACBP4 cDNA fragment by PCR using primers ML849 (5’-AGCTCGAGATGGCTATGCCTAGGGCAAC −3’, XhoI site underlined), and ML850 (5’- CGGAGCTCAATGGCATTACCGGACCAAA −3’, SacI site underlined) and cloning into pGEM-T Easy vector to obtain plasmid pAT361. The XhoI-SacI fragment from pAT361 was then inserted into the XhoI and SacI sites of plant transformation vector pBI-eGFP. For the pACBP5∷GFP construct, a 1.9-kbp and BglII-BamHI fragment derived from plasmid pAT282 was cloned into the BamHI site of pBI-eGFP. The pACBP5∷Red plasmid was generated by insertion of a 2.6-kbp BglII-BglII fragment from plasmid pAT283 into the BamHI site of plant transformation vector pSa13. Plasmid pSa13 is a pBI121 derivative obtained by a 0.5-kbp SalI deletion of plasmid pSa7 (Xu et al., 2004). Since the T-DNAs of plant transformation vectors, pGFP∷ACBP4, pACBP5∷GFP and pACBP5-Red harbor the kanamycin-resistant selectable marker, they were used for the generation of transgenic Arabidopsis plants. The cloning junctions in all constructs were confirmed by nucleotide sequence analysis before further use.
Transient expression in onion epidermal cells
Plasmids encoding autofluorescent protein fusions were introduced into onion epidermal cells by particle gun bombardment using a Biolistic PDS-1000/He system (BioRad). Gold particles (1.0 µm) were coated with the respective plasmid DNA and a helium pressure of 9.3 MPa was employed. About 360 µg of gold particles coated with 0.9 µg DNA was used in one shot. The target distance between the stop screen and onion piece was set at 9 cm. Following incubation in darkness at 22 °C for 15 h, the onion epidermal cells were examined under a Zeiss LSM 510 inverted confocal laser-scanning microscope Zeiss (Jena, Germany). Single optical sections were scanned as resulting images for each transient expression.
Northern blot analysis
Total RNA from plant tissues was isolated using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Thirty micrograms of total RNA were separated on a 1.5% agarose gel containing 6% formaldehyde and transferred to Hybond N membranes (Amersham). The following gene-specific primers were used to generate probes for RNA blot analysis: ACBP4 (ML350 and ML682) and ACBP5 (ML352, 5’-CGGATCCAATGGCTCACATGGTGAGAGCAG −3’ and ML353, 5’-CGAATTCTCATGGGCACTCATGTTTTAGGC −3’). The fragments were labeled with the PCR Digoxigenin Probe Synthesis Kit according to the manufacturer‧s instructions (Roche). Hybridization and detection were performed according to standard procedures as advised by the manufacturer (Roche). The blots were washed under conditions of high stringency (2 × SSC, 0.1% SDS for 2 × 15 min at room temperature; 0.5 × SSC, 0.1% for 2 × 15 min at 68 °C; 0.1 × SSC, 0.1% SDS for 2 × 15 min at 68 °C).
Laser scanning confocal microscopy
A Zeiss LSM 510 inverted confocal laser scanning microscope equipped with helium/neon lasers and multitracking was used for the analysis of GFP and DsRed localizations. GFP fluorescence was excited at 488 nm, filtered through a primary dichroic (UV/488/543), a secondary dichroic of 545 nm and subsequently through BP505-530 nm emission filters to the photomultiplier tube (PMT) detector. DsRed fluorescence was excited at 543 nm, the emission was passed through similar primary and secondary dichroic mirrors and finally through a BP560–615 nm emission filter to the PMT detector. The images were processed using the LSM 510 software (Zeiss, Jena, Germany).
Generation of transgenic plants expressing the autofluorescence-tagged fusions
Transgenic Arabidopsis plant lines expressing GFP∷ACBP4, ACBP5∷GFP and ACBP5∷Red fusions (Fig. 3b) were generated by Agrobacterium-mediated plant transformation (Clough and Bent 1998). Following the initial screening of positive transformants on kanamycin-containing plant growth medium, they were confirmed by PCR analysis by using the CaMV 35S promoter specific primer 35SB (5’-CAATCCCACTATCCTTCGCAAGACC-3’) and gene-specific reverse primers ML850 (for GFP∷ACBP4) and ML852 (for ACBP5∷GFP and ACBP5∷Red). Preparation of mesophyll protoplasts from transgenic Arabidopsis expressing GFP∷ACBP4 and ACBP5∷GFP was performed according to Abel and Theologis (1994).
Fig. 3.
Constructs containing ACBP4- and ACBP5-tagged fusions.
(a) Generation of pACBP4∷DsRed, pACBP5∷DsRed and pGFP∷ACBP5 fusion constructs in which DsRed2 and GFP are translationally fused to amino acids 1 to 668 of ACBP4 and 1 to 648 of ACBP5, respectively.
(b) Generation of pGFP∷ACBP4, pACBP5∷GFP and pACBP5∷Red fusions. GFP from vector pBI-eGFP (Shi et al., 2005) translationally fused to the N-terminus of ACBP4 and the C-terminus of ACBP5 to generate pGFP∷ACBP4 and pACBP5∷GFP, respectively. The ACBP5∷DsRed fragment in (a) was cloned into binary vector pSa13, to generate pSa13-ACBP5∷DsRed (for differentiation, this fusion was designated as ACBP5∷Red).
Identification and complementation of an acbp4 mutant
The acbp4 T-DNA insertion mutant (SALK_040164) was screened from a T-DNA seed pool from SALK Institute Genomic Analysis Laboratory (http://signal.salk.edu/). The T-DNA insertion in the gene was identified by PCR using T-DNA left border primer LBa1 (5’-TTTTTCGCCCTTTGACGTTGGA-3’), ACBP4-specific forward primer ML412 (5’-CAGATCCTGTTGTAGAT-3’) and reverse primer ML418 (5’-TTGCCCGCCAAATATCA-3’). The PCR products were sequenced to confirm the T-DNA insertion site. Individual homozygous T-DNA mutant plants were identified by PCR. Amplification was initiated with denaturation at 95 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min, and another extension at 72 °C for 10 min.
To check for ACBP4 mRNA expression in the acbp4 mutant, total RNA was isolated using TRIzol reagent (Invitrogen, Cat No. 15596-018) from leaves of 3-week-old wild-type and acbp4 mutant plants, followed by reverse transcription-PCR (RT-PCR) analysis using the Superscript™ First-strand synthesis system (Invitrogen, Cat No. 12371-019). Gene-specific primers used in RT-PCR were ML413 (5’-CAACAAGCTGCTGTCTATC-3’) and ML416 (5’-CCATGACAATTTCCCGTAC-3’). The ACTIN control was amplified using primers 5’-CACCGCTTAACCCGAA-3’ and 5’-GTGAGGTCACGACCAG-3‧. PCR amplification was initiated with denaturation at 95 °C for 3 min, followed by 28 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, and an extension at 72 °C for 10 min.
The complementation of the acbp4 mutant was carried out by Agrobacterium-mediated transformation (Clough and Bent, 1998) using plant transformation vector pAT324, which was generated by insertion of a 2.0-kbp XhoI-EcoRI ACBP4 full-length cDNA fragment from pAT181 (Leung et al. 2004) in binary vector pKMB (Mylne and Botella 1998). The putative transformants (designated as cACBP4) were selected on MS medium containing Basta (57.8 µg/mlglufosinate). They were screened by PCR analysis using primer pairs 35SB/ML418, ML412/ML418 and LBa1/ML418 to identify putative cACBP4 transgenic plant lines.
Lipid analysis
Total lipids were extracted from 4-week-old seedlings of wild type, acbp4 mutant and two independent acbp4-complemented transgenic lines (cACBP4 #1 and #2) grown on MS medium containing 2% sucrose under continuous light according to the protocol provided by the Kansas Lipidomics Research Center (www.K-state.edu/lipid/lipidomics). Lipids were dried under nitrogen and polar lipids were analyzed on activated silica gel TLC plates (Merck, Germany) developed with chloroform/methanol/acetic acid/water (170/30/20/7, v/v/v/v) as a solvent system according to Branen et al. (2003). Lipids were visualized by staining with α-naphthol spray reagent and were detected at 120 °C.
For membrane lipid profiling, lipids were extracted as described above, from 5-week-old wild-type (Col-0) and acbp4 mutant Arabidopsis grown in a growth chamber under 16 h light, 23 °C/8 h dark, 21 °C. The solvent was evaporated under nitrogen after extraction and samples were sent by courier service for lipid profiling at the Kansas Lipidomics Research Center.
RESULTS
Prediction on the subcellular localization of ACBP4 and ACBP5
The PSORT WWW server (http://psort.nibb.ac.jp) is a computer program for the prediction of protein subcellular localization (Nakai and Horton 1999). We have found it reliable in predictions on the subcellular localizations of ACBP1, ACBP2 and ACBP3, which were subsequently experimentally verified (Leung et al. 2006; Li and Chye 2003). Using the PSORT WWW server, the predicted subcellular localization scores for ACBP4 and ACBP5 were highest in the cytosol (0.65 and 0.45, respectively). However, these scores for ACBP4 and ACBP5 were lower than those previously predicted for ACBP1 (0.69), ACBP2 (0.81) and ACBP3 (0.85). To investigate if they are targeted to organelles such as the chloroplast (scores of 0.28 for both ACBP4 and ACBP5), peroxisomes (0.36 for ACBP5) or mitochondrial matrix space (0.36 for ACBP5), we searched for the presence of putative targeting signal sequences in ACBP4 and ACBP5 using the TargetP 1.1 Server, but none were located.
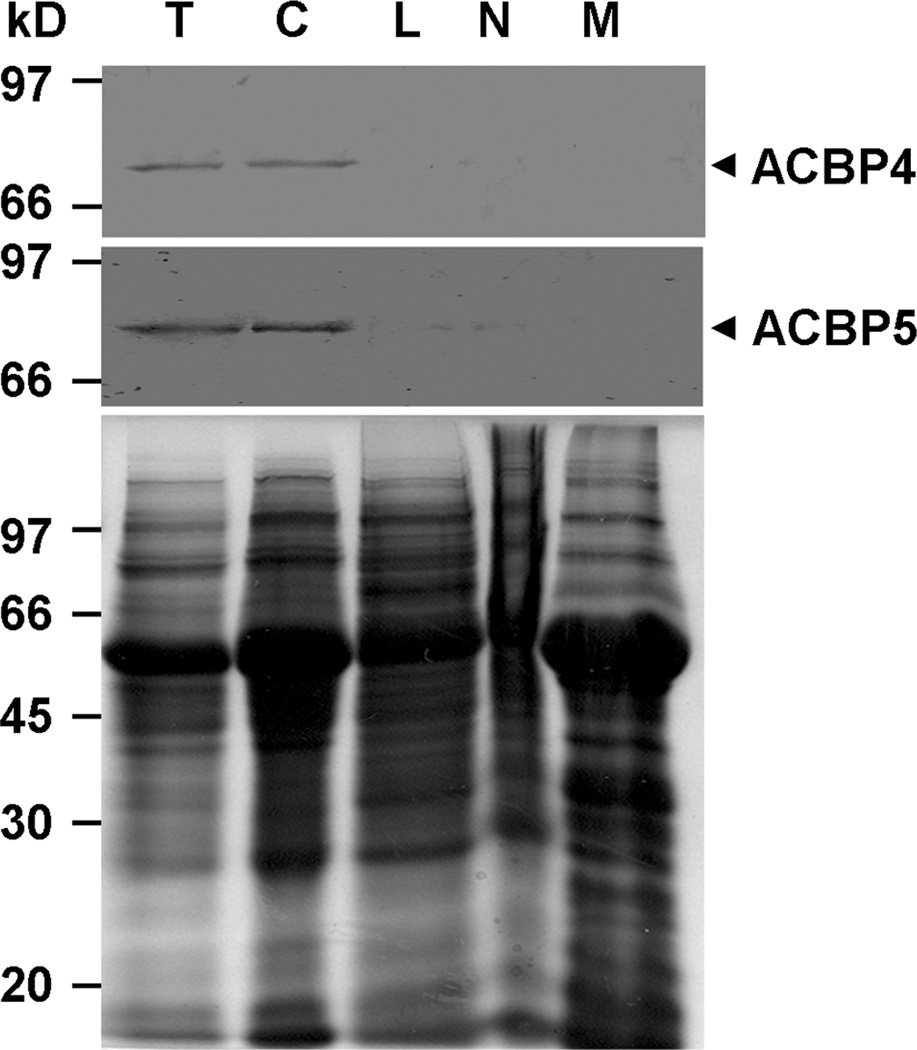
Localization of ACBP4 and ACBP5 proteins in the cytosolic fraction of Arabidopsis Detection of ACBP4 and ACBP5 proteins was investigated by western blot analysis of subcellular protein fractions from 3-week-old wild-type Arabidopsis (Col-0) derived following differential centrifugation. Protein extracts were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blot analysis using ACBP4-specific and ACBP5-specific polyclonal antibodies. Results from western blot analysis revealed that anti-ACBP4 antibodies cross-reacted with a band of apparent molecular mass of 73.1 kD, as predicted for ACBP4, in total protein (Fig. 1, upper panel, lane T) and in the cytosol fraction (Fig. 1, upper panel, lane C). Western blot analysis also showed that the anti-ACBP5 antibodies cross-reacted with a 71-kDa band in total protein (Fig. 1, middle panel, lane T) and in the cytosol fraction (Fig. 1, middle panel, lane C), corresponding well to the predicted mass of ACBP5.
Fig. 1.
Western blot analyses of protein extracts from subcellular fractions of 3-week-old Arabidopsis using anti-peptide antibodies against ACBP4 and ACBP5.
Total whole plant protein (lane T), cytosol (lane C), large particles including mitochondria, chloroplasts and peroxisomes (lane L), nuclei (lane N), and membrane (lane M) subcellular fractions. Bottom, gel identically loaded was stained with Coomassie blue to show the protein amount in each lane.
Both ACBP4 and ACBP5 are immunolocalized in the cytosol of Arabidopsis roots and leaves by electron microscopy
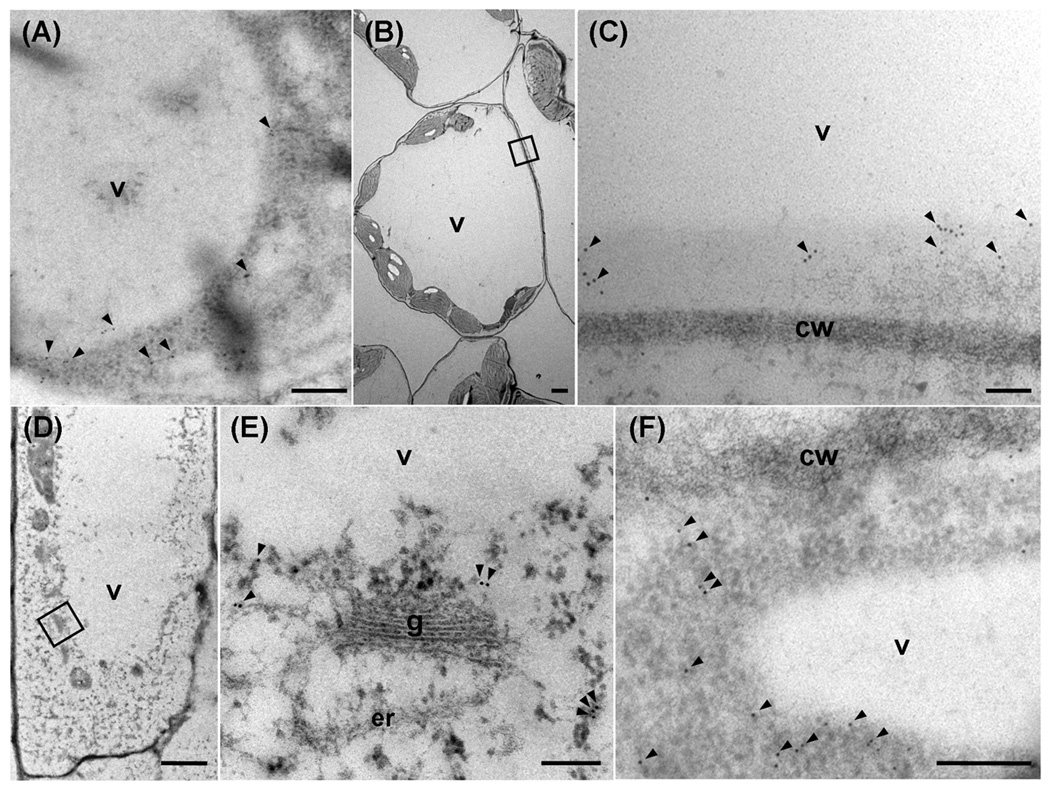
To confirm the subcellular localization of ACBP4 and ACBP5 in the Arabidopsis cytosol, immuno-electron microscopy was carried out using transverse sections of leaves and roots of 2-week-old Arabidopsis germinated and grown in MS medium under a 16 h light/8 h dark regime. In leaves, ACBP4 (Fig. 2a), and ACBP5 (Fig. 2b, c) were detected in the cytosol. In roots, ACBP4 (Fig. 2d, e) and ACBP5 (Fig. 2f) were again detected in the cytosol. In the control, no immuno-gold labeling was seen when the primary antibody was replaced with blocking solution.
Fig. 2.
Comparison of ACBP4 and ACBP5 in Arabidopsis leaf and root cells by immuno-gold labeling using transmission electron microscopy.
Transverse sections were stained with affinity-purified ACBP4-specific antibodies (a, d and e) or ACBP5-specific antibodies (b, c and f).
(a) and (b) Transverse sections of leaves stained with ACBP4-specific antibodies (a) or ACBP5-specific antibodies (b).
(c) Magnification of the boxed area in (b).
(d) and (f) Transverse sections of roots stained with ACBP4-specific antibodies (d) or ACBP5-specific antibodies (f).
(e) Magnification of the boxed area in (d).
Arrowheads, gold particles. v, vacuole; cw, cell wall; g, Golgi apparatus; er, endoplasmic reticulum. Bars in (a), (c), (e) and (f) represent 0.2 µm, and in (b) and (d), 2 µm.
Comparison of the expression of autofluorescence-tagged ACBP4 and ACBP5 in transiently expressed onion epidermal cells
To verify the subcellular localizations of ACBP4 and ACBP5 in vivo, we tagged each of ACBP4 and ACBP5 to the N-terminus of the red fluorescent protein (DsRed2) in vector pRGD (Leung et al. 2006) generating ACBP4∷DsRed and ACBP5∷DsRed fusions (Fig. 3a). ACBP5 was also fused to the C-terminus of the green fluorescent protein (GFP) in vector pGDG (Goodin et al. 2002) generating GFP∷ACBP5 (Fig. 3a). Since plasmids pRGD and pGDG lack antibiotic-resistant marker genes for selection in plants (Leung et al. 2006; Goodin et al. 2002), its derivatives were not applicable for stable plant transformation. Hence, binary vector pBI-eGFP (Shi et al. 2005) was utilised in the generation of GFP∷ACBP4 and ACBP5∷GFP fusions (Fig. 3b), and were subsequently used to produce transgenic Arabidopsis by kanamycin-selection (Fig. 3b). Also, the ACBP5∷DsRed fragment (Fig. 3a) was cloned into binary vector pSa13 (a pBI121 derivative) to generate plasmid pSa13-ACBP5∷DsRed. For differentiation, this fusion was designated as ACBP5∷Red (Fig. 3b).
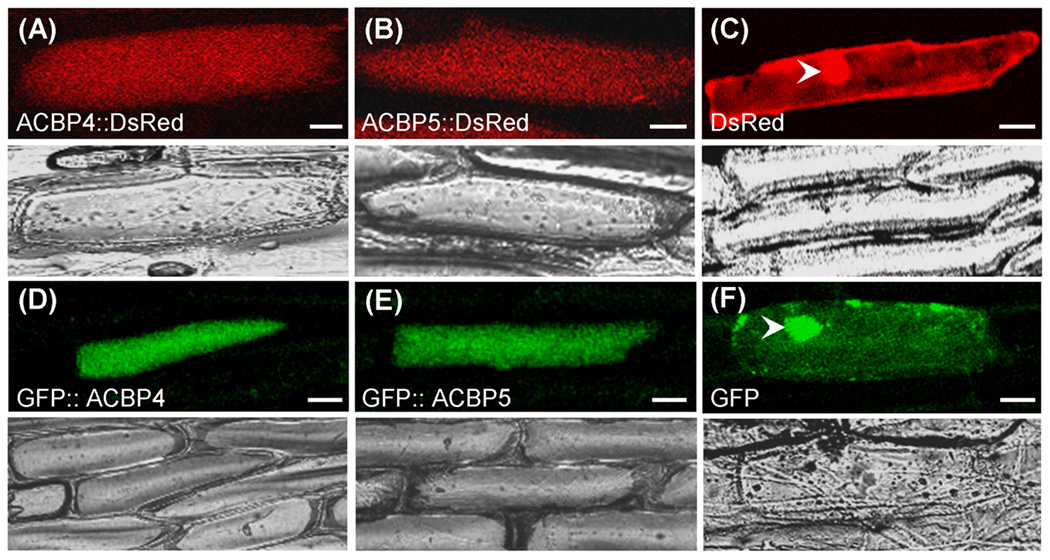
The ACBP4∷DsRed, ACBP5∷DsRed, GFP∷ACBP4 and GFP∷ACBP5 constructs were transiently expressed in onion epidermal cells by particle gun bombardment (Fig. 4), and the images of single optical sections were analyzed by confocal laser scanning microscopy. In onion epidermal cells expressing ACBP4∷DsRed (Fig. 4a) and ACBP5∷DsRed (Fig. 4b), fluorescence was observed throughout the cells, consistent with its predicted cytosolic localization. Onion cells expressing DsRed from vector pRGD showed fluorescence in the cytosol and nuclei, the latter due to diffusion (Fig. 4c). When the C-terminal fusions, GFP∷ACBP4 (Fig. 4d) and GFP∷ACBP5 (Fig. 4e), were transiently expressed in onion epidermal cells, their fluorescence patterns were similar to those of the N-terminal fusions. With GFP expressed from vector pGDG (Fig. 4f) or pBI-eGFP (data not shown), signals were detected in the cytosol and nuclei as expected.
Fig. 4.
Comparison of the expression of ACBP4- and ACBP5-tagged DsRed and GFP fusion proteins transiently expressed in onion epidermal cells.
(a–c) Confocal images showing localization of ACBP4∷DsRed (a) and ACBP5∷DsRed (b) and DsRed control (c). The arrowhead indicates cell nucleus. Bottom, corresponding transmitted light images.
(d–f) Confocal images showing localization of GFP∷ACBP4 (d), GFP∷ACBP5 (e) and GFP control (g). The arrowhead indicates cell nucleus. Bottom, corresponding transmitted light images.
Bars = 20 µm.
Localization of GFP∷ACBP4, ACBP5∷GFP and ACBP5∷Red fusion proteins in transgenic plants
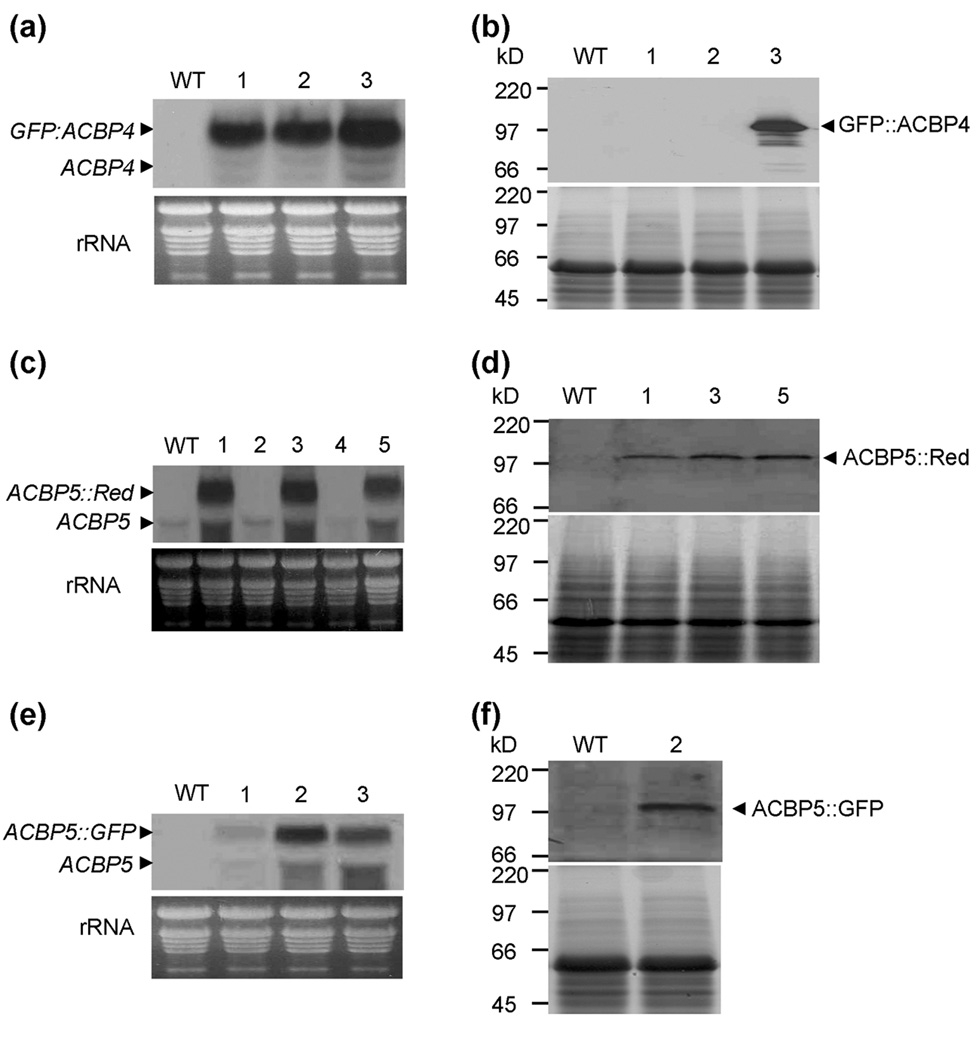
To further confirm the subcellular localization of ACBP4 and ACBP5, transgenic Arabidopsis expressing GFP∷ACBP4, ACBP5∷GFP and ACBP5∷Red (Fig. 3b) were generated by Agrobacterium-mediated transformation. Positive transformants were initially selected using kanamycin-containing plant growth medium and subsequently confirmed by PCR analysis (data not shown) using forward primer 35SB, which is specific to the CaMV 35S promoter, and gene-specific reverse primers ML850 (for GFP∷ACBP4) and ML852 (for ACBP5∷GFP and ACBP5∷Red). The expression of GFP∷ACBP4, ACBP5∷GFP and ACBP5∷Red in T3 stable transgenic plants were examined by northern blot analysis. A 2.7-kbp GFP∷ACBP4 mRNA was detected in all 3 independent transgenic lines tested (Fig. 5a), a 2.6-kbp ACBP5∷Red mRNA band in 3 out of 5 independent transgenic lines tested (Fig. 5c) and a 2.6-kbp ACBP5∷GFP mRNA in all 3 independent transgenic lines (Fig. 5e). In these RNA blots, no hybridizing band was detected in wild-type Arabidopsis. Subsequently, by using anti-GFP or anti-DsRed antibodies, western blot analysis indicated that some transgenic lines were confirmed to produce 100.4-kDa GFP∷ACBP4 (Fig. 5b, line 3), 99.5-kDa ACBP5∷Red (Fig. 5d, lines 1, 3 and 5) and 98.5-kDa ACBP5∷GFP (Fig. 5f, line 2) cross-reacting bands.
Fig. 5.
Identification of transgenic Arabidopsis expressing GFP∷ACBP4, ACBP5∷GFP and ACBP5∷Red fusion proteins by northern blot and western blot analyses.
(a) Northern blot analysis of wild type and 3 independent transgenic plants transformed with GFP∷ACBP4. The 2.7-kbp GFP∷ACBP4 and the 2.0-kbp endogenous ACBP4 mRNAs are marked. Bottom, ethidium bromide-stained rRNA on the gel before blotting indicates the relative amounts of total RNA loaded per lane.
(b) Western blot analysis of wild type and 3 independent transgenic plants transformed with GFP∷ACBP4 show a 100.4-kDa GFP∷ACBP4 cross-reacting band. Bottom, protein gel identically loaded, stained with Coomassie blue.
(c) Northern blot analysis of wild type and 5 independent transgenic plants transformed with ACBP5∷Red. The 2.6-kbp ACBP5∷Red and the 1.9-kbp endogenous ACBP5 mRNAs are marked. Bottom, ethidium bromide-stained rRNA on the gel before blotting indicate the relative amounts of total RNA loaded per lane.
(d) Western blot analysis of wild type and transgenic lines #1, #3 and #5 transformed with ACBP5∷Red show a 99.5-kDa GFP∷ACBP4 cross-reacting band. Bottom, a protein gel identically loaded, stained with Coomassie blue.
(e) Northern blot analysis of wild type and 3 independent transgenic plants transformed with ACBP5∷GFP. The 2.6-kbp ACBP5∷GFP and the 1.9-kbp endogenous ACBP5 mRNAs are marked. Bottom, ethidium bromide-stained rRNA on the gel before blotting indicate the relative amounts of total RNA loaded per lane.
(f) Western blot analysis of wild type and transgenic lines #2 transformed with ACBP5∷GFP show a 98.5-kDa GFP∷ACBP4 cross-reacting band. Bottom, a protein gel identically loaded, stained with Coomassie blue.
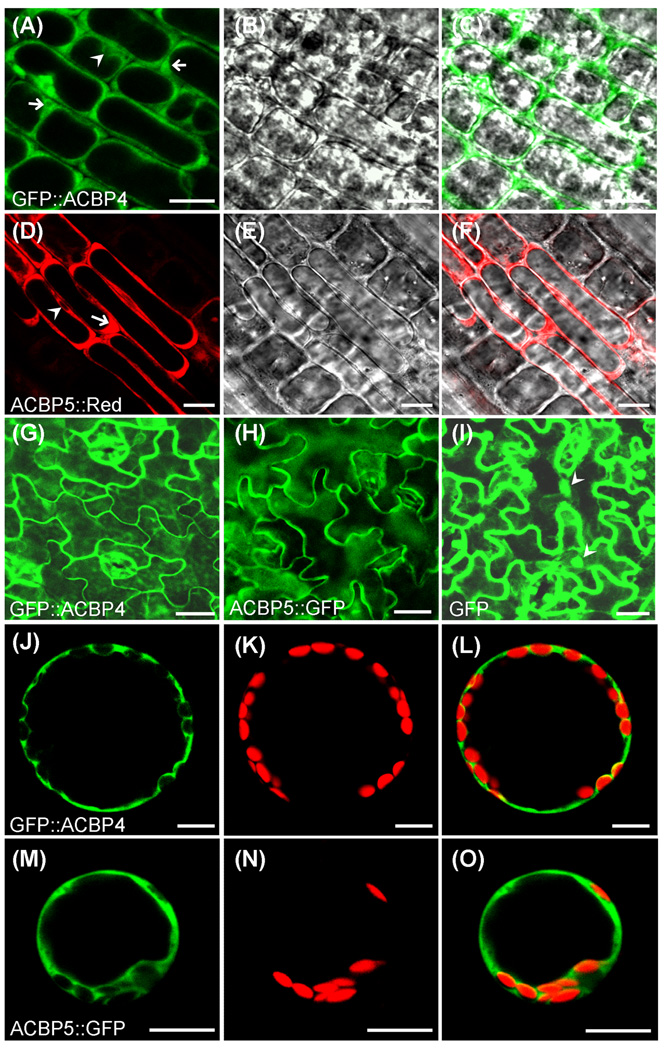
Confocal microscopy on the transgenic lines, previously verified by northern blot and western blot analysis, showed fluorescence of GFP∷ACBP4 and ACBP5∷Red predominately in the cytosol of premature primary root tip cells. Fluorescence of GFP∷ACBP4 (Fig. 6a–c) and ACBP5∷Red (Fig. 6d–f) were restricted to the cytosolic zones surrounding the premature vacuoles (arrows in Fig. 6a, d) and the nuclei (arrowheads in Fig. 6a, d). Detection of fluorescence in the cytosol surrounding the nuclei (Fig. 6a, d) implies that signals are within the cytosol rather than the plasma membrane, despite the presence of vacuoles which occupy much intracellular space.
Fig. 6.
Analysis of transgenic Arabidopsis expressing GFP∷ACBP4 and ACBP5∷GFP/ACBP5∷Red fusion proteins.
(a) and (d) Confocal images showing premature cells of root tips expressing GFP∷ACBP4 (a) and ACBP5∷Red (d). Arrows indicate the cytosol zones adjacent to large vacuoles and arrowheads indicate cell nuclei. Bars = 10 µm.
(b) and (e) Light images of (a) and (d), respectively.
(c) Immerged images of (a) and (b).
(f) Immerged images of (d) and (e).
(g–i) Confocal images of leaf epidermal cells expressing GFP∷ACBP4 (g), ACBP5∷GFP (h) and GFP control (i). Arrowheads indicate cell nuclei. Bars = 20 µm.
(j–l) Confocal image of a mesophyll protoplast showing GFP fluorescence of GFP∷ACBP4 (j), red fluorescence of chloroplasts (k) and their overlapping images (l). Bars = 10 µm.
(m–o) Confocal image of a mesophyll protoplast showing the GFP fluorescence of ACBP5∷GFP (m), red fluorescence of chloroplast (n) and their overlapping images (o). Bars = 10 µm.
Consistently, GFP fluorescence was observed in the cytosol of leaf epidermal cells from transgenic plants expressing GFP∷ACBP4 (Fig. 6g) and ACBP5∷GFP (Fig. 6h). Green fluorescence from the GFP control was observed in both cytosol and nuclei (Fig. 6i). To distinguish localization of these fusions in the cytosol from the plasma membrane, mesophyll protoplasts were isolated from transgenic Arabidopsis expressing GFP∷ACBP4 and ACBP5∷GFP. In protoplasts, GFP∷ACBP4 (Fig. 6j–l) and ACBP5∷GFP (Fig. 6m–o) fluorescence signals were detected in the cytosol and not the plasma membrane or the chloroplasts, which was distinguished by a red autofluorescence due to chlorophyll. Further, green signals detected in the cytosol surrounding the chloroplasts (Fig. 6i–o), confirmed presence of the fusion proteins in the cytosol rather than the plasma membrane. Taken together, our results using various methods of subcellular localization (western blot analyses and immuno-electron microscopy using ACBP4- and ACBP5-specific antibodies and confocal microscopy of autofluorescent-tagged ACBP4 and ACBP5) are consistent with computer predictions on the subcellular localization of both ACBP4 and ACBP5 in the cytosol.
Characterization of an acbp4 knockout mutant
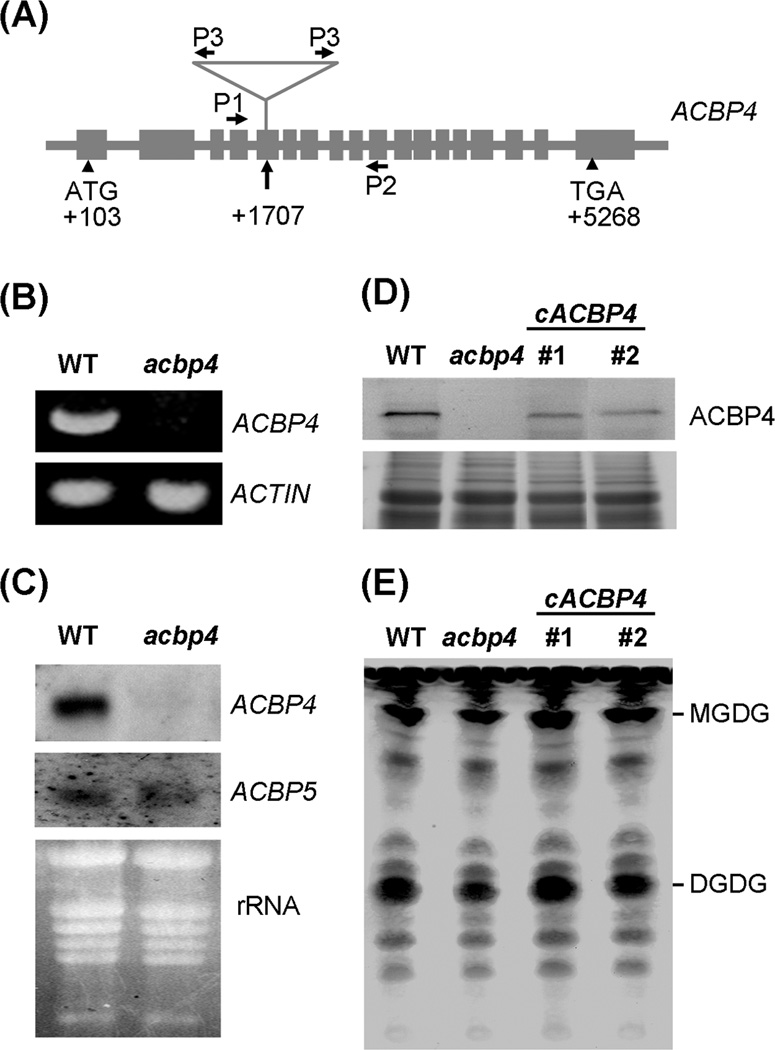
To further examine the function of ACBP4 in lipid metabolism, a T-DNA knock-out mutant of ACBP4 (designated acbp4) was identified from the SALK No. SALK_040164 (ecotype Col-0). PCR analysis of this mutant followed by DNA sequence analysis confirmed that the T-DNA was inserted in the fifth exon of ACBP4 at position +1707 (Fig. 7a). The homozygous mutant plants were backcrossed twice with wild type (Col-0) to clear out other potential insertions. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) (Fig. 7b), northern blot (Fig. 7c) and western blot (Fig. 7d) analyses confirmed the lack of ACBP4 mRNA and protein in the acbp4 mutant, confirming that it was indeed a knockout mutant. The acbp4 mutant did not exhibit an altered phenotype in comparison to wild type when grown under normal conditions (16 h light, 23 °C/8 h dark, 21 °C). Northern blot analyses also indicated that ACBP5 mRNA levels were similar in wild-type and acbp4 mutant plants (Fig. 7c).
Fig. 7.
Characterization of the acbp4 mutant.
(a) Location of T-DNA insertion within ACBP4 (At3g05420) in the acbp4 mutant (SALK_040164). Primers (P1, ML412; P2, ML418; P3, LBa1) used for genotyping the mutant are indicated.
(b) Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis of ACBP4 expression in wild type and acbp4 mutant using primer pair ML413 and ML416 show the presence of a 0.8-kbp band in wild type that is absent in the mutant. Bottom, ACTIN transcript was used as a control in RT-PCR analysis.
(c) Northern blot analyses of ACBP4 and ACBP5 expression in wild type and the acbp4 mutant. RNAs were extracted from leaves of 4-week-old Arabidopsis germinated and grown on MS medium. DIG-labeled ACBP4- and ACBP5-cDNA probes were used in northern blot analysis and the blots were washed under conditions of high stringency.
(d) Western blot analysis on wild type, acbp4 mutant and two independent acbp4-complemented transgenic lines (cACBP4 #1 and #2) using antibodies against ACBP4. The cross-reacting 73.1-kDa ACBP4 band in wild type and cACBP4 transgenic lines was detected using the Amplified Alkaline Phosphatase Assay Kit. Bottom, gel identically loaded and stained with Coomassie blue.
(e) Thin-layer chromatography of leaf lipids extracted from 4-week-old Arabidopsis of wild type, acbp4 mutant and two independent acbp4-complemented transgenic lines (cACBP4 #1 and #2). The leaf tissues (100 mg) of each genotype were weighed before lipid extraction and the lipids were dissolved in 100 µl of chloroform after drying under nitrogen. Equal volumes (30 µl) of lipids were loaded in each lane. The galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) are indicated. The experiment was repeated twice with similar results.
Lipid changes in the acbp4 mutant
Lipid composition in leaves of the acbp4 knockout mutant was compared to wild type. Lipid analyses using thin-layer chromatography suggested that the levels of leaf polar lipids digalactosyldiacylglycerol (DGDG) and monogalactosyldiacylglycerol (MGDG) in the acbp4 mutant were noticeably reduced in comparison to wild type (Fig. 7e). To determine that the lipid changes in the acbp4 mutant were due to knockout of ACBP4, a complementation test was carried out using a construct expressing ACBP4 from the CaMV 35S promoter. Following Agrobacterium-mediated transformation of the acbp4 mutant, two independent transgenic lines were obtained and their genotypes were confirmed by PCR analyses using gene-specific primer pair ML412/ML418 and 35S promoter primer 35SB paired with gene-specific reverse primer ML418. Western blot analyses on these two transgenic lines revealed the ACBP4 protein were expressed upon introduction of ACBP4 cDNA in the acbp4 mutant (Fig. 7d, upper image). As expected, the levels of DGDG and MGDG in both acbp4-complemented transgenic Arabidopsis lines were elevated to wild type (Fig. 7e), suggesting that the lipid changes in the acbp4 mutant could be complemented by the introduction of the ACBP4 cDNA.
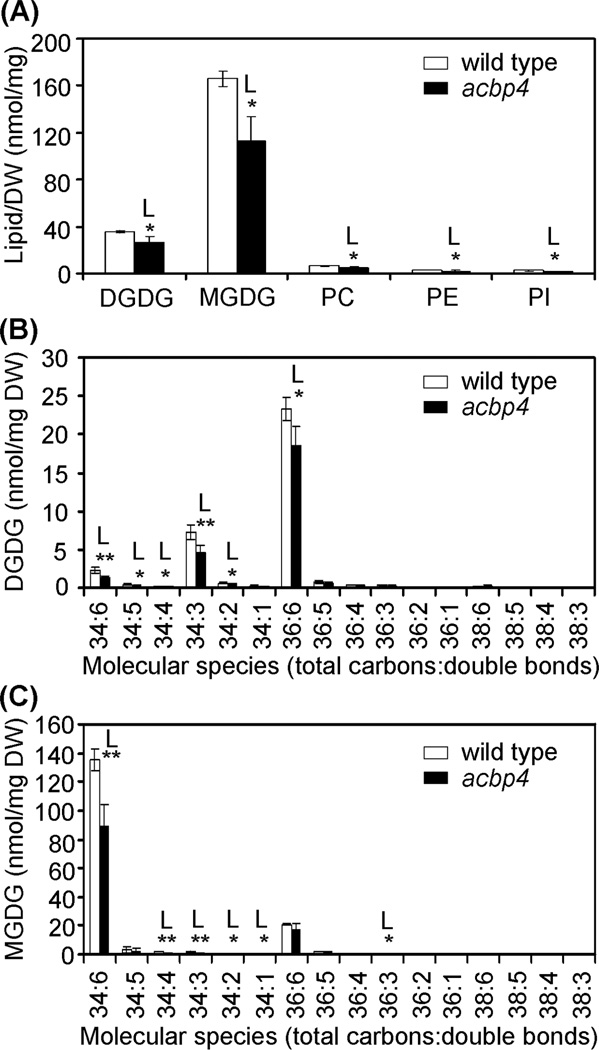
The lipid composition in the leaves of the acbp4 mutant were further analyzed by ESI-MS/MS (Welti et al., 2002) and compared to wild type. Only significant differences in membrane lipids were observed and these are shown in Fig. 8a. The total amounts of galactolipids, digalactosyldiacylglycerol (DGDG) and monogalactosyldiacylglycerol (MGDG), showed significant declines, 25.2% and 31.9% (P < 0.05), respectively, in the acbp4 mutant when compared to wild type. In addition, there were significantly decreases (P < 0.05) in phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylinositol (PI) (Fig. 8a). In particular, when the fatty acid compositions of galactolipids DGDG and MGDG in the leaves of the acbp4 mutant were analyzed, the amounts of 36:6, 34:6, 34:5, 34:4, 34:3 and 34:2 of DGDG (Fig. 8b), and 36:3, 34:6, 34:4, 34:3, 34:2 and 34:1 of MGDG (Fig. 8c) significantly declined in the acbp4 mutant in comparison to wild type (P < 0.05 or P < 0.01).
Fig. 8.
Polar lipid composition (a) and changes of DGDG (b) and MDGD (c) species in leaves of Arabidopsis wild type and acbp4 knockout mutant. Values are means ± SD (nmol/mg DW; n = 4). White and black bars represent WT and acbp4, respectively. “L” indicates that the value of the acbp4 mutant is lower than wild type in the same experiment (*, P < 0.05; **, P < 0.01 by Student’s t-test). DW, dry weight.
Discussion
Subcellular localization of ACBP4 and ACBP5
We have experimentally verified that Arabidopsis ACBP4 and ACBP5 are present in the cytosol by using biochemical fractionation, immuno-electron microscopy, and autofluorescence-tagged fusions. These results are consistent with their predicted subcellular localizations. Given their high affinities in binding oleoyl-CoA ester in vitro (Leung et al. 2004), we suggest that ACBP4 and ACBP5 in the cytosol, are available for the cytosolic maintenance and trafficking of oleoyl-CoA, which may include transfer of oleoyl-CoA from chloroplasts to the ER for lipid biosynthesis via the “eukaryotic pathway”. Results from transient expression of autofluorescence-tagged ACBP4 and ACBP5 in onion epidermal cells and from analyses of stably-transformed Arabidopsis did not show fluorescence in either chloroplast or ER, consistent with the lack of appropriate targeting signals to these subcellular compartments in both ACBP4 and ACBP5.
Functions of ACBP4 and ACBP5 in plant lipid metabolism
It is not clear how acyl-CoAs are exported from chloroplasts to the ER. Palmitoyl-CoA (16:0-CoA) and oleoyl-CoA (18:1-CoA) may be exported by intracellular lipid transporters to the ER for the “eukaryotic pathway” (Ohlrogge and Browse 1995). Acyl-CoAs used in de novo biosynthesis of glycerolipids in the ER may primarily recycle through an acyl editing process (Bates et al. 2007). However, intracellular cytosolic transfer proteins involved in binding and recycling of these acyl-CoAs remain unidentified. Both ACBP4 and ACBP5 bind oleoyl-CoA esters better than palmitoyl-CoA esters in vitro (Leung et al. 2004) and the dissociation constants (Kd values) for recombinant (His)6-ACBP4, (His)6-ACBP5 and (His)6-ACBP6 binding with oleoyl-CoA esters were 5.0 × 10−7 M, 9.3 × 10−7 M and 3.7 × 10−6 M, respectively (M.L. Chye and S. Xiao, unpublished data), implying that ACBP4 and ACBP5 may play a more significant role than ACBP6 in oleoyl-CoA binding and trafficking in the cytosol.
Furthermore, lipid profile analyses of the acbp4 mutant provide direct evidence supporting a role for ACBP4 in plant lipid metabolism. Membrane lipids including galactolipids (MGDG and DGDG) and phospholipids (PC, PE and PI) were significantly reduced in the acbp4 mutant. In the “eukaryotic pathway”, the synthesis of phosphatidic acid (PA) in the ER subsequently gives rise to other phospholipids (PC, PE and PI) which are eventually utilised for the biosynthesis of cell membranes. It appears that a mutation in ACBP4 decreased phospholipid biosynthesis in the ER, consistent with a role for cytosolic ACBP4 in lipid (acyl-CoA) transfer from chloroplast to the ER, as this transfer provides the substrates for the “eukaryotic pathway” of lipid biosynthesis in the ER. Reduction in acyl-CoA import into the ER in the acbp4 mutant, resulted in a decrease in the biosynthesis of phospholipids PC, PE and PI. Subsequently, a deficiency in lipid flow from the ER to the chloroplasts may also follow, affecting the production of galactolipids (MGDG and DGDG). Indeed, ACBP4 and ACBP5 in the cytosol are available to mediate the shuttling of acyl-CoAs between acyl-CoA pools in the chloroplasts and the ER.
There were few significant differences in the decrease between galactolipid species produced by the “eukaryotic pathway” (e.g. 18:3–18:3) and the “prokaryotic pathway” (e.g. 18:3–16:3). From fatty acid compositions of DGDG and MGDG in the leaves of the acbp4 mutant, we observed that the amounts of 36:6, 34:6, 34:5, 34:4, 34:3 and 34:2 of DGDG, and 36:3, 34:6, 34:4, 34:3, 34:2 and 34:1 of MGDG were significantly reduced. This indicates that galactolipid biosynthesis via the “prokaryotic pathway” in the acbp4 mutant was subsequently affected. Possibly, a lack in ACBP4- mediated acyl-CoA transfer between the chloroplasts and the ER may have resulted in an imbalance of acyl-CoA pools or acyl-CoA-dependent lipid biosynthesis, within each of these two subcellular compartments, thereby affecting the “prokaryotic pathway” in galactolipid biosynthesis. Since the trafficking of lipids from the “eukaryotic pathway” to the chloroplasts contributes to most of the DGDG and part of the MGDG content in the chloroplasts, a block in lipid flow from the ER to the chloroplast, that first causes a decrease in galactolipid production via the “eukaryotic pathway”, may subsequently adversely affect galactolipid synthesis via the “prokaryotic pathway”, if these two pathways are closely-linked for thylakoid membrane biogenesis.
Mutation in the genes encoding the chloroplast envelope protein DGD1 involved in the trafficking of lipids, derived from ER into chloroplasts, resulted in a reduction of DGDG in Arabidopsis leaves (Dörmann et al. 1999). Unlike the dgd1 mutant (Dörmann et al. 1999), or the mod1 and mgd1 mutants which are impaired in fatty acid biosynthesis (Kobayashi et al. 2007; Mou et al. 2000), the acbp4 mutant did not exhibit phenotypic changes when grown under normal conditions (16 h light, 23 °C/8 h dark, 21 °C). This can be explained by the presence of a possibly functionally redundant protein, ACBP5, in Arabidopsis. In fact, knockout of ACBP4 in the acbp4 mutant caused about 25.3% reduction in of DGDG content, which is less severe than that (about 85.2%) seen in the dgd1 mutant which is stunted in growth (Dörmann et al. 1999). In comparison, the lack of galactolipids in the Arabidopsis mgd1 mutant completely abrogated both photosynthesis and embryogenesis (Kobayashi et al. 2007).
Although the functional redundancy of ACBP4 and ACBP5, initially proposed based on their similar binding to oleoyl-CoA (Leung et al. 2004), is reflected by findings in this study, based on their subcellular localization, we have yet to characterize an acbp5 knockout mutant to incorporate its effect in association with the acbp4 mutation. Our current findings reinforce our hypothesis that ACBP4 and ACBP5 are likely involved in lipid trafficking in the cytosol. These two larger isoforms of ACBPs may function either alone or with possible protein partners interacting at their kelch motifs (Leung et al. 2004). Screening for their protein interactors by yeast two-hybrid system has already been initiated and the identification of such interactors would be expected to further enhance our knowledge on ACBP4 and ACBP5.
Acknowledgements
We thank Drs. M.M. Goodin (University of California, Berkeley) and W.C. Yang (Institute of Genetics and Developmental Biology, Beijing) for provision of vectors pGDG and pBI-eGFP, respectively, J. Tam for technical assistance in ultra-thin sectioning, Mary Roth (Kansas Lipidomics Research Center) for lipid profiling, Dr. W.K. Yip for provision of the Biolistic PDS-1000/He system, and Dr. S.F. Sin for generation of construct pAT324 used in complementation of the acbp4 mutant. This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region, China (project HKU7504/05M), the University of Hong Kong (grant no. 10208034) and the Croucher Foundation (MLC, HL). SX, JZ and SC were supported by postgraduate studentships from the University of Hong Kong. The Kansas Lipidomics Research Center where lipid analysis was carried out was supported by National Science Foundation (EPS 0236913, MCB 0455318, DBI 0521587), Kansas Technology Enterprise Corporation, Kansas IDeA Network of Biomedical Research Excellence (INBRE) of National Institute of Health (P20RR16475), and Kansas State University.
Abbreviations
- ACBP
acyl-coenzyme-A-binding protein
- DGDG
digalactosyldiacylglycerol
- MGDG
monogalactosyldiacylglycerol
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
References
- Abel S, Theologis A. Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 1994;5:421–427. doi: 10.1111/j.1365-313x.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- Bates PD, Ohlrogge JB, Pollard M. Incorporation of newly-synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J Biol Chem. 2007;282:31206–31216. doi: 10.1074/jbc.M705447200. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Branen JK, Shintani DK, Engeseth NJ. Expression of antisense acyl carrier protein-4 reduces lipid content in Arabidopsis leaf tissue. Plant Physiol. 2003;132:748–756. doi: 10.1104/pp.102.018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana . Biochem J. 1986;235:25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton M, Rose TM, Faergeman NJ, Knudsen J. Evolution of the acyl-CoA binding protein (ACBP) Biochem J. 2005;392:299–307. doi: 10.1042/BJ20050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML. Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Mol Biol. 1998;38:827–838. doi: 10.1023/a:1006052108468. [DOI] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY. Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J. 1999;18:205–214. doi: 10.1046/j.1365-313x.1999.00443.x. [DOI] [PubMed] [Google Scholar]
- Chye ML, Li HY, Yung MH. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol Biol. 2000;44:711–721. doi: 10.1023/a:1026524108095. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dörmann P, Balbo I, Benning C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB. Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana . Arch Biochem Biophys. 1996;331:55–62. doi: 10.1006/abbi.1996.0282. [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA. 2007;104:17216–17221. doi: 10.1073/pnas.0704680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KC, Li HY, Mishra G, Chye ML. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol Biol. 2004;55:297–309. doi: 10.1007/s11103-004-0642-z. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Xiao S, Tse MH, Chye ML. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta. 2006;223:871–881. doi: 10.1007/s00425-005-0139-2. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Mol Biol. 2003;51:483–492. doi: 10.1023/a:1022330304402. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. Arabidopsis acyl-CoA binding protein ACBP2 interacts with an ethylene-responsive element binding protein AtEBP via its ankyrin repeats. Plant Mol Biol. 2004;54:233–243. doi: 10.1023/B:PLAN.0000028790.75090.ab. [DOI] [PubMed] [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. Deficiency in fatty acid synthase leads to remature cell death and dramatic alterations in plant morphology. Plant Cell. 2000;12:405–418. doi: 10.1105/tpc.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J, Botella JR. Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol Biol Rep. 1998;16:257–262. [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell. 2005;17:2340–2354. doi: 10.1105/tpc.105.033563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Krauss MR, Borkird C, Sung ZR. A nuclear-protein associated with cell divisions in plants. Planta. 1988;174:462–472. doi: 10.1007/BF00634474. [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis . Plant J. 2008;54:141–151. doi: 10.1111/j.1365-313X.2008.03402.x. [DOI] [PubMed] [Google Scholar]
- Xu ZF, Teng WL, Chye ML. Inhibition of endogenous trypsin- and chymotrypsin-like activities in transgenic lettuce expressing heterogeneous proteinase inhibitor SaPIN2a. Planta. 2004;218:623–629. doi: 10.1007/s00425-003-1138-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. SDIR1 Is a RING Finger E3 Ligase That Positively Regulates Stress-Responsive Abscisic Acid Signaling in Arabidopsis . Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]