Background: cGMP-dependent protein kinases utilize their leucine zipper (LZ) domains to bind interacting proteins in an isotype-specific manner.
Results: Structural and biophysical analysis reveals residues for the PKG II-Rab11b interaction.
Conclusion: PKG II utilizes an electroneutral surface on the LZ domain to bind Rab11b.
Significance: This is the first structure of PKG bound to one of its interacting proteins.
Keywords: Cyclic GMP (cGMP), Protein Kinase G (PKG), Protein-Protein Interaction, Rab, Second Messenger, Serine/Threonine Protein Kinase, Signal Transduction, G-kinase-interacting Protein (GKIP), Type II cGMP-dependent Protein Kinase (PKG II)
Abstract
cGMP-dependent protein kinase (PKG)-interacting proteins (GKIPs) mediate cellular targeting of PKG isoforms by interacting with their leucine zipper (LZ) domains. These interactions prevent aberrant signaling cross-talk between different PKG isotypes. To gain detailed insight into isotype-specific GKIP recognition by PKG, we analyzed the type II PKG leucine zipper domain and found that residues 40–83 dimerized and specifically interacted with Rab11b. Next, we determined a crystal structure of the PKG II LZ-Rab11b complex. The PKG II LZ domain presents a mostly nonpolar surface onto which Rab11b docks, through van der Waals interactions. Contact surfaces in Rab11b are found in switch I and II, interswitch, and the β1/N-terminal regions. This binding surface dramatically differs from that seen in the Rab11 family of interacting protein complex structures. Structural comparison with PKG Iα and Iβ LZs combined with mutagenic analysis reveals that GKIP recognition is mediated through surface charge interactions.
Introduction
Cyclic guanosine monophosphate (cGMP) is a crucial second messenger that relays extracellular signals to various effectors inside the cell. As the main receptor for cGMP, cGMP-dependent protein kinase (PKG) is a central mediator of the nitric oxide (NO)-cGMP and atrial natriuretic peptide-cGMP pathways. PKG relays these signals by specifically binding to effector proteins and regulating their functions through phosphorylation (1, 2). Because PKG is a broad specificity serine/threonine kinase, cellular targeting of PKG is essential for proper substrate phosphorylation and signal transduction fidelity; this localization is mediated by PKG-interacting proteins (GKIP)2 (3).
Three types of PKG are found in mammalian cells. PKG Iα and Iβ are splice variants of the type I gene and are found mostly in the cytosol, whereas PKG II is transcribed from a separate gene and is associated with the membrane via a myristoylated glycine at the N terminus (4–6). All PKGs share the same domain organization as follows: an N-terminal regulatory domain (R) consisting of a leucine zipper (LZ) domain; an autoinhibitory sequence; two tandem cyclic nucleotide-binding domains; and a C-terminal catalytic domain (C) (Fig. 1A) (1). Functions of PKG I and II are nonredundant, as shown by distinct phenotypes of PKG I and PKG II-deficient mice (7, 8). PKG I regulates smooth muscle tone, vasorelaxation, and platelet aggregation, whereas PKG II is crucial for regulating intestinal secretion, bone growth, renin secretion, and circadian rhythm.
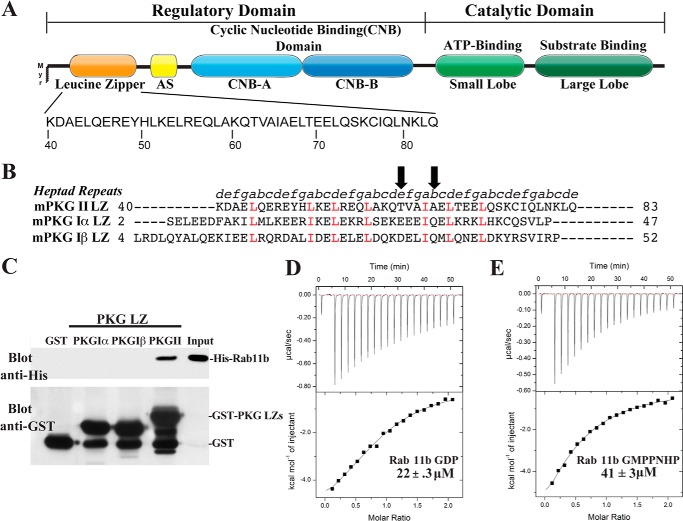
FIGURE 1.
Domain organization and direct binding of PKG II LZ and Rab11b. A, domain organization of PKG II with the sequence of the leucine zipper domain highlighted. AS, autoinhibitory sequence; Myr, myristoylation site. B, sequence alignment of the PKG Iα, PKG Iβ, and PKG II LZ domains. Positions within the heptad repeat (a–g) are shown at the top. The key interface residues, Thr-62 and Ala-66, mutated for disrupting the PKG II LZ and Rab11b interaction are marked with arrows. C, GST or GST-fusion proteins containing the PKG LZs were immobilized on glutathione-Sepharose beads and incubated with purified His-tagged Rab11b as described under “Experimental Procedures.” Beads were washed, and bound proteins were analyzed by SDS-PAGE/immunoblotting. Blots were probed with an antibody specific for the His tag to detect Rab11b binding and GST to demonstrate equal loading of GST and the GST-tagged PKG leucine zippers. D and E, isothermal titration calorimetry experiments demonstrate that PKG II LZ (residues 40–83) has a slightly higher binding preference for Rab11b-GDP (D) than Rab11b bound to the nonhydrolyzable GTP analog GMPPNHP (E).
The three LZ domains share little sequence similarity, except for leucine or isoleucine residues at a and d position of the heptad repeat that form the dimer interface (Fig. 1B). The LZ domains mediate homodimerization of the kinase and have been shown to target PKGs to isoform-specific substrates and binding partners through their interaction with GKIPs. For example, the LZ domain of PKG Iα interacts directly with both the small GTPase RhoA, and the myosin light chain phosphatase (MYPT1). These interactions are essential for mediating phosphorylation of both proteins that contribute to smooth muscle relaxation (9, 10). The PKG Iβ LZ specifically binds the inositol triphosphate receptor-associated PKG substrate. Phosphorylation of the inositol triphosphate receptor-associated PKG substrate by PKG inhibits Ca2+ release from the 1,4,5-inositol triphosphate receptor, which also contributes to smooth muscle relaxation (11, 12). PKG II phosphorylates and activates the cystic fibrosis transmembrane conductance regulator (CFTR) in intact cells (13, 14). CFTR is a good in vitro substrate for both PKG II and PKG Iβ, but PKG Iβ is unable to activate CTFR in intact cells. However, a PKG Iβ chimera containing the membrane targeting domain of PKG II (residues 1–29) activates CFTR, although only 30–40% as effective as PKG II. These results suggest that other regions of PKG II are involved in targeting it to CFTR. Although these data show that each isoform of PKG interacts with isoform-specific GKIPs, which mediates specific subcellular localization and provides a mechanism for substrate specificity, little is known about the details of these interactions due to the lack of structural information (13, 14).
Rab11 is a subfamily of the Ras small GTPases and includes Rab11a and Rab11b, which share 89% sequence identity with each other, and Rab25, which has 61 and 66% sequence identity to Rab11a and Rab11b, respectively (15). Although Rab11a and Rab11b are ubiquitously expressed, Rab25 is found exclusively in epithelial cells (16–18). Rab11 plays a major role in the maintenance of the slow recycling endosome pathway and trafficking cargo to the plasma membrane. Structurally, the small GTPase family has a conserved G protein fold consisting of a six-strand β-sheet flanked by α-helices on each side. Two structurally conserved motifs of the Ras family, the switch I and II regions, play a role in specifically binding downstream effectors and modulating effector affinity by having two distinct functional states. The GTP-bound state is considered the “on” state and has high affinity for downstream effectors; the GDP-bound state is considered the “off” state with low affinity (19, 20). A major focus of research on Rab11 signaling has been to study its interaction with the five members of the Rab11 family of interacting proteins (FIPs) (21). The FIPs form homodimers through a C-terminal leucine zipper, which functions as a Rab11-binding domain (RBD). FIPs preferentially interact with GTP-bound Rab11 (22–25). The interactions between Rab11 and FIPs are essential in regulating recycling endosome trafficking and delivery of cargo to specific locations on the plasma membrane (21).
Reports have shown that Rab11 can interact with several membrane-associated proteins. Specifically, it was shown that PKG II interacts with GDP-bound Rab11b and that this interaction is crucial for their co-localization at the recycling endosome and their subsequent return to the plasma membrane (26). Rab11 has also been reported to interact with other membrane-associated proteins such as the β2-adrenergic receptor, TRPV5 and TRPV6 Ca2+ channels, b-isoform of the thromboxane A2 receptor (TPb), and brain-derived neurotrophic factor-dependent TrkB (TrkB-FL) receptors (27–30).
To understand the molecular details of the PKG II-Rab11b interaction, we identified a PKG II LZ fragment that stably bound Rab11b and determined a crystal structure of their complex. Our structure of the PKG II-Rab11b complex, combined with mutagenic analysis, reveals the molecular details of the PKG II-Rab11b interaction and provides the structural insight into the isotype-specific GKIP-PKG interactions.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
PKG II LZ (residues 40–83) was inserted into the expression vector pQTEV with an N-terminal His6 tag and transformed into BL21 (DE3) Escherichia coli cells (31). Cells were grown at 37 °C and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at A600 = 0.5. The cells continued to grow for 18 h at 18 °C after induction. The cell pellet was suspended in Lysis Buffer A (50 mm Tris (pH 7.5), 200 mm NaCl, 5 mm MgCl2, and 5 mm imidazole) and lysed with Constant Systems TS cell disrupter (Daventry Northants, UK). The lysate was cleared using ultracentrifugation, and the supernatant was loaded onto an IMAC nickel column. The column was washed with Lysis Buffer A, and the sample was eluted in Lysis Buffer A containing 300 mm imidazole. His-tagged tobacco etch virus (TEV) protease was added to cleave the His tag, and the cleaved sample was again loaded onto an IMAC nickel column for TEV separation. The sample was concentrated and loaded onto a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare) equilibrated with 25 mm Tris (pH 7.5), 25 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine.
Using a protocol similar to the one above, Rab11b (residues 8–205) was inserted into the expression vector pQTEV and expressed in BL21 (DE3) E. coli cells (31). Cells were grown at 37 °C, induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at A600 = 0.6, and continued to grow for 4 h at 37 °C after induction. The cell pellet was suspended in Lysis Buffer B (50 mm Tris (pH 7.5), 50 mm NaCl, 5 mm β-mercaptoethanol) and cleared using ultracentrifugation. The supernatant was loaded onto an IMAC nickel column, and the column was washed with Lysis Buffer B, and the sample was eluted with Lysis Buffer B containing 300 mm imidazole. TEV was added to cleave the His tag. Cleaved sample was loaded again onto an IMAC nickel column for TEV separation. The sample was concentrated and loaded onto a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare) equilibrated with 25 mm Tris (pH 7.5), 25 mm NaCl, 1 mm MgCl2, and 0.5 mm tris(2-carboxyethyl)phosphine.
In Vitro Pulldowns
GST and GST-tagged PKG leucine zipper domains were expressed in E. coli and bound to glutathione-Sepharose beads as described (32). Binding reactions contained 10 μg of GST or each GST-tagged leucine zipper incubated with 10 μg of Rab11 in 200 μl of Binding Buffer (50 mm HEPES (pH 7.4), 100 mm NaCl, 1 mm MgCl2, 0.1 mm EDTA, 0.1 mm EGTA, 0.2% BSA, and 0.1% Triton X-100). The reactions were incubated at 4 °C for 1 h. The beads were washed three times with the binding buffer, and bound proteins were analyzed by SDS-PAGE followed by immunoblotting with anti-His and anti-GST antibodies (Santa Cruz Biotechnology).
Crystallization and Structure Solution
Prior to crystallization, the protein complex was formed by mixing an equal molar ratio of Rab11b and PKG II LZ along with 5 mm MgCl2 and 10 mm guanosine diphosphate (GDP) to a final concentration of 3 mm. Crystals were obtained by mixing in a 1:1 ratio of protein and 0.056 m sodium phosphate monobasic monohydrate, 1.344 m potassium phosphate dibasic (pH 8.2), and 10 mm EDTA disodium salt dehydrate. Crystals formed within 2 days at 22 °C. Crystals were harvested, and diffraction data were collected at the Advanced Photon Source (Argonne National Laboratories, IL). A single data set was collected from a crystal that diffracted to a resolution of 2.66 Å and in the primitive tetragonal space group, P43212. Data were processed using Mosflm and scaled using SCALA (33, 34). Phasing was accomplished using the molecular replacement program AutoMR from the PHENIX suite with the previously determined Rab11b structure (PDB code 2F9L) as a search model (35, 36). Two molecules of Rab11b were identified in the initial search, and unambiguous electron density was observed representing the PKG II LZ. Manual building of the model was performed using the program Coot, followed by rounds of refinement using phenix.refine. The final refinement round included restrained TLS refinement parameters (37, 38).
Mutagenesis and ITC Measurements
Mutations were made to the PKG II LZ-pQTEV construct using the QuikChange site-directed mutagenesis kit (Stratagene). The following mutagenic primers were used: 5′-gcagctagctaagcaggaggtggccattgcggagc-3′ (T62E sense) and 5′-gctccgcaatggccacctcctgcttagctagctgc-3′ (T62E antisense); 5′-gcagaccgtggccattcaggagctcaccg-3′ (A66Q sense) and 5′-cggtgagctcctgaatggccacggtctgc-3′ (A66Q antisense). Successful mutagenesis was confirmed by DNA sequencing. Expression and purification of the mutant proteins were performed as described for the wild-type protein.
Nucleotide loading was performed as described (36), and samples were dialyzed into 25 mm HEPES (pH 8.0), 150 mm NaCl, 5 mm MgCl2, 1 mm tris(2-carboxyethyl)phosphine, and 5 mm of either of the nucleotides overnight at 4 °C. Titrations were performed at 35 °C on a MicroCal AutoITC calorimeter system injecting aliquots of 600 μm Rab11b with GDP or GTP into 60 μm PKG II LZ. Calculation of baselines and integration of peak volumes were analyzed using Origin software.
Immunofluorescence Analysis
HeLa cells grown on poly-l-lysine-coated chamber slides were co-transfected with pcDNA3.1-hPKG2-FLAG or pcDNA3.1-hPKG2 T62E/A66Q-FLAG and pcDNA3.1-HA-mRab11b. At 24 h post-transfection, cells were washed twice with phosphate-buffered saline (PBS) and fixed for 20 min in 3.7% formaldehyde. Following sequential washes with PBS, cells were permeabilized for 5 min in 0.1% Triton X-100, washed three times with PBS, and then treated with PBS containing 5% bovine serum albumin for 30 min at room temperature. Cells were subsequently incubated with mouse anti-FLAG M2 IgG (Sigma) or rabbit anti-HA polyclonal antibody (Santa Cruz Biotechnology) overnight at 4 °C. Following three washes with PBS, cells were incubated for 1 h with Alexa Fluor 488 goat anti-mouse IgG or Alexa Fluor 555 goat anti-rabbit IgG (Invitrogen). The slides were washed thoroughly with PBS and mounted in fluorescent mounting medium Vectashield (Vector Laboratories). Fluorescent images were obtained using a Leica TCS-SP5confocal laser-scanning microscope (TCS-SP5; Leica) was used to obtain staining profiles.
RESULTS
PKG II LZ Binds Directly to Rab11b in a GDP-dependent Manner
PKG II was previously found to interact with Rab11b using the yeast two-hybrid system and co-immunoprecipitation from transiently transfected cells (26). Alanine mutations within the dimeric interface of the PKG II LZ were shown to disrupt Rab11b association, suggesting that the PKGII LZ domain mediates the interaction; however, direct binding between the proteins was not shown (26). We analyzed the N-terminal region of PKG II using the program 2ZIP to identify boundaries of the heptad repeat motif common in the LZ domains, which defined the PKG II LZ to be residues 40–83 (Fig. 1B) (39). To test whether Rab11b and the PKG II LZ interact directly, we performed in vitro pulldowns using His-tagged Rab11b (residues 8–205) and GST-tagged PKG Iα, Iβ, or II LZ domains immobilized on glutathione-Sepharose beads. As shown in Fig. 1C, Rab11b bound specifically to the PKG II LZ. Under these conditions, no interaction was seen between Rab11b and GST or the PKG Iα/PKG Iβ LZs (Fig. 2C). Next, we analyzed the PKG II LZ-Rab11b interaction using isothermal titration calorimetry (ITC). ITC measurements showed Kd values ranging 22–41 μm for the PKG II LZ. Interestingly, the Kd value was slightly lower when Rab11b was bound to GDP compared with the GTP (22 μm versus 41 μm) (Fig. 1, D and E). When the PKG Iα or Iβ LZ was mixed with Rab11b, we did not detect a heat exchange signature suggesting that neither PKG I isoform interacts with Rab11b in solution (data not shown).
FIGURE 2.
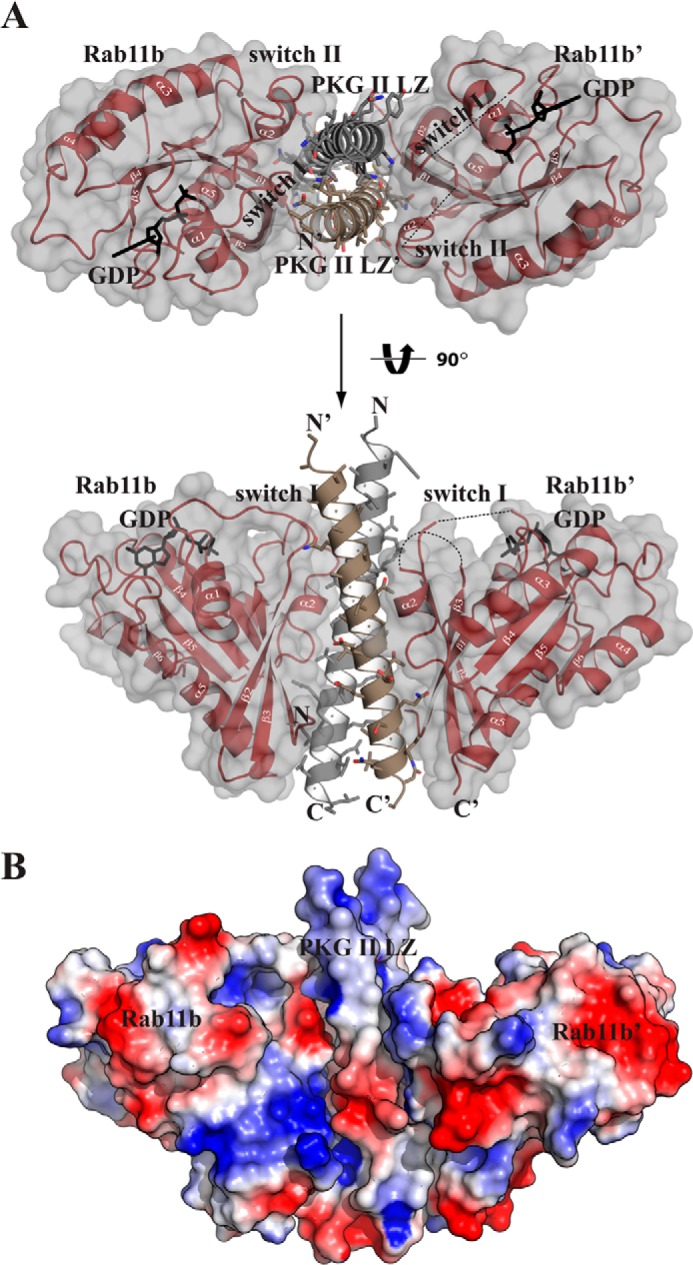
Crystal structure of the PKG II LZ-Rab11b complex. A, structure of the PKGII LZ-Rab11b complex with the secondary structure elements labeled. The disordered switch I and II regions of Rab11b′ are shown as a dotted lines. All structure images were generated using PyMOL (Delano Scientific). B, surface representation of the complex is shown, colored according to its electrostatic potential (red, electronegative; blue, electropositive).
Overall Structure of the PKG II LZ-Rab11b Complex
To gain structural insight into how the PKG II LZ domain interacts with Rab11b, we determined the crystal structure of the PKG II LZ-Rab11b complex (Fig. 2 and Table 1). The structure was determined to 2.66 Å with the asymmetric unit containing two molecules of PKG II LZ (referred as LZ and LZ′) and two molecules of Rab11b (referred to as Rab11b and Rab11b′). The asymmetric unit contains a Rab11b molecule on either side of the PKG II LZ (Rab11b, LZ-LZ′, and Rab11b′). The PKG II LZ mainly interacts with the switch I, II, interswitch, and the β1/N-terminal regions of Rab11b (Fig. 2A).
TABLE 1.
Data and refinement statistics
| PKG II LZ-Rab11b-GDP | |
|---|---|
| Data collection | |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 136, 136, 76.9 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 19.9–2.66 (2.8–2.66)a |
| Rsym (%) | 10.6 (69.5) |
| I/σ (I) | 9.3 (3.5) |
| Completeness (%) | 99.2 (99.5) |
| Redundancy | 14.2 (13.9) |
| Refinement | |
| Resolution | 19.9–2.66 (2.77–2.66) |
| No. of unique reflections | 21,139 (3030) |
| Rwork/Rfreeb | 20.1/25.1 (33.9/38.7) |
| No. of atoms | |
| Protein | 3083 |
| Ligand | 56 |
| Water | 21 |
| B-factors | |
| Protein(chain) | 62.6(A), 73.6(B), 74.1(C), 76.8(D) |
| Ligand | 57.3 |
| Water | 57.9 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.694 |
| Ramachandran plot (%) | |
| Most favorable region | 96.9 |
| Additional allowed region | 3.1 |
| Outliers | 0 |
a Highest resolution shell is shown in parentheses.
b 5.0% of the observed intensities were excluded from refinement for cross-validation purposes.
Overall Structure of the PKG II LZ Domain
The structure shows that the PKG II LZ forms a parallel coiled-coil, with the heptad repeats placing the hydrophobic portion of the amphiphilic helices in the dimerization interface (Figs. 2A and 3). Although the purified protein contained residues 40–83 corresponding to six heptad repeats, the first heptad repeat was not modeled due to lack of electron density. Seven N-terminal residues are missing in one of the chains (LZ, residues 47–83), although two additional residues are missing at either terminus for the other chain (LZ′, residues 49–81).
FIGURE 3.
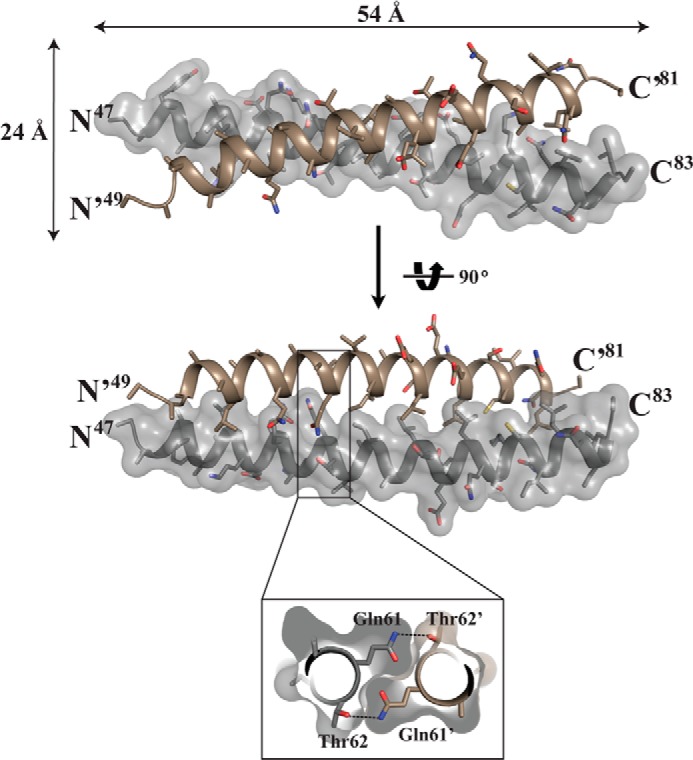
Structure of the PKG II LZ domain. The structure of the PKG II LZ domain showing the overall dimensions in angstroms (Å), with stick representation of the side chains from each monomer. Only one of the chains is shown with surface. Inset, cutaway view looking down the 2-fold axis. This view shows the symmetrical hydrogen bond formed by Gln-61 of one chain and Thr-62 of the other.
With exception of Gln-61 and Lys-75, leucine or isoleucine residues at the a and d positions of heptad repeats of each monomer pack in a “knob-into-holes” manner forming an extensive hydrophobic core with a 735 Å2 surface area. Interestingly, the side chain of Gln-61 at the d position from one monomer forms a hydrogen bond with the side chain of Thr-62′ at the e position of the other monomer (Fig. 3). The interhelical hydrogen bond between Gln-61 and Thr-62′ forms in a symmetrical manner. The d position Lys-75 shows no interhelical salt bridges.
The surface of PKG II LZ domain shows two distinct surfaces: a charged surface with negative and positive patches near the N and C termini, respectively, and a hydrophobic surface at the center of the LZ, which provides most of the docking surface for Rab11b. Superhelical and α-helical parameters were analyzed using Twister showing similar values as the previously reported coiled-coil structures, including PKG Iβ LZ (40, 41). Both strands exhibit higher temperature factors toward the N and C termini, suggesting increased thermal motion or flexibility in those regions, and show minor distortion based on the α-helical parameters by calculated Twister (41). LZ and LZ′, superimpose well with a root mean square deviation of 0.89 Å for 30 equivalent Cα atoms between residues 49 and 81. The N terminus of LZ′ shows a slight kink that may be caused by crystal packing or by its interaction with Rab11b.
Rab11b Structure
As mentioned, the asymmetric unit of the crystal contained two molecules of Rab11b (Rab11b and Rab11b′) both with GDP bound (Fig. 4). Superposition of each molecule with the previously determined Rab11b-GDP structure gives a root mean square deviation of 0.92 Å for Rab11b and 0.69 Å for Rab11b′, with major structural differences found in the 310-helix region located immediately N-terminal of the switch II helix (Fig. 5A) (36, 42). The 310-helix region tilts ∼40° away from the nucleotide binding pocket toward the LZ interface, resulting in major structural rearrangement (Fig. 5B). For Rab11b′, the 310-helix (residues 68–75) and a part of switch I (residues 40–43) show poor electron density, suggesting these regions are flexible and adopt multiple conformations.
FIGURE 4.
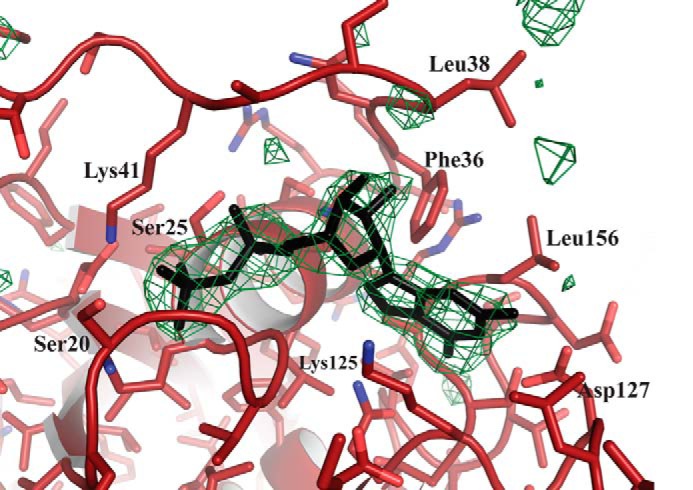
Electron density of GDP bound to Rab11b. Unbiased ligand density is shown for Rab11b; the electron density maps were calculated with coefficients of the form |Fo| − |Fc| determined prior to insertion of GDP in the refinement. The maps are contoured at 4σ, and the side chains of neighboring residues are labeled.
FIGURE 5.
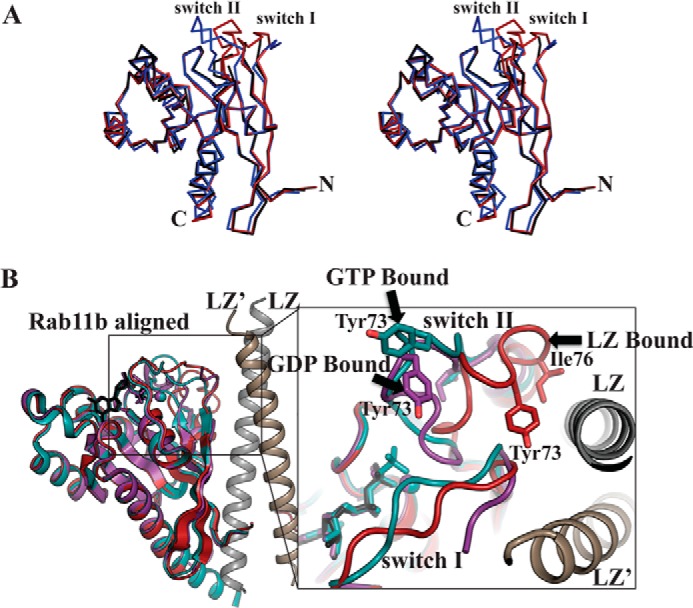
Structure of Rab11b. A, stereo image of the superposition of Rab11b (red), Rab11b′ (black), and the previously solved Rab11b-GDP complex (blue) (PDB code 2F9L) (36). B, structural alignment of the PKG II LZ-Rab11b (red) with the previously determined structures of Rab11b bound to GMPPNHP in teal (PDB code 2F9M) and GDP in purple (PDB code 2F9L) (36). The enlarged panel to the right shows a close-up view of the rearrangement of the 310-helix near the switch II region of Rab11b, which places Tyr-73 and Ile-76 in position to make contacts with the PKG II LZ.
PKG II LZ and Rab11b Complex Interface
The crystal structure of the complex reveals that PKG II uses a total of four heptad repeats to bind Rab11b (Fig. 6A). Residues 54–83 of each PKG II LZ interact with both molecules of Rab11b mainly through hydrophobic interactions. Because of disorder in the switch I and II regions in Rab11b′, the overall interaction surface is not the same between the two chains of the LZ. The total interface surface area between the Rab11b molecule (with the ordered switch regions) and PKG II LZ is 1078 Å2. The interface surface area for the less ordered Rab11b′ is 855.8 Å2. The PKG II LZ docks to multiple regions of Rab11b, including switch I and II, the interswitch, and the β1/N-terminal regions (Fig. 6B).
FIGURE 6.
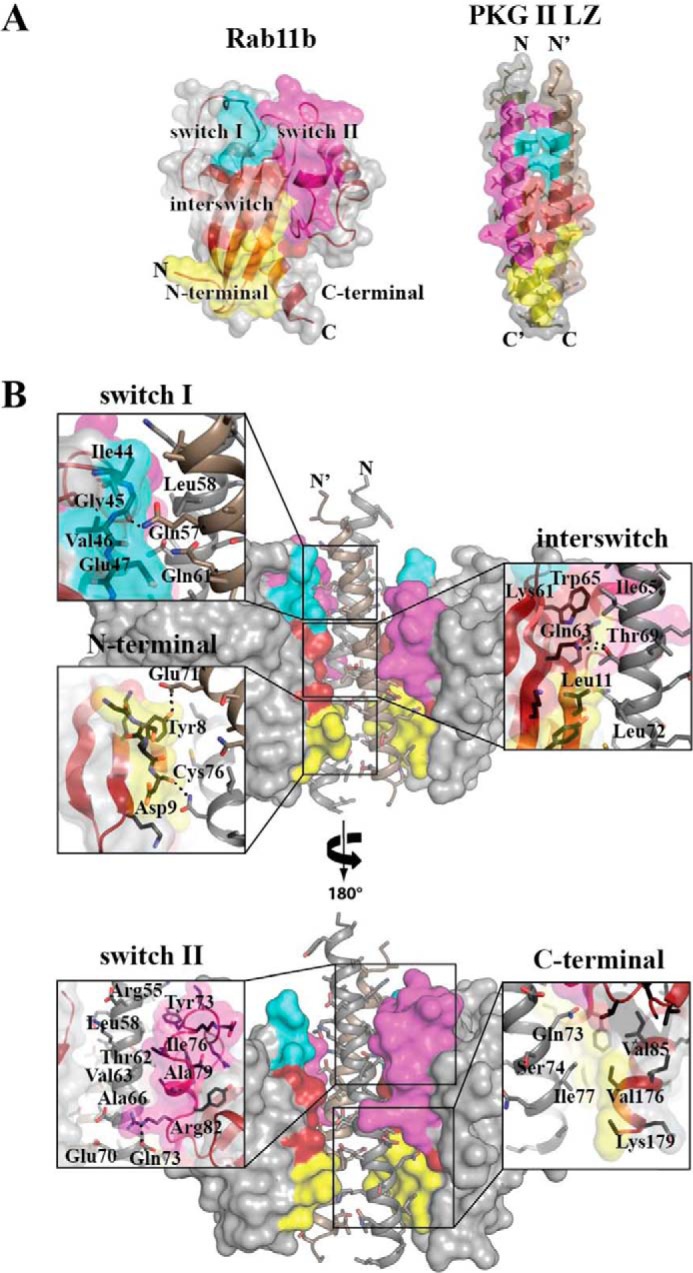
Interface of the PKG II LZ-Rab11b complex. A, color-coded representation of the regions on Rab11b and PKG II that form the interaction interface. The coloration corresponds to the Rab11b surfaces that interact between the two proteins: switch I (teal), switch II (magenta), interswitch (red), and N terminus (yellow). B, PKG II LZ-Rab11b interactions with zoom-in views. Rab11b provides the largest interface at the interswitch/N-terminal region, contributing van der Waals contacts from residues Asp-9 to Phe-12, Phe-48, Lys-58, Lys-61, Trp-65, and Val-85. PKG II LZ provides residues Thr-62, Ile-65, Ala-66, Leu-72, Gln-73, Cys-76, Ile-77, and Lys-81 of PKG II LZ and Gln-61′, Ala-64′, Glu-67′, and Leu-68′ of PKG II LZ′ to this interface. Dashed lines in the insets represent hydrogen bonds and salt bridge interactions.
The first interface forms between the N-terminal region of the PKG II LZ and switch I of Rab11b and, as mentioned above, shows little symmetry in the interaction pattern due to the partially disordered regions in Rab11b′ (upper panel of Fig. 6B). Interestingly, the core residues (Leu-58 from one strand and Gln-61′ from the other at the a and d positions) of the LZ docks to switch I (residues 44–48) mainly through van der Waals (VDW) interactions. A single hydrogen bond forms between Gln-57′ NE2 of PKG II LZ′ and the backbone carbonyl oxygen of Gly-45 of Rab11b at this interface.
The second interface forms between the middle section of the LZ (residues 69–83) and switch II of Rab11b and involves mainly VDW contacts, with the following two exceptions: a salt bridge between side chains of Glu-70 of the PKG II LZ and Arg-82 of Rab11b and a hydrogen bond between the side chain of Gln-73 of LZ and the backbone carbonyl of Gly-83 at the end of switch II (lower panel of Fig. 6B). The hydrophobic interface at this region can be broken into two parts. The first part is the aromatic ring of Tyr-73 of the Rab11b docking to a hydrophobic cleft formed by Arg-55, Leu-58, Ala-59, and Leu-54′ of the PKG II LZ. The second part involves the side chain of Ile-76 and Ala-59 from Rab11b docking onto another hydrophobic patch consisting of Thr-62 and Val-63 of the PKG II LZ. These interactions induce major structural changes in switch II, which involves the 310-helix and α2-helix moving 10 Å away from the core toward the interaction interface (Fig. 4B). The structural rearrangement exposes the buried side chain of Ile-76 for its interaction with PKG II LZ (Fig. 5B).
The third interface forms between the C-terminal end of the LZ and the continuous surface formed between the interswitch, strand β1, and N-terminal regions of Rab11b (Fig. 6B). This region makes up over 50% of the total interface. Although the majority of the contacts are through VDW interactions, four hydrogen bonds are also present at this interface. PKGII Thr-69 contributes two hydrogen bonds through its interaction with the side chain of Gln-63 of Rab11b, and the side chains of PKGII Gln-73 and Asn-80 form hydrogen bonds with the Rab11b backbone amide of Lys-13 and the carbonyl of Asp-9, respectively. Another hydrogen bond was formed between the side chain of PKGII Glu-71 and the hydroxyl group of Tyr-8 of Rab11b. Unlike the asymmetrical interactions at switch I and II, the interactions at the third interface are highly similar for both Rab11b and Rab11b′ molecules in the asymmetric unit and show clear electron density for all contact residues.
T62E or A66Q Mutations in PKG II LZ Disrupt Rab11b Interaction
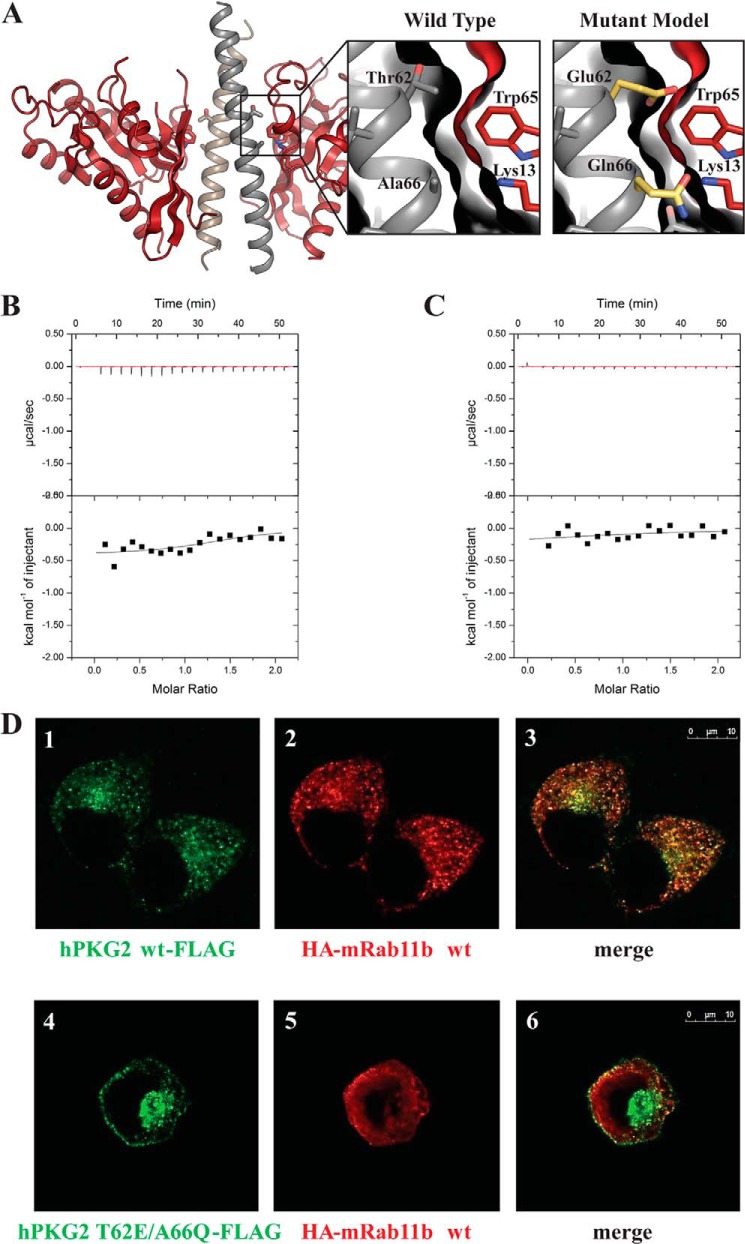
To verify our structural results, we mutated key Rab11b contact residues within the PKG II LZ (Thr-62 and Ala-66) and measured their affinities of the mutants for Rab11b using ITC (Fig. 7). Based on our sequence alignment (Fig. 1B), we mutated Thr-62 and Ala-66 to the corresponding residues (Glu and Gln) in PKG Iα. BecauseThr-62 and Ala-66 are located at the center of the PKG II LZ-Rab11b interface, we reasoned that the mutations would destabilize their interaction by changing the surface contour and the overall charge distribution of the binding surface (Fig. 7A). Indeed, our ITC data showed a lack of exothermal response upon injection indicating that either mutation in the PKG II LZ abolished its interaction with the Rab11b (Fig. 7B).
FIGURE 7.
Representation of mutations made to Rab11b, mutant ITC measurements, and subcellular localizations of PKG II and Rab11b in HeLa cells. A, schematic representation of the PKGII LZ-Rab11b complex with enlarged panels showing the interface seen in the crystal structure (left) and in a modeled structure with PKG II T62E/A66Q mutations (right). B and C, ITC was performed as described under “Experimental Procedures.” No exothermal response was seen when titrating Rab11b into PKG II LZ with a T62E mutation (B) or A66Q mutation (C). D, HeLa cells expressing PKG II-FLAG (panels 1–3) or PKG II T62E/A66Q-FLAG (panels 4–6) together with HA-Rab11b were analyzed by confocal microscopy, as described under “Experimental Procedures.” PKG II WT-FLAG (panel 1, green) and HA-Rab11b (panel 2, red) co-localized in the recycling compartment (panel 3, areas of colocalization are yellow), whereas PKG II T62E/A66Q-FLAG did not co-localize with HA-Rab11b (panels 4–6).
T62E/A66Q Mutation in PKG II Disrupts PKG II and Rab11b Interaction in Vivo
Next, we introduced the T62E/A66Q mutations into full-length PKG II and tested to see whether these mutations would disrupt the interaction with Rab11b in mammalian cells. We co-transfected HeLa cells with Rab11b and full-length wild-type PKG II or T62E/A66Q mutant PKG II and looked for co-localization using immunofluorescence microscopy. In cells with wild-type PKG II, Rab11b and PKG II were co-localized in a punctate staining pattern (Fig. 7D, panels 1–3), consistent with the known association of Rab11b′ with recycling endosomes (43). However, cells transfected with the T62E/A66Q mutant PKGII (43) showed a completely different staining pattern (Fig. 7D, panels 4–6), where PKG II localized to the plasma membrane and pericentriolar region and showed very little co-localization with Rab11b. In addition, Rab11b showed a diffuse staining pattern in PKG II T62E/A66Q mutant-expressing cells, suggesting that the mutant PKG II may influence the distribution of Rab11b. These results demonstrated that the T62E/A66Q mutations in the full-length PKG II abolish its interaction with Rab11b in vivo, further supporting our structural results.
DISCUSSION
Although PKG LZ-GKIP interactions are known to play key roles in mediating PKG signaling, to this point there has been no detailed structural insight into how the PKG isotypes utilize their LZ domains to recognize GKIPs. Our PKG II LZ-Rab11b complex represents the first crystal structure of the LZ domain of PKG II, and the first structure of a PKG-GKIP complex. With this structure, we provide new insight into how the PKG II LZ provides an isotype-specific docking surface for Rab11b and reveal a new protein-protein interaction surface in Rab11b.
Structural comparison with other LZ domains shows that the surface of PKG II LZ is drastically different in its charge distribution and surface contour. Structures of LZs are reported for both PKG Iα (PDB 1ZXA) and Iβ (PDB 3NMD) (40, 44). As seen in the Fig. 8, the PKG II LZ displays a large uncharged surface at the center. In major contrast, the Iα LZ shows both positively and negatively charged regions across its LZ domain, whereas the Iβ LZ shows a largely electronegative surface throughout.
FIGURE 8.
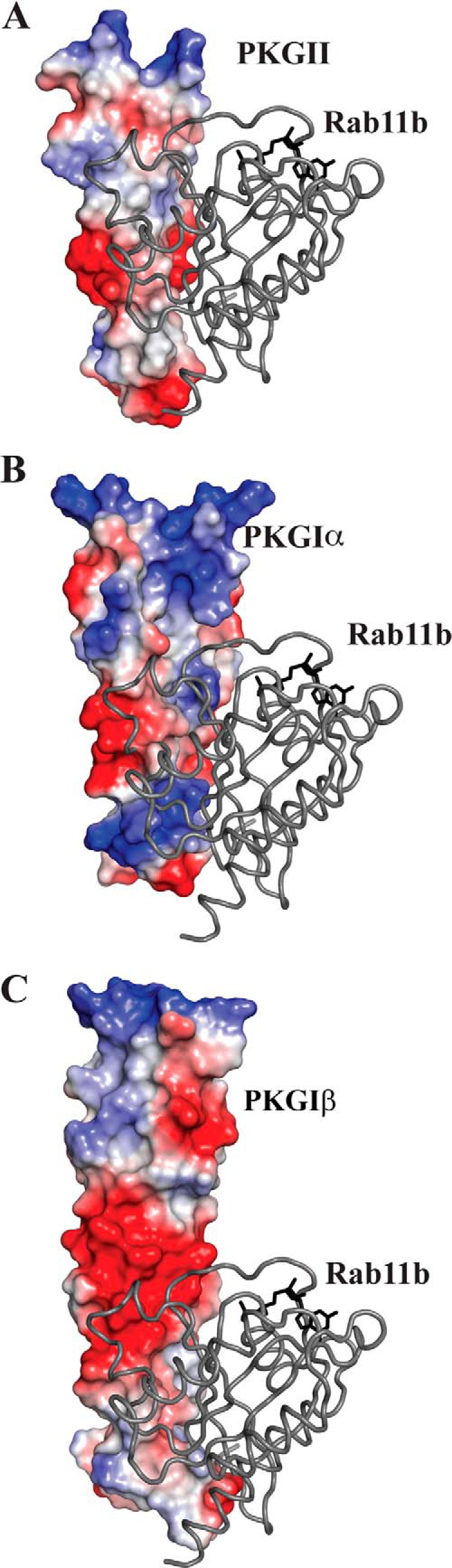
Rab11b modeled to the surface of PKG Iα and PKG Iβ. A, crystal structure of PKG II LZ-Rab11b complex showing the electrostatic surface (electrostatic potential = −85 to 85) of PKG II and Rab11b represented as a ribbon diagram. B, NMR structure of PKG Iα LZ (PDB code 1ZXA) (36); C, crystal structure of PKG Iβ (PDB code 3NMD) (44) were superposed onto the PKG II LZ-Rab11b complex using the sequence alignment from Fig. 1B. Electrostatic surfaces were calculated in PyMOL revealing a charge distribution and surface contour for both PKG Iα and PKG Iβ that would be unfavorable for Rab11b interaction. Electrostatic potential range = −96 to 96 for PKG Iα and electrostatic potential range = −92 to 92 for PKG Iβ.
Alignment of the PKG II LZ with both the PKG I LZs shows that the regions that overlap with the Rab11b docking surface within PKG II LZ display a very different charge distribution and a unique contour suggesting that these surfaces do not provide a compatible surface for the Rab11b binding. We exploited these differences in selecting which PKG II residues to mutate to disrupt the PKG II LZ and Rab11b interaction. Indeed, subsequent mutagenic analysis combined with ITC and co-localization experiments shows that mutating Thr-62 and Ala-66 residues of PKG II LZ disrupt the interaction in vitro and in vivo.
Structural alignments between the PKG LZs, combined with mutagenesis/binding data, suggest that both surface charge distribution and topology are crucial in isotype-specific PKG-GKIP interactions. Based on our structural analysis of the PKG II LZ-Rab11b interaction, we predict that PKG Iα and Iβ will predominantly recognize their isotype-specific GKIPs via hydrogen bond and salt bridge interactions. In support of this, we have previously shown that electrostatic interactions play a major role in mediating PKG Iβ binding to TFII-I and inositol triphosphate receptor-associated PKG substrate (12). In contrast, the structure presented here shows that PKG II binds to Rab11b predominantly through VDW interactions.
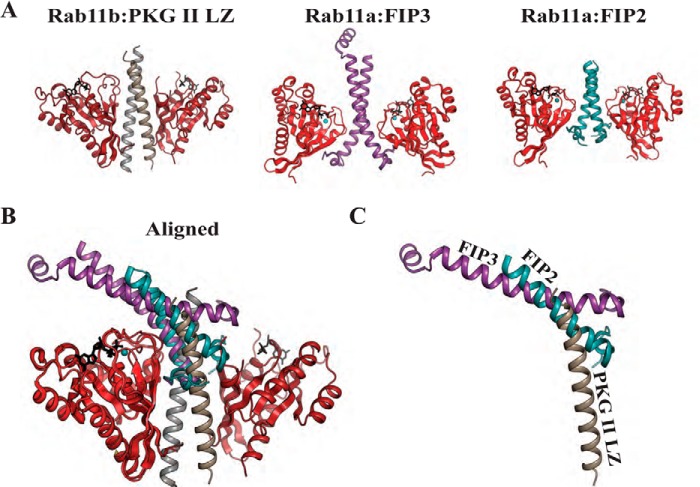
Our PKG II LZ-Rab11b structure also identified a novel surface within Rab11b that recognized the PKG II LZ. As the PKG II LZ was shown to preferentially interact with GDP-bound Rab11b, it was not surprising to find that switches I and II are part of the interface. But we were surprised to see that PKG II utilizes the interswitch and β1/N-terminal regions, which contribute ∼55% of the total interface. This mode of interaction is quite different from the previously reported protein complexes of Rab11 and the RBDs of FIP proteins (22–24). In the Rab11-FIP complexes, the RBD domain forms an interface predominantly with the switch I and II regions of Rab11 (24). When we structurally aligned our complex with the previously determined Rab11-FIP complex, we found that the PKG II LZ and the RBD domains dock to the same side of Rab11b, but with the two very different angles (Fig. 9). Thus, although Rab11 utilizes different interaction surfaces for the recognition of FIPs and PKG II, PKG II and FIPs cannot bind to Rab11 at the same time due to steric clashes.
FIGURE 9.
Structural comparison of the PKG II LZ-Rab11b complex with Rab11a-FIP complexes. A, cartoon representations of the Rab11b-PKG II LZ complex, Rab11a-FIP2 (PBD code 2HV8) (25), and Rab11a-FIP3 (PDB code 2GZD) (24) with PKG II LZ and FIPs aligned. B, complexes aligned with molecules of Rab11. Note that the FIP binding region is mainly along switch I and II of Rab11a, whereas PKG II LZ interacts mainly in the interswitch and N-terminal region of Rab11b. C, view showing only the PKG II LZ, FIP2, and FIP3 highlights the steric clashes.
Structural comparison of PKG II LZ-Rab11b with previously determined structures of Rab11b may explain the 2-fold difference in affinity between the GDP and GTP state of Rab11b. In the GTP-bound state, switch II directly interacts with GTP, and its conformation appears fixed (36). In contrast, in the Rab11b GDP structure, switch II does not interact with GDP, and the structure shows higher temperature factors for the residues in this region suggesting it is flexible (36). This flexibility allows switch II to make a direct interaction with the PKG II LZ. However, in the GTP-bound state, switch II is pulled away from the LZ interface, and it no longer interacts with the LZ domain. The switch II interaction is made up of relatively weak VDW contacts, which contribute little to the overall interface. Nevertheless, these contacts may explain the increased affinity in the GDP-bound state.
In conclusion, we present the first crystal structure of a PKG and GKIP complex. This structure has provided structural insight into the direct interaction between PKG II and Rab11b. Our comparative structural analysis combined with biochemical and co-localization data has shown that the drastically different surface charge distribution of each leucine zipper domain is likely to be one of the major determents for the isotype-specific PKG-GKIP interactions; these interactions are required for PKG subcellular localization and ensure signaling fidelity by limiting cross-talk between different PKG isotypes. It will be interesting to further investigate how other PKG I isoforms use the more electrostatically charged LZ surface to recognize interacting proteins.
Acknowledgments
We thank B. Goud, C. Peters, G. Y. Huang, T. Palzkill, and J. J. Kim for critical reading of the manuscript. We appreciate the technical support, advice, and assistance of D. C. Chow and K. Klerec in the Center for Drug Discovery at Baylor College of Medicine. We also thank S. R. Wasserman (Eli Lilly Beamline, Advanced Photon Source) for assistance with data collection. Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy Office of Science by Argonne National Laboratory, was supported by the United States Department of Energy under Contract DE-AC02-06CH11357. Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Co., which operates the facility.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM090161 (to C. K.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 4OJK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GKIP
- PKG-interacting protein
- LZ
- leucine zipper
- RBD
- Rab11-binding domain
- PDB
- Protein Data Bank
- CFTR
- cystic fibrosis transmembrane conductance regulator
- FIP
- family of interacting protein
- TEV
- tobacco etch virus
- ITC
- isothermal titration calorimetry
- VDW
- van der Waals
- IMAC
- immobilized metal ion affinity chromatography
- GMPPNHP
- guanosine 5′-[β,γ-imido]triphosphate.
REFERENCES
- 1. Hofmann F., Bernhard D., Lukowski R., Weinmeister P. (2009) cGMP-regulated protein kinases (cGK). Handb. Exp. Pharmacol. 191, 137–162 [DOI] [PubMed] [Google Scholar]
- 2. Hofmann F., Feil R., Kleppisch T., Schlossmann J. (2006) Function of cGMP-dependent protein kinases as revealed by gene deletion. Physiol. Rev. 86, 1–23 [DOI] [PubMed] [Google Scholar]
- 3. Francis S. H., Busch J. L., Corbin J. D., Sibley D. (2010) cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol. Rev. 62, 525–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orstavik S., Natarajan V., Taskén K., Jahnsen T., Sandberg M. (1997) Characterization of the human gene encoding the type Iα and type Iβ cGMP-dependent protein kinase (PRKG1). Genomics 42, 311–318 [DOI] [PubMed] [Google Scholar]
- 5. Sandberg M., Natarajan V., Ronander I., Kalderon D., Walter U., Lohmann S. M., Jahnsen T. (1989) Molecular cloning and predicted full-length amino acid sequence of the type Iβ isozyme of cGMP-dependent protein kinase from human placenta. Tissue distribution and developmental changes in rat. FEBS Lett. 255, 321–329 [DOI] [PubMed] [Google Scholar]
- 6. de Jonge H. R. (1981) Cyclic GMP-dependent protein kinase in intestinal brush borders. Adv. Cyclic Nucleotide Res. 14, 315–333 [PubMed] [Google Scholar]
- 7. Pfeifer A., Klatt P., Massberg S., Ny L., Sausbier M., Hirneiss C., Wang G. X., Korth M., Aszódi A., Andersson K. E., Krombach F., Mayerhofer A., Ruth P., Fässler R., Hofmann F. (1998) Defective smooth muscle regulation in cGMP kinase I-deficient mice. EMBO J. 17, 3045–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pfeifer A., Aszódi A., Seidler U., Ruth P., Hofmann F., Fässler R. (1996) Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science 274, 2082–2086 [DOI] [PubMed] [Google Scholar]
- 9. Kato M., Blanton R., Wang G. R., Judson T. J., Abe Y., Myoishi M., Karas R. H., Mendelsohn M. E. (2012) Direct binding and regulation of RhoA protein by cyclic GMP-dependent protein kinase Iα. J. Biol. Chem. 287, 41342–41351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee E., Hayes D. B., Langsetmo K., Sundberg E. J., Tao T. C. (2007) Interactions between the leucine-zipper motif of cGMP-dependent protein kinase and the C-terminal region of the targeting subunit of myosin light chain phosphatase. J. Mol. Biol. 373, 1198–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schlossmann J., Ammendola A., Ashman K., Zong X., Huber A., Neubauer G., Wang G. X., Allescher H. D., Korth M., Wilm M., Hofmann F., Ruth P. (2000) Regulation of intracellular calcium by a signalling complex of IRAG, IP3 receptor and cGMP kinase Iβ. Nature 404, 197–201 [DOI] [PubMed] [Google Scholar]
- 12. Casteel D. E., Boss G. R., Pilz R. B. (2005) Identification of the interface between cGMP-dependent protein kinase Iβ and its interaction partners TFII-I and IRAG reveals a common interaction motif. J. Biol. Chem. 280, 38211–38218 [DOI] [PubMed] [Google Scholar]
- 13. French P. J., Bijman J., Edixhoven M., Vaandrager A. B., Scholte B. J., Lohmann S. M., Nairn A. C., de Jonge H. R. (1995) Isotype-specific activation of cystic fibrosis transmembrane conductance regulator-chloride channels by cGMP-dependent protein kinase II. J. Biol. Chem. 270, 26626–26631 [DOI] [PubMed] [Google Scholar]
- 14. Vaandrager A. B., Smolenski A., Tilly B. C., Houtsmuller A. B., Ehlert E. M., Bot A. G., Edixhoven M., Boomaars W. E., Lohmann S. M., de Jonge H. R. (1998) Membrane targeting of cGMP-dependent protein kinase is required for cystic fibrosis transmembrane conductance regulator Cl− channel activation. Proc. Natl. Acad. Sci. U.S.A. 95, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelly E. E., Horgan C. P., McCaffrey M. W. (2012) Rab11 proteins in health and disease. Biochem. Soc. Trans. 40, 1360–1367 [DOI] [PubMed] [Google Scholar]
- 16. Sakurada K., Uchida K., Yamaguchi K., Aisaka K., Ito S., Ohmori T., Takeyama Y., Ueda T., Hori Y., Ohyanagi H. (1991) Molecular cloning and characterization of a ras p21-like GTP-binding protein (24KG) from rat liver. Biochem. Biophys. Res. Commun. 177, 1224–1232 [DOI] [PubMed] [Google Scholar]
- 17. Lai F., Stubbs L., Artzt K. (1994) Molecular analysis of mouse Rab11b: a new type of mammalian YPT/Rab protein. Genomics 22, 610–616 [DOI] [PubMed] [Google Scholar]
- 18. Goldenring J. R., Shen K. R., Vaughan H. D., Modlin I. M. (1993) Identification of a small GTP-binding protein, Rab25, expressed in the gastrointestinal mucosa, kidney, and lung. J. Biol. Chem. 268, 18419–18422 [PubMed] [Google Scholar]
- 19. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 20. Pfeffer S. R. (2005) Structural clues to Rab GTPase functional diversity. J. Biol. Chem. 280, 15485–15488 [DOI] [PubMed] [Google Scholar]
- 21. Horgan C. P., McCaffrey M. W. (2009) The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 22. Shiba T., Koga H., Shin H. W., Kawasaki M., Kato R., Nakayama K., Wakatsuki S. (2006) Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc. Natl. Acad. Sci. U.S.A. 103, 15416–15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Junutula J. R., Schonteich E., Wilson G. M., Peden A. A., Scheller R. H., Prekeris R. (2004) Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279, 33430–33437 [DOI] [PubMed] [Google Scholar]
- 24. Jagoe W. N., Lindsay A. J., Read R. J., McCoy A. J., McCaffrey M. W., Khan A. R. (2006) Crystal structure of rab11 in complex with rab11 family interacting protein 2. Structure 14, 1273–1283 [DOI] [PubMed] [Google Scholar]
- 25. Eathiraj S., Mishra A., Prekeris R., Lambright D. G. (2006) Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364, 121–135 [DOI] [PubMed] [Google Scholar]
- 26. Yuasa K., Yamagami S., Nagahama M., Tsuji A. (2008) Trafficking of cGMP-dependent protein kinase II via interaction with Rab11. Biochem. Biophys. Res. Commun. 374, 522–526 [DOI] [PubMed] [Google Scholar]
- 27. Parent A., Hamelin E., Germain P., Parent J. L. (2009) Rab11 regulates the recycling of the β2-adrenergic receptor through a direct interaction. Biochem. J. 418, 163–172 [DOI] [PubMed] [Google Scholar]
- 28. van de Graaf S. F., Chang Q., Mensenkamp A. R., Hoenderop J. G., Bindels R. J. (2006) Direct interaction with Rab11a targets the epithelial Ca2+ channels TRPV5 and TRPV6 to the plasma membrane. Mol. Cell. Biol. 26, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamelin E., Thériault C., Laroche G., Parent J. L. (2005) The intracellular trafficking of the G protein-coupled receptor TPβ depends on a direct interaction with Rab11. J. Biol. Chem. 280, 36195–36205 [DOI] [PubMed] [Google Scholar]
- 30. Huang S. H., Wang J., Sui W. H., Chen B., Zhang X. Y., Yan J., Geng Z., Chen Z. Y. (2013) BDNF-dependent recycling facilitates TrkB translocation to postsynaptic density during LTP via a Rab11-dependent pathway. J. Neurosci. 33, 9214–9230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Büssow K., Scheich C., Sievert V., Harttig U., Schultz J., Simon B., Bork P., Lehrach H., Heinemann U. (2005) Structural genomics of human proteins–target selection and generation of a public catalogue of expression clones. Microb. Cell Fact. 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwappacher R., Rangaswami H., Su-Yuo J., Hassad A., Spitler R., Casteel D. E. (2013) cGMP-dependent protein kinase Iβ regulates breast cancer cell migration and invasion via interaction with the actin/myosin-associated protein caldesmon. J. Cell Sci. 126, 1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scapin S. M., Carneiro F. R., Alves A. C., Medrano F. J., Guimarães B. G., Zanchin N. I. (2006) The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform. J. Struct. Biol. 154, 260–268 [DOI] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 39. Bornberg-Bauer E., Rivals E., Vingron M. (1998) Computational approaches to identify leucine zippers. Nucleic Acids Res. 26, 2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Casteel D. E., Smith-Nguyen E. V., Sankaran B., Roh S. H., Pilz R. B., Kim C. (2010) A crystal structure of the cyclic GMP-dependent protein kinase Iβ dimerization/docking domain reveals molecular details of isoform-specific anchoring. J. Biol. Chem. 285, 32684–32688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strelkov S. V., Burkhard P. (2002) Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137, 54–64 [DOI] [PubMed] [Google Scholar]
- 42. Pasqualato S., Senic-Matuglia F., Renault L., Goud B., Salamero J., Cherfils J. (2004) The structural GDP/GTP cycle of Rab11 reveals a novel interface involved in the dynamics of recycling endosomes. J. Biol. Chem. 279, 11480–11488 [DOI] [PubMed] [Google Scholar]
- 43. Schlierf B., Fey G. H., Hauber J., Hocke G. M., Rosorius O. (2000) Rab11b is essential for recycling of transferrin to the plasma membrane. Exp. Cell Res. 259, 257–265 [DOI] [PubMed] [Google Scholar]
- 44. Schnell J. R., Zhou G. P., Zweckstetter M., Rigby A. C., Chou J. J. (2005) Rapid and accurate structure determination of coiled-coil domains using NMR dipolar couplings: application to cGMP-dependent protein kinase Iα. Protein Sci. 14, 2421–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]