Abstract
Here we demonstrate that natural antisense transcripts (NATs), which are abundant in mammalian genomes, can function as repressors of specific genomic loci and that their removal or inhibition by AntagoNAT oligonucleotides leads to transient and reversible upregulation of sense gene expression. As one example, we show that Brain-Derived Neurotrophic Factor (BDNF) is under the control of a conserved noncoding antisense RNA transcript, BDNF-AS, both in vitro and in vivo. BDNF-AS tonically represses BDNF sense RNA transcription by altering chromatin structure at the BDNF locus, which in turn reduces endogenous BDNF protein and function. By providing additional and analogous examples of endogenous mRNA upregulation, we suggest that antisense RNA mediated transcriptional suppression is a common phenomenon. In sum, we demonstrate a novel pharmacological strategy by which endogenous gene expression can be upregulated in a locus-specific manner.
Introduction
Although RNA interference and other technologies have provided useful tools for downregulation of mRNA and subsequently proteins, specific upregulation of individual genes remains challenging. Natural antisense transcripts (NATs) are transcribed from the opposite strand of many protein-coding (sense) genes and overlap in part with sense RNA, promoter region and their regulatory regions12. Here, we demonstrate a potent mechanism by which endogenous NATs suppress transcription of their sense gene counterparts. We show that endogenous gene expression can be upregulated, in a locus-specific manner by the removal or inhibition of the NATs, which are transcribed from most transcriptional units1,3. Our study provides examples of functional ncRNAs that regulate protein output, by altering chromatin structure and we posit that this phenomenon is applicable to many other genomic loci.
Brain-derived Neurotrophic Factor (BDNF) is a member of the "neurotrophin" family of growth factors, essential for neuronal growth, maturation4,5, differentiation and maintenance6. BDNF is also essential for neuronal plasticity and shown to be involved in learning, and memory processes7. The BDNF locus is on chromosome 11 and shows active transcription from both strands, which leads to transcription of a noncoding NATs8. Here, we characterize the regulatory role of this antisense RNA molecule, BDNF-AS that exerts a potent reciprocal and dynamic regulation over the expression of sense BDNF mRNA and protein, both in vitro and in vivo. We also report similar upregulation of Glial-derived Neurotrophic Factor (GDNF) and Ephrin receptor B2 (EphB2), by blocking endogenous NATs. We introduced a novel strategy for upregulation of mRNA expression, using antisense RNA transcript inhibitory molecules, which we term AntagoNATs.
Results
Genomic organization of the human BDNF locus
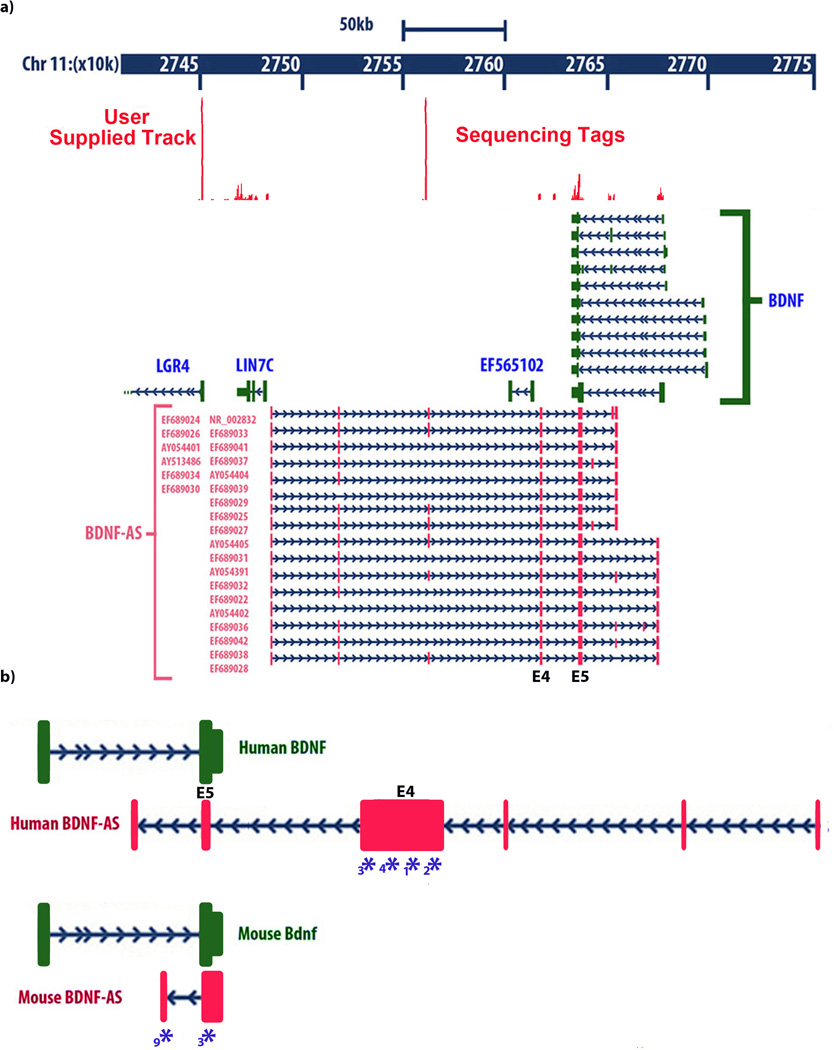
BDNF mRNA as well as BDNF antisense RNA (BDNF-AS, also annotated as BDNF-OS) displays a complex splicing pattern; however, all variants share a common sense-antisense overlapping region8,9. The transcription start site (TSS) of human BDNF-AS is approximately 200 kb downstream from the BDNF promoter and it is located on the positive strand of chromosome-11. Transcription from this site gives rise to 16–25 splice variant long ncRNAs with 6–8 exons8. Exon-5 of BDNF-AS, which contain 225-nucleotides of full complementarity to BDNF mRNA (overlapping) and exon-4 (non-overlapping) are common between all these variants (Fig. 1a). Nucleotide sequence of human BDNF-AS is provided in supplementary data file 1. BDNF mRNA is transcribed from the negative strand of chromosome-11 and shows 11 alternative splicing patterns and one coding exon. All variants of BDNF mRNA also share the 225-nucleotide overlapping region with the BDNF-AS transcript. Our next generation sequencing (deep sequencing) data confirms expression of BDNF-AS transcript in human brain RNA samples (Fig. 1a inset data). Therefore, BDNF-AS has the potential to form an in vivo RNA-RNA duplex with BDNF mRNA through 225 complementary nucleotides overlap.
Figure 1. Genomic organization of the human BDNF locus showing.
(A) genomic location of the BDNF sense and antisense transcripts and their relation to the other neighboring genes on chromosome 11. Solid boxes show exons and arrows show introns and direction of transcription. Different splice variants of BDNF-AS transcript are transcribed from the opposite DNA strand as that of BDNF mRNA. All BDNF-AS splice variants have a common exon that overlaps with 225 bp of all variants of BDNF mRNA. Inset data: Sequence tags generated by next-generation sequencing (RNA deep-seq), derived from human entorhinal cortex are aligned to the UCSC genome browser. Peaks represent nucleotide coverage, indicating reliable detection of BDNF-AS exons.
(B) Identification of mouse Bdnf-AS by RACE experiments followed by sequencing. Proportional drawing representing human and mouse BDNF loci, showing direction of transcription for both BDNF and BDNF-AS transcripts, as well as the potential overlapping region. Mouse Bdnf-AS transcript is shorter than that of human and contains 1–2 exons. The potential overlapping region is bigger in mouse (934 bp compared to 225 bp in human), but the common overlap shows 90% homology across the two species. Real-time PCR (RT-PCR) primers, probes, and siRNAs are designed to target the non-overlapping parts of both transcripts. Blue Asterisks and numbers below each denote target sites of siRNAs used in this study, as well as mBdnf-AntagoNAT3(*3) and mBdnf-AntagoNAT9 (*9).
Identification of mouse Bdnf-AS transcript
Although several human EST’s have been reported to have the potential of forming sense-antisense pairs with BDNF transcripts8, the mouse antisense transcript was not previously identified and thus BDNF-AS was erroneously reported as a primate-specific transcript by others9. Using 5’ and 3’ Rapid amplification of cDNA ends (RACE) experiments; we identified the mouse Bdnf-AS transcript (Fig. 1b). Based on RACE data we designed primers and probes for detection of mouse Bdnf-AS by real-time PCR (RT-PCR) experiments. RACE experiments followed by sequencing and RT-PCR indicate the existence of a conserved noncoding antisense transcript to the mouse Bdnf mRNA. The mouse Bdnf-AS transcript has two splice variants with 1–2 exons and 934-nucleotide complementarity to Bdnf mRNA (Fig. 1b). Nucleotide sequence of mouse Bdnf-AS is provided in Supplementary data file-2. Although the architectures of BDNF-AS in human and mouse are not entirely similar, the 225 bp overlapping region is almost identical between these two species (90% homology across the two species), suggesting the presence of an evolutionarily conserved functional binding domain.
Expression analysis of BDNF and BDNF-AS
We assessed expression of BDNF and BDNF-AS transcripts in various human (adult and embryonic), as well as monkey and mouse tissues, by RT-PCR and RNA fluorescence in situ hybridization (FISH). We also measured the absolute expression of BDNF and BDNF-AS transcript by generating standard curves, using DNA vectors containing cDNA of each transcript (Supplementary Fig. 1). BDNF mRNA levels are generally 10–100 fold higher than BDNF-AS transcript, except in testis, kidney and heart, which contain equal or higher levels of BDNF-AS. Both transcripts are expressed in brain, muscle and embryonic tissues (Supplementary Fig. 2). BDNF mRNA levels were relatively low in all post-natal tissues examined except in brain, bladder, heart and skeletal muscle (Supplementary Fig. 3). We examined the expression pattern of sense and antisense transcripts in rhesus monkey (Supplementary Fig. 4) and mouse tissues by RT-PCR (Supplementary Fig. 5) and RNA FISH (Supplementary Fig. 6). Both transcripts are co-expressed in many tissues, which suggest BDNF-AS potential for regulation of BDNF mRNA.
Knockdown of BDNF-AS increases BDNF in vitro
Transfection of several human and mouse cell lines including HEK293T cells with three independent siRNA molecules that targeted to non-overlapping regions of the BDNF-AS transcript, shown by asterisks in Figure 1b, resulted in over 85% knockdown of BDNF-AS transcript, accompanied by 2 to 6 fold upregulation of the BDNF transcript (Fig. 2a). Sequence information of these siRNAs as well as scrambled controls, AntagoNATs and other oligonucleotides are listed in supplementary Table S1. The upregulation of BDNF was not related to the choice of endogenous controls (Supplementary Fig. 7). BDNF-AS is a very low abundance transcript and therefore, in order to reliably detect the transcript, avoiding false signals from genomic DNA contamination and as control for RT-PCR reactions, we included no reverse transcriptase (NRT) and no template (NTC) controls in our experiments. Additionally, we tested the final PCR products for each set of primers and probe on a gel to ensure that only one product was amplified (Supplementary Fig. 8).
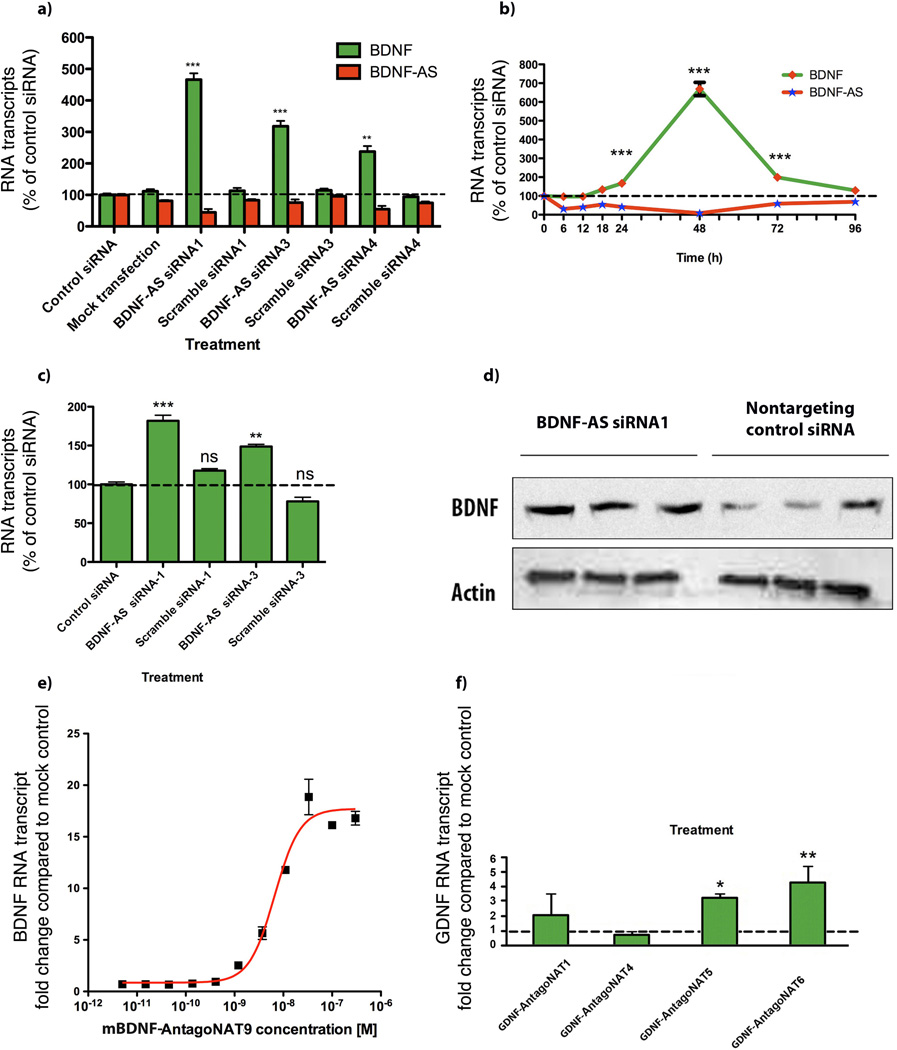
Figure 2. Antisense-mediated regulation of sense mRNA and protein.
(A) Knockdown of brain derived neurotrophic factor (BDNF) natural antisense transcript, BDNF-AS, in HEK293T cells (n=12 per treatment) with each of three unique siRNAs (10 nM) targeting the non-overlapping region of BDNF-AS transcript, caused 2–6 fold upregulation of BDNF (sense) mRNA (n=6 for each data point/treatment ***= P < 0.001, **= P < 0.01). Similar results were obtained from experiments using Human cortical neuron (HCN), glioblastoma (MK059) cells, mouse N2a cells and neurospheres “data not shown”. Scrambled sequences, mock transfection and control siRNAs were used as controls. Control siRNA for this and other experiments is an inert siRNA (CCUCUCCACGCGCAGUACATT) that does not target any known sequence in the mammalian genome. All measurements were normalized to the 18S rRNA and graphed as a percentage of each mRNA to the negative siRNA control sample.
(B) We assessed changes in BDNF and BDNF-AS transcripts over a period of time, following BDNF-AS knockdown (n=6 for each data point/treatment). siRNA knockdown of human BDNF-AS resulted in efficient and consistent downregulation of BDNF-AS, starting at 6 h and continuing on to 72 h. BDNF mRNA levels rose at 18 h, remaining high for more than 72 h, reversing to pre-treatment levels at 96 h. Note that the peak at 48 h is consistent and reproducible. Although BDNF-AS knockdown begins after 6 h, upregulation of BDNF started 18 h post-treatment. This time lag between the depletion of BDNF-AS and the increase of BDNF mRNA shows the sequential order of events indicating that the cells require time to adapt to the removal of the antisense transcript before upregulating BDNF.
(C) siRNA-mediated knockdown of BDNF-AS transcript caused an increase in BDNF protein levels measured by ELISA. Cells were transfected with 10 nM of two active siRNAs for BDNF-AS, scrambled siRNAs or a control siRNA for 48 hours. The supernatants of these cells were concentrated and analyzed for BDNF protein by ELISA, using a commercially available kit. BDNF protein was significantly increased (n=6 per treatment, ***=P < 0.0001, **= P<0.001) with siRNA targeting BDNF-AS transcript.
(D) Western blots confirmed that knockdown of the non-protein-coding BDNF-AS, with BDNF-AS siRNA1, but not control non-targeting siRNA transcript increased BDNF protein levels without changing the levels of beta-actin. Collectively, these data suggest that there is a discordant relationship between the sense and antisense BDNF transcripts in which BDNF-AS suppresses the expression of BDNF mRNA and protein. Removal of this negative regulatory effect, by BDNF-AS knockdown, causes upregulation of BDNF mRNA and protein levels.
(E) Dose-dependent increases in Bdnf following Bdnf-AS depletion: We performed dose-response experiments using 11 different concentrations (1:3 serial dilutions ranging from 300nM to 5pM) of mBdnf-AntagoNAT9 (n=6 per data point/treatment) and we observed a dose-dependent increase in Bdnf mRNA levels at 1–300 nM concentration with an EC50 of 6.6 nM.
(F) Selective knockdown of GDNF-AS increases GDNF mRNA: We treated cells with various AntagoNATs targeting a low abundance noncoding antisense RNA, GDNF-AS. We observed that GDNF-AntagoNAT5 and GDNF-AntagoNAT6 increase the GDNF mRNA by 3–4 fold (n=6 per treatment *= P < 0.05, **= P < 0.01).
To monitor the sequential events after administration of BDNF-AS-targeted siRNA, we performed a time-course study (Fig. 2b) in which we collected HEK-293 cells and assessed the (endogenous) expression of both BDNF and BDNF-AS transcripts at several time points after treatment. We observed downregulation of BDNF-AS at 6 h with maximum efficacy between 24–48 h and it lasting up to 72 h (Fig. 2b). Upregulation of BDNF mRNA only started at 18 h and reached the maximum at 48 h then decreased by 72 h, the expression returning to pre-treatment levels by 96 h. These data show that the observed increase of BDNF commences sequentially after reduction of the BDNF-AS transcript and displays full reversibility.
Moreover, we examined BDNF protein levels following treatment of HEK-293 cells with two active BDNF-AS siRNAs, control siRNA or scrambled siRNAs by ELISA (Fig. 2c). We measured BDNF protein level by western blot following transfection of cells with BDNF-AS siRNA-1 or control siRNA (Fig. 2d). Both ELISA (Fig. 2c) and western blotting (Fig 2d) experiments demonstrated that siRNA targeting BDNF-AS significantly increased the expression of BDNF protein. We observed that the magnitude of BDNF protein upregulation (~2 fold) was somewhat lower than with mRNA upregulation (2–6 fold); this suggests that protein upregulation kinetics lags that of mRNA in these cells or possibly an involvement of post-transcriptional regulatory mechanisms aside from antisense RNA to control BDNF protein output, including, for example, microRNA regulation10–12 (Supplementary Fig. 9).
BDNF-AS did not alter BDNF sense RNA stability
In order to examine the effects of BDNF-AS transcript on the stability of BDNF sense transcript, we depleted BDNF-AS with siRNA then treated cells with α-amanitin. We found that the baseline half-life for BDNF-AS was 15.3 h, nearly 3 h longer than that of the BDNF sense transcript (Supplementary Fig. 10). There was no significant change in BDNF sense RNA stability after reduction of BDNF-AS transcript, suggesting that unlike some other highly abundant antisense transcripts13, BDNF-AS does not alter BDNF sense mRNA stability. Recently it has been suggested that some NATs can produce endogenous siRNAs from the overlapping region between sense and antisense RNAs. Indeed Watanabe et al. published a list of endogenous siRNAs from mouse oocytes14. We examined this list but we were not able to find any endogenous siRNAs originating from the Bdnf locus.
Targeting of BDNF-AS by AntagoNATs
We introduce the term AntagoNAT here to describe single-stranded oligonucleotide molecules that inhibit sense-antisense interactions (with different modifications, see supplementary methods). We hypothesized that use of AntagoNATs should have a similar outcome on BDNF-AS as that observed with BDNF-AS siRNA and designed gapmer single–stranded oligonucleotides, 14 nucleotides in length, with 2’O-Methyl RNA and/or locked nucleic acid (LNA) modifications. Using this strategy, we tiled the entire overlapping region between human BDNF-AS and BDNF transcripts and identified several efficacious AntagoNATs capable of upregulating of BDNF mRNA. hBDNF-AntagoNAT1 and hBDNF-AntagoNAT4, targeting the first part of the overlapping region, produced the largest response (Supplementary Fig. 11). Our data suggests that blockage of BDNF antisense RNA, by single-stranded AntagoNATs, is sufficient in causing an increase in BDNF mRNA.
We then designed single-stranded gapmer LNA-modified15 DNA oligonucleotides (AntagoNATs) 16-nucleotides in length with phosphorothioate backbone, complementary to mouse Bdnf-AS. Two AntagoNATs (mBdnf-AntagoNAT3 and mBdnf-AntagoNAT9) consistently showed a statistically significant increase in Bdnf mRNA levels in mouse N2a cells (Supplementary Fig. 12). In order to establish an optimal dosage for further in vitro studies, we performed concentration-response experiments with 11 different concentrations (1:3 serial dilutions ranging from 300 nM to 5 pM) of mBdnf-AntagoNAT9, measuring fold changes in Bdnf mRNA levels (Fig. 2e). A concentration-dependent increase in Bdnf mRNA levels at 1–300 nM with EC50 of 6.6 nM was determined.
Knockdown of GDNF-AS increases GDNF mRNA in vitro
We selected another low abundance noncoding antisense RNAs that is transcribed from opposite strand of GDNF at chromosome 5 and designed several AntagoNATs targeting the GDNF antisense transcript. We found two AntagoNATs that significantly increase the GDNF mRNA by 3–4 fold (Fig. 2f). Additionally, we show AntagoNAT-mediated upregulation of EphB2, a member of a different gene family (Supplementary Fig. 13). These results suggest that antisense RNA mediated transcriptional suppression is a widespread phenomenon in the mammalian genome.
Bdnf upregulation increases neuronal outgrowth
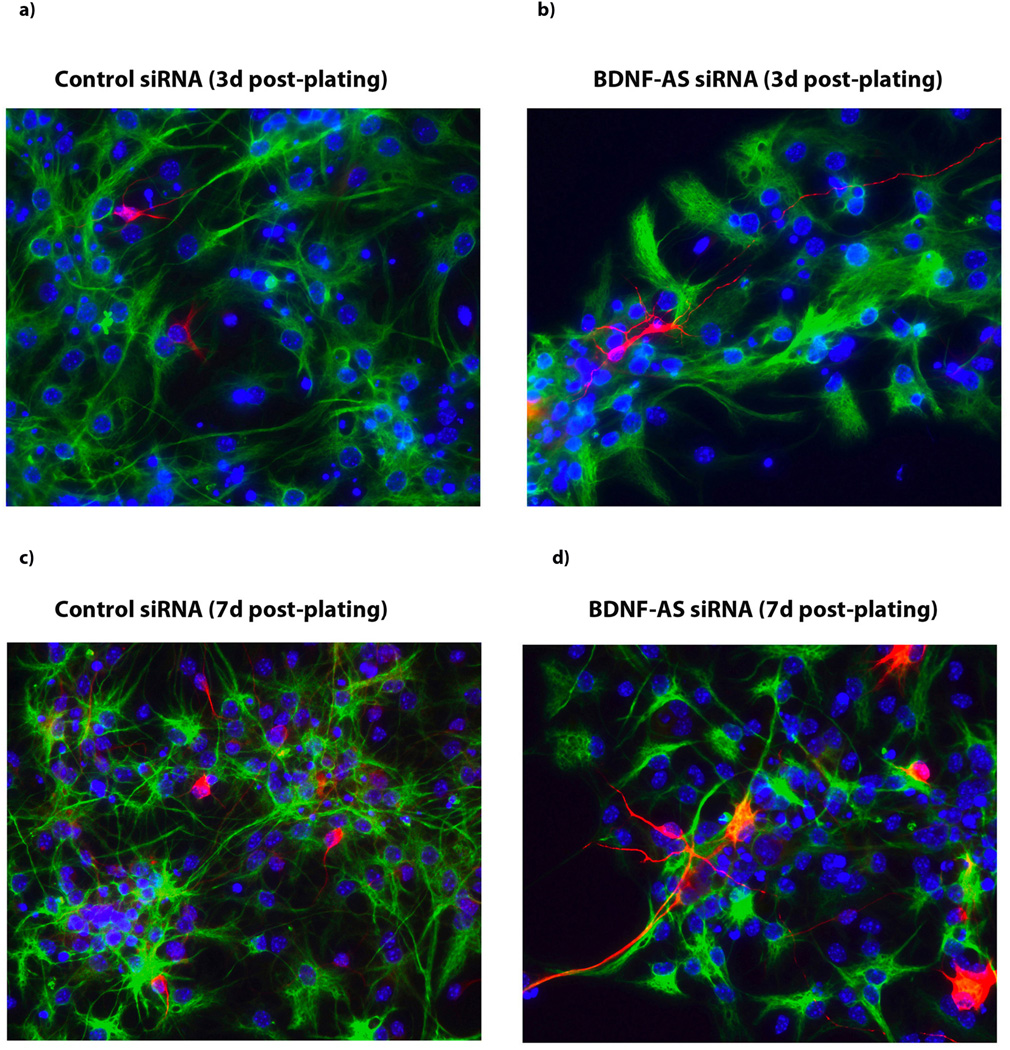
Consistent with many previous reports that indicate stimulatory effects of Bdnf on neuronal outgrowth and adult neurogenesis16–17, we found that an increase in the endogenous Bdnf level due to the knockdown of Bdnf-AS transcript resulted in increased neuronal cell number and in neurite outgrowth and maturation at 3 and 7 days post-plating in neurospheres (Fig. 3 a–d). These data suggest that the upregulation of endogenous Bdnf, due to inhibition of antisense RNA, induces neuronal differentiation in neuronal progenitor cells and might cause a mature phenotype in nascent neurons.
Figure 3. Bdnf upregulation increases neuronal outgrowth.
(A–B) Immunocytochemistry images of hippocampal neurospheres treated with either control siRNA (A) or Bdnf-AS siRNA (B) 3 d post-platting. (C–D) Immunocytochemistry images of neuronal maturation and neurite outgrowth in hippocampal neurospheres treated with either control siRNA (C) or Bdnf-AS siRNA (D) 7 d post-plating. Treatment of cells with siRNA targeting the Bdnf-AS transcript resulted in increased neuronal cell number as well as increase in neurite outgrowth and maturation, both at 3d or 7d post-plating neurospheres. B-tubulin III stained red, GFAP stained green and DAPI stained blue.
Knockdown of Bdnf-AS increases Bdnf in vivo
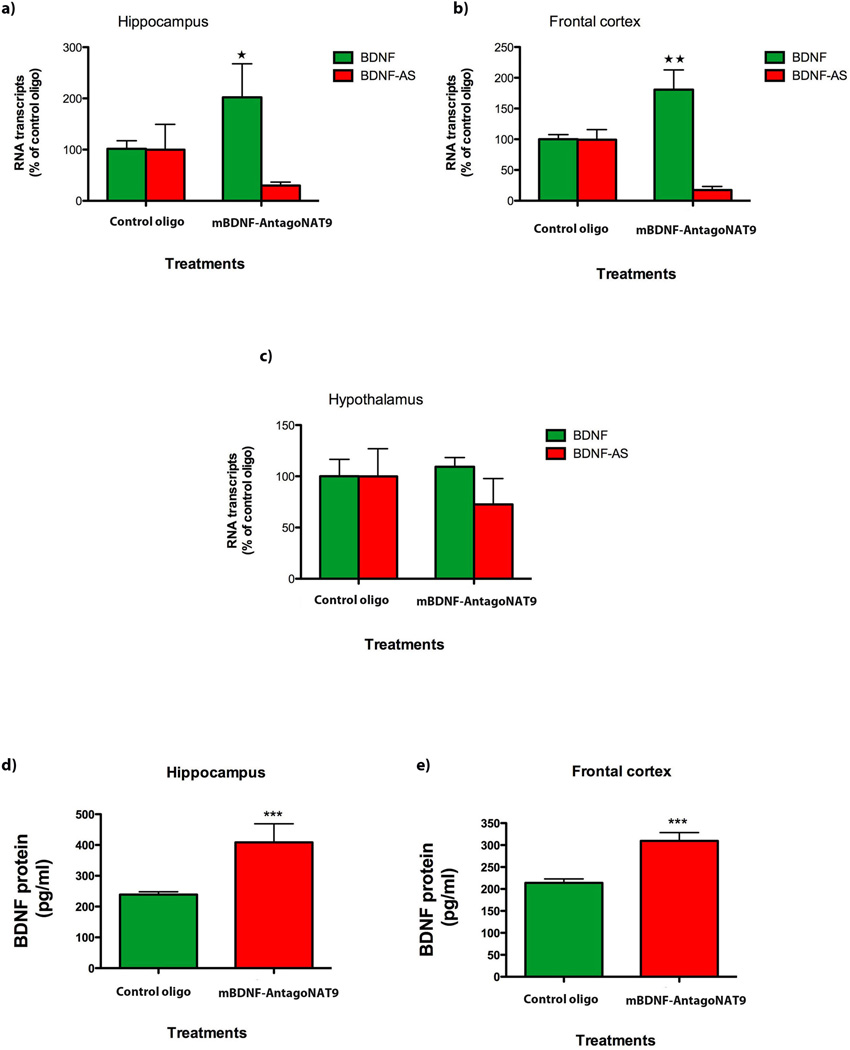
We utilized osmotic mini-pumps for intracerebroventricular (ICV) delivery of mBdnf-AntagoNAT9 to C57BL/6 mice. We selected mBdnf-AntagoNAT9, which is targeting a non-overlapping region of mouse Bdnf-AS, over other active AntagoNATs, based on its high efficacy to increase in Bdnf mRNA in vitro. After 28 days of continuous AntagoNAT infusion, Bdnf mRNA levels were increased across forebrain regions adjacent to the third ventricle in mice treated with mBdnf-AntagoNAT9 as compared to levels unaltered by an inert control oligonucleotide (Fig. 4a,b). Bdnf and Bdnf-AS transcripts were unaltered in the hypothalamus, a structure that is not immediately adjacent to the third ventricle (Fig. 4c). Moreover, we find that AntagoNAT-mediated blockade of Bdnf-AS results in increased Bdnf protein levels (Fig 4 d,e). These findings correspond with the in vitro data described above and indicate that the blockade of Bdnf-AS results in the increase of Bdnf mRNA and protein expression in vivo.
Figure 4. Bdnf-AS regulates Bdnf mRNA and protein in vivo.
(A–C) Using osmotic mini-pumps, we infused mBdnf-AntagoNAT9 (CAACATATCAGGAGCC) or control oligonucleotide (CCACGCGCAGTACATG) constantly over a period of 28 d, into the third ventricle of mouse brain (n=5 per treatment group *= P < 0.05, **= P < 0.01, ***= P < 0.001). mBdnf-AntagoNAT9 directed against Bdnf-AS but not the control oligonucleotide resulted in an increase in Bdnf levels in the hippocampus (A) and frontal cortex (B). In the hypothalamus (C) both transcripts were unchanged, as was expected for a tissue that is not directly connected to the third ventricle of the brain.
(D–E) We assessed BDNF protein levels by ELISA and found that mBdnf-AntagoNAT9 treatment results in an increase in BDNF protein, both in the hippocampus (D) and frontal cortex (E), as compared to control oligonucleotide treated mice.
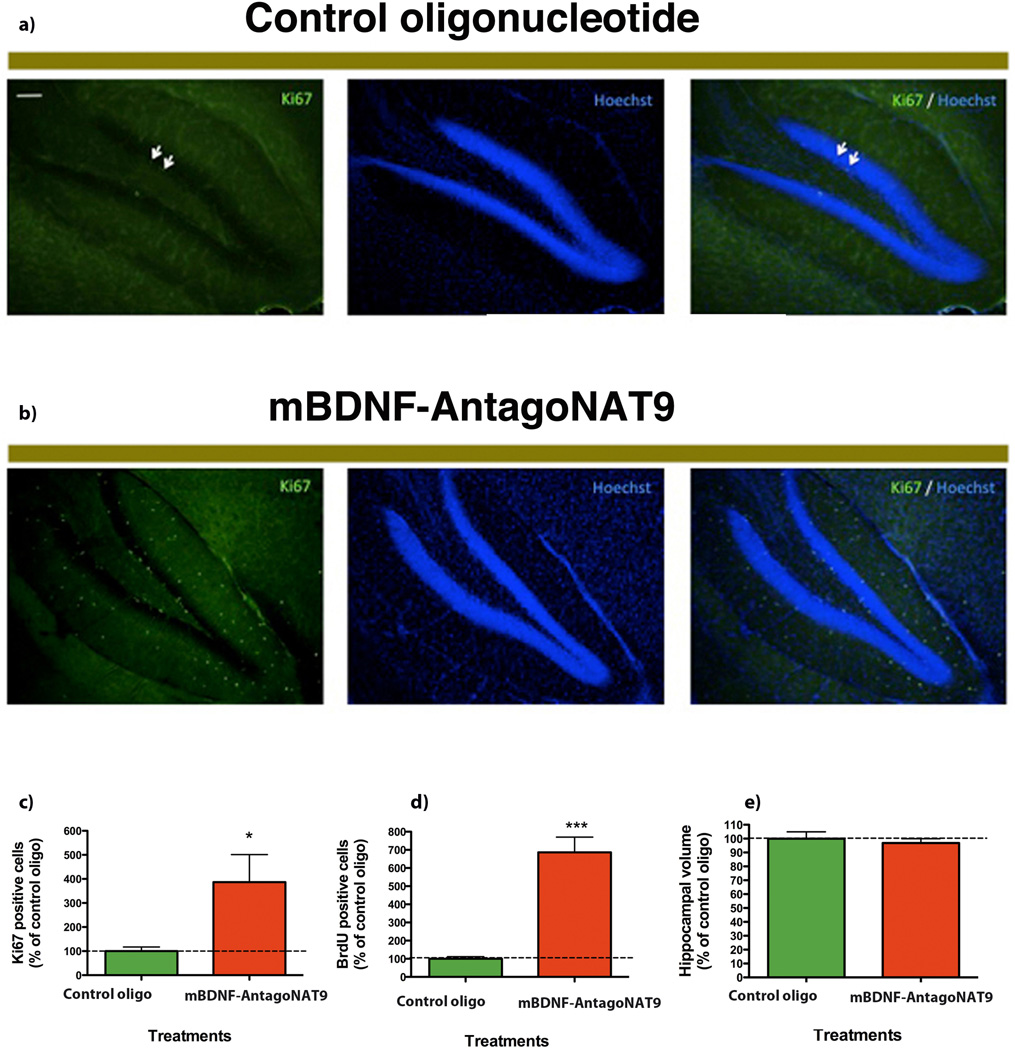
We injected BrdU in the mice treated with mBdnf-AntagoNAT9 in the first week of the study for 5 days. After 28 days of continuous AntagoNAT infusion, we performed histological examination of brain tissues and quantified neuronal proliferation and survival using Ki67 and BrdU markers, respectively. In mice treated with mBdnf-AntagoNAT9, we observed an increase in Ki67 positive (proliferating) cells as compared to control treated mice (Fig 5a,b). We quantified the number of Ki67 positive cells and found a significant increase in cell proliferation in mice treated with mBdnf-AntagoNAT9 compared to control oligonucleotide (Fig. 5c). In mice treated with mBdnf-AntagoNAT9, there was a significant increase in BrdU incorporation (surviving cells) as compared to the control oligonucleotide- treated mice (Fig. 5d). There were no differences in hippocampal volume between control and mBdnf-AntagoNAT9 treated mice (Fig. 5e). These findings demonstrate that Bdnf-AS regulates Bdnf levels in vivo.
Figure 5. Blocking of Bdnf-AS, in vivo, causes an increase in neuronal survival and proliferation.
(A–B) We treated mice with mBdnf-AntagoNAT9 or control oligos. After 28 d of continuous mBdnf-AntagoNAT9 infusion, we performed histological examination of brain tissues, using Ki67. Ki67 is the marker of proliferating cells in hippocampus and we observed an increase in the number of proliferating cells in mice received mBdnf-AntagoNAT treatment compare to mice received control oligos. In mice treated with mBdnf-AntagoNAT9 (B), there was an increase in Ki67 positive cells (proliferating cells), as compared to control treated mice (A).
(C) Mice treated with mBdnf-AntagoNAT9 had a significant increase in the number of Ki67 positive cells as compared to control treated mice.
(D) In mice treated with mBdnf-AntagoNAT9, there was a significant increase in the number of surviving cells (BrdU positive) as compared to control oligonucleotide treated mice.
(E) There were no differences in hippocampal volume between control and mBdnf-AntagoNAT9 treated mice.
Together these data (n=5 per treatment group *= P < 0.05, ***= P < 0.001) demonstrates that Bdnf-AS regulates Bdnf levels in vivo and that blocking Bdnf sense-antisense interactions results in an increase in neuronal lineage, proliferation and survival.
BDNF-AS induces repressive chromatin marks
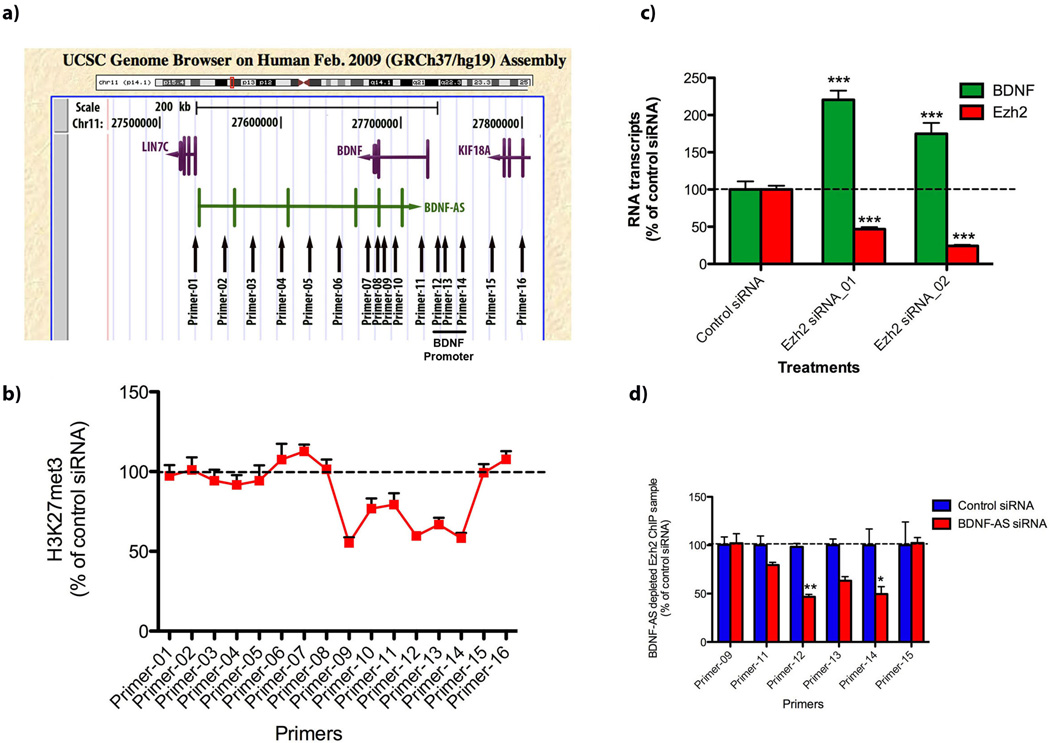
We measured the association of repressive (H3K9met3, H3K27met3) and active (H3K4met3, H3K36met3) chromatin marks to the BDNF genomic locus (Fig. 6a). Treating with siRNA, intracellular BDNF-AS was depleted and chromatin immunoprecipitation (ChIP) assays were performed. DNA was extracted and analyzed using 16 primer sets matching regions along the entire BDNF locus including the sense-antisense (S-AS) overlapping and BDNF promoter region (Fig-6a). Primers were designed to span the entire BDNF locus, at 20 kb increments, covering more than 270 kb and extending through to neighboring genes LIN7C and KIF18A. Initially, we performed RT-PCR on RNA samples and found that the knockdown of BDNF-AS transcript increases BDNF mRNA without any effects on neurotrophic tyrosine kinase, receptor, type 2 (TrkB) or on neighboring genes (LIN7C and KIF18A) in both directions (Supplementary Fig. 14). Next, we studied the immunoprecipitated DNA, using individual primer sets using RT-PCR and found that the siRNA-mediated depletion of the BDNF-AS transcript causes a significant reduction in H3K27met3 association in both the sense-antisense overlapping region and in the upstream BDNF promoter region (Fig. 6b). We found similar reduction in H3K27met3 binding to the promoter of the mouse Bdnf gene upon treatment of N2a ells with mBdnf-AntagoNAT9 (Supplementary Fig. 15). Furthermore, we found that alterations in repressive marks were specific to H3K27met3 as we were not able to detect significant changes in H3K9met3, H3K4met3 and H3K36met3 (Supplementary Fig. 16). These data suggest that BDNF-AS might play a role in the guidance, introduction and maintenance of H3K27met3 at the BDNF locus.
Figure 6. Removal of BDNF-AS resulted in the modification of chromatin marks.
(A) HEK293T cells were treated with control or BDNF-AS siRNA. 48 h post-transfection cells were harvested, fixed with formaldehyde, sonicated and incubated with antibodies against H3K27met3. Immunoprecipitation was performed followed by DNA extraction. DNA samples were analyzed using 16 primer sets covering the entire BDNF gene locus, and the BDNF promoter region, as indicated.
(B) There was a decrease in association of the repressive chromatin marker, H3K27met3, upon treatment of the cells with BDNF-AS siRNA, both at the sense-antisense overlapping (primer-9) and promoter (primer12–14) regions (n=6 for each data point). The observed chromatin modification did not extend toward neighboring genes. These results suggest that local antisense-mediated chromatin modifications are occurring, beginning from the sense-antisense overlapping region and spreading in the 5’ direction towards the BDNF promoter region.
(C) Knockdown of Ezh2, by either one of two different siRNAs, mimics or phenocopies the BDNF-AS knockdown and causes upregulation of BDNF mRNA (n=6 for each data point/treatment ***= P < 0.001).
(D) ChIP assay using an Ezh2 antibody revealed that there was a decrease in Ezh2 association with the BDNF promoter upon depletion of the BDNF-AS transcript by siRNA. Not all 16 primer sets gave detectable PCR signals, possibly due to the lack of direct Ezh2-chromatin binding (n=6 for each data point/treatment **= P < 0.01, *= P < 0.05).
We observed that the ablation of Ezh2 activity using two different siRNAs phenocopied the BDNF-AS knockdown effect and caused the upregulation of BDNF mRNA (Fig. 6c). A ChIP assay was performed using the Ezh2 antibody. ChIP data revealed a significant reduction in Ezh2 binding at the BDNF promoter, upon depletion of BDNF-AS by siRNA (Fig. 6d). However, not all 16 primer sets gave detectable PCR signals, which could be attributed to the lack of direct Ezh2-chromatin binding. Given the reduction of binding of Ezh2 and the loss of the H3K27met3 at the BDNF promoter when BDNF-AS is knocked down, we conclude that BDNF-AS represses chromatin by recruiting Polycomb Repressive Complex 2 (PRC2) to the BDNF promoter region. Removal or inhibition of BDNF-AS could lead to the locus-specific upregulation of BDNF mRNA and protein.
Discussion
The number of ncRNAs in eukaryotic genomes have been shown to increase as a function of developmental complexity18,19 and there is, for example, a great deal of diversity in ncRNAs expressed in the nervous system20,21. Over the past few years, we and others have reported on functional NATs and showed their potential involvement in human disorders, including Alzheimer’s disease13, Parkinson’s disease22 and Fragile X syndrome23. Moreover, previously we showed that upregulation of CD97 sense gene can be attained by knockdown of its antisense RNA transcript1. Upregulation of progesterone receptor (PR), and other endogenous transcripts was reported following targeting of promoter-derived noncoding RNAs24,25. Transcriptional activation of p21 gene26 and Oct4 promoter27 were reported following NATs depletion. Antisense RNA-induced chromatin remodeling seems to be a feasible and dynamic mode of action for many low-copy number NATs2,28. If so, antisense RNA might predominantly exert local effects to maintain or modify chromatin structure, ultimately activating or suppressing sense gene expression.
PCR2 is a protein complex that consists of four core subunits: Eed, Suz12, RbAp48 and the catalytic Ezh2, that catalyzes the trimethylation of histone H3-lysine 27 (H3K27met3)32. Recent studies provide evidence for direct RNA-potein interaction between Ezh2 and many ncRNA transcripts29. Other studies of X inactivation30 and HOX gene cluster31 show RNA transcripts to be involved in the PRC2-mediated induction of H3K27met3, repressive chromatin marks. PRC2 transcriptome profiling has identified over 9,000 PRC2-interacting RNAs in embryonic stem cells, many of them categorized as antisense RNA transcripts29. Epigenetic silencing of p15 and DM1 genes were reported to involve heterochromatin formation by its antisense RNA33,34. The traditional binary division of chromatin into hetero- or eu-chromatin categories might not be complete as recent work has shown that there are five principal chromatin types49 that are more dynamic and flexible than originally believed. Likely applicable to a large number of gene loci, NATs can be manipulated in order to obtain a locus-specific alteration in chromatin modification. As examples, we show here that cleavage (by siRNA) or inhibition (by AntagoNATs) of the antisense transcripts of BDNF, GDNF and EphB2 genes leads to the upregulation of corresponding mRNAs.
Neurotrophins belong to a class of secreted growth factors that enhance the survival, development, differentiation and function of neurons and BDNF is an important molecular mediator of synaptic plasticity35–37. BDNF is suggested to synchronize neuronal and glial maturation38, participate in axonal and dendritic differentiation39 and protect and enhance neuronal cell survival40,41. Neurotrophin expression levels are impaired in neurodegenerative42–45 and in psychiatric and neurodevelopmental disorders46–48. The upregulation of neurotrophins is believed to have beneficial effects on several neurological disorders. AntagoNATs can potentially be used as a therapeutic strategy to inhibit BDNF-AS and consequently enhance neuronal proliferation and survival in a variety of disease states. It cannot be excluded that the herein described approach to upregulate the synthesis of endogenous BDNF molecules, presumed to contain natural modifications and to represent all known splice forms, will prove to be distinct, and perhaps superior, to administrating synthetic BDNF molecules.
Many low abundance noncoding antisense RNAs, which are transcribed on the opposite strands of genomic loci, endogenously suppress corresponding sense gene expression; inhibition or removal of antisense transcript leads to locus-specific upregulation of sense mRNA, protein and function.
Supplementary Material
Acknowledgements
Dr. Qing-Rong Liu from the National Institute of Drug Abuse kindly provided us constructs that contain three splice variants of the human BDNF-AS transcript. We thank Drs. Carlos Coito, Philip Frost, Jane Hsiao and Olga Khorkova at OPKO-CURNA for helpful discussions. The National Institutes of Health (5R01NS063974 and 5RC2AG036596) funded this work.
References
- 1.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasbi A, et al. Calcium signaling cascade links dopamine D1–D2 receptor heteromer to striatal BDNF production and neuronal growth. Proc Natl Acad Sci U S A. 2009;106:21377–21382. doi: 10.1073/pnas.0903676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trzaska KA, et al. Brain-derived neurotrophic factor facilitates maturation of mesenchymal stem cell-derived dopamine progenitors to functional neurons. J Neurochem. 2009;110:1058–1069. doi: 10.1111/j.1471-4159.2009.06201.x. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimura R, Ito K, Endo Y. Differentiation/maturation of neuropeptide Y neurons in the corpus callosum is promoted by brain-derived neurotrophic factor in mouse brain slice cultures. Neurosci Lett. 2009;450:262–265. doi: 10.1016/j.neulet.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Choi DC, et al. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proc Natl Acad Sci U S A. 2010;107:2675–2680. doi: 10.1073/pnas.0909359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu QR, et al. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics. 2007;2:214–222. doi: 10.4161/epi.2.4.5212. [DOI] [PubMed] [Google Scholar]
- 11.Ogier M, et al. Brain-derived neurotrophic factor expression and respiratory function improve after ampakine treatment in a mouse model of Rett syndrome. J Neurosci. 2007;27:10912–10917. doi: 10.1523/JNEUROSCI.1869-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 13.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 15.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hökfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Ørum H, Jacobsen MH, Wengel J. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci (USA) 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park HR, et al. A high-fat diet impairs neurogenesis: involvement of lipid peroxidation and brain-derived neurotrophic factor. Neurosci Lett. 2010;482:235–239. doi: 10.1016/j.neulet.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 17.Im SH, et al. Induction of striatal neurogenesis enhances functional recovery in an adult animal model of neonatal hypoxic-ischemic brain injury. Neuroscience. 2010;169:259–268. doi: 10.1016/j.neuroscience.2010.04.038. [DOI] [PubMed] [Google Scholar]
- 18.Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. J Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 20.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheele C, et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics. 2007;8:74. doi: 10.1186/1471-2164-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile × syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 25.Watts JK, et al. Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters. Nucleic Acids Res. 2010;38:5242–5259. doi: 10.1093/nar/gkq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000258. e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Antal A, et al. Brain-derived neurotrophic factor (BDNF) gene polymorphisms shape cortical plasticity in humans. Brain Stimul. 2010;3:230–237. doi: 10.1016/j.brs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Castillo DV, Escobar ML. A role for MAPK and PI-3K signaling pathways in brain-derived neurotrophic factor modification of conditioned taste aversion retention. Behav Brain Res. 2011;217:248–252. doi: 10.1016/j.bbr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Nikolakopoulou AM, Meynard MM, Marshak S, Cohen-Cory S. Synaptic maturation of the Xenopus retinotectal system: effects of brain-derived neurotrophic factor on synapse ultrastructure. J Comp Neurol. 518:972–989. doi: 10.1002/cne.22258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortega JA, Alcantara S. BDNF/MAPK/ERK-induced BMP7 expression in the developing cerebral cortex induces premature radial glia differentiation and impairs neuronal migration. Cereb Cortex. 20:2132–2144. doi: 10.1093/cercor/bhp275. [DOI] [PubMed] [Google Scholar]
- 39.Chapleau CA, Larimore JL, Theibert A, Pozzo-Miller L. Modulation of dendritic spine development and plasticity by BDNF and vesicular trafficking: fundamental roles in neurodevelopmental disorders associated with mental retardation and autism. J Neurodev Disord. 2009;1:185–196. doi: 10.1007/s11689-009-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu SY, et al. Running exercise protects the substantia nigra dopaminergic neurons against inflammation-induced degeneration via the activation of BDNF signaling pathway. Brain Behav Immun. 2011;25:235–243. doi: 10.1016/j.bbi.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Yang LC, et al. Extranuclear estrogen receptors mediate the neuroprotective effects of estrogen in the rat hippocampus. PLoS One. 2010;5:e9851. doi: 10.1371/journal.pone.0009851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laske C, et al. Higher BDNF serum levels predict slower cognitive decline in Alzheimer's disease patients. Int J Neuropsychopharmacol. 2011;14:1–6. doi: 10.1017/S1461145710001008. [DOI] [PubMed] [Google Scholar]
- 43.Zeng Y, Zhao D, Xie CW. Neurotrophins Enhance CaMKII Activity and Rescue Amyloid-beta-Induced Deficits in Hippocampal Synaptic Plasticity. J Alzheimers Dis. 2010;21:823–831. doi: 10.3233/JAD-2010-100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frade JM, Lopez-Sanchez N. A novel hypothesis for Alzheimer's disease based on neuronal tetraploidy induced by p75(NTR) Cell Cycle. 2010;9:1934–1941. doi: 10.4161/cc.9.10.11582. [DOI] [PubMed] [Google Scholar]
- 45.Massa SM, et al. Small molecule BDNF mimetics activate TrkB signaling and prevent neuronal degeneration in rodents. J Clin Invest. 2010;120:1774–1785. doi: 10.1172/JCI41356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo KR, et al. Differential regulation of neurotrophin S100B and BDNF in two rat models of depression. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:1433–1439. doi: 10.1016/j.pnpbp.2010.07.033. [DOI] [PubMed] [Google Scholar]
- 47.Gonul AS, et al. Association of the brain-derived neurotrophic factor Val66Met polymorphism with hippocampus volumes in drug-free depressed patients. World J Biol Psychiatry. 2011;12:110–118. doi: 10.3109/15622975.2010.507786. [DOI] [PubMed] [Google Scholar]
- 48.Dell'osso L, et al. Associations between Brain-Derived Neurotrophic Factor Plasma Levels and Severity of the Illness, Recurrence and Symptoms in Depressed Patients. Neuropsychobiology. 2010;62:207–212. doi: 10.1159/000319946. [DOI] [PubMed] [Google Scholar]
- 49.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Toledano MA, Shelanski ML. Neurogenic effect of beta-amyloid peptide in the development of neural stem cells. J Neurosci. 2004;24:5439–5444. doi: 10.1523/JNEUROSCI.0974-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.