Abstract
Purpose.
Large copy number variations (CNV) can contribute to increased burden for neurodegenerative diseases. In this study, we analyzed the genome-wide burden of large CNVs > 100 kb in primary open angle glaucoma (POAG), a neurodegenerative disease of the eye that is the largest cause of irreversible blindness.
Methods.
Genome-wide analysis of CNVs > 100 kb were analyzed in a total of 1720 individuals, including an Indian cohort (347 POAG cases and 345 controls) and a Caucasian cohort (624 cases and 404 controls). All the CNV data were obtained from experiments performed on Illumina 660W-Quad (infinium) arrays.
Results.
We observed that for both the populations CNVs > 1 Mb was significantly enriched for gene-rich regions unique to the POAG cases (P < 10−11). In the Indian cohort CNVs > 1 Mb (39 calls) in patients influenced 125 genes while in controls 31 such CNVs influenced only 5 genes with no overlap. In both cohorts we observed 1.9-fold gene enrichment in patients for deletions compared to duplications, while such a bias was not observed in controls (0.3-fold). Overall duplications > 1 Mb were more than deletions (Del/Dup = 0.82) confirming that the enrichment of gene-rich deletions in patients was associated with the disease. Of the 39 CNVs > 1 Mb from Indian patients, 28 (72%) also were implicated in other neurodegenerative disorders, like autism, schizophrenia, sensorineural hearing loss, and so forth. We found one large duplication encompassing CNTN4 gene in Indian and Caucasian POAG patients that was absent in the controls.
Conclusions.
To our knowledge, our study is the first report on large CNV bias for gene-rich regions in glaucomatous neurodegeneration, implicating its impact across populations of contrasting ethnicities. We identified CNTN4 as a novel candidate gene for POAG.
Keywords: glaucoma, POAG, genomics, CNV, CNTN4
This study shows large CNV bias for gene-rich regions for glaucomatous neurodegeneration, implicating its impact across populations of contrasting ethnicities.
Introduction
Copy number variations (CNVs) are known to be one of the common types of genomic variants present in every cell or tissue types. Similar to other types of genetic variants (e.g., single nucleotide polymorphisms [SNPs]) CNVs can be selectively neutral or under selection pressure, and can contribute to disease phenotypes. The CNVs can widely vary in size, from approximately 1 kilobase pair (kbp) to a few megabase pairs (Mbp) and the reliability of detection increases with increasing size.1
Large CNVs, namely, those with sizes > 100 kb have been reported to be associated with several biological traits. In the recent past, a number of studies on several phenotypic traits have shown that the presence of large CNVs (>100 kb) are more common in patients compared to controls.2 Large CNV burden also has been shown to be associated with mortality at old age3 as well as confer higher risk for a number of neurodegenerative diseases.2 The role of larger CNVs in phenotypic diversity and possible deleterious effects were shown in a study on 26 diverse populations from the Indian subcontinent, as well as in the Caucasian population.1,4
Glaucomatous neurodegeneration is the largest cause of irreversible blindness worldwide, with a population prevalence of 1%, and primary open angle glaucoma (POAG) is the most common subtype in most parts of the world, including India. The condition of POAG is a complex multifactorial disease with large heritable components. It usually follows autosomal dominant trait when familial and, so far, 17 loci has been identified to be linked to POAG.5 A number of genes also were reported to have common variants associated significantly with the disease.5 In the last few years, several genome-wide association studies (GWAS) also have been reported from different populations claiming association of various loci to be associated with the disease.6,7
Genome-wide studies also have reported involvement of rare as well as common CNVs with POAG,8,9 and CNVs associated with IOP, a major risk factor for POAG.10 Family-based linkage studies have identified a CNV segregating with normotensive glaucoma (NTG), an important subtype of POAG.11 Studies have explored candidate gene-based CNVs for their possible role in the disease.12,13 However, overlap between these findings is poor, warranting further studies in different populations to decipher the molecular mechanism with better clarity.
The impact of large CNVs in POAG has not been reported so far in the literature. In this study, we specifically looked into the burden of large CNVs (>100 kb) on a genome-wide scale, with a focus for their enrichment in gene-rich regions for POAG in an Indian cohort, and used a publically available Caucasian dataset to check for reproducibility of such burden across ethnic boundaries.
Methods
Study Subjects
This study includes 364 POAG patients and 365 controls recruited from a large population residing in eastern part of India (West Bengal). Inclusion criteria for cases were optic disc cupping or visual field changes in patients with an IOP of greater than 20 mm Hg, and optic disc cupping and visual field changes in patients with an IOP of less than 20 mm Hg. Cases with ocular hypertension but without cupping and/or visual field changes were excluded from the study,14 The samples were recruited after explaining the study to them individually and taken written consent. This study also was approved by the human ethics committee of the organization and followed the tenets of the Declaration of Helsinki.
For validation in an independent population, we included genome-wide CNV data of 866 cases and 495 controls from a POAG cohort of Caucasian background (GLAUGEN: dbGaP Study Accession:phs000308.v1.p1).
DNA Isolation and Genome-Wide Genotyping for CNVs
For samples collected in the Indian cohort, genomic DNA was extracted from peripheral blood leukocytes using a standard salting-out procedure. We performed genome-wide genotyping, using the Infinium Human660W-quad BeadChip (Illumina, Inc., San Diego, CA, USA). We used 200 ng of genomic DNA for each sample, in accordance with the manufacturer's guidelines. The raw data files were processed by the GenomeStudio software package provided by the manufacturers. For the Caucasian cohort, genome-wide data was generated on the same platform and was made available through the dbGAP server, which we have accessed and analyzed (see below).
CNV Quality Control and CNV Call
We removed 25 samples for their close genetic relatedness (IBS test) with other samples in the Indian cohort. Furthermore, we used a threshold of 0.35 for the standard deviation for logR ratio of normalized intensity (LRR) and 12 samples failed to meet this criteria (Supplementary Fig. S1). To call CNVs, we used the PennCNV algorithm15 and applied a stringent criterion of at least 10 consecutive probes to show altered intensity to qualify as a CNV call. The CNVPartition16 was used as an independent tool and 82% concordance was noted with the calls generated by PennCNV. The data presented in the study is the result of PennCNV algorithm. Using the aforementioned filter criteria on Caucasian samples, 306 samples were removed from1334 total samples and, thus, we analyzed data for 1028 samples (624 cases, 404 controls) from this cohort. We focused on individual CNV calls > 100 kb in patient or control groups independently. An independent validation of CNV calls was done on Indian samples by real-time PCR taking representative CNVs and we could validate approximately 70% of the CNV calls (Kaurani et al., unpublished data).
The datasets used for the analyses described in this manuscript were obtained from dbGaP, available in the public domain at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gapthrough dbGaP accession number phs000308.v1.p1.
Results
In this study, we analyzed genome-wide large CNVs (>100 kb) in 1720 individual samples, representing 692 Indians (347 cases, 345 controls) and 1028 Caucasians (624 cases, 404 controls). In addition, we also used CNVs from the database of genomic variants (DGV), which represents more than 11,000 samples. In the Indian POAG group, the mean age of the patients was 49.196 ± 16.45 years and for the controls it was 49.75 ± 11.36 years. The mean IOP for the patients was 22.93 ± 7.74 mm of Hg.
Genomic Landscape of Larger CNVRs in POAG Patients and Controls
We identified a total of 208,929 CNV calls from 692 samples and 453,761 CNV calls from 1028 samples from the Indian and the Caucasian datasets, respectively. Upon applying a minimum size filter of 100 kb in cases and controls separately, we identified 2116 and 2064 individual CNVs in the Indian cohort (Fig. 1, Supplementary Table S1). In the Caucasian cohort, the corresponding numbers were 15,155 and 8582 (Supplementary Fig. S2, Supplementary Table S2). Genome-wide occurrence of deletions and duplications was not significantly different between the cases and the controls in either cohort (Supplementary Fig. 3). Average CNV rate per genome was similar between the cases and controls (6.1 vs. 6.0 in Indian data; 24.3 vs. 21.1 in Caucasian data, respectively), but was different between the two cohorts as depicted in Table 1. Analyzing the standard deviation of the logR ratios, we showed that the higher number of CNVs in the Caucasian data was not due to a technical noise, but was a feature of the specific population (Supplementary Fig. 4). Average CNV size was similar between cases and controls, and was not significantly different between the Indian and the Caucasian cohorts (237.2 kb vs. 237.7 kb in Indian data; 343.3 kb vs. 323 kb in Caucasian data) as depicted in Table 1.
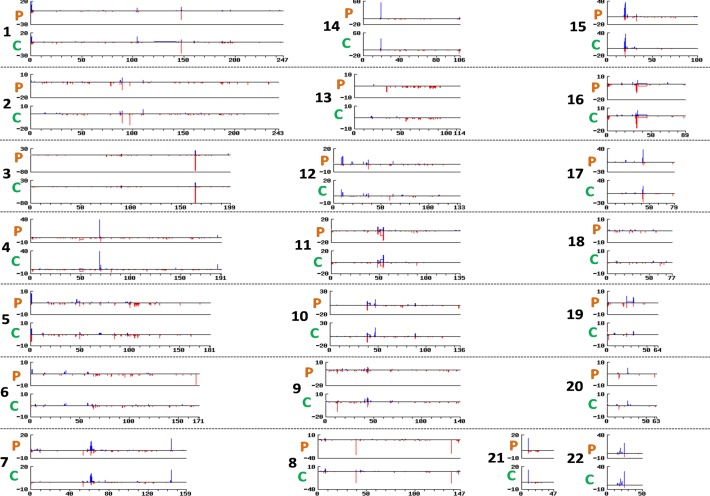
Figure 1.
Genomic landscape of large CNVs in POAG cases and controls. The Figure depicts whole genome distribution of CNVs > 100 kb in POAG cases and controls (Indian cohort). For each chromosome the upper panel represent cases (P) and the lower panel for controls (C). The horizontal axes is for chromosome size (in Mbp), the vertical axes represents segment count. Deletion is depicted in red and duplication is in blue.
Table 1.
Summary of CNVs in the Study Populations
|
Indian Data |
Caucasian Data |
|||
|
Cases,
n
= 347 |
Controls,
n
= 345 |
Cases,
n
= 624 |
Controls,
n
= 404 |
|
| Total CNV calls, >100 kb | 2116 | 2064 | 15154 | 8542 |
| Average CNV per genome, >100 kb | 6.1 | 6.0 | 24.3 | 21.1 |
| Average CNV size in kb (median) | 237.2 (146.1) | 237.7 (144.9) | 343.3 (151.7) | 323 (149.2) |
CNVs >1 Mb Are Significantly Enriched for Genes in the POAG Patients
We segregated the CNVs into four separate size bins, and compared CNVs frequencies in each size bin between the cases and the controls. Inconsistent with literature of large CNVs, we did not find significantly increased burden of large CNVs in POAG samples in either of the cohorts (Table 2).
Table 2.
Size Distribution of CNVs in the Study Populations
|
CNVs Size Bins |
CNVs Indian Data |
CNVs Caucasian Data |
||||||
|
No. of CNVs, Case |
CNVs Freq, Case |
No. of CNVs, Control |
CNVs Freq, Control |
No. of CNVs, Case |
CNVs Freq, Case |
No. of CNVs, Control |
CNVs Freq, Control |
|
| 100–250 kb | 1761 | 0.83 | 1734 | 0.84 | 12070 | 0.8 | 6935 | 0.81 |
| 250–500 kb | 258 | 0.12 | 247 | 0.12 | 2106 | 0.14 | 1123 | 0.13 |
| 500–1000 kb | 58 | 0.03 | 52 | 0.03 | 443 | 0.03 | 237 | 0.03 |
| >1000 kb | 39 | 0.02 | 31 | 0.01 | 535 | 0.03 | 247 | 0.03 |
We analyzed CNVs potentially influencing genes (by overlapping coordinates) in each size bin. Interestingly, for the largest size range (i.e., CNVs > 1 Mb) a total of 39 CNVs in the patients overlapped with 125 genes ,whereas 31 CNVs in the controls overlapped only with 5 genes not found in the patients (P value 1.17 × 10−22, Table 3). In the Caucasian dataset, this also held true (P value 1.33 × 10−11, Table 3). This observation was not due to a few CNVs in the cases as of 39 CNVs 15 overlapped with genes, whereas for the controls only two CNVs were in the genic region. In the Indian patient group, 1.8% of the large CNV calls were >1 Mb and for the controls it was 1.5%, while that for the Caucasian data is 3.5% and 2.9%, respectively. We did not observe this gene enrichment in cases in the three other CNV size bins of sizes less than 1 Mb, for either of the cohorts.
Table 3.
Significant Enrichment of Genic CNVs > 1 Mb
|
CNV Size Range |
Status |
Number of Genes in Each Size Bin |
Total Genes |
P Value (χ2 test) |
| Indian data | ||||
| 100 kb–250 kb | case | 1266 | 1971 | 0.28 |
| control | 1207 | 1776 | ||
| 250 kb–500 kb | case | 498 | 1971 | 0.16 |
| control | 404 | 1776 | ||
| 500 kb–1 Mb | case | 82 | 1971 | 1.6 × 10−8 |
| control | 160 | 1776 | ||
| >1 Mb | case | 125 | 1971 | 1.17 × 10−22 |
| control | 5 | 1776 | ||
| Caucasian data | ||||
| 100 kb–250 kb | case | 6974 | 11399 | 0.18 |
| control | 4710 | 7454 | ||
| 250 kb–500 kb | case | 2902 | 11399 | 0.45 |
| control | 1851 | 7454 | ||
| 500 kb–1 Mb | case | 928 | 11399 | 0.09 |
| control | 664 | 7454 | ||
| >1 Mb | case | 595 | 11399 | 1.33 × 10−11 |
| control | 229 | 7454 | ||
Deletion CNVs > 1 Mb Enriched for Genes Are Overrepresented in the POAG Patients
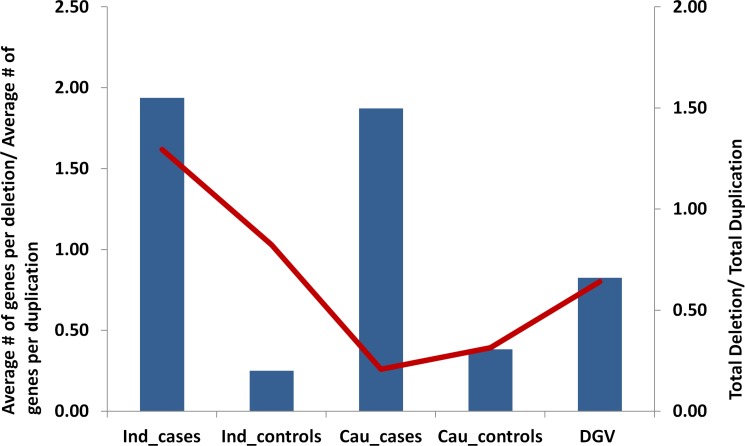
We found deletions to be overrepresented in the CNVs > 1 Mb that overlapped with the genic regions. In the genomic regions harboring functional transcripts (refseq dataset), deletions were found to be approximately 1.9-fold enriched compared to duplications in the POAG patients of Indian and Caucasian origins. Such a bias for large deletions was not observed in the control samples of either population (Fig. 2). For a larger pool of data from the general population, we analyzed CNVs > 1 Mb from the database of genomic variants. This dataset contained a total of 245,487 nonredundant CNV calls from 11,261 samples, of which 251 were unique CNVs of >1 Mb. This dataset also did not show any bias of gene-rich deletions. Interestingly, in the overall dataset of CNV > 1 Mb, the number of deletions was less than duplications (Del/Dup, 0.82) and a poor correlation (r2 = 0.15) indicated that this enrichment was not due to a bias of more deletions in general (Fig. 2, Supplementary Table S3). The bias for gene-rich deletions in cases was not observed for smaller size CNVs (>100 Kb and <1 Mb) (Supplementary Table S4 for Indian data and Supplementary Table S5 for Caucasian data).
Figure 2.
Large deletions of > 1 Mb, enriched in genic regions, are overrepresented in glaucomatous neurodegeneration. Individual CNV calls > 1 Mb are analyzed for their possible impact in the disease. The vertical bars show the ratio of deletions/duplications overlapping with known protein-coding genes. The left vertical axis reflect the values. Note the higher number of genes under deletions specifically in cases. The red line is the ratio for all deletions/all duplications > 1 Mb. The values are reflected at the right vertical axis. Note that, except Indian cases, for all other groups the total number of duplications is higher than that of deletions (ratio < 1). Indian and the Caucasian POAG cases show a much larger fraction (∼2-fold) of deletions possibly disrupting protein-coding genes compared to corresponding controls. The larger dataset of DGV also matches with the trend observed in POAG controls. The source data for this plot is given in Supplementary Table S3.
Genes Influenced by >1 Mb CNVs in POAG Cases Influence Relevant Pathways
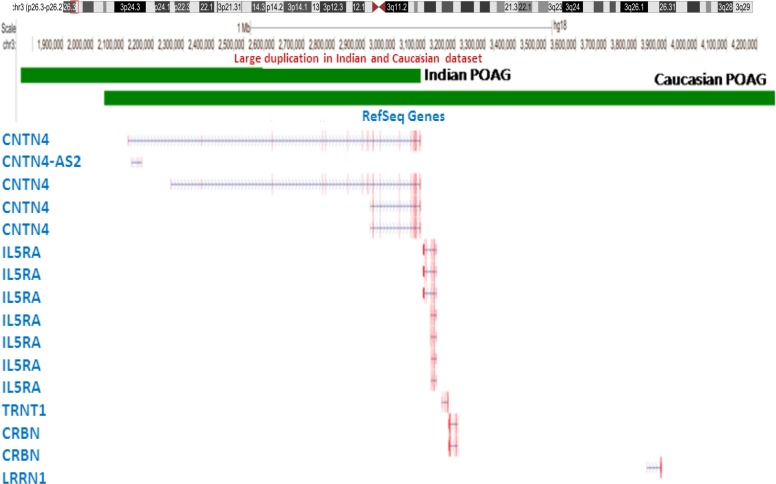
Of the 125 genes found in Indian patients that overlap with CNVs > 1 Mb, 63% (78 genes) overlapped with deletions. These genes are overrepresented in pathways of apoptosis, transcription regulation, and cell death, while cell adhesion pathway genes are overrepresented in the duplication CNVs. The Indian and Caucasian POAG patients shared one duplication >1 Mb unique to the patients on chromosome 3, which potentially can influence three different genes; namely, CNTN4, CNTN4-AS2, and IL5RA. In Indian POAG cases, this was 1.3 Mb duplication, while in the Caucasian dataset it was 2.2 Mb duplication (Fig. 3). The overlapping region of the duplications found in both cohorts completely overlaps only with CNTN4, making it a novel candidate gene for POAG (Fig. 3). This large duplication was not found in either of the control cohorts or in the world populations (DGV). It would be interesting to study these genes in future to evaluate their possible involvement in the disease biology.
Figure 3.
A large duplication involving CNTN4 unique to POAG patients. The scheme displays the genomic perspective of the large duplication on 3p26.3 found in both the cohorts and was absent in all controls, including DGV. The green rectangles represent the regions under individual duplication and below the refseq genes are shown. Note that CNTN4 is the only gene that completely encompasses the overlapping region of the two CNVs. In Indian POAG, this was a 1.3 Mb duplication (chr3:1,761,751- 3,086,458) while in the Caucasian dataset it was a 2.2 Mb duplication (chr3: 2,034,893- 4,258,763). The coordinates are from hg18.
CNVs > 1 Mb Found in POAG Patients Are Common With Other Neurodevelopmental Disorders
As mentioned above, we identified 39 CNVs > 1 Mb in the Indian cohort. When checked for previous reports of association of these 39 large CNVs in other disorders (source, CNVD available in the public domain at http://202.97.205.78/CNVD/), we found 54 diseases, including 14 CNVs for nonsyndromic sensorineural hearing loss (35%), and 14 for autism, bipolar disorder, and schizophrenia together. Thus, 28 CNVs of 39 (72%) were implicated previously in other neurodegenerative diseases. The complete list of other disorders involved with these 39 CNVs is provided as supplementary data (Supplementary Table S6).
Discussion
The importance of large CNV events and their cumulative burden has been highlighted for various diseases by multiple studies and especially in a variety of neurodevelopmental phenotypes. For example, diseases, like autism and intellectual disability, have been shown to have larger CNVs compared to controls.2 We explored the possible role of large CNVs in the POAG subtype of glaucoma, a neurodegenerative disease of the central nervous system (retina). In a cohort collected from the eastern part of India (state of West Bengal), we identified a significant enrichment of gene-rich deletions of CNVs > 1 Mb. In another publically available dataset of POAG cases and controls of Caucasian origin, we found similar bias of large CNVs uniquely in the POAG cases.
It was intriguing to observe the consistent distribution of deletions and duplications in cases and controls within a particular ethnic background vis-à-vis the contrast in average number of CNVs in samples between two ethnic populations analyzed during this study. In the Indian cohort we observed an average of 6.0 CNVs > 100 kb per individual, compared to an average of 22.0 for the Caucasian cohort (24 for cases and 21 for controls). However, as shown in the Results section, this difference in the basal CNV number did not affect the relative frequencies of the larger CNVs in POAG patients with respect to the controls of the same ethnicity. It was reported earlier that the interindividual and interpopulation differences in the copy number of genes (e.g., CCL3L1) can significantly alter the individual and population threshold for susceptibility to diseases (HIV-1 infection).17 In addition, our previous study on 26 diverse populations of India showed widespread differences in average number of CNVs per sample,4 which was related to ethnic and geographic backgrounds. Globally, it has been shown that different populations harbor different average number of CNVs per sample, where admixed populations had higher number of CNVs,18 Interestingly, the GLAUGEN study used for the Caucasian ethnicity had samples of European as well as Hispanic origins, which might contribute to the higher CNV per sample. Some of the samples from the GALUGEN data might have been generated from saliva DNA instead of blood DNA, and the tissue mediated variability also might contribute to different CNV number, but in absence of that information in the dataset accessible to us, we cannot assess this factor. We also have performed a correlation of standard deviation of logR ratio with the number of CNV calls in the Indian and the Caucasian data to check whether samples with higher logR ratio variation show higher CNVs. As shown in Supplementary Figure 4, the correlations were poor (0.009 and 0.002 for the Indian and Caucasian data, respectively), indicating that the differences in CNV numbers in different populations is a biological feature rather than a technical noise.
We investigated each individual CNV calls in the data with a size filter of >100 kb so that rare events with greater impact are not missed. We observed very similar frequencies of CNVs in cases and controls, as well as genes influenced by them for size ranges between 100 and 500 kb. However, when we analyzed even larger CNVs, particularly of size > 1 Mb, we found a distinct bias of CNVs influencing genes in the cases compared to controls and this observation was consistent in both populations. We identified 39 and 31 CNVs > 1 Mb in POAG patients and controls, in the Indian cohort influencing 125 and 5 genes, respectively, This indicated that the CNVs in patients, especially large deletions were enriched for gene-rich regions compared to the controls.
The specificity of gene-rich large deletions in POAG patients was checked further in CNVs > 1 Mb from > 11,000 samples from database of genomic variants, which corroborated our data for the control cohorts. We also ruled out the possibility that this enrichment is due to a larger representation of deletion CNV in the dataset (Fig. 2). The enrichment of gene-rich deletion CNVs was not observed in the CNVs smaller than 1 Mb. We observed 39 CNVs > 1 Mb in 31 patient samples, of which eight harbored more than one event. The majority of singleton CNVs > 1 Mb implied the fact that genetic etiology of individual patients might be unique and different from each other. This was consistent with the recent observations that >80% of the deleterious variants are singletons in the general population.19 In POAG in particular and complex phenotypes in general, rare personalized genetic events might contribute more to the phenotypic perturbation assisted by common variants.20
In our dataset of CNVs > 100 Kb, we identified regions overlapping with nine different GLC1 loci (GLC1B, -C, -D, -H, -I, -J, -K, -M, and -N), but all except GLC1C also were found in the controls. In one patient (GL439), we identified a 212.5 Kb heterozygous deletion in chromosome 3 overlapping with the GLC1C locus. This locus was identified to be linked to POAG in two independent studies.21,22 However, CNVs in this region were observed in the DGV data. For all the nine GLC1 loci, we found CNVs in cases and controls of the Caucasian data. It will be interesting to investigate in larger cohorts whether CNVs in these regions show significant different distribution between the cases and controls to provide insights into the etiology of common CNVs in glaucomatous neurodegeneration.
Two CNVs > 1 Mb were common between the Indian and Caucasian POAG patients, but absent in the matched control sets as well as in the global samples. One of these was duplication in both populations influencing three genes; namely, CNTN4, CNTN4-AS2, and IL5RA. The CNTN4 CNVs have been reported in 3p syndromes characterized by imperfect motor movements and behavior abnormalities.23 At the molecular level, CNTN4 CNVs affects synapse formation and cell adhesion.24 The CNVs in this gene have been implicated in neurodegenerative diseases like ataxia, autism, and so forth.25,26 The CNTN4 encodes a member of the contactin family of immunoglobulins. Contactins are axon-associated cell adhesion molecules that function in neuronal network formation. The encoded protein is a glycosylphosphatidylinositol-anchored neuronal membrane protein that may have a role in the formation of axon connections in the developing nervous system.27 The CNTN4 expression is seen in the retina.28 Duplication in this gene is associated with neurodegenerative diseases, like autism and ataxia.25,26 It has been proposed that the duplicated allele may generate a truncated protein that potentially could interfere with normal CNTN4 function and the same study has found duplication in CNTN4 associated to optic nerve aplasia disease, which causes visual impairment in children, in which the abundance of RGCs is significantly reduced.27 Glaucoma is a neurodegenerative disease, where optic nerve atrophies are crucial clinical factors caused by retinal ganglion cell (RGC) death. Our finding of CNTN4 duplication in both cohorts is interesting for further studies to bring more insights into disease biology.
Complex diseases are known to have overlapping genetic etiologies.29 We checked for the 39 CNVs > 1 Mb found in our POAG patients for their involvement in other complex neurodegenerative disorders. Four diseases; namely, autism, schizophrenia, bipolar disorder, and nonsyndromic sensorineural hearing loss, accounted for 71.8% (28/39) of the CNVs identified in our samples. This indicates overlapping genetic basis for these diseases. Interestingly, as per our data, large CNVs in genic regions are present more in the cases, but in the same genomic region much smaller CNVs are observed in the control samples (Supplementary Table S1). This indicates a negative selection for larger CNVs in the healthy genome, while retaining the polymorphic diversity of variant regions, which might enable the species to adapt and evolve to changing environment.
In summary, we have shown significant bias for deletions >1 Mb in gene-rich regions for POAG patients across population backgrounds and identified CNTN4 as a potential novel candidate for POAG. We also have shown that different populations can have different basal CNV occurrence implying that the relative number of CNVs within a population might be a better disease indicator/marker than the absolute comparison of CNV numbers across populations. Our study provides new insights into the genomic basis of POAG and paves future directions for better understanding of glaucomatous neurodegeneration in the light of selective enrichment of large gene-rich deletions.
Acknowledgments
The authors thank Subhashree Nayak and Pramod Gautam for their help in various technical issues throughout the study, Prajakta Bajad for critical review of the manuscript, the GENEVA Coordinating Center (U01 HG004446) for assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, and the National Center for Biotechnology Information for assistance with data cleaning.
Supported by the Council of Scientific and Industrial Research (CSIR), India (Grants MLP-0016 and BSC-0123), and a senior research fellowship from CSIR-India (LK). The GLAUGEN study was supported by the National Institutes of Health (NIH) Genes, Environment and Health Initiative ([GEI], U01HG004728). The GLAUGEN study is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Genotyping, which was performed at the Broad Institute of the Massachusetts Institute of technology (MIT; Cambridge, MA, USA) and Harvard University (Cambridge, MA, USA), was supported by the NIH GEI (U01 HG04424).
Disclosure: L. Kaurani, None; M. Vishal, None; D. Kumar, None; A. Sharma, None; B. Mehani, None; C. Sharma, None; S. Chakraborty, None; P. Jha, None; J. Ray, None; A. Sen, None; D. Dash, None; K. Ray, None; A. Mukhopadhyay, None
References
- 1. Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009; 84: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Girirajan S, Brkanac Z, Coe BP, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011; 7: e1002334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuningas M, Estrada K, Hsu YH, et al. Large common deletions associate with mortality at old age. Hum Mol Genet. 2011; 20: 4290–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gautam P, Jha P, Kumar D, et al. Spectrum of large copy number variations in 26 diverse Indian populations: potential involvement in phenotypic diversity. Hum Genet. 2012; 131: 131–143 [DOI] [PubMed] [Google Scholar]
- 5. Ray K, Mookherjee S. Molecular complexity of primary open angle glaucoma: current concepts. J Genet. 2009; 88: 451–467 [DOI] [PubMed] [Google Scholar]
- 6. Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011; 43: 574–578 [DOI] [PubMed] [Google Scholar]
- 7. Scheetz TE, Fingert JH, Wang K, et al. A genome-wide association study for primary open angle glaucoma and macular degeneration reveals novel Loci. PLoS One. 2013; 8: e58657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis LK, Meyer KJ, Schindler EI, et al. Copy number variations and primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2011; 52: 7122–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, Gibson J, Wheeler J, et al. GALC deletions increase the risk of primary open-angle glaucoma: the role of Mendelian variants in complex disease. PLoS One. 2011; 6: e27134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nag A, Venturini C, Hysi PG, et al. Copy number variation at chromosome 5q21.2 is associated with intra-ocular pressure. Invest Ophthalmol Vis Sci. 2013; 54: 3607–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fingert JH, Robin AL, Stone JL, et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum Mol Genet. 2011; 20: 2482–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lehmann OJ, Ebenezer ND, Ekong R, et al. Ocular developmental abnormalities and glaucoma associated with interstitial 6p25 duplications and deletions. Invest Ophthalmol Vis Sci. 2002; 43: 1843–1849 [PubMed] [Google Scholar]
- 13. Chanda B, Asai-Coakwell M, Ye M, et al. A novel mechanistic spectrum underlies glaucoma-associated chromosome 6p25 copy number variation. Hum Mol Genet. 2008; 17: 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee D, Bhattacharjee A, Ponda A, Sen A, Ray K. Comprehensive analysis of myocilin variants in east Indian POAG patients. Mol Vis. 2012; 18: 1548–1557 [PMC free article] [PubMed] [Google Scholar]
- 15. Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007; 17: 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsuang DW, Millard SP, Ely B, et al. The effect of algorithms on copy number variant detection. PLoS One. 2010; 5: e14456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005; 307: 1434–1440 [DOI] [PubMed] [Google Scholar]
- 18. Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008; 451: 998–1003 [DOI] [PubMed] [Google Scholar]
- 19. Fu W, O'Connor TD, Jun G, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013; 493: 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012; 148: 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wirtz MK, Samples JR, Kramer PL, et al. Mapping a gene for adult-onset primary open-angle glaucoma to chromosome 3q. Am J Hum Genet. 1997; 60: 296–304 [PMC free article] [PubMed] [Google Scholar]
- 22. Kitsos G, Eiberg H, Economou-Petersen E, et al. Genetic linkage of autosomal dominant primary open angle glaucoma to chromosome 3q in a Greek pedigree. Eur J Hum Genet. 2001; 9: 452–457 [DOI] [PubMed] [Google Scholar]
- 23. Fernandez T, Morgan T, Davis N, et al. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2004; 74: 1286–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansford LM, Smith SA, Haber M, Norris MD, Cheung B, Marshall GM. Cloning and characterization of the human neural cell adhesion molecule, CNTN4 (alias BIG-2). Cytogenet Genome Res. 2003; 101: 17–23 [DOI] [PubMed] [Google Scholar]
- 25. Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009; 459: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka E, Maruyama H, Morino H, Nakajima E, Kawakami H. The CNTN4 c.4256C>T mutation is rare in Japanese with inherited spinocerebellar ataxia. J Neurol Sci. 2008; 266: 180–181 [DOI] [PubMed] [Google Scholar]
- 27. Prasov L, Masud T, Khaliq S, et al. ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum Mol Genet. 2012; 21: 3681–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci. 2012; 32: 14402–14414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci U S A. 2007; 104: 11694–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]