Abstract
Sporadic iron overload in rats has been reported, but whether it is due to genetic or environmental causes is unknown. In the current study, phenotypic analysis of Hsd:HHCL Wistar rats revealed a low incidence of histologically detected liver iron overload. Here we characterized the pathophysiology of the iron overload and showed that the phenotype is heritable and due to a mutation in a single gene. We identified a single male rat among the 132 screened animals that exhibited predominantly periportal, hepatocellular iron accumulation. This rat expressed low RNA levels of the iron regulatory hormone hepcidin and low protein levels of transferrin receptor 2 (Tfr2), a membrane protein essential for hepcidin expression in humans and mice and mutated in forms of hereditary hemochromatosis. Sequencing of Tfr2 in the iron-overloaded rat revealed a novel Ala679Gly polymorphism in a highly conserved residue. Quantitative trait locus mapping indicated that this polymorphism correlated strongly with serum iron and transferrin saturations in male rats. Expression of the Gly679 variant in tissue culture cell lines revealed decreased steady-state levels of Tfr2. Characterization of iron metabolism in the progeny of polymorphic rats suggested that homozygosity for the Ala679Gly allele leads to a hemochromatosis phenotype. However, we currently cannot exclude the possibility that a polymorphism or mutation in the noncoding region of Tfr2 contributes to the iron-overload phenotype. Hsd:HHCL rats are the first genetic rat model of hereditary hemochromatosis and may prove useful for understanding the molecular mechanisms underlying the regulation of iron metabolism.
Abbreviations: Bmp6, bone morphogenetic protein 6; Hfe, hemochromatosis protein; HHC, hereditary hemochromocytosis; Hjv, hemojuvelin; qPCR, qualitative PCR; QTL, quantitative trait locus; SNP, single-nucleotide polymorphism; SSLP, simple sequence-length polymorphism; Tfr2, transferrin receptor 2
Rodents often are used as model systems for studies on mammalian iron metabolism. Although a transporter crucial for intracellular iron trafficking has been identified in both rats and mice,19 mice have been the predominant rodent system used to investigate inherited forms of iron deficiency or overload. The effect of iron overload via dietary means has been evaluated in rats,1,6,8,10,25,38 but genetically defined rat models of inherited iron overload have been unavailable. A retrospective survey of toxicology studies revealed liver-specific iron accumulation in male and female Sprague–Dawley rats beginning at 2 mo of age,33 similar to the iron accumulation in a 7-wk-old female Han Wistar GALAS rat.36 The underlying cause of iron overload was not determined in either report,1,6 although similarities to hereditary hemochromatosis (HHC) were noted.
HHC is a group of inherited human diseases characterized by excessive intestinal iron absorption and progressive tissue iron loading in the setting of normal dietary iron intake.14 The excessive dietary iron absorption results from deficiency of hepcidin, a hormone secreted predominantly by the liver that inhibits uptake of iron from the intestine into the blood. In addition, hepcidin negatively regulates recycling of iron from senescent RBC by macrophages. Deficiency of hepcidin can result from mutations in hepcidin itself, in bone morphogenetic protein 6 (Bmp6) and transferrin (Tf), both of which are stimulators of hepcidin expression, or in any of several other factors required for hepcidin expression, including β2-microglobulin, hemochromatosis protein (Hfe), hemojuvelin (Hjv), transferrin receptor 2 (Tfr2), and neogenin.31 The mechanisms by which these factors influence hepcidin expression remain incompletely understood, although they are believed to mediate Tf- or Bmp6-dependent stimulation of hepcidin expression.
In the current study, routine phenotypic analysis of a closed outbred colony of Wistar rats designated Hsd:HHCL revealed a low incidence of histologic liver iron accumulation. We analyzed serum iron levels, Tf saturations, and liver and spleen nonheme iron content and measured RNA and protein levels of genes associated with iron overload in a cohort of animals from this population. We then performed quantitative trait locus (QTL) mapping to identify genetic loci linked to the measured phenotypes. QTL analysis identified a single locus, which includes rat Tfr2, with a major effect on iron parameters. We selectively bred rats to generate and characterize progeny carrying the candidate causative allele in Tfr2 in hetero- or homozygosity. This effort demonstrated that the iron overload phenotype segregates largely as a codominant Mendelian trait in this population. Overall, our data indicate that a mutation in Tfr2 alters the hepcidin-iron regulatory axis in this subpopulation of Wistar rats and causes the HHC-like phenotype.
Materials and Methods
Animal care and characterization.
All animal procedures were approved by the Harlan Laboratories IACUC. Animals used for this study were Rattus norvegicus and a distinct subpopulation of RccHan:WIST rats (Harlan Laboratories, Indianapolis, IN), designated Hsd:HHCL, in which routine histologic analysis revealed less than 1% incidence of hepatocytic iron accumulation. The Hsd:HHCL stock was maintained by using a 12-section Poiley breeding system to maximize colony heterogeneity.40 Rats were housed according to standards in the Guide for the Care and Use of Laboratory Animals.23 Rats were individually or socially housed to meet standards regarding cage size in open polycarbonate ‘shoebox’ cages on hardwood chip bedding (Sani-Chips 7090, Harlan Teklad, Madison, WI). Rats were provided with municipal tap water filtered to 0.2 μm and chlorinated to 7 to 9 ppm and fed an autoclaved pelleted maintenance diet (2018S Teklad Global 18% Protein Rodent Diet, Sterilizable, Harlan Teklad) ad libitum. A 12:12-h light:dark light cycle was provided. Air was 100% fresh supply at more than 12 air changes hourly. Temperature was 72 ± 4 °F (23.3 ± 2.2 °C) and humidity was 50% ± 30%. Rats originated from a colony free of rat parvoviruses (Kilham rat virus, rat minute virus, rat parvovirus, and Toolan H1 virus), pneumonia virus of mice, rat Theiler virus, reovirus 3, rat coronavirus, Sendai virus, Hantaan virus, lymphocytic choriomeningitis virus, mouse adenovirus 1 and 2, Bordetella bronchiseptica, cilia-associated respiratory bacillus, Clostridium piliforme, Corynebacterium kutscheri, Encephalitozoon cuniculi, Helicobacter spp., Mycoplasma pulmonis, Pasteurella multocida, Pasteurella pneumotropica, Pneumocystis spp., Pseudomonas aeruginosa, Salmonella spp., Streptobacillus moniliformis, Streptococcus pneumoniae, dermatophytes, and endo- and ectoparasites.
Genetic characterization.
For the initial characterization of the Hsd:HHCL stock, 74 male and 58 female rats (age, 7 mo to 1 y) were selected randomly and euthanized by carbon dioxide inhalation. Blood was collected by cardiocentesis and serum prepared and refrigerated for analysis. Spleen, liver, and tail tissue were collected and samples placed in 10% buffered formalin and then in 70% ethanol or were flash-frozen on dry ice. After initial mapping and mutation screening, we identified a Tfr2 A679G polymorphism. To characterize the phenotypic effect of the A679G polymorphism, A679/A679 (homozygous AA at amino acid 679), A679/G679 (heterozygous AG), and G679/G679 (homozygous GG) rats were selected and interbred to generate 9 to 13 male and female rats of each genotype. Harvest and characterization of samples from progeny was performed in similar manner as for the characterization of the larger population.
Sample analysis.
Serum iron levels, total iron-binding capacities, and Tf saturations were determined by using Iron/UIBC Kit (Thermo Scientific, Pittsburgh, PA). Liver and spleen iron levels were determined with a spectrophotometric assay using acid-digested tissues and bathophenanthroline sulfonate as previously described.48 Hepcidin RNA levels depicted in Figure 1 were measured in triplicate from total RNA isolated by TRIzol (Invitrogen, Carlsbad, CA) extraction and isopropanol–ethanol precipitation. Total RNA underwent reverse transcription by using High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, Carlsbad, CA) and thermal cycling. Data collection and analysis was performed on an automated sequencer (model 7900HT, ABI, Foster City, CA) using the ABI SDS analysis software version 2.3 according to manufacturer's instructions. Prussian blue tissue staining of histologic sections was performed as previously described.48 RNA levels depicted in Figure 2 were measured as previously described by quantitative PCR (qPCR) of total RNA48 by using previously published primers for hepcidin,56 β-actin,51 Id1,52 Hfe,18 β2-microglobulin,28 Hfe2,35 neogenin,55 and Tfr2.56 Forward and reverse primers for Bmp6 were 5′ GGC TGA AGT CCG CTC CGC TC 3′ and 5′ CGG GCT CCA GTC CCT CTC CC 3′. Immunoblotting was performed by using rabbit antimouse Tfr2 (Alpha Diagnostics International, San Antonio, TX) and antihuman β-actin antibodies (Cell Signaling, Danvers, MA) as previously described.48
Figure 1.
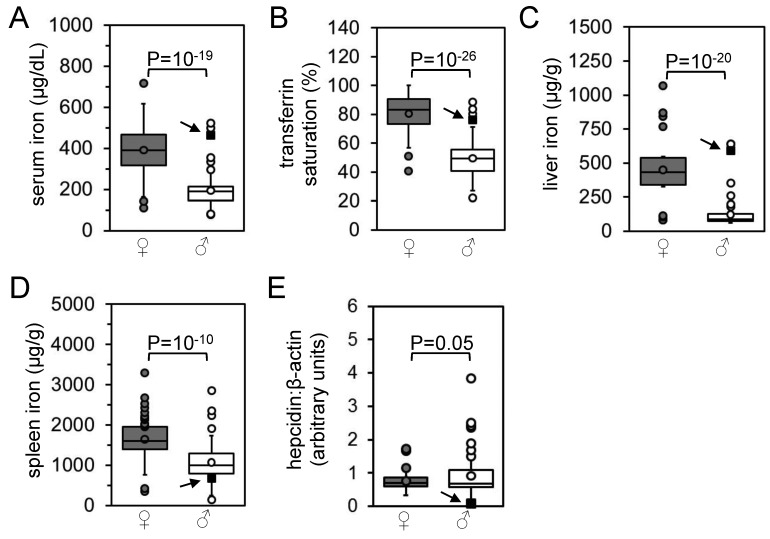
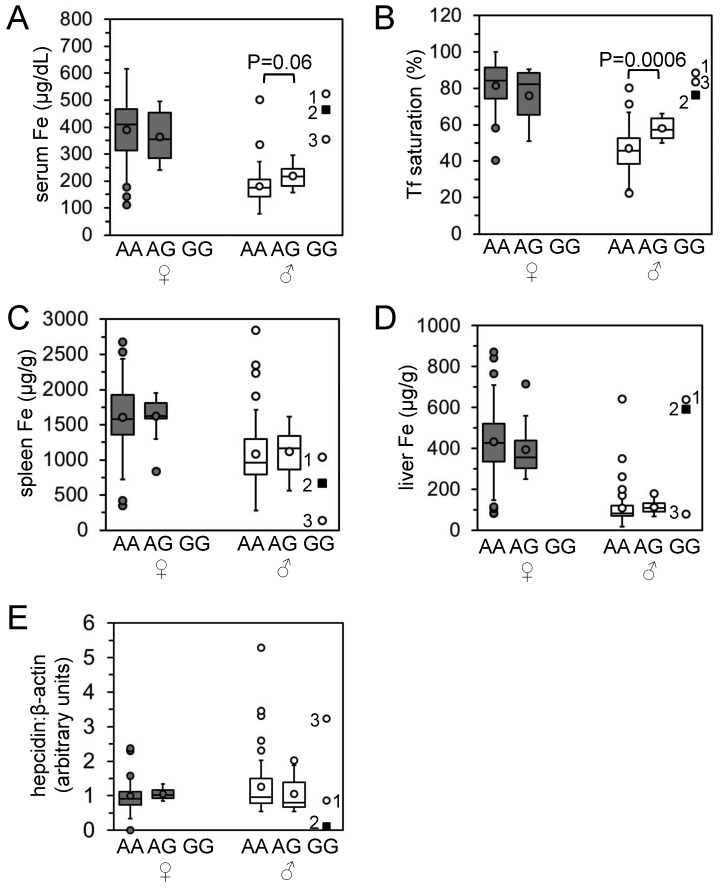
Characterization of iron homeostasis in original rat population. (A) Serum iron levels, (B) transferrin saturations, (C) liver iron levels, (D) spleen iron levels, and (E) RNA levels of liver hepcidin relative to β-actin levels as determined by qPCR were measured in 58 female (dark gray) and 74 male (light gray) rats and represented as boxplots. Circles indicate outlier values (see Materials and Methods for definition of outlier). P values between female and male data sets were calculated by using Student t tests. Arrows and filled black squares indicate the male rat with histologically evident liver iron overload.
Figure 2.
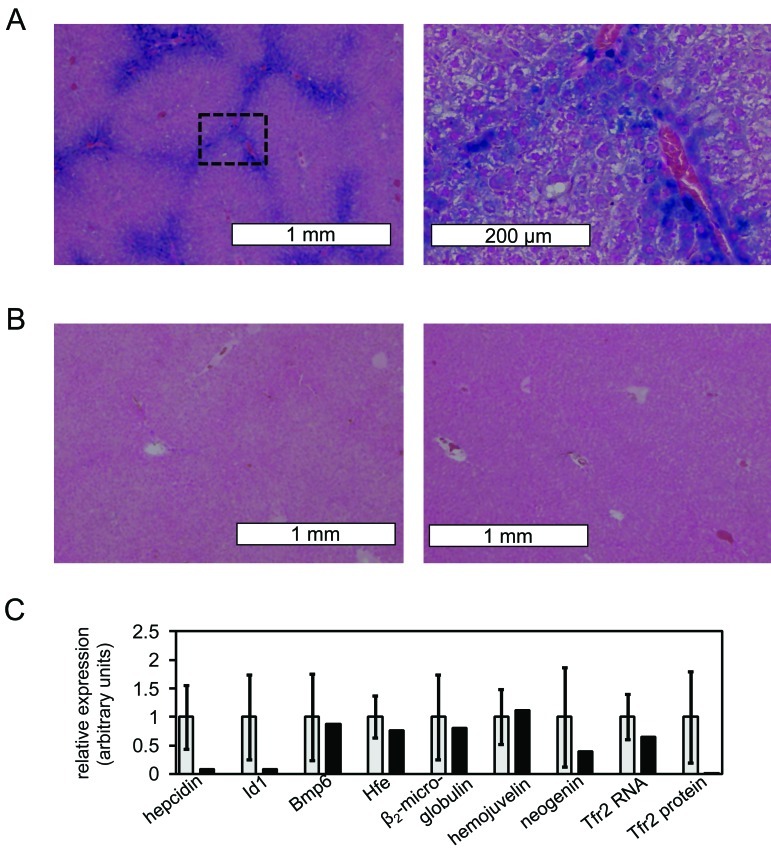
Phenotype of male rat with histologically evident liver iron overload and identification of possible alleles contributing to phenotype. (A) Prussian Blue staining of liver section of affected male rat is shown (magnification, 4× [left], 20× [right]). Blue stain indicates areas of high iron content; the box in the left image indicates the area magnified in the right image. (B) Prussian Blue staining of liver sections of 2 unaffected nonoutlier male rats is shown (magnification, 4×). (C) Liver expression levels (black bars) from affected rat are shown for hepcidin, Id1, BMP6, Hfe, β2-microglobulin, Hjv, and neogenin RNA and Tfr2 RNA and Tfr2 protein. Average expression levels for unaffected male rats (light gray bars) are shown for reference, with error bars indicating 1 SD. Unaffected male rats are defined as those rats with liver iron levels less than 1.5 SD above or below the mean for all male rats; 16 randomly selected unaffected male rats were analyzed. Liver RNA and protein levels were determined by using qPCR and Western blotting and normalized to β-actin levels in each sample. All unaffected male rats carried the Tfr2 A/A genotype.
For QTL mapping, a total of 96 single-nucleotide polymorphisms (SNP) and 9 simple sequence-length polymorphisms (SSLP; Table 1) were used for typing the Hsd:HHCL population. SNP genotypes were determined by using TaqMan (ABI) chemistry with probes and primers designed by using Primer Express version 3.0 (ABI). SSLP length was measured by 4% agarose gel electrophoresis of products from PCR by using genomic DNA and primers from the Rat Genome Database.50 Genomic DNA sequencing was performed using primers designed using Primer3 based on Ensembl rat sequences17,44 and genomic DNA isolated by using DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). Genomic sequence analysis was performed by using Geneious (Biomatters, Auckland, New Zealand). Primary sequence alignments were performed by using Clustal W2.9 Sequences similar to rat Hjv and Tfr2 were identified with NCBI BLAST (www.blast.ncbi.nlm.nih.gov) searches. Molecular modeling was performed by using HHPred and Swiss PDBViewer4,20 and primary sequences for rat Tfr2 (NCBI accession no., NP_001099386.1).
Table 1.
Location (chromosome [Chr], bp) of single nucleotide polymorphisms (SNPs) and simple sequence length polymorphisms (SSLP) used in mapping of qualitative trait loci
| Chr | Location (bp) | SNP | SSLP |
| 1 | 3, 272, 130 | R2783245–Chr1 | |
| 1 | 29, 764, 495 | SNP2783398–SNP | |
| 1 | 43, 994, 504 | SNP2783486–SNP | |
| 1 | 54, 055, 373 | R2783531–Chr1 | |
| 1 | 57, 358, 720 | SNP2783562–SNP | |
| 1 | 73, 244, 382 | SNP2783623–SNP | |
| 1 | 80, 640, 569 | R2783669–Chr1 | |
| 2 | 12, 363, 339 | SNP2785272–SNP | |
| 2 | 14, 167, 043 | SNP2785285–SNP | |
| 2 | 15, 053, 536 | SNP2785289–SNP | |
| 2 | 23, 840, 189 | SNP2785359–SNP | |
| 2 | 33, 524, 941 | SNP2785456–SNP | |
| 2 | 37, 559, 484 | SNP2785491–SNP | |
| 2 | 49, 741, 757 | SNP2785610–SNP | |
| 3 | 3, 409, 637 | SNP2787430–SNP | |
| 3 | 22, 923, 762 | SNP2787617–SNP | |
| 3 | 31, 460, 331 | R2787680–Chr3 | |
| 3 | 35, 627, 977 | SNP2787709–SNP | |
| 3 | 66, 099, 992 | R2787998–Chr3 | |
| 3 | 88, 240, 209 | R2788175–Chr3 | |
| 3 | 9, 472, 233 | SNP2787490–SNP | |
| 4 | 4, 235, 558 | SNP2788847–SNP | |
| 4 | 24, 097, 530 | R2789021–Chr4 | |
| 4 | 29, 420, 955 | SNP2789049–SNP | |
| 4 | 43, 109, 503 | SNP2789147–SNP | |
| 4 | 48, 793, 078 | SNP2789191–SNP | |
| 4 | 82, 005, 094 | SNP2789491–SNP | |
| 5 | 33, 427, 737 | SNP2790580–SNP | |
| 5 | 42, 887, 011 | SNP2790658–SNP | |
| 5 | 137, 257, 797 | R2791522–Chr5 | |
| 5 | 157, 573, 576 | R2791725–Chr5 | |
| 5 | 158, 040, 110 | R2791730–Chr5 | |
| 5 | 158, 085, 094 | R2791731–Chr5 | |
| 6 | 1, 954, 289 | SNP2791870–SNP | |
| 6 | 7, 893, 672 | SNP2791917–SNP | |
| 6 | 46, 447, 474 | SNP2792137–SNP | |
| 6 | 48, 598, 961 | R2792166–Chr6 | |
| 6 | 49, 185, 615 | SNP2792170–SNP | |
| 6 | 55, 888, 289 | R2792230–Chr6 | |
| 6 | 71, 260, 344 | R2792351–Chr6 | |
| 7 | 12, 616, 806 | R2793064–Chr7 | |
| 7 | 13, 726, 199 | R2793079–Chr7 | |
| 7 | 35, 201, 533 | R2793243–Chr7 | |
| 7 | 35, 344, 889 | R2793244–Chr7 | |
| 7 | 72, 199, 760 | SNP2793505–SNP | |
| 7 | 79, 011, 007 | SNP2793558–SNP | |
| 7 | 84, 558, 379 | SNP2793607–SNP | |
| 8 | 130, 343 | SNP2794162–SNP | |
| 8 | 7, 496, 895 | SNP2794197–SNP | |
| 8 | 38, 009, 549 | R2794391–Chr8 | |
| 8 | 53, 106, 706–53, 107, 102 | D8Rat44 | |
| 8 | 56, 172, 391–56, 172, 586 | D8Rat169 | |
| 8 | 58, 386, 194 | SNP2794569–SNP | |
| 8 | 64, 572, 921–64, 573, 148 | D8Rat185 | |
| 8 | 77, 767, 696 | SNP2794733–SNP | |
| 8 | 80, 169, 819 | R2794750–Chr8 | |
| 8 | 88, 129, 945 | SNP2794825–SNP | |
| 8 | 114, 974, 499–114, 974, 688 | D8Rat90 | |
| 8 | 115, 337, 809–115, 338, 082 | D8Rat11 | |
| 9 | 10, 646, 165 | R2795347–Chr9 | |
| 9 | 13, 988, 198 | SNP2795390–SNP | |
| 9 | 24, 414, 293 | SNP2795465–SNP | |
| 9 | 47, 158, 416 | R2795587–Chr9 | |
| 9 | 47, 745, 902 | SNP2795595–SNP | |
| 10 | 8, 131, 820 | SNP2796134–SNP | |
| 10 | 11, 765, 336 | SNP2796173–SNP | |
| 10 | 15, 264, 431 | SNP2796217–SNP | |
| 10 | 18, 460, 765 | SNP2796244–SNP | |
| 10 | 22, 750, 042 | SNP2796293–SNP | |
| 10 | 37, 341, 080 | SNP2796430–SNP | |
| 11 | 23, 197, 858 | SNP2797286–SNP | |
| 11 | 77, 950, 820 | SNP2797738–SNP | |
| 12 | 7, 960, 942 | SNP2797872–SNP | |
| 12 | 10, 191, 946–10, 192, 153 | D12Rat62 | |
| 12 | 15, 087, 719 | SNP2797936–SNP | |
| 12 | 17, 692, 182–17, 692, 411 | D12Mit7 | |
| 12 | 19, 651, 743 | C > G (Tfr2 Ala679Gly) | |
| 12 | 22, 300, 576–22, 300, 992 | D12Rat33 | |
| 12 | 23, 933, 587 | SNP2798012–SNP | |
| 12 | 30, 592, 930 | SNP2798077–SNP | |
| 12 | 34, 919, 871 | SNP2798129–SNP | |
| 12 | 36, 448, 697–36, 448, 841 | D12Rat52 | |
| 13 | 43, 607, 893 | SNP2798538–SNP | |
| 13 | 48, 026, 949 | SNP2798585–SNP | |
| 14 | 7, 428, 232 | SNP2799075–SNP | |
| 14 | 80, 754, 739 | SNP2799705–SNP | |
| 15 | 405, 033 | SNP2799950–SNP | |
| 15 | 17, 803, 205 | SNP2800070–SNP | |
| 15 | 60, 891, 581 | SNP2800310–SNP | |
| 16 | 13, 329, 049 | SNP2800796–SNP | |
| 16 | 15, 598, 493 | SNP2800818–SNP | |
| 16 | 61, 532, 266 | SNP2801097–SNP | |
| 16 | 69, 892, 722 | SNP2801163–SNP | |
| 16 | 79, 128, 540 | SNP2801238–SNP | |
| 17 | 1, 167, 413 | SNP2801323–SNP | |
| 17 | 3, 878, 364 | SNP2801351–SNP | |
| 18 | 13, 028, 236 | SNP2802288–SNP | |
| 18 | 25, 091, 085 | SNP2802416–SNP | |
| 18 | 49, 449, 530 | SNP2802561–SNP | |
| 19 | 21, 706, 839 | SNP2803042–SNP | |
| 19 | 38, 643, 927 | SNP2803159–SNP | |
| 19 | 45, 845, 821 | SNP2803217–SNP | |
| 20 | 13, 595, 941 | SNP2803493–SNP | |
| 20 | 43, 433, 739 | SNP2803725–SNP | |
| 20 | 49, 189, 263 | SNP2803779–SNP |
Cell culture.
pExpress-1 vector carrying wild-type rat Tfr2 cDNA was obtained from Source BioScience (Nottingham, UK). Intact coding sequence was confirmed by sequencing. The Gly679 substitution was introduced by using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). HEK293T, HepG2, and Hepa1-6 cells were transiently transfected by using Lipofectamine 2000 (Invitrogen) and rat Tfr2 cDNA expression vectors. Tfr2 and β-actin protein levels in cells lysed in RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) were measured as described above.
Statistical analysis.
Basic statistical analysis was performed by using Excel (Microsoft, Redmond, WA) and Student 2-tailed t tests with unequal variance. A P value of less than 0.05 was used to define statistical significance. In boxplots, the lower and upper borders of box indicate the first and third quartiles, respectively; lines and circles within boxes indicate medians and averages, respectively. The upper and lower bars indicate the high and low control limits, respectively, with control limits defined as median ± 1.5 times the difference between the first and third quartiles. Outliers are those values greater than the high control limit or less than the low control limit.49 A subpopulation of nonoutlier rats, consisting of 13 female and 16 male rats, was selected randomly from the initial population depicted in Figure 1. None of the parameters depicted in Figure 1 differed significantly between all rats, all nonoutlier rats, and the nonoutlier subpopulation in either male or female groups (data not shown). Furthermore, differences between female and male nonoutlier subpopulations were maintained in this smaller group (data not shown), indicating that the selected group of nonoutlier rats accurately represented the entire population of nonoutlier rats. To map QTL, we performed a genome-wide scan of all 74 male and 58 female rats by using Outbred Lines, a program developed for genetic mapping of crosses derived from outbred parents.37 We tested for correlation between genotype and phenotype distribution by using nonparametric tests, the Kolmogorov–Smirnov and Wilcoxon–Mann–Whitney ranking tests. We computed genome-wide significance through random reassignment (permutation) of phenotype to typed subjects. The procedure is appropriate for an outbred rat population like Hsd:HHCL, because no diversity assumptions are used to evaluate statistical significance—for example, we do not assume the presence of just 2 haploid genomes, which is assumed to be the case in crosses of isogenic lines. Familial relationships were not tracked and were not used for this analysis. The power of this approach is due in part to the limited diversity of the colony and the prolonged duration of haplotypes within it; the few SSLP tested were biallelic at most. Initial mapping was performed separately for male and female rats because of large differences in phenotype distributions.
Results
Routine histologic analysis of a subpopulation of Wistar rats designated Hsd:HHCL revealed a low incidence of liver hepatocellular iron accumulation (data not shown). To characterize iron homeostasis in this rat population, we analyzed samples from 58 female and 74 male Hsd:HHCL rats, all of which were randomly selected. Serum iron levels, total iron-binding capacities, Tf saturations, liver and spleen iron levels, and liver:spleen iron ratios were significantly (P < 0.05) greater in female than male rats (Figure 1 A through D; data not shown). There was a wide range of values in all parameters for both sexes. Despite the significant differences in iron parameters between female and male rats, hepcidin RNA levels (normalized to β-actin RNA levels) did not differ between female and male rats (Figure 1 E). Prussian blue staining of liver sections revealed a single rat with histologically characteristic liver iron accumulation in a predominantly periportal, hepatocellular distribution (Figure 2 A). In this male rat, serum iron level, total iron-binding capacity, Tf saturation, liver iron level, and liver:spleen iron ratio were increased and spleen iron level, liver hepcidin RNA level, and hepcidin:liver iron ratio were decreased relative to those of the rest of the population. All of these values differed from their respective means by at least 1 SD (arrows and square points, Figure 1; data not shown).
To determine whether the expression of iron metabolism genes was abnormal in the affected rat, we randomly selected a group of 13 female and 17 male nonoutlier rats as a comparison subpopulation. None of the parameters presented in Figure 1 differed significantly between all rats, all nonoutlier rats, and the nonoutlier subpopulation in either male or female groups (data not shown). Furthermore, differences between female and male nonoutlier subpopulations were maintained in this smaller group (data not shown). These findings indicated that the selected group of nonoutlier rats accurately represented the whole population of nonoutlier rats. None of the nonoutlier rats displayed liver iron accumulation as measured by Prussian blue staining (Figure 2 B, data not shown).
We compared expression levels of several genes involved in mammalian iron metabolism in the nonoutlier rat subpopulations and the affected male rat. We examined Bmp6, Hfe, β2-microglobulin, Hjv, neogenin, and Tfr2, because mice carrying mutations in these genes exhibit hepcidin deficiency and hepatocellular liver iron overload14,31 similar to that of the affected rat. We also analyzed Id1, because the expression of both Id1 and hepcidin is stimulated by iron overload through a BMP–SMAD signaling pathway.26 In addition, we measured Tfr2 protein levels, which correlate with liver iron content in rats.41 The liver hepcidin and Id1 RNA levels and liver Tfr2 protein levels in the male rat with histologically obvious liver iron accumulation were more than 1 SD below the respective mean values (Figure 2 B). This result suggested that a primary defect in Tfr2 resulted in decreased Tf or Bmp6 signaling (or both) and that hepcidin expression could account for the iron overload phenotype in the male rat.
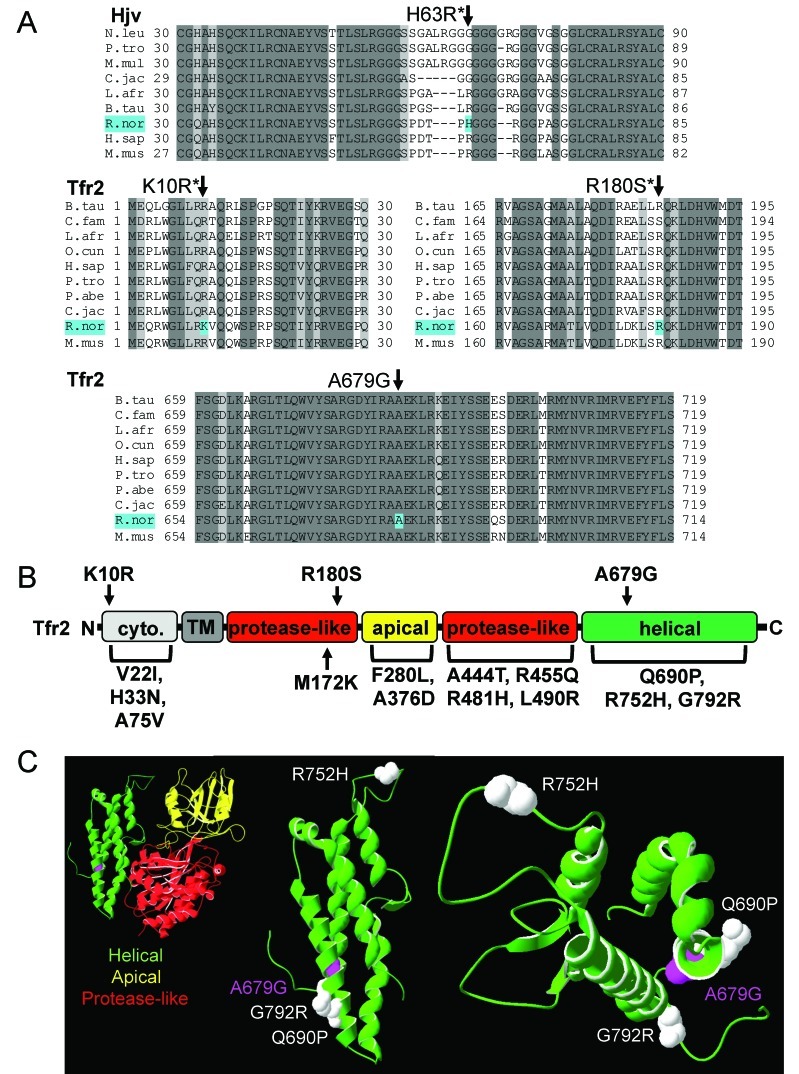
To determine whether the affected rat carried any sequence polymorphisms in genes involved in iron metabolism, we sequenced exons and exon–intron junctions in Bmp6, Hfe, β2-microglobulin, Hjv, neogenin, Tfr2, hepcidin, Tf, and ferroportin, which encodes a cellular iron exporter required for dietary iron absorption and macrophage iron efflux.10 All sequences corresponded to the reported wildtype rat sequences,17 except for those of Hjv and Tfr2. An H63R polymorphism, previously noted in Ensembl, affected a poorly conserved residue in Hjv (Figure 3 A).12 K10R, R180S, and A679G polymorphisms were identified in Tfr2. The first 2 polymorphisms had been described previously,13 but A679G, which alters a highly conserved residue in a highly conserved region of the protein (Figure 3 A), had not. The K10R, R180S, and A679G polymorphisms reside in the cytoplasmic, protease-like, and helical domains, respectively, of Tfr2 (Figure 3 B). Putative and causative mutations have been noted in each of these domains in human patients with HHC2,3,5,45,21,22,27,32,34,42,43,46 (Figure 3 B). Because all of the rats in the tested population were homozygous for the K10R and R180S alleles, we concluded that they were not responsible for the differences in iron metabolism. However, the A679G allele segregated as a minor allele in the population.
Figure 3.
Alignment of Hjv and Tfr2 sequences; schematic of Tfr2 with rat Tfr2 polymorphisms and hemochromatosis mutations. (A) Alignment of rat hemojuvelin (Hjv) and transferrin receptor 2 (Tfr2) and related primary sequences from other species, constructed by using ClustalW 2.0. Species and accession numbers include: Nomascus leucogenys (N.leu), XP_003268155.1; Pan troglodytes (P.tro), XP_001154526.1 and XP_003318698.1; Macaca mulatta (M.mul), XP_001092987.1; Callithrix jacchus (C.jac), XP_002759852.1 and XP_002744042.1; Loxodonta africans (L.afr), XP_003422013.1 and XP_003422538.1; Bos taurus (B.tau), NP_001193843.1 and NP_001171212.1; Rattus norvegicus (R.nor), NP_001012080.1 and NP_001099386.1; Homo sapiens (H.sap), Q8N7M5.1 and NP_003218.2; Mus musculus (M.mus) NP_081402.3 and NP_056614.3; Canis familaris (C.fam), XP_546952.2; Oryctolagus cuniculus (O.cun), XP_002711959.1; and Pongo abelii (P.abe) XP_002817816.1. Identical and similar residues are highlighted in dark and light gray, respectively. Residues affected by polymorphisms in affected rat (H63R, K10R, R180S, and A679G) are highlighted in cyan. Asterisks indicate polymorphisms previously documented in Ensembl. (B) Schematic of Tfr2 protein structure with cytoplasmic region (cyto.), transmembrane domain (TM), protease-like, apical, and helical domains adapted from human Tfr1 structure. Rat Tfr2 polymorphisms K10R, R180S, and A679G are indicated above the protein; TFR2 mutations in patients with hereditary hemochromatosis are shown below the sequence. (C) Models of extracellular regions of rat Tfr2 protein, constructed by using rat Tfr2 primary sequence (NCBI accession no., NP_001099386.1), HHPred, and SwissPDBViewer. Protease-like, apical, and helical domains are shown in red, yellow, and green, respectively. Residues affected by the A679G polymorphism are indicated as magenta space-filling side chains; residues affected by Tfr2 hemochromatosis mutations Q690P, R752H, and G792R are indicated as white space-filling side chains. The helical domain is shown as part of the larger extracellular Tfr2 fragment consisting of helical, apical, and protease-like domains (left image) or as a separate domain (middle and right images; 2 different viewing angles).
To gauge the potential effect of the A679G polymorphism on Tfr2 structure and function, we mapped the rat Tfr2 and human TFR2 sequences onto the crystal structure of the extracellular domain of human TFR1.29 The rat A679 residue is located in the helical domain, which resides at the interface of the 2 Tfr2 monomers in the mature homodimer. A679 also lies in the vicinity of 2 HHC disease-associated residues, G690P and G792R (Figure 3 C). These modeling data suggest that the A679G polymorphism could have structural effects relevant to Tfr2 function.
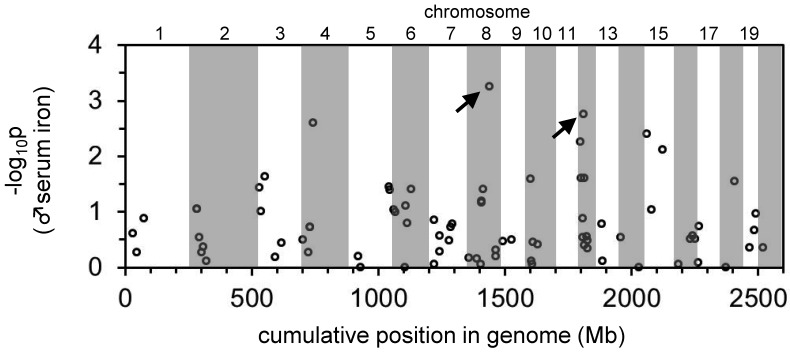
To identify chromosomal loci associated with traits related to iron metabolism in an unbiased manner, we genotyped the initial cohort of 132 rats for 96 SNP spanning the rat genome (Table 1). Although the use of these 96 SNP did not provide complete coverage of the rat genome, Wilcoxon and Kolmogorov–Smirnov tests did indicate significant associations between 2 SNP on chromosome 8 and 12 and serum iron and Tf saturations in male rats (data not shown). To better localize these QTL, we genotyped the entire cohort for an additional 9 SSLP on chromosomes 8 and 12 and repeated the analysis, including the A679G genotype (Table 1). The peak associated with the chromosome 8 QTL localized to 88.13 Mb (Figure 4). Two genes of interest lie near this peak, the Tf locus at 108.20 to 108.24 Mb and the neogenin locus at 62.68 to 62.79 Mb. Neogenin is required for hepcidin expression in mice but its specific function is unclear.11,31 Notably, the SSLP at 64 Mb had a lower logarithm-of-odds score than did the SSLP at 88 Mb, suggesting that the gene of interest lay distal to the 88 Mb SSLP. We were unable to identify any SSLP or SNPs mapping to 88 to 108 Mb that were polymorphic in the cohort, thus limiting our ability to further delineate this locus. The A679G polymorphism in Tfr2 defined the peak logarithm-of-odds score of the chromosome 12 QTL (Figure 4). Repeating the QTL mapping analysis by using the Tfr2 A679G polymorphism as a covariate rendered the chromosome 8 locus nonsignificant (data not shown). This result is not surprising because genotypes are highly correlated across chromosomes in this rat strain, thus suggesting a selection effect that we are currently investigating. To look for a second significant locus, we first performed a simple linear model in which the only explanatory variable was the Tfr2 A679G genotype. We used the result of the linear fit to compute the residual phenotype, that is, the original phenotype minus the value attributed to the Tfr2 A679G genotype. A genome scan using the residual phenotype did not produce any significant linkage. Therefore, no independent locus other than A679G contributes significantly to the phenotype of the affected male rate.
Figure 4.
Manhattan plot demonstrating the positions in the rat genome of quantitative trait loci for serum iron levels in male rats. The entire rat genome (approximately 2.6 Gb total) is represented along the x axis from chromosome 1 (left end of axis) to 20 (right end of axis). Regions of the genome corresponding to individual chromosomes are indicated as alternating white and gray shading. Numbers for most chromosomes are indicated above the graph. Negative logarithms of genome-wide P values are shown on the y axis. The plot represents the results of allele tests; results of genotype tests were similar (data not shown). Arrows indicate the locations of the 2 mostly highly significant quantitative trait loci for serum iron levels in male rats.
Because the mRNA and protein expression, DNA sequence, and QTL mapping strategies all suggested a primary defect in Tfr2, we focused on Tfr2 and the A679G polymorphism as the causative genetic factor related to aberrant iron metabolism in the Hsd:HHCL rat stock. To refine the phenotypic analysis, we reassessed the rats depicted in Figure 1 according to their A679G genotype. We found that 3 male and no female rats carried the G679/G679 (GG) genotype (Figure 5). As might be predicted from the QTL analysis, serum iron levels and Tf saturations of all homozygous male GG rats were greater than the average values for homozygous A679/A679 (AA) and heterozygous A679/G679 (AG) rats. AG male rats had significantly (P < 0.05) or near-significantly greater serum iron levels and Tf saturations than did AA male rats (Figure 5 A and B). Spleen iron levels in all male GG rats were lower than the average values for AA and AG male rats (Figure 5 C). Two (rats 1 and 2) of the 3 GG rats had increased liver iron levels and decreased hepcidin levels, whereas rat 3 had mildly decreased liver iron levels and increased hepcidin levels relative to the average values for AA and AG male rats (Figure 5 E). Correspondingly, Prussian blue staining of liver sections revealed a diffuse iron-rich pattern for rat 1; a predominantly periportal, hepatocellular pattern for rat 2; and a relative paucity of stainable iron for rat 3 (data not shown). Rat 2 is the affected male rat of interest and is indicated by arrows in Figure 1 and characterized in Figure 2.
Figure 5.
Characterization of iron homeostasis in rat population according to A679G genotype. Data from Figure 1 are shown here according to the Tfr2 A679G genotypes of 70 female rats (43 homozygous AA, 27 heterozygous AG, and 0 homozygous GG) and 72 male rats (60 AA, 9 AG, and 3 GG). (A) Serum iron levels, (B), transferrin saturations, (C) spleen iron levels, (D) liver iron levels, and (E) hepcidin RNA levels relative to β-actin RNA levels are shown.
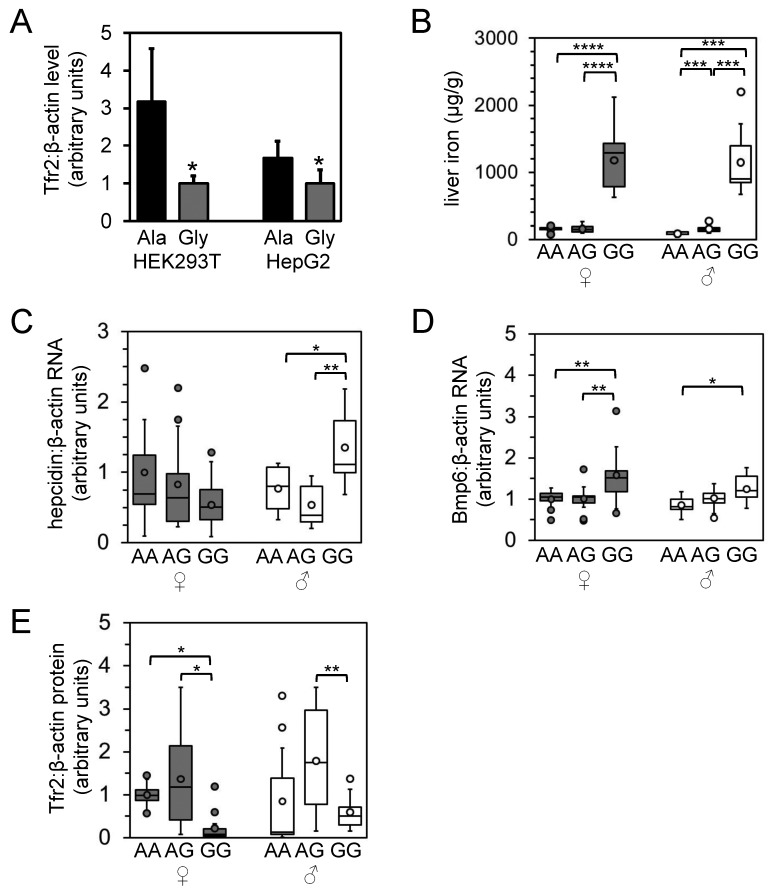
To assess the effect of the A679G polymorphism on Tfr2 structure and function, we transiently transfected expression constructs for the A679 or the G679 Tfr2 variant into HEK293T and HepG2 cell lines and measured steady-state Tfr2 protein levels by immunoblot. Expression levels of the G679 variant were significantly decreased relative to that of the A679 variant (Figure 6 A), suggesting that the A679G polymorphism reduces stability of the Tfr2 protein.
Figure 6.
In vitro characterization of Tfr2 A679G polymorphism and characterization of iron homeostasis in derived rat population according to A679G genotype. (A) HEK293T and HepG2 cells were transiently transfected with cDNA expression vectors encoding the Ala679 (Ala) or Gly679 (Gly) Tfr2 variant; protein levels of Tfr2 and β-actin then were measured by immunoblotting and densitometry. Asterisks indicate significantly different (Student t test, P < 0.05) values. Rats carrying the A679G allele were bred to generate 9 to 13 male and female rats of the homozygous AA, heterozygous AG, and homozygous GG phenotypes. F1 rats were analyzed for (B) liver iron levels, (C) hepcidin levels, (D) Bmp6 RNA levels relative to β-actin RNA levels as measured by qPCR, and (E) liver Tfr2 protein levels relative to β-actin protein levels. Data are presented as in Figure 5. Asterisks indicate statistical significance as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To characterize the effect of the A679G polymorphism on iron metabolism in a larger population, we selectively bred rats of specific A679G genotypes and characterized the phenotypes of the progeny (Figure 6 B through F). Although the original population was analyzed at 7 to 12 mo of age, this second population was analyzed at 1 mo of age. Notably, tissue iron levels were lower (P < 0.05) in this younger population (Figure 6 B; data not shown) than in the older population depicted in Figure 1. A similar trend in spleen iron levels has been noted in female rats and was shown to be hormone-dependent.7 Nevertheless, even at this young age, liver nonheme iron levels and ratios of liver:spleen nonheme iron were significantly (P < 0.05) higher in female and male GG than in AG or GG rats (Figure 6 B; data not shown). Although spleen iron levels in male rats did not vary greatly, levels decreased (P < 0.05) or trended toward decreasing in female rats carrying 1 or 2 G679 alleles (data not shown). Prussian blue staining of livers revealed histologically apparent iron overload in GG rats in a pattern similar to that shown in Figure 2 A (data not shown). Hepcidin and Id1 mRNA levels did not differ in female rats of different genotype, whereas levels in male GG rats were greater (P < 0.05) than those in AA and AG rats (Figure 6 C; data not shown). Bmp6 RNA levels were increased in female and male GG rats (Figure 6 E), but Tfr2 RNA levels did not differ between genotypes of either sex (data not shown). Tfr2 protein levels, relative to β-actin protein levels, were decreased (P < 0.05) in GG female rats relative to AA and AG female rats, whereas Tfr2 protein levels were decreased (P < 0.05) in GG male rats relative to AG male rats only (Figure 6 F).
Discussion
Although rodent models of aberrant metal metabolism are typically established in mice, the study of inbred rat strains has contributed greatly to the study of metal biology. Belgrade rats exhibit microcytic, hypochromic anemia and aberrant reticulocyte iron uptake and dietary iron absorption. Characterization of these rats revealed a mutation in divalent metal transporter 1,15 a cellular iron importer that is mutated in the inbred mk mouse strain16 and in specific cases of human anemia24 as well. Long–Evans cinnamon rats harbor a mutation in the rat homolog of ATP7B, a copper-transporting ATPase mutated in Wilson disease, a human disease of tissue copper toxicity caused by impaired hepatobiliary copper excretion.47,53,54 The current study is the first to identify a mutation in a hemochromatosis gene in rats. Although multiple mouse models of inherited iron overload already exist, a rat model of this disease has several possible advantages. First, mouse models of hemochromatosis do not always recapitulate characteristics of the human disease, such as hepatic fibrosis.30 We have yet to explore the long-term sequelae of iron overload in our rat population, but the Hsd:HHCL rat model may more closely mimic the human disease than do current mouse models. Second, because rats are used often for drug testing, a rat model of inherited iron overload would allow for such studies in an animal model of gross iron overload or predisposition to iron overload. Testing in a model of increased risk of iron overload may be particularly attractive given the high prevalence of hemochromatosis alleles in humans.39
We propose that the iron overload in our rat population results, at least in part, from a polymorphism in Tfr2. Our data suggest that A679G substitution contributes to the iron overload observed in our rat population, but we cannot exclude the possibility that a polymorphism in a noncoding region of Tfr2 plays a role in the phenotype. Nevertheless, we hypothesize that the A679G polymorphism leads to decreased hepatic Tfr2 protein levels, decreased Tfr2-mediated stimulation of hepcidin expression, increased dietary iron absorption and, ultimately, tissue iron overload. Notably, we observed considerable variability in this phenotype. In addition to the sex- and age-related differences, the variability within each sex may reflect that fact that colony is not inbred and that other alleles exist within the population that modify the hemochromatosis phenotype. For example, whereas liver Bmp6 RNA levels were increased in female homozygous rats, hepcidin levels were not increased, suggesting that Bmp6 levels may not be stimulated adequately by iron overload in these rats. However, adequate stimulation of Bmp6 in this population cannot be defined until AA rats are iron overloaded by chemical or dietary means for comparison. Similarly, a single homozygous rat (no. 3 in Figure 5) exhibited moderate liver iron and abundant hepcidin levels. The fact that homozygous rats with moderate liver iron levels did not occur in the group of rats in Figure 6 may reflect the fact that the rats in Figure 6 were generated from only a few breeders selected solely in light of their A679G genotype. Identification of loci contributing to the phenotypic variability in homozygous rats may permit us to establish a rat model with more severe hemochromatosis and identify novel genes required for iron metabolism.
Acknowledgments
We gratefully acknowledge the contributions of the following animal, technical, and caretaking staff at Harlan Laboratories: Ms Lisa Mitchell, Ms Sarah Garmon, Ms Joyce Cox, Ms Missie Graham, Ms Pauline Muncy, Ms Jessica Payne, Mr Jeff Schutte, Ms Rebecca Ransdell, and Lotena Hatter for their husbandry and animal care expertise; Ms April Pruet, Ms Amber Taylor, and Ms Jessica Payne for their technical assistance with tissue and blood collection. We further acknowledge Dr Klaus Weber and Ms Kellie Martel of Harlan Laboratories and Dr Craig Franklin of MURADIL for their scientific expertise and involvement with study coordination. This work was supported by National Institutes of Health grants K99DK084122 (TBB) and R01DK080011 (MDF).
References
- 1.Awai M, Narasaki M, Yamanoi Y, Seno S. 1979. Induction of diabetes in animals by parenteral administration of ferric nitrilotriacetate. A model of experimental hemochromatosis. Am J Pathol 95:663–673 [PMC free article] [PubMed] [Google Scholar]
- 2.Biasiotto G, Belloli S, Ruggeri G, Zanella I, Gerardi G, Corrado M, Gobbi E, Albertini A, Arosio P. 2003. Identification of new mutations of the HFE, hepcidin, and transferrin receptor 2 genes by denaturing HPLC analysis of individuals with biochemical indications of iron overload. Clin Chem 49:1981–1988 [DOI] [PubMed] [Google Scholar]
- 3.Biasiotto G, Camaschella C, Forni GL, Polotti A, Zecchina G, Arosio P. 2008. New TFR2 mutations in young Italian patients with hemochromatosis. Haematologica 93:309–310 [DOI] [PubMed] [Google Scholar]
- 4.Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res 34:W335–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bittencourt PL, Marin MLC, Couto CA, Cançado ELR, Carrilho FJ, Goldberg AC. 2009. Analysis of HFE and nonHFE gene mutations in Brazilian patients with hemochromatosis. Clinics (Sao Paulo) 64:837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonkovsky HL, Healey JF, Lincoln B, Bacon BR, Bishop DF, Elder GH. 1987. Hepatic heme synthesis in a new model of experimental hemochromatosis: studies in rats fed finely divided elemental iron. Hepatology 7:1195–1203 [DOI] [PubMed] [Google Scholar]
- 7.Borràs M. 1998. Hormone dependency of splenic iron stores in the rat: effect of oestrogens on the recuperation of reserves in ferrodeficient subjects. Lab Anim 32:290–297 [DOI] [PubMed] [Google Scholar]
- 8.Brown KE, Knudsen CA. 1998. Oxidized heme proteins in an animal model of hemochromatosis. Free Radic Biol Med 1998 24:239–244 [DOI] [PubMed] [Google Scholar]
- 9.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabbagh AJ, Mannion T, Lynch SM, Frei B. 1994. The effect of iron overload on rat plasma and liver oxidant status in vivo. Biochem J 300:799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enns CA, Ahmed R, Zhang AS. 2012. Neogenin interacts with matriptase-2 to facilitate hemojuvelin cleavage. J Biol Chem 287:35104–35117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensembl Genome Browser 69. [Internet]. 2012. Rattus norvegicus—cDNA sequence—transcript: Hfe2 (ENSRNOT00000028781). [Cited 19 November 2012]. Available at: www.ensembl.org.
- 13.Ensembl Genome Browser 69. [Internet]. 2012. Rattus norvegicus—cDNA sequence—transcript: Tfr2 (ENSRNOT00000001905). [Cited 19 November 2012]. Available at: www.ensembl.org.
- 14.Fleming MD. 2008. The regulation of hepcidin and its effects on systemic and cellular iron metabolism. Hematolo Am Soc Hematol Educ Program 2008:151–158 [DOI] [PubMed] [Google Scholar]
- 15.Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. 1998. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA 95:1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming MD, Trenor CC, 3rd, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. 1997. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet 16:383–386 [DOI] [PubMed] [Google Scholar]
- 17.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Gordon L, Hendrix M, Hourlier T, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Larsson P, Longden I, McLaren W, Overduin B, Pritchard B, Riat HS, Rios D, Ritchie GRS, Ruffier M, Schuster M, Sobral D, Spudich G, Tang YA, Trevanion S, Vandrovcova J, Vilella AJ, White S, Wilder SP, Zadissa A, Zamora J, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJP, Parker A, Proctor G, Vogel J, Searle SMJ. 2011. Ensembl 2011. Nucleic Acids Res 39:D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambling L, Czopek A, Andersen HS, Holtrop G, Srai SKS, Krejpcio Z, McArdle HJ. 2009. Fetal iron status regulates maternal iron metabolism during pregnancy in the rat. Am J Physiol Regul Integr Comp Physiol 296:R1063–R1070 [DOI] [PubMed] [Google Scholar]
- 19.Garrick MD, Dolan KG, Horbinski C, Ghio AJ, Higgins D, Porubcin M, Moore EG, Hainsworth LN, Umbreit JN, Conrad ME, Feng L, Lis A, Roth JA, Singleton S, Garrick LM. 2003. DMT1: a mammalian transporter for multiple metals. Biometals 16:41–54 [DOI] [PubMed] [Google Scholar]
- 20.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss–PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 21.Hofmann WK, Tong XJ, Ajioka RS, Kushner JP, Koeffler HP. 2002. Mutation analysis of transferring receptor 2 in patients with atypical hemochromatosis. Blood 100:1099–1100 [DOI] [PubMed] [Google Scholar]
- 22.Hsiao PJ, Tsai KB, Shin SJ, Wang CL, Lee ST, Lee JF, Kuo KK. 2007. A novel mutation of transferrin receptor 2 in a Taiwanese woman with type 3 hemochromatosis. J Hepatol 47:303–306 [DOI] [PubMed] [Google Scholar]
- 23.Institute for Laboratory Animal Research. 1996. Guide for the care and use of laboratory animals. Washington (DC): National Academies Press.
- 24.Iolascon A, De Falco L. 2009. Mutations in the gene encoding DMT1: clinical presentation and treatment. Semin Hematol 46:358–370 [DOI] [PubMed] [Google Scholar]
- 25.Kang JO, Jones C, Brothwell B. 1998. Toxicity associated with iron overload found in hemochromatosis: possible mechanism in a rat model. Clin Lab Sci 11:350–354 [PubMed] [Google Scholar]
- 26.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang R-H, Deng C, Vaulont S, Mosser J, Coppin H, Roth M-P. 2008. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood 112:1503–1509 [DOI] [PubMed] [Google Scholar]
- 27.Koyama C, Wakusawa S, Hayashi H, Suzuki R, Yano M, Yoshioka K, Kozuru M, Takayamam Y, Okada T, Mabuchi H. 2005. Two novel mutations, L490R and V561X, of the transferrin receptor 2 gene in Japanese patients with hemochromatosis. Haematologica 90:302–307 [PubMed] [Google Scholar]
- 28.Langnaese K, John R, Schweizer H, Ebmeyer U, Keilhoff G. 2008. Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model. BMC Mol Biol 9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence CM, Ray S, Babyonyshev M, Galluser R, Borhani DW, Harrison SC. 1999. Crystal structure of the ectodomain of human transferrin receptor. Science 286:779–782 [DOI] [PubMed] [Google Scholar]
- 30.Lebeau A, Frank J, Biesalski HK, Weiss G, Srai SKS, Simpson RJ, McKie AT, Bahram S, Gilfillan S, Schümann K. 2002. Long-term sequelae of HFE deletion in C57BL/6 × 129/O1a mice, an animal model for hereditary haemochromatosis. Eur J Clin Invest 32:603–612 [DOI] [PubMed] [Google Scholar]
- 31.Lee DH, Zhou LJ, Zhou Z, Xie JX, Jung JU, Liu Y, Xi CX, Mei L, Xiong WC. 2010. Neogenin inhibits HJV secretion and regulates BMP-induced hepcidin expression and iron homeostasis. Blood 115:3136–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee PL, Halloran C, West C, Beutler E. 2001. Mutation analysis of the transferrin receptor 2 gene in patients with iron overload. Blood Cells Mol Dis 27:285–289 [DOI] [PubMed] [Google Scholar]
- 33.Masson R, Roome NO. 1997. Spontaneous iron overload in Sprague–Dawley rats. Toxicol Pathol 25:308–316 [DOI] [PubMed] [Google Scholar]
- 34.Mendes AI, Ferro A, Martins R, Picanço I, Gomes S, Cerqueira R, Correia M, Nunes AR, Esteves J, Fleming R, Faustino P. 2009. Non-classical hereditary hemochromatosis in Portugal: novel mutations identified in iron metabolism-related genes. Ann Hematol 88:229–234 [DOI] [PubMed] [Google Scholar]
- 35.Merle U, Theilig F, Fein E, Gehrke S, Kallinowski B, Riedel HD, Bachmann S, Stremmel W, Kulaksiz H. 2007. Localization of the iron-regulatory proteins hemojuvelin and transferrin receptor 2 to the basolateral membrane domain of hepatocytes. Histochem Cell Biol 127:221–226 [DOI] [PubMed] [Google Scholar]
- 36.Miyata K, Sukata T, Kushida M, Ogata K, Suzuki M, Ozaki M, Ozaki K, Uwagawa S. 2009. Spontaneous iron accumulation in hepatocytes of a 7-week-old female rat. J Toxicol Pathol 22:199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niaré O, Markianos K, Volz J, Oduol F, Touré A, Bagayoko M, Sangaré D, Traoré SF, Wang R, Blass C, Dolo G, Bouaré M, Kafatos FC, Kruglyak L, Touré YT, Vernick KD. 2002. Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science 298:213–216 [DOI] [PubMed] [Google Scholar]
- 38.Park CH, Bacon BR, Brittenham GM, Tavill AS. 1987. Pathology of dietary carbonyl iron overload in rats. Lab Invest 57:555-563 [PubMed] [Google Scholar]
- 39.Pietrangelo A.2010. Hereditary hemochromatosis: pathogenesis, diagnosis, and treatment. Gastroenterology 139: 393–408.
- 40.Poiley S. 1960. A systematic method of breeder rotation for non-inbred laboratory animals colonies. Proc Anim Care Panel 10:159–166 [Google Scholar]
- 41.Robb A, Wessling-Resnick M. 2004. Regulation of transferrin receptor 2 protein levels by transferrin. Blood 104:4294–4299 [DOI] [PubMed] [Google Scholar]
- 42.Roetto A, Daraio F, Alberti F, Porporato P, Calì A, De Gobbi M, Camaschella C. 2002. Hemochromatosis due to mutations in transferrin receptor 2. Blood Cells Mol Dis 29:465–470 [DOI] [PubMed] [Google Scholar]
- 43.Roetto A, Totaro A, Piperno A, Piga A, Longo F, Garozzo G, Calì A, De Gobbi M, Gasparini P, Camaschella C. 2001. New mutations inactivating transferrin receptor 2 in hemochromatosis type 3. Blood 97:2555–2560 [DOI] [PubMed] [Google Scholar]
- 44.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 45.Rueda Adel C, Grande NC, Fernández EA, Enríquez de Salamanca R, Sala LA, Jiménez MJ. 2011. Mutations in HFE and TFR2 genes in a Spanish patient with hemochromatosis. Rev Esp Enferm Dig 103:379–382 [DOI] [PubMed] [Google Scholar]
- 46.Santos PCJL, Cançado RD, Pereira AC, Schettert IT, Soares RAG, Pagliusi RA, Hirata RDC, Hirata MH, Teixeira AC, Figueiredo MS, Chiattone CS, Krieger JE, Guerra-Shinohara EM. 2011. Hereditary hemochromatosis: mutations in genes involved in iron homeostasis in Brazilian patients. Blood Cells Mol Dis 46:302–307 [DOI] [PubMed] [Google Scholar]
- 47.Sasaki N, Hayashizaki Y, Muramatsu M, Matsuda Y, Ando Y, Kuramoto T, Serikawa T, Azuma T, Naito A, Agui T, Yamashita T, Miyoshi I, Takeichi N, Kasai N. 1994. The gene responsible for LEC hepatitis, located on rat chromosome 16, is the homolog to the human Wilson disease gene. Biochem Biophys Res Commun 202:512–518 [DOI] [PubMed] [Google Scholar]
- 48.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. 2008. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab 7:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tukey J. 1977. Exploratory data analysis. Reading (MA): Addison-Wesley. [Google Scholar]
- 50.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. 2007. The Rat Genome Database, update 2007—easing the path from disease to data and back again. Nucleic Acids Res 35:D658–D662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, Du F, Qian ZM, Ge XH, Zhu L, Yung WH, Yang L, Ke Y. 2008. Lipopolysaccharide induces a significant increase in expression of iron regulatory hormone hepcidin in the cortex and substantia nigra in rat brain. Endocrinology 149:3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiercinska E, Wickert L, Denecke B, Said HM, Hamzavi J, Gressner AM, Thorikay M, ten Dijke P, Mertens PR, Breitkopf K, Dooley S. 2006. Id1 is a critical mediator in TGFβ-induced transdifferentiation of rat hepatic stellate cells. Hepatology 43:1032–1041 [DOI] [PubMed] [Google Scholar]
- 53.Wu J, Forbes JR, Chen HS, Cox DW. 1994. The LEC rat has a deletion in the copper transporting ATPase gene homologous to the Wilson disease gene. Nat Genet 7:541–545 [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi Y, Heiny ME, Shimizu N, Aoki T, Gitlin JD. 1994. Expression of the Wilson disease gene is deficient in the Long–Evans cinnamon rat. Biochem J 301:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang AS, Anderson SA, Meyers KR, Hernandez C, Eisenstein RS, Enns CA. 2007. Evidence that inhibition of hemojuvelin shedding in response to iron is mediated through neogenin. J Biol Chem 282:12547–12556 [DOI] [PubMed] [Google Scholar]
- 56.Zhang AS, Xiong S, Tsukamoto H, Enns CA. 2004. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood 103:1509–1514 [DOI] [PubMed] [Google Scholar]