Abstract
Stem cell fate decisions are controlled by a molecular network in which transcription factors and miRNAs are of key importance. To systemically investigate their impact on neural stem cell (NSC) maintenance and neuronal commitment, we performed a high-throughput mRNA and miRNA profiling and isolated functional interaction networks of involved mechanisms. Thereby, we identified an E2F1–miRNA feedback loop as important regulator of NSC fate decisions. Although E2F1 supports NSC proliferation and represses transcription of miRNAs from the miR-17∼92 and miR-106a∼363 clusters, these miRNAs are transiently up-regulated at early stages of neuronal differentiation. In these early committed cells, increased miRNAs expression levels directly repress E2F1 mRNA levels and inhibit cellular proliferation. In mice, we demonstrated that these miRNAs are expressed in the neurogenic areas and that E2F1 inhibition represses NSC proliferation. The here presented data suggest a novel interaction mechanism between E2F1 and miR-17∼92 / miR-106a∼363 miRNAs in controlling NSC proliferation and neuronal differentiation.
INTRODUCTION
Stem cells are characterized by their ability to self-renew indefinitely and to differentiate into committed progeny (1). During embryonic development, stem cells give rise to all tissues and organs where some of them remain as multi-potent somatic stem cells. In adult tissues, these somatic stem cells are mostly found in homeostasis balancing between self-renewal and differentiation. Consequently, key features of stem cells, including proliferation, differentiation, migration, polarity and death are controlled by a tightly regulated network of signalling pathways (2).
For a long time, the ‘no new neuron’ dogma, which stated that brain tissue is quiescent and does not undergo notable cell turnover, was widely accepted. However, multiple reports confuted this dogma. Studies in songbirds (3), mice (4), monkeys (5) and humans (6,7) accentuated neurogenesis and neuron replacement in adult brains. Thereby, the existence of adult neural stem cells (NSCs) was proven. In the adult mammalian brain, the main niches of adult NSCs are the subventricular zone (SVZ) of the lateral ventricle and the subgranular zone of the dentate gyrus within the hippocampus (8).
Like other stem cells, NSCs bear the capacity to self-renew and to differentiate into more committed progeny (9–11). Under maintenance conditions, NSCs are characterized by a very low degree of epigenetic silencing, suggesting that a high amount of genes has to be activated in order to ensure the self-renewing state (12). In good agreement, only a small number of microRNAs (miRNAs) have been identified in proliferating NSCs. Therefore, a decreased amount of miRNA-regulated post-transcriptional inhibition was suggested for self-maintaining NSCs (13). In contrast, neuronal differentiation of NSCs induces drastic changes in the transcriptomic profile. These changes include the activation of numerous miRNAs. For instance in differentiating neurons, miR-9/9* represses the transcription factors Foxg1, Nr2e1, Gsh2 and Meis2, which are important for stem cell maintenance (14–19). However, the knowledge about the molecular events governing stem cell fate decisions is still fragmentary. Moreover, a systemic analysis, integrating gene regulation by miRNAs and highlighting the functional network and interplay of involved molecules and pathways is currently not available.
In this study, we used an mRNA–miRNA-based systemic analysis in order to identify and characterize molecular interaction networks implicated in the maintenance of self-renewing NSCs as well as in the induction of neuronal differentiation. By that means, we show evidence of an E2F1–miRNA feedback loop regulating NSC proliferation. In this system, E2F1 promotes proliferation of NSCs while directly repressing the expression of multiple miRNAs belonging to the miR-17∼92 and miR-106a∼363 clusters. On the other hand, upon induction of neuronal differentiation, the expression levels of these miRNAs are strongly increased, which represses E2F1 expression levels as well as cell proliferation rates. Interestingly, we demonstrate that these miRNAs, generally annotated as proliferation inducers, are only transiently up-regulated during the course of neuronal differentiation and strongly decrease in more mature neurons. Together, our data shed light on the complex molecular mechanism of NSC maintenance and neuronal differentiation and underline the modulating effect of miRNAs on neural stem cell fate decisions.
MATERIALS AND METHODS
Cell culture
Mouse NSCs were cultivated in a niche-independent cell culture system as described by Conti et al. (9,10). In brief, primary NSCs were kept on polystyrene poly-d-lysine (Sigma-Aldrich)-coated 10-cm dishes (Greiner) in DMEM HAM’s F12 medium (PAA) supplemented with Epidermal Growth Factor (EGF) (Peprotech), Fibroblast Growth Factor-basic (bFGF) (Peprotech), N2 (Invitrogen), l-glutamine (Invitrogen) and penicillin/streptomycin (Invitrogen). Neuronal differentiation was induced by exchanging 50% of the maintenance medium by Neurobasal medium (Gibco), supplemented with N2, B27 (Invitrogen), l-glutamine and penicillin/streptomycin. After 3 days of neuronal differentiation, we observed ∼30% of TuJ1-positive cells; this value increases to ∼50% after 5 days. With this protocol, only a low level of glial differentiation is detectable.
Neuroblastoma cells; Neuro-2a cells (N2A) cells and NIH3T3 cells are kept on polystyrene poly-d-lysine-coated 10-cm dishes in DMEM medium (Sigma-Aldrich) supplemented with 10% FCS (PAA), l-glutamine and penicillin/streptomycin.
C2C12 (muscle precursor cells) are kept in DMEM medium supplemented with 15% FCS, l-glutamine and penicillin/streptomycin. Differentiation was induced by switching the medium to differentiation medium composed of DMEM medium supplemented with l-glutamine, penicillin/streptomycin and 2% of Horse serum (Sigma Aldrich).
Microarray analysis
MRNA was extracted from NSCs and derived neurons using the RNAeasy kit (Quiagen) following manufacturer’s recommendations. mRNA quantity and purity were determined by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Additional quality check was performed by the Agilent Bioanalyzer (Agilent). Gene expression profiles were generated using MouseGene 1.0ST arrays according to manufacturer’s recommendations (Affymetrix).
MiRNA was extracted from NSCs and derived neurons using the miRNAeasy kit (Quiagen) following manufacturer’s recommendations. Gene expression profiles were generated using miRXplore Microarrays according to manufacturer’s recommendations (Miltenyi Biotech). Data processing of digitalized array intensity files was performed using the Mayday software (20).
Software
The used software is listed in Supplemental Data.
Immunohistochemistry
For immunohistochemical analysis, cells were fixed with 4% paraformaldehyde in 120 mM phosphate buffer, pH 7.4 (PBS) (4%PFA/1PBS), permeabilized with 0.05% Triton X-100 in PBS, blocked with 10% goat serum in PBS and subjected to immunohistochemistry staining with primary and secondary antibodies diluted in the blocking solution.
For immuno-labelling the following antibodies were used: anti-Nestin (BD Bioscience), anti-TuJ1 (Covance), anti-GFAP (Sigma-Aldrich), anti-E2F1 (Gene Tex), anti-Ki67 (Vector Labs), anti-phospho-Histone 3 (New England Biolabs), anti-PCNA (BD Biosciences), anti-Pax7 (Neuromics) and anti-Myosin (DSHB). For immunofluorescence staining, secondary Alexa-fluorophore-conjugated antibodies (Invitrogen) were used. DNA was stained using Hoechst 33258 (Invitrogen).
Transfection assays
N2A and NIH3T3 cells were transfected with 5 µg of indicated vectors. Transfection has been performed using Turbofect (Fermentas), following manufacturer’s recommendations. NSCs were electroporated with 5 µg of indicated vectors using the Amaxa mouse neural stem cell Nucleofector Kit (Lonza) by means of the Amaxa Nucleofector II Device (Lonza). Used plasmids are listed in Supplemental Data.
Knock-down of E2F1 was achieved with shRNA-expressing vectors shE2F1_A and shE2F1_B (21). As control, the same vector backbone leading to expression of scrambled sequence (shSCR) was used. Additionally in several experiments for controls the pEGFP-N1 vector (Clontech) was used.
Chromatin immunoprecipitation
N2A cells were transfected with 5 µg of E2F1-HA-expressing vectors. Transfection has been performed using Turbofect (Fermentas), following manufacturer’s recommendations. The chromatin immunoprecipitation (ChIP) assay was realized with the Imprint® Chromatin Immunoprecipitation Kit (Sigma-Aldrich). For each ChIP reaction, 250 000 cells has been used following manufacturer’s recommendations. Chromatin was sonicated using the Bioruptor® Water cooler (Diagenode). The mean fragment size of sonicated DNA was 400 bp. Bound E2F1-HA was immunoprecipitated using an anti-HA antibody (Roche). Quantification of precipitated DNA was performed by PCR reaction. Used primer pairs are listed in Supplemental Data.
RTq-PCR
For quantification analysis, total RNA (mRNAs and miRNAs) was extracted from NSCs, NSC-derived neurons, NIH3T3 and N2A cells by the miRNAeasy kit (Quiagen) following manufacturer’s recommendations. E2F1 expression levels were evaluated by the SYBR-Green Jump Start Taq Ready Mix (Sigma-Aldrich) following manufacturer’s recommendations. E2F1-related intensity levels were evaluated upon normalization with GAPDH levels. Used primers are listed in Supplemental Data.
Expression levels of miRNA extracted from NSCs, derived neurons or N2A cells were evaluated by microRNA LNA™ PCR primer sets (Exiqon) and normalized relative to miR-184 expression levels.
Luciferase assay
N2A cells were seeded to reach 80% confluence and transfected with 1.2 μg plasmid DNA and 4.0 μl polyethylenimine in a 24-well plate. Each transfection contained 50 ng pRL-SV40 Renilla expression construct for normalization purposes, 150 ng pcDNA3.1D-Firefly expression construct including the 3′-UTR of interest and 1 μg expression plasmid for the miRNA of interest. After 48 h, cells were washed with 500 μl PBS and lysed in 150 μl Passive Lysis Buffer (Promega). Twenty microlitre lysate was subsequently analysed using 50 μl LARII substrate or Stop&Glow (1:5 dilutions) to determine Firefly and Renilla luciferase activity, respectively (Dual Luciferase Reporter Assay System; Promega). Each sample was analysed in quadruplicates, and each transfection was carried out independently at least three times. Firefly activity was normalized to Renilla activity, and mean values plus standard error of mean are depicted. Reporter constructs encoding luciferase fused to a 3′-UTR with mismatches in the seed region served as negative controls due to their lack of complementarity with the miRNA. Mutagenesis of miRNA-binding sites in luciferase vectors was performed by QuikChange PCR using Pfu Ultra AD (Invitrogen) followed by a Dpn I digest (Fermentas) and transformation into MACH1 chemocompetent Escherichia coli cells as described previously (22,23). Used primer pairs are listed in the Supplemental Data.
Mice
Mice were kept under standard conditions according to governmental rules and regulations. All experiments involving mice have been conducted according to German Animal Welfare Act and have been approved by the responsible authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen).
Stereotactic injection
For stereotactic injections, surgery animals were under deep anaesthesia (intraperitoneal injection of 0.017 ml of 2.5% Avertin per gram of body weight) and fixed into a stereotactic frame (Kopf). Three microlitres concentrated lentivirus was injected into the lateral ventricle over 5 min using a Hamilton 7005KH 5 µl syringe. Following stereotactic co-ordinates in relation to bregma were used: anteroposterior: 1.4 mm, mediolateral: ±0.84 mm, dorsoventral: −2.5 mm below skull.
Lentiviral vector particle production
Lentiviral vector particles were produced using a three-plasmid transient transfection protocol. The day before transfection, 2 × 106 HEK293T cells were seeded in a 10 cm2 tissue culture dish in Dulbecco’s modified Eagle’s medium high glucose GlutaMAX (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin. Prior to transfection, medium was changed to fresh DMEM without serum and antibiotics. pGIPZ plasmid (pGIPZ-shScr, pGIPZ-shE2F1A or pGIPZ-shE2F1B) were mixed with a second-generation packaging plasmid (pCMV-dR8.2 dvpr) and a plasmid encoding the glycoprotein G of vesicular stomatitis virus (pCMV-VSV-G) in OptiMEM I Reduced Serum Media (Gibco). Fugene HD (Roche Applied Biosciences) was added, the solution was incubated for 15 min at room temperature and added drop wise to the cells. Next day, the supernatant was discarded and fresh DMEM supplemented with 30% FBS and 1% penicillin/streptomycin was added. Days 2 and 3 post transfection, supernatants were harvested and media were replaced. Supernatants were cleared through filtering, and vector particles were concentrated by low-speed centrifugation (4 h, 26 000g) at 4°C. Pellets were resuspended in an appropriate amount of DMEM supplemented with polybrene (Sigma-Aldrich) resulting in a 500-fold concentration, and stored at −80°C.
Perfusion, sectioning and immunohistochemistry
Eight days after virus injection, animals under deep anaesthesia were intracardially perfused with 50 ml 1× PBS following 50 ml 4% PFA /1 PBS solution. After dissection, isolated brains were post-fixed in 4% PFA /1 PBS solution over night at 4°C. By the use of a Vibratom (Leica VT 1200 S), 40 µm sagittal brain sections were made; free-floating sections were permeabilized in TBS 0.1 M Tris, 150 mM NaCl, pH 7.4 / 0.5% Triton-X 100 / 0.1% Na-Azide / 0.1% Na-Citrate / 5% normal goat serum (TBS+/+/+) for at least 1 h. The primary anti-Ki67 antibody (Vector Labs) was diluted in TBS+/+/+ and incubated for 48 h on a shaker at 4°C. For immunofluorescence staining, secondary Alexa-fluorophore-conjugated antibodies (Invitrogen) and 33258 (Invitrogen) were used. Sections were analysed with a Zeiss LSM 710 confocal microscope.
RESULTS
A systemic analysis of mRNA and miRNA profiles reveals numerous pathways involved in neural stem cell maintenance and neuronal differentiation
To identify key molecules and mechanisms implicated in NSC maintenance and neuronal differentiation, we made use of a primary mouse NSC in vitro culture system (9,10). The hallmarks of stem cells, i.e. maintenance capacity and multi-lineage differentiation potential, were verified by positive staining for the stem cell marker Nestin under maintenance conditions, positive staining for the neuronal marker TuJ1 after induction of neuronal differentiation and positivity for the astrocyte marker GFAP after induction of glial differentiation (Supplementary Figure S1A).
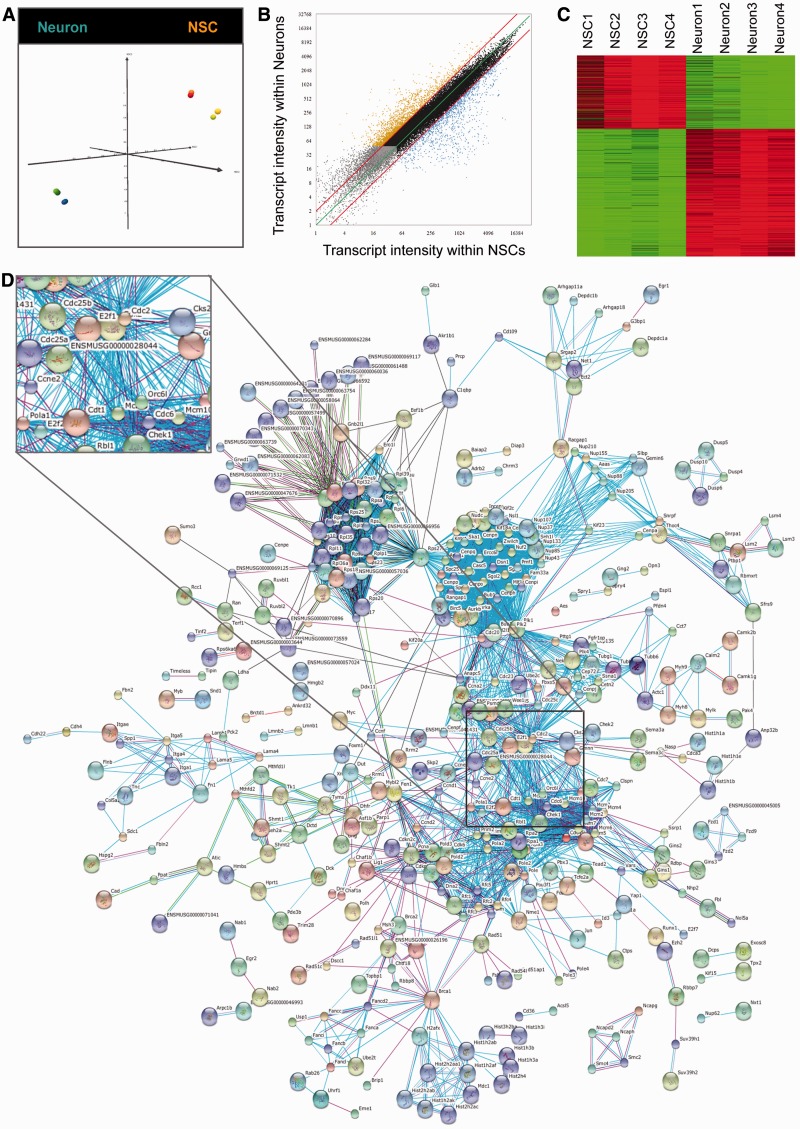
The mRNA expression signatures of NSCs and NSC-derived young neurons (2 days of neuronal differentiation) were determined by using whole-transcriptome expression analysis MouseGene 1.0ST arrays from Affymetrix. Firstly, we performed a multi-dimensional scale test (MDS) to check for outlier samples. Interestingly, the result of this analysis highlighted that most accentuated transcriptomic differences are found between the cell types, i.e. NSCs and neurons (Figure 1A). This MDS analysis was performed on the 500 transcripts with the highest standard deviation observed throughout the eight samples analysed. To isolate transcripts that are regulated during the process of neuronal differentiation, we compared expression intensities of NSCs and derived neurons in a scatter blot (Figure 1B). Genes characterized by expression intensities close to background signal for both cell types (Figure 1B, grey dots), or genes with low transcriptional variations (Figure 1B; black dots) were discarded from further analysis. Only transcripts with a >4-fold up- or down-regulation were scored as regulated during neuronal differentiation and used for further analysis. (Figure 1C and Supplementary Table S1).
Figure 1.
Systemic analysis of neural stem cells and differentiated neurons. (A) MDS analysis based on the 500 transcripts with the highest standard deviation within the eight samples analysed (NSC, yellow–red; neurons, green–blue). (B) mRNA scatter blot of analysed NSC and neurons. Four fold cut-off lines are represented in red. Gray dots represent mRNAs scored as absent in NSCs and in neurons; black dots represent mRNAs scored as not regulated during differentiation; orange dots represent mRNAs scored as up-regulated during neuronal differentiation and blue dots represent mRNAs scored as up-regulated in self-renewing NSCs. (C) Heat-map of differentially expressed genes isolated in (B). Red colour shows high expression levels and green colour shows low expression levels. (D) Functional network of genes/proteins with specific high expression levels in self-renewing NSCs. The network was generated with the STRING tool (http://string.embl.de/). The black square indicates enlargement of the network around E2F1. Network subclusters are presented in Figure S1N–R.
The differential expression profiles of regulated transcripts were functionally analysed using the GSEA (Gene Set Enrichment Analyzer) software (24). NSCs grown under maintenance conditions were characterized by a strong association with the activated cell cycle, p53 signalling pathway, DNA replication and repair mechanisms, as well as with the expression of histone-coding genes located in the chromosomal region 6p22 (Supplementary Figure S1B–G). Interestingly, by this approach, we could further identify that these 6p22 histone-coding genes share the consensus site of a yet unknown transcription factor that is probably important for the transcription for most of them (Supplementary Figure S1H). On the other hand, this in silico analysis revealed that the early stages of neuronal differentiation are associated with an up-regulation of neuroactive ligand receptor interactions, calcium signalling and activation of numerous genes downstream of NRSF (Rest, RE1-silencing transcription factor), SF1 and CDPCR3HD (Cux1, cut-like homeobox 1) (Supplementary Figure S1I–M).
To investigate which genes and pathways are important either for NSC maintenance or for neuronal differentiation, we decided to perform a systemic analysis of the obtained transcription profiles. To conduct this kind of analysis, we made use of the STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins 8.3; http://string-db.org/) (25). This database allowed us to highlight known as well as predicted direct and functional gene and protein interactions (Figure 1D). In agreement with the GSEA data (Supplementary Figure S1B–M), the network analysis underlined the importance of cell cycle components, DNA replication genes and histones within self-renewing NSCs (Figure 1D and Supplementary Figure S1N–P). Additionally, we identified functional subclusters for genes involved in ribosome biosynthesis and extracellular matrix composition (Supplementary Figure S1Q and R).
The functional network of genes highly expressed in young neurons was characterized by a lower degree of complexity (Supplementary Figure S1S). Nevertheless, multiple functional subclusters were identified. According to this analysis, numerous genes coding for interferon alpha signalling, olfactory receptors, glutamate receptors, voltage-dependent calcium channels, FGF-signalling, gap junctions and integrins are important for the induction of neuronal differentiation (Supplementary Figure S1T–Z).
In summary, by combining gene expression profiling and systemic network analysis, it was possible to attribute specific functional clusters and molecular processes either to NSC self-renewal or to neuronal commitment.
The majority of miRNAs is up-regulated during neuronal differentiation
Analogous to the expression profiling of mRNAs in NSCs and young NSC-derived neurons, we also profiled the expression of miRNAs in these two cell types. Also similar to the mRNA profiling analysis, we compared the expression levels of miRNAs from both sample groups by a scatter blot analysis. (Supplementary Figure S2A). MiRNAs that showed a >4-fold up- or down-regulation were considered as differentially regulated. Interestingly, nearly all miRNAs that were classified as regulated were up-regulated during neuronal differentiation. Out of these neuron-specific miRNAs, we selected the 40 transcripts with the highest standard deviation for further analysis (Supplementary Table S2).
To predict the target genes of selected miRNAs, we used the target gene finder tool miRDB (26). By this approach, a total of 2686 genes with a minimal target score of 70 (reflecting a high degree of confidence) were identified (Supplementary Table S3). These predicted target genes were used to generate a second functional network (Supplementary Figure S2B). This network represents functional gene groups that are potentially down-regulated during neuronal differentiation in an miRNA-dependent way.
The transcription factor E2F1 is highly expressed in self-renewing neural stem cells
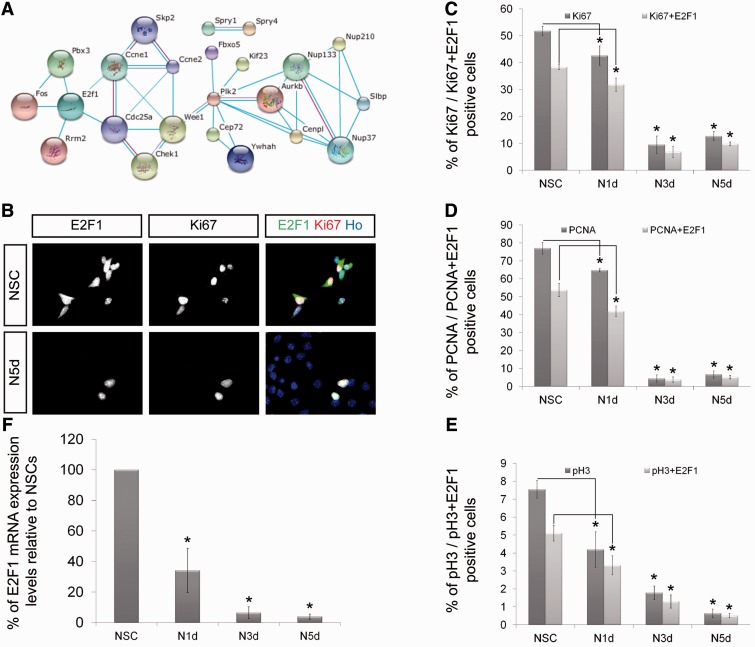
In the next step, we focused on the identification of genes and pathways that are important for NSC maintenance and whose activity is down-regulated in an miRNA-dependent manner during neuronal differentiation. Therefore, we superimposed the network of genes that are highly expressed in NSCs (Figure 1D) with the network of the predicted neuron-specific miRNA target genes (Supplementary Figure S2B). This superimposition resulted in a third network including the transcription factor E2F1 (Figure 2A). This transcription factor is of special interest because it already previously has been described to be involved in stem cell maintenance (27). Furthermore, E2F1 regulates stem cell self-renewal together with miR-223 in a negative feedback loop (28,29). Based on these published findings and the results from our bioinformatic analysis, E2F1, E2F1-regulated miRNAs as well as E2F1-regulating miRNAs are promising candidates for a function during NSC maintenance and neuronal differentiation.
Figure 2.
E2F1 is strongly associated with proliferation of NSCs. (A) Congruent network of genes simultaneously up-regulated in self-renewing NSCs (Figure 1D) and potentially controlled by neuron-specific miRNAs (Figure S2B). (B) NSCs and derived neurons after 5 days of neuronal differentiation (N5d) labelled with indicated markers. NSCs expressing high levels of Ki67 mostly express E2F1. After 5 days of neuronal differentiation, most cells exit the cell cycle; the few proliferating Ki67-positive cells also express E2F1 (scale bar, 20µm). (C–F) Quantitative analysis indicating the percentage of cells positively labelled for the proliferation markers Ki67 (C), pospho-Histone 3 (D) and PCNA (E) with or without E2F1 co-labelling (n ≥ 100; N ≥ 3; mean ± SEM; *P < 0.05; U-Test). (F) E2F1 RT-qPCR in self-renewing NSC and derived neurons after 1 day (N1d), 3 days (N3d) and 5 days (N5d) of neuronal differentiation. E2F1 expression levels in self-renewing NSCs are set to 100%; after 1 day of differentiation, relative E2F1 expression levels are reduced to 35% and tend to a low expression level plateau already after 3 days of differentiation (N ≥ 3; mean ± SEM; *P < 0.05; U-Test).
To characterize whether E2F1 regulates the cell cycle of NSCs, we performed immunofluorescence co-stainings for E2F1 and for the cell cycle markers Ki67 and PCNA as well as the mitosis marker phospho-Histone 3 (Figure 2B–E). As expected, the majority of self-renewing NSCs were positive for Ki67. In fact, most of these cells also displayed high levels of E2F1. With ongoing neuronal differentiation, the majority of NSCs stop to proliferate and become negative for the three proliferation markers tested. Three and five days after induction of neuronal differentiation, we observed a significant decrease in the number of proliferating cells (Figure 2C–E). However, virtually all of the few cells that still proliferate even 5 days after induction of differentiation express high levels of E2F1 (Figure 2B–E). Finally, we wanted to quantitatively evaluate this differentiation-dependent down-regulation of E2F1 at the mRNA level. Therefore, we performed RT-qPCR measurements on mRNA isolated from NSCs and from NSC-derived neurons 1, 3 and 5 days after induction of differentiation (Figure 2F and Supplementary Figure S2C–D). This analysis confirmed high E2F1 expression levels within NSCs and a strong down-regulation upon neuronal differentiation (Figure 2F). Additionally, this experiment revealed that the down-regulation of E2F1 is very fast. Already at the first day of differentiation, the level of the E2F1 mRNA is reduced by >50% (Figure 2F). From these data, we conclude that the expression of E2F1 strongly correlates with neural stem cell maintenance and proliferation.
Inhibition of E2F1 blocks cell cycle transition in NSCs
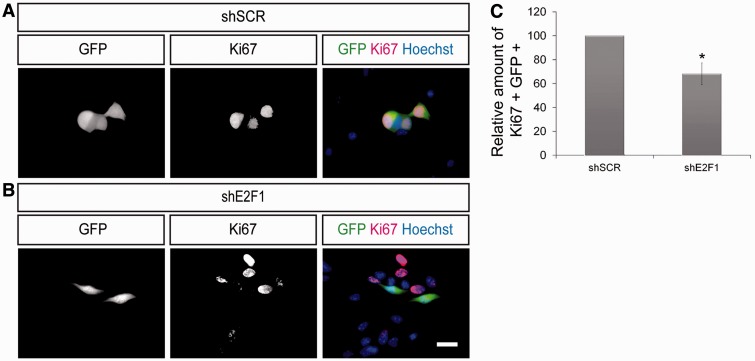
Based on our so far presented gene expression data and immunofluorescence results, we assume that E2F1 activity is associated to stem cell maintenance characteristics like cell cycle progression. Accordingly, diminished expression of E2F1 should inhibit NSC proliferation. To experimentally address this hypothesis, we made use of vectors coding for shRNA sequences directed against E2F1 (shE2F1) leading to an efficient knock-down of E2F1 (Supplementary Figure S3). NSCs were electroporated either with these shE2F1 vectors or with control vectors against a scrambled sequence (shSCR, negative control). All vectors additionally code for GFP under the control of an independent promoter. Two days after transfection the proliferation rate of electroporated NSCs (GFP positive) was analysed by immunofluorescence staining for Ki67. Under these conditions, the fraction of GFP–Ki67 double-positive NSCs is significantly decreased upon knock-down of E2F1 (Figure 3A–C). These data demonstrate that E2F1 is necessary for NSC proliferation.
Figure 3.
E2F1 is important for the proliferation of self-renewing NSCs. (A and B) NSCs were electroporated with shSCR or with shE2F1 coding vectors (both expressing GFP) and labelled with indicated markers. Analysis of electroporated NSCs (GFP positive) showed that transfection with shE2F1 resulted in a strong decrease of proliferating NSC (GFP–Ki67 double positive cells) when compared to the control situation (scale bar, 20 µm). (C) Relative amount of GFP–Ki67 double positive NSCs after electroporation with shE2F1 when compared to control treatment (n ≥ 100; N ≥ 3; mean ± SEM; *P < 0.05; U-Test).
MiRNAs mediate the down-regulation of E2F1 during neuronal differentiation
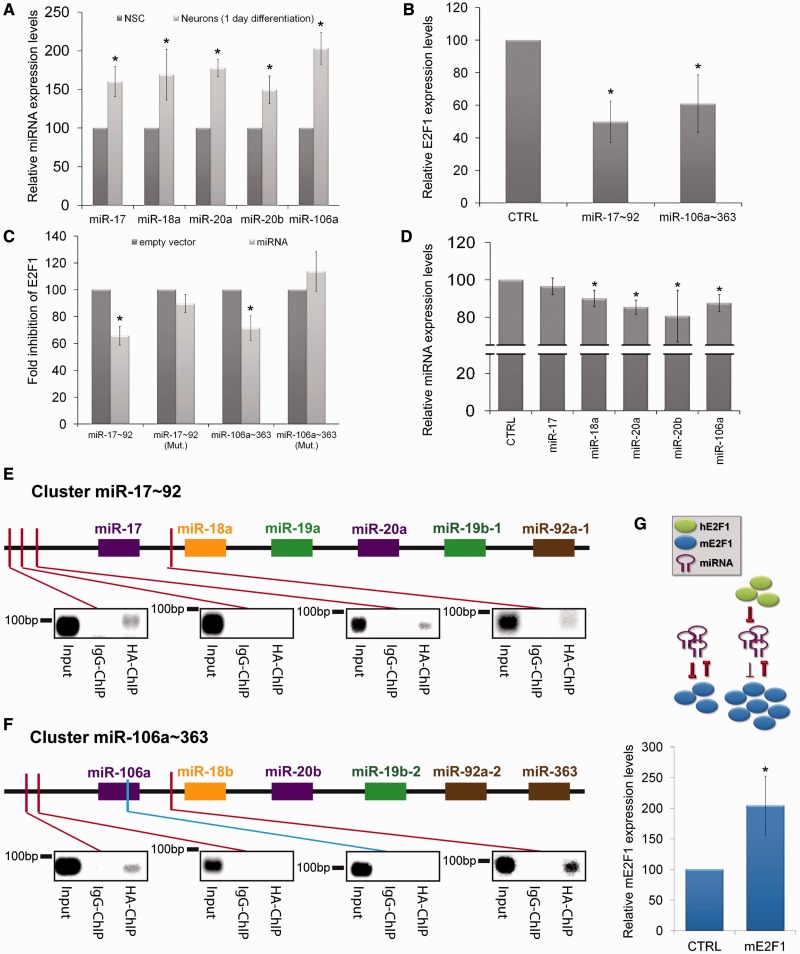
Based on the network superimposition (Figure 2A), we predict that the down-regulation of E2F1 during neuronal differentiation is mediated by miRNAs. This prediction is supported by the observation that among the miRNAs that are up-regulated during neuronal differentiation, several have corresponding seed sequences in the E2F1 3′UTR. Interestingly, among these, the miRNAs miR-17, miR-18a, miR-20a, miR-20b and miR-106a also have E2F1 binding consensus sequences in their promoter region. Consequently, these miRNAs potentially do not only regulate E2F1 expression but are themselves regulated in their expression levels by E2F1 (representing regulation in a classical feedback loop). Strikingly, this group of five miRNAs belongs to only two genomic miRNA clusters (miR-17∼92 and miR-106a∼363), which both show a high degree of similarity (Supplementary Figure S4).
Firstly, we wanted to confirm that these five miRNAs are up-regulated early during neuronal differentiation. Because the down-regulation of E2F1 is already strong only 1 day after induction of differentiation (Figure 2F), we decided to measure the levels of miR-17, miR-20a, miR-20b, miR-106a and miR-18a at this point of differentiation. As expected, the expression levels of all six analysed miRNAs significantly increased rapidly upon initiation of neuronal differentiation (Figure 4A).
Figure 4.
miRNAs act with E2F1 in a feedback loop. (A) RT-qPCR comparing the relative expression levels of indicated miRNAs extracted from self-renewing NSCs (normalized to 100%) and neurons after 1 day of differentiation (N ≥ 3; mean ± SEM; *P < 0.05; U-Test). (B) RT-qPCR measuring the relative E2F1 expression levels within N2A cells transfected with indicated miRNA constructs (N ≥ 3; mean ± SEM; *P < 0.05; U-Test). (C) Luciferase assay measuring the fold inhibition of E2F1 by indicated miRNA constructs. Mutations (Mut.) were introduced on the miRNA seed sequences located on the E2F1 3′UTR. (N ≥ 3; mean ± SEM; *P < 0.05; U-Test). (D) RT-qPCR comparing the relative expression levels of indicated miRNAs extracted from N2A cells either transfected with a control construct or with an E2F1 over-expressing construct (N ≥ 3; mean ± SEM; *P < 0.05; U-Test). (E and F) Chromatin immunoprecipitation performed on N2A cells transfected with HA-E2F1. Binding of E2F1 on predicted consensus sites (red bars) in the miR-17∼92 (E) and miR-106a∼363 (F) genomic clusters was analysed by PCR (Input, non immunoprecipitated DNA; Negative Control of reaction: miR-106a ORF—blue bar). (G) RT-qPCR comparing the relative expression levels of endogenous mouse E2F1. Upper part: Experimental set-up: mouse cells were either transfected with a control vector (CTRL) or with a vector expressing human E2F1 (hE2F1). Lower part: relative expression levels of endogenous mouse E2F1 (mE2F1) (N ≥ 3; mean ± SEM; P < 0.05; U-Test).
In the next step, we assessed whether these miRNAs indeed have the ability to down-regulate E2F1 expression levels. Therefore, we transfected the neural progenitor cell line N2A with vectors either expressing EGFP (CTRL) or the miRNA clusters miR-17∼92 or miR-106a∼363. The expression of the miRNA cluster miR-17∼92 or miR-106a∼363 led to a significant down-regulation of E2F1 (Figure 4B). To delineate whether E2F1 is directly targeted by the miRNAs of both clusters, we placed the 3′UTR of the E2F1 mRNA downstream to a luciferase-coding sequence and transfected N2A cells with this construct. In this assay, we observed a significant reduction in the luciferase signal upon presence of miR-17∼92 or miR-106a∼363 (Figure 4C). This direct interaction of miR-17∼92 and miR-106a∼363 with the E2F1 3′UTR was further strengthened by the absence of down-regulation when the miRNA-specific seed sequences were mutated. Taken together, these results suggest that expression of the miRNA clusters miR-17∼92 and miR-106a∼363 directly mediate the down-regulation of E2F1 upon induction of neuronal differentiation.
The investigated miRNAs (miR-17, miR-20a, miR-20b, miR-106a and miR-18a) were selected not only because they were significantly up-regulated upon induction of neuronal differentiation or because of their predicted ability to directly down-regulate E2F1 (confirmed in Figure 4B and C) but also because they have E2F1 consensus binding sequences in their promoter region. If there is a regulatory feedback loop between E2F1 and these miRNAs, we would expect that the expression of the miRNAs in turn is negatively regulated by E2F1. To experimentally address this question, we transfected N2A cells with vectors expressing EGFP (control) or E2F1. Twenty hours after transfection, the expression levels of the five miRNAs were measured by RT-qPCR. In agreement with our prediction, over-expression of E2F1 was able to significantly down-regulate the levels of four out of the five miRNAs (Figure 4D).
To investigate whether E2F1 indeed mediates this transcriptional repression by direct binding to regulatory elements within the genomic miR-17∼92 and miR-106a∼363 loci, we performed ChIP experiments. Potential E2F1 consensus sites were identified using the software ‘TFSEARCH’ (30) and ‘TFBIND’ (31). The investigated regions of both genomic miRNA clusters spanned the 3000 bp upstream to the first miRNA till the end of the first intron. ChIPs were performed from N2A cells transfected with an E2F1-HA expression vector. Binding to the predicted sites was analysed by subsequent PCR with specific primer sets. In total, we analysed seven potential E2F1-binding sites as well as the miR-106a ORF region as negative control (Figure 4E and F). Interestingly, in addition to binding to several of the predicted consensus sites located within the promoter regions, we were able to demonstrate that E2F1 also directly binds within the first intron of both miRNA clusters.
So far our results lead to a model in which E2F1 inhibits the expression of miRNAs from the 17∼92 and the 106a∼363 cluster. Furthermore, miRNAs from these clusters repress the expression of E2F1. If this model is correct, over-expression of exogenous E2F1 should inhibit the expression of the endogenous miRNAs from these clusters, consequently the inhibition of endogenous E2F1 should be abrogated and more of this endogenous E2F1 should be expressed. Indeed, when we transfected murine cells with a vector coding for human E2F1 (hE2F1) and then used murine-specific primers to measure the levels of endogenous E2F1 (mE2F1) we were able to detect a >2-fold increase of mE2F1 mRNA levels (Figure 4G).
Transient up-regulation of miRNAs from the miRNA clusters miR-17∼92 and miR-106a∼363 induce cell cycle exit in neural stem cells
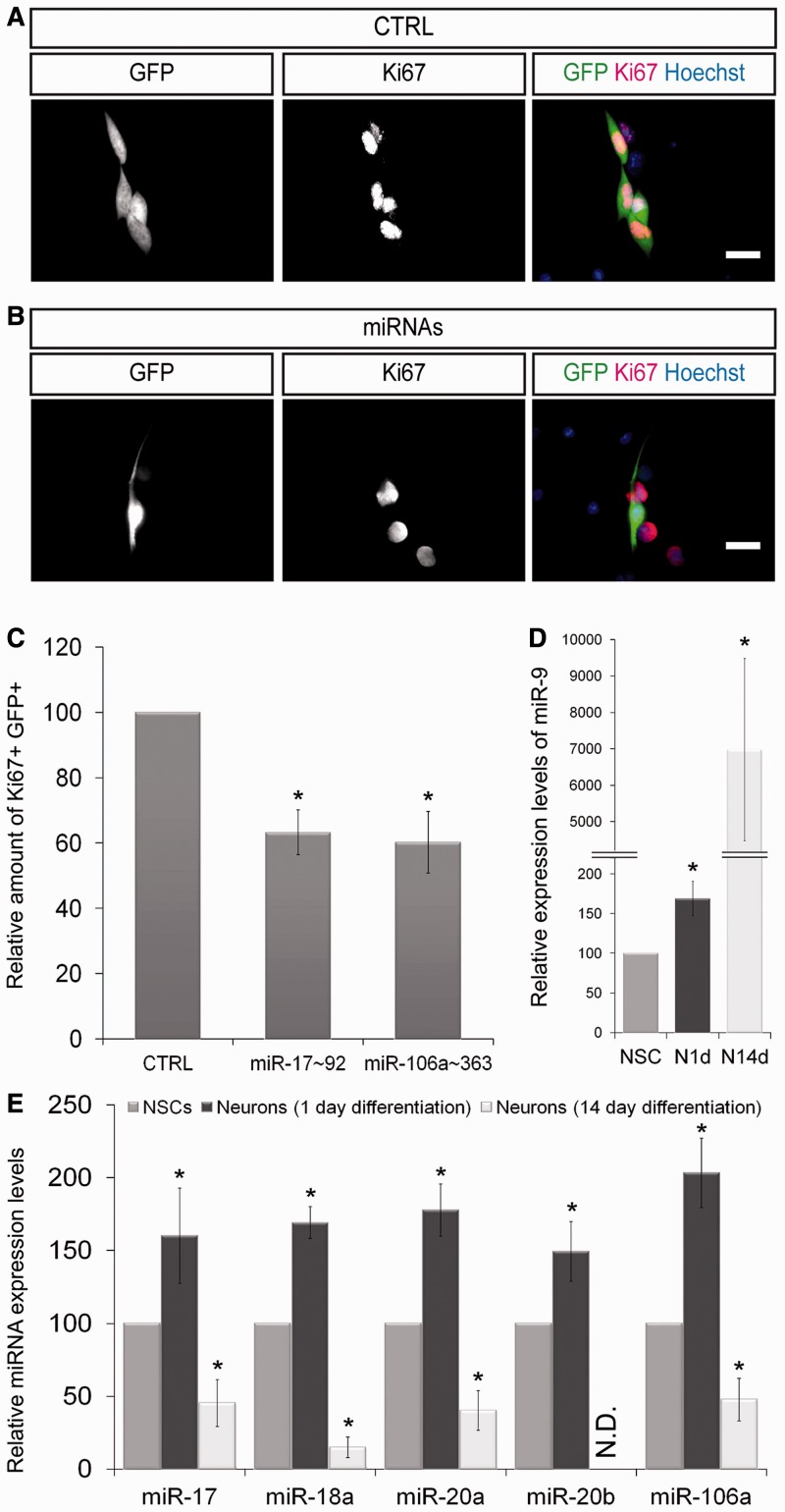
Based on our results indicating that miRNAs from the miRNA clusters miR-17∼92 and miR-106a∼363 induce a direct down-regulation of E2F1, their over-expression might, similarly to an E2F1 knock-down, inhibit the self-renewal and proliferation abilities of NSCs. Thereby, these miRNAs would be able to induce the fate transition from NSCs into neurons. To experimentally address this question, we electroporated NSCs with vectors either expressing GFP alone (control; CTRL) or in combination with the miRNA clusters miR-17∼92 or miR-106a∼363. Two days after transfection, the proliferation of electroporated NSCs (GFP positive) was analysed by immunofluorescence staining for Ki67. In agreement with our prediction, the amount of miRNA expressing Ki67-positive NSCs was strongly reduced. We observed a decrease of proliferating NSCs to 50 and 61% upon over-expression of miR-17∼92 and of miR-106a∼363, respectively (Figure 5A–C). This effect is similar to the before-observed reduction of proliferation after direct knock-down of E2F1 (Figure 3B–D). Finally, we verified that the observed decreased proliferation rate was not due to increased apoptosis (data not shown). In contrast to our results, previous studies reported a down-regulation of miRNAs from the clusters miR-17∼92 and miR-106a∼363 upon stem cell differentiation. However, these studies compare the expression levels of the miRNAs at the extremes of cell fate determination, i.e. they compare self-renewing stem cells and fully differentiated mature cells (32). To compare our results with these previously published data, we investigated the expression levels of miR-9, miR-17, miR-18a, miR-20a, miR-20b and miR-106a (miR-9 was included as a bona fide miRNA marker for neuronal differentiation) in NSCs and NSC-derived neurons 1 day and 14 days after induction of differentiation (Figure 5D and E). This analysis clearly revealed that the investigated miRNAs are transiently up-regulated directly after induction of differentiation, while they are significantly down-regulated in terminally differentiated neurons. This transient up-regulation during differentiation is in good agreement with a modulating function of miRNAs during cell fate transitions.
Figure 5.
Transient up-regulation of miRNAs induces early neuronal differentiation. (A and B) NSCs were electroporated with GFP or with the miRNA cluster miR-17∼92 and miR-106a∼363 coding vectors (in combination with GFP) and labelled with indicated markers. Analysis of electroporated NSCs (GFP positive) showed that expression of the indicated miRNAs resulted in a strong decrease in proliferating NSC (GFP–Ki67 double positive cells) when compared to the control transfections. (C) Relative amount of GFP–Ki67 double positive NSCs after electroporation with indicated miRNAs when compared to control transfections (n ≥ 100; N ≥ 3; mean ± SEM; *P < 0.05; U-Test). (D) RT-qPCR measuring the relative expression levels of the neuron-associated miRNA miR-9 under maintenance conditions (NSC) and after 1 (N1d) and 14 days (N14d) of neuronal differentiation (N ≥ 3; mean ± SEM; P < 0.05; U-Test). (E) Transient up-regulation of indicated miRNAs at the early time points of neuronal differentiation. miRNA expression levels strongly decrease after 14 days of neuronal differentiation (N ≥ 3; mean ± SEM; *P < 0.05; U-Test). N.D.: Not Detectable.
Inhibition of E2F1 blocks cell cycle transition in NSCs in vivo
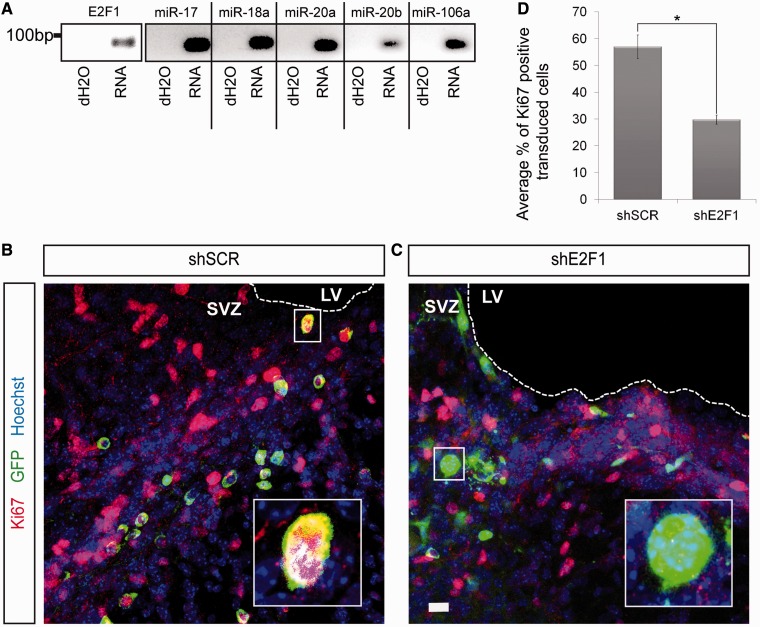
To investigate the function of E2F1 on NSC proliferation in vivo, we focussed on the SVZ, which is the largest neurogenic niche within the adult mouse brain. By performing a PCR on RNA extracted from the SVZ, we confirmed that not only E2F1 but also the miRNAs miR-17, miR-18a, miR-20a, miR-20b and miR-106a are expressed in this neurogenic area (Figure 6A).
Figure 6.
E2F1 is important for the NSC proliferation in the subventricular zone. (A) PCR performed on RNA extracted from the SVZ of adult mice brain. (B and C) Proliferation rate within the mouse SVZ of 5 treated and 5 control mice was analysed. Cells were transduced by lentiviruses containing constructs either targeting a scrambled sequence (shSCR) or E2F1 (shE2F1). Viruses were stereotactically injected in the SVZ of adult mice brain. Additionally, these vectors contained a GFP-coding gene under the control of an independent promoter. Transduced cells are GFP positive, Ki67 is labelled in red and nuclei are labelled with Hoechst (blue). While control animals show high amounts of proliferating transduced cells, the amount of dividing cells with E2F1 knock-down is strongly decreased (SVZ: subventricular zone; LV: lateral ventricle) (Scale bars: B and C, 20 µm. (D) Quantitative analysis highlighting the average percentage of proliferating cells transduced with the control (shSCR) or with the construct directed against E2F1 (shE2F1). The relative amount of proliferating SVZ cells is significantly decreased when E2F1 knocked-down. (N ≥ 5; n ≥ 100; mean ± SEM; *P < 0.05; U-Test).
To address the function of E2F1 in SVZ neural progenitor cells, we stereotactically injected lentiviruses carrying a plasmid coding for either an shRNA directed against a scrambled sequence (shSCR) or against E2F1 (shE2F1). Both vectors further coded the GFP gene under the control of an independent promoter. Eight days after injection, proliferation of transduced cells was analysed by immunofluorescence stainings with an anti-Ki67 antibody. Figures 6B and C show that the SVZ is densely populated by dividing Ki67-positive cells. Importantly, the number of transduced and proliferating (GFP–Ki67 double positive) cells was significantly reduced when E2F1 was knocked-down (Figure 6C–D). Taken together, these results clearly demonstrate that E2F1 is necessary for proliferation of neural progenitor cells in vivo.
In summary, our data indicate that a regulatory feedback loop between E2F1 and specific miRNAs is important for the maintenance of self-renewal and proliferation abilities of NSCs. In particular, up-regulation of these miRNAs during the early stages of neuronal differentiation induces a direct down-regulation of E2F1, leading to the exit from the cell cycle. In contrast, during stem cell maintenance E2F1 represses the expression of miR-17, miR-18a, miR-20a, miR-20b and miR-106a and thereby ensures NSC self-renewal and proliferation (Supplementary Figure S5).
DISCUSSION
Recently, multiple studies demonstrated the possibilities to maintain, reprogram, transform and differentiate embryonic and somatic stem cells. However, the knowledge about the exact molecular mechanisms important for such processes is very incomplete. Moreover, the massive transcriptomic differences between stem cells and their differentiated progeny suggest that large and complex networks of interacting proteins and pathways are involved. One efficient tool to delineate these molecular networks is the use of systemic analysis approaches. Such an approach has been used previously (33) to construct a functional interaction network centred on the transcription factor Oct4. In contrast to this approach, in our study, we asked which part of the expression profile is unique in NSCs in comparison with early committed neurons. The identification of differentially expressed genes allowed us to determine the molecular fingerprint of NSCs and to bring it into a functional context. Our systemic analysis associated functional subclusters containing components of the cell cycle machinery, DNA replication, ribosome synthesis, histone synthesis and extracellular matrix composition with proliferating NSCs. On the other hand, functional subclusters specifically associated with differentiating neurons highlighted numerous genes coding for olfactory receptors, interferon alpha proteins, glutamate receptors, voltage-dependent calcium channels, members of the FGF pathway, integrin–Reelin interaction genes and genes coding for different units of gap junctions. Strikingly, for all these processes, implications in neurogenesis have been shown previously (34–39).
Transcription factors as well as miRNAs are known to play important roles in stem cell fate decisions (40). MiRNAs are non-coding RNAs with regulatory functions on mRNA stability and translation rate, and thus, are direct control elements of protein production levels (41). Within NSCs, miR-9/9*, together with miR-124, have been shown to be of major importance for governing their differentiation towards neurons by completing the switch from the neural-progenitor-specific BRG1/BRM-associated factor (BAF) complex composition to the neuron-specific BAF complex (42). In agreement with these observations, expression of miR-9/9* and miR-124 induces the direct conversion of human fibroblasts to neurons. This effect is even more pronounced by additional expression of the transcription factors ASCL1 and MYT1L (18). Interestingly, besides the importance of miRNAs in determining the cellular identity of committed cells, their cooperation with transcription factors has an enhanced effect on cellular fate decisions. In our study, we demonstrated that the transcription factor E2F1 is highly expressed in NSCs while it is strongly down-regulated upon initiation of differentiation. E2F1 plays a major role in promoting the cell cycle. During the G1 phase, this transcription factor binds its consensus sequences in the promoter regions of cell cycle genes such as c-myc, B-myb, cdc2, DNA polymerase A, DHFR, TK, cyclinD and cyclinE, activates their expression and ensures the progression from the G1 to the S phase (43). In agreement with these findings and our results, Arai and collaborators demonstrated that short G1 phases are found in self-renewing stem cells, while lengthening of the G1 phase is associated with the transition into a more differentiated state (44). Accordingly, higher expression levels of E2F1 promote the G1/S transition (44,45) and therefore inhibit the induction of differentiation. When NSCs start to differentiate, E2F1 levels decrease, consequently length of the G1 phase increases and differentiation is favoured. To evaluate whether the E2F1 influence on stem cell proliferation is specific for NSCs, we quantitatively analysed E2F1 mRNA expression levels within the muscle precursor cell line C2C12. Also in these cells, E2F1 is significantly decreased after 3 and 5 days of differentiation (Supplementary Figure S6). However, the significant up-regulation at the first day of differentiation suggests that the regulatory mechanisms controlling E2F1 expression levels are different in this cell type.
Beside its role in promoting the cell cycle, our data suggest that E2F1 further inhibits expression of certain miRNAs, including miR-17, miR-18a, miR-20a, miR-20b and miR-106a. This implies that E2F1 can simultaneously act as transcription activator (for cell cycle genes) and transcription repressor (for the mentioned miRNAs). Previously, the direct transcription repressor function of E2F1 has been described for Interferon regulatory factor 3 (IRF-3) (46). Additionally, it has been shown that E2F1 is able to influence the differentiation program of granulocytes by inhibiting the expression of miR-223 (29). In the here presented study, we show that E2F1 inhibits the transcription of the miRNA clusters miR-17∼92 and miR-106a∼363 in self-renewing NSCs. Additionally, for the miRNAs miR-17, miR-18a, miR-20a, miR-20b and miR-106a, we demonstrate an early up-regulation during the fate transition from stem cell identity into neuronal fate. In good agreement with the well described function of miRNAs during cell fate transitions (47), this up-regulation is transient and has a modulator effect on neural stem cell fate decisions.
Interestingly, previous studies focusing on the role of miR-17∼92 and miR-106a∼363 miRNAs showed that these miRNAs are important activators of tumour cell proliferation (48). Moreover, the few studies based on physiological stem cells presented indications that E2F1 acts as an activator on miR-17∼92 and miR-106a∼363 miRNAs (49). Furthermore, it was reported that these miRNAs exhibit lower expression levels in the more differentiated progeny (32). On the first glance, these observations seem to be contradictory to the here reported results. However, usually those studies investigate the start and endpoint of a differentiation process. They compare the expression profile of an undifferentiated stem cell to a mature fully differentiated somatic cell. By investigating only these endpoints, functions that transcription factors and miRNAs might have during cell fate transitions may be overlooked. In the present study, we demonstrate that directly during cell fate transition from neural stem cell identity to neuronal identity, expression of specific miRNAs belonging to the miR-17∼92 and miR-106a∼363 clusters first need to increase in order to down-regulate the expression of cell cycle activators (like E2F1). With ongoing differentiation, these miRNAs diminish to levels even below the ones measured in undifferentiated stem cells. Because we clearly observe that these miRNAs are not absent within self-renewing NSCs, it is conceivable that under stem cell maintenance conditions, these miRNAs target cell cycle inhibitors like p21 (CDKN1A) (50). Therefore, we suggest that during neuronal differentiation, miRNAs from the miR-17∼92 and miR-106a∼363 clusters exert their modulating effect on NSC fate decisions by acting, beyond other cell cycle activators, on E2F1, and in this manner support the transcriptomic transition towards the expression profile of a fate-committed neuron. This post-transcriptional down-regulation may act together with a micro-RNA-independent transcriptional down-regulation of E2F1. The importance of E2F1 in stem cell fate decisions is further supported by the fact that increased expression of E2F1 has been found in embryonic stem cells and has been associated with the transcriptional regulatory network governing stem cell maintenance (51). Furthermore, transgenic mice over-expressing E2F1 show inhibited chondrocyte differentiation, which is reflected by delayed bone formation (52). On the other hand, loss of E2F1 in transgenic mice led to decreased neurogenic activity (53).
Whether E2F1 functions as activator or as inhibitor of transcription might depend on its binding to different regulatory elements either in the promoter or the intron of the regulated gene or miRNA cluster. Accordingly, Brosh and collaborators (54) demonstrated that in proliferating cells, E2F1 binds to numerous promoter regions. In contrast, in the here present study, we additionally demonstrated binding of E2F1 into the intronic regions of both miRNA clusters. This intronic binding may account for its repressor activity. A similar molecular behaviour was previously observed in reprogrammed pluripotent cells that need to down-regulate the Xist [X (inactive)-specific transcript] gene in order to acquire pluripotency. The pluripotency factors, Nanog, Oct4 and Sox2, which usually activate transcription, ensure this down-regulation by binding to the first intron of Xist (55).
If E2F1 has a higher affinity for its promoter binding sites compared with intronic binding regions, the following model would be conceivable: Under conditions of stem cell maintenance, high levels of E2F1 are present, consequently both sites (promoter as well as intron) would be bound by E2F1. This would lead to moderate expression of the miRNAs. They in turn regulate expression of E2F1, leading to stable steady state levels of both. Upon the induction of differentiation, E2F1 levels decrease. Consequently only the promoter binding sites would be bound by E2F1, which would lead to an activation of the respective miRNAs. If this process of differentiation continues, levels of E2F1 would be further decreased. Consequently also the activating promoter binding sites would not be bound by E2F1 anymore. Therefore, at later stages of differentiation, levels of the microRNAs decrease even below the levels present in NSCs.
Overall, our data strongly suggest that transcription factors, miRNAs and their regulatory interplay are of outstanding importance for stem cell fate decisions. Here, we demonstrated that at early time points of neuronal differentiation, the mutual control of the transcription factor E2F1 and the miRNAs miR-17, miR-18a, miR-20a, miR-20b and miR-106a play an important role in the modulation of proliferative characteristics of NSCs and thus on their maintenance and differentiation characteristics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6 and Supplementary References [21,22].
FUNDING
Marie Curie Fellowship (to T.P.); Münster Graduate Program for Cell Dynamics and Disease (CEDAD) (to A.L.H. and L.B.); German Research Foundation [DFG: Emmy Noether Program, SCHW1392/2-1; SFB629 and SPP1356, SCHW1392/4-1], Schram-Stiftung [T287/21795/2011], Else Kröner-Fresenius-Stiftung [2011_A94] and the Boehringer Ingelheim Foundation (to J.C.S.); fund ‘Innovative Medical Research’ of the University of Münster Medical School [SC120901 and SC411003]; Interdisciplinary Center for Clinical Research (IZKF) Münster [SchwJ3/001/11]. Funding for open access charge: German Research Foundation (DFG).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sandra Stelzer, Inga Werthschulte and Anna-Lena Schön for excellent technical assistance.
REFERENCES
- 1.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blank U, Karlsson G, Karlsson S. Signaling pathways governing stem-cell fate. Blood. 2008;111:492–503. doi: 10.1182/blood-2007-07-075168. [DOI] [PubMed] [Google Scholar]
- 3.Goldman SA, Nottebohm F. Neuronal production, migration, and differentiation in a vocal control nucleus of the adult female canary brain. Proc. Natl Acad. Sci. USA. 1983;80:2390–2394. doi: 10.1073/pnas.80.8.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 5.Bunk EC, Stelzer S, Hermann S, Schafers M, Schlatt S, Schwamborn JC. Cellular organization of adult neurogenesis in the Common Marmoset. Aging Cell. 2011;10:28–38. doi: 10.1111/j.1474-9726.2010.00639.x. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat. Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 7.Godfraind C, Friedrich VL, Holmes KV, Dubois-Dalcq M. In vivo analysis of glial cell phenotypes during a viral demyelinating disease in mice. J. Cell Biol. 1989;109:2405–2416. doi: 10.1083/jcb.109.5.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 9.Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat. Rev. Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- 10.Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillje AL, Worlitzer MM, Palm T, Schwamborn JC. Neural stem cells maintain their stemness through protein kinase c zeta-mediated inhibition of TRIM32. Stem Cells. 2011;29:1437–1447. doi: 10.1002/stem.687. [DOI] [PubMed] [Google Scholar]
- 12.Massirer KB, Carromeu C, Griesi-Oliveira K, Muotri AR. Maintenance and differentiation of neural stem cells. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:107–114. doi: 10.1002/wsbm.100. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS. MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 2010;30:14931–14936. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell. 2012;22:1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Palm T, Bahnassawy L, Schwamborn J. miRNAs and neural stem cells: a team to treat Parkinson's disease? RNA Biol. 2012;9:720–730. doi: 10.4161/rna.19984. [DOI] [PubMed] [Google Scholar]
- 17.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao M, Janson Lang AM. Bromodeoxyuridine infused into the cerebral ventricle of adult mice labels nigral neurons under physiological conditions–a method to detect newborn nerve cells in regions with a low rate of neurogenesis. J. Neurosci. Methods. 2009;184:327–331. doi: 10.1016/j.jneumeth.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Battke F, Symons S, Nieselt K. Mayday–integrative analytics for expression data. BMC Bioinformatics. 2010;11:121. doi: 10.1186/1471-2105-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moiola C, De Luca P, Gardner K, Vazquez E, De Siervi A. Cyclin T1 overexpression induces malignant transformation and tumor growth. Cell Cycle. 2010;9:3119–3126. doi: 10.4161/cc.9.15.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc. Natl Acad. Sci. USA. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeo HC, Beh TT, Quek JJ, Koh G, Chan KK, Lee DY. Integrated transcriptome and binding sites analysis implicates E2F in the regulation of self-renewal in human pluripotent stem cells. PLoS One. 2011;6:e27231. doi: 10.1371/journal.pone.0027231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han BW, Feng DD, Li ZG, Luo XQ, Zhang H, Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, et al. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum. Mol. Genet. 2011;20:4903–4915. doi: 10.1093/hmg/ddr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 32.Iwaniuk KM, Schira J, Weinhold S, Jung M, Adjaye J, Muller HW, Wernet P, Trompeter HI. Network-like impact of MicroRNAs on neuronal lineage differentiation of unrestricted somatic stem cells from human cord blood. Stem Cells Dev. 2011;20:1383–1394. doi: 10.1089/scd.2010.0341. [DOI] [PubMed] [Google Scholar]
- 33.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forster E, Tielsch A, Saum B, Weiss KH, Johanssen C, Graus-Porta D, Muller U, Frotscher M. Reelin, disabled 1, and beta 1 integrins are required for the formation of the radial glial scaffold in the hippocampus. Proc. Natl Acad. Sci. USA. 2002;99:13178–13183. doi: 10.1073/pnas.202035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imbeault S, Gauvin LG, Toeg HD, Pettit A, Sorbara CD, Migahed L, DesRoches R, Menzies AS, Nishii K, Paul DL, et al. The extracellular matrix controls gap junction protein expression and function in postnatal hippocampal neural progenitor cells. BMC Neurosci. 2009;10:13. doi: 10.1186/1471-2202-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lahti L, Saarimaki-Vire J, Rita H, Partanen J. FGF signaling gradient maintains symmetrical proliferative divisions of midbrain neuronal progenitors. Dev. Biol. 2011;349:270–282. doi: 10.1016/j.ydbio.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Moriyama M, Fukuhara T, Britschgi M, He Y, Narasimhan R, Villeda S, Molina H, Huber BT, Holers M, Wyss-Coray T. Complement receptor 2 is expressed in neural progenitor cells and regulates adult hippocampal neurogenesis. J. Neurosci. 2011;31:3981–3989. doi: 10.1523/JNEUROSCI.3617-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinheiro PS, Mulle C. Presynaptic glutamate receptors: physiological functions and mechanisms of action. Nat. Rev. Neurosci. 2008;9:423–436. doi: 10.1038/nrn2379. [DOI] [PubMed] [Google Scholar]
- 39.Sheng ZH, Westenbroek RE, Catterall WA. Physical link and functional coupling of presynaptic calcium channels and the synaptic vesicle docking/fusion machinery. J. Bioenerg. Biomembr. 1998;30:335–345. doi: 10.1023/a:1021985521748. [DOI] [PubMed] [Google Scholar]
- 40.Tiscornia G, Izpisua Belmonte JC. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 2010;24:2732–2741. doi: 10.1101/gad.1982910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 42.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin XQ, Barsoum J. Differential cell cycle effects induced by E2F1 mutants. Oncogene. 1997;14:53–62. doi: 10.1038/sj.onc.1200809. [DOI] [PubMed] [Google Scholar]
- 44.Arai Y, Pulvers JN, Haffner C, Schilling B, Nusslein I, Calegari F, Huttner WB. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M, Wu S, Jia J, May WS. JAZ mediates G1 cell cycle arrest by interacting with and inhibiting E2F1. Cell Cycle. 2011;10:2390–2399. doi: 10.4161/cc.10.14.16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu HG, Ren W, Zou L, Wang Y, Jin R, Zhou GP. Direct repression of the human IRF-3 promoter by E2F1. Immunogenetics. 2011;63:189–196. doi: 10.1007/s00251-010-0505-5. [DOI] [PubMed] [Google Scholar]
- 47.Perruisseau-Carrier C, Jurga M, Forraz N, McGuckin CP. miRNAs stem cell reprogramming for neuronal induction and differentiation. Mol. Neurobiol. 2011;43:215–227. doi: 10.1007/s12035-011-8179-z. [DOI] [PubMed] [Google Scholar]
- 48.De Brouwer S, Mestdagh P, Lambertz I, Pattyn F, De Paepe A, Westermann F, Schroeder C, Schulte JH, Schramm A, De Preter K, et al. Dickkopf-3 is regulated by the MYCN-induced miR-17-92 cluster in neuroblastoma. Int. J. Cancer. 2011;130:2591–2598. doi: 10.1002/ijc.26295. [DOI] [PubMed] [Google Scholar]
- 49.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 52.Scheijen B, Bronk M, van der Meer T, Bernards R. Constitutive E2F1 overexpression delays endochondral bone formation by inhibiting chondrocyte differentiation. Mol. Cell. Biol. 2003;23:3656–3668. doi: 10.1128/MCB.23.10.3656-3668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper-Kuhn CM, Vroemen M, Brown J, Ye H, Thompson MA, Winkler J, Kuhn HG. Impaired adult neurogenesis in mice lacking the transcription factor E2F1. Mol. Cell. Neurosci. 2002;21:312–323. doi: 10.1006/mcne.2002.1176. [DOI] [PubMed] [Google Scholar]
- 54.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol. Syst. Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.