Genome integrity of meiotic initial cells is essential for haploid spore formation and consolidates the ploidy stability of successive generations. This study demonstrates that nonlethal defects in Arabidopsis cytokinesis (e.g., that alter callose or sterol synthesis) lead to premeiotic endomitosis and generate tetraploid meiocytes, constituting a route for sexual polyploidization in plants.
Abstract
In sexually reproducing plants, the meiocyte-producing archesporal cell lineage is maintained at the diploid state to consolidate the formation of haploid gametes. In search of molecular factors that regulate this ploidy consistency, we isolated an Arabidopsis thaliana mutant, called enlarged tetrad2 (et2), which produces tetraploid meiocytes through the stochastic occurrence of premeiotic endomitosis. Endomitotic polyploidization events were induced by alterations in cell wall formation, and similar cytokinetic defects were sporadically observed in other tissues, including cotyledons and leaves. ET2 encodes GLUCAN SYNTHASE-LIKE8 (GSL8), a callose synthase that mediates the deposition of callose at developing cell plates, root hairs, and plasmodesmata. Unlike other gsl8 mutants, in which defects in cell plate formation are seedling lethal, cytokinetic defects in et2 predominantly occur in flowers and have little effect on vegetative growth and development. Similarly, mutations in STEROL METHYLTRANSFERASE2 (SMT2), a major sterol biosynthesis enzyme, also lead to weak cytokinetic defects, primarily in the flowers. In addition, SMT2 allelic mutants also generate tetraploid meiocytes through the ectopic induction of premeiotic endomitosis. These observations demonstrate that appropriate callose and sterol biosynthesis are required for maintaining the ploidy level of the premeiotic germ lineage and that subtle defects in cytokinesis may lead to diploid gametes and polyploid offspring.
INTRODUCTION
In order to generate haploid spores, sexually reproducing organisms need to control the genomic ploidy level of their gamete-producing cell lineages by maintaining them in their diploid state. This premeiotic diploid consistency suggests that plants essentially suppress all potential ploidy increasing mechanisms in their reproductive cell lineage. However, despite its biological relevance and the large set of molecular factors regulating cell proliferation and endopolyploidization in somatic tissue, little is known about the molecular mechanism(s) maintaining the basic diploid ploidy level in meiotic precursor cells.
In both animals and plants, the cell-specific multiplication of the nuclear DNA content, also termed endopolyploidization, generally results from two alternative cell cycle processes; namely, endomitosis or endoreduplication (Joubès and Chevalier, 2000). Although both mechanisms are considered alterations of the mitotic cell cycle, there is an essential difference. In contrast with endoreduplication, in which mitotic chromosome condensation and subsequent division are completely absent (Lee et al., 2009), endomitotic cells enter but do not fully complete the mitotic M-phase. As these cells proceed through anaphase but lack nuclear division and cytokinesis (D’Amato, 1984), endomitosis typically causes a doubling of the absolute chromosome number. By contrast, endoreduplication generates polytenal chromosomes, without affecting the total number of chromosomes (Edgar and Orr-Weaver, 2001).
Besides alterations in mitotic cell cycle regulation, endomitotic events are generally induced by a loss of cell wall formation. In plant cell division, cytokinesis comprises four main steps: (1) the construction of the phragmoplast, (2) directed transport and fusion of vesicles at the phragmoplast, (3) centrifugal outgrowth of the cell plate, and (4) fortification of the cell plate into a cell wall (Samuels et al., 1995; Assaad, 2001; Van Damme et al., 2008). Several reports have demonstrated that the alteration of one of these processes induces the formation of multinucleate cells, which, upon nuclear fusion, often yield endomitotic polyploid cells (Mayer et al., 1999; Müller et al., 2002; Strompen et al., 2002). Functional loss of KNOLLE or KEULE, for example, two proteins involved in cytokinetic vesicle fusion (Lauber et al., 1997), causes severe defects in embryonic cell plate formation and consequently generates high numbers of multinucleate and endomitotic cells (Lukowitz et al., 1996; Assaad et al., 2001). However, since the functional loss of most proteins involved in cytokinesis and cell plate formation typically induce seedling lethality, potential effects on further plant development (e.g., reproductive tissues) could not be analyzed.
Callose, a β-1,3-glucan polymer with β-1,6-branches, is synthesized in both sporophytic and gametophytic plant tissues and plays an important role in several developmental processes (Chen and Kim, 2009). In dividing plant cells, callose is deposited at the lumen of the tubulo-vesicular phragmoplast matrix and promotes the maturation of the tubular network into a fenestrated cell plate, hence stabilizing the newly formed cell plate. Moreover, as phragmoplastin is one of the subunits of the callose synthase complex (Hong et al., 2001b), callose-driven membrane dynamics are thought to be directed by an associated squeezing of phragmoplastin polymers (Verma and Hong, 2001). Callose also plays an important role in microspore maturation, exine patterning, and pollen tube formation (Dong et al., 2005) and accumulates in plasmodesmata, where it regulates intercellular communication by regulating the symplasmic movement of signaling macromolecules (Guseman et al., 2010). Additionally, in the case of mechanical injury, pathogen attack, and metal toxicity, the deposition of callose protects the plant by sealing sieve plates in dormant phloem or by forming physical plugs (e.g., papillae) at affected sites (Jacobs et al., 2003).
In Arabidopsis thaliana, like in other plants, callose is synthesized from its precursor UDP-Glc by a class of enzymes, termed callose synthases or glucan synthase-like enzymes. In the Arabidopsis genome, 12 genes encoding putative callose synthase have been identified (At-GSL1 to At-GSL12) (Hong et al., 2001a). Although most callose synthases are essential for pollen development or fertilization (Dong et al., 2005; Enns et al., 2005; Nishikawa et al., 2005; Töller et al., 2008), the normal sporophytic growth phenotype of the corresponding single mutants suggests a high level of genetic redundancy during sporophytic development (Chen et al., 2009). In mutant forms of GLUCAN SYNTHASE-LIKE8 (GSL8), however, severe defects in somatic cell plate formation and patterning were observed, indicating that GSL8 is essential for sporophytic plant development. Indeed, genetic studies demonstrated that GSL8 enables the phragmoplast-directed synthesis and deposition of callose at the cell equator and thereby stabilizes the establishment of the newly formed cell plate (Chen et al., 2009; Thiele et al., 2009). Moreover, GSL8 is also required for the deposition of callose at symplasmic plasmodesmatal channels and the concomitant regulation of cell-to-cell communication (Guseman et al., 2010).
In this study, we identified an Arabidopsis mutant, named et2, which produces diploid spores through the ectopic induction of premeiotic endomitosis. We demonstrate that et2 has a mutation in GSL8 and consequently exhibits defects in stomatal distribution, epidermal patterning, and cell wall formation. However, in contrast with previously reported gsl8 mutants, which are all seedling lethal, developmental defects in et2 are relatively mild and allow the plants to reach the reproductive phase. Using cytological approaches, we demonstrate that defects in et2 cell wall establishment are most prominent in flower organs and lead to endomitotic polyploidization in petals and other reproductive tissues, including the premeiotic germ cells. Similar developmental alterations in cell plate formation and premeiotic ploidy stability were also observed in Arabidopsis mutants frill1 and cvp1-3, carrying allelic mutations in STEROL METHYL TRANSFERASE (SMT2). The double sterol biosynthesis mutant frill1 smt3-1 shows more severe cytokinesis defects and elevated endoploidy levels in its reproductive structures, reinforcing a tight correlation between the ploidy consistency of the reproductive system and appropriate control over cytokinesis.
RESULTS
The Arabidopsis Mutant et2 Produces Diploid Male and Female Spores
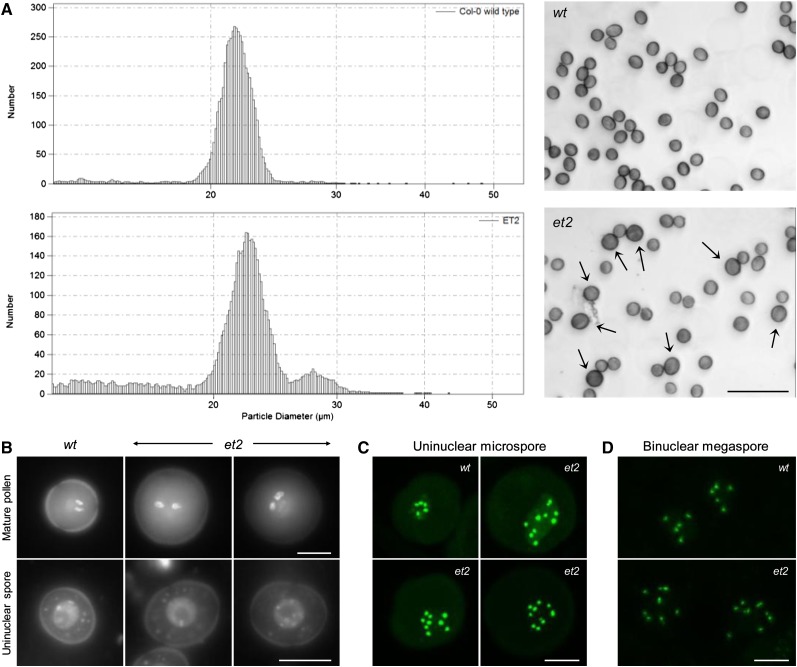
In the search for genes that regulate the haploid genome dosage of male gametes, 4800 ethyl methanesulfonate (EMS)–mutagenized Arabidopsis M2 plants were screened for the production of diploid and/or polyploid pollen. Based on the correlation between pollen size and gametophytic ploidy level (Altmann et al., 1994), individual plants were monitored for enlarged pollen formation. Mutant enlarged tetrad2 (et2) showed a significant number of larger pollen grains at a frequency of 5.3% ± 0.4% (Figure 1A). To verify the nuclear configuration, pollen DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Enlarged and normal sized et2 pollen exhibited the typical wild type–like tricellular nuclei configuration with one vegetative nucleus and two highly condensed sperm cells (Figure 1B). However, in contrast with normal sized pollen, enlarged spores clearly showed more intensively stained, enlarged nuclei, suggesting an increased gametophytic DNA content. Similar observations were made at earlier stages of spore development (Figure 1B), indicating that the increase in DNA content occurred prior to or early in male sporogenesis.
Figure 1.
The Arabidopsis Mutant et2 Produces Diploid Male and Female Gametes.
(A) Volume-based pollen size distribution and microscopy analysis of wild-type (wt) and et2 mature pollen grains. Larger et2 pollen are indicated by arrows. Bar = 100 µm.
(B) DAPI staining of wild-type and et2 spores at the mature tricellular pollen stage and the uninuclear microspore stage. Bars = 10 µm.
(C) In vivo quantification of the gametophytic chromosome number in wild-type and et2 uninuclear microspores using ProWOX2-CENH3-GFP–mediated centromere labeling. Bar = 5 µm.
(D) ProWOX2-CENH3-GFP–mediated chromosome quantification in wild-type and et2 ovules at the binuclear megaspore stage. Bar = 5 µm.
[See online article for color version of this figure.]
Similarly to other Arabidopsis 2n pollen-producing mutants (e.g., jason and atps1), et2 showed a bimodal pollen size distribution (Figure 1A), with the low and the high value peak corresponding to haploid and diploid spores, respectively (De Storme and Geelen, 2011). To quantify the gametophytic chromosome number in et2 enlarged spores, we introgressed the recombinant green fluorescent protein (GFP) marker ProWOX2CENH3-GFP that specifically labels the centromeres in nuclei of developing gametes (De Storme and Geelen, 2011). In diploid control plants harboring the centromere marker, uninuclear microspores generally contain five centromeric dots (86.3% ± 9.3%; n = 383), reflecting their haploid genome content (Figure 1C). In accordance, regular-sized et2 spores showed the haploid-specific number of five centromeric dots (89.0% ± 8.1%; n = 473). For enlarged et2 spores, on the other hand, the gametophytic chromosome number appeared highly variable and ranged from six to 19 centromeric dots per microspore (n = 183) (Figure 1C), indicating an increased number of chromosomes. A substantial part of these enlarged spores (34.5% ± 12.6%) contained the euploid number of 10 centromeric dots, allowing us to conclude that et2 generates a significant level of diploid spores.
In the female germ lineage, expression of the centromeric GFP marker is generally lower and only allows for a distinct observation of centromeric dots at the postmeiotic binuclear stage. In this stage, wild-type ovules typically contain two nuclei that contain five centromeres each (n = 34). In et2, on the other hand, besides the presence of haploid ovules, also female gametophytes having diploid (2 × 10 centromeres) or aneuploid chromosome numbers were observed (Figure 1D).
These observations indicate that the gametophytic ploidy level in both et2 male and female gametogenesis is affected, with the prevalent formation of diploid gametes. Moreover, as both et2 selfing and reciprocal backcrosses (with diploid wild type) resulted in the spontaneous formation of polyploid offspring (Table 1), both male and female diploid gametes were considered functionally viable and able to confer sexual polyploidization events.
Table 1. Frequency of Polyploid Progeny in et2 Selfing and Crosses as a Female Donor Plant.
| Progeny Plants (%) |
|||||
|---|---|---|---|---|---|
| Line | n | 2× | 3× | 4× | Aneuploid |
| et2 selfing | 84 | 94.0 | 2.4 | 1.2 | 2.4 |
| et2 × wild type | 49 | 95.9 | 4.1 | 0.0 | 0.0 |
Tetraploid Meiocytes in et2 Are Indicative of Premeiotic Endomitosis
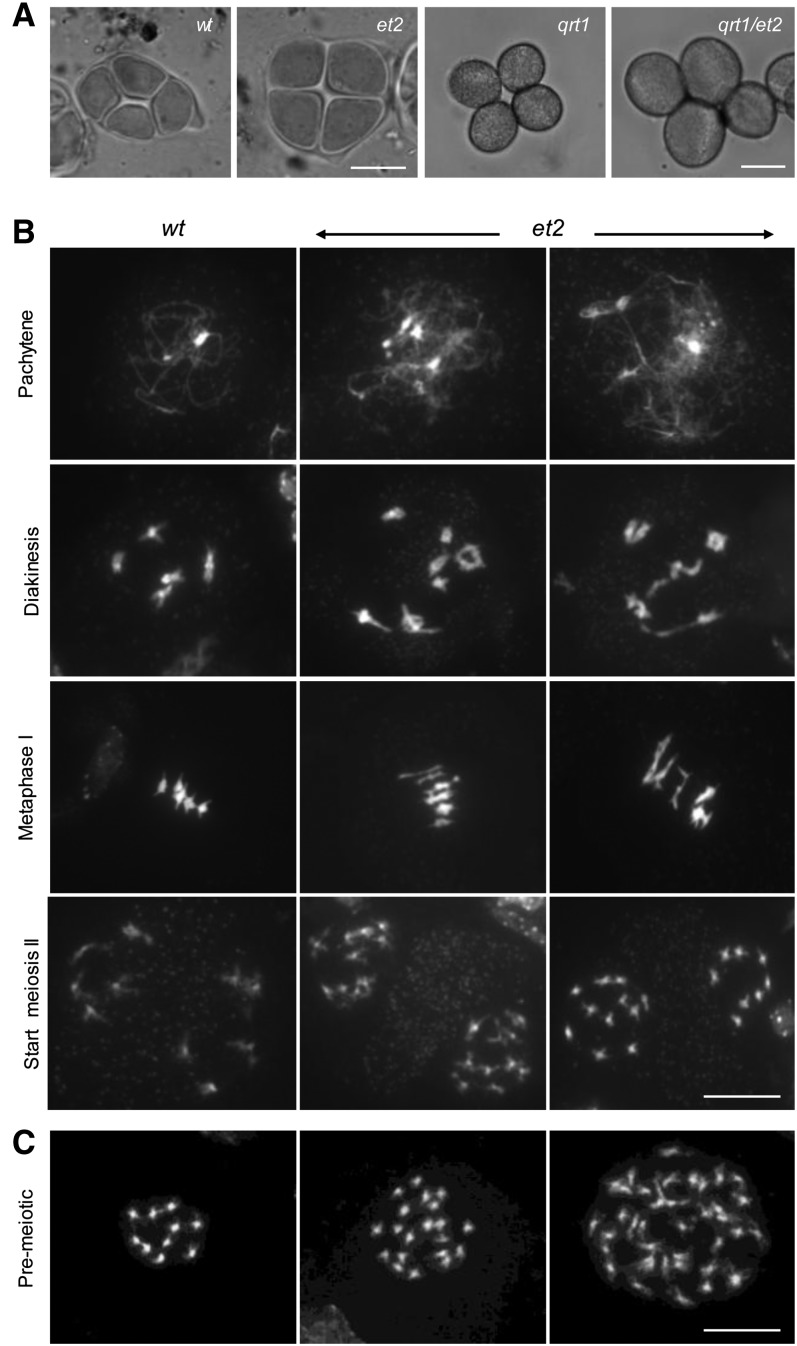
Because diploid gametes are mostly formed through a restitution of the meiotic cell cycle (Brownfield and Köhler, 2011), we analyzed the outcome of male meiosis in et2. In wild-type male sporogenesis, tetrad stage meiotic products are highly uniform and always contain four equally sized haploid microspores (100%, n > 250) (Figure 2A). In et2, tetrads appeared highly similar to the wild type, except for a subpopulation (9.0% ± 5.4%, n = 455) that was substantially larger, equaling the size of tetrads generated by tetraploid Arabidopsis plants (Figure 2A). Based on this finding, we presumed that enlarged et2 tetrads contain polyploid spores and result from a genome duplication event occurring either before or during meiosis.
Figure 2.
Cytological Analysis of et2 Male Meiosis.
(A) Tetrad stage meiocytes in wild-type (wt) and et2 male sporogenesis (bar = 10 µm) and mature pollen of the wild type and et2 in the qrt1-1−/− mutant background (bar = 20 µm).
(B) DAPI-stained chromosome spreading of wild-type and et2 male meiosis shows the presence of tetraploid meiocytes in different stages of et2 meiosis. Bar = 10 µm.
(C) Chromosome spreads of premeiotic cells in wild-type and et2 flower buds. The presence of polyploid cells with condensed chromosomes indicates endomitotic polyploidization. Bar = 10 µm.
To assess the potential occurrence of postmeiotic genome duplication in the formation of et2 diploid gametes, et2 was crossed into the qrt1-1−/− mutant background. QUARTET1 (QRT1) encodes a pectin methylesterase that is involved in the degradation of the pollen mother cell wall at the end of meiosis. As qrt1-1−/− mutants do not degrade this cell wall, microspores are kept together in their meiotic configuration and mature pollen are released in groups of four (Figure 2A) (Francis et al., 2006). In qrt1-1−/−et2−/− male sporogenesis, two types of tetrads were observed: regular-sized tetrads (∼90%) and enlarged tetrads (∼10%) (Figure 2A). In both types of tetrads, all four spores appeared equally sized, and tetrads with only a partial number of enlarged spores were not observed. Based on these data, we conclude that et2 diploid gametes do not result from postmeiotic genome duplication but rather from ectopic doubling during or before meiosis.
To determine the developmental stage at which et2 sporogenesis performs ectopic genome duplication (meiotic or premeiotic), DAPI-stained chromosome counts were performed at all stages of male meiocyte development. Similar to the wild type, et2 showed a normal progression and chromosome behavior throughout meiotic cell division (Figure 2B). However, besides the predominant presence of diploid meiocytes, tetraploid cells also were observed. Indeed, in contrast with the normal set of 10 chromosomes typically observed at the start of meiosis II (2 × 5 chromosome), 10.7% of et2 MII meiocytes displayed 20 chromosomes, indicating that the meiocyte’s chromosomes had undergone duplication. Additionally, in meiosis I, a similar frequency of meiocytes having the tetraploid number of 20 chromosomes was observed (7.4% in diakinesis and 12.5% in metaphase I) (Figure 2B). Quantitative analysis demonstrated that the average frequency of tetraploid meiocytes in all stages of et2 male meiosis was highly similar and did not increase, indicating that the reproductive genome doubling events in et2 occur prior to meiosis, in the pollen mother cell–producing archesporal cell lineage. Moreover, as endoreduplication does not duplicate the absolute number of chromosomes, but endomitosis does, tetraploid meiocytes were suggested to result from a premeiotic endomitotic polyploidization event. This was confirmed by the presence of endomitotic cells (polyploid cells with condensed chromosomes) in somatic tissue of premeiotic flower buds (Figure 2C).
Defects in et2 Cell Wall Establishment Lead to Ectopic Endomitosis Primarily in Floral Organs
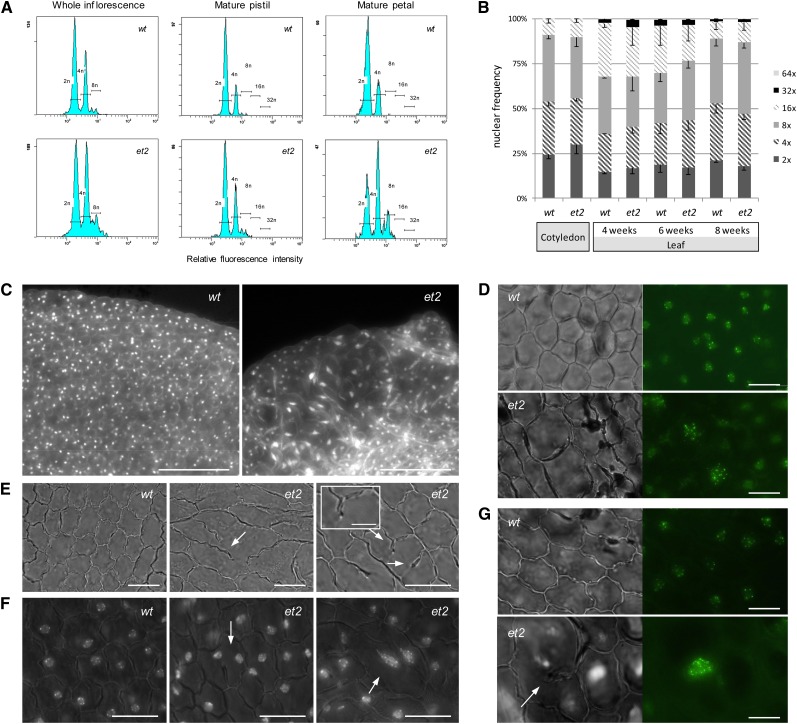
The presence of polyploid cells in premeiotic reproductive tissues prompted us to assess potential polyploidization events in other et2 flower organs. In whole inflorescences, only a small, yet significant increase in polyploidy was observed in the et2 mutant. Indeed, in contrast with wild-type inflorescences, which only contain diploid and tetraploid cells (Galbraith et al., 1991), et2 showed an increased level of tetraploid nuclei and an additional subset of octaploid nuclei (Figure 3A). Organ-specific analyses demonstrated that this increase in et2 flower endopolyploidy is mainly attributed to the accumulation of tetraploid and octaploid cells in the petals. In other floral organs (e.g., pistils), we did not observe a significant change in ploidy, although some of the samples analyzed contained more polyploid cells.
Figure 3.
Endoploidy Analysis of et2 Flower Organs and Other Somatic Tissues.
(A) Flow cytometric analysis of endoploidy levels in wild-type (wt) and et2 flower organs.
(B) Comparison of endopolyploidy distribution in wild-type and et2 cotyledons (of 7-d-old seedlings) and somatic leaf tissue.
(C) DAPI staining of mature petals shows distinct regions of polyploid cell clusters in et2 petals, particularly at the distal end. Bars = 100 µm.
(D) Somatic ploidy determination of single nuclei in wild-type and et2 petals, using transgenic ProSD-CENH3-GFP–mediated centromere labeling. Bars = 10 µm.
(E) Bright-field microscopy of epidermal cell wall morphology in mature petals. Bars = 20 µm. Inset shows a cell wall stub, indicative of cytokinetic defects. Bar = 5 µm.
(F) Nuclear DNA staining (DAPI) of wild-type and et2 petals at anthesis. Bars = 20 µm.
(G) Centromere labeling using ProSDS-CENH3-GFP in mature wild-type and petals. Note that the endomitotic polyploid cell in et2 shows defects in cell plate formation (arrow). Bars = 20 µm.
[See online article for color version of this figure.]
To monitor potential increases in et2 leaf endoploidy, DNA flow cytometry was performed on 10-d-old cotyledons and 4-, 6-, and 8-week-old leaves. In all stages of plant development, the endoploidy profile of et2 somatic tissue appeared highly similar to the wild type (Figure 3B), indicating that ectopic polyploidization did not occur or was undetectable in the constitutive endopolyploidy background of the rosette leaves. Based on these data, we conclude that ectopic polyploidization in et2 is more prominent in the reproductive flower organs and less abundant, or completely absent, in somatic tissues.
To verify whether the endoploidy increase in et2 petals is also attributed to ectopic endomitosis, we analyzed ProSDSCENH3-GFP expression in the et2 background. As this centromere-labeling construct is expressed throughout all plant organs, fluorescent GFP detection allows for the in vivo quantification of the absolute number of chromosomes in various types of cells and organs, including petals. In wild-type petals, nuclei always show the presence of seven to 10 centromeric dots, reflecting a basic diploid status (Figure 3D; see Supplemental Figure 1 online). In et2, on the other hand, petal nuclei occasionally display significantly more centromeric dots (ranging from 14 to 24), indicative of tetraploid and/or polyploid cells (Figure 3D; see Supplemental Figure 1 online). Moreover, based on the notion that endoreplication does not alter the cell-specific number of centromeres (Lermontova et al., 2006), we conclude that polyploid et2 petal cells originate from endomitotic polyploidization that typically involves a doubling and associated separation of centromeres. Besides petals, other floral organs (e.g., stamens and sepals) also displayed cells that contain an increased number of fluorescent CENH3-GFP signals (see Supplemental Figure 2 online), indicating that basically all et2 flower organs undergo ectopic induction of endomitosis.
To determine the spatial patterning of et2 endomitotic polyploidization events, petals were cleared and stained with DAPI to visualize the nuclei. In contrast with wild-type petals, which always display homogeneously sized cells that contain equally sized nuclei (diploid consistency), et2 petals show the additional presence of enlarged cells containing enlarged, polyploid nuclei (Figure 3C). In most cases, these polyploid cells form variably sized cell clusters stochastically spread along the petal leaf. Interestingly, inspection of mature et2 anthers also showed that polyploid spores were spatially clustered in the anther locule (see Supplemental Figure 3 online), suggesting that endomitotic initials were formed early in development and consequently created a clustered polyploid cell lineage upon maturation. In support of this cell lineage hypothesis, in vivo centromere labeling using the ProSDS-CENH3-GFP construct revealed that early premeiotic et2 flower buds often contained single enlarged endomitotic cells scattered along the petal leaf, whereas in later flower stages, large areas of clustered polyploid cell were found (see Supplemental Figure 4 online).
In search of a cytological defect that induces ectopic endomitosis, we found that et2 petals exhibit distinct alterations in cell plate formation and cross-wall establishment. Indeed, in contrast with the fully enclosed cell walls in wild-type epidermal petal tissue, et2 occasionally showed the presence of incomplete cell walls and cell wall stubs (Figure 3E). These cell wall aberrations were rather rare (<1% of all cells) and showed a higher prevalence at the apical leaf margin. In addition, we found several et2 petal cells that formed multinuclear syncytia containing three or even more diploid nuclei (Figure 3F). Moreover, since uninuclear endomitotic polyploid petal cells often exhibited partial cell walls and cell wall stubs (Figure 3G), we concluded that endomitotic events in et2 originate from defects in cell plate formation and subsequent fusion of syncytial nuclei.
Cell Wall Defects in et2 Are Caused by a Splice Site Mutation in GSL8
The endomitosis-causing mutation in et2 was identified through a chromosome landing approach followed by SHOREmap analysis. After crossing et2 (Columbia-0 [Col-0]) into the Landsberg erecta background, resulting F1 plants were selfed to generate an F2 mapping population. Phenotypic pollen analysis demonstrated that the F1 plants produced normal pollen, whereas the subsequent F2 mapping population showed a 1:3 segregation of the mutant phenotype, indicating that the et2 allele is sporophytic monogenic recessive.
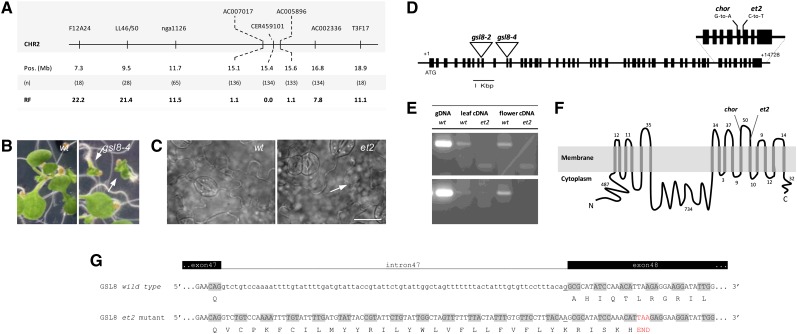
Cumulative genotyping of increasing numbers of F2+ (et2 phenotype positive) plants confined the et2 locus to a region of 0.5 Mb (15.1 to 15.6 Mb) on the bottom arm of chromosome II (Figure 4A). In a similar way, OFFSHORE sequencing of a pooled set of 500 F2+ plants also located the mutation on the bottom arm of chromosome II with a calculated maximum likelihood at 15.4 Mb (see Supplemental Figure 5 online). Using SHOREmap analysis, we identified four mutations in the 15.1 to 15.6 Mb interval on chromosome 2 (see Supplemental Table 1 online) with the C-to-T base pair switch in the GSL8 gene closest to the maximum likelihood peak. As the other three mutations occurred either in an untranslated gene sequence or caused a conservative amino acid switch, we considered the mutation in GSL8, which disrupts the splice acceptor site at intron 47, as the prime candidate for the described defects in et2.
Figure 4.
Positional Cloning of ET2.
(A) Positional mapping of the ET2 locus on the bottom arm of chromosome II. Locations of molecular markers, the number of recombinants (n) and the corresponding recombination frequency (RF) are indicated.
(B) Morphology of 7-d-old seedlings in the wild type (wt) and heterozygous gsl8-4 mutant background (arrows indicate seedling lethal phenotype).
(C) Cell wall pattern and stomatal distribution in wild-type and et2 7-d-old cotyledons. Note the dimeric stomatal cluster and the aberrant cell wall (arrow) in the et2 background. Bar = 20 µm.
(D) GSL8 structure with introns (lines) and exons (boxes) and the position of T-DNA insertions (triangles) and relevant EMS-induced mutations (i.e., et2 and chorus).
(E) RT-PCR analysis using intron-specific primers to monitor the putative retention of intron 47 in et2 cDNA. Three biological replicates were performed and gave a similar result. Sequences of primers used are listed in Supplemental Table 4 online, and the corresponding genomic regions in the GSL8 open reading frame are graphically represented in Supplemental Figure 10 online. gDNA, genomic DNA.
(F) Topology of the GSL8 protein.
(G) Predicted alteration of GSL8 cDNA splicing through the retention of intron 47 in et2 causes the premature establishment of a STOP-codon (red) and results in a truncated GSL8 protein with altered C-terminal amino acid composition.
[See online article for color version of this figure.]
We next characterized the phenotype of available T-DNA insertion alleles for both GSL8 and the other three candidate genes. For all candidates, except for GSL8, homozygous T-DNA lines showed a normal pollen phenotype and a wild-type-like petal cell wall patterning, indicating that they do not underlie the et2-specific defects. Strikingly, for GSL8, mature homozygous T-DNA lines were never retrieved (see Supplemental Table 2 online). As this suggested that gsl8 is gametophytically or sporophytically lethal, we phenotypically analyzed progeny of heterozygous gsl8 T-DNA lines and found that all these lines exhibit a seedling lethal phenotype that segregates at a 1:3 ratio (Figure 4B; see Supplemental Table 3 online). Based on these data, we concluded that GSL8 is essential for sporophytic growth and that loss of GSL8 functionality induces a high level of seedling lethality. Indeed, as demonstrated in previous reports, loss of GSL8 leads to severe defects in cytokinesis and cell wall establishment in young developing seedlings (e.g., cotyledons) and consequently induces premature growth arrest and seedling abortion (Chen et al., 2009; Thiele et al., 2009). Strikingly, in the epidermal pavement cell layer of et2 cotyledons, we occasionally observed cells with incomplete cell walls (Figure 4C, arrow), similar to what has been described for the GSL8-deficient mutant chorus (Guseman et al., 2010) and other gsl8 mutants. Moreover, as we additionally observed a high number of endomitotic cells in the roots and cotyledons of the GSL8 chorus mutant (see Supplemental Figure 6 online), we concluded that the defects in cell wall formation and the associated induction of ectopic endomitosis in et2 is caused by the earlier described, splice site–specific mutation in GSL8.
Loss of GSL8 function reduces the deposition of callose in root hair tips and plasmodesmata and induces correlated aberrations in root hair morphology and epidermal patterning (e.g., stomatal clustering), respectively (Guseman et al., 2010). In accordance, we found that et2 root hairs show a lack of callose staining with associated defects in branching (see Supplemental Figure 7 online) and additionally noticed that et2 displays stomatal clustering and an enhanced proliferation of meristemoid cells in cotyledons and first leaves, indicative of a reduced level of callose at the plasmodesmata (see Supplemental Figure 8 online).
Arabidopsis GSL8/MASSUE/CHORUS spans a genomic region of 14,433 bp, consisting of 50 exons with a coding sequence region of 5712 bp, and encodes a large integral membrane protein of 1904 amino acids with 16 predicted transmembrane helices (TMHs). GSL8 bears an IPR003440 glycosyl transferase domain and catalyzes the formation of (1,3)-β-glucan polymers (e.g., callose) from the precursor UDP-Glc. In the et2 mutant, GSL8 shows a C-to-T base pair shift at the splice acceptor site at position 13,929 (Figure 4D), inducing the specific retention of intron 47 in posttranscriptional splicing (Figure 4E). This change in GSL8 transcript processing is predicted to cause a frameshift in the open reading frame leading to the establishment of a premature stop codon (TAA) 17 bp downstream of the end of intron 47 (Figure 4G). As a result, the deduced protein sequence starting from the end of exon 47 (197 amino acids in the wild type) is replaced by a shortened version of 33 amino acids, producing a truncated protein that lacks the five terminal TMHs (encoded by exons 48 to 50) (Figure 4F). Strikingly, the GSL8 mutant allele chorus was also predicted to produce a truncated protein lacking the same terminal TMHs, but additionally in chorus a major part of the sixth extracytoplasmic hydrophilic loop is also deleted (Figure 4F). Quantitative PCR expression analysis demonstrated that GSL8 expression was unaltered in et2, indicating that defects in cell wall formation were not caused by an altered GSL8 transcript level, but rather by a structural change in the C-terminal part of the protein. Moreover, in comparison to the extreme dwarfism in chorus seedlings, et2 plants showed normal growth and development, indicating that the et2-specific alterations in the GSL8 transcript sequence only cause a mild change in GSL8 activity and/or stability.
et2 Shows Homeotic Flower Alterations and Defects in Petal Vasculature
In addition to the ectopic formation of polyploid cell regions, et2 flowers often exhibited an altered morphology, with serrated petal margins, premature bud opening, and clear aberrations in petal vasculature (Figure 5). In contrast with wild-type petals, in which vascular patterning always displays four fully enclosed loops of xylem strands emanating from the central midvein, vascular loops in et2 petals were not fully closed and consequently displayed alterations in vein continuity.
Figure 5.
Morphological Analysis of et2 Flower Structure and Petal Vasculature.
Morphological analysis of et2 flower structures shows frilled petal margins and homeotic flower defects, similar as in the SMT2 allelic mutant frill1. Bar = 500 µm.
[See online article for color version of this figure.]
Besides these petal-specific defects, et2 flowers also displayed homeotic organ transformations and alterations in the spacing of floral organs. Unlike wild-type flowers, in which floral organs are uniformly spaced within the four concentric whorls, et2 petals and anthers often appeared improperly localized, showing regions with overlapping and regions with no flower structures (Figure 5). In addition, in late developmental stages, we found that et2 flowers display defects in organ identity and show homeotic transformations, such as extra petals or fused stamens.
Cell Wall Alterations in smt2 Sterol Biosynthesis Mutants Also Induce Ectopic Endomitosis
Based on the close correlation between the endomitotic cells and serrated margins in et2 petals, we presume that loss of diploid consistency in Arabidopsis flowers is easily detected at the macroscopic level by assessing petal morphology. In search of other factors that regulate flower-specific ploidy maintenance, both serrated petal margins and premature bud opening were also retrieved in the Arabidopsis mutants frill1 and cvp1-3 (Figure 5). Both mutants are defective in SMT2, a key enzyme of the sterol biosynthesis pathway. Together with the functionally redundant SMT3, SMT2 acts at a branching point promoting the synthesis of free membrane sterols (sitosterol and stigmasterol) and negatively regulating brassinosteroid (BR) biosynthesis (Carland et al., 2002).
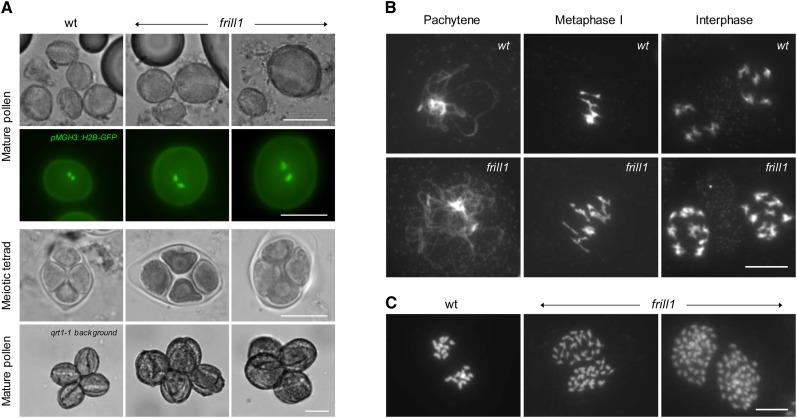
Based on the phenotypic petal similarity to et2, both smt2 mutants were screened for the presence of diploid gametes. Microscopy analysis demonstrated that both frill1 and cvp1-3 produce a significant number of enlarged pollen, containing substantially larger nuclei compared with haploid control spores (Figure 6A). Using the gametophytic ploidy marker ProWOX2-CENH3-GFP, we found that enlarged spores in frill1 contain an increased number of chromosomes (ranging from 6 to 19; 43.5% ± 6.8% has 10 centromeric dots), indicating that frill1 produces both diploid and aneuploid spores (see Supplemental Figure 9 online). Furthermore, in accordance with the formation of polyploid gametes, both frill1 and cvp1-3 generate enlarged tetrads and tetraploid meiocytes (Figures 6A and 6B), indicating that gametophytic polyploidization in smt2 mutants is the result of premeiotic endomitosis (Figure 6C).
Figure 6.
Cytological Analysis of frill1 Male Sporogenesis.
(A) Bright-field microscopy of mature pollen isolated from wild-type (wt) and frill1 flowers at anthesis and fluorescent visualization of sperm cells in mature pollen grains using the ProMGH3-H2B-GFP construct. Analysis of the male meiotic products in wild-type and et2 anthers through cytological tetrad stage meiocyte examination and mature pollen grain observations in the qrt1-1−/− mutant background. Bars = 20 µm.
(B) DAPI-stained meiotic chromosome spreads of wild-type and frill1 male sporogenesis at different stages of the meiotic cell cycle. Bar = 10 µm.
(C) Chromosome spreading of somatic cells in premeiotic frill1 flower buds shows the ectopic formation of endomitotic polyploid cells. Bar = 5 µm.
[See online article for color version of this figure.]
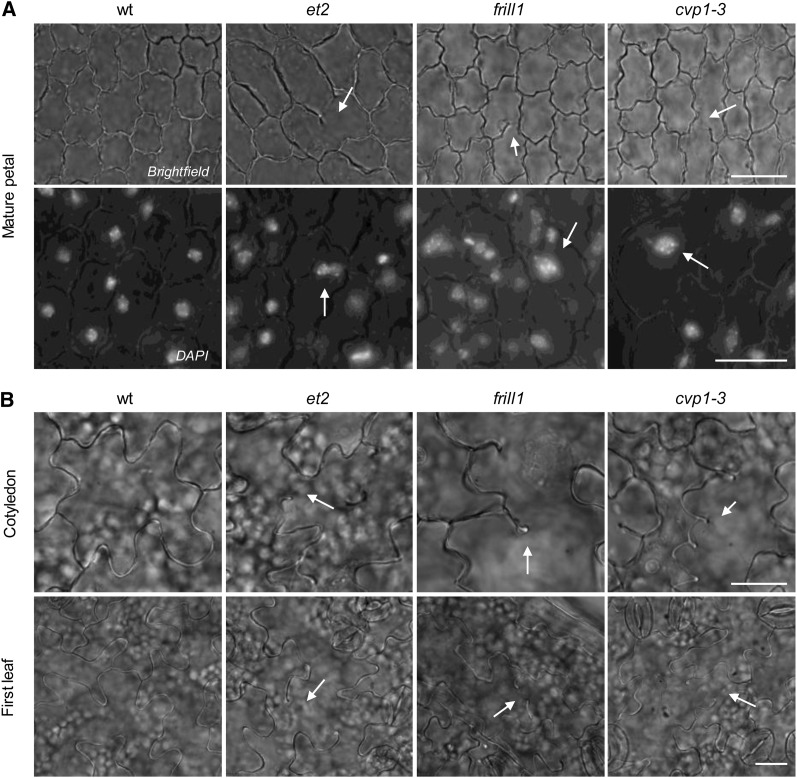
Similarly to et2, frill1 and cvp1-3 flower organs (e.g., petals and sepals) also display endomitotic polyploid cells and associated defects in cell wall establishment (Figure 7A). Moreover, as enlarged polyploid cells in SMT2-deficient flowers occasionally show impartial cell walls, endomitotic cells are presumed to originate from the spontaneous fusion of syncytial nuclei in cell plate–defective multinucleate cells. Defects in cell plate formation were also detected in the epidermal cell layer of smt2 cotyledons and leaves (Figure 7B), albeit at a lower frequency (<1%). In conclusion, these observations indicate that SMT2 and the associated synthesis of sterols are essential for cellular cross-wall formation in several stages of plant development and play an important role in the maintenance of the ploidy stability of the reproductive germ lineage.
Figure 7.
Cytological Analysis of Epidermal Cell Walls in et2 and smt2 Petals, Cotyledons, and Leaves.
(A) Epidermal cell wall morphology and nuclear DNA staining in mature petals of the wild type (wt), et2, and the SMT2 mutant alleles frill1 and cvp1-3. Arrows indicate aberrations in cell wall formation and the ectopic presence of polyploid nuclei. Bars = 10 µm.
(B) Epidermal cell wall morphology in cotyledons of 7-d-old seedlings and in emerging leaves of 14-d-old seedlings. Arrows indicate defects in cell wall formation, such as partial cell walls and cell wall stubs. Bars = 10 µm.
The GSL8 Mutant et2 Shows Normal Levels of Free Sterols
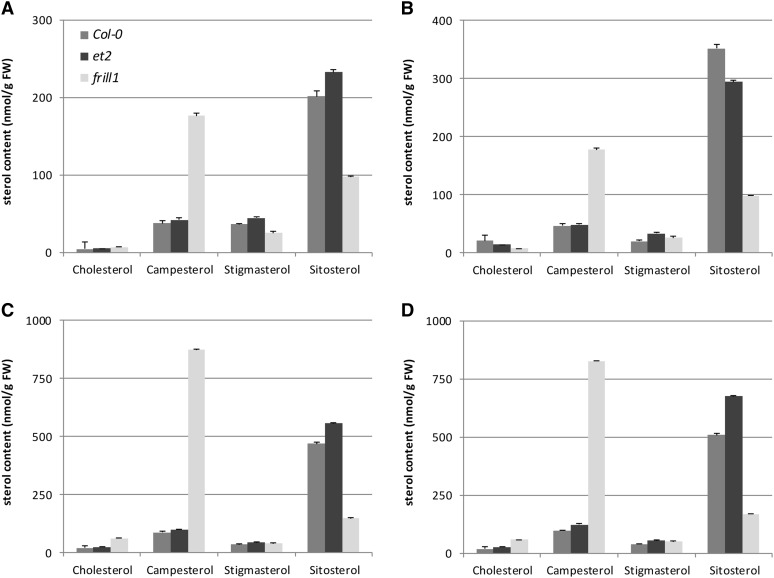
Both frill1 and cvp1-3 demonstrate that an altered sterol profile induces endomitotic polyploidization in several flower organs, similar as in the GSL8 hypomorph et2. Although GSL8 has not been reported to be involved in sterol metabolism, we investigated whether the endomitotic polyploidization events in et2 flowers are caused by an indirect change in free sterols. As a reference, we analyzed sterol levels in frill1 and wild-type seedlings, rosette leaves, whole inflorescences, and mature flowers and compared the cholesterol, campesterol, stigmasterol, and sitosterol content with that of et2. In frill, we observed a strong increase in campesterol together with a significant decrease of sitosterol in all tissues analyzed (Figure 8). Sterol levels in et2, on the other hand, did not substantially deviate from the wild type. The slight increase in C24-ethyl sterols together with the concomitant increase in campesterol indicates that the minor differences in the et2 sterol profile are caused by an alteration of the total sterol content and not by a shift in sterol composition. In conclusion, these results indicate that ectopic endomitosis in et2 flowers, unlike frill1 and cvp1-3, is not caused by an alteration of the plant’s sterol profile.
Figure 8.
Quantification of Sterol Levels
Quantitative analysis of free sterols in 7-d-old seedlings (A), rosette leaves of 3-week-old plants (B), whole inflorescences (C), and mature flowers (D) in wild-type control (Col-0), et2, and frill1. For each sample, three biological repeats were analyzed and error bars represent sd. FW, fresh weight.
Double smt2 smt3 Sterol Mutants Have an Increased Gametophytic Ploidy Level
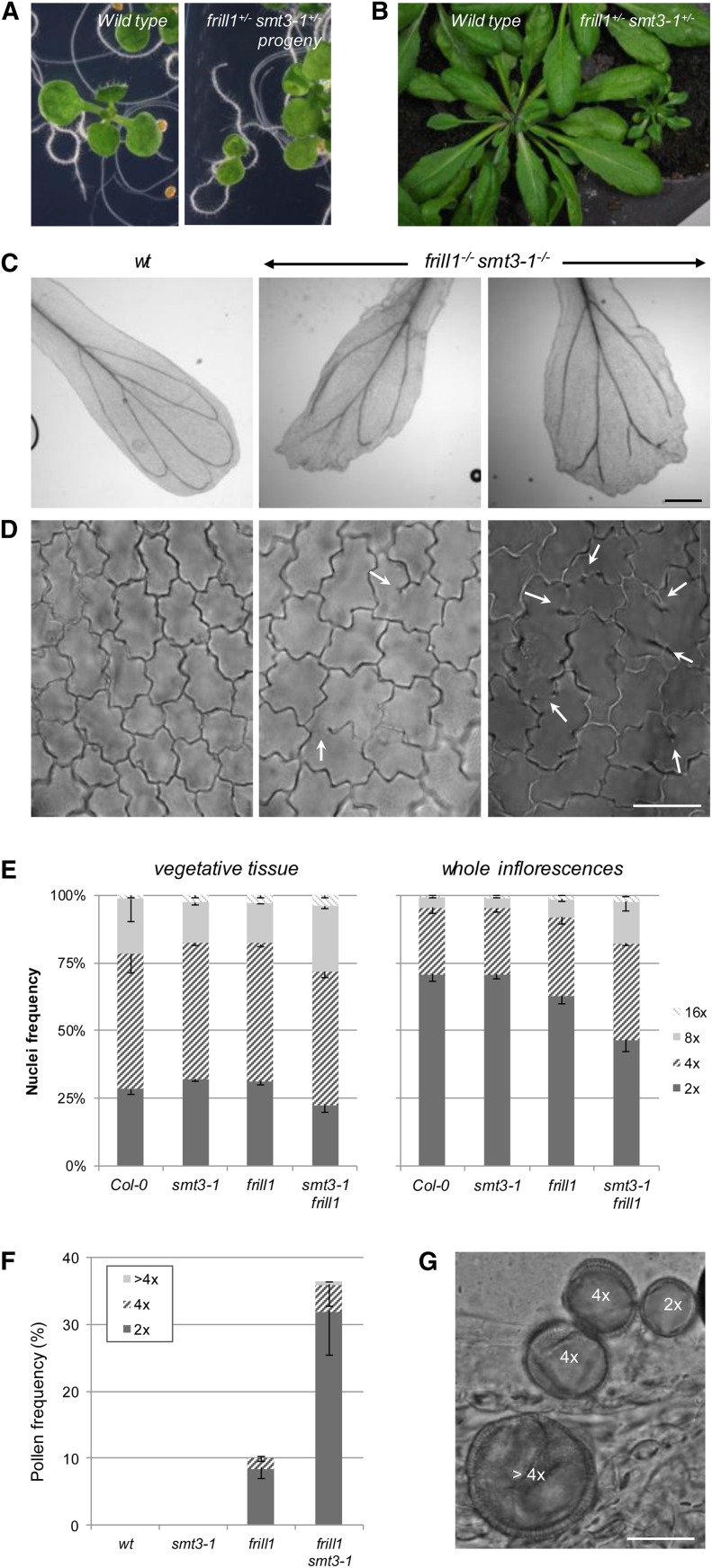
Both et2 and the SMT2 sterol mutants suggest that mild defects in cytokinesis may lead to endomitotic polyploidization and that reproductive structures are particularly sensitive to cytokinetic aberrations, typically inducing diploid and polyploid gamete formation. To confirm this, we next assessed both the somatic and gametophytic ploidy level of double frill1 smt3-1 mutants and compared this with the corresponding single mutants. Since SMT3 acts redundantly with SMT2 at the phytosterol-BR branching point to promote sterol biosynthesis (Carland et al., 2002), the combinatorial loss of SMT2 and SMT3 substantially reduces the main phytosterol content compared with the single mutants (Carland et al., 2010). As a result, smt2 smt3 double mutants show enhanced morphological defects, such as discontinuous cotyledon vein patterning, reduced root growth, loss of apical dominance, and enhanced homeotic flower transformations. In line with this, we found that frill1 smt3-1 plants are significantly smaller and display distinct alterations in morphology, including a reduced stature and irregular-shaped leaves (Figures 9A and 9B). In addition, frill1 smt3-1 petals appeared highly disorganized with extreme serrated leaf margins and broad areas of enlarged cells (Figure 9C). Cytological analysis of the epidermal cell layer of frill1 smt3-1 leaves and petals revealed an increased incidence of cytokinetic alterations, as observed from the enhanced level of cell wall stubs (±20% of epidermal cells) and enlarged cells (Figure 9D). Similar as in et2 and frill1, flow cytometric ploidy analysis demonstrated that frill1 smt3-1 inflorescences have an enhanced endoploidy pattern, showing more tetraploid and octaploid nuclei compared with the wild type (Figure 9E). Interestingly, the endoploidy pattern in frill1 smt3-1 inflorescences appeared substantially higher compared with the single mutants, indicating that the enhanced level of cytokinetic defects correlates with an increased endoploidy level. In line with this, frill1 smt3-1 mutants also showed a significant increase in larger pollen grain formation (Figures 9F and 9G), indicating that polyploidization is also enhanced in the gametophytic germ lineage. Strikingly, in comparing the somatic endoploidy level (e.g., in the rosette leaf), both frill1 smt3-1 single and double mutants showed a ploidy pattern similar to the wild type (Figure 9E). Thus, in contrast with the endoploidy pattern in reproductive flower tissues, the overall ploidy distribution of vegetative tissues is less or not affected by defects in cell plate formation.
Figure 9.
Phenotypic Characterization of Double frill1−/− smt3-1−/− Mutants.
(A) and (B) General morphology of wild-type and frill1−/− smt3-1−/− 7-d-old seedlings and 4-week-old plants.
(C) Cytological analysis of petal vasculature and distal margin morphology in the wild type (wt) and frill1−/− smt3-1−/−. Bar = 250 µm.
(D) Epidermal cell wall morphology in wild-type and frill1−/− smt3-1−/− petals. Arrows indicate cell wall stubs and incomplete cell walls. Bar = 20 µm.
(E) Quantitative analysis of the endoploidy pattern of vegetative leaf tissue and whole inflorescences of 6-week-old plants. For each sample, three biological replicates were analyzed and error bars represent sd.
(F) Quantitative analysis of larger pollen grain formation in flowers of 6-week-old plants. For each sample, three biological replicates were analyzed and error bars represent sd.
(G) Normal sized diploid (2x), enlarged tetraploid (4x), and jumbo polyploid (>4x) pollen grains in frill1−/− smt3-1−/− flowers. Bar = 20 µm.
[See online article for color version of this figure.]
DISCUSSION
et2 Is a Nonlethal GSL8 Allele That Induces Endomitosis through Defects in Cytokinesis
In this report, we document the identification of an Arabidopsis mutant, named et2, which produces diploid spores through the ectopic induction of premeiotic endomitosis. Endomitotic polyploidization events were not only observed in the premeiotic germ lineage, but also frequently occur in all other et2 flower organs and sporadically in other somatic tissues (e.g., roots). Using cytological approaches, we additionally demonstrate that endomitotic cells in both somatic and reproductive et2 organs originate from defects in cell plate formation or cell wall establishment.
The molecular identification of et2 as a novel allelic variant of Arabidopsis GSL8 is in strong agreement with the observed defects in cell wall formation. GSL8 encodes GLUCAN SYNTHASE-LIKE8, a callose synthase that catalyzes the local biosynthesis of callose. As callose is a major intermediate component of developing cell plates during plant cytokinesis (Samuels et al., 1995; Verma, 2001), loss of GSL8 functionality has been shown to induce defects in cell plate and cell wall formation (Chen et al., 2009). Indeed, previously identified gsl8 mutants (e.g., chorus and massue) exhibit distinct aberrations in cross-wall and cell plate formation (Thiele et al., 2009; Guseman et al., 2010). However, in contrast with chorus and massue, in which cell wall aberrations are severe and typically induce seedling lethality, cytokinetic defects in et2 are relatively mild and allow plantlets to continue development onto the fully matured, seed-setting state. As such, et2 allows for the functional analysis of GSL8 in all stages of plant development. Based on the observed cell wall defects and the associated formation of endomitotic cells in et2 (e.g., in flowers and germ lineage), we conclude that GSL8-driven callose deposition plays a prominent role in de novo cell plate formation and genomic ploidy maintenance in all stages of plant development, particularly in the reproductive tissues. The phenotypic aberrations in the GSL8 hypomorph et2 strongly correlate with the AtGenExpress (Schmid et al., 2005) and RT-PCR expression data, which indicate that GSL8 is widely expressed in most organs throughout plant development, with transcript maxima in actively dividing cells, such as those in root tips and floral buds (Chen et al., 2009).
The basis for the lethality in previously isolated gsl8 knockout mutants is not entirely clear. Since main organs are formed and a bipolar axis established (Chen et al., 2009; Thiele et al., 2009), the growth arrest may be attributed to a defect in the cell’s ability to perform chromosome condensation and segregation in combination with an altered cell-to-cell communication and cellular patterning. Indeed, successive divisions without cytokinesis generally lead to cells carrying a highly polyploid nucleus for which the disentanglement of the replicated daughter strands becomes too complex (Sugimoto-Shirasu and Roberts, 2003). As such, successive defects in cell plate formation (e.g., in seedling-lethal gsl8 mutants) may induce a premature arrest of proliferative cell division. Interestingly, in et2, the majority of ploidy increases consist of a single nuclear genome duplication and only sporadically generate octaploid cells, suggesting that successive events of defective cytokinesis are rare. As such, cytokinetic defects in et2 are rather mild compared with other gsl8 mutants and most presumably lies at the basis of the nonlethality of the GSL8-specific et2 mutant.
Cell Wall Alterations in SMT2 Sterol Mutants Also Lead to Endomitotic Endopolyploidy
Several reports have demonstrated a role for sterols in the regulation of somatic cytokinesis and cell wall formation. For example, Schrick et al. (2004) demonstrated that mutations in the major Arabidopsis sterol biosynthesis genes FACKEL, HYDRA1, and SMT1, despite distinct alterations in the sterol profile, all induce similar cytokinetic defects, such as incomplete cell walls, cell wall stubs, and aberrant cell wall thickenings in embryonic and postembryonic tissues. Because of the severity of cytokinetic defects, functional loss of sterol biosynthesis enzymes upstream of 24-methylene lophenol (i.e., the common BR and sterol precursor) has been shown to be seedling lethal. By contrast, here, we show that functional loss of SMT2, which is involved in the biosynthesis of free sterols downstream of 24-methylene lophenol, causes much weaker defects in cell wall establishment and plant growth, allowing plants to complete their life cycle up until the seed setting stage. We additionally found that cytokinetic defects in smt2 occasionally induce ectopic endomitosis in both vegetative and reproductive flower tissues and thereby generate alterations in flower morphology, including vasculature defects and serrated petals. Although Hase et al. (2005) postulated that the serrated petal phenotype is caused by ectopic endoreduplication, generating large amorphous cell clusters, here, we demonstrate that endopolyploid cells in frill1 petals are endomitotic and result from stochastic aberrations in cross-wall formation.
The ectopic induction of endopolyploidy is a general feature of cytokinesis-defective cells and has been observed in several organisms and cell types (Hatzfeld and Buttin, 1975; Thompson and Lindl, 1976; Karess et al., 1991; Castrillon and Wasserman, 1994; Neufeld and Rubin, 1994; Liu et al., 1997; Spielman et al., 1997; Lordier et al., 2008; Pampalona et al., 2012; Serres et al., 2012) and under certain cell plate–destructive conditions (e.g., caffeine and cold) (Roper and Roper, 1977; De Storme et al., 2012). In mutant forms of the Arabidopsis KNOLLE and TSO1, for example, cell plate–defective embryos and flower organs exhibit enlarged cells with both multinucleate and polyploid nuclei (Lukowitz et al., 1996; Liu et al., 1997). However, despite the ubiquitous presence, the exact cytological mechanism underlying nuclear fusion in cell plate–defective cells has not been revealed yet. In theory, the ectopic formation of polyploid nuclei in multinucleate cells can occur via two different processes: (1) either through an active migration and colocalization of syncytial nuclei, followed by an endocytosis-like fusion of nuclear membranes during G1- or S-phase; or (2) through the coallocation of all syncytial chromosomes in one metaphase spindle figure during the subsequent M-phase. In support of the first mechanism, we occasionally observed colocalization of noncondensed nuclei in cell wall–defective multinucleate et2 and smt2 cells. However, to fully elucidate the cytological mechanism underlying nuclear fusion, more in-depth studies need to be performed.
The Ploidy Integrity of the Arabidopsis Reproductive System Is More Sensitive to Cytokinetic Defects Compared with Somatic Tissues
In both the GLS8 hypomorph et2 and the sterol smt2 mutants, flower organs (e.g., petals and sepals) were found to contain large clusters of endomitotic polyploid cells, whereas ectopic polyploidization events were not or only rarely detected in cotyledon and leaf tissues. As defects in cell plate formation occur in both types of tissues, these findings suggest that the ploidy integrity of flower organs is more sensitive to cytokinetic alterations compared with other somatic tissue types. This was confirmed by the analysis of the more severe cytokinesis-defective frill1 smt3-1 double mutant. Whereas single smt2 mutants only show a moderate reduction in sterol content, the combinatorial loss of SMT2 and SMT3 causes almost a complete loss of phytosterols (Carland et al., 2010). In agreement with a role for sterols in cell wall formation, double smt2 smt3 mutants show an aggravated cytokinesis phenotype, both in the somatic and reproductive structures, causing morphological defects, such as reduced stature and irregular-shaped leaves and petals. Correlated with the enhanced cytokinetic defects, we found that smt2 smt3 inflorescences show an increased level of endomitotic polyploidization compared with single mutants. The somatic endoploidy level, on the other hand, appeared unchanged. These findings confirm that the ploidy stability of Arabidopsis flower organs is more sensitive to cytokinetic alterations compared with the ploidy pattern of other somatic tissues and that the increase in inflorescence endoploidy is positively correlated with the severity of cell wall aberrations.
A putative hypothesis for the discrepancy in cytokinesis-defective ploidy alterations between vegetative and reproductive structures is based on the difference in the underlying cell cycle pattern. Indeed, in contrast with most plants, Arabidopsis vegetative tissues show a high level of endoreduplication, typically resulting in an endoploidy pattern consisting of diploid, tetraploid, and octaploid nuclei. By contrast, Arabidopsis flower organs do not show endoreduplication, but instead proliferate through mitotic cell cycles, typically resulting in a ploidy pattern consisting of diploid and tetraploid (dividing cells in S- or G2-phase) cells (Galbraith et al., 1991). Based on these endoploidy differences, it is plausible to assume that ectopically formed endomitotic nuclei (e.g., those formed through defects in cell wall formation) are easier to detect in nonreplicative flower structures compared with the endoreplicative vegetative tissues. Moreover, since several authors have postulated the existence of an organ size checkpoint in Arabidopsis (Sugimoto-Shirasu and Roberts, 2003; Tsukaya, 2003), ectopic increases in vegetative endoploidy (e.g., through extra endomitotic events) may be systematically blocked or masked by a reduced level of endoreduplication. Alternatively, the frequency of nuclear fusion in terminally differentiated leaf cells may possibly be much lower compared with that in proliferative flower tissues, eventually leading to an increased flower endoploidy pattern without significant changes in the vegetative ploidy distribution.
Premeiotic Endomitosis in Cell Wall–Defective Flowers Leads to Polyploid Meiocytes
Besides an increased endoploidy pattern, both et2 and smt2 inflorescences also produce a significant population of tetraploid meiocytes through the ectopic induction of premeiotic endomitosis. As such, these mutants generate a subset of viable diploid male and female gametes that enable the formation of polyploid offspring (sexual polyploidization). In a broader context, these findings suggest that alterations in callose synthesis, sterol biosynthesis, and most likely also other cytokinesis-related processes in the premeiotic germ cell lineage may lead to 2n gamete formation and hence constitute an important route for sexual polyploidization. In line with this, diploid gametes in certain interspecific Brassica hybrids were also found to result (partially) from tetraploid meiocytes, most presumably originating from premeiotic chromosome doubling events (Mason et al., 2011). Moreover, since Mason et al. (2011) demonstrated that adverse temperature conditions significantly increase 2n male gamete frequency, sexual polyploidization in plants may possibly be stimulated by stress-induced defects in premeiotic cell plate formation and associated endomitosis.
In vertebrates, the production of diploid gametes through premeiotic endomitosis has already been documented in several parthenogenetically reproducing species (Kobel and Dupasquier, 1986). For example, in oogonia of natural loach clones and hybrid fish species, the ectopic induction of premeiotic endomitosis leads to 2n eggs (Shimizu et al., 2000; Liu et al., 2004; Itono et al., 2006). In all these cases, however, the underlying cytological defect is unknown and may not relate to defects in cytokinesis. Here, in Arabidopsis et2 and smt2, endomitotic polyploidization in the reproductive archesporal cells originates from defects in cell plate formation and subsequent fusion of syncytial nuclei. Although previously reported cytokinesis mutants (e.g., knolle, keule, cyt1, hinkel, runkel, and pilz also show occasional endomitotic polyploidization, these lines have been mostly found to be embryo or seedling lethal and thus are not able to induce polyploidy in the reproductive germ lineage (Mayer et al., 1991, 1999; Assaad et al., 1996; Nickle and Meinke, 1998; Strompen et al., 2002; Krupnova et al., 2009). By contrast, in some Arabidopsis cytokinesis-defective mutants (e.g., scd1-1, pleiade, hyade, cyd1, and tso1), cellular defects are relatively mild or tissue specific, allowing plants to develop into the flowering stage (Liu et al., 1997; Yang et al., 1999; Müller et al., 2002; Falbel et al., 2003). Although in these lines the ploidy of the reproductive organs (e.g., meiocytes and pollen) was not assessed, the presence of larger pollen grains in both tso1 (Andersen et al., 2007) and cyd1 (Yang et al., 1999) suggests that flower-specific cytokinetic defects in these lines, similar as in et2 and smt2, lead to premeiotic endomitosis and the associated induction of diploid and polyploid spore formation. Hence, it can be stated that moderate defects in cell wall formation constitute an important route for diploid and/or polyploid gamete production and subsequent sexual polyploidization in plants.
Besides defects in cell wall establishment, endomitotic polyploidization can also be induced by alterations of the mitotic cell cycle. For example, Iwata et al. (2011) demonstrated that loss of GIG1/OSD1, a plant-specific inhibitor of the APC/C ubiquitin ligase complex, induces ectopic endomitosis in Arabidopsis cotyledons. Similarly, Weingartner et al. (2004) postulated that the expression of a nondegradable cyclin B1 significantly affects exit from mitosis and consequently generates multinucleate and endomitotic cells. Although these reports clearly demonstrate the ectopic induction of endomitosis in vegetative tissues, the ectopic occurrence of premeiotic endomitosis and the associated formation of polyploid meiocytes were not reported.
Conclusions
Based on observations in the Arabidopsis mutants et2 and smt2, we conclude that stochastic defects in cell wall formation and the associated formation of endomitotic cells in vegetative tissues do not affect the somatic endoploidy level and only have a minor effect on Arabidopsis growth and development. In the reproductive flower tissues, on the other hand, GSL8-mediated callose deposition and SMT2-mediated sterol synthesis are critically required for maintaining the basic ploidy level in all organ types, particularly in the gametophytic meiotic precursor cells. Indeed, upon loss of GSL8 or SMT2 function, both male and female sporogenesis generate a subset of polyploid meiocytes, which eventually develop into viable diploid or polyploid gametes. In a broader developmental context, these findings suggest that the ploidy integrity of the reproductive tissues is more sensitive to cytokinetic defects compared with other tissues and may therefore constitute an important pathway for diploid gamete formation and sexual polyploidization in plants.
METHODS
Plant Materials
Mutant et2 was isolated out of an EMS-mutagenized M2 Arabidopsis thaliana Col-0 seed stock (Lehle Seeds). The following mutants and transgenic lines were previously described: frill1 (Hase et al., 2005), cvp1-3 (Carland et al., 2002), smt3(-1) (Carland et al., 2010), qrt1-1 (Preuss et al., 1994), ProMGH3-H2B-GFP (Brownfield et al., 2009), chorus (Guseman et al., 2010), and ProWOX2-CENH3-GFP (De Storme and Geelen, 2011).
The ProSDS-CENH3-GFP cassette was constructed using the Multisite-Gateway cloning system according to the manufacturer’s instructions (Invitrogen). The verified plasmid was transformed into the Agrobacterium tumefaciens strain GV3130 and used to generate transgenic Arabidopsis Col-0 lines. Single T-DNA transgenic progeny plants were selected based on kanamycin selection and stable GFP expression. Recombinant reporter constructs were introduced into et2 and frill1 via genetic crosses.
Arabidopsis plants were grown under the same conditions as reported by De Storme and Geelen (2011). For analysis of seedling morphology, plantlets were kept on the plate in the growing chamber.
Genetic Screen
A large population of 4800 EMS-mutagenized M2 plants was screened for the production of larger pollen grains. For each individual plant, extraction of pollen and analysis of pollen size distribution was performed as described previously (De Storme and Geelen, 2011).
Low-Resolution Mapping and SHOREmap Sequencing
Plants homozygous for et2 (Col-0) were crossed to the Landsberg erecta strain to generate a recombinant F2 mapping population that segregates the et2 phenotype. Homozygous mutant F2 segregants were identified based on the production of larger pollen grains and used as a source of DNA for genetic mapping. To map the nontagged EMS mutation, two alternative approaches were used: marker-assisted positional cloning and sequencing-based SHOREmapping (Schneeberger et al., 2009).
Flow Cytometric Ploidy Assessment
The somatic ploidy level of leaves, flower buds, and flower organs was determined using DNA flow cytometry (Epics Altra; Beckman Coulter) with the nuclear preparation protocol of Galbraith et al. (1983).
Cytology and Histology
Nuclear DNA staining of developing and mature spores and the analysis of male meiosis were performed according to protocols described earlier (De Storme and Geelen, 2011). To monitor the nuclear size in somatic tissue, plant organs were fixed in ethanol:acetic acid (3:1) fixative for at least 1 h, cleared in 70% ethanol, and dyed with 2 µg/mL DAPI solution.
For histochemical callose staining, plant material was infiltrated with 0.1% (m/v) Aniline Blue fluorochrome in 0.033% (m/v) K3PO4 for 10 min and washed in distilled water. Visual assessment of vascular patterning in both cotyledons and petal leaves was performed by the method of Carland et al. (2002).
The analysis of the epidermal cell layer was performed according to Guseman et al. (2010) with minor modifications. Twelve-day-old seedlings were fixed in ethanol:acetic acid (3:1) for at least 1 h, washed in 70% ethanol, and used for microscopy analysis.
Microscopy
Bright-field and fluorescence imaging were performed using an Olympus IX81 inverted fluorescence microscope equipped with an X-Cite Series 120Q UV lamp and an XM10 camera. Image processing and z-stack projections were conducted using ImageJ.
Sterol Analysis
Quantification of sterol contents was performed using the quantitative time-of-flight method, previously described by Wewer et al. (2011).
RNA Extraction and RT-PCR
Total plant RNA was prepared using the RNeasy plant mini kit with additional on-column DNaseI treatment (Qiagen). First-strand cDNA was synthesized using the RevertAid H− cDNA synthesis kit (Fermentas) according to the manufacturer’s guidelines. Both quantitative RT-PCR and RT-PCR experiments were performed as described earlier (De Storme and Geelen, 2011). Gene-specific primers are listed in Supplemental Table 4 online. The genomic location of the primers used to detect the retention of intron 47 in GSL8 cDNA is shown in Supplemental Figure 10 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GSL8 (At2g36850), NM_179940; SMT2 (At1g20330), NM_101884; and SMT3 (At1g76090), NM_106258.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Ectopic Presence of Endomitotic Nuclei in et2 Petal Cells.
Supplemental Figure 2. Endomitotic Nuclei in et2 Anther Filaments.
Supplemental Figure 3. Clusters of Larger Pollen Grains in et2 Mature Anthers.
Supplemental Figure 4. Presence of Endomitotic Cells in et2 Petal Development.
Supplemental Figure 5. SHOREmap Analysis Localizes et2 on the Bottom Arm of Chromosome 2.
Supplemental Figure 6. Endomitotic Nuclei in the Roots of et2 and chorus Seedlings.
Supplemental Figure 7. Loss of Callose Deposition and Ectopic Branching in et2 Root Hairs.
Supplemental Figure 8. Stomatal Clusters and Excessive Cell Proliferation in et2 Leaves and Cotyledons.
Supplemental Figure 9. Male Gametophytic Ploidy Analysis in frill1 Using ProSDS-CENH3-GFP.
Supplemental Figure 10. GSL8 Gene Structure and Position of et2 Mutation and Transcript Analysis Primers Used.
Supplemental Table 1. Candidate Genes and Corresponding Mutations Resulting from et2 SHOREmap Analysis.
Supplemental Table 2. Candidate Gene Analysis for ET2 Gene Identification.
Supplemental Table 3. Segregation of Seedling Phenotype in Two Heterozygous gls8 Lines.
Supplemental Table 4. Sequences of Primers Used and Corresponding PCR Settings.
Acknowledgments
We thank Monica Höfte and David De Vleesschauwer (University of Ghent) for technical support regarding quantitative RT-PCR. We thank Susan Armstrong (University of Birmingham) for demonstrating the meiotic chromosome-spreading technique and for helpful comments. We also thank Keiko Torii (University of Washington) for providing the chorus seeds and Ellen Van Gysegem and Christophe Petit (University of Ghent) for technical help with reporter constructs and plant growing.
AUTHOR CONTRIBUTIONS
N.D.S. and D.G. designed the research project. N.D.S. carried out most experiments. J.D.S. and W.V.C. performed the OFFSHORE mapping of ET2. V.W. and P.D. contributed by analyzing sterol contents. N.D.S. and D.G. wrote the article.
Glossary
- EMS
ethyl methanesulfonate
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- Col-0
Columbia-0
- TMH
transmembrane helix
- BR
brassinosteroid
References
- Altmann T., Damm B., Frommer W.B., Martin T., Morris P.C., Schweizer D., Willmitzer L., Schmidt R. (1994). Easy determination of ploidy level in Arabidopsis thaliana plants by means of pollen size measurement. Plant Cell Rep. 13: 652–656 [DOI] [PubMed] [Google Scholar]
- Andersen S.U., Algreen-Petersen R.G., Hoedl M., Jurkiewicz A., Cvitanich C., Braunschweig U., Schauser L., Oh S.A., Twell D., Jensen E.O. (2007). The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J. Exp. Bot. 58: 3657–3670 [DOI] [PubMed] [Google Scholar]
- Assaad F.F. (2001). Plant cytokinesis. Exploring the links. Plant Physiol. 126: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F.F., Huet Y., Mayer U., Jürgens G. (2001). The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J. Cell Biol. 152: 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F.F., Mayer U., Wanner G., Jürgens G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253: 267–277 [DOI] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Borg M., Sidorova A., Mori T., Twell D. (2009). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Köhler C. (2011). Unreduced gamete formation in plants: Mechanisms and prospects. J. Exp. Bot. 62: 1659–1668 [DOI] [PubMed] [Google Scholar]
- Carland F., Fujioka S., Nelson T. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F.M., Fujioka S., Takatsuto S., Yoshida S., Nelson T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D.H., Wasserman S.A. (1994). Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 120: 3367–3377 [DOI] [PubMed] [Google Scholar]
- Chen X.Y., Kim J.Y. (2009). Callose synthesis in higher plants. Plant Signal. Behav. 4: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.Y., Liu L., Lee E., Han X., Rim Y., Chu H., Kim S.W., Sack F., Kim J.Y. (2009). The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 150: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato, F. (1984). Role of polyploidy in reproductive organs and tissues. In Embryology of Angiosperms, B.M. Johri, ed (New York: Springer-Verlag), pp. 519–566. [Google Scholar]
- De Storme N., Copenhaver G.P., Geelen D. (2012). Production of diploid male gametes in Arabidopsis by cold-induced destabilization of postmeiotic radial microtubule arrays. Plant Physiol. 160: 1808–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Geelen D. (2011). The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol. 155: 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.Y., Hong Z.L., Sivaramakrishnan M., Mahfouz M., Verma D.P.S. (2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Orr-Weaver T.L. (2001). Endoreplication cell cycles: More for less. Cell 105: 297–306 [DOI] [PubMed] [Google Scholar]
- Enns L.C., Kanaoka M.M., Torii K.U., Comai L., Okada K., Cleland R.E. (2005). Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 58: 333–349 [DOI] [PubMed] [Google Scholar]
- Falbel T.G., Koch L.M., Nadeau J.A., Segui-Simarro J.M., Sack F.D., Bednarek S.Y. (2003). SCD1 is required for cytokinesis and polarized cell expansion in Arabidopsis thaliana [corrected]. Development 130: 4011–4024 [DOI] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Copenhaver G.P. (2006). Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142: 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Knapp S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z.L., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Hase Y., Fujioka S., Yoshida S., Sun G.Q., Umeda M., Tanaka A. (2005). Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J. Exp. Bot. 56: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Hatzfeld J., Buttin G. (1975). Temperature-sensitive cell cycle mutants: a chinese hamster cell line with a reversible block in cytokinesis. Cell 5: 123–129 [DOI] [PubMed] [Google Scholar]
- Hong Z.L., Delauney A.J., Verma D.P.S. (2001a). A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z.L., Zhang Z.M., Olson J.M., Verma D.P.S. (2001b). A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itono M., Morishima K., Fujimoto T., Bando E., Yamaha E., Arai K. (2006). Premeiotic endomitosis produces diploid eggs in the natural clone loach, Misgurnus anguillicaudatus (Teleostei: Cobitidae). J. Exp. Zoolog. A Comp. Exp. Biol. 305: 513–523 [DOI] [PubMed] [Google Scholar]
- Iwata E., Ikeda S., Matsunaga S., Kurata M., Yoshioka Y., Criqui M.C., Genschik P., Ito M. (2011). GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23: 4382–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A.K., Lipka V., Burton R.A., Panstruga R., Strizhov N., Schulze-Lefert P., Fincher G.B. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15: 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J., Chevalier C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43: 735–745 [DOI] [PubMed] [Google Scholar]
- Karess R.E., Chang X.J., Edwards K.A., Kulkarni S., Aguilera I., Kiehart D.P. (1991). The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65: 1177–1189 [DOI] [PubMed] [Google Scholar]
- Kobel H.R., Dupasquier L. (1986). Genetics of polyploid Xenopus. Trends Genet. 2: 310–315 [Google Scholar]
- Krupnova T., et al. (2009). Microtubule-associated kinase-like protein RUNKEL needed [corrected] for cell plate expansion in Arabidopsis cytokinesis. Curr. Biol. 19: 518–523 [DOI] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jürgens G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.O., Davidson J.M., Duronio R.J. (2009). Endoreplication: Polyploidy with purpose. Genes Dev. 23: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Schubert V., Fuchs J., Klatte S., Macas J., Schubert I. (2006). Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18: 2443–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Sun Y.D., Zhang C., Luo K.K., Liu Y. (2004). Production of gynogenetic progeny from allotetraploid hybrids red crucian carp x common carp. Aquaculture 236: 193–200 [Google Scholar]
- Liu Z.C., Running M.P., Meyerowitz E.M. (1997). TSO1 functions in cell division during Arabidopsis flower development. Development 124: 665–672 [DOI] [PubMed] [Google Scholar]
- Lordier L., Jalil A., Aurade F., Larbret F., Larghero J., Debili N., Vainchenker W., Chang Y.H. (2008). Megakaryocyte endomitosis is a failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling. Blood 112: 3164–3174 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Mayer U., Jürgens G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- Mason A.S., Nelson M.N., Yan G.J., Cowling W.A. (2011). Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biol. (online) 11: doi: 10.1186/1471-2229-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U., Herzog U., Berger F., Inzé D., Jürgens G. (1999). Mutations in the pilz group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78: 100–108 [DOI] [PubMed] [Google Scholar]
- Mayer U., Ruiz R.A.T., Berleth T., Misera S., Jurgens G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353: 402–407 [Google Scholar]
- Müller S., Fuchs E., Ovecka M., Wysocka-Diller J., Benfey P.N., Hauser M.T. (2002). Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 130: 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., Rubin G.M. (1994). The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77: 371–379 [DOI] [PubMed] [Google Scholar]
- Nickle T.C., Meinke D.W. (1998). A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15: 321–332 [DOI] [PubMed] [Google Scholar]
- Nishikawa S.I., Zinkl G.M., Swanson R.J., Maruyama D., Preuss D. (2005). Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona J., Frías C., Genescà A., Tusell L. (2012). Progressive telomere dysfunction causes cytokinesis failure and leads to the accumulation of polyploid cells. PLoS Genet. 8: e1002679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Roper W., Roper S. (1977). Centripetal wall formation in roots of Vicia faba after caffeine treatment. Protoplasma 93: 89–100 [Google Scholar]
- Samuels A.L., Giddings T.H., Jr., andStaehelin L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130: 1345–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.E., Weigel D., Andersen S.U. (2009). SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schrick K., Fujioka S., Takatsuto S., Stierhof Y.D., Stransky H., Yoshida S., Jürgens G. (2004). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38: 227–243 [DOI] [PubMed] [Google Scholar]
- Serres M.P., Kossatz U., Chi Y., Roberts J.M., Malek N.P., Besson A. (2012). p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J. Clin. Invest. 122: 844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Shibata N., Sakaizumi M., Yamashita M. (2000). Production of diploid eggs through premeiotic endomitosis in the hybrid medaka between Oryzias latipes and O. curvinotus. Zoolog. Sci. 17: 951–958 [Google Scholar]
- Spielman M., Preuss D., Li F.L., Browne W.E., Scott R.J., Dickinson H.G. (1997). TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124: 2645–2657 [DOI] [PubMed] [Google Scholar]
- Strompen G., El Kasmi F., Richter S., Lukowitz W., Assaad F.F., Jürgens G., Mayer U. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12: 153–158 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Thiele K., Wanner G., Kindzierski V., Jürgens G., Mayer U., Pachl F., Assaad F.F. (2009). The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant J. 58: 13–26 [DOI] [PubMed] [Google Scholar]
- Thompson L.H., Lindl P.A. (1976). A CHO-cell mutant with a defect in cytokinesis. Somatic Cell Genet. 2: 387–400 [DOI] [PubMed] [Google Scholar]
- Töller A., Brownfield L., Neu C., Twell D., Schulze-Lefert P. (2008). Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J. 54: 911–923 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2003). Organ shape and size: A lesson from studies of leaf morphogenesis. Curr. Opin. Plant Biol. 6: 57–62 [DOI] [PubMed] [Google Scholar]
- Van Damme D., Inzé D., Russinova E. (2008). Vesicle trafficking during somatic cytokinesis. Plant Physiol. 147: 1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D.P.S. (2001). Cytokinesis and building of the cell plate in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 751–784 [DOI] [PubMed] [Google Scholar]
- Verma D.P.S., Hong Z.L. (2001). Plant callose synthase complexes. Plant Mol. Biol. 47: 693–701 [DOI] [PubMed] [Google Scholar]
- Weingartner M., Criqui M.C., Mészáros T., Binarova P., Schmit A.C., Helfer A., Derevier A., Erhardt M., Bögre L., Genschik P. (2004). Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wewer V., Dombrink I., vom Dorp K., Dörmann P. (2011). Quantification of sterol lipids in plants by quadrupole time-of-flight mass spectrometry. J. Lipid Res. 52: 1039–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Nadeau J.A., Zhao L.M., Sack F.D. (1999). Characterization of a cytokinesis defective (cyd1) mutant of Arabidopsis. J. Exp. Bot. 50: 1437–1446 [DOI] [PubMed] [Google Scholar]