Abstract
Owing to their role in controlling the efflux of toxic compounds, transporters are central players in the process of detoxification and elimination of xenobiotics, which in turn is related to cancer risk. Among these transporters, ATP-binding cassette B1/multidrug resistance 1 (ABCB1/MDR1), ABCC2/multidrug resistance protein 2 (MRP2), and ABCG2/breast cancer resistance protein (BCRP) affect susceptibility to many hematopoietic malignancies. The maintenance of regulated expression of these transporters is governed through the activation of intracellular “xenosensors” like the nuclear receptor 1I2/pregnane X receptor (NR1I2/PXR). SNPs in genes encoding these regulators have also been implicated in the risk of several cancers. Using a tagging approach, we tested the hypothesis that common polymorphisms in the transporter genes ABCB1, ABCC2, ABCG2, and the regulator gene NR1I2 could be implicated in lymphoma risk. We selected 68 SNPs in the 4 genes, and we genotyped them in 1,481 lymphoma cases and 1,491 controls of the European cases-control study (EpiLymph) using the Illumina™ GoldenGate assay technology.Carriers of the SNP rs6857600 minor allele in ABCG2, was associated with a decrease in risk of B-cell lymphoma (B-NHL) overall (p<0.001). Furthermore, a decreased risk of chronic lymphocytic leukemia (CLL) was associated with the ABCG2 rs2231142 variant (p=0.0004), which could be replicated in an independent population. These results suggest a role for this gene in B-NHL susceptibility, especially for CLL.
Keywords: Lymphoma, multidrug resistance 1 (MDR1), multidrug resistance protein 2 (MRP2), breast cancer resistance protein (BCRP), pregnane X receptor (PXR)
Introduction
Lymphomas are a heterogeneous group of malignancies arising in the lymphoid tissue, 1, 2 whose known risk factors include acquired or congenital immune system deficiency 3, familial history of a first degree relative with a hematological malignancy 4, and viral infections including Epstein Barr and hepatitis C virus infection. 5 Additionally, an increasing number of publications reported on the association between genetic variants and lymphoma risk, indicating the relevance of genetic variability in lymphomagenesis. 6, 7
There is considerable epidemiological evidence that the process of detoxification and elimination of xenobiotics has implications for cancer risk. 8–11 Membrane transporters are central players in this task, owing to their role of controlling the efflux of toxic compounds and reducing the local cellular burden of xenobiotics. Transporter proteins reduce the entrance of harmful substances (“phase 0 metabolism”) and increase the excretion of their detoxification products (“phase III metabolism”). 12 Among several players in this detoxifying/elimination process three ABC (ATP binding cassette) transporters ATP-binding cassette B1/multidrug resistance 1 (ABCB1/MDR1), ATP-binding cassette C2/multidrug resistance protein 2 (ABCC2/MRP2) and ATP-binding cassette G2/breast cancer resistance protein (ABCG2/BCRP), have a key role in protecting the organism in various tissues, including peripheral blood. 13–17
The maintenance of regulated expression of both drug transporters and metabolizing enzymes is governed through the activation of intracellular “xenosensors”. These ligand-activated transcription factors sense the intracellular level of xenobiotics and, upon ligand binding, translocate into the nucleus and transcriptionally activate genes involved in xenobiotic metabolism and transport. One of the best characterized of these sensors is nuclear receptor 1I2 (NR1I2), also known as pregnane X receptor (PXR), whose target genes include ABCB1, ABCC2 and ABCG2. 18
Some polymorphisms in the ABC transporter genes and their regulators have a demonstrated effect on gene expression and/or protein function. 19, 20 Several have also been implicated in the risk of various cancers and they are thought to have a major impact on the genetic susceptibility to hematopoietic malignancies. However, results have been inconsistent thus far. 21–26
To verify the impact of polymorphisms in genes related to xenobiotic metabolism on lymphomagenesis, using a tagging approach, we studied common genetic variability of ABCB1, ABCC2, ABCG2 and NR1I2. The selection of the genes was driven by the abundance of reports on how polymorphisms in the selected transporters can modify the protein function in comparison to other genes in the same family that were left out. We selected 68 single nucleotide polymorphisms (SNPs) which tag all frequent polymorphisms in the four genes, and we typed them in 1,481 lymphoma cases and 1,491 controls in the context of the EpiLymph study.
Design and Methods
Study population
Detailed information about the EpiLymph study is given elsewhere. 27 Briefly, this is a multi-centric case-control study carried out in six European countries (Spain, France, Italy, Germany, Ireland and Czech Republic) between 1998 and 2004. Incident cases with a diagnosis of a lymphoid malignancy were included in the study and categorized according to the WHO classification. 2 In Germany and Italy the corresponding controls were randomly sampled from population registries as described elsewhere and matched by age (± 5 years), sex and area of residence. 27 In the other study centers corresponding controls were recruited from the same hospitals as the cases, but had to have a diagnosis other than cancer, infectious diseases and immunodeficiency disorders. The participation rate was 88% in cases, 81% and 57% in hospital and population controls, respectively. Informed consent was sought from the 2,348 cases and 2,462 controls. All participants were asked to provide a blood sample for DNA extraction and took part in a standardized face-to-face interview requesting information on sociodemographic characteristics, familial and medical history, smoking and alcohol habits, a complete occupational history and data on leisure and occupational sun exposure. In each country the study was approved by the responsible ethics committee.
In this study 1,481 cases for which DNA was available were included and consisted of Hodgkin’s Lymphoma (HL, n=272), T-cell non-Hodgkin Lymphoma (T-NHL, n=95), and B-cell non-Hodgkin Lymphoma (B-NHL, n=1,114). Among the B-NHL different entities were present: diffuse large B-cell lymphoma (DLBCL, n=408), follicular lymphoma (FL, n=200), chronic lymphocytic leukemia (CLL, n=340) including small lymphocytic lymphoma (SLL), marginal zone B-cell lymphoma (MZL, n=112), and mantle cell lymphoma (MCL, n=54). The corresponding EpiLymph controls (n=1,491) were frequency-matched by age, sex and study center. Details regarding the main characteristics of the study population used for the statistical analysis and the distribution of cases are presented in Table 1.
Table 1.
Characteristics of the study population.
| Variable | Cases | Control subjects |
|---|---|---|
| N (%)* | N (%)* | |
| Age | ||
| <30 | 147 (10) | 129 (9) |
| 30–39 | 147 (10) | 153 (11) |
| 40–49 | 175 (12) | 183 (13) |
| 50–59 | 269 (19) | 274 (19) |
| 60–69 | 390 (28) | 397 (27) |
| =70 | 286 (21) | 311 (21) |
| Study Centre | ||
| Spain | 301 (21) | 318 (22) |
| France | 149 (11) | 152 (10) |
| Germany | 518 (37) | 530 (37) |
| Italy | 106 (7) | 110 (8) |
| Ireland | 105 (7) | 102 (7) |
| Czech Republic | 235 (17) | 235 (16) |
| Sex | ||
| Male | 787 (56) | 802 (55) |
| Female | 627 (44) | 645 (45) |
| Lymphoma subtype | ||
| Hodgkin lymphoma (HL) | 259 (18) | - |
| T-cell lymphoma (T-NHL) | 90 (6) | - |
| B-cell lymphoma (B-NHL) | 1114 (76) | - |
| Diffuse large B-cell lymphoma (DLBCL) | 391 (28) | - |
| Follicular lymphoma (FL) | 195 (14) | - |
| Chronic lymphocytic leukaemia (CLL) | 321 (23) | - |
| Mantle cell lymphoma (MCL) | 48 (3) | - |
| Marginal zone B-cell lymphoma (MZL) | 110 (8) | - |
| Removed samples† | 67 | 44 |
| 1481 | 1491 |
Percentage is based on the final population used for statistical analysis.
65 cases and 44 controls were removed due to low performance of the genotyping (call-rate below 0.95); additionally, 2 cases were excluded due to missing information on age.
Selection of tagging single nucleotide polymorphisms
We aimed at surveying the entire set of common genetic variants in the ABCB1, ABCC2, ABCG2 and NR1I2 genes. All polymorphisms in the candidate gene regions (including 5 kb upstream of the first known exon and 5 kb downstream of the last known exon of each gene), with minor allele frequency (MAF) ≥5% in Caucasians from the International HapMap Project (version 22; http://www.hapmap.org), were included. Tagging SNPs were selected with the use of the Tagger program within Haploview (http://www.broad.mit.edu/mpg/haploview/;http://www.broad.mit.edu/mpg/tagger/; 28, 29) using pairwise SNP selection with a minimum r2 threshold of 0.8. This resulted in a selection of 15 tagging SNPs for ABCG2, with a mean r2 of the selected SNPs with the SNPs they tag of 0.963, 23 tagging SNPs for ABCB1, with a mean r2 of 0.956, 12 tagging SNPs for ABCC2, with a mean r2 of 0.981, 14 tagging SNPs for PXR, with a mean r2 of 0.984. Therefore, the selected SNPs capture over 95% of the known common variability in these genes. Considering that the genomic regions of the three genes are characterized by high levels of linkage disequilibrium (LD), we postulate that such SNPs are also likely to tag any hitherto unidentified common SNPs in the respective genes. Since rs2725256 showed a poor Illumina score (0.76) we added rs1564481, which is in almost complete LD with the previous one (r2=1). Moreover rs2231142 was added, due to its demonstrated functional relevance. 30 Online Supplementary Table S1 shows details of the SNPs included in the study.
Sample preparation and genotyping
DNA was extracted from blood clots (Gentra Puregene Blood Kit, Qiagen, Hilden, Germany). The order of DNA samples of cases and controls was randomized on PCR plates in order to ensure that an equal number of cases and controls could be analyzed simultaneously. Genotyping was carried out using the Illumina™ GoldenGate technology (San Diego, CA, USA), according to the protocol specified by the manufacturer. For quality control 3% of the samples were replicated.
Data filtering and statistical analysis
Samples where more than 4% of the SNPs had failed were dropped from the analyses (65 cases and 44 controls), 2 additional cases were excluded from the analysis due to missing age information. We then filtered data to remove poorly performing SNPs: all SNPs that failed on at least 5% of the samples (n=5) were set to missing. Statistically significant (p<10−5) deviation from Hardy-Weinberg equilibrium (HWE) among controls was tested as well, but it did not lead to elimination of any SNP. After data filtering for quality control, 63 SNPs were analyzed in 1414 lymphoma cases and 1447 controls.
The association between genetic variants and risk of HL, T-NHL and B-NHL, as well as with the most common B-NHL entities was estimated by calculating Odds Ratios (OR), the corresponding 95% confidence intervals (CI) and p-values using unconditional logistic regression. The dominant, co-dominant and log-additive model of inheritance were used, and the homozygous genotype of the common allele was set as reference. All calculations were adjusted for age (continuous), sex and study centre.
In order to take into account the large number of tests performed in this candidate gene approach, we calculated for each gene the number of effective independent variables, Meff, by use of the SNP Spectral Decomposition approach (simpleM method). 31 We obtained a gene-wise Meff value for each gene and also a study-wise Meff value, by adding up the gene Meff’s. We calculated Meff values for each candidate gene separately and for the whole study (by adding the individual gene Meff values). The study-wise Meff was 51. We therefore used a study-wise significance p-value threshold of 0.05/51=0.001.
As a sensitivity analysis for B-NHL we performed stratified analysis by gender and age, while the cut point for age was set at 50 years, according to previous publications by Wang et al. 4 Among the HL cases, the cut point was set at the age of 40years, due to the typical bimodal age distribution. To evaluate heterogeneity across countries, we used the likelihood ratio test to compare the models with and without an interaction term between country and SNP.
All statistical analyses were performed using SAS 9.2 (SAS Institute Inc. Cary, NC).
Meta-analysis
For replication of the strongest associations identified in the present study population (EpiLymph), information on CLL risk was obtained from a NHL GWAS (SF1; 211 CLL cases and 750 controls) 7 and information of a GWAS on CLL (Mayo-GEC; 407 CLL cases and 296 controls)32. To compare the three studies, allelic odds ratios were estimated. Furthermore, a meta-analysis was performed using the “meta” and “rmeta” packages of the statistical software environment R (2.13.0) 33. We used the random effects model for combining the study estimates. The individual study results and the combined odds ratios and 95% CIs were presented in forest plots.
Haplotype reconstruction
Haplotype blocks were constructed from the control genotyping data using SNPtool (http://www.dkfz.de/de/molgen_epidemiology/tools/SNPtool.html, 34) and the algorithm implemented in Haploview based on confidence bounds by Gabriel et al. 35 The following cut-off values were used: MAF>5%, HWE p≥0.01 and 75% of non-missing genotypes. Maximum likelihood estimates of the haplotype frequencies were generated with an expectation-maximization based algorithm implemented in the PROC HAPLOTYPE procedure of SAS. Unconditional logistic regression adjusted for age (continuous), sex and study centre was used to calculate risk estimates. The most frequent haplotype was set as the reference, whereas haplotypes with a frequency below 0.05 were declared as rare haplotypes and combined.
Gene-gene interactions
The nonparametric Multifactor Dimensionality Reduction (MDR) approach was selected to complement logistic regression for the analysis of gene–gene interactions. The details of MDR are described elsewhere. 36 Briefly, MDR is a data reduction approach that seeks to identify combinations of multilocus genotypes and discrete environmental factors that are associated with either high risk or low risk of disease. MDR defines a single variable that incorporates information from several loci and/or environmental factors. This new variable can be evaluated for its ability to classify and predict outcome risk status using cross-validation and permutation testing. The MDR software is available from http://www.epistasis.org.
Results
Main effects of genotyped single nucleotide polymorphisms
Twelve SNPs showed statistically significant associations at the conventional p-value of 0.05, with B-NHL (overall) and HL considering the three different inheritance models (dominant, co-dominant and log-additive). Online Supplementary Table S2 shows the corresponding results for all analyzed SNPs in all histological subtypes.
One SNP, rs6857600, situated in the ABCG2 gene, was study-wise significantly associated with a 24% risk reduction for B-NHL. Carriers of at least one A allele were associated with a decreased risk of B-NHL (OR=0.75, 95%CI 0.64–0.90, p=0.001). Furthermore several other associations approached study-wise significance as detailed in Table 2, where SNPs associated with B-NHL, HL or CLL risk (p<0.01) are presented.
Table 2.
Odds Ratios (OR) and 95% Confidence Intervals (CI) for transporter gene related SNPs with B-NHL, HL and CLL in samples of the Epilymph study. Only SNPs with at least an association showing p<0.01 are presented. Significant associations after correction for multiple testing (p≤0.001) are highlighted in bold).
| Genotype | Controls* | B-NHL | CLL | Controls* | HL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | N (%) | OR† (95% CI) | p | N (%) | OR (95%CI) | p | N (%) | N (%) | OR (95%CI) | p | |
| ABCC2 rs3740067 | |||||||||||
| CC | 450 (38) | 418 (39) | Reference | 129 (40) | Reference | 417 (36) | 115 (44) | Reference | |||
| CG | 562 (47) | 493 (46) | 0.95 (0.79–1.13) | 0.5 | 145 (45) | 0.90 (0.68–1.18) | 0.4 | 569 (49) | 105 (41) | 0.63 (0.46–0.87) | 0.004 |
| GG | 183 (15) | 151 (14) | 0.90 (0.69–1.16) | 0.4 | 46 (14) | 0.89 (0.60–1.31) | 0.6 | 170 (5) | 39 (15) | 0.7 (0.45–1.09) | 0.1 |
| CG/GG | 0.93 (0.79–1.11) | 0.4 | 0.90 (0.69–1.16) | 0.4 | 0.65 (0.48–0.87) | 0.004 | |||||
| p-trend | 0.4 | p-trend | 0.5 | p-trend | 0.02 | ||||||
| ABCG2 rs1481012 | |||||||||||
| AA | 959 (80) | 902 (85) | Reference | 284 (88) | Reference | 923 (80) | 224 (86) | Reference | |||
| AG | 228 (19) | 155 (15) | 0.71 (0.57–0.90) | 0.004 | 33 (10) | 0.50 (0.33–0.74) | 0.0005 | 220 (19) | 33 (13) | 0.61 (0.40–0.93) | 0.02 |
| GG | 9 (1) | 10 (1) | 1.15 (0.47–2.86) | 0.8 | 4 (1) | 1.57 (0.47–5.26) | 0.5 | 13 (1) | 2 (1) | 0.51 (0.10–2.53) | 0.4 |
| AG/GG | 0.73 (0.59–0.91) | 0.005 | 0.54 (0.37–0.78) | 0.001 | 0.60 (0.40–0.91) | 0.02 | |||||
| p-trend | 0.01 | p-trend | 0.005 | p-trend | 0.02 | ||||||
| ABCG2 rs1564481 | |||||||||||
| GG | 501 (42) | 407 (38) | Reference | 121 (38) | Reference | 473 (41) | 95 (37) | Reference | |||
| GA | 564 (47) | 496 (46) | 1.08 (0.90–1.29) | 0.4 | 145 (45) | 1.08 (0.82–1.42) | 0.6 | 554 (48) | 128 (49) | 1.26 (0.92–1.74) | 0.2 |
| AA | 131 (11) | 164 (15) | 1.54 (1.18–2.00) | 0.002 | 55 (17) | 1.85 (1.26–2.71) | 0.002 | 129 (11) | 36 (14) | 1.43 (0.90–2.28) | 0.1 |
| GA/AA | 1.17 (0.98–1.38) | 0.08 | 1.22 (0.94–1.58) | 0.1 | 1.30 (0.96–1.75) | 0.09 | |||||
| p-trend | 0.002 | p-trend | 0.007 | p-trend | 0.08 | ||||||
| ABCG2 rs2231142 | |||||||||||
| CC | 957 (80) | 898 (84) | Reference | 284 (88) | Reference | 921 (80) | 224 (86) | Reference | |||
| CA | 229 (19) | 158 (15) | 0.73 (0.58–0.91) | 0.005 | 33 (10) | 0.49 (0.33–0.73) | 0.0004 | 221 (19) | 33 (13) | 0.60 (0.40–0.92) | 0.02 |
| AA | 10 (1) | 11 (1) | 1.15 (0.49–2.74) | 0.7 | 4 (1) | 1.42 (0.43–4.65) | 0.6 | 14 (1) | 2 (1) | 0.49 (0.10–2.43) | 0.4 |
| CA/AA | 0.75 (0.60–0.93) | 0.009 | 0.53 (0.36–0.77) | 0.001 | 0.59 (0.39–0.90) | 0.01 | |||||
| p-trend | 0.02 | p-trend | 0.004 | p-trend | 0.01 | ||||||
| ABCG2 rs2725256 | |||||||||||
| AA | 498 (42) | 403 (38) | Reference | 119 (37) | Reference | 469 (41) | 94 (36) | Reference | |||
| AG | 562 (47) | 496 (46) | 1.09 (0.91–1.30) | 0.4 | 146 (45) | 1.10 (0.84–1.45) | 0.5 | 555 (48) | 129 (50) | 1.29 (0.94–1.77) | 0.1 |
| GG | 136 (11) | 168 (16) | 1.52 (1.17–1.98) | 0.002 | 56 (17) | 1.82 (1.25–2.66) | 0.002 | 132 (11) | 36 (14) | 1.40 (0.88–2.23) | 0.2 |
| AG/GG | 1.17 (0.99–1.39) | 0.07 | 1.24 (0.96–1.60) | 0.10 | 1.31 (0.97–1.77) | 0.08 | |||||
| p-trend | 0.005 | p-trend | 0.007 | p-trend | 0.08 | ||||||
| ABCG2 rs6857600 | |||||||||||
| GG | 696 (58) | 693 (65) | Reference | 211 (66) | Reference | 681 (59) | 153 (59) | Reference | |||
| GA | 421 (35) | 319 (30) | 0.76 (0.64–0.91) | 0.003 | 86 (27) | 0.63 (0.47–0.84) | 0.002 | 409 (35) | 84 (32) | 0.94 (0.69–1.30) | 0.7 |
| AA | 79 (7) | 55 (5) | 0.71 (0.49–1.01) | 0.06 | 24 (7) | 0.94 (0.57–1.55) | 0.8 | 66 (6) | 22 (8) | 1.37 (0.77–2.42) | 0.3 |
| GA/AA | 0.75 (0.64–0.90) | 0.001 | 0.68 (0.52–0.88) | 0.004 | 1.01 (0.75–1.36) | 1.0 | |||||
| p-trend | 0.002 | p-trend | 0.03 | p-trend | 0.6 | ||||||
| NR1I2 rs11917714 | |||||||||||
| GG | 822 (69) | 684 (64) | Reference | 215 (67) | Reference | 800 (69) | 164 (63) | Reference | |||
| GA | 333 (28) | 327 (31) | 1.18 (0.98–1.42) | 0.07 | 91 (28) | 1.03 (0.78–1.36) | 0.9 | 319 (28) | 82 (32) | 1.29 (0.94–1.79) | 0.1 |
| AA | 40 (3) | 55 (5) | 1.65 (1.08–2.51) | 0.02 | 15 (5) | 1.44 (0.77–2.70) | 0.3 | 37 (3) | 13 (5) | 1.35 (0.67–2.76) | 0.4 |
| GA/AA | 1.23 (1.03–1.47) | 0.02 | 1.07 (0.82–1.40) | 0.6 | 1.30 (0.96–1.77) | 0.09 | |||||
| p-trend | 0.007 | p-trend | 0.4 | p-trend | 0.1 | ||||||
1196 controls were taken into account for the risk analysis with the B-cell lymphoma cases and 1153 controls were taken into account for the risk analysis with the Hodgkin lymphoma cases.
Odds Ratios were adjusted for the matching variables: age (continuous), sex and study center (categorical); the common homozygote was used as reference group for each SNP.
We have also analyzed the SNPs in each histological subtypes of B-NHL. Two SNPs belonging to the ABCG2 gene showed a study-wise association with disease risk. Heterozygous carriers of the SNP rs1481012 (ORAG=0.50, 95%CI 0.33–0.74, p=0.0005; ORAG/GG=0.54, 95%CI 0.37–0.78, p=0.001), and heterozygous carriers of the rs2231142 SNP (ORCA=0.49, 95%CI 0.33–0.73, p=0.0004, ORCA/AA =0.53, 95%CI 0.36–0.77, p=0.001) were associated with a decreased CLL risk The two SNPs are in high linkage disequilibrium with each other (r2=0.989).
For all the other subtypes no association was found at the study-wise significance threshold, but several were significant at the canonical p-value of 0.05, as shown in Online Supplementary Table S2.
We also stratified the analysis by age and gender, but we did not find any further statistical study-wise association in the different strata. Results are shown in Online Supplementary Table S4 and S5.
Results of the replication and meta-analysis
For the purpose of validation of SNPs significantly associated with B-NHL or CLL risk, allelic odds ratios were calculated with genotyping information of two GWAS on NHL and CLL 7, 32. In the Mayo-GEC study two ABCG2 SNPs showed a statistically significant association with decreased risk of CLL, with ORs (rs1481012: OR=0.69, 95% CI 0.49–0.97, p=0.03; rs2231142: OR 0.67, 95% CI 0.50–0.98, p=0.03) of a similar magnitude as those of our study. In the smaller SF1 study neither of the SNPs showed any significance with the disease risk. We also performed a meta-analysis between the three studies. A significant association with decreased CLL risk was observed for the non-synonymous rs2231142 SNP (OR=0.74, 95% CI 0.55–0.99, p=0.04). Figure 1 shows the forest plot of the individual study results and the combined estimate for rs2231142. Supplementary figure 1 illustrates the results for the remaining SNPs included in the meta-analysis.
Figure 1. Association between rs2231142 (ABCG2) and CLL risk.
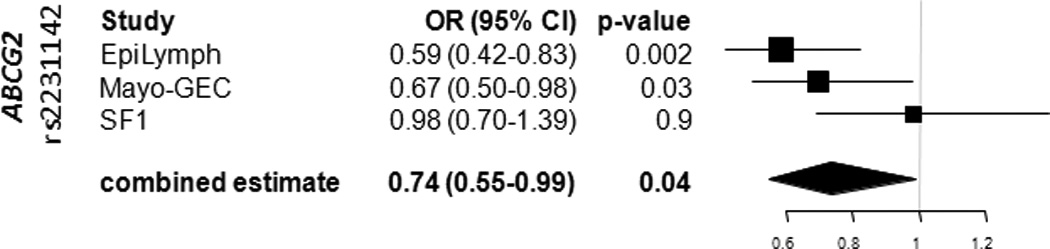
Relative samples sizes are represented by size of symbols. Horizontal lines indicate the 95% confidence intervals for the respective allelic odds. The combined estimate is indicated by a diamond. For the Mayo-GEC population the OR calculation was based on imputed information.
Haplotypes and interaction analysis
Haplotype analysis was performed for each gene in the study. Two ABCG2 haplotypes showed a study-wise significant association. Haplotype (rs13120400-rs1481012-rs2725256-rs2231142-rs1564481: A_A_A_C_G) (OR=0.78; 95%CI 0.67–0.91; p=0.001) and Haplotype (rs13120400-rs1481012-rs2725256-rs2231142-rs1564481: A_g_A_a_G) (OR=0.70; 95%CI 0.56–0.87; p=0.002) were associated with a decreased risk of B-NHL. None of the other haplotypes showed a study-wise statistically significant association with lymphoma risk, although haplotype (rs11917714-rs6785049-rs3732360-rs1054190: a_g_g_G) of the NR1I2 gene showed a suggestive association with increased risk of B-NHL, which however did not reach study-wise significance (OR=1.22; 95%CI 1.05–1.43; p=0.009). Table 3 shows the distribution of the ABCG2 and NR1I2 haplotypes and their associations with lymphoma risk. Online Supplementary Table S3 shows the haplotypes of each gene and their association with lymphoma risk. The structure of the haplotype blocks of ABCG2 is presented in Figure 2. Online Supplementary Figure 2 shows the LD blocks of the ABCB1, ABCC2 and NRI12 genes.
Table 3.
Odds Ratios and 95% Confidence Intervals for estimated Haplotypes of the ABCG2 and NR1I2 genes associated with B-NHL and subtypes.
| B-NHL | DLBCL | FL | CLL | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Structure | Freq. | OR* | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p |
| ABCG2 | Block2† | |||||||||||||
| A-A-g-C-a | 0.36 | Reference | Reference | Reference | Reference | |||||||||
| g-A-A-C-G | 0.28 | 0.95 | (0.82–1.10) | 0.5 | 0.99 | (0.80–1.21) | 0.9 | 0.93 | (0.70–1.22) | 0.6 | 0.89 | (0.71–1.11) | 0.3 | |
| A-A-A-C-G | 0.25 | 0.78 | (0.67–0.91) | 0.001 | 0.80 | (0.64–0.99) | 0.04 | 0.79 | (0.60–1.05) | 0.10 | 0.74 | (0.59–0.94) | 0.01 | |
| A-g-A-a-G | 0.09 | 0.70 | (0.56–0.87) | 0.002 | 0.70 | (0.51–0.96) | 0.03 | 0.86 | (0.58–1.28) | 0.5 | 0.53 | (0.37–0.77) | 0.001 | |
| NR1I2 | Block2‡ | |||||||||||||
| G-A-A-G | 0.60 | Reference | Reference | Reference | Reference | |||||||||
| a-g-g-G | 0.19 | 1.22 | (1.05–1.43) | 0.009 | 1.26 | (1.03–1.55) | 0.03 | 1.39 | (1.06–1.83) | 0.02 | 1.13 | (0.90–1.43) | 0.3 | |
| G-g-A-a | 0.13 | 1.09 | (0.90–1.30) | 0.4 | 0.90 | (0.69–1.18) | 0.4 | 1.28 | (0.92–1.77) | 0.1 | 1.20 | (0.91–1.57) | 0.2 | |
| G-g-g-G | 0.08 | 0.82 | (0.66–1.01) | 0.07 | 0.83 | (0.62–1.13) | 0.2 | 0.56 | (0.34–0.91) | 0.02 | 0.97 | (0.70–1.33) | 0.8 | |
Abbreviations are explained in Table 2.
Odds Ratios were adjusted for the matching variables: age (continuous), sex and study center (categorical); the common haplotype was used as reference group.
SNP order: rs13120400-rs1481012-rs2725256-rs2231142-rs1564481. Uppercase letters indicate major allele, lowercase letters denote minor allele
SNP order: rs11917714-rs6785049-rs3732360-rs1054190. Uppercase letters indicate major allele, lowercase letters denote minor allele
Figure 2. Haplotype blocks of the ABCG2 gene.
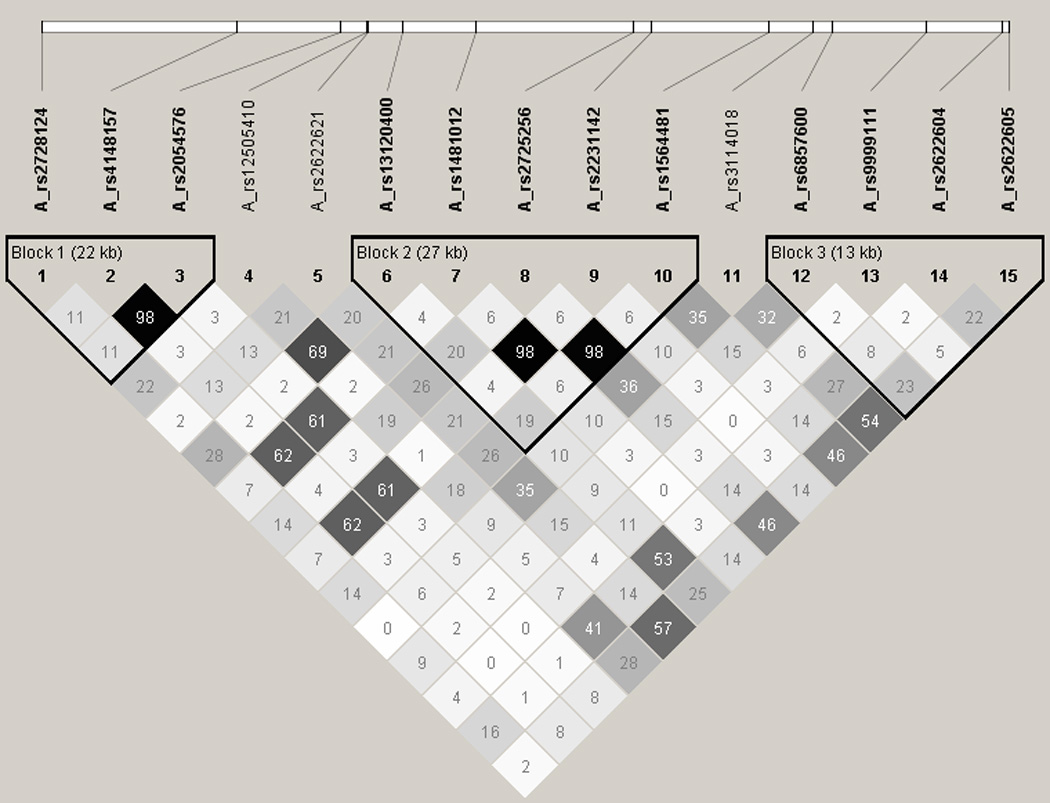
Figure 2 shoes the estimates of the correlation coefficient r2 in the ABCG2 gene, derived from the genotypes in the underlying study population in the Haploview software28. The LD relationship between each pair of SNP is indicated by the values in the squares. The shading indicates the extent of LD, a greater LD is represented by darker shading.
The rs1481012 and rs2231142 are in high LD (r2=0.989), and the rs2775256 is in high LD with the rs1564481 (r2=0.982).
Furthermore we analyzed all the possible pair-wise interactions between SNPs using the MDR method. No interactions between genes emerged with a study-wise significant p-value.
Discussion
Detoxification of xenobiotics, including toxins, carcinogens, and drugs, is the main task of many metabolizing enzymes. Transporter proteins are centrally involved in xenobiotic defence. Export pumps are the gatekeepers for all cells and organelles, controlling uptake and efflux of crucial compounds such as sugars, amino acids, nucleotides, inorganic ions and drugs and reducing the local cellular burden of toxic compounds giving the individual cell protection against toxic effects. The importance of the genes that code for these proteins is underlined by the fact that it is generally assumed that at least 5% (>2,000) of all human genes are transport-related. 14
In this study, we thoroughly captured common genetic variation across ABCB1, ABCC2, ABCG2 and NR1I2 genes and, to our knowledge, this is the most comprehensive evaluation of common variation in this crucial biological mechanism in relation with lymphoma risk. The intensive SNP tagging approach used provides a close to exhaustive analysis of associations of lymphoma risk with common polymorphic variants known for each of the loci studied. Moreover the analyses of haplotypes and gene-gene interactions give a comprehensive picture of the common genetic variability of the 4 genes in relation with HL, B-NHL and its more common subgroups. In this study we had sufficient power (over 0.80 for a codominant model) to detect OR=1.29 at alpha=0.001 (study-wise significance p-threshold) for a SNP with a MAF of 0.30 when considering all the study population as well as the B-NHL subgroup. For analyses in the HL sample set the power to detect an OR=1.5 for a SNP with a MAF of 0.30 was 0.80 for the codominant genetic model.
ABCG2 expression and its polymorphisms have been found, alone or in combination with polymorphisms of other transporters, to be associated with risk or prognosis of various malignant diseases and hematological diseases. 13, 24, 37–40 We found three SNPs (rs6857600, rs2231142 and rs1481012) on the ABCG2 locus associated with risk of B-NHL overall, and with CLL in particular. One of these polymorphisms, rs6857600, emerged at study-wise level of significance threshold of p=0.001 with a decreased risk of B-NHL. This association was consistent also in the CLL subgroup although the association was not statistically significant after correction for multiple testing (p=0.004).
Two SNPs, rs1481012 and rs2231142, emerged at study-wise significance threshold of p=0.001 with a decreased risk of CLL, (p=0.0005 for rs1481012 and p=0.0004 for rs2231142). These two polymorphisms are in close linkage disequilibrium with each other (r2=0.92) and therefore they have to be considered as a single association. To validate our strongest associations (rs11917714, rs1481012, rs15644814, rs2231142, rs2725256, rs6857600) we used the data from 2 recent GWAS 7, 32 and performed a meta-analysis. We observed that the rs2231142 SNP showed a statistically significant association (p=0.04) in the meta-analysis. On the other hand, rs1481012 did not show a statistically significant association with CLL risk in the meta-analysis, although the ORs and 95% CI calculated within the CLL GWAS (Mayo-GEC) were very similar to those ascertained in the EpiLymph study, while the results of the SF1 were different. It is worth mentioning that this polymorphism (rs1481012) was imputed in SF1 study and genotyped in the Mayo-GEC population, moreover the power to detect the association observed in the EpiLymph population was limited in the SF1 sample (67%). Hu and colleagues (2007) found that carriers of the A allele of the SNP rs2231142 run an increased risk of diffuse large B-cell lymphoma (DLBCL) in a Chinese population. 24 In our study we found that carriers of C allele of rs2231142 were associated with decreased risk of CLL and B-NHL, although they were not associated with DLBCL risk.
The reason of this discrepancy in the findings may be due to various factors, including the different ethnicity of the two populations, different environmental risk factors or the difference in the sample size of the two studies, the Chinese study being rather small compared to ours.
SNP rs2231142, which has been associated with risk of various diseases 41–44, lies in the fifth exon of the ABCG2 gene and is a non synonymous SNP which results from an amino acid change from glutamine to lysine. This nonsynonymous substitution has been proposed by several studies to affect ABCG2 protein expression, membrane surface translocation, efflux activity, or ATPase activity. 30, 45–48 The majority of these reports point to the possible decreased activity of the mutant (A) allele. Therefore, our finding that the polymorphism is associated with decreased risk would seem to be inconsistent with the functional evidence. Nevertheless it is worth noting that the results of these studies vary, showing inconsistencies in the effect of this polymorphism on ABCG2 protein activity. Additionally, several of the functional studies were performed in cell lines and therefore might not reflect the physiological situation in the organism.
None of the SNPs emerged as significant in this report were listed among the most significant results of the three recently conducted genome wide association studies (GWAS) on CLL. 32, 49, 50 We also analyzed haplotypes and SNP-SNP interactions, but the results did not explain more of the genetic susceptibility to the disease than the three SNPs alone. In this report we focused our attention on 4 genes where profound a priori knowledge of the influence of genetic variability on protein function exists. However other players (such as ABCC1 and ABCC3) were left out and we cannot exclude possible interaction between the latter ones and the ABCB1, ABCC2, ABCG2 and NR1I2 genes. In conclusion, we have found strong associations between three common variants of the ABCG2 gene, which has an important role in the elimination process of toxic agents in the organism, and the risk of B-NHL and CLL. We replicated the association of rs2231142 SNP in an independent population and after performing a meta-analysis the association remained statistically significant. Considering the three populations together, results of 939 cases and 2242 controls are presented, making it the largest investigation on genetic susceptibility of CLL. As transporter and well as metabolic genes affect risk of lymphoma only in presence of their carcinogenic substrate, further analyses shall be conducted to test the interaction between such substrates and the transporter and metabolic gene polymorphisms. This finding if further replicated may be a useful tool to identify a sub-group of individuals with an increased risk of CLL.
Supplementary Material
The association of the three studies included in the meta-analysis are presented in forest plots for selected ABCG2 (rs1481012, rs1564481, rs2725256, rs6857600) and NR1I2 (rs11917714) variants.
The reconstructed haplotype structures of the ABCB1, ABCC2, and NR1I2 genes based on the genotyping information of the genotyped controls are shown.
Details of the SNPs selected for the study. Columns in this table show: gene name; NCBI dbSNP rs number; the two possible alleles; the gene chromosome; the SNP position on the chromosome, relative location in the gene (intron, coding, 3UTR, flanking 3UTR, 5UTR), MAF in the CEU population; the minor allele; the major allele; HWE value for each SNP in the controls; the genotyping success rate in the controls; the genotyping success rate in the cases.
Main effect of 63 SNPs successfully genotyped. Columns in this table show: gene name; NCBI dbSNP rs number; odds ratios, with 95% confidence interval, for heterozygotes; odds ratios, with 95% confidence interval for homozygotes for the rare allele, relative p-value (referred to the homozygotes for the common allele), relative p-value; odds ratios, with 95% confidence interval, for Dominant model and p-value of the trend test. Each sheet in the file is referred to a specific subtype: B-NHL, DLBCL, FL, CLL, MZL, MCL, HL.
Results of haplotype analysis. Columns in this table show: gene name number of haplotype block in the gene; reconstructed haplotype; Haplotype frequency, odds ratios, with 95% confidence interval and related p-value for B-NHL, T-NHL and subtypes (sheet 1) and HL (sheet 2).
Effect of the 63 SNPs stratified for age. Columns in this table show: gene name; NCBI dbSNP rs number; odds ratios, with 95% confidence interval, for heterozygotes; odds ratios, with 95% confidence interval for homozygotes for the rare allele, relative p-value (referred to the homozygotes for the common allele), relative p-value; odds ratios, with 95% confidence interval, for Dominant model and p-value of the trend test. The two sheets in the file are referred to B-NHL and HL.
Effect of the 63 SNPs stratified for gender. Columns in this table show: gene name; NCBI dbSNP rs number; odds ratios, with 95% confidence interval, for heterozygotes; odds ratios, with 95% confidence interval for homozygotes for the rare allele, relative p-value (referred to the homozygotes for the common allele), relative p-value; odds ratios, with 95% confidence interval, for Dominant model and p-value of the trend test. The two sheets in the file are referred to B-NHL and HL.
Acknowledgements
The authors would like to thank our study subjects for their willingness to participate in the study. We are grateful to Bettina Ehret and Marco Uhrig for their help in sample preparation, Matthias Schick and Roger Fischer in the Genomics & Proteomics Core Facility for their great support with genotyping and Dr. Bernhard Korn for the possibility to perform the genotyping in his lab. For statistical support we thank Dorothee Zoller, Helena Schock and Dr. Sabine Behrens.
We would like to thank all the contributors of the GEC Consortium, namely Sara J Achenbach, Timothy G Call, Neil E Caporaso, James R Cerhan, Julie M Cunningham, Martha Glenn, Lynn R Goldin, Curtis A Hanson, Neil E Kay, Mark C Lanasa, Laura F Leis, Brian K Link, Gerald E Marti, Vicki A Morrison, Kari G Rabe, Susan L Slager, Logan G Spector, Sara S Strom, Celine M Vachon, and J Brice Weinberg.
Funding
This work was supported by the German José Carreras Leukemia Foundation (DJCLS_R04/08), EC 5th Framework Program Quality of Life grant No. QLK4-CT-2000-00422; by the EC 6th Framework Program grant No. FP6-2003-FOOD-2-B; by the Federal Office for Radiation Protection grants No. StSch4261 and StSch4420 (Germany); by the Spanish Ministry of Health grants CIBERESP (06/02/0073), FIS 08-1555 1 and Marato V3 (051210) (Spain); by La Fondation de France; by the Compagnia di San Paolo di Torino, Programma Oncologia; and by the Health Research Board, Ireland.
Glossary
Abbreviations
- ABC
ATP-binding cassette
- MDR1
multidrug resistance 1
- MRP2
multidrug resistance protein 2
- BCRP
breast cancer resistance protein
- NR1I2
nuclear receptor 1I2
- PXR
pregnane X receptor
- SNP
single nucleotide polymorphism
- WHO
World Health Organization
- HL
Hodgkin’s Lymphoma
- T-NHL
T-cell non-Hodgkin Lymphoma
- B-NHL
B-cell non-Hodgkin Lymphoma
- DLBCL
diffuse large B-cell lymphoma
- FL
follicular lymphoma
- CLL
chronic lymphocytic leukemia
- MZL
marginal zone B-cell lymphoma
- MCL
mantle cell lymphoma
- MAF
minor allele frequency
- LD
linkage disequilibrium
- HWE
Hardy-Weinberg Equilibrium
- MDR
Multifactor Dimensionality Reduction
- OR
odds ratio
- 95%CI
95% confidence interval
- GWAS
genome wide association study
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Turner JJ, Morton LM, Linet MS, Clarke CA, Kadin ME, Vajdic CM, Monnereau A, Maynadie M, Chiu BC, Marcos-Gragera R, Costantini AS, Cerhan JR, et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO Classification (2008): update and future directions. Blood. 2010 doi: 10.1182/blood-2010-06-289561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe ESHNL, Stein H, Vardiman JW. Pathology and Genetics of Tumours of Heamatopoietic and Lymphoid Tissuesed. Lyon: IARC PRess; 2001. [Google Scholar]
- 3.Alexander DD, Mink PJ, Adami HO, Chang ET, Cole P, Mandel JS, Trichopoulos D. The non-Hodgkin lymphomas: a review of the epidemiologic literature. Int. J. Cancer. 2007;120(Suppl 12):1–39. doi: 10.1002/ijc.22719. [DOI] [PubMed] [Google Scholar]
- 4.Wang SS, Slager SL, Brennan P, Holly EA, De Sanjose S, Bernstein L, Boffetta P, Cerhan JR, Maynadie M, Spinelli JJ, Chiu BC, Cocco PL, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;109:3479–3488. doi: 10.1182/blood-2006-06-031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Sanjose S, Benavente Y, Vajdic CM, Engels EA, Morton LM, Bracci PM, Spinelli JJ, Zheng T, Zhang Y, Franceschi S, Talamini R, Holly EA, et al. Hepatitis C and non-Hodgkin lymphoma among 4784 cases and 6269 controls from the International Lymphoma Epidemiology Consortium. Clin. Gastroenterol. Hepatol. 2008;6:451–458. doi: 10.1016/j.cgh.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skibola CF, Bracci PM, Nieters A, Brooks-Wilson A, de Sanjose S, Hughes AM, Cerhan JR, Skibola DR, Purdue M, Kane E, Lan Q, Foretova L, et al. Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph Consortium. Am. J. Epidemiol. 2010;171:267–276. doi: 10.1093/aje/kwp383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, Nieters A, Slager SL, Brooks-Wilson A, Agana L, Riby J, Liu J, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat. Genet. 2010;42:661–664. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agundez JA. Polymorphisms of human N-acetyltransferases and cancer risk. Curr. Drug Metab. 2008;9:520–531. doi: 10.2174/138920008784892083. [DOI] [PubMed] [Google Scholar]
- 9.Leslie EM, Deeley RG, Cole SP. Toxicological relevance of the multidrug resistance protein 1, MRP1 (ABCC1) and related transporters. Toxicology. 2001;167:3–23. doi: 10.1016/s0300-483x(01)00454-1. [DOI] [PubMed] [Google Scholar]
- 10.Sheweita SA, Tilmisany AK. Cancer and phase II drug-metabolizing enzymes. Curr. Drug Metab. 2003;4:45–58. doi: 10.2174/1389200033336919. [DOI] [PubMed] [Google Scholar]
- 11.Singh MS, Michael M. Role of xenobiotic metabolic enzymes in cancer epidemiology. Methods Mol. Biol. 2009;472:243–264. doi: 10.1007/978-1-60327-492-0_10. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa T. The ATP-dependent glutathione S-conjugate export pump. Trends Biochem. Sci. 1992;17:463–468. doi: 10.1016/0968-0004(92)90489-v. [DOI] [PubMed] [Google Scholar]
- 13.Abbott BL. ABCG2 (BCRP) expression in normal and malignant hematopoietic cells. Hematol. Oncol. 2003;21:115–130. doi: 10.1002/hon.714. [DOI] [PubMed] [Google Scholar]
- 14.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 15.Huls M, Russel FG, Masereeuw R. The role of ATP binding cassette transporters in tissue defense and organ regeneration. J. Pharmacol. Exp. Ther. 2009;328:3–9. doi: 10.1124/jpet.107.132225. [DOI] [PubMed] [Google Scholar]
- 16.Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol. Appl. Pharmacol. 2005;204:216–237. doi: 10.1016/j.taap.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr. Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- 19.Robey RW, To KK, Polgar O, Dohse M, Fetsch P, Dean M, Bates SE. ABCG2: a perspective. Adv Drug Deliv Rev. 2009;61:3–13. doi: 10.1016/j.addr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wada M. Single nucleotide polymorphisms in ABCC2 and ABCB1 genes and their clinical impact in physiology and drug response. Cancer Lett. 2006;234:40–50. doi: 10.1016/j.canlet.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 21.Campa D, Pardini B, Naccarati A, Vodickova L, Novotny J, Forsti A, Hemminki K, Barale R, Vodicka P, Canzian F. A gene-wide investigation on polymorphisms in the ABCG2/BRCP transporter and susceptibility to colorectal cancer. Mutat. Res. 2008;645:56–60. doi: 10.1016/j.mrfmmm.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Gardner ER, Ahlers CM, Shukla S, Sissung TM, Ockers SB, Price DK, Hamada A, Robey RW, Steinberg SM, Ambudkar SV, Dahut WL, Figg WD. Association of the ABCG2 C421A polymorphism with prostate cancer risk and survival. BJU Int. 2008;102:1694–1699. doi: 10.1111/j.1464-410X.2008.07913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattori H, Suminoe A, Wada M, Koga Y, Kohno K, Okamura J, Hara T, Matsuzaki A. Regulatory polymorphisms of multidrug resistance 1 (MDR1) gene are associated with the development of childhood acute lymphoblastic leukemia. Leuk. Res. 2007;31:1633–1640. doi: 10.1016/j.leukres.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Hu LL, Wang XX, Chen X, Chang J, Li C, Zhang Y, Yang J, Jiang W, Zhuang SM. BCRP gene polymorphisms are associated with susceptibility and survival of diffuse large B-cell lymphoma. Carcinogenesis. 2007;28:1740–1744. doi: 10.1093/carcin/bgm113. [DOI] [PubMed] [Google Scholar]
- 25.Jamroziak K, Mlynarski W, Balcerczak E, Mistygacz M, Trelinska J, Mirowski M, Bodalski J, Robak T. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur. J. Haematol. 2004;72:314–321. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 26.Jamroziak K, Robak T. Pharmacogenomics of MDR1/ABCB1 gene: the influence on risk and clinical outcome of haematological malignancies. Hematology. 2004;9:91–105. doi: 10.1080/10245330310001638974. [DOI] [PubMed] [Google Scholar]
- 27.Besson H, Brennan P, Becker N, Nieters A, De Sanjose S, Font R, Maynadie M, Foretova L, Cocco PL, Staines A, Vornanen M, Boffetta P. Tobacco smoking, alcohol drinking and non-Hodgkin's lymphoma: A European multicenter case-control study (Epilymph) Int. J. Cancer. 2006;119:901–908. doi: 10.1002/ijc.21913. [DOI] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 30.Morisaki K, Robey RW, Ozvegy-Laczka C, Honjo Y, Polgar O, Steadman K, Sarkadi B, Bates SE. Single nucleotide polymorphisms modify the transporter activity of ABCG2. Cancer Chemother. Pharmacol. 2005;56:161–172. doi: 10.1007/s00280-004-0931-x. [DOI] [PubMed] [Google Scholar]
- 31.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 32.Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, Lanasa MC, Spector LG, Rassenti LZ, Leis JF, Camp NJ, Glenn M, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2010 doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Team RDC. R: a language and environment for statistical computing. Austria: R Foundation for Statistical Computing Vienna; 2008. [Google Scholar]
- 34.Chen B, Wilkening S, Drechsel M, Hemminki K. SNP_tools: A compact tool package for analysis and conversion of genotype data for MS-Excel. BMC Res. Notes. 2009;2:214. doi: 10.1186/1756-0500-2-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 36.Moore JH. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Review of Molecular Diagnostics. 2004;4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- 37.Cortez MA, Scrideli CA, Yunes JA, Valera ET, Toledo SR, Pavoni-Ferreira PC, Lee ML, Petrilli AS, Brandalise SR, Tone LG. mRNA expression profile of multidrug resistance genes in childhood acute lymphoblastic leukemia. Low expression levels associated with a higher risk of toxic death. Pediatr. Blood Cancer. 2009;53:996–1004. doi: 10.1002/pbc.22220. [DOI] [PubMed] [Google Scholar]
- 38.Kim JE, Singh RR, Cho-Vega JH, Drakos E, Davuluri Y, Khokhar FA, Fayad L, Medeiros LJ, Vega F. Sonic hedgehog signaling proteins and ATP-binding cassette G2 are aberrantly expressed in diffuse large B-cell lymphoma. Mod. Pathol. 2009;22:1312–1320. doi: 10.1038/modpathol.2009.98. [DOI] [PubMed] [Google Scholar]
- 39.Plasschaert SL, van der Kolk DM, de Bont ES, Kamps WA, Morisaki K, Bates SE, Scheffer GL, Scheper RJ, Vellenga E, de Vries EG. The role of breast cancer resistance protein in acute lymphoblastic leukemia. Clin. Cancer Res. 2003;9:5171–5177. [PubMed] [Google Scholar]
- 40.Szczuraszek K, Materna V, Halon A, Mazur G, Wrobel T, Kuliczkowski K, Maciejczyk A, Zabel M, Drag M, Dietel M, Lage H, Surowiak P. Positive correlation between cyclooxygenase-2 and ABC-transporter expression in non-Hodgkin's lymphomas. Oncol. Rep. 2009;22:1315–1323. doi: 10.3892/or_00000570. [DOI] [PubMed] [Google Scholar]
- 41.Dehghan A, Kottgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC, Benjamin EJ, van Duijn CM, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korenaga Y, Naito K, Okayama N, Hirata H, Suehiro Y, Hamanaka Y, Matsuyama H, Hinoda Y. Association of the BCRP C421A polymorphism with nonpapillary renal cell carcinoma. Int. J. Cancer. 2005;117:431–434. doi: 10.1002/ijc.21187. [DOI] [PubMed] [Google Scholar]
- 43.Phipps-Green AJ, Hollis-Moffatt JE, Dalbeth N, Merriman ME, Topless R, Gow PJ, Harrison AA, Highton J, Jones PB, Stamp LK, Merriman TR. A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Maori, case and control sample sets. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq412. [DOI] [PubMed] [Google Scholar]
- 44.Tabara Y, Kohara K, Kawamoto R, Hiura Y, Nishimura K, Morisaki T, Kokubo Y, Okamura T, Tomoike H, Iwai N, Miki T. Association of four genetic loci with uric acid levels and reduced renal function: the J-SHIPP Suita study. Am. J. Nephrol. 2010;32:279–286. doi: 10.1159/000318943. [DOI] [PubMed] [Google Scholar]
- 45.Imai Y, Nakane M, Kage K, Tsukahara S, Ishikawa E, Tsuruo T, Miki Y, Sugimoto Y. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol. Cancer Ther. 2002;1:611–616. [PubMed] [Google Scholar]
- 46.Kondo C, Suzuki H, Itoda M, Ozawa S, Sawada J, Kobayashi D, Ieiri I, Mine K, Ohtsubo K, Sugiyama Y. Functional analysis of SNPs variants of BCRP/ABCG2. Pharm. Res. 2004;21:1895–1903. doi: 10.1023/b:pham.0000045245.21637.d4. [DOI] [PubMed] [Google Scholar]
- 47.Mizuarai S, Aozasa N, Kotani H. Single nucleotide polymorphisms result in impaired membrane localization and reduced atpase activity in multidrug transporter ABCG2. Int. J. Cancer. 2004;109:238–246. doi: 10.1002/ijc.11669. [DOI] [PubMed] [Google Scholar]
- 48.Sparreboom A, Loos WJ, Burger H, Sissung TM, Verweij J, Figg WD, Nooter K, Gelderblom H. Effect of ABCG2 genotype on the oral bioavailability of topotecan. Cancer Biol. Ther. 2005;4:650–658. doi: 10.4161/cbt.4.6.1731. [DOI] [PubMed] [Google Scholar]
- 49.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, Ruiz-Ponte C, Enjuanes A, Rosenquist R, Carracedo A, Jurlander J, Campo E, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, Sullivan K, Vijayakrishnan J, Wang Y, Pittman AM, Sunter NJ, Hall AG, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The association of the three studies included in the meta-analysis are presented in forest plots for selected ABCG2 (rs1481012, rs1564481, rs2725256, rs6857600) and NR1I2 (rs11917714) variants.
The reconstructed haplotype structures of the ABCB1, ABCC2, and NR1I2 genes based on the genotyping information of the genotyped controls are shown.
Details of the SNPs selected for the study. Columns in this table show: gene name; NCBI dbSNP rs number; the two possible alleles; the gene chromosome; the SNP position on the chromosome, relative location in the gene (intron, coding, 3UTR, flanking 3UTR, 5UTR), MAF in the CEU population; the minor allele; the major allele; HWE value for each SNP in the controls; the genotyping success rate in the controls; the genotyping success rate in the cases.
Main effect of 63 SNPs successfully genotyped. Columns in this table show: gene name; NCBI dbSNP rs number; odds ratios, with 95% confidence interval, for heterozygotes; odds ratios, with 95% confidence interval for homozygotes for the rare allele, relative p-value (referred to the homozygotes for the common allele), relative p-value; odds ratios, with 95% confidence interval, for Dominant model and p-value of the trend test. Each sheet in the file is referred to a specific subtype: B-NHL, DLBCL, FL, CLL, MZL, MCL, HL.
Results of haplotype analysis. Columns in this table show: gene name number of haplotype block in the gene; reconstructed haplotype; Haplotype frequency, odds ratios, with 95% confidence interval and related p-value for B-NHL, T-NHL and subtypes (sheet 1) and HL (sheet 2).
Effect of the 63 SNPs stratified for age. Columns in this table show: gene name; NCBI dbSNP rs number; odds ratios, with 95% confidence interval, for heterozygotes; odds ratios, with 95% confidence interval for homozygotes for the rare allele, relative p-value (referred to the homozygotes for the common allele), relative p-value; odds ratios, with 95% confidence interval, for Dominant model and p-value of the trend test. The two sheets in the file are referred to B-NHL and HL.
Effect of the 63 SNPs stratified for gender. Columns in this table show: gene name; NCBI dbSNP rs number; odds ratios, with 95% confidence interval, for heterozygotes; odds ratios, with 95% confidence interval for homozygotes for the rare allele, relative p-value (referred to the homozygotes for the common allele), relative p-value; odds ratios, with 95% confidence interval, for Dominant model and p-value of the trend test. The two sheets in the file are referred to B-NHL and HL.


