Abstract
Age-dependent leaf senescence and cell death in Arabidopsis (Arabidopsis thaliana) requires activation of the transcription factor ORESARA1 (ORE1) and is not initiated prior to a leaf age of 28 d. Here, we investigate the conditional execution of events that regulate early senescence and cell death in senescence-associated ubiquitin ligase1 (saul1) mutants, deficient in the PLANT U-BOX-ARMADILLO E3 ubiquitin ligase SAUL1. In saul1 mutants challenged with low light, the switch of age-dependent cell death was turned on prematurely, as indicated by the accumulation of ORE1 transcripts, induction of the senescence marker gene SENESCENCE-ASSOCIATED GENE12, and cell death. However, ORE1 accumulation by itself was not sufficient to cause saul1 phenotypes, as demonstrated by double mutant analysis. Exposure of saul1 mutants to low light for only 24 h did not result in visible symptoms of senescence; however, the senescence-promoting transcription factor genes WRKY53, WRKY6, and NAC-LIKE ACTIVATED BY AP3/PI were up-regulated, indicating that senescence in saul1 seedlings was already initiated. To resolve the time course of gene expression, microarray experiments were performed at narrow intervals. Differential expression of the genes involved in salicylic acid and defense mechanisms were the earliest events detected, suggesting a central role for salicylic acid in saul1 senescence and cell death. The salicylic acid content increased in low-light-treated saul1 mutants, and application of exogenous salicylic acid was indeed sufficient to trigger saul1 senescence in permissive light conditions. Double mutant analyses showed that PHYTOALEXIN DEFICIENT4 (PAD4) but not NONEXPRESSER OF PR GENES1 (NPR1) is essential for saul1 phenotypes. Our results indicate that saul1 senescence depends on the PAD4-dependent salicylic acid pathway but does not require NPR1 signaling.
Plant senescence is an age-dependent phenomenon that closely correlates with cell death. Leaf senescence is developmentally well defined to guarantee the recycling of resources from senescing leaves into young leaves or seeds, thus optimizing the growth and reproductive capacity of plants. This process becomes visible as yellowing of leaves due to chlorophyll degradation. Senescence is genetically associated with aging and involves the regulated expression of senescence-associated genes (SAGs; Yoshida, 2003; Lim et al., 2007). Microarray studies on adult Arabidopsis (Arabidopsis thaliana) plants indicated large changes in gene expression patterns during age-dependent senescence (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Wagstaff et al., 2009). In Arabidopsis, a feed-forward regulatory switch of age-dependent cell death during senescence has recently been demonstrated (Kim et al., 2009). The NAC transcription factor ORESARA1 (ORE1; also named ANAC092) triggers aging-associated cell death in leaves that are at least 28 d old. In young leaves, ORE1 accumulation is prevented by miRNA164 (miR164), which targets ORE1 mRNA for cleavage. This negative regulation of ORE1 expression by miR164 is released during aging through the down-regulation of miR164 (Kim et al., 2009).
In addition to ORE1, other components of the transcription factor network regulating gene expression during senescence have been identified. High expression of the WRKY6 transcription factor gene leads to leaf necrosis (Robatzek and Somssich, 2002). It has been shown that WRKY6 activates the promoter of SENESCENCE-INDUCED RECEPTOR KINASE1 encoding senescence-induced receptor kinase, which is specifically induced in senescent leaves. Premature senescence has been observed in WRKY53-overexpressing plants, whereas wrky53 mutants showed delayed senescence (Miao et al., 2004). Similarly, overexpression of the NAC-LIKE ACTIVATED BY AP3/PI (AtNAP)/ANAC029 gene encoding a NAC family transcription factor results in premature senescence (Guo and Gan, 2006). In contrast, senescence was delayed in two independent atnap mutant alleles. The NAC transcription factor ORE1 SISTER1/ANAC059 is also a positive regulator of senescence (Balazadeh et al., 2011). The hierarchy within the network of transcription factors during senescence is still not known.
Besides regulation at the level of transcription, protein turnover represents a highly important molecular mechanism for control of the onset and progression of senescence and cell death. On the one hand, bulk protein turnover during senescence occurs in autophagic vesicles involving ubiquitin-like conjugation pathways (Thompson and Vierstra, 2005; Bassham, 2007; Phillips et al., 2008; Yoshimoto et al., 2009). On the other hand, selective protein turnover is mediated by the ubiquitin/proteasome pathway (Sullivan et al., 2003; Smalle and Vierstra, 2004). The F-box protein ORE9 has been suggested to be a positive regulator of leaf senescence, because mutants lacking ORE9 expression are delayed in senescence (Woo et al., 2001). Generally, F-box proteins are part of SCF-type E3 complexes with ubiquitin ligase activity and recruit target substrates to such complexes (Lechner et al., 2006). It has recently been shown that ORE9 indeed interacts with the core SCF subunits ARABIDOPSIS SKP1 HOMOLOGUE1 and CULLIN1 in planta (Stirnberg et al., 2007). The arginyl-tRNA:protein arginyltransferase ATE1, which is a component of the N-end rule pathway within the ubiquitin-dependent proteolytic system, also positively regulates senescence. Knockout of the ATE1 gene in delayed senescence1 mutant plants resulted in delayed leaf senescence (Yoshida et al., 2002). In contrast to ORE9 and ATE1, the RING-type ubiquitin ligase NITROGEN LIMITATION ADAPTATION (NLA) and the PLANT U-BOX (PUB)-ARMADILLO (ARM) E3 ubiquitin ligase SENESCENCE-ASSOCIATED UBIQUITIN LIGASE1 (SAUL1; At1g20780) are negative regulators of plant senescence (Peng et al., 2007; Raab et al., 2009). A mutation in NLA results in early senescence when growing the mutants in nitrogen-limiting conditions. The ubiquitin ligase activity of NLA has indirectly been demonstrated through interaction with the Arabidopsis ubiquitin conjugase AtUBC8 (Peng et al., 2007). Similarly, saul1 mutant plants lacking any expression of SAUL1 show early senescence in low-light conditions. Senescence can be prematurely induced in saul1 seedlings by transfer to low light. Thus, saul1 mutants have been established as a low-light-inducible and age-independent model system for senescence (Raab et al., 2009).
In plant and animal cells, the ubiquitin/proteasome pathway has critical functions for cell survival and repair (Vernace et al., 2007; Vierstra, 2009). It is the major proteolytic pathway that mediates regulated protein degradation. The age-dependent decline of this process leads to the accumulation of aberrant proteins and has been correlated with certain human diseases (Grune et al., 2004; Hyun et al., 2004). It has also been suggested that the inhibition of proteasome function induces morphological symptoms of plant programmed cell death (Kim et al., 2003). Generally, plant programmed cell death occurs during normal development, for example, embryo formation and leaf senescence, but also during the hypersensitive response following pathogen attack to trigger cell death around infection sites and restrict pathogen spread. In the pathogen response, regulatory functions have been assigned to components of the ubiquitin/proteasome pathway, including many PUB proteins (Vierstra, 2009; Yee and Goring, 2009). Most of these belong to the subfamily of PUB-ARM proteins that contain ARM repeats, named after the Drosophila segment polarity gene armadillo, in addition to the U-box (Nüsslein-Volhard and Wieschaus, 1980; Riggleman et al., 1989). PUB-ARM proteins carry distinct numbers of ARM repeats where each repeat consists of three α-helices, yielding a conserved three-dimensional structure that is implicated to function as a protein interaction domain (Peifer et al., 1994; Huber et al., 1997; Coates, 2003). Tobacco (Nicotiana tabacum) CMPG1 and Avr9/Cf-9 Rapidly Elicited276, rice (Oryza sativa) SPOTTED LEAF11, and Arabidopsis AtPUB17, AtPUB22, AtPUB23, and AtPUB24 are PUB-ARM proteins and have been implicated in cell death control during pathogen responses (Zeng et al., 2004; González-Lamothe et al., 2006; Trujillo et al., 2008). The PUB-ARM protein SAUL1 has a role in the regulation of senescence and is localized to the plasma membrane (Drechsel et al., 2011). This localization may render SAUL1 a regulatory component that, by modifying signaling components through ubiquitination at the plasma membrane, is positioned very high in the hierarchy of senescence and cell death control.
To further define the function of SAUL1 during the onset and progression of senescence and cell death, we asked whether the feed-forward regulatory switch of age-dependent cell death during senescence involving ORE1 induction was turned on in very young saul1 seedlings. In addition, we aimed to reveal the time course of gene expression changes in saul1 mutants after transfer to low light on a genomic scale to resolve the underlying signaling events. We show that, indeed, ORE1 expression and cell death are rapidly induced in saul1 seedlings after transfer to low light. However, additional knockout of ORE1/ANAC092 in saul1-1/anac092-1 double mutants was not sufficient to suppress saul1 phenotypes. Microarray analyses identified the timing of responses in saul1 cell death and senescence and indicated that salicylic acid (SA) pathways may play a central role in saul1 senescence. Abolishing SA pathways in saul1-1/pad4 but not in saul1-1/nonexpresser of PR genes1 (npr1) double mutants was sufficient to suppress saul1 phenotypes. Our results suggested that specifically the PHYTOALEXIN DEFICIENT4 (PAD4)-mediated SA pathway is important for saul1 senescence.
RESULTS
The Switch for Age-Dependent Cell Death Is Prematurely Turned on in saul1 Seedlings
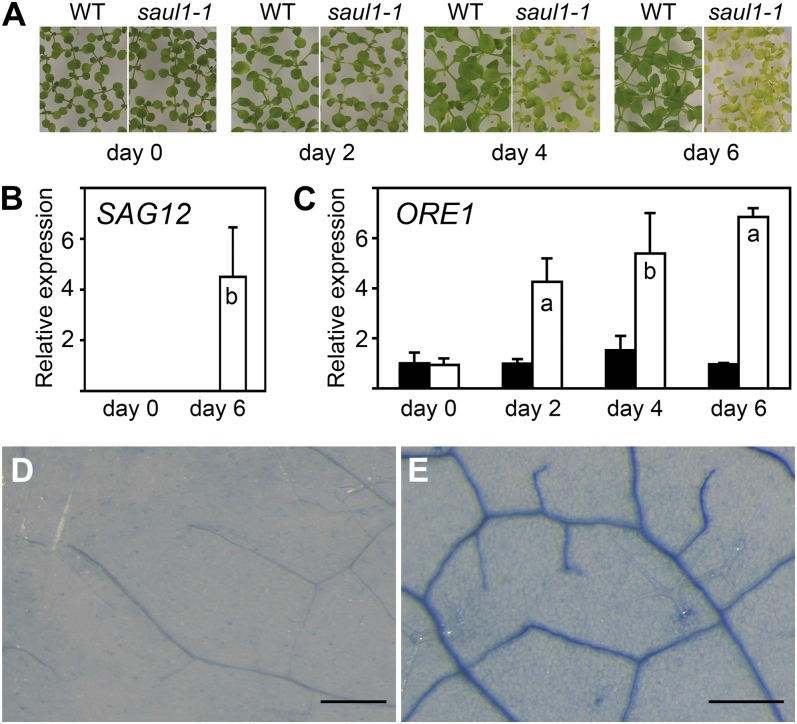
Recent mutant analyses in Arabidopsis showed that mutant plants, which do not express SAUL1 encoding an E3 ubiquitin ligase of the PUB protein family, exhibit early senescence (Raab et al., 2009). When growing in low light (photon flux density less than 30 µmol m−2 s−1), saul1 mutants show premature aging of leaves younger than 10 d (Fig. 1A). In permissive light conditions (photon flux density greater than 60 µmol m−2 s−1), saul1 seedlings are indistinguishable from the wild type, but senescence can be induced rapidly at any developmental stage by transfer to low light. Expression of the SAG12 gene encoding a Cys protease is strictly associated with senescent tissues, and SAG12 expression is used as a highly reliable marker of age-dependent senescence (Weaver et al., 1998; Noh and Amasino, 1999). Here, we show that at the molecular level senescence in saul1 mutants becomes manifested in SAG12 expression that is normally absent from young seedlings (Fig. 1B). This implies a premature turning on of a regulatory switch in saul1 mutants leading to SAG12 expression that is suppressed by SAUL1 in wild-type plants.
Figure 1.
Premature turning on of the switch for age-dependent cell death in saul1 seedlings. A, Senescence phenotype of saul1-1 mutants. Wild-type (WT) and saul1-1 plants were grown for 12 d at 60 µmol m−2 s−1 photon flux density followed by growth for 2, 4, and 6 d at 20 µmol m−2 s−1 photon flux density. Yellowing of leaves appears as a visible symptom of senescence. B, SAG12 transcript level in young saul1-1 seedlings. Bars show relative expression of the senescence marker gene SAG12 in wild-type (black bars) and saul1-1 (white bars) plants at the indicated time points (mean ± sd; n = 3) as determined by qPCR. Note that SAG12 transcripts are absent from wild-type seedlings and saul1-1 mutant seedlings without the senescence phenotype. C, Increased ORE1 transcript levels in saul1 seedlings. Bars show relative expression determined by qPCR of the senescence regulatory gene ORE1 in growth kinetics as in A in wild-type (black bars) and saul1-1 (white bars) seedlings at the indicated time points (mean ± sd; n = 3). D and E, Premature cell death in saul1 mutants. Trypan blue staining indicated dead or dying cells in saul1-1 mutant (E) but not in wild-type (D) seedlings. Assayed leaves (n = 16) were less than 20 d old, and identical leaves and leaf areas were chosen for comparison. Bars = 250 µm.
To identify the status of the feed-forward regulatory switch of age-dependent cell death during senescence (Kim et al., 2009) in saul1 mutants after transfer to low light, we tested for ORE1 expression through real-time reverse transcription-PCR (qPCR) experiments and for symptoms of cell death. Indeed, ORE1 transcript levels were up-regulated in saul1-1 leaves with leaf ages below 10 d counted from the time of leaf emergence (Fig. 1C). Accumulation of ORE1 transcripts was followed by cell death in saul1 mutants, as expected and indicated by the presence of trypan blue-stained cells (Fig. 1, D and E; Salt et al., 2011). These findings suggest that the feed-forward regulatory switch of age-dependent cell death has prematurely been turned on in saul1 mutant seedlings.
High Salinity Severely Affects the Growth and Development of saul1 Mutants
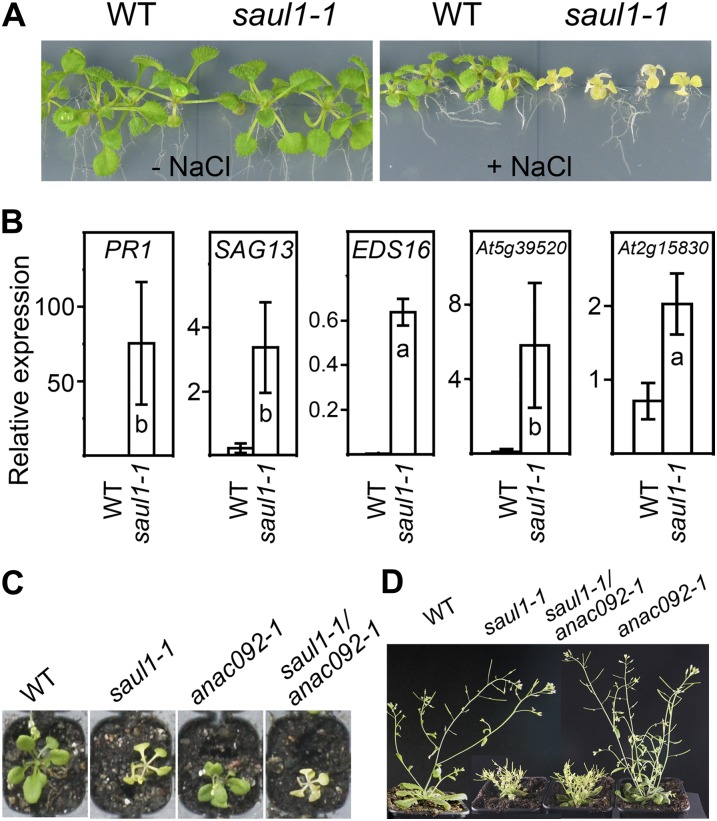
A common abiotic stress that may cause senescence and cell death in plants is high salinity. Next to its role in age-dependent cell death, ORE1/ANAC092 has been shown to mediate salt-induced senescence and cell death, too. Compared with the wild type, anac092-1 mutants were more tolerant to salt (Huh et al., 2002; Balazadeh et al., 2010). To study whether SAUL1 is also involved in the salt response, wild-type and saul1-1 mutant seedlings were grown in permissive light conditions for 5 d, transferred to agar plates containing 0 or 150 mm NaCl, and kept in permissive light. Whereas saul1-1 mutants were indistinguishable from wild-type plants in the absence of salt, saul1-1 mutants but not wild-type plants showed leaf yellowing as visible symptoms of senescence in the presence of NaCl (Fig. 2A). The expression of selected genes that are known to be induced by salt or regulated by ORE1 (Balazadeh et al., 2010), namely PATHOGENESIS-RELATED GENE1 (PR1), SAG13, ENHANCED DISEASE SUSCEPTIBILITY16 (EDS16), At5g39520, and At2g15830, also increased upon NaCl treatment in saul1-1 in comparison with wild-type plants (Fig. 2B). These data suggested that SAUL1 is a regulatory factor integrating not only low-light-induced, but also salt-stress-triggered, senescence and cell death.
Figure 2.
Hypersensitivity of saul1 mutants to high salt concentrations. A, Wild-type (WT) and saul1-1 mutant seedlings were grown in permissive light conditions (80 µmol m−2 s−1) for 5 d and then transferred to 0 or 100 mm NaCl to investigate the effect of salt on growth and development in permissive light conditions. In contrast to the wild type, saul1-1 mutant seedlings showed leaf yellowing on high salt. B, Relative expression levels were determined by qPCR to demonstrate salt induction of PR1, SAG13, EDS16, At5g39520, and At2g15830 in saul1-1 seedlings. Values represent means ± sd from three independent experiments. C, Leaf yellowing was monitored in wild-type, saul1-1, anac092-1, and saul1-1/anac092-1 seedlings that were grown at 100 µmol m−2 s−1 on soil. D, Growth defects in adult plants were observed in saul1-1 and saul1-1/anac092-1 but not in anac092-1 plants.
The Absence of ORE1/ANAC092 Is Not Sufficient to Suppress saul1 Phenotypes
We were aiming to test whether the knockout of ORE1/ANAC092 may suppress saul1 senescence and cell death, because this important switch during age-dependent cell death was turned on in low-light-treated saul1 mutants (Fig. 1). By crossing saul1-1 mutants to anac092-1 mutants, saul1-1/anac092-1 double knockout mutants were generated and studied with respect to the occurrence of leaf yellowing. In growth conditions that led to saul1-1 senescence and cell death, saul1-1/anac092-1 double mutants also showed yellowing of leaves, indicating that additional knockout of ORE1/ANAC092 did not suppress saul1 senescence and cell death phenotypes (Fig. 2C). The growth defect of saul1-1 mutants at later developmental stages (Raab et al., 2009) was also not suppressed in these double mutants (Fig. 2D).
Regulatory Events in saul1 Seedlings Precede the Occurrence of a Visible Phenotype and the Induction of ORE1 Expression
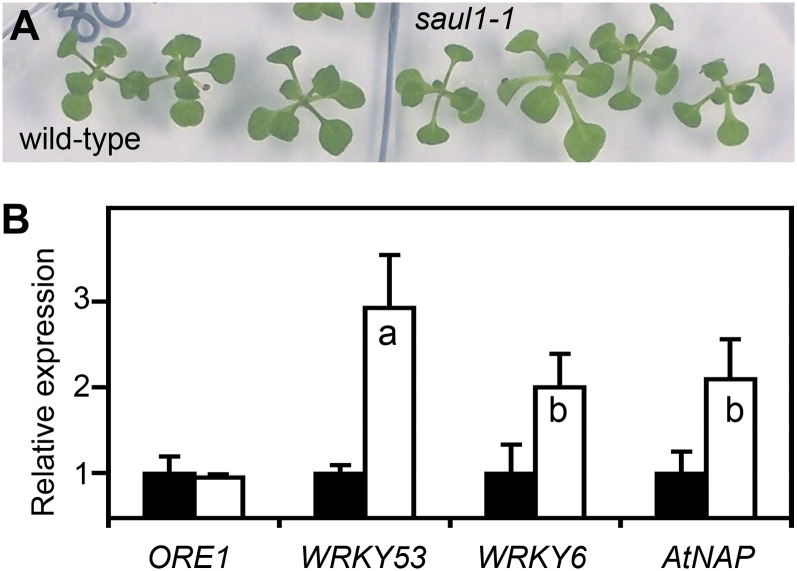
To investigate whether senescence in saul1 seedlings was initiated at the molecular level in the absence of visible symptoms and before the induction of ORE1 expression, plants were grown in permissive light conditions for 14 d and then transferred to low light. One day after transfer, saul1-1 seedlings still looked like the wild type (Fig. 3A). At this time, ORE1 expression was identical in saul1-1 mutant and wild-type plants (Fig. 3B). We tested for the expression of other well-known senescence regulatory genes encoding transcriptional regulators of the NAC and WRKY transcription factor families. Interestingly, the WRKY53, WRKY6, and AtNAP transcript levels were significantly increased in saul1-1 seedlings already 24 h after transfer to low light (Fig. 3B). This line of evidence demonstrates that changes in the regulatory network of transcription took place even before the occurrence of visible symptoms and before the increase of ORE1 expression. These observations implied that important regulatory events at the molecular level were established in the first 24 h after transfer to low light. In wild-type plants, the plasma membrane-localized ubiquitin ligase SAUL1 functions to prevent these expression changes. This prompted us to use a genomics approach to resolve the time course of gene expression changes that result in the onset and progression of saul1 senescence and cell death.
Figure 3.
Regulatory events at the molecular level in the absence of visible symptoms of senescence. A, Wild-type and saul1-1 mutant seedlings were grown for 14 d in permissive light (60 µmol m−2 s−1) followed by growth for 24 h in low-light conditions (20 µmol m−2 s−1). Note that wild-type and saul1-1 mutant seedlings were indistinguishable. B, Induction of transcription factor gene expression. Relative expression levels for ORE1, WRKY53, WRKY6, and AtNAP were determined by qPCR from three independent experiments. Means ± sd (n = 3) are shown for wild-type (black bars) and saul1-1 mutant (white bars) seedlings. Whereas ORE1 transcript levels were indistinguishable in wild-type and saul1-1 seedlings, the expression of WRKY53, WRKY6, and AtNAP was induced significantly.
Microarray Analyses Reveal Different Levels of Response in Low-Light-Induced saul1 Senescence and Cell Death
To study gene expression changes, saul1-1 mutant and wild-type seedlings were grown at permissive light (75 µmol m−2 s−1) for 11 d and then challenged with low light (20 µmol m−2 s−1) for 6, 24, or 48 h. Differential expression profiles of genes in saul1-1 compared with wild-type seedlings were determined by using Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays. Microarray hybridizations, data acquisition, and bioinformatics analysis of the microarray experiments were performed as described (Deeken et al., 2006; Duy et al., 2007) with adaptations (see “Materials and Methods”). For each time-point, T0 (low-light challenge: time = 0 h, just before transfer to low light), T6 (6 h), T24 (24 h), and T48 (48 h), the fold changes and adjusted P values for genes differentially expressed were calculated from three replicate saul1 mutant samples versus three replicate wild-type samples. On the basis of fold changes (log2) greater than 0.585 or (log2) less than −0.585 that met the significance criterion of adjusted P < 0.01, we found 24 up-regulated genes and one down-regulated gene at T0, 224 up-regulated and 134 down-regulated genes at T6, 2,031 up-regulated and 1,994 down-regulated genes at T24, and 2,737 up-regulated and 3,281 down-regulated genes at T48 (Supplemental Tables S1 and S2).
Previously, we determined the low-light-induced accumulation of abscisic acid (ABA) in saul1 mutant plants (Raab et al., 2009). Therefore, we checked for the regulation of ABA-dependent genes in low-light-grown saul1 mutants. Frequently used ABA marker genes were not regulated before T24 (ABI1 [At4g26080] and ATHB7 [At2g46680]) or T48 (ATHB12 [At3g61890] and RD26 [At4g27410]), suggesting that the ABA regulation of gene expression was not initiated at T6 (Supplemental Table S1). To confirm this suggestion, we screened the differential expression of genes that we previously identified to be responsive to ABA (Hoth et al., 2002) for fold changes (log2) greater than 0.379 or less than −0.379 meeting the significance criterion of an adjusted P < 0.05 in our microarray experiments. Of the 598 up- and 617 down-regulated ABA-responsive genes that were also present on the array, 31.4% and 37.3% were also up- and down-regulated in low-light-challenged saul1 plants, respectively (Supplemental Table S3). However, induction or repression did not appear until 24 h or even 48 h after transfer to low light (with very few exceptions), indicating that ABA control (like ORE1 regulation) of gene expression was not the earliest event.
To elucidate the events leading to saul1 cell death and senescence, we scanned the genes that were differentially expressed at T6 already. Strikingly, among the 224 up-regulated genes at T6, we found many genes that have previously been related to pathogen defense and the response to SA in Arabidopsis, including PR1, PR5, EDS1, EDS16, and PAD4 (Supplemental Table S1). Therefore, we compared the genes that were differentially expressed at T6 with gene expression changes upon pathogen or SA treatment. Of the 224 induced genes, 66% were also up-regulated in response to powdery mildew, indicating substantial overlap with biotic stress responses (Supplemental Table S4; Nishimura et al., 2003). Almost one-quarter of the 224 genes (22%) were also induced upon SA treatment (Supplemental Table S4; Blanco et al., 2009). Plant defense involves SA-mediated inhibition of auxin signaling by repressing 21 genes related to auxin signal transduction (Wang et al., 2007). With two exceptions, these genes were repressed at T24 in saul1 mutants (Supplemental Table S5). These data pointed to a participation of defense mechanisms and SA in the response of saul1 mutants to low light.
The PAD4-Dependent SA Pathway Is Required for saul1 Senescence and Cell Death
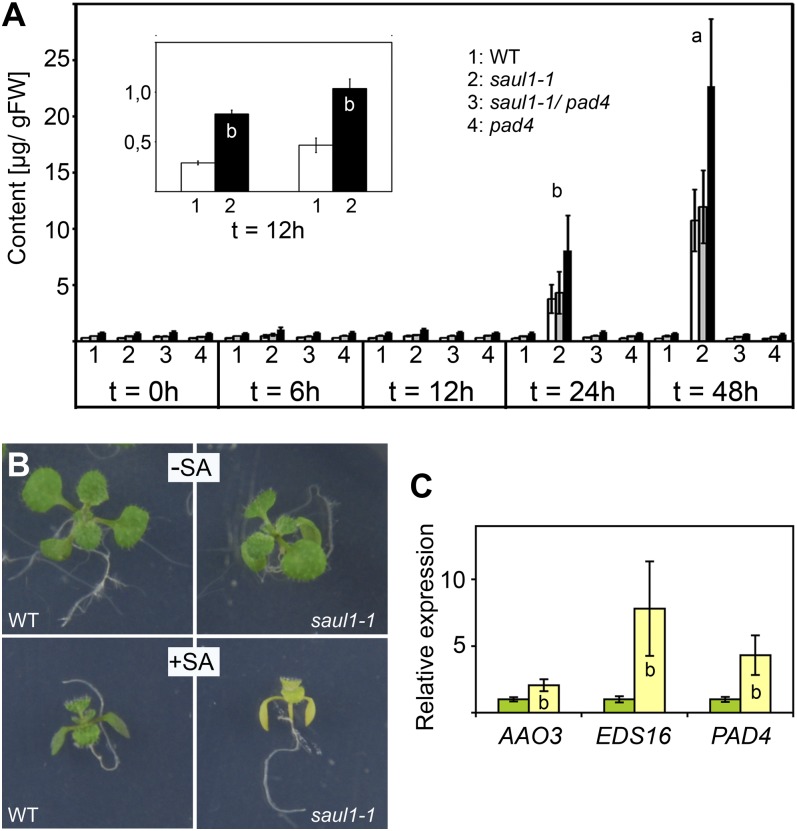
To further substantiate the role of SA in saul1 mutant phenotypes, we measured SA and SA glucoside contents in the wild type and saul1-1 mutants at 6, 12, 24, and 48 h after transfer to low light. Whereas no increase was observed in wild-type seedlings, SA and SA glucoside (and thus total SA) contents increased considerably in saul1-1 mutant seedlings at 24 and 48 h (Fig. 4A). However, increased contents could be detected already at 12 h (Fig. 4A, inset). In the next step, we studied the growth of mutant seedlings in the presence of SA in permissive light conditions. As expected from our gene expression data, saul1-1 mutant seedlings were indeed more sensitive to SA treatment than wild-type seedlings and showed yellowing of leaves (Fig. 4B). We monitored the expression of SA-responsive genes in the presence of SA in saul1-1 mutants compared with wild-type seedlings. Indeed, SA induction of EDS16 and PAD4 expression was much stronger in saul1-1 plants (Fig. 4C). The expression of AAO3 was also slightly changed. Taken together, our data suggested that SA signaling is an important event in low-light-induced saul1 cell death and senescence. To test this hypothesis, we crossed saul1-1 to pad4 plants that are defective in SA responses and isolated saul1-1/pad4 double mutants. In contrast to saul1-1 mutant seedlings showing growth arrest and yellowing of leaves in low light, these saul1-1/pad4 plants were indistinguishable from wild-type plants (Fig. 5A). We confirmed this also at the molecular level by analyzing AAO3, WRKY6, and EDS16 expression. Whereas transcript levels of all three genes were highly increased in saul1-1 mutants, their expression resembled wild-type levels in saul1-1/pad4 double mutants (Fig. 5B). The increase of SA and SA glucoside that was observed in low-light-treated saul1-1 mutants was also absent in saul1-1/pad4 double mutants (Fig. 4A). Disruption of the PAD4-dependent SA pathway was thus sufficient to suppress the low-light-induced saul1 phenotype. In contrast, disruption of the major SA signaling component NPR1 or of EDS5, which is required for SA biosynthesis, in saul1-1/npr1-1 or saul1-1/eds5-1 double mutants, respectively, did not suppress saul1 phenotypes (Supplemental Fig. S1, A and B). These data suggested that saul1 senescence specifically requires the PAD4-dependent SA pathway.
Figure 4.
Determination of SA content and SA-induced saul1 senescence. A, SA (white bars), SA glucoside (gray bars), and total SA (black bars) contents at the indicated times are shown for wild-type (WT), saul1-1, saul1-1/pad4, and pad4 plants. Means ± se (n = 6) are shown. Significant differences in SA content and total SA already at 12 h were only detected in saul1-1 mutants (inset). Significance differences were determined by Student’s t test analysis (a, P < 0.01; b, P < 0.05). FW, Fresh weight. B, When growing in the absence of SA in permissive light conditions, wild-type (top left) and saul1-1 mutant (top right) seedlings developed indistinguishably. SA treatment resulted in growth arrest of both wild-type (bottom left) and saul1-1 (bottom right) seedlings, but only saul1-1 mutants showed symptoms of senescence, namely yellowing of leaves. C, Relative expression levels for AAO3, EDS16, and PAD4 in the presence of SA were determined by qPCR. In comparison with the wild type, the expression of these genes was significantly increased in saul1-1 mutants. Means ± sd (n = 3) are shown for wild-type (green bars) and saul1-1 (yellow bars) plants.
Figure 5.
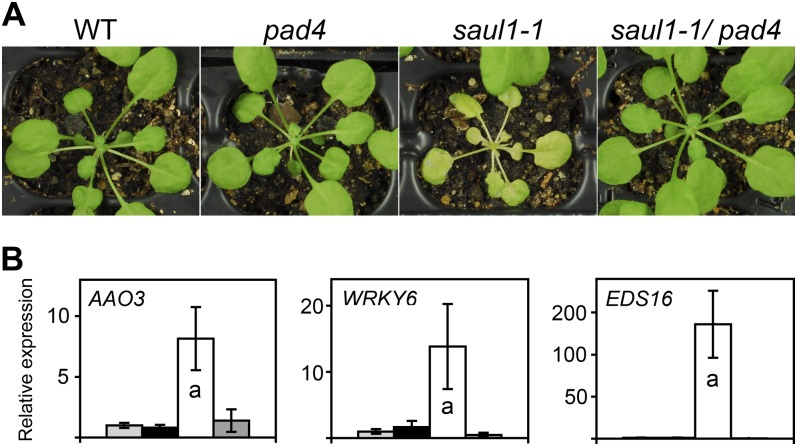
Rescue of the saul1 senescence and molecular phenotypes in saul1/pad4 double mutants. A, The phenotypes of wild-type (WT), saul1-1 and pad4 single mutant, and saul1-1/pad4 double mutant plants, which were grown for 4 weeks in permissive light, were studied 3 d after transfer to low light. The saul1 mutants but not the saul1/pad4 double mutants exhibited yellowing of leaves and reduced growth. B, Induction of gene expression of senescence- and SA-related genes. Relative expression levels for AAO3, WRKY6, and EDS16 were determined by qPCR from three independent experiments. Means ± sd (n = 3) are shown for wild-type (light gray bars), pad4 (black bars), saul1-1 (white bars), and saul1-1/pad4 double mutant (dark gray bars) seedlings. Induction of all three genes was only seen in saul1-1 seedlings. This induction was prevented in saul1-1/pad4 double mutants.
DISCUSSION
In this study, we demonstrate that saul1 mutants lacking expression of the E3 ubiquitin ligase gene SAUL1 appear to misjudge their developmental age and turn on a regulatory switch of age-dependent cell death in the young seedling stage. In wild-type Arabidopsis, this age-dependent switch involving ORE1, miR164, and EIN2 ensures that aging triggers cell death in leaves (Kim et al., 2009). During leaf development, down-regulation of miR164 allows for ORE1 accumulation. This age-dependent down-regulation requires ETHYLENE INSENSITIVE2, encoding a putative transport protein in the endoplasmic reticulum membrane, which is involved in ethylene signaling and leaf senescence (Oh et al., 1997; Alonso et al., 1999; Bisson et al., 2009). A critical developmental turning point occurs between leaf ages of 26 and 28 d. At this time, ORE1 accumulation apparently becomes sufficient to induce SAG12 expression and cell death (Kim et al., 2009). In saul1 seedlings challenged with low light, the turning point leading to cell death is reached at leaf ages below 10 d. Accordingly, ORE1 accumulation, SAG12 expression that is normally strictly associated with age-dependent senescence, and cell death can be detected prematurely (Fig. 1). In wild-type plants, SAUL1 function is thus crucial to prevent cell death under low-light conditions. However, through genetic analyses, we could show that disruption of ORE1/ANAC092 in saul1-1/anac092-1 double mutants did not result in the suppression of saul1 phenotypes. Apparently, enhanced ORE1 expression by itself is not sufficient to cause senescence and cell death in saul1 plants.
Obviously, our data raise an important question: is the onset of senescence and cell death in saul1-1 seedlings established at the molecular level before symptoms such as growth arrest and yellowing of leaves are visible? ORE1 accumulation in saul1 seedlings was observed after 2 d in low-light conditions, but ORE1 transcript levels in saul1 mutants were still indistinguishable from those in wild-type plants after 1 d of low-light treatment. In contrast, WRKY53, WRKY6, and AtNAP transcript levels were already significantly increased after 1 d (Fig. 3). Expression of these transcription factor genes has previously been shown to induce plant senescence (Robatzek and Somssich, 2002; Miao et al., 2004; Guo and Gan, 2006). These data suggest that WRKY53, WRKY6, and AtNAP are activated prior to ORE1 in saul1 mutants. It will be necessary to find out the hierarchy in the regulatory network of transcription during low-light-induced senescence and cell death in saul1 mutants compared with age-dependent processes in wild-type plants.
The microarray analyses indicated that SA-dependent signaling represents the first detectable regulatory event in low-light-challenged saul1-1 mutant seedlings. The SA biosynthetic genes EDS16 and EDS5 and the SA signaling components PAD4, EDS1, and WRKY70 that are crucial for SA signaling were present at higher levels already at T6. Various presumed defense effector genes were also activated, including PR1, PR2, and PR5 (Supplemental Table S1; Glazebrook, 2005). In line with the predominant association of SA with resistance to biotrophic pathogens, we not only found an overlap of gene expression changes in saul1 mutants with SA-triggered expression changes but also a high degree of overlap with expression changes upon treatment with powdery mildew (Nishimura et al., 2003; Wang et al., 2007). Full activation of SA and defense marker genes requires the signaling component NPR1 and involves transcriptional regulators such as TGACG MOTIF-BINDING FACTOR5 (TGA5) and TGA6 (Kim and Delaney, 2002; Kesarwani et al., 2007; Trujillo and Shirasu, 2010). The NPR1 gene and the genes encoding the transcriptional regulators TGA5 and TGA6 were indeed increased with a small delay at T24 (Supplemental Table S1). At T6, many genes with functions in the control of host defense against pathogens and of cell death, which is not essentially required for defense gene expression, were also regulated (Supplemental Table S1). It has been shown previously that SA causes the repression of auxin genes (Wang et al., 2007). This repression, which is thought to be an important aspect of SA-dependent defense responses, was observed in low-light-grown saul1 mutants at T24/T48 and may partly be responsible for the growth defects of saul1 seedlings in low light (Supplemental Table S5). At this point in time, and thus also following SA action, transcript levels of ABA-regulated genes were changed (Supplemental Table S3). At the gene expression level, an overlap between pathogenicity responses and ABA application has previously been described (de Torres-Zabala et al., 2007). In addition, genetic and molecular analyses of constitutive expressor of PR genes22 (cpr22) mutants showed that elevated SA content resulted in alterations of ABA signaling (Mosher et al., 2010).
The importance of SA pathways for saul1 phenotypes was supported by physiological and genetic analyses. Increased SA content was determined in low-light-grown saul1-1 mutants, exogenous application of SA was sufficient to trigger saul1 senescence (Fig. 4), and disruption of PAD4 in saul1-1/pad4 double mutants was sufficient to fully suppress saul1 defects (Fig. 5). However, disruption of NPR1 and EDS5 in saul1-1/npr1-1 and saul1-1/eds5-1 double mutants did not suppress saul1 phenotypes, thus pointing to a specific requirement of PAD4. The PAD4 gene encodes a protein with similarity to triacyl glycerol lipases, but PAD4 function could not be experimentally shown yet (Jirage et al., 1999). Initially, pad4 mutant alleles were identified in a screen for enhanced disease susceptibility (Glazebrook et al., 1996), and additional alleles were found when searching for mutants with reduced R gene-mediated resistance to Peronospora parasitica-specified resistance (Feys et al., 2001). When infected with the virulent bacterial pathogen Pseudomonas syringae pv maculicola ES4326, these pad4 mutants show typical properties, such as lower SA content, reduced expression of PR1, and lower content of the phytoalexin camalexin. Application of SA rescues many of the pad4 mutant phenotypes. Together with the putative lipase EDS1, PAD4 is thought to be part of an amplification loop that generates increased SA levels sufficient for SA signaling and defense (Wiermer et al., 2005). Direct interactions have been detected between PAD4 and EDS1, between EDS1 and SAG101, a well-known positive regulator of senescence, and between all three proteins in a ternary complex (He and Gan, 2002; Rietz et al., 2011; Zhu et al., 2011). It has been shown that PAD4 and EDS1 participate not only in resistance to pathogens but also in the transduction of photooxidative stress signals resulting in cell death and growth inhibition. Cell death and growth inhibition phenotypes of lsd1, acd6-1, cpr1, and cpr6 mutants require the presence of PAD4 and EDS1, as demonstrated by the rescue of mutant phenotypes in double mutants with pad4 or eds1 (Jirage et al., 2001; Rustérucci et al., 2001; Mateo et al., 2004; Ochsenbein et al., 2006; Mühlenbock et al., 2008; Ng et al., 2011). The SA amplification loop is also crucial for cell death and senescence, increased SA content, and gene expression changes in saul1 mutants challenged by low light (Fig. 5). Such a role of SA in the regulation of gene expression in cell death during the final stage of leaf senescence has previously been suggested from microarray analyses (Morris et al., 2000).
From the presented data, a timing of events for low-light-induced saul1 senescence and cell death can be deduced. The E3 ubiquitin ligase SAUL1 is associated with the plasma membrane (Drechsel et al., 2011). We hypothesize that target proteins are either modulated by monoubiquitination or subjected to degradation by polyubiquitination through SAUL1 at the plasma membrane. This regulation of target proteins is important to prevent cell death and senescence in response to low light or salt stress. In the absence of SAUL1 in saul1 mutants, the regulation of genes involved in defense and/or the SA responses was the first event (T6) that we resolved after transfer to low light. In line with a role of SA in saul1 senescence and cell death, SA content increased in saul1 mutants in low light and application of SA triggered growth arrest and yellowing of leaves in saul1 mutant seedlings growing in permissive light (Fig. 5). However, SA application did not result in yellowing of leaves but only in growth arrest in wild-type seedlings at this time. This indicated that SA is not sufficient to trigger yellowing of leaves in the presence of functional SAUL1 as easily as in the absence of SAUL1 in saul1 mutants, and thus that other changes in saul1 mutants are required for the complete SA response. This may depend on the activation of PAD4, because saul1 phenotypes were suppressed in saul1-1/pad4-1 double mutants. Following regulation of defense and SA genes, repression of auxin genes and regulation of ABA genes were detected at T24. This might be due to increased SA activity, because SA effects on the expression of auxin and ABA signaling components have been published previously (Wang et al., 2007; Mosher et al., 2010).
We have shown that during senescence and cell death, plasma membrane-associated SAUL1 is important in suppressing prodeath events. In animals, extrinsic signaling leading to programmed cell death has recently been shown to involve ubiquitin-dependent steps at the plasma membrane, too (Jin et al., 2009). Cell death control at the plasma membrane thus represents another facet of ubiquitination. The Arabidopsis RING1 E3 ligase that has a function in pathogen-induced programmed cell death is also associated with the plasma membrane in lipid rafts (Lin et al., 2008). Recently, it has been shown that the pepper (Capsicum annuum) plasma membrane-associated CaRING1 is part of the SA-dependent defense responses (Lee et al., 2011). Regulated ubiquitin/proteasome-dependent processing at membranes has been suggested as a possible mechanism of transcription factor regulation and thus gene expression control (Hoppe et al., 2001). Future research will help to identify in vivo targets that are modified by SAUL1 at the plasma membrane.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
Prior to sowing on petri dishes containing Murashige and Skoog salts, pH 5.7, 0.05% MES, and 0.8% phytoagar (Duchefa Biochemie), Arabidopsis (Arabidopsis thaliana) wild-type and saul1 mutant seeds were surface sterilized and stratified in the dark at 4°C for 3 d. Plants were grown in long-day conditions (16 h of light, 8 h of dark) at 21°C and photon flux densities of 20 to 100 µmol m−2 s−1 (as indicated in the text and/or figure legends) for the indicated numbers of days starting with transfer from dark to light after stratification. Photon flux densities were determined using a LI-250A light meter in combination with a Quantum sensor (LI-COR Biosciences). For SA treatment, SA was added to the medium to a final concentration of 0.5 mm. For studying NaCl-dependent gene expression, wild-type and saul1-1 plants were grown on agar for 8 d and then transferred to a hydroponics system containing 150 mm NaCl or control solution for 2 d prior to harvesting the samples. To provide independent biological replicates, three wild-type samples and three saul1-1 samples were collected for all treatments, and each sample consisted of five individual seedlings.
qPCR Analysis of Transcript Levels
qPCR analysis was performed as described (Raab et al., 2006). Samples were standardized to ACTIN mRNA levels. Gene expression levels were normally given relative to the expression levels in the wild type (set to 1) at the respective control conditions. Significances of differences were determined by Student’s t test analysis. The respective columns were labeled with letters a (P < 0.01) and b (P < 0.05) in the figures. Primers for PCR amplification were 5′-CTTACCATGGAAGGCTAAGATGGG-3′ and 5′-TTCCAATAACCGGCTTCTGTCG-3′ for ORE1 (At5g39610), 5′-TCCTTACAAAGGCGAAGACGCTAC-3′ and 5′-AACCGGGACATCCTCATAACCTG-3′ for SAG12 (At5g45890), 5′-CGCATCCTCACCGAGCGTACAAC-3′ and 5′-GGTGTTCTTGTGATCGCCTGC-3′ for WRKY53 (At4g23810), 5′-GGGAAGAGACTTGGGCGTG-3′ and 5′-GCAACGGATGGTTATGGTTTC-3′ for WRKY6 (At1g62300), 5′-GCCATTCACAGCGGTTCAAG-3′ and 5′-CAACAAATGAGCCAGCGAAC-3′ for AtNAP (At1g69490), 5′-CTCCGCCATGGTCTCTACTCATAC-3′ and 5′-ACTTAGGGAAGAAAGCCGATGATG-3′ for At2g15830, 5′-ACGACCATTAGCTTCGTGCTTCC-3′ and 5′-AAATCGCATTGGCCATGTTTCTCC-3′ for At5g39520, 5′-GCTTGGCTAGCACAGTTACAGC-3′ and 5′-CACTGCAGACACCTAATTGAGTCC-3′ for EDS16 (At1g74710), 5′-CGTGCTCATATCCTCTGCTGC-3′ and 5′-CCGTCAACGCAAATGGTCTG-3′ for SAG13 (At2g29350), 5′-CTCGTAATCTCAGCTCTTATTTG-3′ and 5′-CACTACACTCAAGTTGTTTGGAG-3′ for PR1 (At2g14610), 5′-GAGCATTTGTTCAAGGCATCGG-3′ and 5′-GCAAAGAGACTTCACCACAGGC-3′ for AAO3 (At2g27150), and 5′-AGATACGCGAGCACAACGCAAG-3′ and 5′-TTTCTCGCCTCATCCAACCACTC-3′ for PAD4 (At3g52430).
Trypan Blue Staining
To visualize dying cells, leaves were detached and submerged in lactophenol-trypan blue solution (0.03% trypan blue, 33% [w/v] lactic acid, 33% water-saturated phenol, and 33% glycerol). Samples were incubated at 99°C for 1 min followed by incubation at room temperature for 24 h, washed in chloral hydrate solution (2.5 g mL−1) to reduce background staining, and photographed using a Leica MZLIII stereomicroscope (Leica Microsystems).
Determination of SA and SA Glucoside Contents
Free SA and SA glucoside were extracted and analyzed as described with minor modifications (Voll et al., 2012). Per sample, four to seven seedlings (28–38 mg fresh weight) that were grown for 13 d in 100 µmol m−2 s−1 followed by low-light challenge (20 µmol m−2 s−1) for 0, 6, 12, 24, and 48 h were harvested, supplemented with 250 ng of o-anisic acid (Acros Organics, Thermo Fisher Scientific) as an internal standard, and then extracted once with 600 µL of 70% methanol at 65°C for 1 h and once with 600 µL of 90% methanol at 65°C for 1 h. The solvent of the combined extracts was then evaporated with a vacuum concentrator, followed by a precipitation step with 500 µL of 5% (w/v) TCA. Free phenols were partitioned two times against 600 µL of cyclohexane:ethyl acetate (1:1). The combined organic phases were evaporated and resuspended in 400 µL of 20% acetonitrile in 25 mm KH2PO4 (pH 2.6). The remaining aqueous phase was acidified with 1 volume of 8 m HCl and incubated at 80°C for 1 h. After partitioning as above, the organic phase was supplemented with 20 µL of water to prevent sublimation of SA, evaporated, and resuspended in 400 µL of 20% acetonitrile in 25 mm KH2PO4 (pH 2.6; i.e. the HPLC starting mobile phase). HPLC separation of SA and o-anisic acid was performed on a Dionex Summit system (P680, ASI-100, TCC-100, RF-2000) equipped with a Phenomenex Luna Security Guard C18 column (4.0 × 3.0 mm) followed by a 5-µm Luna C18(2) reverse-phase column (250 × 4.6 mm) as described by Voll et al. (2012).
Microarray Analysis, Data Preprocessing, and Differential Gene Expression Analysis
Total RNA for microarray analysis was isolated in two consecutive steps using Trizol reagent (Invitrogen) and the Plant RNeasy extraction kit (Qiagen) from wild-type and saul1-1 mutant seedlings grown on petri dishes in permissive light (60 µmol m−2 s−1) for 11 d and then challenged with low light (20 µmol m−2 s−1) for 6, 24, or 48 h. To provide biological replicates, three wild-type and three saul1-1 mutant samples were harvested for each time point. Each sample consisted of at least 15 individual seedlings. For microarray analysis, 5 µg of RNA was processed and hybridized to Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays using the Affymetrix One-Cycle Labeling and Control (Target) kit according to the manufacturer’s instructions. Raw signal intensity values were computed from the scanned array images using the Affymetrix GeneChip Command Console 3.0. Statistical gene expression analysis was performed analogously to the analysis described by Deeken et al. (2006). Data from gene expression microarrays were analyzed using the statistical software R (R Development Core Team; www.R-project.org) and add-on packages for microarray analysis from Bioconductor (Gentleman et al., 2004). Raw data were normalized with a variance stabilization algorithm (Huber et al., 2002). To summarize individual probes of an Affymetrix probe set, the median polish algorithm from the Robust Multichip Average normalization was used (Irizarry et al., 2003). Differentially expressed genes were assessed with a linear model approach implemented in the R package LIMMA (Smyth, 2004). Lists of differentially expressed genes were corrected for multiple testing with a false discovery rate (Benjamini and Hochberg, 2000), yielding an adjusted P value for each gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of saul1/npr1-1 and saul1/eds5-1 double mutants.
Supplemental Table S1. Up-regulated genes in low-light-treated saul1-1 mutant seedlings.
Supplemental Table S2. Down-regulated genes in low-light-treated saul1-1 mutant seedlings.
Supplemental Table S3. Regulation of ABA-induced genes in low-light-treated saul1-1 mutant seedlings.
Supplemental Table S4. Regulation of genes up-regulated at T6 in low-light-treated saul1-1 mutant seedlings by powdery mildew and SA.
Supplemental Table S5. Regulation of auxin-related genes, which were identified to be modulated by SA, in low-light-treated saul1-1 mutant seedlings.
Supplementary Material
Acknowledgments
We thank Tanja Bender for technical assistance, Magdalena Weingartner for discussions, Barbara Niemeyer for comments on the manuscript, and Salma Balazadeh and Bernd Müller-Röber for supplying seeds of anac092-1 mutants.
Glossary
- SA
salicylic acid
- qPCR
real-time reverse transcription-PCR
- ABA
abscisic acid
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B. (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Bassham DC. (2007) Plant autophagy: more than a starvation response. Curr Opin Plant Biol 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (2000) On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat 25: 60–83 [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G. (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6 [DOI] [PubMed] [Google Scholar]
- Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L. (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Coates JC. (2003) Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol 13: 463–471 [DOI] [PubMed] [Google Scholar]
- Deeken R, Engelmann JC, Efetova M, Czirjak T, Müller T, Kaiser WM, Tietz O, Krischke M, Mueller MJ, Palme K, et al. (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18: 3617–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Bergler J, Wippel K, Sauer N, Vogelmann K, Hoth S. (2011) C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J Exp Bot 62: 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wirén N, Soll J, Philippar K. (2007) PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19: 986–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD. (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker K, Davies KJ. (2004) Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol 36: 2519–2530 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S. (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- He Y, Gan S. (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S. (2001) Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol 13: 344–348 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH. (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. (1997) Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90: 871–882 [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics (Suppl 1) 18: S96–S104 [DOI] [PubMed] [Google Scholar]
- Huh GH, Damsz B, Matsumoto TK, Reddy MP, Rus AM, Ibeas JI, Narasimhan ML, Bressan RA, Hasegawa PM. (2002) Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J 29: 649–659 [DOI] [PubMed] [Google Scholar]
- Hyun DH, Gray DA, Halliwell B, Jenner P. (2004) Interference with ubiquitination causes oxidative damage and increased protein nitration: implications for neurodegenerative diseases. J Neurochem 90: 422–430 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. (2009) Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137: 721–735 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J. (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J 26: 395–407 [DOI] [PubMed] [Google Scholar]
- Kesarwani M, Yoo J, Dong X. (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Delaney TP. (2002) Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J 32: 151–163 [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn JW, Jin UH, Choi D, Paek KH, Pai HS. (2003) Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem 278: 19406–19415 [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. (2006) F-box proteins everywhere. Curr Opin Plant Biol 9: 631–638 [DOI] [PubMed] [Google Scholar]
- Lee DH, Choi HW, Hwang BK. (2011) The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol 156: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin SS, Martin R, Mongrand S, Vandenabeele S, Chen KC, Jang IC, Chua NH. (2008) RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J 56: 550–561 [DOI] [PubMed] [Google Scholar]
- Mateo A, Mühlenbock P, Rustérucci C, Chang CC, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S. (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 136: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. (2010) The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol 152: 1901–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng G, Seabolt S, Zhang C, Salimian S, Watkins TA, Lu H. (2011) Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189: 851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Noh YS, Amasino RM. (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Göbel C, Imboden A, Feussner I, Apel K. (2006) The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J 47: 445–456 [DOI] [PubMed] [Google Scholar]
- Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12: 527–535 [DOI] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76: 789–791 [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD. (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S. (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59: 39–51 [DOI] [PubMed] [Google Scholar]
- Raab S, Toth Z, de Groot C, Stamminger T, Hoth S. (2006) ABA-responsive RNA-binding proteins are involved in chloroplast and stromule function in Arabidopsis seedlings. Planta 224: 900–914 [DOI] [PubMed] [Google Scholar]
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Vlot AC, Feys BJ, Niefind K, Parker JE. (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Riggleman B, Wieschaus E, Schedl P. (1989) Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev 3: 96–113 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Aviv DH, Holt BF, III, Dangl JL, Parker JE. (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt JN, Yoshioka K, Moeder W, Goring DR. (2011) Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS ONE 6: e21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW. (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4: 948–958 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD. (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K. (2010) Ubiquitination in plant immunity. Curr Opin Plant Biol 13: 402–408 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernace VA, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. (2007) Aging and regulated protein degradation: who has the UPPer hand? Aging Cell 6: 599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Voll LM, Zell MB, Engelsdorf T, Saur A, Wheeler MG, Drincovich MF, Weber APM, Maurino VG. (2012) Loss of cytosolic NADP-malic enzyme 2 in Arabidopsis thaliana is associated with enhanced susceptibility to Colletotrichum higginsianum. New Phytol 195: 189–202 [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Yang TJ, Stead AD, Buchanan-Wollaston V, Roberts JA. (2009) A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 57: 690–705 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Yoshida S. (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Callis J, Nishida I, Watanabe A. (2002) A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J 32: 129–137 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P. (2011) SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog 7: e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.