Abstract
P-element vectors are commonly used to make transgenic Drosophila and generally insert in the genome in a nonselective manner. However, when specific fragments of regulatory DNA from a few Drosophila genes are incorporated into P-transposons, they cause the vectors to be inserted near the gene from which the DNA fragment was derived. This is called P-element homing. We mapped the minimal DNA fragment that could mediate homing to the engrailed/invected region of the genome. A 1.6 kb fragment of engrailed regulatory DNA that contains two Polycomb-group response elements (PREs) was sufficient for homing. We made flies that contain a 1.5kb deletion of engrailed DNA (enΔ1.5) in situ, including the PREs and the majority of the fragment that mediates homing. Remarkably, homing still occurs onto the enΔ1. 5 chromosome. In addition to homing to en, P[en] inserts near Polycomb group target genes at an increased frequency compared to P[EPgy2], a vector used to generate 18,214 insertions for the Drosophila gene disruption project. We suggest that homing is mediated by interactions between multiple proteins bound to the homing fragment and proteins bound to multiple areas of the engrailed/invected chromatin domain. Chromatin structure may also play a role in homing.
Introduction
P-element mediated transformation has been used to generate transgenic Drosophila for about 30 years [1]. In large-scale screens, P-element based vectors have been used for mutagenesis and for enhancer-detection [2]–[6]. In these cases, P-element vectors were found to insert in the genome in a relatively non-selective manner, with the exception of some hotspots for insertion. However, in a few specific cases, when particular fragments of regulatory DNA are included in a P-element vector, they dramatically alter the insertional specificity of that vector. The first case of this was described by Hama, Ali, and Kornberg in 1990 [7]; regulatory DNA from the engrailed (en) gene, when included in a P-element vector (P[en]), caused P[en] to insert in the vicinity of the en gene at a high frequency. They named this phenomenon P-element homing. Since that time, regulatory DNA fragments from the linotte gene, bithorax complex, and the even-skipped (eve) gene have been found to mediate homing [8]–[10]. For homing mediated by en, bithorax, and eve DNA, insertions are not site-specific but are regional; that is, they occur in the vicinity of the endogenous gene but can be distributed over a large genomic region. For homing by P[en], insertions occur over about a 300 kb region, including en, inv and flanking genes [7], [11], [12].
The segmentation gene en exists in a gene complex with the related gene invected (inv). en and inv encode homeodomain-containing proteins, are co-expressed, and share regulatory DNA [13]. en and inv are regulated by the Polycomb group genes (PcG) [14]. In tissue culture cells and embryos, H3K27me3, the distinctive PcG chromatin modification, covers a 100kb region that includes en and inv, but not flanking genes [15]. In chromatin immunoprecipitation experiments in tissue culture cells and adults, there are three regions of en/inv that associate with PcG proteins, one just upstream on the en promoter, one coincident with an inv promoter, and one 6 kb upstream of inv [15], [16]. These fragments of DNA act as Polycomb group response elements (PREs) in transgenic Drosophila [17]–[19]. The PRE located upstream of the en gene is a complex element, including 2 minimal PREs [18], [20].
We have been using P-element homing by en DNA as a tool for our experiments for about 20 years. For example, we used a homing P-element to insert a mosquito en cDNA onto an en mutant chromosome to show it can substitute for the Drosophila en protein coding region [11]. More recently, we used P[en] homing to generate a large number of P[en] insertions near en and study the long-range action and promoter specificity of en enhancers [12]. Here we took two experimental approaches in order to further understand the mechanism of P[en] homing: 1) we dissected the en homing fragment and 2) we deleted the homing fragment from the genome, and asked whether homing could still occur. Our data show that the minimal fragment of DNA that can mediate homing is 1.6kb. Since we could not get a smaller fragment of DNA to mediate homing, we suggest that homing is not caused by a single protein or protein complex. Second, we found that homing still occurs, albeit at a reduced frequency, into genomic DNA that lacks the majority of the homing fragment. Therefore, homing is not solely dependent on interactions between the genomic and P[en] homing fragments. Our data suggest that homing is mediated by multiple protein-protein interactions between proteins bound to the homing fragment in P[en] and proteins bound to multiple regions of the en/inv genes. Since homing occurs in the germ line, we suggest that the en/inv genomic region is bound by many regulatory proteins and has a specific chromatin structure in germ cells.
Results
Identifying a minimal homing fragment
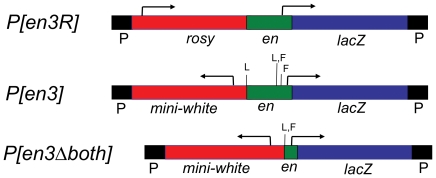
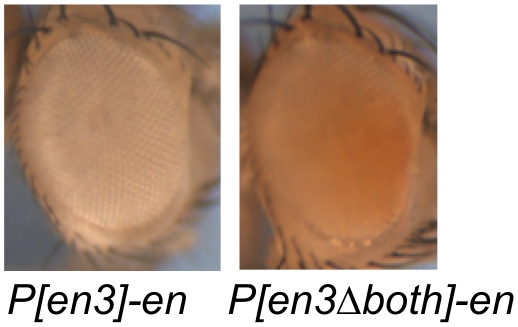
In previous experiments, we found that a 2.6 kb DNA fragment, extending from −2.4 kb to +188 bp was sufficient to mediate homing to the vicinity of the en/inv region of the genome at a frequency of 5% (7 out of 131 insertions; [21]) (Construct P[en3R], Fig. 1, called P[en1] in [21]). Since that time, whenever this en fragment was included in a P-element vector we localized the P-insertions in the genome. In 2002, we studied the pairing-sensitive silencing properties of the 2.6kb fragment, and generated 43 lines, using mini-white as the reporter gene to recognize transgenic flies [17]. Insertions into the vicinity of en/inv region occurred at a frequency of 4.6% (2/43; Table 1). In another study, we generated 21 lines using P[en3] (Fig. 1), and obtained two insertions in the vicinity of en/inv (a frequency of 10%; [18]). In P[en3], the reporter gene used to recognize transgenic flies is mini-white. Notably, flies that contained P[en3] inserted 6kb upstream of en (P[en3]-en) had white eyes (Fig. 2). This line was recovered fortuitously; when using inverse PCR to localize the site of a P[en3] insertion, we identified two insertions in one transgenic fly, one on chromosome 3R, and the other at en (P[en3]-en). The white eye color of P[en3]-en is due to the repression of mini-white by the PREs in the 2.6kb fragment, since deletion of both PREs by Cre recombinase and Flp recombinase to generate P[en3Δboth] gives flies with colored eyes (Fig. 2; no eye color was seen when either one of the PREs was present). In contrast, flies with P[en3] inserted into tou, the gene just upstream of en, have light orange eyes (not shown). Using constructs similar to P[en3], but with different reporter genes (an en-lacZ fusion protein, construct F in [22]; the mosquito en cDNA [11]) we know that insertions into the inv intron and just downstream of the en transcription unit give flies with colored eyes. Nevertheless, because mini-white silencing by the PREs would lead us to miss insertions in the vicinity of the en promoter, the homing frequency obtained in experiments using P[en3]-mini-white vectors is probably an underestimation of the true homing frequency.
Figure 1. P-constructs used in homing experiments.
Arrow shows the direction and start of transcription. L, loxP site. F, FRT site. P[en3R] is from [21] (called P[en1] in that study). P[en3] and P[en3Δboth] are from [18].
Table 1. Insertions near en/inv on wild-type chromosomes.
| Construct | Linea | Positionb | Genec | Locationd | Reference |
| 2.6lacZ | 2A | −751 bp | en | 7416139 | 17 |
| 2.6up | 4A | −87 kb | tou | 7492660 | 17 |
| P[walter] | 48A | −67 kb | tou | 7482784 | 26 |
| P[walter] | 4A | −86 kb | tou | 7501690 | This study |
| P[en3] | en | −6 kb | en | 7421628 | 18 |
| P[en3R] | 1A | −472 bp | en | 7415760 | This study |
| P[en3R] | 11A | −72 kb | tou | 7487611 | This study |
| P[en3R] | +52kb | +52 kb | inv | 7363188 | 12 |
| P[en3R] | −87kb | −87 kb | tou | 7502613 | 12 |
Line names
The number of kilobases or base pairs upstream (negative numbers) or downstream (positive numbers) of the major en transcription start site (at 7415388) [35].
Closest gene.
Nucleotide insertion site (genome version R5.1, FlyBase).
Figure 2. PREs silence mini-white expression in P[en3]-en eyes.
Eyes of 5-day old male flies are shown. w; P[en3]-en/+ (left) and w; P[en3Δboth]-en/+ (right).
Experiments designed to test PRE activity lead us to use the vector pUZ [23]. In this vector, the mini-white promoter is separated from the PRE by the lacZ transcription unit (Fig. 3). We made a pUZ construct containing a 2 kb en fragment, extending from −2.4 to −0.4 kb upstream of the en transcription unit (P[enHSP1] [12]; Fig. 3). Notably, when P[enHSP1] was inserted in the genome just upstream of en, the flies had colored eyes (not shown). The homing frequency of P[enHSP1] was 15% (7/46 lines; one line had two insertions, one in en and one in inv, this line was only counted once). Similar to the previous experiments, P[enHSP1] insertions were not site specific but occurred over a 149kb region with two insertions in en, two in inv, three in tou, and one in E(Pc) [12].
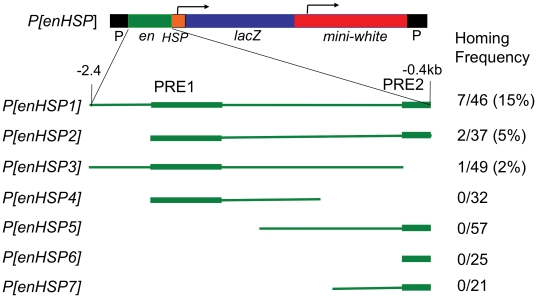
Figure 3. P[enHSP] constructs used to dissect the homing fragment.
The P[enHSP] construct is shown on top with the extent of the en DNA present shown below by the green line. The PREs are shown as thicker areas of the green line. The homing frequency is shown on the left; the number of insertions in the en/inv region/the total number of transgenic lines recovered is shown on the right side. P[enHSP1], P[enHSP2], and P[enHSP3] are from [12].
We used the pUZ vector to narrow down the DNA fragment required for homing. Deletion of the 181bp en PRE decreased the homing ability of this fragment (P[enHSP3]) when compared with P[enHSP1] (2% versus 15%, P = 0.0274 by a Fisher's exact test). Therefore we conclude that the 181bp fragment contributes to homing efficiency. Can the 181bp-en PRE alone mediate homing? Over the years we have generated 166 lines with the 181-bp fragment cloned upstream or downstream of mini-white in various constructs ([17], [20] and P[enHSP6] in Fig. 3) and mapped their insertions sites using either recombination mapping, hybridization to polytene chromosomes, or inverse PCR, and never recovered an insertion near the en gene. Also, in this study, P[enHSP5] and P[enHSP7] contain the 181-bp fragment and we did not obtain any inserts of these two constructs into the vicinity of en/inv. Thus we conclude that the 181-bp fragment is not sufficient for homing to en.
Data from a 1994 study [20] led us to test the ability of the fragments in P[enHSP4] and P[enHSP5] to home. In that study, one P-insertion from a pCaSpeR construct with sequences extending from −1184 to −944 bp was localized to polytene band 48A on salivary chromosomes, out of 4 insertion events (its exact location was never determined and the line was lost many years ago). Second, we obtained an insertion of a pCaSpeR construct containing sequence −1944 to −1166 bp into E(Pc), out of a total of 13 insertion events. Both P[enHSP4] and P[enHSP5] contain the fragment extending from −1184 to −944 bp. Out of 89 lines obtained, none was inserted in the vicinity of en. P[enHSP4] contains the sequences −1944 to −1166 bp. We did not obtain any insertions into en in the 32 lines recovered. Thus, our results suggest that if these fragments cause homing, it is at a very low frequency.
In this study we did find one sub-fragment that mediates homing. P[enHSP2] contains a 1.6kb fragment that includes the two PREs. Two out of 37 insertions of this construct were at en (5%). The homing frequency of P[enHSP2] is not significantly different from that obtained with P[enHSP1] (P = 0.2867 by a Fisher's exact test). Therefore, we consider the 1.6kb fragment to be the minimal homing fragment.
Homing occurs in flies that lack the genomic homing fragment
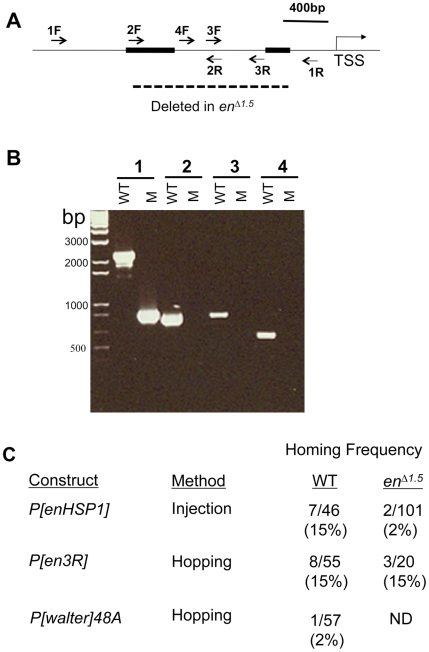
We generated a 1.5kb genomic deletion that removes both PREs upstream of the en transcription start site (Fig. 4A, B), enΔ1.5kb. These flies are both viable and fertile, and have a loss-of-function wing phenotype similar to that of enΔ530, which contains a smaller deletion in the same region [18]. P[enHSP1] was injected into enΔ1.5kb homozygous embryos; two insertions (out of 102) were in the en/inv chromosomal region; one insertion was into the en promoter, and one was in the inv promoter (Table 2, Fig. 4C). The frequency of homing into enΔ1.5 flies was reduced as compared to wild-type (2% vs. 15%; P = 0.0041 by a Fisher's exact test), however, it is notable that it occurred at all, given that majority of the fragment required for P-element homing was deleted from the endogenous en gene in these flies.
Figure 4. Characterization of and homing to the enΔ1.5 chromosome.
(A) Thin line represents DNA upstream and including the transcription start site (TSS) of en. PRE1 and PRE2 are depicted by rectangular boxes. The primers used for PCR reactions are indicated. The extent of the enΔ1.5 deletion is indicated by the dashed line. (B) PCR reactions using wild type (WT) or enΔ1.5 DNA (M) and the following primer sets (1) 1F and 1R (2) 2F and 2R (3) 3F and 1R (4) 4F and 3R. The M PCR product from primer set 1 was sequenced and shows that 1462 bases are deleted in enΔ1.5. (C) Homing frequency onto wild type and enΔ1.5 chromosomes. Homing frequency is the number of insertions near the en/inv region/the total number of transgenic lines. ND, not done.
Table 2. Insertions near en/inv on chromosomes carrying enΔ1.5.
| Construct | Linea | Positionb | Locationc |
| P[en3R] | inv | +52 kb | 7363188 |
| P[en3R] | en | −216 bp | 7415604 |
| P[en3R] | CG9005 | −105 kb | 7520040 |
| P[enHSP1] | inv | +52 kb | 7363272 |
| P[enHSP1] | en | −26 bp | 7415414 |
Lines are the named by the closest gene. All lines were generated for this study.
The number of kilobases or base pairs upstream (negative numbers) or downstream (positive numbers) of the major en transcription start site (at 7415388) [35].
Nucleotide insertion site (genome version R5.1, FlyBase).
As an additional test as to whether homing could occur onto enΔ1.5, we mobilized P[en3R]-87kb, an insertion in the tou gene on a CyO chromosome with P(Δ2,3) transposase [24] and screened for transposition off the CyO chromosome onto the enΔ1.5 chromosome. Of 16 transposition events, two occurred into the vicinity of en/inv; one 1.2kb downstream of the inv promoter, and another insertion 20kb upstream of tou (a homing frequency of 12.5%) (Table 2). Mobilization of P[en3R]+52kb, inserted in inv on a CyO chromosome also generated an insertion on enΔ1.5 near en/inv, upstream of CG9005 (out of 4 transposition events, Table 2, Fig. 4C). As a control we mobilized P[en3R]-87kb with P(Δ2,3) transposase in a wild-type 2nd chromosome background and obtained one insertion into the tou gene and another insertion 55 bases upstream of the en transcription start site (2/21 transposition events, a frequency of 9.5%, Table 1; Fig. 4C). In another experiment we mobilized a different P[en3R] insertion in tou to generate 34 transposition events in a wild-type 2nd chromosome background and 6 were in the vicinity of en/inv (17%). Thus, by P-element mobilization, the frequency of homing is similar onto a wild-type and enΔ1.5 chromosome.
P-elements are known to transpose locally, and, in one study, it was reported that there was also preferential transposition of P-elements to a corresponding region of the homologous chromosome [25]. Therefore we wondered whether the high frequency of transposition events obtained from P[en3R] inserted into tou and inv were the result of local transposition or representative of homing to the en/inv region. Of note, CyO does not disrupt homolog pairing in the vicinity of en/inv, thus, local transposition was a distinct possibility. We tested whether there is preferential transposition to a corresponding region of the homologous chromosome in the vicinity of en/inv using a P[walter] element inserted in tou (Table 1). P[walter]-48A contains a modified mini-white gene used to detect gene conversion events [26] and does not contain a PRE. Thus, by analogy with P[en3]Δboth, mini-white would not be expected to be silenced if P[walter] is inserted in the vicinity of en/inv. We looked for transposition of P[walter]-48A off a wild-type chromosome onto either a CyO or a chromosome carrying the marker Sp, but wild type for the en/inv region. 52 P[walter] transposition events were found, and only one was near en, inserted in tou, a frequency of 1.9% (1/52). This is much lower than rates obtained by transposition of P[en3R] onto either a wild-type (8/55) or enΔ1.5 chromosomes (3/20) (P = 0.0126 by a Fisher's exact test compared to wild-type; P = 0.0516 compared to enΔ1.5). We suggest that the high rate of insertion into the en/inv region via transposition from P[en3R] inserted in tou or inv is largely the result of homing and not local transposition.
Are there other preferred targets for P[en] constructs?
To address the question of whether constructs containing the homing fragment have additional preferred targets, we analyzed the insertion sites of all 40 P[enHSP1] insertions generated by injection into enΔ1.5 embryos that mapped to chromosome 2 (Table 3). We reasoned that the enΔ1.5 chromosome should not alter the chromatin structure of areas outside the en/inv region of the chromosome. In this experiment, 40 of 102 P[enHSP1] insertions were on chromosome 2, including one at the en and one at the inv promoter (Table 3). We recovered four insertions into lola, a hotspot for P-element insertions, two into bun, another P-element hotspot, showing that P[enHSP1] behaves like other P-elements with respect to P-element hotspots [3]. In addition, we obtained two insertions into the gene Vha16-1 (Table 3). Vha16-1 is not a reported hotspot for P-element insertions although there are 29 P-element inserts in this vicinity recorded in FlyBase. From these data we conclude that P[enHSP1], aside from it's homing to en/inv region, behaves like most other P-elements in its target site selection.
Table 3. P[enHSP1] insertions on chromosomes carrying enΔ1.5.
| Linea | Locationb | Genec |
| 5A | 2L:109235 | Sam-S |
| f-14-3 | 2L:11057544 | Samuel |
| f-11 | 2L:11173008 | Vm32E |
| 16A | 2L:12011113 | Rha5, CG6734 |
| m-55-L | 2L:12539996 | bun * |
| m-58 | 2L:12540005 | bun * |
| 4E | 2L:14234118 | smi35A |
| f-29V | 2L:14490554 | noc |
| m-55-S | 2L:14997589 | mol |
| f-89 | 2L:16485712 | dac |
| f-14-1 | 2L:16685386 | grp * |
| f-28-1 | 2L:20382385 | CG16798 |
| 10A | 2L:20972144 | CG42238 |
| 2B | 2L:21618794 | CG2201 |
| 4D | 2L:21828581 | tsh |
| 2A | 2L:22019207 | CR33987 |
| 6A | 2L:5108441 | Msp-300 |
| 30f1 | 2L:5981758 | eIF-4A |
| 14C | 2L:8416598 | Akap200 |
| m-98 | 2R:12468461 | Cdk4 |
| m-69 | 2R:12744739 | Dek |
| 2C | 2R:1632293 | ap |
| f-23 | 2R:18100371 | ari-2 |
| m-3-1 | 2R:19757865 | ken |
| m-53-L | 2R:20289791 | slik |
| 12A | 2R:20857342 | CG3776 |
| f-28-2 | 2R:221423 | Gprk1 |
| m-47 | 2R:2518935 | Vha16-1 |
| m-45 | 2R:2519424 | Vha16-1 |
| m-74 | 2R:2839867 | mim |
| f-58 | 2R:2993199 | esn |
| 14E | 2R:4061646 | CG14757 |
| 15C | 2R:6421820 | lola * |
| 14B | 2R:6422854 | lola * |
| 3A | 2R:6429207 | lola * |
| 15A | 2R:6429232 | lola * |
| inv | 2R:7363272 | inv |
| en | 2R:7415414 | en |
| 6B | 2R:7779622 | E1alpha48D |
| f-12 | 2R:8481109 | Amph |
Name of line.
Nucleotide insertion site (genome version R5.1).
Nearest gene(s).
*Hotspot for P-element insertions [3]. PcG targets are shown in bold.
It has been suggested that P-elements that contain PREs insert in the genome near PcG-regulated genes (reviewed in [27]). Therefore, we examined if any of the P[enHSP1] insertion sites were in PcG targets. For PcG targets we used the class I high confidence target genes from Schwartz et al., 2010 [28]. Insertions of P[enHSP1] were found in 6 PcG target genes: CR33987, apterous, teashirt, dachshund, en, and inv. Notably, these genes are not hotspots for P-element insertions.
It was recently reported that Polycomb-target genes are cold spots for P-elements and other transposons [4]. In that study, the sites of 18,214 unselected P{EPgy2} insertions were determined [4]. Of these 18,214 insertions, only 156 were into class I high-confidence PcG targets, including 62 in apt, and 20 in esg, both P-element hotspots. There are no P{EYgy2} insertions into en, inv, or dachshund. The number of P[enHSP1] insertions in PcG targets (6/102) is significantly different than the number of P{EYgy2} (156/18,214) (P<0.0001, chi-square test). Thus, our data is consistent with a model that P-elements that contain PREs have an increased frequency of insertion into PcG-regulated genes.
Discussion
A 1.6kb fragment of en DNA can mediate homing
Our previous results indicated that a 2.6 kb fragment of en DNA, extending from −2.4 kb upstream through +188bp of the en transcription unit could mediate P-element homing to the en/inv domain [21]. Here we show that a 1.6kb fragment that extends from −2.0 kb through −0.4 kb is sufficient for homing. We had hoped to identify a small fragment of en DNA that could mediate homing, but this was not the case. Thus we suggest that homing is mediated by a complex array of proteins and/or chromatin structure.
PcG proteins are thought to mediate long-range chromatin interactions at the Bithorax complex [29], between the Bithorax and Antennapedia complexes [30], and also between PcG targets on the same chromosome arm [31]. We note that one study suggests that the interactions at the Bithorax complex are not mediated by PREs, but by closely associated insulator elements [32]. Biochemical studies show that PcG protein complexes can interact in vitro [33]. Our results suggest that PREs play a role in P[en] homing: 1) deletion of the 181-bp PRE in the transgene decreases the homing frequency and 2) P[enHSP1] insertions occur in PcG-regulated genes at a higher frequency than P{EYgy2} insertions. We note that both the eve and Bithorax homing fragments are thought to be insulator elements [9], [10]. The en homing fragment is located just upstream of the en promoter and we consider it unlikely that it is an insulator. However, the insulator proteins GAGA Factor, CTCF, and Mdg4 are associated with this DNA in embryos [15]. Therefore, it is possible that the homing fragment has some of the same properties as insulators.
In our previous study [21], we found that embryonic lacZ expression from P[en3R] (called P[en1] in that study) occurred in stripes at a much higher frequency than with the enhancer trap P[lacW]. Our hypothesis was that P[en3R] caused selective insertion of P[en3R], not just to en/inv, but also to many genes expressed in stripes. We know now that both the en promoter and en PREs (or sequences closely associated with them) mediate interactions with distant enhancers [12], [18]. Thus, one reason for the enriched number of lacZ stripe patterns with P[en3R] could be its ability to work with distant enhancers. In support of this, when P[en3R] is inserted up to 140 kb and 5 transcription units away from the nearest en stripe enhancer (either upstream or downstream), P[en3R]-encoded lacZ is still expressed in en-like stripes [12]. In contrast, when P[lacW] is inserted about 45kb upstream of the nearest en stripe enhancer, into tou, the gene adjacent to en, P[lacW]-encoded lacZ is not expressed in stripes [12]. In fact, the PREs in P[en3] facilitate long-distance interactions with enhancers in many different regions of the genome [18]. We suggest that the high percentage of striped lacZ expression from P[en3R] insertions is due both to the ability of the en promoter and PREs to act with distant enhancers and also to increased insertion into PcG-regulated genes, many of which are developmental regulators and expressed in stripes.
Homing in enΔ1.5
We generated a 1.5 kb deletion of en DNA in situ, including the two PREs. Surprisingly, enΔ1.5 flies are homozygous viable and fertile. En expression appears normal in these flies (data not shown). We suggest that these PREs are redundant with inv PREs and that the en/inv H3K27me3 domain is not disrupted in enΔ1.5 flies. Interestingly, P[en] homing still occurs in enΔ1.5 flies. These data suggest that homing is not mediated solely by self-self interactions between the homing fragment in P[en] and the genomic homing fragment.
A model for P[en] homing
Fig. 5 shows a model for P-element homing. We suggest that P[en] homing is mediated by the interaction of multiple proteins bound to the en fragment within P[en] and proteins bound to the en genomic region, and that these interactions are facilitated by the H3K27me3 mark characteristic of PcG target genes. Note that since P[en] insertions into the en/inv target occur much more frequently than into other PcG-target genes, protein-protein interactions, specific for the en/inv region must be involved in homing. The smallest fragment that could mediate homing was 1.6kb, a size capable of binding many proteins. This suggests that P[en] homing is not caused by a binding of a single protein or protein complex. However, it is also possible that the 1.6 kb fragment is needed to form the chromatin structure that facilitates homing. Finally, we suggest that P[en] interacts with multiple proteins bound to the en/inv domain, since homing still occurs in enΔ1.5, where the majority of the genomic homing fragment has been deleted.
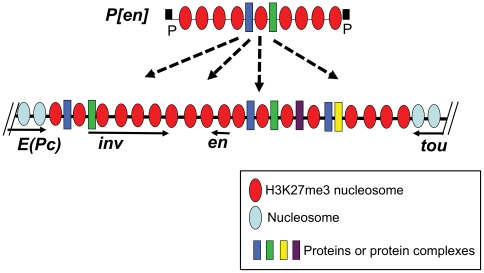
Figure 5. A model for P[en] homing.
P[en] and the en/inv DNA are packaged in H3K27me3 nucleosomes (red ovals) [15]. Nucleosomes associated with tou and E(Pc) do not have the H3K27me3 mark (blue ovals) [15]. Specific proteins bound to both P[en] and en/inv DNA are represented by different colored rectangular boxes. We suggest that the en DNA in P[en] adopts the same chromatin structure and has the same proteins bound to it as in the genomic context. (P[en] is not drawn to scale). Since the inv promoter is co-regulated with en, and also has a PRE associated with it, we draw its structure as identical to the en promoter. We suggest that P-element homing occurs through specific interactions between P[en] and multiple proteins bound to the genomic en/inv region and that chromatin structure also plays a role. These interactions cause P[en] to be concentrated in the vicinity of en/inv and then transposition occurs into en/inv and genes nearby.
P-element homing occurs in germ cells. En is not expressed in these cells. Recent results indicate that the H3K27me3 modification is present at many developmental loci in germ cells (reviewed in [34]). P[en] homing suggests that, in addition to the H3K27me marks, there are specific proteins bound to en DNA in the germ cells. These proteins could be present to keep en silenced, or perhaps they are there to facilitate rapid initiation of en transcription in the embryo.
Materials and Methods
Construction of plasmids
P[enHSP] constructs were generated by inserting PCR fragments with SpeI and NotI ends into pUZ [23]. Inserts were generated by PCR with the following en primers. P[enHSP1] (−2.407 to −0.395 kb): P1-GGGGCGGCCGCGAATTCCGTTGATATGAT and P2-GCGACTAGTGCATGCTGGAGCTGTCAG; P[enHSP2] (-1.945 to -0.395kb): P3-GCGGCCGCGAAAGTGTGTAGGGGAAT and P2; P[enHSP3] (-2.407 to -0.579kb): P1 and GCGACTAGTCCACAGACACTTTTC [12]; P[enHSP4] (-1.945 to -0.847kb) P3 and GCGACTAGTGAGGCCTTCAATTAACCA; P[enHSP5] (-1.495 to -0.395kb): GCGGCCGCGCGCATAAAAATTGA and P2; P[enHSP6] (-0.576 to -0.395kb): GCGGCGGCCGCGAGATGGCATGTGGCTCTC and P2; P[enHSP7] (-0.941 to -0.415kb): GCGGCGGCCGCCGATGGGCAATATAAATTAAATG and GCGACTAGTGGTTGACAACTGTGTCCCCAGCG. The PCR-amplified fragments were cut with both Spe1 and Not1 and ligated into pUZ. The resulting clones were sequenced in and around the cloned PCR fragment to ensure sequence fidelity.
Transgenic lines
P[enHSP] lines were generated by injections into w1118 embryos by Genetic Services (Sudbury, MA, USA). For P[enHSP] inserts that mapped to chromosome 2, recombination mapping was used to determine whether they were in the vicinity of en. For P[en3R] mobilization, P[en3R]-87kb or P[en3R]+52kb CyO/Sp or enΔ1.5; P(Δ2,3)99B Sb ry506/ry506 males were crossed to ry506 females. ry+ non-CyO progeny represented transposition events. All P[en3R] insertions on the second chromosome were localized by inverse PCR as described [12]. For P[walter]-48A mobilization, w*; P[walter]-48A/Sp or CyO; P(Δ2,3)99B Sb ry506/+ males were crossed to y Df(1)w67c2 females and w+ Sp or CyO males were selected and mapped with respect to chromosome. Insertions sites on the CyO chromosome were determined by inverse PCR [12]. Insertions on the Sp chromosome were mapped via recombination mapping to determine whether they were tightly linked with en.
Generation of enΔ1.5
enΔ1.5 was obtained in an experiment designed to recover a gene conversion event replacing wild-type en DNA (Fig. S1). It arose as the result of an imprecise excision of a P-element inserted 412bp upstream of the major transcription start site of en. The primer sequences used to characterize the deletion are: 1F: CAGTGCGACAATTGAGTTG, 1R: GCTTGTTAGGCAGCAAT, 2F:GGAAAGTGTGTAGGGGAAT, 2R:GAATCTGTTCGATGTGA, 3F:TCACATCGAACAGATTCG, 3R: ATCGATTTGCCAGACGAG. 4F: TTCAAGTCCATTGATC. The enΔ1.5 deletion includes 1462 bp of genomic sequence from 7415804 to 7417265 (genome release 5.36), and it leaves 32bp (CATGATGAAATTATGTTAATAACATAATAATTA) in this genomic location.
Supporting Information
Generation of enΔ1.5 . (A) enΔ1.5 was generated by a crossing scheme designed to obtain a gene conversion event by P-element excision [26]. The recipient chromosome was marked with Sp and contained a ry+ P-element inserted 412 bp upstream of the en transcription start site. The donor chromosome contained a 16kb deletion of en DNA (enΔJ83B) from −15 kb upstream through the first intron of en (generated in our lab), and the donor P-element that contained LoxP sites and FRT sites flanking (indicated by vertical lines in the en DNA) the PREs (P[en2] from [18]). We were trying to get a gene conversion event that would put LoxP and FRT sites into the genome. We set up 400 vials of Cross 1 and 300 vials of Cross 2. From this, we obtained 2 potential deletions (enΔ1.5 and one other) and one potential gene conversion event. The 2 deletions were recovered and balanced from Cross 3 but the gene conversion event was not recovered. (B) Schematic representation of the PCR reactions used to detect the gene conversion event. L(LoxP), F(FRT). A, B are approximate primer locations.
(TIF)
Acknowledgments
We thank Bill Engels for the P[walter]-48A flies; Jeff Americo for inverse PCR on P[walter]-48A transposition lines; Sarah DeVido for making some of the P[enHSP] constructs; and Elissa Lei, Jim Kennison, and Payal Ray for discussion and comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Child Health and Human Development (NIH, NICHD). Diane Mucci was supported by a grant from the Cystic Fibrosis Foundation. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Rubin GM, Spradling, AC Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 2.Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, et al. P-element-mediated enhancer detection-A versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 3.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, et al. The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 6.O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hama C, Ali Z, Kornberg TB. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- 8.Taillebourg E, Dura JM. A novel mechanism for P element homing in Drosophila. Proc Natl Acad Sci U S A. 1999;96:6856–6861. doi: 10.1073/pnas.96.12.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- 10.Fujioka M, Wu X, Jaynes JB. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development. 2009;136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiteley M, Kassis JA. Rescue of Drosophila engrailed mutants with a highly divergent mosquito engrailed cDNA using a homing, enhancer-trapping transposon. Development. 1997;124:1531–1541. doi: 10.1242/dev.124.8.1531. [DOI] [PubMed] [Google Scholar]
- 12.Kwon D, Mucci D, Langlais KK, Americo JL, Devido SK, et al. Enhancer-promoter communication at the Drosophila engrailed locus. Development. 2009;136:3067–3075. doi: 10.1242/dev.036426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustavson E, Goldsborough AS, Ali Z, Kornberg TB. The Drosophila engrailed and invected genes: Partners in regulation, expression, and function. Genetics. 1996;142:893–906. doi: 10.1093/genetics/142.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moazed D, O'Farrell PH. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development. 1992;116:805–810. doi: 10.1242/dev.116.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Négre N, Hennetin J, Sun LV, Lavrov S, Bellis M, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biology. 2006;4(6):e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Americo J, Whiteley M, Brown JL, Fujioka M, Jaynes JB, et al. A complex array of DNA binding proteins required for pairing-sensitive silencing by a Polycomb-group response element from the Drosophila engrailed gene. Genetics. 2002;160:1561–1571. doi: 10.1093/genetics/160.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeVido S, Kwon D, Brown JL, Kassis, JA The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development. 2008;135:669–676. doi: 10.1242/dev.014779. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham MD, Brown JL, Kassis JA. Characterization of the Polycomb group response elements of the Drosophila melanogaster invected locus. Mol Cell Biol. 2010;30:820–828. doi: 10.1128/MCB.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassis JA. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassis JA, Noll E, Vansickle EP, Odenwald WF, Perrimon N. Altering the insertional specificity of a Drosophila transposable element. Proc Natl Acad Sci U S A. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassis JA. Spatial and temporal control elements of the Drosophila engrailed gene. Genes Dev. 1990;4:433–443. doi: 10.1101/gad.4.3.433. [DOI] [PubMed] [Google Scholar]
- 23.Lyko R, Brenton JD, Surani MA, Paro R. An imprinting element from the mouse H19 locus functions as a silencer in Drosophila Nat Genet. 1997;16:171–173. doi: 10.1038/ng0697-171. [DOI] [PubMed] [Google Scholar]
- 24.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Betz WK, et al. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tower J, Kurapati R. Preferential transposition of a Drosophila P element to the corresponding region of the homologous chromosome. Mol Gen Genet. 1994;244:484–490. doi: 10.1007/BF00583899. [DOI] [PubMed] [Google Scholar]
- 26.Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P-element-induced gap repair. Science. 1991;263:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 27.Kassis JA. Pairing-sensitive silencing, Polycomb group response elements, and transposon homing in Drosophila. Adv. Genetics. 2002;46:421–438. doi: 10.1016/s0065-2660(02)46015-4. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, et al. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet. 2010;6(1):e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 30.Bantignies F, Roure V, Comet I, Leblanc G, Schuettengruber, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 31.Tolhius B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, et al. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7(3):e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H-B, Müller M, Bahechar IA, Kyrchanova O, Ohno K, et al. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–625. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavigne M, Francis NJ, King IFG, Kingston RE. Propagation of silencing: recruitment and repression of naïve chromatin in trans by Polycomb repressed chromatin. Mol Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- 34.Eun SH, Gan Q, Chen X. Epigenetic regulation of germ cell differentiation. Cur Opin Cell Biol. 2010;22:737–743. doi: 10.1016/j.ceb.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soeller WC, Poole SJ, Kornberg T. In vitro transcription of the Drosophila-engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of enΔ1.5 . (A) enΔ1.5 was generated by a crossing scheme designed to obtain a gene conversion event by P-element excision [26]. The recipient chromosome was marked with Sp and contained a ry+ P-element inserted 412 bp upstream of the en transcription start site. The donor chromosome contained a 16kb deletion of en DNA (enΔJ83B) from −15 kb upstream through the first intron of en (generated in our lab), and the donor P-element that contained LoxP sites and FRT sites flanking (indicated by vertical lines in the en DNA) the PREs (P[en2] from [18]). We were trying to get a gene conversion event that would put LoxP and FRT sites into the genome. We set up 400 vials of Cross 1 and 300 vials of Cross 2. From this, we obtained 2 potential deletions (enΔ1.5 and one other) and one potential gene conversion event. The 2 deletions were recovered and balanced from Cross 3 but the gene conversion event was not recovered. (B) Schematic representation of the PCR reactions used to detect the gene conversion event. L(LoxP), F(FRT). A, B are approximate primer locations.
(TIF)