Abstract
The ability to alter nutrient partitioning within plants cells is poorly understood. In Arabidopsis (Arabidopsis thaliana), a family of endomembrane cation exchangers (CAXs) transports Ca2+ and other cations. However, experiments have not focused on how the distribution and partitioning of calcium (Ca) and other elements within seeds are altered by perturbed CAX activity. Here, we investigate Ca distribution and abundance in Arabidopsis seed from cax1 and cax3 loss-of-function lines and lines expressing deregulated CAX1 using synchrotron x-ray fluorescence microscopy. We conducted 7- to 10-μm resolution in vivo x-ray microtomography on dry mature seed and 0.2-μm resolution x-ray microscopy on embryos from lines overexpressing deregulated CAX1 (35S-sCAX1) and cax1cax3 double mutants only. Tomograms showed an increased concentration of Ca in both the seed coat and the embryo in cax1, cax3, and cax1cax3 lines compared with the wild type. High-resolution elemental images of the mutants showed that perturbed CAX activity altered Ca partitioning within cells, reducing Ca partitioning into organelles and/or increasing Ca in the cytosol and abolishing tissue-level Ca gradients. In comparison with traditional volume-averaged metal analysis, which confirmed subtle changes in seed elemental composition, the collection of spatially resolved data at varying resolutions provides insight into the impact of altered CAX activity on seed metal distribution and indicates a cell type-specific function of CAX1 and CAX3 in partitioning Ca into organelles. This work highlights a powerful technology for inferring transport function and quantifying nutrient changes.
Transporters play fundamental roles in life, namely, the selective import or efflux of molecules through biological membranes. The vast majority of transporters are secondary, energized by the proton gradient and membrane potential. The plant cation/H+ exchangers (CAXs) are part of the ensemble of transporters that may coordinate the redistribution of various cations, including Ca2+, in exchange for the protons generated by H+ pumps (Hirschi, 2004; McAinsh and Pittman, 2009). Elucidating CAX, and other transporter functions, is challenging because it is difficult to discern phenotypes among a multitude of transporters with similar putative functions.
The Arabidopsis (Arabidopsis thaliana) genome contains six CAX open reading frames, and the transporters predominantly reside on the vacuole (Shigaki et al., 2006; Martinoia et al., 2007). CAX1 is a high-affinity Ca2+/H+ transporter and is expressed strongly in the leaves (Cheng et al., 2003). The cax1 loss-of-function mutants have subtle phenotypes, and microarray studies indicate compensatory expression of other transporters in the mutant (Cheng et al., 2005; Conn et al., 2011). The absence of strong phenotypes prompted the development of dominant gain-of-function CAX variants, namely a deregulated, N-terminally truncated, or short, sCAX1 (Diener and Hirschi, 2000). Recently, a role for CAX1 in controlling apoplastic Ca2+ concentrations has been delineated (Conn et al., 2011). CAX3 is 77% similar to CAX1, and CAX3 can compensate for the loss of CAX1 function (Cheng et al., 2005; Conn et al., 2011). When both CAX1 and CAX3 are perturbed, stunting and leaf chlorosis are observed; however, experimental tools to discern functional differences among these related transporters have not been readily available.
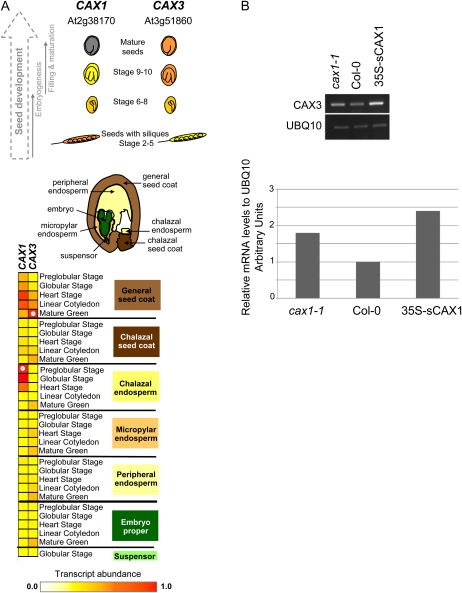
CAX1 and CAX3 are coexpressed only at specific stages during the Arabidopsis life cycle (Zhao et al., 2009; Conn et al., 2011) and may interact to form a transporter with distinct substrate transport characteristics (Cheng et al., 2005). Both genes are expressed during seed development. Figure 1A shows the relative expression levels of CAX1 and CAX3 during seed development using data from the Harada-Goldberg Arabidopsis LCM GeneChip data set (on the Seedgenenetwork Web site: http://estdb.biology.ucla.edu/seed/; Gene Expression Omnibus accession series no. GSE12404). These expression data were normalized to the highest expression value set to 1 and visualized using Genesis software (Sturn et al., 2002). CAX1 expression peaks during the earlier stages of seed development, whereas CAX3 expression is higher in the later stages. CAX1 is expressed in the chalazal endosperm at the preglobular stage, and CAX3 is expressed in the mature green seed coat. The expression of both CAX1 and CAX3 during seed development and germination provides a compelling rationale for examining the roles these transporters play in seed nutrient partitioning.
Figure 1.
A, Expression of CAX1 and CAX3 genes during seed development (Harada-Goldberg Arabidopsis LCM GeneChip data set, Seedgenenetwork Web site, http://estdb.biology.ucla.edu/ seed/; Gene Expression Omnibus accession series no. GSE12404). These expression data were normalized to the highest expression value set to 1.0 and visualized using Genesis software (Sturn et al., 2002). A color scale is used to represent variations in transcript abundance for each gene, in which red represents the highest expression and yellow represents the lowest expression. B, Semiquantitative RT-PCR measurement of CAX3 transcript level in the mature dry seeds of cax1-1, Col-0 wild type, and 35S-sCAX1. Top, semiquantitative RT-PCR gel image; bottom, relative expression level to UBQ10 as determined by relative densitometry analysis.
Evidence suggests diversity in the substrate range and function of the CAX transporters. In addition to calcium (Ca), CAXs may transport cadmium (Cd), manganese (Mn), and zinc (Zn; Hirschi et al., 2000; Kamiya and Maeshima, 2004). Studies have primarily focused on CAXs as drivers of Ca2+ accumulation in plant tissues, and engineering CAX is thought to have great potential in biofortification (Hirschi, 2009); however, how the distribution and partitioning of Ca and other elements in plant tissues are altered by perturbed CAX activity has not been systematically addressed.
The study of metal ion homeostasis in plants can benefit from spatially resolved metal analysis techniques, such as particle-induced x-ray emission (Bhatia et al., 2003; Isaure et al., 2006) and nanosecondary ion mass spectroscopy (Moore et al., 2010; Smart et al., 2010). Synchrotron x-ray fluorescence (SXRF) microscopy has been used to characterize gene function in plants (Kim et al., 2006). SXRF can be used to show the quantitative elemental characteristics of plant tissues, frequently without sample preparation, on a submicrometer scale. This can be informative for characterizing genes that facilitate metal ion movement in and out of specific cells or organelles, such as those that encode for membrane transport proteins. The simultaneous multielemental information in SXRF is particularly useful in less-characterized gene families, where imaging can show tissue- or cell-level elemental localization and thereby focus searches for metal-responsive genes expressed in those tissues. Likewise, multielemental analysis can reveal unexpected elemental coassociations, and a high level of coassociation can provide candidate elements for binding partners of metals of interest, which can be investigated using complementary techniques such as x-ray absorption spectroscopy. SXRF microtomography allows the collection of elemental information from intact samples in the form of virtual cross-sections (tomograms), removing the need for physical sectioning and chemical preservation, and is well suited to small samples such as Arabidopsis seed.
In this study, we used two synchrotron microprobes with different spatial resolutions to collect elemental images from seed of lines with altered CAX expression. Images suggested that overexpression of a CAX1 with an N-terminal truncation in the regulatory region (sCAX1) caused a disruption of selective Ca accumulation by cell types, rendering all layers equally Ca rich, and that deletion of both CAX1 and CAX3 caused a disruption in Ca storage within the cell. The use of two different spatial resolutions (micrometer and submicrometer) not only demonstrates the capabilities of SXRF for the analysis of biological tissue but also allows for the comparison between sample preparation techniques. It also adds to the developing body of literature on elemental distribution within the plant cell, which finds application in the manipulation of membrane transporters for various purposes, such as biofortification and the exclusion of toxic metals from edible plant parts.
RESULTS
Volume-Averaged Analysis via Inductively Coupled Plasma Mass Spectroscopy
In order to gain a first approximation of the role of CAX transporters in mineral seed content, bulk analysis of soil-grown seed was performed on lines altered in CAX expression, via inductively coupled plasma mass spectroscopy (ICP-MS; Table I). Notable alterations in elemental composition were disrupted bulk Ca levels; statistically, there were higher Ca concentrations in cax1 and lower Ca in 35S-sCAX1: 22% higher and 14% lower, respectively, than ecotype Columbia (Col-0). Among the micronutrient elements analyzed, Mn showed the greatest changes as a result of CAX gene disruption, mirroring the trend seen in Ca, with higher Mn in cax1 (32% higher than Col-0) and lower Mn in 35S-sCAX1 (14% lower than Col-0). This trend was also observed in bulk concentrations of copper (Cu). Iron (Fe) and Zn appeared minimally perturbed by CAX gene disruption.
Table I. Mean ± sd elemental concentrations of Arabidopsis seed grown under standard soil conditions (μg g−1 dry weight), where n = 8 to 16.
Pairwise comparison of means used the Tukey-Kramer honestly significant difference statistical test. Means not showing the same letter are significantly different (P < 0.05).
| Genotype | K | Ca | Mg | Mn | Fe | Zn | Cu |
| Col-0 | 11,024c± 962 | 3,593b± 242 | 3,097bc± 168 | 48.67b± 4.1 | 46.8bc± 2.8 | 51.1b± 0.66 | 6.4c± 0.6 |
| cax1-1 | 11,241bc± 571 | 4,393a± 516 | 3,124ab± 98 | 63.82a± 4.5 | 41.5d± 1.2 | 59.9a± 4.44 | 8.8a± 0.5 |
| cax3-1 | 12,081a± 481 | 3,60b± 68 | 3,238a± 111 | 46.83bc± 5.9 | 44.3cd± 1.1 | 59.7a± 1.81 | 7.6b± 0.3 |
| cax1cax3 | 11,860ab± 331 | 3,647b± 410 | 2,980c± 12 | 51.11b± 2.2 | 48.7ab± 3.3 | 58.4a± 1.19 | 8.4a± 0.22 |
| 35S-sCAX1 | 10,592c± 316 | 3,088c± 193 | 2,741d± 91 | 41.77c± 1.0 | 51.9a± 4.2 | 48.7c± 0.75 | 6.1c± 0.3 |
Semiquantitative Reverse Transcription-PCR Gene Expression Analysis
As determined by semiquantitative reverse transcription (RT)-PCR, the transcript levels of CAX3 in 35S-sCAX1 and cax1-1 are about 0.5- and 1-fold, respectively, higher than that of Col-0 wild type (Fig. 1B). For CAX1, the transcript was at low levels in all the cDNA samples and was undetectable after 30 cycles of amplification (data not shown). Even after 35 cycles, CAX1 bands were weak (data not shown). At 35 cycles, nonspecific amplification also becomes a factor that severely decreases the accuracy of measurement of very low levels of transcripts of interest. Thus, no conclusion was drawn regarding the relative levels of CAX1 transcripts in cax3-1, Col-0 wild type, and 35S-sCAX1.
SXRF Microtomography
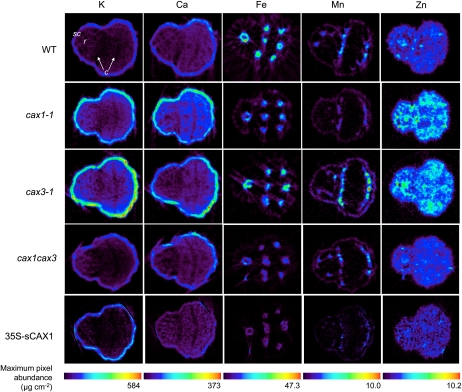
We conducted SXRF microtomography to analyze the impact of CAX1 and CAX3 disruption on the spatial distribution of elements within mature seeds (Fig. 2). Microtomography of intact seed showed no distributional anomalies at the tissue level of either macronutrients (potassium [K] and Ca) or micronutrients (Fe, Mn, and Zn), nor was elemental allocation between seed coat and embryo disrupted (Supplemental Fig. S1). There were differences in elemental abundances between lines, with higher K, Ca, and Zn in cax1 and cax3 and lowest abundances of these elements in 35S-sCAX1.
Figure 2.
Quantified SXRF tomograms of Arabidopsis seed, Col-0 wild type (WT), cax1-1, cax3-1, and cax1cax3 mutants, and transgenic line 35S-sCAX1, collected with a 7- × 10-μm beam. Tomograms for each element are scaled to the highest maximum pixel abundance, shown on the color bar, expressed as μg cm−2; the minim is zero for all elements. c, Cotyledons; r, radicle; sc, seed coat (within the embryo). Embryonic cotyledons are arranged with the adaxial (upper) surfaces facing one another.
Volume-averaged seed analysis via ICP-MS (Table I) and spatially resolved analysis via SXRF microtomography (Fig. 2) agreed on the following trends: K and Ca were highest in cax1-1 and cax3-1, Mn was lowest in 35S-sCAX1, whereas Zn was highest in cax1-1 and cax3-1 and lowest in 35S-sCAX1. However, even if expressed in the same units, absolute values between these two data sets differ, and the extent to which K, Ca, and Zn differ is less pronounced in volume-averaged data. We offer several explanations for this. First, volume-averaged data are the average elemental concentration of a large number of seeds (there are approximately 4,000–5,000 seeds per 50-mg aliquot) and consider the whole seed volume. This is compared with the elemental composition of a 10-μm-thick slice through a single Arabidopsis seed. In volume-averaged data, averaging over a large number of seeds will reduce between-seed elemental variability, whereas in spatially resolved data, the inherently low replicate number enhances variability. Additionally, spatially resolved data are presented as maximum pixel abundance, indicating only the upper limit of the data, rather than the average.
For this reason, we have conducted region of interest (ROI) analysis on identified tissue layers to derive their mean elemental abundances. These tissues consisted of seed coat (Table II) and embryo (Table III), showing macronutrients K and Ca and the micronutrients Fe, Mn, Cu, and Zn. Table II shows that Ca abundances in the seed coat of cax1-1, cax3-1, and cax1cax3 are higher than the wild type and lower in 35S-sCAX1. The high sd values of Fe and Mn in the embryo are due to their discrete distribution in specific tissue layers, which were analyzed separately (Table IV). Fe localizes to the endodermal cells (radicle) continuous with the bundle sheath cells (cotyledons) at abundances in the range of 4 to 17 μg cm−2. Mn localizes to the subepidermal layer of the abaxial side of the cotyledons, with abundances in the range of 0.6 to 3.4 μg cm−2 (Table IV). High sd values (60%–70% of the mean) for Mn- and Fe-enriched cell layers suggest that the spatial resolution of this ROI analysis is not optimal, strongly indicating subcellular heterogeneity, consistent with varying numbers and sizes of vacuole.
Table II. Elemental concentration of the seed coat of Arabidopsis seed from in vivo SXRF microtomography analysis, as defined by ROI analysis.
Values are means ± sd of each user-defined region, expressed as μg cm−2.
| Genotype | K | Ca | Mn | Fe | Zn | Cu |
| Col-0 | 99.29± 16.86 | 70.00± 33.05 | 0.54± 0.37 | 1.42± 1.09 | 1.12± 0.70 | 0.88± 0.55 |
| cax1-1 | 205.15± 75.17 | 137.79± 54.61 | 0.53± 0.32 | 1.76± 1.07 | 1.61± 0.86 | 0.93± 0.37 |
| cax3-1 | 301.93± 110.91 | 129.29± 55.67 | 1.03± 0.62 | 1.60± 1.18 | 1.83± 1.08 | 1.04± 0.44 |
| cax1cax3 | 100.79± 37.29 | 81.98± 36.05 | 0.48± 0.39 | 0.72± 0.99 | 1.08± 0.67 | 0.61± 0.30 |
| 35S-sCAX1 | 160.49± 73.82 | 46.00± 22.07 | 0.34± 0.28 | 0.45± 0.54 | 1.19± 0.82 | 0.34± 1.15 |
Table III. Elemental concentration of the embryos of Arabidopsis seed from in vivo SXRF microtomography analysis, as defined by ROI analysis.
Values are means ± sd of each user-defined region, expressed as μg cm−2.
| Genotype | K | Ca | Mn | Fe | Zn | Cu |
| Col-0 | 52.92± 13.74 | 36.09± 11.76 | 0.61± 0.83 | 3.85± 6.42 | 2.56± 0.67 | 0.51± 0.20 |
| cax1-1 | 104.12± 38.07 | 85.88± 25.43 | 0.46± 0.46 | 3.03± 4.02 | 3.93± 1.05 | 0.68± 0.24 |
| cax3-1 | 127.08± 43.14 | 55.05± 19.68 | 1.04± 1.39 | 3.81± 6.12 | 3.89± 0.92 | 0.69± 0.28 |
| cax1cax3 | 50.09± 20.33 | 37.88± 14.44 | 0.52± 0.74 | 1.82± 2.79 | 2.35± 0.56 | 0.53± 0.16 |
| 35S-sCAX1 | 69.25± 35.60 | 34.13± 13.11 | 0.30± 0.30 | 1.01± 1.67 | 1.97± 0.71 | 0.23± 0.49 |
Table IV. Abundance of Fe in the endodermis-bundle sheath tissue layer, and Mn in the abaxial subepidermis, of Arabidopsis seed from in vivo SXRF microtomography analysis, as defined by ROI analysis.
Values are means ± sd of a user-defined region, expressed as μg cm−2.
| Genotype | Mn | Fe | |
| Radicle | Cotyledon | ||
| Col-0 | 1.8± 1.3 | 16.7± 28.7 | 14.3± 24.6 |
| cax1-1 | 1.1± 0.7 | 10.4± 19.2 | 8.9± 16.4 |
| cax3-1 | 3.4± 2.0 | 17.1± 29.8 | 12.5± 21.8 |
| cax1cax3 | 8.5± 11.6 | 1.8± 1.2 | 6.3± 8.6 |
| 35S-sCAX1 | 0.6± 0.4 | 4.8± 8.1 | 4.1± 7.1 |
High-Resolution SXRF Mapping of Embryo Sections
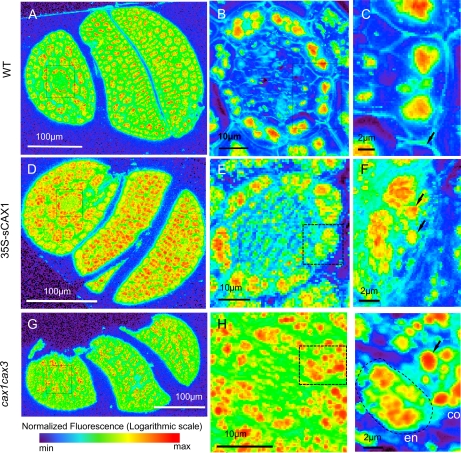
We conducted higher resolution (submicrometer) imaging to fully investigate the elemental distribution of subcellular compartments. High-resolution SXRF mapping (0.2-μm2 beam) of the Ca distribution of whole embryo thick sections of the wild type, 35S-sCAX1, and cax1cax3 mutants are shown in Figure 3, A, D, and G, respectively. We chose the double knockout and lines expressing deregulated CAX1 (35S-sCAX1) with the assumption that elemental differences would be more pronounced than in the single mutants (Conn et al., 2011).
Figure 3.
High-resolution SXRF elemental maps of Ca in Col-0 wild type (WT), 35S-sCAX1, and cax1cax3 embryo sections. Data are expressed as normalized fluorescence on a logarithmic scale, with each image individually scaled. A, D, and G, Whole embryo sections (0.2-μm beam, 0.3-μm step) oriented with the hypocotyl on the left and the cotyledons to the right. Boxes indicate the positions from which higher resolution maps were collected. B, E, and H, Endodermal layers of the hypocotyl. C, F, and I, Single endodermal cells of the hypocotyl showing subcellular localization of Ca in vacuoles.
Ca was present in all cells, both enclosed within numerous organelles at higher pixel abundances and as a component of the cell walls at several orders of magnitude lower abundance (Fig. 3C, arrow). The Ca abundance range typically spanned several orders of magnitude; therefore, normalized fluorescence is shown on a logarithmic scale. The Ca abundance of the epidermal cell layer of the embryo was lower than that of the internal cell layers. This distribution was also observed for phosphorus (P; Supplemental Fig. S2). In wild-type embryos, Ca abundance in cells of the endodermis (Fig. 3B) was comparatively greater than in neighboring layers: the pericycle, protoxylem, and protophloem on the interior and cortical cells on the exterior (Fig. 3B). The endodermal cells contain storage vacuoles rich in Fe (Roschzttardtz et al., 2009), which corresponded to enrichment of Fe, Ca, Mn, and Zn. This tissue-level Ca gradient was less pronounced in 35S-sCAX1 (Fig. 3, D and E) and absent in cax1cax3 (Fig. 3H). Given the Ca enrichment of the endodermis, and the likelihood that endodermal organelles are storage vacuoles, we chose to further analyze this layer for CAX phenotypes.
Looking closely at individual endodermal cells (Fig. 3, C, F, and I), Ca appeared to be highly localized within the lumen of a subcellular organelle in the wild type. This is indicated by the increased Ca abundance at the center of the body in comparison with the lower abundance at the margin. This contrasted with lines expressing deregulated CAX1 (35S-sCAX1), where Ca was localized at higher abundances at the margin (Fig. 3F, arrows). In this line, Ca appeared to be associated with numerous bodies of generally smaller size (Fig. 3F) as well as with irregularly shaped masses that could indicate its presence outside of the organelle. The cax1cax3 endodermal cell image (Fig. 3I) was conducted at an identical analytical resolution (0.5-s dwell, 0.15-μm step), and yet distinct organelles within the endodermal cells were difficult to discern. In cax1cax3, elements were localized in regions without a distinct margin and did not resemble the large bodies seen in Col-0. Figure 3I shows both an endodermal cell and a cortical cell, and organelles are clearly visible in the cortical cell (Fig. 3I, arrow). This suggests that the diffuse elemental distribution in the endodermis is not attributable to insufficient analytical resolution, (although insufficient fixation of tissue cannot completely be ruled out), because organelles with distinct margins are evident in neighboring cortical cells. This indicates that these elements were not confined within organelles in cax1cax3 endodermal cells in the same manner as in wild-type cells.
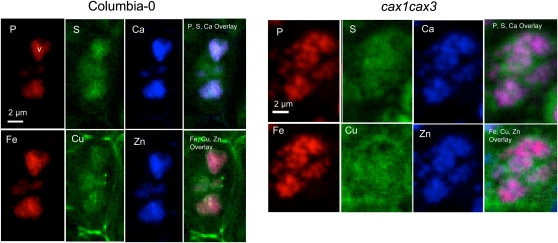
High-resolution images were conducted at an energy (10 keV) and a resolution that made it possible to see other elements of biological interest within the cell, such as P, sulfur (S), Fe, and Cu. Given the novelty of subcellular resolution images of the embryo we examined the characteristic elemental distributions in these sections. We found common distributions of elements; namely, that certain elements were either located within the lumen of subcellular organelles (e.g. in the wild type; P, Fe, and Ca), outside of organelles (e.g. S), or associated with the cell wall or cell membrane (e.g. Cu and Ca; Fig. 4).
Figure 4.
High-resolution SXRF elemental maps showing normalized fluorescence of P, S, Ca, Fe, Cu, Zn, and macronutrient and micronutrient overlay distribution in the endodermal cells of the embryonic hypocotyl in Col-0 wild-type and cax1cax3 lines.
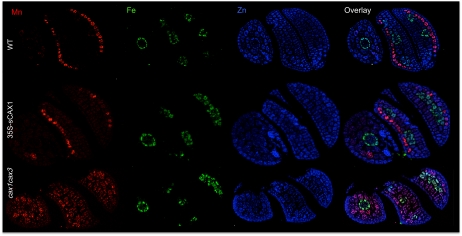
High-resolution images were also collected of whole seed sections to gain an understanding of elemental association with certain tissue types. Recent studies suggest that only certain cells have the ability to accumulate Ca (Conn et al., 2011), but this has not previously been imaged at this resolution in Arabidopsis seed for a range of biologically relevant elements. Whole embryo high-resolution images of Ca in Col-0, 35S-sCAX1, and cax1cax3 (Fig. 3, A, D, and G) show that Ca is present at maximal abundances (i.e. red on the color bar) in the majority of cells expressing deregulated CAX1, in comparison with the wild type. In the double mutant, it is the cytosol and cell membrane regions that dominate the Ca abundance. Given the changes we observed in Mn bulk and spatially resolved concentrations, we wanted to look at Mn in greater detail. We found in our earlier studies that Mn was strongly localized to a subepidermal layer of the cotyledons (Kim et al., 2006), whereas high-resolution mapping of cax1cax3 whole embryo sections (Fig. 5) showed a shallow Mn gradient. Fe was distributed solely within organelles of the endodermal cells of the radicle and around the vasculature of the cotyledons (Fig. 5). Zn was distributed uniformly throughout the cells of the embryo, strongly colocalizing with Ca.
Figure 5.
High-resolution SXRF maps of micronutrient distribution within whole Arabidopsis seed, showing Mn, Fe, Zn, and overlays in Col-0 wild-type (WT), 35S-sCAX1 overexpressor, and cax1cax3 lines. Images are individually scaled.
DISCUSSION
Planting the Seeds of SXRF in Plant Science
Nutrient storage within cells is often compartmentalized, but the mechanisms underpinning this compartmentalization are not fully appreciated (Leegood, 2008; Conn et al., 2011). Ionomic measurements have fostered the concept of ion homeostasis networks (Salt, 2004), but this technology has yet to fully address the spatial distribution of macronutrient and micronutrient elements within the various plant organs. Here, we have used two SXRF microprobes to collect high-resolution spatially resolved elemental images of how CAX transporters impact nutrient distribution within seeds.
For technical reasons, we focused our SXRF analysis on seeds. Seed is naturally dehydrated and has an extended stability during analysis; therefore, it is ideally suited to x-ray imaging studies. Numerous experiments have used transgenic approaches to alter rice nutrient content (Lee et al., 2009), and SXRF can now be used to analyze the spatial distribution of nutrients within seeds.
Some of ours observations in seeds may not be applicable to other parts of the plant (Vreugdenhil et al., 2004). For example, in seeds, some nutrients may be in a complex with the P-containing compound phytate. Elemental distribution patterns may vary among tissues, and it is likely that the correlations observed here would not be found in other tissues.
A Closer Look at CAX1 and CAX3
Imaging the cellular and subcellular distribution of Ca and other elements within seed of CAX1 and CAX3 mutants has shown changes in seed Ca distribution as a result of CAX gene disruption. On the cellular level, deletion of CAX1 and CAX3 has caused relative increases in Ca outside organelles (Fig. 3H); tentatively, different types of storage vacuoles and the expression of deregulated CAX1 disrupted the selective Ca accumulation abilities of certain cell types, rendering all layers Ca rich (Fig. 3E) and suggesting that CAX1 is only operating in certain cell types. On the tissue level, significant increases in Ca as a result of CAX1 and CAX3 deletion, observed in both volume-averaged and spatially resolved analytical techniques, are counterintuitive. They suggest either an overcompensation of subsequently up-regulated CAX3 in cax1-1 or the enhanced expression of other Ca transport systems. Recent studies also suggest that altering CAX activity can alter shoot-to-root phosphate signaling (Liu et al., 2011); however, this does not appear to drastically alter the P levels in the seeds.
The distributional changes observed (increases in Ca outside of organelles) are consistent with an altered ability for effective Ca storage. Furthermore, high-resolution imaging of individual cells indicates that an overexpression of a deregulated version of CAX1 and a deletion of cax1cax3 disrupts the normal elemental distribution on the cellular level. Cells expressing the deregulated CAX1 transporter appear replete, with more numerous, smaller organelles (in comparison with wild-type cells) that have comparatively greater Ca abundance at the margin than the lumen. By contrast, images of a cell in which both CAX1 and CAX3 have been deleted showed no readily discernible vacuoles (Fig. 3I), despite an identical analytical resolution. In these cells, elements appeared diffusely distributed. Future studies are needed to clarify these observations and survey other cell layers for evidence of similar mislocalization with greater replication.
Our study demonstrates how physiological changes in the plant can impact the concentration and distribution of multiple elements. Although the seed ionome of CAX mutants has not previously been measured, the vegetative phase shoot ionome of soil-grown cax1, cax3, and cax1cax3 via ICP-MS shows some perturbation of elements in cax1 and cax3, which corresponded with our measurements in the seed (Cheng et al., 2005), in particular, the increases in Mn and Zn concentration in seeds of cax1, cax3, and cax1cax3 (Fig. 2).
Recent studies have established CAX1 and CAX3 as key regulators of apoplastic Ca levels (Conn et al., 2011). Ca moves apoplastically through the transpiration stream and is generally present at low levels in seeds (White and Broadley, 2003). Although there is no active transpiration stream within cells of the mature embryo, nutrient transfer between maternal and filial tissue is restricted to the apoplast (Patrick and Offler, 2001); therefore, changes in apoplastic Ca levels of the maternal plant may be reflected in the mature embryo or seed coat. During seed development, the seed coat is derived from maternal tissues (the integuments of the ovule), and in this study, differences in Ca concentration of the embryo and the seed coat were found in CAX mutants.
The semiquantitative RT-PCR provides a snapshot of the transcript levels of CAX1 and CAX3 in mature dry seed. At this stage, the level of CAX1 transcript is low, which agrees with the Harada-Goldberg Arabidopsis LCM GeneChip data set (Fig. 1A). At the dry seed stage, CAX3 transcript was more abundant than CAX1 and was enhanced in both the cax1-1 and 35S-sCAX1 lines. This fits previous reports showing that CAX3 expression is enhanced in cax1-1 lines (Cheng et al., 2003); however, in 35S-sCAX1 lines, the marginal increase in CAX3 expression was unexpected. Since there is evidence that CAX1 and CAX3 function as dimers (Zhao et al., 2009), there may be coupled regulation of gene expression. The mRNA transcript levels reported here may not reflect the transport activity of CAX1 and CAX3 at this stage of development. CAX transcripts at earlier developmental periods may influence the transport activity that mediates the nutrient distribution patterns shown here (Fig. 2).
The Future of SXRF in Plant Sciences
It is clear from this and other studies (Kim et al., 2006; Carey et al., 2010, 2011; Moore et al., 2010) that spatially resolved elemental mapping can be a powerful technique for analyzing transporter phenotypes. While some shared elemental network components can be predicted by binding and transport properties, in practice, living systems are so complex that most relationships cannot be predicted easily from chemical or biological principles and must be determined experimentally (Baxter, 2009). For SXRF to become a widely used experimental platform among plant biologists, certain limitations must be addressed. It remains necessary to stabilize hydrated tissue such as leaves and roots, which require extended analysis times to image micronutrient elements present at very low abundances.
Microtomographic analysis of the low atomic number (Z) elements (the macronutrient elements P, S, and Ca) is hindered by self-absorption effects, where the low-energy x-rays emitted by these elements are easily absorbed by the air path between the sample and detector and by the sample itself. In this study, we added an additional silicon drift detector at 180° geometry to the existing GE multielement array. With this new setup, as soon as self-absorption effects begin to dominate for one detector, the opposite detector is ideally placed to receive fluorescent x-rays from the sample, and count rates are doubled. This geometry effectively eliminates self-absorption effects and enhances the detection of low-abundance elements. Tomograms shown in Figure 2 were collected using this new configuration.
As SXRF develops under the continued presence of users from the life sciences, there is the potential to use elemental imaging to explore gene × environment interactions by imaging samples with altered gene expression under various environmental stresses. With advances in detector technology, the collection of in vivo data will remove the need for intrusive sample preparation in the majority of instances. However, even with faster detectors, it is likely that radiation damage from x-rays will limit the possibility of in vivo imaging. Currently, SXRF remains a static image of a dynamic system, and consideration of time is particularly crucial in elements like Ca, where concentration gradients and spatial distribution patterns are known to change rapidly in response to stimuli (White and Broadley, 2003).
Evidence has shown that nutrient localization is crucial in normal growth and development (Kim et al., 2006); therefore, our working hypothesis is that plant growth and nutrient bioavailability are determined by the correct cellular and subcellular localization of macronutrient and micronutrient elements. For endeavors such as biofortification, it is necessary also to increase the bioavailable form of the nutrient rather than simply increasing the bulk amount. Our long-term goal is to combine ionomic and imaging approaches to identify the relationships between nutrient distribution and subsequent changes in the chemical forms of nutrients in the plant cell.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used in this study. Seeds from wild-type and transgenic plants were surface sterilized, germinated, grown on full-strength B5 medium containing 0.5% Suc, and solidified with 0.8% agar. All plates were sealed with paper surgical tape and incubated at 22°C under continuous cool-fluorescence illumination. Seedlings were transplanted to soil (3:1:1, Pro-Mix:vermiculite:perlite) and grown under a 16-h-light/8-h-dark cycle. Plants were frequently watered with nutrient solution [5 mm KNO3, 50 mm KPO4, 2 mm MgSO4, 2 mm Ca(NO3)2, 50 mm FeEDTA, 70 μm H3BO3, 14 μm MnCl2, 0.5 mm CuSO4, 1 μm ZnSO4, 0.2 mm NaMoO4, 10 mm NaCl, and 0.001 mm CoCl2].
Volume-Averaged Elemental Analysis via ICP-MS
cax1-1, cax3-1, cax1cax3, 35S-sCAX1, and wild-type plants were grown as described above, in the same growth chamber, and harvested on the same day. Three replicates of approximately 50-mg aliquots of dry seed were digested in 2 mL of Optima HNO3 in Teflon vessels using a MARS5 EXPRESS microwave-assisted reaction system (CEM Corporation). A standard reference material (NIST 1573a, Tomato Leaves; National Institute of Standards and Testing) and a HNO3 blank were included after every fifth sample. Sample volume was brought to 10 mL with deionized water. The vessels were heated to 180°C in 10 min and held at that temperature for a further 10 min. After the samples had cooled, they were brought up to approximately 15-mL volume with deionized water. Samples were analyzed for trace element concentrations using an Agilent 7500cx ICP-MS system operating in collision mode at the Trace Element Analysis Core facility of Dartmouth College.
Semiquantitative RT-PCR Gene Expression Analysis
CAX1 and CAX3 transcript levels were measured in the mature dry seeds of cax1-1, cax3-1, Col-0 wild type, and 35S-sCAX1. Total RNA was extracted from about 50 mg of seeds for each sample with the Spectrum Plant Total RNA kit (Sigma-Aldrich). First-strand cDNA was synthesized from 1 μg of RNA with the SuperScript First-Strand Synthesis system for RT-PCR (Invitrogen) with oligo(dT)12–18. The cDNA samples were then diluted to the equivalent of 20 ng RNA μL−1. PCR was performed with the following thermal program: 94°C for 5 min to denature DNA; then in each cycle, 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s; and 72°C for a 7-min final extension. UBQ10 was used as the internal reference gene. The number of cycles used in PCR for UBQ10, CAX1, and CAX3 were 21, 30, and 35, respectively. These cycle numbers were determined to be within the exponential phase of PCRs for these three genes for the mature dry seed cDNA samples. For CAX1, the forward primer was 5′-TTCCGGCCATTCCTGCCG-3′ and the reverse primer was 5′-CACCGCGGTTTCTTGCTCC-3′. For CAX3, the forward primer was 5′-ACAACTACGGTCGTCCGTT-3′ and the reverse primer was 5′-GCGACATTTTGTTAATCATAGAGGTCG-3′. For UBQ10, the forward primer was 5′-GATCTTTGCCGGAAAACAATTGGAG-3′ and the reverse primer was 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAG-3′. All amplicon sizes were around 500 bp. The DNA gel was imaged by the Bio-Rad Gel Doc XR+ system with exposure time of 2 s. The relative densitometry of the bands was analyzed with Bio-Rad Image Lab software.
SXRF Microspectroscopy of Intact Seed
Tomograms were collected at the bending magnet Beamline X26A at the National Synchrotron Light Source, Brookhaven National Laboratory. X-ray fluorescence measurements were conducted using a 12-keV monochromatic x-ray beam. Due to spectral overlap with the abundant macronutrient K, fluorescence data could not be collected for Cd at the 12-keV excitation energy. Monochromatic x-rays were tuned using a Si(111) channel-cut monochromator and focused to a beam size of 5 × 8 μm using rhodium-coated, silicon Kirkpatrick-Baez microfocusing mirrors. Incident beam energy was monitored using an ion chamber upstream of the focusing optics. X-ray fluorescence spectra were collected with a Vortex-EX silicon-drift detector (SII Nanotechnology) with an active area of 50 mm2. X-ray transmission through the sample was recorded simultaneously using a p-type, intrinsic, n-type photodiode.
Individual mature, dry, and unsectioned seeds were attached to a 100-μm-diameter silica fiber using Devcon 5-min epoxy resin, with the micropyle uppermost. The fiber was inserted into a Huber 1001 goniometer, mounted on a xyzθ stage (where xyzθ represents the axes through which the stage moves: x, left and right; y, up and down; z, back and forth; and θ, angle of rotation), and centered. Tomograms were collected from the midpoint of the seed. During fluorescence microtomography, the seed samples were translated horizontally through the focused x-ray microbeam in step sizes ranging from 5 to 7 μm and then rotated at intervals of between 0.8° and 1.1° angular steps, repeating the translation through a total of 180°. Full energy-dispersive spectra were collected at each pixel, with a dwell time of 2 s per pixel. Two-dimensional sinograms (plot of intensity against θ) were computationally reconstructed using fast Fourier transform-based Gridrec software developed by Brookhaven National Laboratory (Dowd et al., 1999), which is controlled by the Interactive Data Language programming software (Research Systems) to provide images of the cross-sectional internal metal distribution.
Abundance Quantification and Postprocessing of SXRF Tomography Data
Elemental abundances (percentage weight fraction) were calculated for the fluorescence measurements, adapted from a description by McNear et al. (2005). Briefly, a thin-film standard reference material (SRM 1833) was measured prior to the collection of each data set to establish elemental sensitivities (counts per second per μg cm−2) for Fe. We used an assumed object density of 1.2 g cm−3 for Arabidopsis seed, a measured voxel size of 3.887 × 10−8 cm3 (reconstructed pixel area × beam height), and the average Fe response from the sample to calculate the Fe content of a whole tomogram. This Fe abundance was used as a fixed value for input into the Naval Research Laboratory X-Ray Fluorescence program (Criss, 1977), from which abundances for K, Ca, Mn, Fe, nickel, Cu, and Zn were calculated. The concentration precision is typically ±15% and ±10% (1σ) for individual and mean values, respectively.
Following quantification, ROI analysis was carried out to investigate differences in elemental abundance and concentration of specific tissues. This was conducted using the freehand ROI capability of the beamline-specific imaging software (muplot_alpha4 running in the Interactive Data Language Virtual Machine, © ITTVisual Information Solutions). Separation of the seed from the background or air surrounding the sample prior to any data inquiries was conducted as standard. ROI analysis was used to calculate nutrient allocation within the various tissues, expressed as the percentage of the total in the tomogram allocated to the seed coat or embryo. Concentration was expressed in μg cm−2, using seed weights determined for each line (weight of 100 seeds per 100, measured for each line).
Preparation of Samples for High-Resolution Two-Dimensional SXRF Raster Scanning
The beamline setup at the Advanced Photon Source 2-ID-D (Argonne National Laboratory), in particular the helium enclosure around the sample stage and detector, required that seed be sectioned for analysis, which necessitated a resin-embedding sample preparation step. Arabidopsis seed was imbibed on moist filter paper for 2 d to allow removal of the seed coat and release the embryo. This was carried out to ensure optimal infiltration of resin into the embryo cells. Embryos were placed in fixative solution (a mixture of 3% glutaraldehyde and 4% paraformaldehyde in 0.4 m sodium cacodylate) under a gentle vacuum overnight. Embryos were rinsed in a solution of 0.2 m sodium cacodylate and 2.5 mm CaCl2, pH 7.2, followed by distilled water. Embryos were then dehydrated in an ethanol series (30%, 50%, 70%, 95%, and 100% for 30 min each), with the final step repeated three times over the course of 1 h. Embryo samples were then immersed in three changes of 100% ethanol for 10 min each, followed by LR White resin:ethanol mixtures of 1:3, 1:2, and 1:1 (twice) for 1 h each, after which they were stored at 4°C in 1:1 LR White resin:ethanol overnight. Samples were warmed to room temperature and moved to a 2:1 LR White resin:ethanol solution over 4 h, before immersing in two changes of 100% LR White solution for 1 h each. Samples were stored at 4°C overnight and then warmed to room temperature the following day before immersing in three changes of 100% LR White resin over a 4-h period. Embryos were transferred to flat embedding molds using a toothpick to achieve the correct orientation, before polymerizing for 24 h.
Embedding molds were constructed following the methods described by Palmieri and Kiss (2005). Custom molds were created to provide flat embedding chambers that exclude oxygen from contacting the embedding medium. We chose 0.254-mm and 0.381-mm polycarbonate films (McMaster-Carr) because both satisfied our size requirements, were easy to work with, were low cost, and withstood exposure to LR White. These polycarbonate films were cut using a scalpel to fit standard 25 × 3 × 75 mm2 glass microscope slides. The slides were precleaned with 100% ethanol and wiped with lint-free Kim wipes. To facilitate specimen removal, the slide was treated with an antistick agent (Fluoroglide spray; Electron Microscopy Sciences) prior to adhering the gasket to the slide. This was applied three times and then polished to remove lubricity. Silicone adhesive was then added to one side of the gasket and pressed onto the slide so an airtight seal was formed. Silicone adhesive was cured at room temperature for 24 h or in a 60°C oven for 1 h and allowed to cool before use.
LR White and embryo specimens were added until the chamber was slightly overfilled and the liquid formed a convex surface, and then an Aclar strip (Ted Pella) was cut to a slightly larger width than the slide and placed on top of the chamber to shield the LR White from oxygen. One end of the Aclar strip was placed onto the edge of the chamber and the rest was rolled down onto the resin so that any excess resin spilled over the side and prevented air bubbles from being trapped underneath. The specimens were polymerized at 60°C for 24 h. After polymerization, the Aclar was removed and specimens were excised with a razor blade while the slide was still warm. Initial samples were cut to between 1 and 5 μm thick with a microtome and a glass knife and allowed to adhere to silicone nitride windows.
High-Resolution SXRF Microscopy
Scanning x-ray fluorescence microscopy was performed at Beamline 2-ID-D of the Advanced Photon Source at the Argonne National Laboratory (Cai et al., 2000). Incident x-rays of 10 keV were chosen to excite elements from P to Zn. A Fresnel zone plate focused the x-ray beam to a spot size of 0.2 × 0.2 μm on the sample, which was raster scanned (Yun et al., 1999) at resolutions of 1.25 μm step in the whole seed images, 0.5 μm step in whole endodermis layer images, and 0.15 μm step in the single cell images, with dwell times ranging from 0.5 to 1 s per pixel. X-ray fluorescence from the sample was captured with an energy-dispersive silicon drift detector. Data were collected over a number of scheduled beamtime experiments, using samples that were prepared and sectioned separately, and therefore are expressed as normalized fluorescence counts, expressed on individual scales.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT2G38170 (CAX1) and AT3G51860 (CAX3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Elemental allocation between seed coat and embryo determined via SXRF region of interest analysis.
Supplemental Figure S2. High-resolution SXRF elemental maps of phosphorus in Arabidopsis seed wild type, 35S-sCAX1, and cax1cax3.
Acknowledgments
We thank Dr. Helene Zuber for work on Figure 1.
References
- Baxter I. (2009) Ionomics: studying the social network of mineral nutrients. Curr Opin Plant Biol 12: 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia NP, Orlic I, Siegele R, Ashwath N, Baker AJM, Walsh KB. (2003) Elemental mapping using PIXE shows the main pathway of nickel movement is principally symplastic within the fruit of the hyperaccumulator Stackhousia tryonii. New Phytol 160: 479–488 [DOI] [PubMed] [Google Scholar]
- Cai Z, Lai B, Yun W, Ilinski P, Legnini D, Maser J, Rodrigues W. (2000) A hard x-ray scanning microprobe for fluorescence imaging and microdiffraction at the Advanced Photon Source. Am Inst Physics Proc 507: 472–477 [Google Scholar]
- Carey AM, Norton GJ, Deacon C, Scheckel KG, Lombi E, Punshon T, Guerinot ML, Lanzirotti A, Newville M, Choi Y, et al. (2011) Phloem transport of arsenic species from flag leaf to grain during grain filling. New Phytol 192: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AM, Scheckel KG, Lombi E, Newville M, Choi Y, Norton GJ, Charnock JM, Feldmann J, Price AH, Meharg AA. (2010) Grain unloading of arsenic species in rice. Plant Physiol 152: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N-H, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD. (2003) The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N-H, Pittman JK, Shigaki T, Lachmansingh J, LeClere S, Lahner B, Salt DE, Hirschi KD. (2005) Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol 138: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Giliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng N-H, Stancombe MA, Hirschi KD, Webb AAR, et al. (2011) Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criss JW. (1977) NRLXRF, A FORTRAN Program for X-Ray Fluorescence Analysis. Naval Research Laboratory, Washington, DC [Google Scholar]
- Diener AC, Hirschi H. (2000) Heterologous expression for dominant-like gene activity. Trends Plant Sci 5: 10–11 [DOI] [PubMed] [Google Scholar]
- Dowd BA, Campbell GH, Marr RB, Nagarkar VV, Tipnis SV, Axe L, Siddons DP. (1999) Developments in synchrotron x-ray computed microtomography at the National Synchrotron Light Source. Proc SPIE 3772: 224–236 [Google Scholar]
- Hirschi KD. (2004) The calcium conundrum: both versatile nutrient and specific signal. Plant Physiol 136: 2438–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD. (2009) Nutrient biofortification of food crops. Cousins R, Bier D, Bowman B, Dean L, , Annual Reviews of Nutrition, Vol 29. Annual Reviews, Palo Alto, CA, pp 401–421 [DOI] [PubMed] [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ. (2000) Expression of Arabidopsis CAX2 in tobacco: altered metal accumulation and increased manganese tolerance. Plant Physiol 124: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaure MP, Fraysse A, Devès G, Le Lay P, Fayard B, Susini J, Bourguignon J, Ortega R. (2006) Micro-chemical imaging of cesium distribution in Arabidopsis thaliana plant and its interaction with potassium and essential trace elements. Biochimie 88: 1583–1590 [DOI] [PubMed] [Google Scholar]
- Kamiya T, Maeshima M. (2004) Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J Biol Chem 279: 812–819 [DOI] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Lee S, Jeon US, Lee SJ, Kim Y-K, Persson DP, Husted S, Schjørring JK, Kakei Y, Masuda H, Nishizawa NK, et al. (2009) Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci USA 106: 22014–22019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC. (2008) Roles of the bundle sheath cells in leaves of C3 plants. J Exp Bot 59: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Liu T-Y, Aung K, Tseng C-Y, Chang T-Y, Chen Y-S, Chiou T-J. (2011) Vacuolar Ca2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis. Plant Physiol 156: 1176–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- McNear DH, Jr, Peltier E, Everhart J, Chaney RL, Sutton S, Newville M, Rivers M, Sparks DL. (2005) Application of quantitative fluorescence and absorption-edge computed microtomography to image metal compartmentalization in Alyssum murale. Environ Sci Technol 39: 2210–2218 [DOI] [PubMed] [Google Scholar]
- Moore KL, Schröder M, Lombi E, Zhao F-J, McGrath SP, Hawkesford MJ, Shewry PR, Grovenor CRM. (2010) NanoSIMS analysis of arsenic and selenium in cereal grain. New Phytol 185: 434–445 [DOI] [PubMed] [Google Scholar]
- Palmieri M, Kiss JZ. (2005) A novel technique for flat-embedding cryofixed plant specimens in LR white resin. Microsc Res Tech 68: 80–84 [DOI] [PubMed] [Google Scholar]
- Patrick JW, Offler CE. (2001) Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52: 551–564 [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S. (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE. (2004) Update on plant ionomics. Plant Physiol 136: 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki T, Rees I, Nakhleh L, Hirschi KD. (2006) Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J Mol Evol 63: 815–825 [DOI] [PubMed] [Google Scholar]
- Smart KE, Smith JAC, Kilburn MR, Martin BGH, Hawes C, Grovenor CRM. (2010) High-resolution elemental localization in vacuolate plant cells by nanoscale secondary ion mass spectrometry. Plant J 63: 870–879 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Vreugdenhil D, Aarts MGM, Koornneef M, Nelissen H, Ernst WHO. (2004) Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ 2004: 828–839 [Google Scholar]
- White PJ, Broadley MR. (2003) Calcium in plants. Ann Bot (Lond) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun W, Lai B, Cai Z, Maser J, Legnini D, Gluskin E, Chen Z, Krasnoperova AA, Vladimirsky Y, Cerrina F, et al. (1999) Nanometer focusing of hard x-rays by phase zone plates. Rev Sci Instrum 70: 2238–2241 [Google Scholar]
- Zhao J, Shigaki T, Mei H, Guo YQ, Cheng N-H, Hirschi KD. (2009) Interaction between Arabidopsis Ca2+/H+ exchangers CAX1 and CAX3. J Biol Chem 284: 4605–4615 [DOI] [PubMed] [Google Scholar]