Abstract
PhoB is a two-component response regulator that activates transcription by interacting with the σ70 subunit of the E. coli RNA polymerase in promoters in which the −35 σ70-recognition element is replaced by the pho box. The crystal structure of a transcription initiation subcomplex that includes the σ4 domain of σ70 fused with the RNA polymerase β subunit flap tip helix, the PhoB effector domain and the pho box DNA reveals how σ4 recognizes the upstream pho box repeat. As with the −35 element, σ4 achieves this recognition through the N-terminal portion of its DNA recognition helix, but contact with the DNA major groove is less extensive. Unexpectedly, the same recognition helix contacts the transactivation loop and helices α2 and α3 of PhoB. This result shows a simple and elegant mechanism for polymerase recruitment to pho box promoters in which the lost −35 element contacts are compensated by new ones with the activator. In addition, σ4 is reoriented, thereby suggesting a remodelling mechanism for transcription initiation.
Keywords: PhoB, RNA polymerase σ70, transcription activation, two-component signal transduction, X-ray crystal structure
Introduction
Bacterial RNA synthesis is carried out by a multisubunit RNA polymerase (RNAP) of 450 kDa, which consists of a stable catalytic core that contains two α, β, β′ and ω subunits and a transiently bound σ factor, which capacitates RNAP to recognize the DNA promoters and initiate transcription (Burgess, 1969; Zhang et al, 1999). The catalytic core structure is conserved from bacteria to eukaryotes (Cramer et al, 2001). In bacteria, the RNAP core bound to a σ factor constitutes the RNA polymerase holoenzyme (RNAPH). In E. coli, the primary σ factor, σ70, controls the transcription of housekeeping genes during bacterial exponential growth. The structure of the σ70-related σA primary factor of Thermus aquaticus shows three all α-folded domains (σ2, σ3 and σ4) separated by long linkers (Campbell et al, 2002). The structure of RNAPH (Murakami et al, 2002b; Vassylyev et al, 2002) shows that the σ factor lies at one side of the crab claw-shaped RNAP core, making extensive contacts with the RNAP β and β′ subunits; in particular, σ2 and σ4 domains contact a coiled-coil in β′ and a flap in β, respectively. The typical σ70 DNA promoter bears two conserved sequence elements, the −10 and the −35 hexamers, separated by a spacer of 17 bp (Helmann and Chamberlin, 1988). The structure of RNAPH bound to a DNA promoter shows that the σ factor makes all the contacts with the DNA, including the −10 and −35 elements (Murakami et al, 2002a). In particular, σ2 subregion 2.4 binds the −10 element while subregion 2.3 melts the DNA around it. Subregion 3.2, located in the linker between σ3 and σ4, contacts RNAP by blocking the exit of the nascent RNA (linker σ R3-4; Murakami et al, 2002b) and two subregions located in σ4—subregions 4.1 and 4.2—bind the RNAP β-flap tip helix and the −35 element DNA, respectively (Gardella et al, 1989; Campbell et al, 2002; Kuznedelov et al, 2002; Murakami et al, 2002b).
In E. coli and closely related bacteria, the histidine kinase PhoR and the response regulator PhoB are part of a two-component system that activates inorganic phosphate (Pi) uptake metabolic pathways when low concentrations of environmental Pi are present (Hsieh and Wanner, 2010). PhoB presents two structural motifs connected by a flexible linker: a conserved N-terminal regulatory domain that folds in an α/β arrangement and contains a conserved aspartate, Asp53, to which a phosphoryl group from PhoR His215 is transferred (Makino et al, 1989; Volz, 1993; Solà et al, 1999); and a C-terminal effector domain (PhoBE) that presents a winged-helix fold with specific DNA binding and transactivation properties turned on upon PhoBN phosphorylation (Makino et al, 1996). However, if the regulatory domain is removed, PhoBE can freely bind DNA and act as a constitutive activator (Ellison and McCleary, 2000). The cellular response after PhoB phosphorylation is transcription activation of the Pho Regulon, a group of nearly 40 genes distributed in 5 operons and 4 independent genes (Wanner, 1996; Kim et al, 2000; Murray and Conway, 2005). In particular, PhoB binds to specific promoters that substitute the canonical −35 sequence recognized by the bacterial transcription machinery. The PhoB-specific promoters contain one to three copies of a pho box, a sequence that comprises two 11-bp direct repeats. In each repeat, the first 7 bp is more conserved than the last 4, which are rich in AT. PhoB binds as a head-to-tail dimer to a pho box, each monomer contacting one direct repeat (Makino et al, 1996; Blanco et al, 2002). Mutational and deletion studies have shown that PhoB activates transcription by an interaction with the σ4 domain of the σ subunit within RNAPH (Makino et al, 1993; Kumar et al, 1994).
Here, we describe the crystal structure of a transcription initiation subcomplex that includes the σ4 domain of the E. coli σ70 RNAP factor fused with the RNAP β subunit flap tip helix, a PhoBE tandem dimer and a DNA pho box. The structure reveals how σ4 domain is recruited to the pho box promoters by PhoB and shows a reoriented σ4 domain with respect to its binding to the canonical −35 promoter sequence. These observations suggest that PhoB enhances transcription initiation by remodelling the RNAPH complex.
Results
A strategy for preparing a transcriptional initiation subcomplex
After DNA binding, PhoB triggers the activation of transcription. Mutations and C-terminal deletions of σ70 showed the involvement of the σ4 subdomain in the transcriptional activation mediated by PhoB (Makino et al, 1993; Kumar et al, 1994; Kim et al, 1995). We decided to characterize the interactions between PhoB, σ4 and a PhoB promoter by crystallographic methods. However, protein production of the σ4 domain resulted very difficult because of poor expression or precipitation during purification. A thorough analysis of genetic studies (Kuznedelov et al, 2002) and two RNAPH crystal structures (Murakami et al, 2002a; Vassylyev et al, 2002) indicated that σ4 has a hydrophobic surface that interacts with one side of the RNAP β-flap, an essential interaction for holoenzyme formation (Geszvain et al, 2004). This finding inspired us to design a chimera by fusing σ4 with the β-flap tip helix through an artificial linker (see Figure 2B) with the aim of obtaining a soluble and stable globular domain. Once purified, the σ4-β-flap construct was incubated with the PhoBE-pho box DNA complex and subjected to size exclusion chromatography (Supplementary Figure 1A). SDS–PAGE of the eluting fractions showed that the ternary complex was stable (Supplementary Figure 1B). The purified complex was subsequently used for structural analysis.
σ4-β-Flap tip helix chimera binds to pho box DNA in the presence of PhoBE
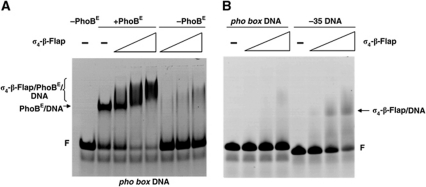
The interaction of the σ4-β-flap tip helix chimera with the pho box sequence was assayed by electrophoretic mobility shift analysis (EMSA). The addition of increasing amounts of purified σ4-β-flap tip helix chimera to a pho box/PhoBE complex resulted in a progressive reduction in electrophoretic mobility (as indicated with a bracket in Figure 1A, lanes 3–5). This change in mobility clearly reflected the formation of the σ4-β-flap tip helix/PhoBE/pho box DNA ternary complex since the σ4-β-flap tip helix chimera did not stably interact with the pho box DNA in the absence of PhoB (Figure 1A, lanes 6–8; Figure 1B, lanes 2–4). In contrast, interaction of the σ4-β-flap tip helix chimera with a conventional −35 DNA sequence was observed in the same conditions (Figure 1B, lanes 6–8), as expected (Campbell et al, 2002).
Figure 1.
σ4-β-Flap tip helix chimera binding to DNA. EMSA of the σ4-β-flap tip helix/PhoBE/pho box DNA ternary complex. (A) Increasing amounts of σ4-β-flap tip helix chimera (0, 0.055, 0.11 and 0.18 nmol, respectively) were added to a pre-formed PhoBE/pho box DNA complex (lanes 2–5) or to a free pho box (lanes 6–8). The positions of the PhoBE/pho box DNA complex and the σ4-β-flap/PhoBE/pho box DNA are indicated (arrow and bracket, respectively). F, free DNA probe. (B) σ4-β-Flap tip helix interaction with a −35 DNA probe. Increasing amounts of σ4-β-flap tip helix (0, 0.055, 0.11 and 0.18 nmol, respectively) were added either to pho box DNA (lanes 1–4) or to a DNA probe containing the canonical −35 motif (see Materials and methods). The position of the σ4-β-flap tip helix/−35 DNA probe is indicated (arrow). F, free DNA probe.
Structure of the transcriptional initiation subcomplex σ4-β-flap/PhoBE/pho box DNA
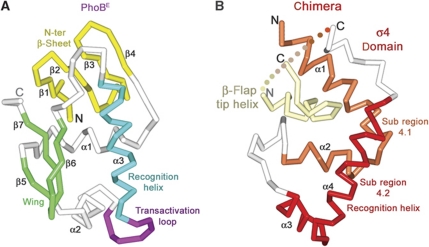
The crystal structure shows two PhoBE protomers and one σ4-β-flap tip helix chimera bound to a 26-mer phoA pho box encoding DNA (Figure 3). The PhoBE dimer binds to the 26-mer double-stranded oligonucleotide in tandem, an arrangement very similar to that previously reported by our group using a different oligonucleotide (Blanco et al, 2002). A PhoBE protomer consists of an N-terminal four-stranded antiparallel β-sheet (strands β1–β4) followed by a compact three α-helix bundle (helices α1–α3) packed against a small C-terminal β-sheet, which includes a β-hairpin or ‘wing’ (Figure 2A). PhoBE has a winged-helix motif for DNA binding, where helix α3 is the recognition helix. Within the modified helix-turn-helix motif between helices α2 and α3 there is a seven-residue loop which has been named the transactivation loop since it was postulated to interact with the σ70 subunit to activate transcription (Makino et al, 1996; Blanco et al, 2002). A tandem of two protomers arranged head-to-tail sits at one side of the DNA molecule. In each protomer, the recognition helices α3 lay along the major groove while the wing tips contact the downstream minor groove. The DNA is bent about 29°, which is slightly less than in the previous binary complex structure.
Figure 2.
Overview of protein–DNA structures. Cα traces of transcriptional activator PhoBE (A) and σ4-β-flap tip helix chimera (B) highlighting important structural elements in different colours.
In the ternary complex, the σ4 domain of the E. coli σ70 factor presents a fold consisting of a C-shaped four-α-helix bundle with an exposed hydrophobic core in its central region (Figure 2B). The last three α helices show a spatial arrangement identical to those found in the structure of σ4 bound to its regulator Rsd (Patikoglou et al, 2007). This fold is similar to that described for σA counterparts from T. aquaticus and T. thermophilus (Campbell et al, 2002; Vassylyev et al, 2002). Minor differences between structures are the angles between helices α1 and α2, and one turn less in helix α4 in our E. coli structure, as a result of the presence of Pro601, which interrupts the helix. Helices α1 and α2 comprise the subregion 4.1, and helices α3 and α4 comprise subregion 4.2 and constitute a helix-turn-helix motif, in which α4 is the recognition helix (Figure 2B). As explained above, the E. coli σ4 domain was expressed in fusion with the helix from the tip of the β-flap of the RNAP β subunit. In the three-dimensional structure, this β-flap tip helix is buried in a deep hydrophobic crevice between helices α1 and α2 that would otherwise be exposed (Figure 3C). The β-flap tip helix surface, which covers the σ4 hydrophobic crevice, exposes hydrophilic residues (Figure 3C, right panel), thus explaining the increased solubility and stability of the chimera during protein production versus native σ4 production. Furthermore, the fitting between the β-flap tip helix and the σ4 subunit shows a good superposition with the RNAPH structures available (data not shown).
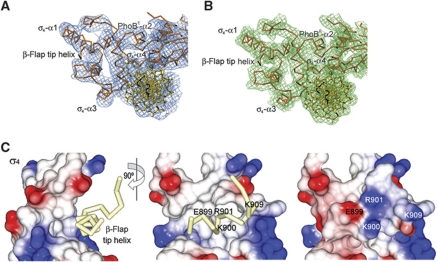
Figure 3.
Electron density maps of the σ4-β-flap/PhoBE/pho box DNA complex and details of the σ4-β-flap tip helix chimera. (A) Experimental σA-weighted electron density map of the ternary complex at 6.5 Å after density modification (see Materials and methods), contoured at 1σ. The final model is shown fitted in the density. (B) Final refined σA-weighted electron density map of the ternary complex at 4.3 Å. (C) Electrostatic potential surface representation of σ4 showing the β-flap tip helix as a Cα trace in pale yellow (left and middle panels) in two views 90° apart. The β-flap tip helix fits in a hydrophobic crevice of the σ4 surface. At the right panel (same view as the middle panel), the electrostatic potential surface representation includes the attached β-flap tip helix.
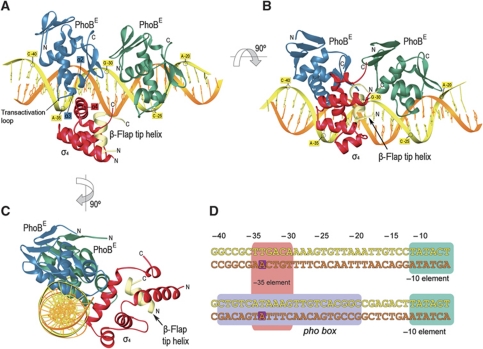
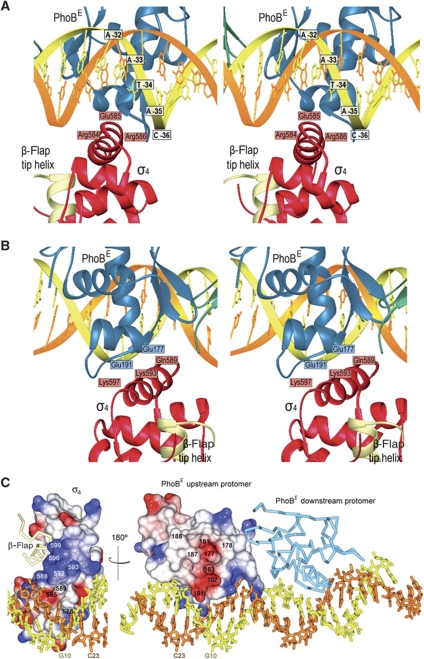
In the crystal asymmetric unit, there are two ternary complexes that show a σ4-β-flap chimera anchored to the major groove of the pho box DNA, at the −35-bp position, and contacting the upstream PhoBE protomer (Figure 4A–C). The position of the PhoB tandem is not modified by the presence of the σ4-β-flap tip helix chimera even though the upstream PhoBE contacts the chimera. Both polypeptides are bound to the same stretch of the DNA double helix but at different sides. The alignment of the DNA sequences of the phoA promoter covered by our σ4 chimera and that of the σ-dependent promoter present in the structure of the RNAPH-DNA complex (Murakami et al, 2002a) shows that the σ4 domains are located exactly at the same position from the transcription origin +1 in both structures (Figure 4D). No σ4 domain was found in contact with the downstream PhoBE protomer, thereby indicating that σ discriminates between the two protomers in the PhoBE-DNA complex and binds only to the one near the −35 position. σ4 Contacts the DNA through the N-terminal end of the recognition helix α4 (Figure 5A). In addition, the central part of the recognition helix interacts with PhoBE, in particular with helices α2 and α3 and the transactivation loop of the activator (see below and Figures 4 and 5B).
Figure 4.
Structure of the σ4-β-flap tip helix chimera/PhoBE/pho box DNA complex. (A) Ribbon diagrams of the ternary complex. The upstream and the downstream protomers of PhoBE are shown in blue and green, respectively; the coding and template DNA strands are coloured in yellow and orange, respectively; the chimera is shown as σ4 in red and the β-flap tip helix in pale yellow. Note that the σ4 subunit interacts only with the upstream PhoBE. (B, C) Different orientations of the ribbon representation in (B) to show the relative positioning of the different multisubunits of the ternary complex. (D) Alignment of an RNAPH promoter containing the canonical −35 and −10 elements (upper sequence) and the phoA promoter containing the pho box (lower sequence). In the ternary complex reported here, the PhoBE dimer covers the whole pho box sequence (light violet rectangle), while σ4 covers the sequence from −35 to −30 (pink rectangle) which coincides with the binding site of σ4 onto the canonical −35 element (upper sequence, pink rectangle). Two conserved adenines at position −34 in both sequences are highlighted in dark violate squares (see text).
Figure 5.
Interactions within the ternary complex. (A) Top: ribbon stereo plot showing the interactions between σ4 (red) and the pho box DNA. Interacting amino acids and DNA bases are indicated. (B) Stereo plot showing the interactions of σ4 (red) with the PhoBE upstream protomer (blue). The view rotated 180° about the horizontal axis relative to (A). (C) Electrostatic potential surface representation of the σ4 domain bound to the DNA (left) or to the upstream protomer of the PhoBE tandem (right) within the ternary complex. The σ4/PhoBE complex has been ‘opened’ by swinging the σ4 domain 180° to show the electrostatic potential of the interaction surface. The residues from each subunit that confront the residues from the other subunit in a contacting distance have been indicated. A base from each DNA strand is also shown to make the 180° turn of the structure clearer. The β-flap tip helix (pale yellow) and the downstream PhoBE protomer (light blue) are shown as Cα traces.
The downstream pho box repeat contains a nearly −35 consensus sequence TTGTCA at positions −29 to −24, which could, in principle, be recognized by σ4. However, if bound to that stretch of the DNA, it would not be correctly placed to establish the interaction observed between the upstream PhoBE and σ4. In order to have a similar interaction with the downstream PhoBE protomer, σ4 would have to be placed over the promoter stretch −25 to −20, which has the sequence CACGGC, far from the −35 consensus (Figure 4D). Note that the position of PhoBE dimer is fixed, and in tandem, as observed in all crystal structures solved so far.
Protein–protein and protein–DNA interactions within the ternary complex
The PhoBE upstream protomer and σ4 contact each other through an interface of 300 Å2. Although the resolution of the X-ray diffraction data does not allow a precise definition of interacting atoms, it is clear that this interaction has a strong electrostatic character, as deduced from the amino-acid content of opposing surfaces at appropriate distances (Figure 5B and C). An acidic patch on the PhoB surface delineated by the transactivation loop and the N-terminus of helix α2 faces a patch of basic residues from σ4 helix α4. This observation supports the postulated transactivation role for the loop as mutations on this segment or nearby (Trp184Arg, Gly185Arg, Val190Met and Asp192Gly) cancel PhoB transcription activation activity (Makino et al, 1996). Therefore, the interaction occurs through a small electrostatic interface, thus suggesting low affinity between components in a transient complex (Nooren and Thornton, 2003). It was suggested that σ4 contacted PhoB by loop α2–α3 and helix α3, because mutations on the corresponding residues (Asp570Gly, Glu575Lys, Tyr571Ala, Thr572Leu, Val576Thr, Lys578Glu, and Phe580Val) cancel PhoB transcription activation (Makino et al, 1993; Kim et al, 1995). However, these segments are located at the opposite side of the σ surface that contacts the upstream PhoBE protomer, and although they face the downstream protomer, they are too far to establish an interaction with it. One possible explanation is that this area is involved in downstream transcription events.
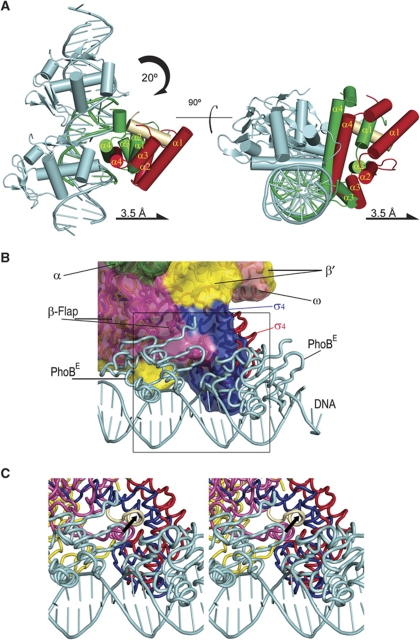
Interestingly, superposition of the DNA phosphates of our ternary complex onto the σ4-DNA complex of T. aquaticus (Campbell et al, 2002) reveals that σ4 with the β-flap tip helix is displaced from the DNA in our structure and reoriented by a 20° clockwise rotation that permits the N-terminal portion of the recognition helix to contact the PhoBE upstream protomer (Figure 6A). This reorientation is not observed in the activating ternary subcomplex of σ4, DNA and the transcription factor λCI (Jain et al, 2004), where σ4 presents the same positioning as in the σ4-DNA structure of T. aquaticus (Murakami et al, 2002a). In our complex structure, the σ4 domain contacts the DNA through the N-terminal residues of its recognition helix α4 (Figure 5A). In particular, the Thr583 (like Thr408 in T. aquaticus) side chain is close to the 5-methyl group of the thymine (T) base located at position −34 of the leading strand; Arg586 (Arg409 in T. aquaticus) is close to the −34T and/or −35T phosphates; and the Glu585 (Glu410 in T. aquaticus) side chain faces the DNA major groove near adenine (A) −34 in the template strand. Importantly, this adenine base is conserved in the phoA pho box and in the −35 element (Figure 4D). Arg554 from helix α2 (Arg387 in T. aquaticus) is at an appropriate distance and orientation for contacting the phosphate groups of the DNA leading strand. A comparison of all these interactions with those observed in the crystal structure of T. aquaticus σ4 bound to the −35 sequence (Campbell et al, 2002) shows that many are conserved, in particular those at the recognition helix α4. However, in our complex the reorientation of σ4 displaces the N- and C-termini of helices α1 and α3 from the DNA, respectively (Figure 6A). This movement causes a considerable loss of protein–DNA interactions, thus decreasing the interface from 600 Å2 in the T. aquaticus σ4-DNA binary complex to 300 Å2 in our ternary complex. The contacts established by helices α2, α3 and α4 and by the equivalent residues to Glu585, Gln589 and Arg584 are missing. However, the loss of interactions between σ4 and the DNA resulting from the σ4 reorientation are compensated by new protein–protein interactions with the PhoBE upstream protomer.
Figure 6.
Displacement of σ4 upon binding to the pho box. (A) Two views of the superposition of the ternary complex reported here and the complex σ4-DNA (1KU7; Campbell et al, 2002); superposition was done by matching the DNA phosphates from the pho box (our structure) and the −35 element (1KU7). Schematic representations of the σ4 domains of σ70 and σA are depicted in red and green, the β-flap tip helix in cream and the PhoBE dimer in cyan. The σ4 domain of σ70 is partly displaced from the DNA as a result of the presence of PhoBE. (B) Fitting of the PhoB-DNA and σ4-β-flap tip helix chimera structure against the RNAPH structure. Colour codes for σ4, the β-flap tip helix and the PhoBE dimer are as in (A) for our ternary complex. The σ4 domain as it is in the RNAPH is shown in blue. RNAPH subunits are differentiated by colours and labelled. The superposition shows that the upstream PhoBE protomer contacts and reorientates the σ4 domain. (C) Close-up stereo view of the area indicated as a square in (B). The σ4 movement would drag the β-flap tip helix, thus facilitating the exit of nascent RNA. The black arrow indicates the movement of the β-flap tip helix, from the position observed in the RNAPH structure (β-flap tip helix in lilac) to the position observed in the present structure, when PhoBE is bound at the Pho promoter.
PhoB in the RNAPH-DNA scenario
Superposition of the DNA phosphates of our structure onto those of the T. aquaticus σ4-DNA complex evidences the different orientations of σ4. In addition, superposition of the σ4 domain of the RNAPH structure (Vassylyev et al, 2002) onto σ4 of the T. aquaticus structure puts our σ4 in the context of the RNA polymerase holoenzyme (Figure 6B and C). As described, the PhoBE upstream protomer contacts the σ4 domain, which holds the β-flap tip helix. Surprisingly, upon superposition, the downstream PhoBE protomer gets close to the bottom of the RNAPH β-flap, which traps the linker between the σ70 domains 3 and 4 against the RNAP core (Figure 6B and C). In this simple superposition, the downstream PhoBE protomer and RNAPH are close, located at the same side of the DNA. This observation suggests that, in addition to the contact with σ70, a direct contact between the PhoBE and the β-subunit of RNAPH can occur.
Discussion
RNAPH recruitment and remodelling mediated by PhoB
In general, in the multistep process of transcription initiation, RNAPH—per se or helped by transcription factors—first engages the promoter DNA to yield an RNAPH-promoter closed complex. In a second step, RNAPH unwinds and melts about 14 bp of DNA surrounding the transcription start site to yield an RNAPH-promoter open complex. In a third step, RNAPH typically stacks into multiple abortive cycles of synthesis and releases short RNA products (about 9–11 nucleotides), failing to escape from the promoter (Revyakin et al, 2006; Goldman et al, 2009). Finally, only when a newly synthesized RNA molecule grows enough to cross the exit channel, located next to the β-flap, does RNAP leave the promoter as an elongation complex to enter productive RNA synthesis (Murakami et al, 2002b; Kapanidis et al, 2006). A mechanistic view of transcription initiation described from kinetic studies distinguishes between two activation modes: (i) a transcription activator ‘recruits’ RNAPH by establishing only a mere adherent interaction, without any conformational change of the latter; and (ii) an activator ‘remodels’ an inactive RNAPH bound to the promoter DNA (Record et al, 1991; Busby and Ebright, 1994; Ptashne and Gann, 1997). In recruiting, the activator favours the formation of the RNAPH-DNA open complex. This mechanism has been described for activators that contact either αCTD, like CRP (Busby and Ebright, 1994; Ptashne and Gann, 1997), or activators that contact σ4, like λcI (Hawley and McClure, 1982; Nickels et al, 2002; Jain et al, 2004). Available structural data show only small interaction surface between the DNA-bound transcription factors CRP or λcI and their targeted αCTD or σ4 subunits of the transcription machinery, without any changes in respective orientations (Benoff et al, 2002; Jain et al, 2004).
The crystal structure of the ternary complex presented here shows that the RNAPH σ70 subunit binds the −35 position in pho box promoters with bound PhoB. The DNA recognition helix of σ4 is displaced and simultaneously binds the DNA major groove and PhoB. In vitro DNA protection studies showed no protection against RNAPH unless PhoB was present, which suggests that the transcription factor, bound to the promoter, recruits RNAPH. The polymerase was unable to recognize the binding site on its own as a result of sequence discrepancy between the pho box and the −35 element (Makino et al, 1993). Thus, the binding of σ to the pho box depended on the DNA-PhoB complex. Our structure shows that, when σ4 is bound to the pho box, a number of contacts that σ4 performed with the −35 sequence DNA are lost but are replaced by new contacts with the DNA-bound PhoB, that is, those of residues Lys593, Gln589 and partly Glu585.
Our analysis also presents an additional feature, namely the σ4 domain in a swung orientation when compared with the structure in complex with the bacterial canonical −35 DNA element (Campbell et al, 2002). The orientation of σ4 bound to a canonical −35 sequence is not possible in the pho box because the PhoBE upstream protomer hinders the full entrance of the σ4 recognition helix into the DNA major groove. Within the full-length σ, this reorientation of σ4 may represent remodelling. Therefore, in addition to recruitment, a remodelling event may eventually lead to transcription initiation. These two events, recruiting and remodelling, are not incompatible. They have been described for CRP which, in a first step, recruits an αCTD protomer and, in a second step, indirectly remodels RNAPH by bridging the αCTD subunit to σ4 (Chen et al, 2003). In PhoB, the combination of the two mechanisms would consist first of recruitment of RNAPH (in agreement previous results of Makino and co-workers) and, subsequently, remodelling of σ by rotating σ4, as in our ternary complex structure (Figure 6A). Remodelling of σ4 would drag the β-flap tip helix closer to the RNA exit channel, by means of hydrophobic interactions like those observed in the chimera, thus opening it and favouring the exit of the nascent RNA (Figure 6B and C).
The interaction of σ70 with PhoB and other transcription factors
The σ4 domain is a specialized platform designed to contact a wide range of transcription factors through specific residues, integrating each input to a concrete response. Moreover, some residues, like Gln589 and Lys593 and possibly Glu585, at subregion 4.2, show multiple roles and have the capacity to contact the DNA backbone (Campbell et al, 2002) or transcription activators like λcI (Jain et al, 2004) or PhoB (this work). Mutagenesis studies have shown that residues located in subregion 4.2 are involved in the binding of activators like λcI, CRP and FNR (Kuldell and Hochschild, 1994; Lonetto et al, 1998). Accordingly, this subregion provides RNAPH with a surface for the recruitment of activators, which we propose to name SAP1 (σ70 activating patch 1). Furthermore, the structure of λcI contacting σ4 of the T. aquaticus σA factor shows salt bridge interactions between basic (Arg413, Lys418 and Arg421) and acidic residues of λcI (Jain et al, 2004). The basic residues of the σ4 domain in σA are conserved in σ70 and are close to acidic residues of PhoB (Glu177, Glu191 and Asp192). This observation points to a common charge-based code among transcription activators, which, despite not showing sequence conservation or the same fold, use acidic patches to interact with σ4 SAP1.
Concluding remarks
There is increasing interest in elucidating the transcription initiation steps that involve RNA synthesis. Three regions of RNAPH have been reported as key players in the first events of transcription initiation, namely the lid, the σ R3-4 linker and the β-flap. The lid is an essential motif to initiate RNA synthesis but not in the RNA transcription elongation stage (Toulokhonov and Landick, 2003); the σ R3-4 linker blocks the RNA exit channel and its deletion cancels RNA abortive transcription (Murakami et al, 2002b); and the β-flap is located next to the RNA exit channel and its mutations are related to various RNAP conformational states (Kuznedelov et al, 2006). In particular, those mutations that strengthen the interaction with σ4 prevent RNA elongation (Nickels et al, 2005). The crystal structure of the complex of the σ4-β-flap tip helix, PhoBE and pho box DNA presented here shows that the σ4 domain contacts the PhoBE upstream protomer, at the −35-bp position. Remarkably, superposition with the RNAP holoenzyme shows that σ4 is reoriented. This reorientation would pull the σ R3-4 linker out of the aperture of the RNA exit channel, thus facilitating RNA release and thus reducing the probability of abortive transcription. This notion leads us to propose that in addition to recruiting the polymerase, the ultimate action of PhoB as a transcription activator is to facilitate transcript release by remodelling σ4.
Materials and methods
Protein and nucleic acid preparation
PhoBE protein expression, purification and DNA complex formation were performed as previously described (Blanco et al, 2002). After the unsuccessful design of E. coli σ70 factor constructs, which resulted in non-soluble protein, we designed a chimeric protein containing σ4(533–613), an artificial linker (Gly-Ser-Ser-Gly-Ser-Gly) and the β-flap tip helix (889–898). The chimera was expressed in E. coli BL21 (DE3) cells for 6 h at 37°C. Protein purification included a heparine Sepharose column chromatography step (GE Biosciences), eluted with buffer 20 mM BisTris (pH 6.5), 1 mM EDTA and gradient buffer 20 mM BisTris (pH 6.5), 1 M NaCl, 1 mM EDTA, followed by a size exclusion HiLoad 26/60 Superdex 75 column (GE Biosciences), previously equilibrated with buffer C (20 mM BisTris (pH 6.5) and 200 mM NaCl). Ternary complex formation was obtained first by mixing PhoBE with an annealed DNA duplex containing the pho box sequence and then adding the σ4-β-flap chimera. The ternary complex appeared in one single elution peak in a Superdex 75 10/30 (GE Biosciences) column in buffer containing 10 mM BisTris (pH 6.5) and 50 mM KCl.
EMSA experiments
We developed a non-radioactive detection system based on fluorescein labelling at the 5′ end of one of the strands of the dsDNA used. In a final volume of 10 μl, EMSA samples were incubated in binding buffer containing 20 mM BisTris (pH 6.5), 100 mM NaCl, 10 mM MgCl2, 100 μg/ml BSA and 5% glycerol. The σ4-β-flap tip helix/PhoBE/pho box DNA complex was assayed using 0.1 nmol of fluorescein labelled-pho box dsDNA and 0.089 nmol of PhoBE in the presence of 0.3 nmol of a non-specific 22 bp-competitor dsDNA, also previously annealed. After 30 min incubation at room temperature, increasing amounts of σ4-β-flap tip helix chimera (0.055, 0.11 and 0.18 nmol, respectively) were added to these samples and incubated for 30 min more. Samples were finally loaded onto 10% polyacrylamide gels (7 cm long, 10 cm wide and 1.5 mm thick) prepared in buffer containing 20 mM Tris (pH 7.9), 10 mM acetate and 0.1 mM EDTA, and run at 120 V at room temperature. The direct interaction of the chimera with pho box DNA and −35 DNA was assayed in the same conditions without the second incubation period. Pictures were taken with an Eugenius gel-recording apparatus (Invitrogen).
The oligonucleotides used were
pho box_leading 5′-F-GAGCTGTCATAAAGTTGTCACGG-3′;
pho box_template 5′-GCCGTGACAACTTTATGACAGCT-3′;
−35_leading 5′-F-GCCGCTTGACAAAAGTGTTAA-3′;
−35_template 5′-TTAACACTTTTGTCAAGCGGC-3′;
Competitive_leading 5′-TCGGCGACTTTTCGGCGACTTT-3′;
Competitive_template 5′-AAAGTCGCCGAAAAGTCGCCGA-3′.
Crystallization and X-ray diffraction data collection
Crystals of the ternary complex σ4-β-flap/PhoBE/pho box DNA were obtained by vapour diffusion from sitting drops by mixing 2 μl of ternary complex solution at 11 mg/ml of protein concentration with 2 μl of crystallization solution containing 50 mM MES (pH 6.0), 100 mM KCl, 10 mM MgCl2 and 8–10% (v/v) PEG 4000. Several oligonucleotides, differing in length and sequence, were tested, the best diffracting crystals being those prepared with a 26-mer DNA duplex with two overhanging bases at each end (5′-TGGCTGTCATAAAGTTGTCACAAAAG-3′/3′-CGACAGTATTTCAACAGTGTTTTCAC-5′). Initially, the crystals diffracted to only 7 Å resolution. However, after dehydration by successive soakings in solutions with a 5% increase in PEG 4000, the resolution improved to ∼4 Å. These crystals, cryo-protected with 20% ethylene glycol, were cryo-cooled in liquid nitrogen and diffraction data were collected at ESRF beamline ID14-4 (Table I).
Table 1. Data collection and phasing statistics.
aOverall/outermost resolution shell.
bRmerge=(Σhkl Σi∣Ii(hkl)−〈Ii(hkl)〉∣/Σhkl Σi Ii(hkl)) × 100 where Ii is the ith measurement of reflection hkl.
Crystals of the ternary complex were very sensitive to heavy atom derivatization. Nevertheless, a crystal was derivatized with Ta6Br12 by first harvesting it in 50 mM MES (pH 6.0), 200 mM KCl, 10 mM MgCl2 and 15% (v/v) PEG 4000 and afterwards adding to the solution 5 mM Ta6Br12 and incubating the crystals for 7 h. This procedure eventually yielded a crystal suitable for MAD data collection (6.5 Å resolution). Data were collected at three wavelengths, 1.2542 Å (peak), 1.2554 Å (inflection) and 1.2498 Å (remote) at ID29 in the ESRF (Grenoble, France; Table I).
Structure determination
The ternary complex was solved by MAD using a Ta4Br13 derivative. MAD analysis SHELXD (Sheldrick et al, 2001) using the three-wavelength data up to 7.4 Å (including the highly redundant peak data with multiplicity 8.76) showed six strong sites. Positions were refined with SHARP (Vonrhein et al, 2007) using data up to 6.5 Å (peak data). Five additional weaker sites were found and initial SHARP phases were improved with SOLOMON and DM (Abrahams and Leslie, 1996; Cowtan, 1999). From the 6.5 Å resolution map, all protein subunit boundaries were easily distinguishable from the solvent. DNA density was also clear and allowed identification of major and minor grooves. Phase extension from 6.5 to 4.3 Å followed, using solvent flattening and averaging algorithms as implemented in Pirate (Cowtan, 2000). The resulting map showed almost all secondary structure features of the proteins and DNA (Figure 3A and B; Supplementary Figure 2). The structure of PhoBE in complex with a 26-mer DNA, previously solved at 2.8 Å resolution, and a σ4-β-flap tip helix chimera model based on the structure of E. coli σ4 (Patikoglou et al, 2007) and the β-flap tip helix from the RNAP structure of T. thermophilus (Vassylyev et al, 2002) were fitted in the electron density map either manually or using MOLREP (Vagin and Teplyakov, 1997). Subsequently, rigid-body refinement was performed with combined phases using Hendrickson–Lattman coefficients (Hendrickson and Lattman, 1970), as implemented in REFMAC5 (Murshudov et al, 1997). Further rigid-body refinement cycles with CNS (Brünger et al, 1998) were combined with manual model building using Turbo (Carranza et al, 1999) or Coot (Emsley and Cowtan, 2004). In the final model, the electron density is continuous for all polypeptide chains, except for the Gly-Ser engineered linker between σ4 the β-flat tip helix, which is probably flexible (Table I; Figure 3B). Atomic coordinates have been deposited with the protein data base under accession code 3T72.
Supplementary Material
Acknowledgments
This study was supported by the Ministerio de Ciencia e Innovación (Grants BFU2008-02372/BMC and CONSOLIDER CSD 2006-23 to MC and BFU2009-07134 to MS), the Generalitat de Catalunya (Grants SGR2009-1309 to MC and SGR2009-1366 to MS) and the European Commission (Spine2-Complexes LSHG-CT-2006-031220 and Rapid FP7-PEOPLE-2011-ITN-290246). Synchrotron data collection was supported by the ESRF and the EC. Crystallization screening and preliminary X-ray analysis were performed at the Automated Crystallography Platform, Barcelona Science Park.
Author contributions: AGB and MC conceived and designed the study; AGB carried out complex preparation and crystallization experiments; AGB collected X-ray data and solved the structure with the assistance of MS and MC; AGB and AC refined the structure; JB performed the EMSA experiments; AGB and MC wrote the manuscript with significant contributions from AC, MS and JB; MC supervised the study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abrahams JP, Leslie AG (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr 52: 30–42 [DOI] [PubMed] [Google Scholar]
- Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH (2002) Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297: 1562–1566 [DOI] [PubMed] [Google Scholar]
- Blanco AG, Solà M, Gomis-Rüth FX, Coll M (2002) Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10: 701–713 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burgess RR (1969) Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem 244: 6168–6176 [PubMed] [Google Scholar]
- Busby S, Ebright RH (1994) Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79: 743–746 [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA (2002) Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell 9: 527–539 [DOI] [PubMed] [Google Scholar]
- Carranza C, Inisan A-G, Mouthuy-Knoops E, Cambillau C, Roussel A (1999) Turbo-Frodo. In AFMB Activity Report 1996–1999, pp 89–90. Marseille: CNRS-UPR 9039 [Google Scholar]
- Chen H, Tang H, Ebright RH (2003) Functional interaction between RNA polymerase alpha subunit C-terminal domain and sigma70 in UP-element- and activator-dependent transcription. Mol Cell 11: 1621–1633 [DOI] [PubMed] [Google Scholar]
- Cowtan K (1999) Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr D Biol Crystallogr 55: 1555–1567 [DOI] [PubMed] [Google Scholar]
- Cowtan K (2000) General quadratic functions in real and reciprocal space and their application to likelihood phasing. Acta Crystallogr D Biol Crystallogr 56: 1612–1621 [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Ellison DW, McCleary WR (2000) The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J Bacteriol 182: 6592–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gardella T, Moyle H, Susskind MM (1989) A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol 206: 579–590 [DOI] [PubMed] [Google Scholar]
- Geszvain K, Gruber TM, Mooney RA, Gross CA, Landick R (2004) A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates sigma(70) region 4 function. J Mol Biol 343: 569–587 [DOI] [PubMed] [Google Scholar]
- Goldman SR, Ebright RH, Nickels BE (2009) Direct detection of abortive RNA transcripts in vivo. Science 324: 927–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DK, McClure WR (1982) Mechanism of activation of transcription initiation from the lambda PRM promoter. J Mol Biol 157: 493–525 [DOI] [PubMed] [Google Scholar]
- Helmann JD, Chamberlin MJ (1988) Structure and function of bacterial sigma factors. Annu Rev Biochem 57: 839–872 [DOI] [PubMed] [Google Scholar]
- Hendrickson WA, Lattman EE (1970) Representation of phase probability distributions for simplified combination of independent phase information. Acta Cryst B26: 136–143 [Google Scholar]
- Hsieh YJ, Wanner BL (2010) Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13: 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Nickels BE, Sun L, Hochschild A, Darst SA (2004) Structure of a ternary transcription activation complex. Mol Cell 13: 45–53 [DOI] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314: 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Kimura S, Shinagawa H, Nakata A, Lee KS, Wanner BL, Makino K (2000) Dual transcriptional regulation of the Escherichia coli phosphate-starvation-inducible psiE gene of the phosphate regulon by PhoB and the cyclic AMP (cAMP)-cAMP receptor protein complex. J Bacteriol 182: 5596–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Makino K, Amemura M, Nakata A, Shinagawa H (1995) Mutational analysis of the role of the first helix of region 4.2 of the sigma 70 subunit of Escherichia coli RNA polymerase in transcriptional activation by activator protein PhoB. Mol Gen Genet 248: 1–8 [DOI] [PubMed] [Google Scholar]
- Kuldell N, Hochschild A (1994) Amino acid substitutions in the −35 recognition motif of sigma 70 that result in defects in phage lambda repressor-stimulated transcription. J Bacteriol 176: 2991–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Grimes B, Fujita N, Makino K, Malloch RA, Hayward RS, Ishihama A (1994) Role of the sigma 70 subunit of Escherichia coli RNA polymerase in transcription activation. J Mol Biol 235: 405–413 [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Lamour V, Patikoglou G, Chlenov M, Darst SA, Severinov K (2006) Recombinant Thermus aquaticus RNA polymerase for structural studies. J Mol Biol 359: 110–121 [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K (2002) A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science 295: 855–857 [DOI] [PubMed] [Google Scholar]
- Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C (1998) Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J Mol Biol 284: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M (1996) DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol 259: 15–26 [DOI] [PubMed] [Google Scholar]
- Makino K, Amemura M, Kim SK, Nakata A, Shinagawa H (1993) Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev 7: 149–160 [DOI] [PubMed] [Google Scholar]
- Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A (1989) Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol 210: 551–559 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002a) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA (2002b) Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 296: 1280–1284 [DOI] [PubMed] [Google Scholar]
- Murray EL, Conway T (2005) Multiple regulators control expression of the Entner-Doudoroff aldolase (Eda) of Escherichia coli. J Bacteriol 187: 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nickels BE, Dove SL, Murakami KS, Darst SA, Hochschild A (2002) Protein-protein and protein-DNA interactions of sigma70 region 4 involved in transcription activation by lambdacI. J Mol Biol 324: 17–34 [DOI] [PubMed] [Google Scholar]
- Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A (2005) The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci U S A 102: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooren IM, Thornton JM (2003) Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol 325: 991–1018 [DOI] [PubMed] [Google Scholar]
- Patikoglou GA, Westblade LF, Campbell EA, Lamour V, Lane WJ, Darst SA (2007) Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J Mol Biol 372: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A (1997) Transcriptional activation by recruitment. Nature 386: 569–577 [DOI] [PubMed] [Google Scholar]
- Record MT Jr, Ha JH, Fisher MA (1991) Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol 208: 291–343 [DOI] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR (2006) Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314: 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G, Hauptmann H, Weeks C, Miller R, Usón I (eds). (2001) International Tables for Crystallography F. Dordrecht: Kluwer Academic Publishers [Google Scholar]
- Solà M, Gomis-Rüth F-X, Serrano L, González A, Coll M (1999) Three-dimensional crystal structure of the transcription factor PhoB Receiver domain. J Mol Biol 285: 675–687 [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Landick R (2003) The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell 12: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Volz K (1993) Structural conservation in the CheY superfamily. Biochemistry 32: 11741–11753 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wanner BL (1996) Phosphorous assimilation and control of the phosphate regulon. In Escherichia coli and Salmonella Typhimurium Cellular and Molecular Biology, Neidhardt FC, Curtiss RI, Ingraham JL, Lin EC, Low KBJ, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger HE (eds). pp 1357–1381. Washington, DC: American Society for Microbiology [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98: 811–824 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.