Pathogen-specific Foxp3+ T reg cells can be identified on the basis of cytokine production, are detected in naive T cell populations, and exhibit suppressive ability toward effector T cells with the same antigen specificity.
Abstract
Foxp3+ CD4 regulatory T cells (T reg cells) are important in limiting immunopathology in infections. However, identifying pathogen-specific epitopes targeted by these cells has been elusive. Using MHC class II/peptide tetramers and intracellular cytokine staining, we identify T reg cells recognizing two virus-specific CD4 T cell epitopes in the coronavirus-infected central nervous system as well as naive T cell precursor pools. These T reg cells are detected at the same time as effector T cells (T eff cells) exhibiting the same specificity and can suppress T eff cell proliferation after stimulation with cognate peptide. These virus-specific T reg cells may be especially effective in inhibiting the immune response during the peak of infection, when virus antigen is maximal. Furthermore, these T reg cells express both IL-10 and IFN-γ after peptide stimulation. IFN-γ expression is maintained during both acute and chronic phases of infection. Identification of T reg cell target epitopes by cytokine production is also applicable in autoimmune disease because myelin oligodendrocyte glycoprotein–specific Foxp3+ T reg cells express IL-10 and IL-17 at the peak of disease in mice with experimental autoimmune encephalomyelitis. These results show that pathogen epitope-specific Foxp3+ T reg cells can be identified on the basis of cytokine production.
Regulatory CD4 T cells (T reg cells), which are characterized by expression of Foxp3, are major contributors to the antiinflammatory response in the settings of autoimmune disease, cancer, and infections (Vignali et al., 2008). In infectious settings, they help to resolve the proinflammatory response, thereby minimizing bystander damage but also potentially delaying or preventing pathogen clearance (Belkaid, 2007). T reg cells may also be required for efficient immune cell entry into sites of infection (Lund et al., 2008). Pathogen-specific T reg cells have been detected in a few studies using whole pathogen preparations as targets, without identification of specific target epitopes. In mice infected with Leishmania major or Toxoplasma gondii, parasite-specific T reg cells were demonstrated, but the target antigen or epitope was not further identified (Suffia et al., 2006; Oldenhove et al., 2009). In another study, T reg cells specific for a Mycobacteria tuberculosis epitope (Mtb) were studied using Mtb-specific TCR Tg cells, but T reg cells specific for this epitope were not found in the natural infection and no Mtb-specific T reg cell target epitope has thus far been identified (Shafiani et al., 2010). Identification of pathogen-specific epitopes recognized by T reg cells would facilitate their study and therapeutic use in disease settings.
The neurotropic rJ2.2 strain of mouse hepatitis virus, a coronavirus, causes mild acute encephalitis and chronic demyelination (Fleming et al., 1986). We previously showed that T reg cells have a critical role in diminishing immunopathological disease in rJ2.2-infected mice. Thus, adoptive transfer of T reg cells from naive spleens (natural T reg cells) into infected mice decreased weight loss, clinical scores, and demyelination (Trandem et al., 2010). However, whether any of these T reg cells are virus specific and whether virus-specific T reg cells are immunosuppressive in a cognate epitope-dependent manner are not known.
In this paper, we identify two epitopes (M133 and S358) targeted by T reg cells in the rJ2.2-infected central nervous system (CNS). Both epitopes are also recognized by effector T cells (Haring et al., 2001). These virus epitope-specific T reg cells express both IL-10 and IFN-γ, suppress proliferation of virus-specific effector cells after stimulation with cognate peptide, and are present throughout the acute and resolution phases of the infection. Virus-specific T reg cells appear to arise, at least in part, from natural T reg cell pools because they are detected in naive Foxp3gfp mice and mice retrogenic (Rg) for an M133-specific T cell receptor (TCR) and express the transcription factor helios.
RESULTS AND DISCUSSION
Virus-specific effector CD4 T cells and T reg cells are detected at the same time after infection in the brains of rJ2.2-infected mice
Foxp3gfp mice were infected with rJ2.2 and examined for the presence of Foxp3+ T reg cells. The frequency of Foxp3+ T reg cells was 20–30% at 3 and 5 d after infection, decreasing to <10% of total CD4 T cells in the brain and spinal cord by 7 d after infection (Fig. 1 A). The decrease in frequency by day 7 did not reflect a lower rate of proliferation because similar fractions of T reg cells and non–T reg CD4 T cells incorporated BrdU at 6 d after infection (Fig. 1 B). Rather, it likely reflects recruitment of virus-nonspecific T reg cells into the CNS at early stages of the infection.
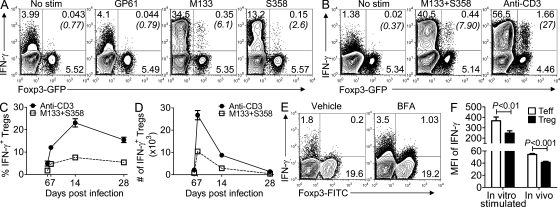
Figure 1.
Identification of epitope M133-specific T reg cells using MHC class II tetramer. (A) CNS-derived lymphocytes from rJ2.2-infected Foxp3gfp mice were analyzed for Foxp3+ CD4 T cells at the indicated times after infection (n = 5 mice for each time point, in at least two independent experiments). (B) BrdU incorporation by CNS-derived CD4 T cells at 6 d after infection is shown. Data shown are representative of six mice analyzed in three independent experiments. (C and D) Foxp3gfp mice were infected with rJ2.2 (C) or rJ2.2.MY135Q (D) and sacrificed at the indicated times after infection. Brain and spinal cord (SC)–derived lymphocytes were stained with I-Ab/M133 tetramers and representative plots gated on CD4+ T cells are shown. (E) Frequency of T reg cells that are I-Ab/M133 tetramer-positive is shown. Data are from four to seven mice per time point in two to five independent experiments. Error bars in A and E indicate mean ± SEM.
We previously showed that virus-specific effector CD4 and CD8 T cells were detected in the infected brain at 6–7 d after infection (Haring et al., 2001). MHC class II/peptide tetramers are available for detecting epitope M133-specific CD4 T cells, and using these tetramers, we also detected M133-specific T reg cells in the brain and spinal cord at 7 d after infection (Fig. 1, C and E). Furthermore, M133-specific T reg cells were present in the infected CNS until at least 49 d after infection. M133-specific T reg cells were not routinely detected above background in draining cervical LNs (CLNs) or spleen (unpublished data). Staining with M133 tetramer was specific because no positive cells were detected in mice infected with an rJ2.2 variant in which the epitope was mutated, abrogating recognition (rJ.2.2.MY135Q; Fig. 1 D; Anghelina et al., 2006).
rJ2.2-specific T reg cells express IFN-γ and IL-10
To further characterize these virus-specific T reg cells, we next assessed whether they produced cytokines in response to peptide stimulation. Remarkably, ∼4–6% of all CNS (pooled brain and spinal cord)-derived T reg cells expressed IFN-γ, after peptide M133 stimulation at 14 d after infection (Fig. 2 A). We also identified IFN-γ+ T reg cells specific for a second rJ2.2-specific CD4 T cell epitope, S358 in the CNS. In experiments described later in this paper, cells were stimulated with M133 and S358 peptides together (M133 + S358) because virus-specific T reg cells were present in low numbers in the infected CNS (Fig. 2 B). Virus-specific IFN-γ+ T reg cells were detected as early as 6 d after infection, at the same time that virus-specific effector CD4 T cells could be identified (Haring et al., 2001), and were present at approximately the same frequency in both the acute and chronic stages of the infection (Fig. 2, C and D). In addition, after stimulation with anti-CD3 mAb, the fraction of T reg cells expressing IFN-γ increased two- to fourfold, suggesting that M133 + S358–specific T reg cells accounted for only a fraction of all IFN-γ+ T reg cells in the CNS (Fig. 2, B–D). Notably, the mean fluorescence intensity of IFN-γ expression was lower in M133 + S358–specific T reg cells compared with effector CD4 T cells, indicating less expression on a per-cell basis (Fig. 2 F). To verify that T reg cells produce IFN-γ in vivo, we treated B6 mice with brefeldin A (BFA) at 6 d after infection (Fig. 2 E; Liu and Whitton, 2005). In the absence of BFA, some Foxp3− CD4 T cells were IFN-γ+, consistent with results showing that cells in the infected brain express IFN-γ in the absence of ex vivo stimulation (Fig. 2 A). The frequency of Foxp3− and Foxp3+ cells expressing IFN-γ increased twofold and fivefold, respectively, after in vivo BFA treatment, confirming in situ IFN-γ expression by T reg cells. Consistent with the in vitro peptide stimulation analyses, the mean fluorescence intensity of IFN-γ expression was lower in T reg cells compared with effector CD4 T cells after in vivo BFA treatment (Fig. 2 F).
Figure 2.
Expression of IFN-γ by rJ2.2-specific T reg cells. (A–D) rJ2.2-infected Foxp3gfp mice were sacrificed at the indicated times after infection and CNS-derived lymphocytes analyzed for IFN-γ production after peptide stimulation. Representative experiments for 14 d after infection are shown in A and B. (A) M133- and S358-specific T reg cells express IFN-γ. Stimulation with lymphocytic choriomeningitis virus GP61 or no peptide served as controls. (B) T reg cells express IFN-γ after stimulation with M133 + S358 peptides or anti-CD3 mAb. Numbers in parentheses represent percentage of IFN-γ+ T reg cells. (C and D) Mean frequency (C) and number (D) of T reg cells that express IFN-γ after stimulation with peptides M133 + S358 or anti-CD3 mAb. Data shown are representative of 3–20 single or pooled brain samples per time point in 3–10 independent experiments. (E) rJ2.2-infected B6 mice were treated with BFA or vehicle at 6 d after infection. After 6 h, CNS-derived lymphocytes were analyzed for IFN-γ expression. Plots shown were gated on CD4 T cells. Data are representative of six mice in two independent experiments. 5.1 ± 0.5 and 4.6 ± 0.3% of Foxp3− and Foxp3+ cells, respectively, expressed IFN-γ after in vivo BFA treatment. (F) IFN-γ is expressed at higher levels in effector CD4 T cells than in T reg cells after peptide M133 + S358 stimulation (data from 10 single or pooled brain samples analyzed at 7, 14, or 28 d after infection are combined) or after in vivo BFA treatment (six mice in two independent experiments). MFI, mean fluorescence intensity. Error bars in C, D, and F indicate mean ± SEM.
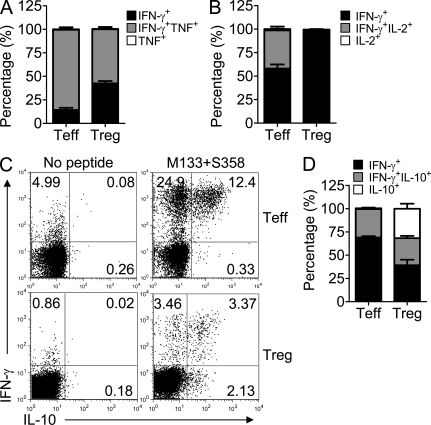
A majority of these virus-specific IFN-γ+ T reg cells expressed TNF but virtually none expressed IL-2 (Fig. 3, A and B). IL-10 production by T reg cells has been previously documented (Vignali et al., 2008) and as shown in Fig. 3 (C and D), ∼50% of IFN-γ+ T reg cells also expressed IL-10. Additionally, a substantial percentage only produced IL-10. T reg cells at sites of Th1-type inflammation express the transcription factor T-bet (Koch et al., 2009). Consistent with the proinflammatory nature of the rJ2.2-infected brain, 35% of all T reg cells also expressed this protein (Fig. S1, A and B). All IFN-γ+ M133 + S358–specific T reg cells were T-bet positive (Fig. S1 C).
Figure 3.
Expression of TNF and IL-10, but not IL-2, by rJ2.2-specific T reg cells. CNS-derived lymphocytes were pooled from rJ2.2-infected Foxp3gfp mice at 14 d after infection and analyzed for IFN-γ, TNF, IL-10, and IL-2 production after peptide stimulation. Cells were gated on GFP− (T eff) or GFP+ (T reg) CD4 T cells. (A and B) A majority of IFN-γ–expressing M133 + S358–specific T reg cells also produced TNF but none expressed IL-2. (C and D) Dual expression of IL-10 and IFN-γ by M133 + S358–specific T reg cells was detected. (A, B, and D) Pooled data from 6–10 pooled brain samples analyzed in three to five independent experiments are shown. Error bars indicate mean ± SEM.
These assays all used Foxp3gfp mice to detect virus epitope-specific T reg cells. Next, we confirmed concordant labeling of GFP and Foxp3 detected with anti-Foxp3 antibody in both naive CLNs and infected brains (Fig. S2 A). To show that IFN-γ virus-specific T reg cells were also detected in WT mice, we infected B6 mice with rJ2.2 and analyzed CNS-derived cells at 7 d after infection. A similar percentage of M133 + S358–specific T reg cells expressed IFN-γ as was detected in infected Foxp3gfp mice (Fig. S2 B).
M133-specific T reg cells suppress proliferation of cognate epitope-specific effector CD4 T cells
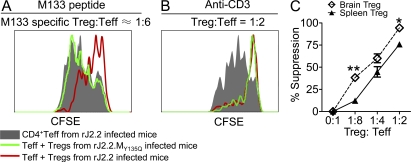
To determine whether M133-specific T reg cells were immunosuppressive, we isolated GFP+ and GFP− CD4 T cells from the brains of rJ2.2-infected Foxp3gfp mice at day 7 after infection and used them in direct ex vivo suppression assays. As a control, we isolated GFP+ T reg cells from mice infected with rJ2.2.MY135Q, which does not express epitope M133. As shown in Fig. 4 A, T reg cells harvested from rJ2.2, but not from rJ2.2.MY135Q-infected mice, suppressed proliferation of brain-derived effector T cells after M133 peptide stimulation. In contrast, T reg cells from brains infected with either virus were suppressive after stimulation with anti-CD3 (Fig. 4 B). To determine the relative suppressive ability of brain-derived T reg cells, we compared them to T reg cells isolated from naive spleens in proliferation assays as described in Materials and methods. In these assays, responder cells were harvested from uninfected spleens and stimulated with anti-CD3 mAb. The brain-derived T reg cells were more suppressive than splenic T reg cells (Fig. 4 C), possibly because T reg cells from sites of inflammation are more activated than those derived from naive mice. Consistent with this, brain-derived T reg cells expressed higher levels of CD69, ICOS, and CTLA-4 directly ex vivo and of IL-10 after stimulation with anti-CD3 antibody or PMA and ionomycin (Fig. S3).
Figure 4.
Suppression mediated by M133-specific T reg cells. (A and B) Foxp3gfp mice were infected with rJ2.2 or rJ2.2.MY135Q. GFP+ (T reg cell) and GFP− (responder, only from rJ2.2-infected mice) CD4 T cells were isolated from infected brains at 7 d after infection and used in T cell proliferation assays. (A) T reg cells from rJ2.2 but not rJ2.2.MY135Q-infected mice suppressed proliferation of responder cells after peptide M133 stimulation. (B) T reg cells from both rJ2.2 or rJ2.2.MY135Q-infected mice suppressed proliferation of responder cells after anti-CD3 mAb stimulation. Data in A and B are representative of three independent experiments, with cells pooled from 5–10 mice/group in each experiment. (C) T reg cells harvested from rJ2.2-infected brains (pools of 10 mice) or naive spleens were used in T cell proliferation assays with CFSE-labeled naive splenic CD4 T cells as responders. Cells were stimulated with anti-CD3 mAb. The percentage of suppression by T reg cells is shown. **, P < 0.01; *, P < 0.05. P = 0.08 at 1:4. Data from three pooled brain samples or naive spleens in three individual experiments are shown. Error bars indicate mean ± SEM.
M133-specific T reg cells are detected in naive M133-specific CD4 T cell populations and express helios
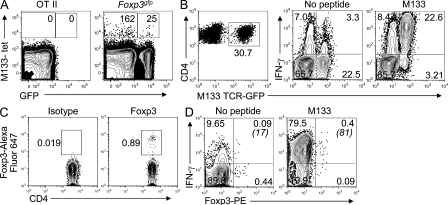
Detection of M133-specific effector CD4 T cells and T reg cells at 6 d after infection was consistent with the notion that M133-specific T reg cells were present in the natural T reg cell pool. To address this, we used MHC/peptide tetramers and magnetic bead technology, which facilitates the identification of epitope-specific precursor T cells in naive mice (Moon et al., 2009). Using this approach and I-Ab/M133 tetramers, we detected 163 ± 10 Foxp3− and 25 ± 2 Foxp3+ M133-specific CD4 T cells per three naive Foxp3gfp mice (n = 3; Fig. 5 A). To confirm these results in mice with a higher M133-specific precursor frequency, we developed Rg mice expressing an M133-specific T cell receptor, as described in Materials and methods (M133 TCR Rg). 0.2–5% of splenic CD4 T cells were M133 specific in uninfected M133 TCR Rg mice and, of these cells, ∼0.5–1% were Foxp3+ (Fig. 5 C). To determine whether IFN-γ+ M133-specific T reg cells were detectable in infected M133 TCR Rg mice, we infected mice that contained <1% Rg CD4 T cells with rJ2.2. The frequency of M133-specific CD4 T cells as assessed by IFN-γ expression (Fig. 5 B) was modestly higher than in B6 mice (Fig. 2 A; Zhao et al., 2009), but most of the cells were M133 Rg cells. A small fraction of these cells were Foxp3+ and most expressed IFN-γ after M133 peptide stimulation (Fig. 5 D).
Figure 5.
Detection of M133-specific T reg cells in naive Foxp3gfp and M133 TCR Rg mice. (A) Lymphocytes were pooled from the spleens and LNs of three naive Foxp3gfp or two OT-II mice and analyzed for tetramer M133–positive T reg cells. Numbers of Foxp3− and Foxp3+ M133-specific CD4 T cells/3 mice are indicated. Representative data from one of three independent experiments are shown. (B–D) M133 TCR Rg mice were infected with rJ2.2 and sacrificed at 7 d after infection (B and D) or were not infected (C). (B) M133-specific (GFP+) CD4 T cells accumulated in the infected brain and expressed IFN-γ after M133 peptide stimulation. (C) GFP+ CD4 T cells from spleens of uninfected mice were analyzed for Foxp3 expression. (D) Lymphocytes from the brains of rJ2.2-infected mice were stimulated with peptide M133 and assayed for Foxp3 and IFN-γ expression after gating on GFP+ CD4 T cells. Numbers in parentheses represent percentage of IFN-γ+ T reg cells. Data shown in B–D are representative of two independent experiments containing three mice each.
To further investigate whether these T reg cells were derived from natural T reg cells, we measured expression of the helios, a transcription factor. Helios has been reported to be expressed by natural but not peripherally converted T reg cells (Thornton et al., 2010). Some M133 + S358–specific IFN-γ+ T reg cells in the infected brain expressed high level of helios, consistent with an origin in natural T reg cell populations (Fig. S4). However, many of the IFN-γ+ cells were helios intermediate, making it uncertain as to whether all of these cells were thymically derived.
IL-10+ and IL-17+ myelin oligodendrocyte glycoprotein (MOG)35-55–specific T reg cells are present in the brains of mice with experimental autoimmune encephalomyelitis (EAE)
Our results from rJ2.2-infected mice showed that virus-specific T reg cells expressed IFN-γ and IL-10 in the inflamed brain. To determine whether this was also true in autoimmune settings, we examined cytokine production by MOG35-55-specific T reg cells in Foxp3gfp mice with EAE (Fig. S5). In these mice, ∼12 and 32% CD4 T cells in the inflamed brain were Foxp3+ at 15 and 32 d after immunization, respectively (Fig. S5 C). To determine whether these T reg cells expressed cytokines in response to peptide stimulation, we exposed cells directly ex vivo to MOG35-55 and measured IL-10, IFN-γ, and IL-17 production. IL-10 was expressed by the majority of cytokine-producing MOG35-55-specific T reg cells, whereas small percentages expressed IL-17 (Fig. S5 D) at day 15. By day 32, only a small fraction of T reg cells expressed either of these cytokines (Fig. S5 E).
Identification of T reg cell–specific target epitopes in infected nontransgenic animals has proven difficult. Our results demonstrate the presence of virus-specific Foxp3+ T reg cells in the rJ2.2-infected CNS but also show that they are present at a low frequency (Figs. 1 and 2). Detection was initially facilitated by the availability of M133-specific MHC class II tetramers. However, we previously showed that this tetramer was not as efficient as cytokine expression in identifying M133-specific effector CD4 T cells (Zhao et al., 2009). Subsequently, we identified a greater frequency of M133-specific T reg cells by cytokine staining for IL-10 and IFN-γ after peptide stimulation. IL-10 expression may be particularly useful for detecting epitope-specific T reg cells because IL-10 was produced in virally infected mice and in mice with EAE after peptide stimulation (Fig. 3 and Fig. S5). IFN-γ production by T reg cells was previously demonstrated in a lethal toxoplasmosis infection (Oldenhove et al., 2009), but our results indicate that IFN-γ is also expressed in the context of a nonlethal viral infection and continues even as the inflammatory response subsides (Fig. 2). The continued expression of IFN-γ by virus-specific T reg cells likely occurs because virus antigen persists in the infected CNS (Bergmann et al., 2006; Zhao et al., 2009). Epitope M133-specific T reg cells suppressed proliferation of effector CD4 T cells responding to the same epitope, suggesting that if IFN-γ expression diminishes T reg cell suppressive function, its effect is limited. rJ2.2-specific T reg cells were detected in the CNS and not draining CLNs or spleen (Fig. 1 and not depicted), suggesting that they function by directly inhibiting T cell function or diminishing antigen presentation by microglia or other antigen-presenting cells at the site of inflammation.
Do virus-specific T reg cells generally play a critical role in diminishing immunopathology? In several infectious diseases, including patients infected with West Nile virus or Dengue fever virus, higher T reg cell frequencies correlated with improved outcomes (Lühn et al., 2007; Lanteri et al., 2009). Relevant for our results, Mtb-specific TCR Tg T reg cells were more potently suppressive than Mtb-nonspecific TCR Tg T reg cells in infected mice (Shafiani et al., 2010). Furthermore, although transfer of natural T reg cells diminished disease in the setting of autoimmune disease, suppression was estimated to be ∼20–50-fold more potent if T reg cells were specific for an epitope at a site of inflammation (Tang et al., 2004; Tarbell et al., 2004). Thus, islet-specific T reg cells prevented diabetes in NOD mice, whereas nonspecific T reg cells at the same dosage could not. If true for rJ2.2-specific T reg cells, these cells may be especially potent in diminishing immunopathology in mice with encephalomyelitis and would be most activated and, thereby, suppressive during acute infection when viral antigen is maximal (Bergmann et al., 2006).
Our results suggest that rJ2.2-specific T reg cells originate, at least in part, from natural T reg cell populations. First, virus-specific T reg cells are detected at 6–7 d after infection at the same time as effector CD4 T cells. If these cells arose by peripheral conversion from effector cells, this process would need to occur concomitantly with priming. Second, M133-specific T reg cells can be detected in the spleens and LNs of naive Foxp3gfp and Rg M133 TCR mice (Fig. 5), showing that they are present in the naive T reg cell pool. Third, some M133 + S358–specific T reg cells expressed high levels of helios, a transcription factor expressed by thymus-derived but not induced T reg cells (Fig. S4; Thornton et al., 2010).
In summary, we demonstrate the presence of IFN-γ– and IL-10–expressing virus-specific T reg cells in the CNS of infected mice and show that these T cells suppress proliferation of cognate epitope-specific CD4 T cells. Detection of epitope-specific T reg cells by cytokine expression may be especially useful because MHC class II/peptide tetramers are not widely available and may detect only high-affinity cells (Sabatino et al., 2011). In addition, previous efforts using T reg cells therapeutically to diminish excessive inflammatory responses have used adoptive transfer of in vitro propagated T reg cells (Tang et al., 2004; Tarbell et al., 2004). Identification of virus-specific T reg cells raises the possibility that an alternative therapeutic approach will be to augment expansion of T reg cells specific for a virus or pathogenic self epitope at sites of inflammation.
MATERIALS AND METHODS
Mice.
Specific pathogen-free 6-wk-old C57BL/6 mice (B6) were purchased from the National Cancer Institute (Bethesda, MD). Foxp3gfp mice on a B6 background were provided by A. Rudensky (University of Washington, Seattle, WA). OT-II mice, transgenic for expression of a TCR specific for epitope OVA323-339, were obtained from T. Griffith (University of Iowa, Iowa City, IA). All animal studies were approved by the University of Iowa Animal Care and Use Committee.
For M133 TCR Rg mice, to identify M133-specific T cell receptors, splenocytes were harvested 7 d after infection from mice infected i.p. with the neurovirulent JHM strain of mouse hepatitis virus and were stimulated with 5 µM of peptide M133. After stimulation, epitope-specific CD4 T cells were captured using a mouse IFN-γ secretion assay (Miltenyi Biotec) as previously described (Pewe et al., 2004). IFN-γ–expressing CD4 T cells were sorted with a FACSDiva (BD). RNA was isolated using an RNeasy Mini kit (Invitrogen) and used for cDNA synthesis. Vβ8-specific PCR products were synthesized because Vβ8 is the most commonly used Vβ element in the M133-specific response (Haring et al., 2001). PCR products were cloned into plasmid vector pCR2.1-TOPO (Invitrogen) and sequenced.
TCR Rg mice were engineered as previously described (Holst et al., 2006). An M133-specific TCR-β (Vβ8.3, TRBV13-1) identified in three of three mice was produced synthetically (Geneart) and then cloned into a retroviral vector containing an IRES-GFP cassette to allow identification of transfected cells by GFP expression. Retroviral producer cell lines expressing this TCR-β were produced and used to transduce B6 donor bone marrow before reconstitution of lethally irradiated B6 recipient mice. After reconstitution, these Rg mice were infected with rJ2.2. The majority of CD4 T cells in the infected CNS were GFP+. I-Ab/M133 tetramer+ GFP+ cells were sorted from the infected CNS and analyzed for TCR-α chain expression. Two TCR-α chains (Vα2.3 and TRAV14-1), differing by a single amino acid in the CDR3 region, were detected using RT-PCR–based cloning. One of these TCR-α was synthesized and cloned into the same retroviral vector described in this section. In this vector, the TCR-α and TCR-β chains are linked via the 2A protease, facilitating cleavage. Because the two chains are linked on the same molecule, they are produced in stoichiometric amounts. TCR Rg mice expressing both M133-specific TCR-α and TCR-β were produced. Epitope M133 specificity was confirmed by analysis of GFP+ cells in the infected CNS using M133-specific tetramer and IFN-γ expression after peptide stimulation.
Viruses and infection.
A recombinant rJ2.2 in which the M133 epitope was mutated to abolish binding to the I-Ab antigen (rJ2.2.MY135Q) was constructed as described previously (Anghelina et al., 2006). rJ2.2 and rJ2.2.MY135Q were propagated in mouse 17Cl-1 cells and titered on HeLa-MHVR cells (Pewe et al., 2005). 6–7-wk-old WT B6 and Foxp3gfp mice were inoculated intracerebrally with 600 PFU rJ2.2 or 2,000 PFU rJ2.2.MY135Q in 30 µl DME.
Antibodies.
The following antibodies were used in this study: CD4-FITC, -PE, or -PerCP-Cy5.5 (RM 4–5), CD69-PE (H1.2F3), IL-2-PE (JES6-5H4; BD); purified functional anti-CD3, CD3-PerCP-Cy5.5 (145-2C11), CD4–Alexa Fluor 700 (RM 4–5), CD16/CD32-biotin or -PerCP-Cy5.5 (93), Foxp3-FITC, -PE, or –Alexa Fluor 647 (FJK-16s), T-bet–PE (eBio4B10), IFN-γ–PE or –allophycocyanin (APC; XMG1.2), TNF-PE (MP6-XT22), CTLA-4–PE (UC10-4B9), ICOS-PE (7E.17G9), eFluor 450–conjugated anti-CD11b (M1/70), CD11c (N418), F4/80 (BM8), B220 (RA3-6B2), and CD49b (DX5) mAbs (eBioscience); and IL-10-APC (JES5-16E3) and Helios-PE (22F6; BioLegend).
Isolation of mononuclear cells from CNS tissue.
Brains and spinal cords harvested after PBS perfusion were dispersed and digested with 1 mg/ml collagenase D (Roche) and 0.1 mg/ml DNase I (Roche) at 37°C for 30 min. Dissociated CNS tissue was passed through a 70-µm cell strainer, followed by Percoll gradient (70/37%) centrifugation. Mononuclear cells were collected from the interphase, washed, and resuspended in culture medium for further analysis.
MHC class II tetramer staining.
CNS-derived mononuclear cells were stained with 8 µg/ml PE-conjugated I-Ab/M133 tetramers (obtained from the National Institutes of Health/National Institute of Allergy and Infectious Diseases MHC Tetramer Core Facility, Atlanta, GA) in RP10 media for 1 h at 37°C. Cells were then incubated with anti–CD16/CD32-biotin and anti–CD4-PerCP-Cy5.5, followed by Avidin-APC. Cells were analyzed using a FACSCalibur (BD). Results are shown after gating on CD16/CD32-negative cell populations.
Measurement of BrdU incorporation.
Foxp3gfp mice were injected i.p. with 2 mg BrdU (BD) at 6 d after infection. BrdU incorporation by brain CD4 T cells was examined 16 h after injection according to manufacturer protocols (APC BrdU flow kit; BD).
Intracellular cytokine staining.
To detect cytokine production by antigen-specific CD4+ effector T cells (T eff cells) and T reg cells, mononuclear cells isolated from the brain and spinal cord were stimulated with 5 µM of peptides M133, S358, M133 + S358 (rJ2.2 infection), or MOG35-55 (in EAE) in complete RPMI 1640 medium for 6 h at 37°C, in the presence of 1 µl/ml Golgiplug (BD) and antigen-presenting cells (CHB3 cells, B cell line, H-2Db, I-Ab). The rJ2.2-specific epitopes encompass residues 133–147 of the transmembrane (M) protein and residues 358–372 of the surface (S) glycoprotein (Xue and Perlman, 1997; Haring et al., 2001). Intracellular expression of IFN-γ, IL-10, TNF, IL-2, or IL-17 was assayed. For some samples, cells were pooled from two to four mice. In some experiments, infected mice were treated with 0.25 mg BFA (Sigma-Aldrich) or vehicle (5% DMSO) intravenously. 6 h later, CNS-derived lymphocytes were analyzed directly ex vivo for IFN-γ expression as described previously (Liu and Whitton, 2005). Only infected mice were analyzed because virtually no T cells are detected in the uninfected brain.
In vitro suppression assay.
CD4+ T cells were enriched by negative magnetic selection from rJ2.2- or rJ2.2.MY135Q-infected brains (pooled from 10 mice) or naive spleens of Foxp3gfp mice using a CD4+ T cell isolation kit and AutoMACS (both from Miltenyi Biotec) or a mouse CD4+ T cell enrichment kit and Purple EasySep Magnet (both from STEMCELL Technologies). After enrichment of CD4+ T cells, CD4+GFP− responder and CD4+GFP+ regulatory T cells were then sorted using a FACSDiva or FACSAria cell sorter (BD). For the suppression assay, 3 × 104 CFSE-labeled (2.5 µM; Invitrogen) responder T cells from rJ2.2-infected brains or naive spleens were co-cultured with the indicated number of T reg cells and 105 irradiated CHB3 cells (2,500 rad) per well, in the presence of 5 µM M133 peptide or 1 µg/ml anti-CD3 mAb in a 96-well round bottom plate. After 66 h, cells were harvested and stained with anti–CD4-PE and anti–Foxp3–Alexa Fluor 647. CD4+Foxp3− responder cells were analyzed for CFSE dilution by flow cytometry. The division index (DI) was obtained using FlowJo software (Tree Star). The percentage of suppression by T reg cells was calculated as follows: % suppression = 100% × [1 − (DI of responders plus T reg cells/DI of responders only)].
Detection of M133-specific T reg cells in naive T cell pool.
M133-specific T reg cells in the naive T cell pool were identified using a previously described method (Moon et al., 2009) with modifications. In brief, single cell suspensions were prepared from spleens and cervical, axillary, brachial inguinal, and mesenteric LNs of uninfected 10-wk Foxp3gfp and OT-II mice. CD4 T cells were isolated by negative selection using a CD4 T cell isolation kit II. CD4 T cells were then stained with anti-CD16/32 mAb (2.4G2) and PE-conjugated I-Ab/M133 tetramers as described in the previous section (except tetramer was used at 4 µg/ml to reduce background). After washing, cells were exposed to anti-PE magnetic microbeads and separated on an MS column (Miltenyi Biotec). Cells were then stained with anti–CD3-PerCP, anti–CD4-Alexa Fluor 700, and non–T cell lineage (dump) markers (eFluor 450–conjugated anti-CD11b, anti-CD11c, anti-F4/80, anti-B220, and anti-CD49b). Cells were then stained with Hoechst 33258 to gate out dead cells, and analyzed with a LSRII cytometer (BD).
Induction of EAE.
EAE was induced in Foxp3gfp mice using a previously described method (Wang et al., 2006). In brief, mice were immunized with 0.4 ml of an emulsion containing 300 µg MOG35-55 (>95% purity; AnaSpec) in PBS and an equal volume of CFA containing 1 mg Mycobacterium tuberculosis H37Ra (Difco Laboratories) by s.c. inoculation at four flank sites. 200 ng pertussis toxin (List Biological Laboratories Inc.) was administered in 0.2 ml PBS i.v. on the day of immunization and 48 h later. Mice were weighed and assessed for clinical symptoms daily with the following scoring system: 0, no clinical signs; 1, limp tail; 2, paraparesis (weakness, incomplete paralysis of one or two hind limbs); 3, paraplegia (complete paralysis of two hind limbs); 4, paraplegia with fore limb weakness or paralysis; and 5, moribund or dead.
Statistical analysis.
Two-tailed unpaired Student’s t tests were used to analyze differences in mean values between groups. All data are presented as mean ± SEM. Differences with values of P < 0.05 were considered significant.
Online supplemental material.
Fig. S1 shows T-bet expression by rJ2.2-specific T reg cells. Fig. S2 shows IFN-γ expression by virus-specific T reg cells in the brains of infected B6 mice. Fig. S3 shows expression of activation markers and IL-10 by T reg cells derived from naive spleens and rJ2.2-infected brains. Fig. S4 shows helios expression by IFN-γ–expressing virus-specific T reg cells in the brains of infected B6 mice. Fig. S5 shows IL-10 and IL-17 expression by MOG35-55-specific T reg cells in the CNS of mice with EAE. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110236/DC1.
Acknowledgments
We thank Drs. John Harty, Steven Varga, and Jonathan Trujillo for stimulating discussion and critical review of the manuscript, the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases Tetramer Core Facility (Atlanta, GA) for providing tetramers, Dr. D. Vignali (St. Jude’s Research Hospital) for reagents and guidance as we developed M133-specific TCR Rg mice, and the University of Iowa Flow Cytometry facility for help with cell sorting and analysis.
This work was supported in part by grants from the NIH (NS36592) and National Multiple Sclerosis Society. K.T. was supported by an NIH training grant (T32 AI007511-14).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- APC
- allophycocyanin
- BFA
- brefeldin A
- CLN
- cervical LN
- CNS
- central nervous system
- EAE
- experimental autoimmune encephalomyelitis
- MOG
- myelin oligodendrocyte glycoprotein
- Rg
- retrogenic
- T eff cell
- effector T cell
- T reg cell
- regulatory T cell
References
- Anghelina D., Pewe L., Perlman S. 2006. Pathogenic role for virus-specific CD4 T cells in mice with coronavirus-induced acute encephalitis. Am. J. Pathol. 169:209–222 10.2353/ajpath.2006.051308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y. 2007. Regulatory T cells and infection: a dangerous necessity. Nat. Rev. Immunol. 7:875–888 10.1038/nri2189 [DOI] [PubMed] [Google Scholar]
- Bergmann C.C., Lane T.E., Stohlman S.A. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121–132 10.1038/nrmicro1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J.O., Trousdale M.D., el-Zaatari F.A., Stohlman S.A., Weiner L.P. 1986. Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies. J. Virol. 58:869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring J.S., Pewe L.L., Perlman S. 2001. High-magnitude, virus-specific CD4 T-cell response in the central nervous system of coronavirus-infected mice. J. Virol. 75:3043–3047 10.1128/JVI.75.6.3043-3047.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J., Szymczak-Workman A.L., Vignali K.M., Burton A.R., Workman C.J., Vignali D.A. 2006. Generation of T-cell receptor retrogenic mice. Nat. Protoc. 1:406–417 10.1038/nprot.2006.61 [DOI] [PubMed] [Google Scholar]
- Koch M.A., Tucker-Heard G., Perdue N.R., Killebrew J.R., Urdahl K.B., Campbell D.J. 2009. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10:595–602 10.1038/ni.1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanteri M.C., O’Brien K.M., Purtha W.E., Cameron M.J., Lund J.M., Owen R.E., Heitman J.W., Custer B., Hirschkorn D.F., Tobler L.H., et al. 2009. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J. Clin. Invest. 119:3266–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Whitton J.L. 2005. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J. Immunol. 174:5936–5940 [DOI] [PubMed] [Google Scholar]
- Lühn K., Simmons C.P., Moran E., Dung N.T., Chau T.N., Quyen N.T., Thao T.T., Van Ngoc T., Dung N.M., Wills B., et al. 2007. Increased frequencies of CD4+CD25high regulatory T cells in acute dengue infection. J. Exp. Med. 204:979–985 10.1084/jem.20061381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J.M., Hsing L., Pham T.T., Rudensky A.Y. 2008. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 320:1220–1224 10.1126/science.1155209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.J., Chu H.H., Hataye J., Pagán A.J., Pepper M., McLachlan J.B., Zell T., Jenkins M.K. 2009. Tracking epitope-specific T cells. Nat. Protoc. 4:565–581 10.1038/nprot.2009.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenhove G., Bouladoux N., Wohlfert E.A., Hall J.A., Chou D., Dos Santos L., O’Brien S., Blank R., Lamb E., Natarajan S., et al. 2009. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 31:772–786 10.1016/j.immuni.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L.L., Netland J.M., Heard S.B., Perlman S. 2004. Very diverse CD8 T cell clonotypic responses after virus infections. J. Immunol. 172:3151–3156 [DOI] [PubMed] [Google Scholar]
- Pewe L., Zhou H., Netland J., Tangudu C., Olivares H., Shi L., Look D., Gallagher T., Perlman S. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335–11342 10.1128/JVI.79.17.11335-11342.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino J.J., Jr, Huang J., Zhu C., Evavold B.D. 2011. High prevalence of low affinity peptide–MHC II tetramer–negative effectors during polyclonal CD4+ T cell responses. J. Exp. Med. 208:81–90 10.1084/jem.20101574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiani S., Tucker-Heard G., Kariyone A., Takatsu K., Urdahl K.B. 2010. Pathogen-specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. J. Exp. Med. 207:1409–1420 10.1084/jem.20091885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffia I.J., Reckling S.K., Piccirillo C.A., Goldszmid R.S., Belkaid Y. 2006. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J. Exp. Med. 203:777–788 10.1084/jem.20052056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Henriksen K.J., Bi M., Finger E.B., Szot G., Ye J., Masteller E.L., McDevitt H., Bonyhadi M., Bluestone J.A. 2004. In vitro–expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 199:1455–1465 10.1084/jem.20040139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbell K.V., Yamazaki S., Olson K., Toy P., Steinman R.M. 2004. CD25+ CD4+ T cells, expanded with dendritic cells presenting a single autoantigenic peptide, suppress autoimmune diabetes. J. Exp. Med. 199:1467–1477 10.1084/jem.20040180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton A.M., Korty P.E., Tran D.Q., Wohlfert E.A., Murray P.E., Belkaid Y., Shevach E.M. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184:3433–3441 10.4049/jimmunol.0904028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trandem K., Anghelina D., Zhao J., Perlman S. 2010. Regulatory T cells inhibit T cell proliferation and decrease demyelination in mice chronically infected with a coronavirus. J. Immunol. 184:4391–4400 10.4049/jimmunol.0903918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali D.A., Collison L.W., Workman C.J. 2008. How regulatory T cells work. Nat. Rev. Immunol. 8:523–532 10.1038/nri2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hong J., Sun W., Xu G., Li N., Chen X., Liu A., Xu L., Sun B., Zhang J.Z. 2006. Role of IFN-gamma in induction of Foxp3 and conversion of CD4+ CD25- T cells to CD4+ Tregs. J. Clin. Invest. 116:2434–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Perlman S. 1997. Antigen specificity of CD4 T cell response in the central nervous system of mice infected with mouse hepatitis virus. Virology. 238:68–78 10.1006/viro.1997.8819 [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. 2009. De novo recruitment of antigen-experienced and naive T cells contributes to the long-term maintenance of antiviral T cell populations in the persistently infected central nervous system. J. Immunol. 183:5163–5170 10.4049/jimmunol.0902164 [DOI] [PMC free article] [PubMed] [Google Scholar]