Abstract
In young Arabidopsis seedlings, retrograde signaling from plastids regulates the expression of photosynthesis-associated nuclear genes in response to the developmental and functional state of the chloroplasts. The chloroplast-located PPR protein GUN1 is required for signalling following disruption of plastid protein synthesis early in seedling development before full photosynthetic competence has been achieved. Recently we showed that sucrose repression and the correct temporal expression of LHCB1, encoding a light-harvesting chlorophyll protein associated with photosystem II, are perturbed in gun1 mutant seedlings.1 Additionally, we demonstrated that in gun1 seedlings anthocyanin accumulation and the expression of the “early” anthocyanin-biosynthesis genes is perturbed. Early seedling development, predominantly at the stage of hypocotyl elongation and cotyledon expansion, is also affected in gun1 seedlings in response to sucrose, ABA and disruption of plastid protein synthesis by lincomycin. These findings indicate a central role for GUN1 in plastid, sucrose and ABA signalling in early seedling development.
Key words: ABA, ABI4, anthocyanin, chloroplast, GUN1, retrograde signalling, sucrose
Arabidopsis seedlings develop in response to light and other environmental cues. In young seedlings, development is fuelled by mobilization of lipid reserves until chloroplast biogenesis is complete and the seedlings can make the transition to phototrophic growth. The majority of proteins with functions related to photosynthesis are encoded by the nuclear genome, and their expression is coordinated with the expression of genes in the chloroplast genome. In developing seedlings, retrograde signaling from chloroplasts to the nucleus regulates the expression of these nuclear genes and is dependent on the developmental and functional status of the chloroplast. Two classes of gun (genomes uncoupled) mutants defective in retrograde signalling have been identified in Arabidopsis: the first, which comprises gun2–gun5, involves mutations in genes encoding components of tetrapyrrole biosynthesis.2,3 The other comprises gun1, which has mutations in a nuclear gene encoding a plastid-located pentatricopeptide repeat (PPR) protein with an SMR (small MutS-related) domain near the C-terminus.4,5 PPR proteins are known to have roles in RNA processing6 and the SMR domain of GUN1 has been shown to bind DNA,4 but the specific functions of these domains in GUN1 are not yet established. However, GUN1 has been shown to be involved in plastid gene expression-dependent,7 redox,4 ABA1,4 and sucrose signaling,1,4,8 as well as light quality and intensity sensing pathways.9–11 In addition, GUN1 has been shown to influence anthocyanin biosynthesis, hypocotyl extension and cotyledon expansion.1,11
gun1 Seedling Development is Hypersensitive to ABA and Sucrose
Norflurazon (an herbicide that results in photo-oxidation of chlorophyll) and lincomycin (an antibiotic that inhibits plastid translation) lead to a loss of photosynthesis-associated nuclear gene expression in wild-type Arabidopsis seedlings. The gun mutants were identified by their ability to express photosynthesis-associated nuclear genes when grown in the presence of norflurazon,12 but only gun1 retained the ability to express photosynthesis-associated nuclear genes when grown in the presence of lincomycin, linking the GUN1 signaling pathway to disruption of chloroplast translation.7 The target genes of this signaling pathway have been shown to contain an ACGT motif, which is core to both the abscisic acid (ABA) response element (ABRE) and the light-responsive G-box.4 Screens of ABA-deficient and ABA-insensitive mutants identified aba insensitive4 (abi4) as having a weak gun phenotype.4 Overexpression of ABI4, which encodes an APETALA 2-type transcription factor, suppressed the gun1 phenotype, suggesting that ABI4 functions downstream of GUN1.4 ABI4 has been shown to regulate RBCS gene expression in response to sucrose and ABA, via an S-box motif in association with the light-responsive G-box element.13 However, most ABA signalling mutants, including abi4, have been identified by their insensitivity to ABA, which mediates seedling developmental arrest in wild type, whereas gun1 does not fall into this category. gun1 has a hypersensitive response to ABA and displays greater developmental arrest compared to wild-type seedlings.1,4
abi4 and many other ABA-insensitive mutants are also sugar insensitive, establishing links between sugar and ABA signalling. However, we showed that gun1 displays sucrose hypersensitivity: sucrosemediated developmental arrest was more severe for gun1 than for wild-type seedlings, predominantly at the stage of cotyledon expansion.1 Sugar developmental-arrest screens have identified many insensitive mutants but very few that are hypersensitive,14 and fewer still that are also hypersensitive to ABA.
The interactions between ABA and sugar signalling in developing seedlings are complex. Although both ABA and sucrose can arrest seedling development, sucrose can rescue seedlings from ABA-mediated developmental arrest.15 This observation led to the hypothesis that ABA inhibits germination and post-germinative growth by inhibiting mobilization of seed lipid reserves.15 However, active lipid mobilization in response to ABA, leading to high levels of endogenous sucrose, has been demonstrated.16 Thus, it is not clear why sucrose should rescue ABA-induced developmental arrest, but one possible explanation is that endogenous and exogenous sugars are perceived and affect developmental signals differently. Exogenous sugars may potentially be perceived by young seedlings as the product of photosynthesis and serve to override ABA signalling. But sugar concentrations may also play a part: 14 mM sucrose rescues ABA developmental arrest,15 whereas 330 mM sucrose arrests seedling development.17 We showed that medium containing 2% sucrose (85 mM) arrests seedling development and LHCB1 expression in wildtype seedlings and to a greater extent in gun1.1 However, sugar sensitivity in gun1 seedlings appears to change during development; the addition of 7% glucose (300 mM) to the growth medium was shown to reduce LHCB1 expression significantly when applied to 3-day-old4 and 3-week-old8 wild-type seedlings, but not when applied to gun1 seedlings.
Temporal Photosynthesis Gene Expression and Anthocyanin Accumulation are Differentially Affected by Sucrose in Wild-Type and gun1 Seedlings
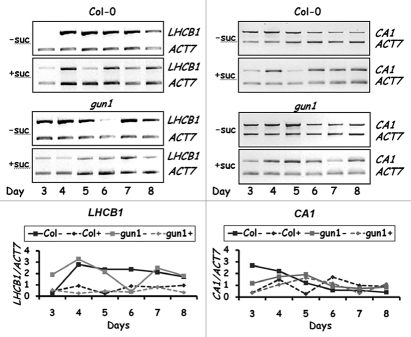
We showed that the expression of LHCB1 fluctuates during early seedling development (3–8 days old) and is different in gun1 seedlings in comparison to wild-type. 1 Sucrose represses expression of both nuclear and plastid genes encoding photosynthetic components18,19 and disrupts the temporal expression profile of LHCB1 in both wild-type and gun1 seedlings. In general, sucrose results in a decrease in LHCB1 transcripts of around 70% in both genotypes.1 The addition of lincomycin resulted in a further reduction of LHCB1 transcripts in wild type, but partially released LHCB1 from sucrose repression in gun1.1 This would imply that GUN1 is required for sucrose repression of LHCB1 expression in the absence of functional chloroplasts. To determine if a similar temporal response was observed with other nuclear genes encoding photosynthesis components, we examined the expression of the CA1 gene (At3g01500) encoding a plastidic carbonic anhydrase, which catalyses the reversible hydration of CO2 to form HCO3- and is thought to supply CO2 for Rubisco. Transcriptome profiling identified transcripts of CA1 as the most responsive to lincomycin treatment in wild-type plants, whereas CA1 transcripts remained unchanged in lincomycin-treated gun1 seedlings.5 Analysis of CA1 transcripts in wild-type and gun1 seedlings over a 6-day period in the absence or presence of 2% sucrose is shown in Figure 1. The temporal patterns of CA1 expression and the effects of sucrose differed between wild-type and gun1 seedlings. In wild-type seedlings, the temporal patterns were markedly different in the absence and presence of sucrose; sucrose repressed CA1 expression in 3–5-day-old seedlings, but had little effect in 6–8-day-old seedlings. In contrast, in gun1 seedlings, sucrose had little effect on CA1 transcript accumulation over the 6-day period. There was a slight repression of CA1 expression by sucrose in 3- and 4-day-old seedlings, but in 5–8-day-old seedlings there was relatively little effect. This is different to LHCB1 expression, which is markedly repressed by sucrose in both wild-type and gun1 seedlings throughout the 6-day period (Fig. 1). These observations imply that the temporal sucrose response of CA1, unlike LHCB1, is almost entirely mediated by GUN1.
Figure 1.
Time course of LHCB1 and CA1 transcript abundance in wild-type and gun1 seedlings. RT-PCR s were performed on RNA extracted from 3- to 8-day-old wild-type (Col-0) and gun1-1 seedlings grown on 0.5x MS-agar medium, ±2% sucrose (suc). The PCR products separated by electrophoresis in 1% agarose gels are shown in the upper part of the figure. The amounts of LHCB1 and CA1 PCR products normalized to those of ACT7 are shown in the lower part of the figure.
Sucrose also induces anthocyanin accumulation in young seedlings in a distinct temporal fashion.20 We showed that anthocyanins reach a peak 1 day later in gun1 seedlings in comparison to wild-type, and that the amounts accumulated were lower in gun1.1 Lincomycin and norflurazon treatments disrupted both the temporal profile and decreased sucrose-induced anthocyanin levels in both genotypes. We demonstrated that GUN1-mediated regulation of anthocyanin biosynthesis was via transcriptional control of the “early” anthocyanin biosynthesis genes (PAL, CHS, CHI and F3H). Whereas ABA-mediated seedling developmental arrest can be rescued by sucrose, ABA has been shown to have a synergistic effect on sucrose-induced anthocyanin accumulation in wild-type Arabidopsis seedlings.21 This appears not to involve ABI4, as 3-day-old abi4 seedlings accumulate more anthocyanin in response to sucrose than wild-type seedlings and CHS transcripts were shown to be increased.22 ABI4 may have a role in negatively regulating “early” anthocyanin biosynthesis genes, in contrast to GUN1 which appears to have a positive regulatory role.
If ABI4 regulates photosynthesis-associated nuclear gene expression in response to GUN1-mediated retrograde signalling, it is not clear how it manifests contrasting seedling developmental phenotypes of gun1 in response to ABA and sucrose. A weak gun phenotype has been reported for 4-day-old abi4 seedlings on medium containing sucrose and lincomycin.4 However, this phenotype would appear to be transient, as 7-day-old abi4 seedlings do not display a gun phenotype, whereas 7-day-old gun1 seedlings do (Fig. 2). Lincomycin treatment completely abolished CA1 expression in wild-type and abi4 seedlings, but had no effect on CA1 transcripts in gun1 seedlings. However, because of the fluctuations in the amounts of LHCB1 and CA1 transcripts during early seedling development and because sucrose disrupts this temporal expression (see Fig. 1), observations at a single specific time point on medium containing sucrose may be potentially misleading. ABI4 may act downstream of GUN1 in response to sucrose, but chloroplast translation-related signaling may act, at least in part, independently of ABI4.
Figure 2.
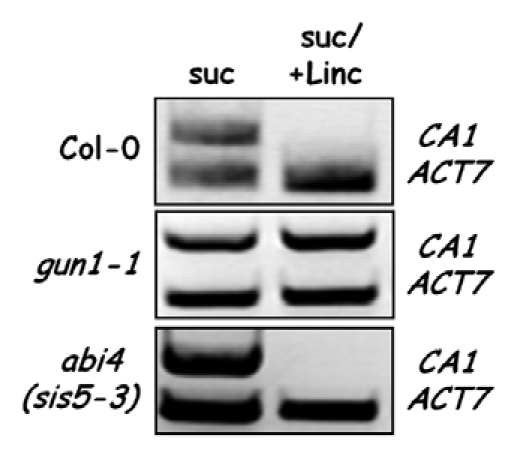
Effect of sucrose and lincomycin on CA1 transcripts in wild-type, gun1 and abi4 seedlings. RT-PCR s for carbonic anhydrase 1 (CA1) and actin (ACT7) transcripts were performed on RNA extracted from 7-day-old wild-type (Col-0), gun1-1 and abi4-1 seedlings grown on 0.5x MS-agar medium, 2% sucrose (suc) and ±0.5 mM lincomycin (Linc). The PCR products were separated by electrophoresis in a 1% agarose gel.
The greatest difference in sucrose-induced anthocyanin accumulation between gun1 and wild-type seedlings was observed on day 4 in the presence of lincomycin, when gun1 contained only 47% of wild-type levels.1 This coincides with a distinct developmental difference between the genotypes: gun1 had expanded cotyledons and extended hypocotyls, whereas wild-type seedlings did not. This phenotype was seen in seedlings grown on medium containing lincomycin with or without sucrose, but was not observed in seedlings grown on norflurazon or on media without inhibitors. This provides further evidence that signaling of the chloroplast developmental/functional status and sucrose signaling may be operating at least partially independently through GUN1. Sucrose inhibited cotyledon opening in gun1 seedlings but not in the absence of functional chloroplasts, whereas in wild-type seedlings both sucrose and decreased chloroplast translation delayed cotyledon opening.
It would appear that GUN1 regulates expression of photosynthesis-related nuclear genes, seedling development and anthocyanin biosynthesis in young seedlings in response to developmental and environmental stimuli before and up until photosynthetic competence has been established. However, in gun1 seedlings these responses are not usually completely abolished, but they are diminished or delayed, suggesting that GUN1 may serve to optimise the timing of a crucial developmental event in seedlings.
GUN1 may Optimize the Switch to Photoautotrophic Growth
In the developing Arabidopsis seedlings, lipid reserves in the form of triacylglycerol (TAG) are mobilized in the form of sucrose; this requires the bypass of the decarboxylative reactions of the tricarboxylic acid (TCA) cycle and is achieved by post-transcriptional regulation of NAD+-isocitrate dehydrogenase.23 Carbon flow through the full TCA cycle is delayed until the seedling is photosynthetically competent and no longer dependent on stored reserves. In the absence of exogenous sugars, the switch from autotrophic to phototrophic growth occurs in 2–3-day-old seedlings,23 but in the presence of sucrose, lipid mobilization is retarded24 and metabolic switching may be delayed. However, the effects of sucrose on lipid mobilization are not seen if sucrose is applied after 3 days.24 Several of the components of GUN1 signaling appear to affect target photosynthesis-related nuclear genes only at an early stage of seedling development. Inhibitors of plastid translation, such as lincomycin and chloramphenicol, are effective in regulating photosynthesis genes only if applied to seedlings younger than 3 days old,25,26 and responsiveness to sucrose changes in older seedlings (see Fig. 1).
These changes in responsiveness appear to coincide with attainment of photosynthetic competence. GUN1 may function in the timing of this event, possibly influenced by inputs from sugar sensing, which may be able to distinguish between sugars generated from lipid mobilization and those produced by photosynthesis. GUN1 also appears to orchestrate seedling development to coincide with the transition to phototrophic growth; this necessitates cotyledon opening to maximise photosynthesis and anthocyanin accumulation to protect new and developing chloroplasts.
Abbreviations
- ABA
abscisic acid
- CHI
gene encoding chalcone isomerase
- CHS
gene encoding chalcone synthase
- F3H
gene encoding flavanone 3-hydroxylase
- GUN1
GENOMES UNCOUPLED1
- LHCB1
gene encoding light-harvesting chlorophyll protein associated with photosystem II
- PAL1
gene encoding phenylalanine ammonia lyase
- RBCS
gene encoding the small subunit of Rubisco
- RT-PCR
reverse transcriptase-polymerase chain reaction
References
- 1.Cottage A, Mott EK, Kempster JA, Gray JC. The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot. 2010;61:3773–3786. doi: 10.1093/jxb/erq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- 4.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 5.Cottage AJ, Mott EK, Wang JH, Sullivan JA, MacLean D, Tran L, et al. GUN1 (GENOMES UNCOUPLED1) encodes a pentatricopeptide repeat (PPR) protein involved in plastid protein synthesis-responsive retrograde signaling to the nucleus. In: Allen JF, Gantt E, Golbeck JH, Osmond B, editors. Photosynthesis. Energy from the Sun; 14th International Congress on Photosynthesis; Springer-Verlag, Berlin. 2008. pp. 1205–1211. [Google Scholar]
- 6.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D. Coordination of plastid and nuclear gene expression. Phil Trans R Soc Lond. 2003;358:135–145. doi: 10.1098/rstb.2002.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang ZW, Yuan S, Xu F, Yang H, Zhang NH, Cheng J, et al. The plastid hexokinase pHXK: a node of convergence for sugar and plastid signals in Arabidopsis. FEBS Lett. 2010;584:3573–3579. doi: 10.1016/j.febslet.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 9.McCormac AC, Terry MJ. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signaling pathways during deetiolation. Plant J. 2004;40:672–685. doi: 10.1111/j.1365-313X.2004.02243.x. [DOI] [PubMed] [Google Scholar]
- 10.Ruckle ME, DeMarco SM, Larkin RM. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell. 2007;19:3944–3960. doi: 10.1105/tpc.107.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruckle ME, Larkin RM. Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol. 2009;182:367–379. doi: 10.1111/j.1469-8137.2008.02729.x. [DOI] [PubMed] [Google Scholar]
- 12.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 13.Acevedo-Hernandez GJ, Leon P, Herrera-Estrella LR. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 2005;43:506–519. doi: 10.1111/j.1365-313X.2005.02468.x. [DOI] [PubMed] [Google Scholar]
- 14.Leon P, Sheen J. Sugar and hormone connections. Trends Plant Sci. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 15.Garciarrubio A, Legaria JP, Covarrubias AA. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta. 1997:203182–203187. doi: 10.1007/s004250050180. [DOI] [PubMed] [Google Scholar]
- 16.Pritchard SL, Charlton WL, Baker A, Graham IA. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002;31:639–647. doi: 10.1046/j.1365-313x.2002.01376.x. [DOI] [PubMed] [Google Scholar]
- 17.Laby RJ, Kincaid MS, Kim D, Gibson SI. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2001;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- 18.Pego JV, Kortstee AJ, Huijser C, Smeekens SC. Photosynthesis, sugars and the regulation of gene expression. J Exp Bot. 2000;51:407–416. doi: 10.1093/jexbot/51.suppl_1.407. [DOI] [PubMed] [Google Scholar]
- 19.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 20.Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs WB, Ausubel FM. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell. 1992;4:1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- 22.Dijkwel PP, Huijser C, Weisbeek PJ, Chua NH, Smeekens SC. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falk KL, Behal RH, Xiang C, Oliver DJ. Metabolic bypass of the tricarboxylic acid cycle during lipid mobilization in germinating oilseeds. Regulation of NAD+-dependent isocitrate dehydrogenase versus fumarase. Plant Physiol. 1998;117:473–481. doi: 10.1104/pp.117.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To JP, Reiter WD, Gibson SI. Mobilization of seed storage lipid by Arabidopsis seedlings is retarded in the presence of exogenous sugars. BMC Plant Biol. 2002;2:4. doi: 10.1186/1471-2229-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. Expression of nuclear genres as affected by treatments acting on plastids. Planta. 1986;168:482–492. doi: 10.1007/BF00392267. [DOI] [PubMed] [Google Scholar]
- 26.Gray JC, Sornarajah R, Zabron AA, Ducket CM, Khan MS. Chloroplast control of nuclear gene expression. In: Mathis P, editor. Photosynthesis, from Light to Biosphere. Kluwer: Dordrecht; 1995. pp. 543–550. [Google Scholar]