Abstract
Arabidopsis (Arabidopsis thaliana) suppressor of npr1-1, constitutive1 (snc1) contains a gain-of-function mutation in a Toll/interleukin receptor-nucleotide binding site-leucine-rich repeat Resistance (R) protein and it has been a useful tool for dissecting R-protein-mediated immunity. Here we report the identification and characterization of snc4-1D, a semidominant mutant with snc1-like phenotypes. snc4-1D constitutively expresses defense marker genes PR1, PR2, and PDF1.2, and displays enhanced pathogen resistance. Map-based cloning of SNC4 revealed that it encodes an atypical receptor-like kinase with two predicted extracellular glycerophosphoryl diester phosphodiesterase domains. The snc4-1D mutation changes an alanine to threonine in the predicted cytoplasmic kinase domain. Wild-type plants transformed with the mutant snc4-1D gene displayed similar phenotypes as snc4-1D, suggesting that the mutation is a gain-of-function mutation. Epistasis analysis showed that NON-RACE-SPECIFIC DISEASE RESISTANCE1 is required for the snc4-1D mutant phenotypes. In addition, the snc4-1D mutant phenotypes are partially suppressed by knocking out MAP KINASE SUBSTRATE1, a positive defense regulator associated with MAP KINASE4. Furthermore, both the morphology and constitutive pathogen resistance of snc4-1D are partially suppressed by blocking jasmonic acid synthesis, suggesting that jasmonic acid plays an important role in snc4-1D-mediated resistance. Identification of snc4-1D provides us a unique genetic system for analyzing the signal transduction pathways downstream of receptor-like kinases.
Receptor-like kinases (RLKs) are a large group of kinases with a variable extracellular domain and a cytoplasmic kinase domain linked by a single transmembrane motif. RLKs have been shown to play diverse roles in regulating plant innate immunity as well as growth and development (Morillo and Tax, 2006). The extracellular domains of RLKs are believed to bind directly to ligands to perceive extracellular signals, whereas the cytoplasmic kinase domains transduce these signals into the cell. There are over 600 RLKs (Shiu and Bleecker, 2001) in Arabidopsis (Arabidopsis thaliana). The biological functions of most RLKs are unknown.
Several RLKs have been identified to be receptors of microbe-associated molecular patterns (MAMPs). FLS2 and EFR are two well-characterized RLKs with extracellular Leu-rich repeats (LRRs) that recognize bacterial flagellin and translation elongation factor EF-Tu, respectively (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006). BAK1 is also an RLK with extracellular LRRs. BAK1 seems to function as an adaptor protein for multiple RLKs including BRI1, FLS2, and BIR1 (Li et al., 2002; Nam and Li, 2002; Chinchilla et al., 2007; Heese et al., 2007; Gao et al., 2009). Interestingly, knocking out BIR1 activates cell death and defense responses mediated by another RLK, SOBIR1 (Gao et al., 2009). Recently, the rice (Oryza sativa) RLK Xa21 was also suggested to be a MAMP receptor. Xa21 recognizes a peptide derived from the secreted effector protein AvrXa21, which is conserved among different Xanthomonas species (Lee et al., 2009). Unlike FLS2, EFR, and Xa21, the putative receptor for chitin is an RLK with three extracellular LysM domains instead of LRRs that are required for the perception of chitin as well as resistance against bacterial pathogens (Miya et al., 2007; Wan et al., 2008; Gimenez-Ibanez et al., 2009).
Perception of MAMPs by receptors leads to the rapid activation of mitogen-activated protein (MAP) kinases including MAP KINASE3 (MPK3), MPK4, and MPK6 (Boller and Felix, 2009). MAP KINASE SUBSTRATE1 (MKS1) was identified as an MPK4-interacting protein that positively regulates defense responses (Andreasson et al., 2005). Silencing of MKS1 compromises basal resistance to Pseudomonas syringae pv tomato DC3000, whereas overexpression of MKS1 leads to enhanced pathogen resistance. Activation of MAMP receptors also induces a number of responses such as oxidative burst, callose deposition, and increased salicylic acid synthesis (Boller and Felix, 2009). Defense responses induced by different MAMPs seem similar, suggesting that they may share common signaling components. Identification of the signaling components downstream of RLK receptors remains a major task in understanding MAMP-triggered immunity.
Here we report the identification and characterization of suppressor of npr1-1, constitutive4-1D (snc4-1D), a gain-of-function mutant of an atypical RLK that is autoactivated by a mutation in its kinase domain. The snc4-1D mutant plants constitutively express defense maker genes PR1, PR2, and PDF1.2, and display enhanced resistance to Hyaloperonospora arabidopsidis (H. a.) Noco2. Epistasis analysis showed that snc4-1D-mediated defense responses are dependent on multiple factors including NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1), MKS1, and OPR3.
RESULTS
Identification and Characterization of snc4-1D
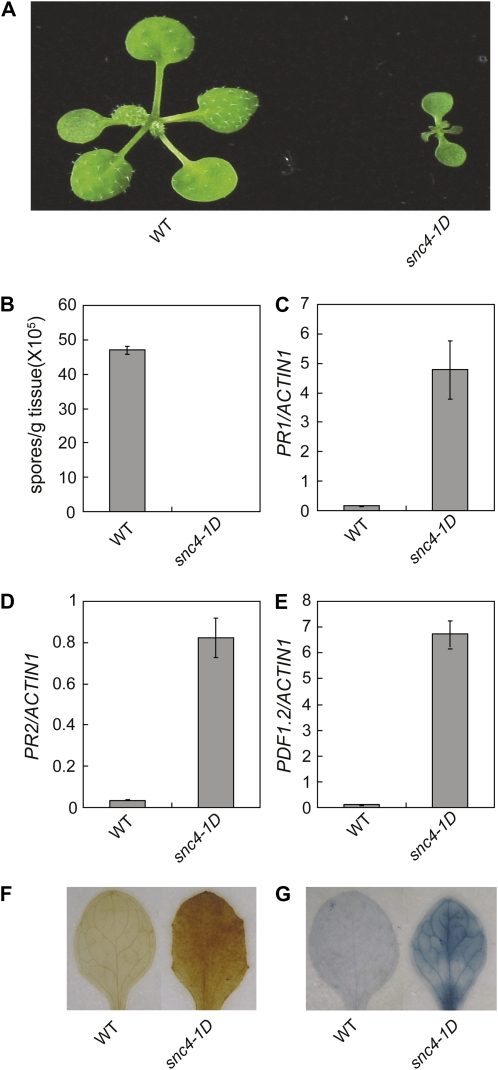
Our lab has been interested in identifying novel negative regulators of plant immunity (Gao et al., 2008). Loss-of-function mutations in these negative regulators would yield mutants with enhanced pathogen resistance and dwarfism that is usually associated with strong resistance phenotypes. Very rarely, gain-of-function mutations in positive regulators in the same pathways would yield mutants with similar phenotypes, such as in snc1 (Li et al., 2001). When we screened an ethyl methanesulfonate (EMS)-mutagenized population in the Columbia-0 (Col-0) background for mutants with enhanced resistance against the oomycete pathogen H. a. Noco2, one of the mutants, designated as snc4-1D, exhibited snc1-like dwarfism (Fig. 1A) and strong resistance to the virulent pathogen H. a. Noco2 (Fig. 1B). Plants heterozygous for snc4-1D have intermediate sizes, suggesting the mutant is semidominant and the mutation is most likely a gain-of-function mutation in a positive regulator of defense. Real-time reverse transcription (RT)-PCR analysis showed snc4-1D constitutively expresses defense marker genes PR1 (Fig. 1C), PR2 (Fig. 1D), and PDF1.2 (Fig. 1E). These data suggest that defense responses are constitutively activated in snc4-1D.
Figure 1.
Characterization of the snc4-1D mutant. A, Morphology of wild type (WT) and snc4-1D. Plants were grown on soil and photographed 3 weeks after planting. B, Growth of H. a. Noco2 on wild-type and snc4-1D mutant plants. Two-week-old seedlings were sprayed with H. a. Noco2 spores (5 × 104 spores/mL). Infection was scored 7 d later by counting the number of conidia spores with a hemocytometer. The values presented are averages of three replicates ± sds. C to E, Expression of PR1 (C), PR2 (D), and PDF1.2 (E) in wild-type and snc4-1D mutant seedlings. Total RNA was extracted from 2-week-old seedlings grown on one-half-strength Murashige and Skoog medium at 23°C. Relative levels of PR1, PR2, and PDF1.2 were determined by real-time PCR. Values were normalized to the expression of ACTIN1. Error bars represent sd from three measurements. F, DAB staining of true leaves of wild-type and mutant plants. G, Trypan blue staining of true leaves of wild-type and mutant plants.
Mutants with enhanced disease resistance often accumulate high levels of hydrogen peroxide (H2O2) and exhibit spontaneous cell death. To test whether snc4-1D accumulates H2O2, 3, 3′-diaminobenzidine (DAB) staining was performed on the snc4-1D mutant seedlings. Strong staining was observed in snc4-1D mutant seedlings (Fig. 1F), indicating that snc4-1D accumulated high levels of H2O2 compared to wild type. In addition, microscopic cell death was also observed in the snc4-1D mutant from trypan blue staining (Fig. 1G).
Map-Based Cloning of snc4-1D
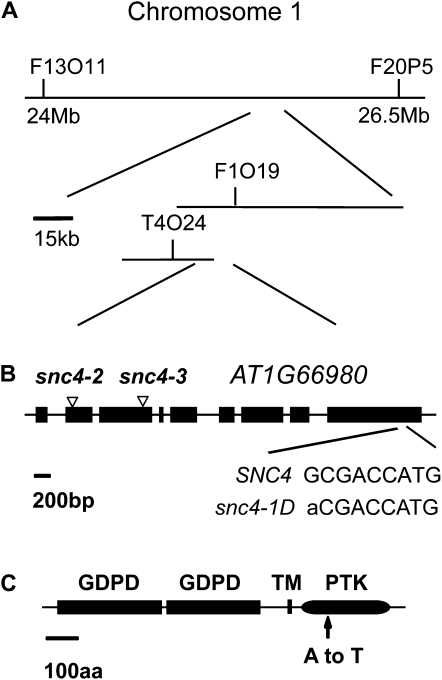
To map the snc4-1D mutation, snc4-1D was crossed with Landsberg erecta (Ler). Plants with snc4-1D morphology were selected from the F2 progeny and used for crude mapping. The mutation was initially mapped to a region between F13O11 and F20P5 on chromosome 1 (Fig. 2A). Further analysis of approximately 1,100 F2 plants flanked the mutation to a 35-kb region between marker T4O24 and F1O19. The coding sequence of genes in the region were amplified by PCR from the genomic DNA of snc4-1D and sequenced. A single G to A mutation was found in one of the sequenced genes, At1g66980 (Fig. 2B). Previously it was reported that a knockout mutant of At1g66980 displayed wild-type morphology (Hayashi et al., 2008), suggesting that the mutant phenotypes observed in snc4-1D is not likely caused by loss of function of the protein encoded by At1g66980.
Figure 2.
Map-based cloning of snc4-1D. A, Map of the snc4-1 locus. B, Exon/intron structure of SNC4. The coding regions are indicated with boxes. snc4-2, Salk_122292; snc4-3, WiscDsLox444C5. C, Predicted protein structure of SNC4. TM, Transmembrane motif; PTK, protein Tyr kinase. The locations of the mutations in SNC4 are as indicated. aa, Amino acids.
At1g66980 encodes a predicted RLK (Fig. 2C). The extracellular region of this protein contains two putative glycerophosphoryl diester phosphodiesterase (GDPD) domains. The protein also has a predicted N-terminal signal peptide and a single transmembrane domain between the cytoplasmic kinase domain and the extracellular GDPD domains. Originally identified in Escherichia coli, GDPD catalyzes the hydrolysis of a glycerophosphodiester to an alcohol and glycerol 3-P (Larson et al., 1983). GDPD activity was also detected in cell wall fractions of carrot (Daucus carota; Van Der Rest et al., 2004). Blast analysis failed to identify any other RLK with extracellular GDPD domains in Arabidopsis and other plants, suggesting that At1g66980 probably evolved recently through combining its GDPD and kinase domains. To confirm that the GDPD and kinase domains are encoded in the same gene, the size of the SNC4 cDNA is determined by RT-PCR (Supplemental Fig. S1A). In addition, western-blot analysis showed that the SNC4-GFP fusion protein expressed in transgenic plants has a mass of about 150 kD, close to the predicted size of the fusion protein (Supplemental Fig. S1B).
The SNC4-GFP fusion protein localized to the plasma membrane (Supplemental Fig. S2), suggesting that SNC4 is a plasma membrane protein as predicted. The mutation in snc4-1D changes the Ala 850 to Thr in the kinase domain of the protein, which does not change the localization of the protein (Supplemental Fig. S2).
Complementation
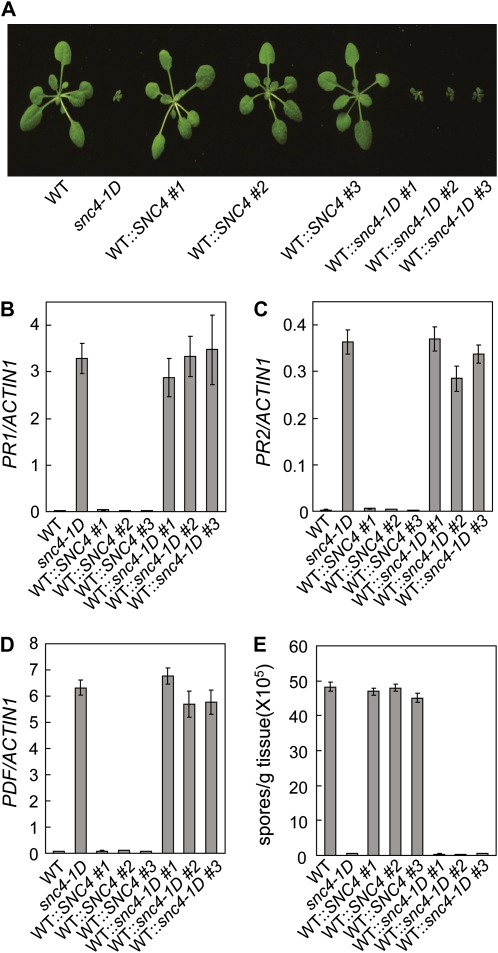
Since T-DNA knockout mutants of At1g66980 did not display snc4-1D phenotypes, we deduced that the Ala-850 to Thr mutation is most likely a gain-of-function mutation in At1g66980. If that is the case, transforming the mutant version of At1g66980 in wild-type background would yield plants with snc4-1D phenotypes. When we transformed constructs expressing the wild-type or mutant protein of At1g66980 under the control of its native promoter into wild-type plants, all transgenic lines obtained for the wild-type SNC4 construct displayed wild-type morphology. In contrast, about half of the transgenic lines with the mutant snc4-1D construct exhibit snc4-1D-like morphology (Fig. 3A). Real-time PCR analysis of defense marker genes PR1, PR2, and PDF1.2 showed that these marker genes were constitutively expressed in the transgenic plants carrying the mutant snc4-1D gene but not in the transgenic plants expressing the wild-type SNC4 gene (Fig. 3, B–D). In addition, transgenic plants expressing the mutant At1g66980 displayed enhanced resistance to H. a. Noco2 (Fig. 3E). Progeny from a representative line expressing the wild-type or mutant gene were further analyzed for the accumulation of H2O2 and cell death. As shown in Supplemental Figure S3, strong DAB and trypan blue staining was observed in the transgenic line carrying the mutant snc4-1D transgene but not in the line with the wild-type SNC4 transgene. Thus, all the snc4-1D mutant phenotypes can be recaptured by expressing the At1g66980 mutant gene in wild-type plants, indicating that SNC4 is At1g66980 and the mutation in snc4-1D is a gain-of-function mutation that activates downstream defense responses.
Figure 3.
Complementation analysis of snc4-1D. A, Morphology of wild-type (WT), snc4-1D, WT::SNC4 (wild type transformed with a SNC4 genomic clone), and WT::snc4-1D (wild type transformed with a snc4-1D genomic clone) transgenic lines. All plants were grown on soil and photographed when they were 3 weeks old. B to D, Expression of PR1 (B), PR2 (C), and PDF1.2 (D) in wild-type, snc4-1D, WT::SNC4, and WT::snc4-1D transgenic lines. Values were normalized to the expression of ACTIN1. Error bars represent sd from averages of three measurements. E, Growth of H. a. Noco2 on wild-type, snc4-1D, WT::SNC4, and WT::snc4-1D transgenic lines. Infection was performed and scored as in Figure 1B. The values presented are averages of three replicates ± sd. [See online article for color version of this figure.]
Analysis of Intragenic Suppressors of snc4-1D
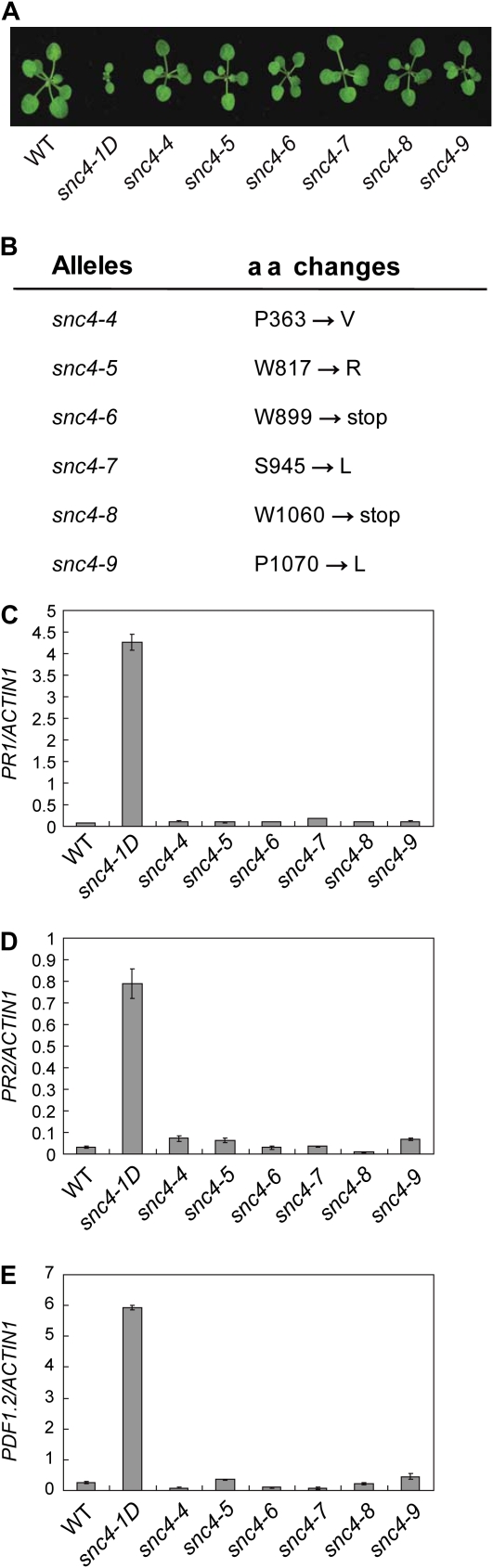
To identify mutants that suppress the snc4-1D mutant phenotypes, we mutagenized snc4-1D seeds with EMS and screened for mutants with wild-type morphology in the M2 population. Twenty-one snc4-1D suppressor mutants were obtained from the screen. Because snc4-1D seems to be a gain-of-function mutation, we hypothesized that some of the mutants recovered may be intragenic suppressors of snc4-1D. Sequence analysis of the SNC4 locus identified six mutants containing mutations in snc4-1D (Fig. 4A). Among these mutations, one is a missense mutation in one of the GDPD domains, two are missense mutations in the kinase domain, and the rest are nonsense mutations that cause early truncations of the protein (Fig. 4B). Real-time RT-PCR analysis showed that suppression of the snc4-1D mutant morphology correlates with reduced expression of PR1, PR2, and PDF1.2 (Fig. 4, C–E). These data further confirm that At1g66980 is SNC4 and snc4-1D is a gain-of-function mutation.
Figure 4.
Characterization of the intragenic suppressors of snc4-1D. A, Morphology of wild-type (WT), snc4-1D, and the intragenic suppressor mutant plants. Plants were grown on soil at 23°C and photographed 3 weeks after planting. B, Mutations identified in the intragenic suppressor mutant alleles. C to E, Expression of PR1 (C), PR2 (D), and PDF1.2 (E) in wild type, snc4-1D, and the suppressor mutants of snc4-1D. Values were normalized to the expression of ACTIN1. Error bars represent sd from averages of three measurements. aa, Amino acid. [See online article for color version of this figure.]
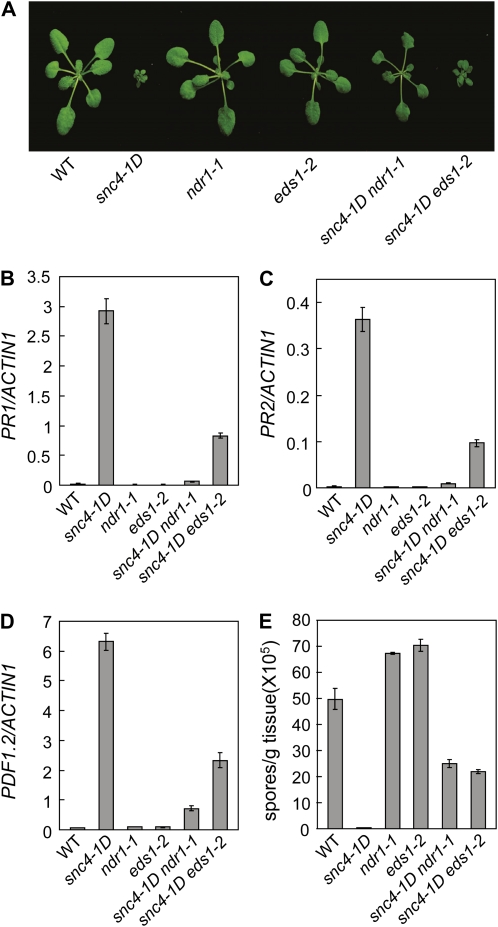
NDR1 Is Required for the Mutant Phenotypes of snc4-1D
Arabidopsis NDR1 is generally required for resistance mediated by coiled coil-nucleotide binding site (NB)-LRR Resistance (R) proteins, whereas EDS1 is required for resistance mediated by many Toll/interleukin receptor-NB-LRR R proteins (Century et al., 1995; Parker et al., 1996; Aarts et al., 1998). To test whether snc4-1D activates NDR1 or EDS1-dependent signaling pathways, snc4-1D was crossed with ndr1-1 or eds1-2 (backcrossed into Col background). As shown in Figure 5A, the snc4-1D mutant morphology was largely suppressed by ndr1-1 but not by eds1-2. Analysis of PR1 and PR2 expression in the double mutants showed that the constitutive expression of both PR1 and PR2 in snc4-1D was largely blocked by ndr1-1. In contrast, PR1 and PR2 expression were only partially affected by the eds1-2 mutation (Fig. 5, B and C). The elevated PDF1.2 expression in snc4-1D is partially suppressed by ndr1-1 and to a lesser extent by eds1-2 (Fig. 5D). We further tested whether resistance to H. a. Noco2 in snc4-1D is affected in the double mutants. As shown in Figure 5E, resistance to H. a. Noco2 was reduced in both snc4-1D ndr1-1 and snc4-1D eds1-2.
Figure 5.
Analysis of the snc4-1D ndr1-1 and snc4-1D eds1-2 double mutants. A, Morphology of 3-week-old plants of the indicated genotypes. B to D, Expression of PR1 (B), PR2 (C), and PDF1.2 (D) in the indicated genotypes. Values were normalized to the expression of ACTIN1. Error bars represent sd from averages of three measurements. E, Growth of H. a. Noco2 on the indicated genotypes. Infection was performed and scored as in Figure 1B. The values presented are averages of three replicates ± sd. [See online article for color version of this figure.]
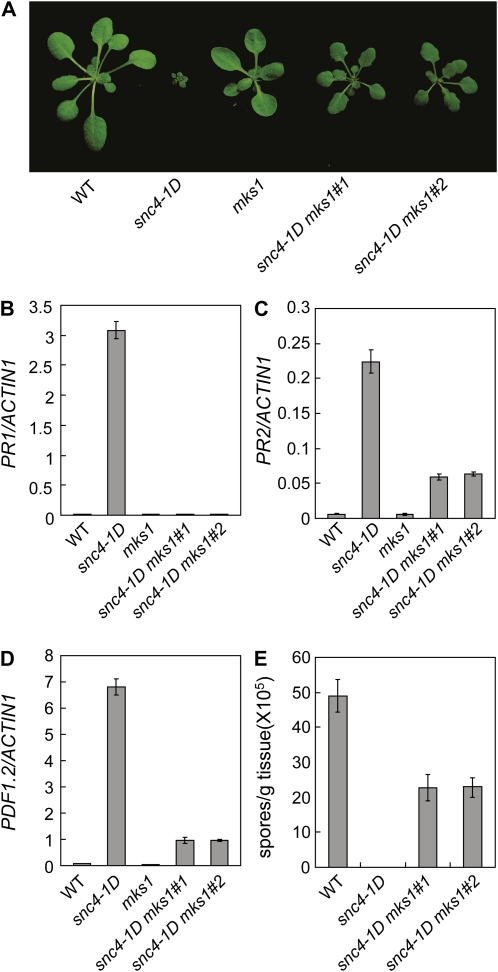
MKS1 Is Required for snc4-1D-Mediated Resistance Responses
RLKs that serve as MAMP receptors, such as FLS2 and EFR, signal through MAP kinase cascades (Boller and Felix, 2009). If snc4-1D signals through a similar cascade, one would expect that mutation in a positive regulator of the MAP kinase pathway would block snc4-1D-mediated immunity. Arabidopsis MKS1 is a positive regulator of defense responses that interacts with MPK4 (Andreasson et al., 2005). When we made the snc4-1D mks1-1 double mutant, the morphological phenotypes of snc4-1D were largely suppressed by mks1-1 (Fig. 6A). Real-time RT-PCR analysis showed that the elevated expression of PR1 in the snc4-1D single mutant was blocked in the snc4-1D mks1-1 double mutant plants (Fig. 6B). In contrast, the expression of PR2 and PDF1.2 expression in snc4-1D is only modestly affected by mks1-1 (Fig. 6, C and D). Analysis of resistance to H. a. Noco2 showed that the enhanced resistance observed in snc4-1D was largely suppressed by mks1-1 (Fig. 6E).
Figure 6.
Analysis of snc4-1D mks1-1 double mutant. A, Morphology of 3-week-old wild type (WT), mks1-1, snc4-1D, and two representative lines for the snc4-1D mks1-1 double mutant. B to D, Expression of PR1 (B), PR2 (C), and PDF1.2 (D) in wild-type, mks1-1, snc4-1D, and snc4-1D mks1-1 seedlings. Values were normalized to the expression of ACTIN1. The values presented are averages of three replicates ± sd. E, Growth of H. a. Noco2 on wild-type, snc4-1D, and snc4-1D mks1-1 plants. Infection was performed and scored as in Figure 1B. The values presented are averages of three replicates ± sd. [See online article for color version of this figure.]
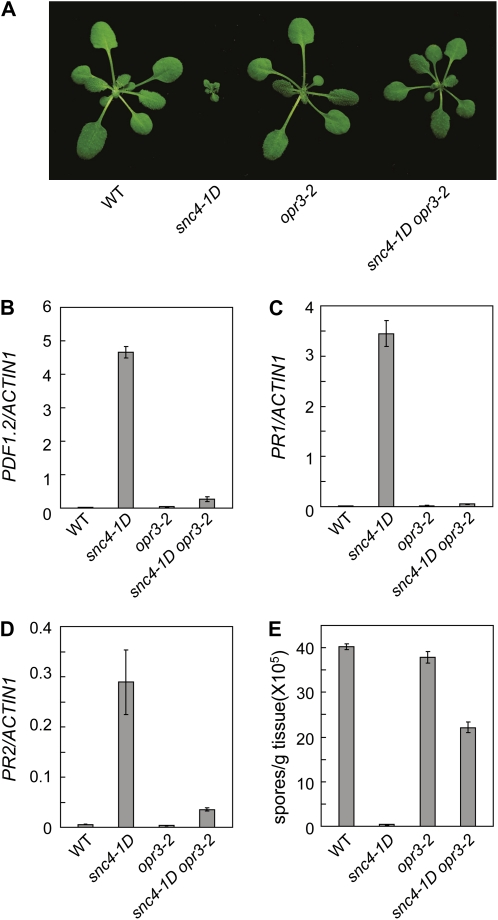
JA Is Required for snc4-1D-Mediated Resistance Responses
Since snc4-1D constitutively expresses PDF1.2, a defense marker gene of the jasmonic acid (JA) pathway, we tested whether JA is required for the snc4-1D mutant phenotypes by crossing snc4-1D into opr3-2, a loss-of-function mutant of 12-oxo-phytodienoic acid reductase 3 that is required for jasmonate biosynthesis (Stintzi and Browse, 2000). As shown in Figure 7A, the snc4-1D opr3-2 double mutant has intermediate size compared to the wild-type and snc4-1D mutant plants, suggesting that opr3-2 partially suppresses the snc4-1D mutant phenotype. PDF1.2 expression in snc4-1D was dramatically lowered by opr3-2 (Fig. 7B). Surprisingly, the expression of PR1 (Fig. 7C) and PR2 (Fig. 7D) in snc4-1D is also largely suppressed by opr3-2. When snc4-1D opr3-2 plants were challenged by H. a. Noco2, the pathogen grew much better on the double mutant than on snc4-1D, but at a lower level than on wild-type plants and opr3-2, suggesting that JA is required for pathogen resistance activated by snc4-1D (Fig. 7E).
Figure 7.
Characterization of snc4-1D opr3-2 double mutants. A, Morphology of wild-type (WT), snc4-1D, opr3-2, and snc4-1D opr3-2 mutant plants. Plants were grown on soil and photographed when they were 3 weeks old. B to D, Expression of PDF1.2 (B), PR1 (C), and PR2 (D) in wild-type, snc4-1D, opr3-2, and snc4-1D opr3-2 mutant seedlings. Error bars represent sd from averages of three measurements. Values were normalized to the expression of ACTIN1. E, Growth of H. a. Noco2 on wild-type, snc4-1D, opr3-2, and snc4-1D opr3-2 double mutant plants. Infection was performed and scored as in Figure 1B. The values presented are averages of three replicates ± sd. [See online article for color version of this figure.]
DISCUSSION
Previous studies have identified several gain-of-function mutations in NB-LRR R genes that lead to autoactivation of defense responses (Shirano et al., 2002; Zhang et al., 2003a; Noutoshi et al., 2005; Igari et al., 2008). One of the mutants, snc1, has been used to identify components involved in R-protein activation (Palma et al., 2005, 2007; Zhang et al., 2005; Zhang and Li, 2005; Goritschnig et al., 2007, 2008; Cheng et al., 2009). The snc4-1D mutation reported here results in activation of an atypical RLK with an unusual extracellular region consisting of two GDPD domains. SNC4 appears to be a unique RLK with no clear orthologs in other plant species. Its kinase domain is most similar to RLKs in the LRK10L-2 subfamily (Shiu and Bleecker, 2001). Functions of RLKs in this subfamily are unknown. The mutation in snc4-1D is located in an unconserved region within subdomain III of the kinase. No similar gain-of-function mutations in other RLKs have been previously identified to constitutively activate downstream signaling pathways. The mechanism on how the mutation in snc4-1D activates defense responses remains to be determined.
In Arabidopsis, there are six GDPD domain-containing proteins that are closely related to the extracellular domain of SNC4 (Supplemental Fig. S4). One of the proteins, SHV3, is involved in the cell wall organization (Hayashi et al., 2008). Sequence analysis showed that some key residues involved in binding to substrates and calcium ions in the E. coli GDPD are not conserved in SHV3 and related proteins (Hayashi et al., 2008). These residues are not conserved in SNC4 either. It suggests that SHV3, SNC4, and related proteins may not have the same enzymatic activity as the GDPD in E. coli.
The GDPD domain of SNC4 could be involved in perception of lipid-derived molecules from microbial pathogens or damage responses. Interestingly, one of the intragenic suppressors of snc4-1D (snc4-4) contains a mutation in the GDPD domain. Western-blot analysis showed that this mutation does not affect the stability of the protein (Supplemental Fig. S5), suggesting that the GDPD domain may be important for the function of SNC4. It is unclear how the snc4-4 mutation affects the function of the protein. Additional mutagenesis analysis of the GDPD domain of SNC4 will help us better understand its function.
To analyze downstream signaling pathways activated in snc4-1D, we carried out epistasis analysis between snc4-1D and several known mutants involved in pathogen resistance. One unexpected finding is that the mutant morphology as well as activation of defense gene expression and pathogen resistance in snc4-1D is suppressed by ndr1-1, a mutant previously known to be defective in resistance mediated by a number of coiled coil-NB-LRR R proteins (Century et al., 1995). In contrast, the eds1-2 mutation does not affect the mutant morphology and PR2 expression in snc4-1D. However, resistance to H. a. Noco2 and expression of PR1 are reduced in the snc4-1D eds1-2 double mutant. Since EDS1 is also required for basal resistance (Parker et al., 1996), reduced resistance to H. a. Noco2 could be caused by lowered basal resistance by eds1-2.
Another mutation that partially suppresses the snc4-1D mutant phenotypes is mks1. MKS1 has previously been shown to be a positive regulator of defense responses, where it is phosphorylated in response to bacterial pathogen infections (Andreasson et al., 2005). Our results suggest that MKS1 functions downstream of the RLK SNC4, suggesting that MAP kinase signaling could participate in snc4-1D-mediated immunity. It will be interesting to determine whether snc4-1D activates MKS1 through a MAP kinase cascade that functions downstream of the known MAMP receptors.
Epistasis analysis showed that JA-dependent resistance pathways are activated in snc4-1D. Blocking JA synthesis suppresses the morphological phenotypes as well as constitutive expression of PDF1.2, PR1, and PR2 in snc4-1D. While it is known that expression of PDF1.2 is regulated by JA, activation of PR1 and PR2 are often associated with salicylic acid-induced defense response. How JA modulates the expression of PR1 and PR2 in snc4-1D remains to be determined.
Based on our data, a working model is proposed (Supplemental Fig. S6). SNC4 probably encodes a pathogen-associated molecular pattern or damage-associated molecular pattern receptor candidate. Perception of the pathogen-associated molecular pattern or damage-associated molecular pattern signal leads to activation of SNC4 and downstream defense pathways. Since mutations in either NDR1 or OPR3 partially suppress the mutant phenotypes of snc4-1D and these mutations have different effects on the expression of defense marker genes, NDR1 and OPR3 may function in parallel downstream of SNC4. The stronger effect of ndr1-1 on PR1 and PR2 expression than mks1-1 suggests that NDR1 may function in earlier events of the signaling pathways comparing to MKS1. We tentatively placed NDR1 upstream of the MAP kinase cascade. Additional studies are required to identify the upstream signal of SNC4 and to better define the relationships between NDR1, OPR3, MKS1, and MAP kinase cascades activated in snc4-1D.
In conclusion, we have identified a unique gain-of-function mutant of an atypical RLK. Epistasis analysis showed that snc4-1D activates multiple downstream resistance pathways. Our results suggest that snc4-1D could be utilized as a powerful tool for genetic analysis of resistance pathways activated by RLKs. Further characterization and cloning of the snc4-1D extragenic suppressors may lead to better understanding of signal transduction downstream of RLKs.
MATERIALS AND METHODS
Plant Growth Conditions and Mutant Screens
All plants were grown under 16 h light at 23°C and 8 h dark at 20°C. To screen for mutants with snc1-like constitutive defense responses, wild-type Col-0 seeds were mutagenized with EMS and the M2 progeny were analyzed for enhanced resistance to H. a. Noco2. snc4-1D was one of mutants identified. To search for suppressors of snc4-1D, seeds of snc4-1D were obtained by growing the plants at 28°C and mutagenized with EMS. About 12,000 M2 plants were screened to identify mutants that can grow to maturity and set seeds at 22°C.
Mutant Characterization
Two-week-old seedlings grown on one-half-strength Murashige and Skoog medium were used for gene expression analysis, trypan blue, and DAB staining. For gene expression analysis, RNA was extracted using Takara RNAiso reagent (Takara) and RT was performed using the Takara M-MLV RTase cDNA synthesis kit. Real-time PCR was carried out with the SYBR Premix Ex (Takara). The primers used for amplification of Actin1, PR1, and PR2 were described previously (Zhang et al., 2003b). The primers used for amplification of PDF1.2 are PDF1.2-F (5′-atcacccttatcttcgctgc-3′) and PDF1.2-R (5′-tgctgggaagacatagttgc-3′). Trypan blue and DAB staining were carried out as previously described (Parker et al., 1996; Thordal-Christensen et al., 1997). Analysis of resistance to H. a. Noco2 was performed by spraying 2-week-old seedlings with H. a. Noco2 spores (5 × 104 spores/mL). For each genotype, seedlings grown in three different pots (15 seedlings per pot) were inoculated. Seven days post inoculation, plants from each pot were collected as one sample and total number of conidia spores for each sample was counted with a hemocytometer.
Map-Based Cloning of snc4-1D
Positional cloning of snc4-1D was performed according to procedures previously described (Zhang et al., 2007). The markers used to map snc4-1D were derived from insertion-deletion and single nucleotide polymorphisms identified from the genomic sequences of Col-0 and Ler ecotypes provided by Monsanto (Jander et al., 2002). The primer sequences for F13O11 are 5′-cacatcttcagacactgccac-3′ and 5′-gcttgaatctgtcgttgcttc-3′. Primers for F20P5 are 5′-agtgccttgttcatcgtctc-3′ and 5′-tggaactgggcgtcttgtcg-3′. These two markers are based on insertion-deletion polymorphisms. For marker T4O24 that detects single nucleotide polymorphism, primers 5′-ctttctaagctcagtaacagg-3′ and 5′-gaccgttatgcacggtaatgt-3′ were used to detect the presence of the Col-0 allele and primers 5′-ctttctaagctcagtaacagg-3′ and 5′-gaccgttatgcacggtaatgc-3′ were used to detect the presence of the Ler allele. For marker F1O19, DNA was amplified by PCR using primers 5′-cttctgttgtctcagccgtg-3′ and 5′-tttggagccggtatcaatgg-3′ and sequenced to detect the polymorphisms in Col-0 and Ler.
Plasmid Construction and Arabidopsis Transformation
For complementation analysis, a 6.6-kb genomic DNA fragment containing SNC4 or snc4-1D was amplified by PCR using primers 5′-acgcacgcgtcgacgcttgacacaagaggtacag-3′ and 5′-cggggatccgagctcgaatctttagtgggccttgg-3′ and cloned into the binary vector pGREEN229 (Hellens et al., 2000). To confirm that the phenotypes observed in snc4-1D were caused by the mutation in snc4-1D, pGREEN229-SNC4 and pGREEN-snc4-1D were transformed into Agrobacterium and subsequently into the wild-type plants by floral dipping (Clough and Bent, 1998). Transgenic plants were identified by Basta selection and confirmed by PCR.
Localization Assay in Protoplast
For localization analysis, the full-length cDNA fragments of SNC4 and snc4-1D were amplified by PCR using primers SNC4-GF (5′-cggggatccgagctcatgaattcccagcaaagtac-3′) and SNC4-GR (5′-acgcacgcgtcgactccttctgtgtagacagagac-3′) and cloned into a modified pCAMBIA1300 vector containing an in-frame GFP tag at the C terminus. The pCAMBIA1300-GFP-SNC4 and pCAMBIA1300-GFP-snc4-1D were transformed into protoplast as described (Sheen, 2001) and examined by confocal microscopy.
Creating the Double Mutants
Since snc4-1D plants cannot grow to maturity, the snc4-1D/SNC4 heterozygous plants were crossed with homozygous opr3-2, eds1-2, ndr1-1, and mks1-1 (GT_5_108403) to generate various double mutants. F1 plants heterozygous for snc4-1D were identified by PCR. All the double mutant plants were identified in the F2 progeny by PCR. eds1-2 and ndr1-1 were described previously (Century et al., 1995; Parker et al., 1996). The eds1-2 line used was introgressed into Col by multiple backcrossing and was kindly provided by Dr. Jane Parker. opr3-2 was kindly provided by Dr. Jianmin Zhou, and it contains a mutation in OPR3 that creates an early stop codon in the cDNA. The mks1-1 mutant is in Ler background and it is closely linked to the RPP5 locus. The snc4-1D mks1-1 double mutants were therefore identified for the presence of the RPP5 locus (from Ler) and two independent lines with RPP5 absent were used for H. a. Noco2 infection experiments.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number HM461758.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the size of SNC4 cDNA and SNC4-GFP fusion protein.
Supplemental Figure S2. Localization of SNC4-GFP and snc4-1D-GFP fusion protein.
Supplemental Figure S3. Trypan blue staining (top section) and DAB staining (bottom section) of true leaves of wild type, snc4-1D, WT::SNC4 line 1, and WT::snc4-1D line 1.
Supplemental Figure S4. Sequence alignment of the extracellular domain of SNC4 and six GDPD domain-containing proteins in Arabidopsis.
Supplemental Figure S5. Western-blot analysis of SNC4 protein levels in wild type (WT), snc4-1D, and snc4-4 using a polyclonal anti-SNC4 antibody generated against a fragment of the SNC4 kinase domain.
Supplemental Figure S6. A working model for SNC4-mediated resistance.
Supplementary Material
Acknowledgments
We thank Dr. Jianmin Zhou for the opr3-2 mutant and Dr. Jane Parker for eds1-2 in Col-0. We also thank Fang Cheng for help with the mapping of snc4-1D and Patrick Gannon for careful reading of the manuscript.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, Wirthmueller L, Despres C, Parker JE, Zhang Y, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Goritschnig S, Weihmann T, Zhang Y, Fobert P, McCourt P, Li X. (2008) A novel role for protein farnesylation in plant innate immunity. Plant Physiol 148: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritschnig S, Zhang Y, Li X. (2007) The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J 49: 540–551 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ishii T, Matsunaga T, Tominaga R, Kuromori T, Wada T, Shinozaki K, Hirayama T. (2008) The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol 49: 1522–1535 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Igari K, Endo S, Hibara K, Aida M, Sakakibara H, Kawasaki T, Tasaka M. (2008) Constitutive activation of a CC-NB-LRR protein alters morphogenesis through the cytokinin pathway in Arabidopsis. Plant J 55: 14–27 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TJ, Ehrmann M, Boos W. (1983) Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem 258: 5428–5432 [PubMed] [Google Scholar]
- Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC. (2009) A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X. (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SA, Tax FE. (2006) Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol 9: 460–469 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K. (2005) A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Palma K, Zhang Y, Li X. (2005) An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol 15: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W, Zhang Y, Li X. (2007) Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev 21: 1484–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shirano Y, Kachroo P, Shah J, Klessig DF. (2002) A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J. (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Van Der Rest B, Rolland N, Boisson AM, Ferro M, Bligny R, Douce R. (2004) Identification and characterization of plant glycerophosphodiester phosphodiesterase. Biochem J 379: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng YT, Bi D, Palma K, Li X. (2005) MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr Biol 15: 1936–1942 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Glazebrook J, Li X. (2007) Identification of components in disease-resistance signaling in Arabidopsis by map-based cloning. Methods Mol Biol 354: 69–78 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X. (2003a) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X. (2005) A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X. (2003b) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.