Abstract
Proteins bearing a SET domain have been shown to methylate lysine residues in histones and contribute to chromatin architecture. Methylation of histone H3 at lysine 9 (H3K9) has emerged as an important player in the formation of heterochromatin, chromatin condensation, and transcriptional repression. Here, we have characterized a previously undescribed member of the histone H3K9 methyltransferase family named CLLD8 (or SETDB2 or KMT1F). This protein contributes to the trimethylation of both interspersed repetitive elements and centromere-associated repeats and participates in the recruitment of heterochromatin protein 1 to centromeres. Consistently, depletion in CLLD8/KMT1F coincides with a loss of CENP proteins and delayed mitosis, suggesting that this protein participates in chromosome condensation and segregation. Altogether, our results provide evidence that CLLD8/KMT1F is recruited to heterochromatin regions and contributes in vivo to the deposition of trimethyl marks in concert with SUV39H1/KMT1A.
Keywords: Cell/Mitosis, Chromatin, Chromatin/ Modification, Chromatin/Structure, Chromatin/Epigenetics, Chromosomes/Chromatin Remodeling
Introduction
Deciphering the pathways that regulate chromatin architecture has been a major goal in the dissection of the histone code predicting that different modifications of specific amino acids in the tails of the core histones (H2A, H2B, H3, and H4) are translated into distinct information (1). These tails that protrude from the nucleosome octamer are subject to various covalent post-translational modifications such as acetylation, phosphorylation, ubiquitination, ADP-ribosylation, and methylation catalyzed by specific enzymes. These signatures provide epigenetically heritable information that regulates transcription, replication, repair, and chromosome condensation through cell divisions. Among them, methylation is now recognized as a major change associated with both repression and activation of transcription (2, 3). Of the nine lysine positions that can be modified in the histone H3 amino terminus, five can display methylation. Methylation of H3K9 and H3K27 are marks of repressive chromatin, whereas methylation of H3K4, H3K36, and H3K79 are found at transcriptionally active sites. The H3K9 and H3K27 residues can be mono, di-, or trimethylated, thereby extending the complexity of the epigenetic code associated with histone modifications. In general, H3K9 trimethylation is linked to chromosome condensation and is important for binding of heterochromatin protein 1 (HP1)6 to discrete regions, thereby regulating gene expression and heterochromatin spreading through self-association properties and association with the SUV39H histone methyltransferase (HMTase). The translation of this code can be affected by another adjacent modification on the same histone tail and the level of cytosine methylation of the underlying DNA. Thus, the combinatory use of different marks participates in the topology of the chromatin fiber and gives enormous potential for the variability of the biological response. In addition, the primary function of a particular modification within a single nucleosome might be modulated by another adjacent residue. For example, a cooperation between H3 and H4 methylation participates in the epigenetic regulation of many processes that include heterochromatin formation, X inactivation, imprinting, and transcriptional regulation. On the contrary, the phosphorylation of serine 10 alters the binding of HP1 to methylated H3K9, thereby regulating the dissociation of HP1 from chromatin during the M phase (4, 5).
Proteins implicated in either the establishment or the translation of this code have been the focus of intensive research. A common feature for HMTases is the presence of an evolutionarily conserved SET domain initially identified in the Drosophila suppressor of position effect variegation (Su(var)3–9), the polycomb group enhancer of zeste, and the trithorax protein. The homology between this SET domain and a plant enzyme, Rubisco large subunit lysine methyltransferase, further led to the characterization of a large family of proteins containing this motif. This family is divided into different subgroups according to amino acid identity within their SET domain (6, 7). In mammals, several HMTases able to methylate lysine 9 of histone H3 have been characterized. These enzymes, SUV39H1 and 2 (KMT1A and KMT1B), G9a (KMT1C), and ESET/SETDB1 (KMT1E) possess a SET domain and two adjacent cysteine-rich regions (pre-SET and post-SET) (7). In addition to these enzymes, a database search revealed that other proteins with SET motifs but devoid of one or both of the flanking regions also exist in eukaryotes (6). Only the SET domain is required for HMTase activity, and the function of the adjacent pre- and post-SET domains might be replaced by other motifs conferring specificity (8, 9).
Among these HMTases, KMT1F (or SETDB2 or CLLD8, named hereafter KMT1F), a candidate protein for B-cell chronic lymphocytic leukemia, has not been characterized so far. The corresponding gene, located on chromosome 13q14.3, spans 49 kb and consists of 15 exons. The coding region encodes a protein of 719 amino acids with a predicted molecular mass of 81.9 kDa (see Fig. 1A) (10). This putative protein contains a SET domain and the adjacent pre-SET cysteine-rich regions, together with a methyl-binding domain (MBD) also found in protein with affinity to methylated DNA. The SET domain is interrupted by a 218-amino acid insertion in the middle of the motif and presents 73% of similarity with the bifurcated ESET/SETDB1 domain (11). This evolutionarily conserved insertion suggests that the SET domain possesses functionally separable domains. The pre-SET, also known as the SAC domain (SET domain-associated cysteine-rich domain), might be involved in DNA binding (12), whereas the post-SET might be implicated in substrate binding and catalysis.
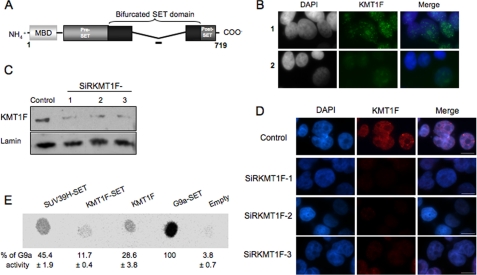
FIGURE 1.
Characterization of the KMT1F protein. A, structure of the KMT1F protein and its predicted protein domains. The KMT1F SET domain contains a central insertion and is described as a “bifurcated SET domain.” The peptide used for the production of a rabbit anti-KMT1F polyclonal antibody within a nonconserved part of the bifurcated SET domain is indicated as a bold line. B, indirect immunofluorescence in 293T cells using anti-KMT1F antibody in the presence of a control peptide (row 1) or the KMT1F-derived peptide (row 2) confirms specificity of the signal. C, the anti-KMT1F antibodies were further characterized by immunoblot on nuclear extracts of 293T cells transiently transfected with control siRNAs (Control) or three distinct small interfering RNA against KMT1F (SiRKMT1F-1, -2, or -3). The level of endogenous KMT1F protein is significantly decreased after transfection of the different siRNAs. D, transfection of the KMT1F siRNAs decreases the intensity of KMT1F staining compared with a control siRNA. Control 1 contained interphase nuclei. E, we investigated the catalytic activity of the KMT1F protein. Therefore, SET domains of SUV39H1, KMT1F and G9a, and full-length KMT1F fused to a FLAG immunoprecipitated from 293T cells were incubated in the presence of S-adenosyl-[methyl-14C]-l-methionine as a donor of methyl groups and a lysine 9-dimethylated H3 peptide (NH2-ARTKQTARK(2Me)STGGKAPRKQLC-COOH). Reaction mixtures were spotted onto an Immobilon membrane and exposed to a PhosphorImager screen, and the spots were quantified using the ImageQuant software. The percentage of signal was determined by comparing the intensity of each spot to G9A. The results from three different experiments ±S.D. are indicated. KMT1F can transfer methyl groups to lysine 9-dimethylated H3 peptide. No significant activity was observed with unmodified or H3-K9me H3 peptides.
Alterations of the SET proteins are associated with a wide range of pathologies. For instance, the mammalian homolog of trx, mll/HRX, is frequently translocated in leukemia, resulting in a fusion protein that lacks the carboxyl-terminal SET domain (13). NSD1 has been associated with Wolf-Hirschhorn syndrome (14), whereas expression of ESET/SETDB1 is altered in Huntington disease (15). Interestingly, the 13q14.3 locus encoding CLLD8/KMT1F is frequently deleted in B-cell chronic lymphocyte leukemia, suggesting a role for KMT1F in pathology.
In this report, we investigated the biological function of KMT1F in the methylation of histone H3K9 residues in human cells. We show that this member of the histone methyltransferase family participates in the distribution of H3K9 trimethylation (H3K9me3) marks and contributes to chromosome segregation during mitosis, suggesting that this previously uncharacterized protein might regulate the level of H3K9 trimethylation and genome stability in human cells.
EXPERIMENTAL PROCEDURES
Cell Culture
The conditions of the culture and transfection of the human embryonic kidney (293T) and epithelial cervix adenocarcinoma HeLa cell lines are detailed in the supplemental information.
Immunoblotting
Anti-KMT1F antibody was produced in collaboration with Abcam®. The peptide sequence DITKYREETPPRSR, corresponding to a short region of the bifurcated SET domain, was selected after a BLAST comparison of SET motifs in mouse, human, and rabbit for its immunogenicity potential. For detection of KMT1F and Lamins A/C, nuclear and cytoplasmic fractions (Nuclear Extract KitTM, Active Motif) were separated by SDS-PAGE, transferred to nitrocellulose membranes, blocked, and incubated with anti-KMT1F (1/200) or anti-Lamins A/C (1/1000, H110; Santa Cruz), probed with horseradish peroxidase-conjugated anti-rabbit and anti-mouse secondary antibodies (Sigma), detected with the ECL chemiluminescent kit, and visualized with Fujifilm LAS-3000 (Multi Gauge; Fujifilm, Tokyo, Japan).
Immunofluorescence
After 2–6 days, the cells grown on coverslips were washed and fixed with 4% paraformaldehyde for 10 min (16, 17), permeabilized, and incubated with rabbit anti-KMT1F (1/50), rabbit anti-monomethyl H3K9 (1/50), anti-dimethyl-H3K9 (1/100), and rabbit anti-trimethyl-H3K9 (1/600; kindly provided by T. Jenuwein) for 1 h at 37 °C. A secondary antibody directed against the rabbit primary antibody conjugated with Alexa Fluor 488 or 555 (Molecular Probes) were used for detection. For antigen competition, anti-KMT1F was preincubated for 2 h either with a KMT1F or a control peptide at a final concentration of 50 μg/ml. The nuclei were counterstained with DAPI (Sigma) for fluorescence analysis and TOTO-3 iodide (Molecular Probes) at 1–2 μm for confocal imaging, mounted in Vectashield (Vector Laboratories), and observed on an Axioplan-2 Imaging Zeiss fluorescent microscope (Carl Zeiss, Le Pecq, France) equipped with Apochromat 63× and 100×/1.4 oil differential interference contrast objectives, Coolsnap HQ CCD camera, and Metamorph 7.5 software (Molecular Devices) for conventional microscopy and on a confocal Axioplan 2 equipped with Neofluar 100×/1.4 oil Ph3 objective using the LSM 510 v 3.2 software for confocal acquisitions. Z stacks were processed through Imaris 4.2 software (Bitplane AG, Zurich, Switzerland) for three-dimensional analyses. For CENP A and CENP B detection, 48 h after transfection, the cells were pre-extracted in 0.25% Triton X-100 buffered in phosphate-buffered saline for 2 min and fixed for 10 min in 3.5% paraformaldehyde. Indirect immunofluorescence was performed as previously described using either mouse anti-CENP A (1/250; Abcam), rabbit anti-CENP B (1/100; Abcam), or mouse anti-CENP C (1/250; Abcam) for 2 h. Secondary anti-mouse and anti-rabbit antibodies were used for detection as previously described. The cells were counterstained with DAPI (Sigma). Imaging was performed using conventional microscopy. Stacks of 20 images every 0.2 μm were collected and analyzed in 30–50 mitosis cells. Student's t tests were performed using S-PLUS® 6.2 (Lucent Technologies).
Live Cell Four-dimensional Confocal Microscopy
HeLa cells were transfected by the calcium phosphate procedure with pBOS-H2B-GFP (BD PharMingen) 16 h before KMT1F siRNA (SiRKMT1F-3) transfection. Imaging was performed on a PerkinElmer Life Sciences UltraView RS Nipkow-Disk system attached to a Zeiss Axiovert 200M microscope equipped with a times 63/1.4NA objective. Stacks of 14 images every 1.5 μm were collected every 90 s with a cooled CCD Hamamatsu ORCA II ER camera, and maximum intensity Z projections were done for each time point using an ImageJ grouped Z projector plug-in.
Chromatin Immunoprecipitation and Quantitative PCR
In vivo protein DNA cross-linking was carried out as described (18). Antibodies specific for total H3 (Abcam), H3K9me3 (Upstate), or IgG (Sigma) were incubated with protein A-clarified chromatin overnight at 4 °C with gentle rocking. DNA samples were quantified using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Real-time PCR experiments were performed with a LightCycler and Stratagene systems and using either the Fast Start DNA Master SYBR Green I mix (Roche Diagnostics) or a QuantiTect SYBR Green PCR kit (Qiagen) according to the manufacturer's instructions. Fold enrichment of target sequence from antibody-bound chromatin DNA (immunoprecipitation (IP)) compared with input (Ref) was calculated using the equation, 2ΔCt(IP)−Ct(Ref). The sequence of the primers are available upon request.
RESULTS
KMT1F Is a Nuclear Protein Associated with DAPI-intense Regions
The classification of KMT1F as a member of the histone H3K9 HMTase family (6) prompted us to test its function on the methylation of histone H3 in human cells. For this purpose, we raised a polyclonal antibody by immunizing rabbits with a peptide corresponding to the nonconserved region of the bifurcated SET motif (Fig. 1A). Indirect immunofluorescence experiments performed with this antibody in 293T interphase cells reveal a diffuse nuclear signal with intense foci as well as larger and more diffuse ones (Fig. 1B) that overlap with DAPI-intense regions in interphase nuclei but are not associated with chromosomes at mitosis (Fig. 1D and supplemental Figs. S1A and S3B). The absence of intense foci when immunofluorescence was performed in the presence of the peptide used for the immunization confirms the specificity of the antibody (Fig. 1B) and the enrichment of KMT1F at nuclear foci. Furthermore, Western blot analysis revealed that the antibody recognizes a 84-kDa product corresponding to the expected size of the endogenous KMT1F protein (Fig. 1C, lane 1) and a transiently expressed flagged form of KMT1F (supplemental Fig. S1B).
To investigate the biological function of KMT1F, we designed three small interfering duplex RNAs targeting different parts of the KMT1F mRNA (supplemental Fig. S1C). The levels of KMT1F mRNA (Fig. S1D) and protein (Fig. 1C, rows 1–3 for cells transfected with SiRKMT1F and supplemental Fig. S1B) were significantly decreased after transient transfection of these siRNA in 293T cells. The siRNA at position 1851 (SiRKMT1F-3) gives a reduction of 75–80% in the level of KMT1F mRNA (supplemental Figs. S1D and S2A) and was selected for further analysis. Also, transfection of the different siRNAs decreases the signal corresponding to the endogenous KMT1F protein after immunofluorescent labeling (Fig. 1D), showing that KMT1F is a detectable nuclear protein that can be efficiently depleted to be further analyzed by different methods using the antibody that we developed.
Inhibition of KMT1F Alters the Distribution of Histone H3 Trimethylation
We next investigated the involvement of KMT1F in the distribution of H3K9 methylation. The 293T cells were transiently transfected with either a negative control siRNA or the SiRKMT1F-3, and the distribution of H3K9 methylation was analyzed by indirect immunofluorescence (Fig. 2, A and B) and Western blot (supplemental Fig. S2B). KMT1F depletion does not affect mono- and dimethylation of H3K9 residues as seen by indirect immunofluorescence at different stages of the cell cycle (Fig. 2A) or by Western blot (supplemental Fig. S2B). In cells transfected with control siRNA (Fig. 2B), H3K9me3 can be detected as a diffuse signal with a few intense foci in interphase cells (Fig. 2B) that overlap with DAPI-intense regions during mitosis. In cells transfected with KMT1F siRNA (Fig. 2B), H3K9me3 staining remains diffuse during interphase, whereas it is significantly reduced upon KMT1F depletion in metaphasic cells (Fig. 2B). This effect is reproducibly observed with each of the three KMT1F-specific siRNAs by Western blotting, ruling out an off target effect of the siRNA transfection. These results indicate that KMT1F depletion is not associated with global changes in the level of H3K9 methylation (mono- and di-) but rather specifically implicated in the trimethylation of histone H3K9 residues with a more pronounced effect for metaphase chromosomes. Confirming the prediction made from sequence analysis of SET proteins (6), in vitro experiments indicate that the full-length KMT1F or the SET domain immunoprecipitated from 293T cells can catalyze the trimethylation of purified substrate (Fig. 1E) and suggest that KMT1F/SETDB2/CLLD8 is a functional member of the HMTase family.
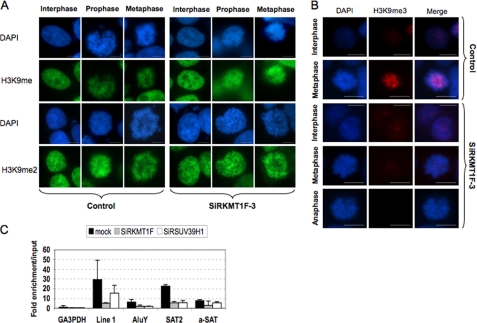
FIGURE 2.
H3K9 trimethylation is lost at pericentric regions upon KMT1F depletion. A and B, post-translational modifications of H3K9 were detected by indirect immunofluorescence with anti-monomethylated (H3K9me), anti-dimethylated (H3K9me2), or anti-trimethylated (H3K9me3) antibody (kindly provided by T. Jenuwein) in 293T cells transfected either with a control siRNA (Control) or a siRNA against KMT1F (SiRKMT1F-3) at different stages of the cell cycle. The cells were counterstained with DAPI, and the merged signals are shown in B. The scale bars represent 10 μm. C, the 293T cells were transiently transfected either with a control siRNA, a siRNA against KMT1F (SiRKMT1F), or SUV39H1 (SiRSUV39H). Chromatin immunoprecipitation was performed 3 days after a second transfection with anti-H3K9me3. Enrichment of the immunoprecipitated DNA fraction compared with input DNA was determined for different primer pairs (GA3PDH promoter, line 1 repeats, AluY subclass, Satellite 2 (SAT2), and α-satellite (a-Sat). The values were normalized using the histone H4 promoter as an internal standard. Each data point is the average of two independent experiments with the standard deviation shown by error bars.
To further investigate the biological function of this protein and compare it with SUV39H1, another HMTase involved in chromosome condensation, we transiently reduced the expression of either KMT1F or SUV39H1 by transient transfection of different siRNA. The efficiency of SUV39H1 depletion was determined by reverse transcription real-time PCR detection and reaches 75% but does not affect KMT1F expression (supplemental Fig. S2A). Depletion in any or both of these two proteins decreases the level of H3K9me3 (supplemental Fig. S2B) but has little effect on mono- and dimethylation, suggesting that both are implicated in the trimethylation of H3K9. We then evaluated the distribution of H3K9me3 for different repetitive DNA elements by chromatin immunoprecipitation assays (Fig. 2C). In control cells, as expected, we observed a significant enrichment of histone trimethylation for different classes of DNA repeats such as interspersed (LINE 1, AluY subclass), pericentric (Satellite 2), and centromeric repeats (α-Satellite). This enrichment is clearly decreased at pericentromeres and interspersed LINE 1 sequences when the expression of KMT1F or SUV39H1 is impaired, whereas the effect is more moderate for the other sequences analyzed (Fig. 2D). Therefore, we propose that KMT1F plays a role in defining the higher order chromatin structure, together with SUV39H1. However, consistent with the changes of the methylation pattern of repetitive sequences dispersed throughout the genome (AluY and LINE 1), our results do not rule out a broader activity at euchromatic loci.
The H3K9me3 is a constitutive mark of condensed chromatin and regulates the distribution of the heterochromatin proteins HP1 (19–21). Therefore, we hypothesized a role for KMT1F in the formation of heterochromatin and examined the colocalization of KMT1F with HP1α foci by immunofluorescence staining and confocal microscopy (supplemental Fig. S3A). In control cells, we observed that large HP1 foci (>1 μm) are always associated with KMT1F staining (average of 3.7 colocalizations/cell in >1-μm foci, 50 cells), suggesting that both proteins might be targeted to constitutive heterochromatin. The overexpression of KMT1F (pKMT1F) results in an intense diffuse nuclear signal for both HP1 and KMT1F, whereas inhibition of either KMT1F or SUV39H1 expression disorganizes HP1 dots, suggesting a close connection between these two proteins and HP1 in interphase (supplemental Fig. S3A). Thus, at this step, we conclude that KMT1F participates in the distribution of H3K9me3 on repetitive DNA and recruitment of HP1, thereby contributing to chromatin organization.
KMT1F Depletion Affects Mitosis
The transient accumulation of KMT1F at a DAPI-intense region and the effect of KMT1F depletion on histone methylation of different classes of repeated elements, especially at centromeres and pericentromeres, suggest that this protein might participate in chromosome condensation and consequently influence mitosis. Indeed, the constitutive heterochromatin formed at centromeres epigenetically regulates their function, and perturbation of proper condensation of these regions can lead to aberrant gene expression and defective chromosome segregation (22). To address this hypothesis, we first analyzed the distribution of the centromeric specific proteins CENP A, B, and C using CREST antibody and H3K9me3 after knockdown of either KMT1F, SUV39H1, or both in 293T cells (supplemental Fig. S3B). Inhibition of the two SET proteins results in a massive loss of the CREST signal, suggesting that inhibition of histone H3 methylation affects the assembly of centromeric heterochromatin and kinetochore. We then used different antibodies against individual CENP proteins and showed that CENP A, B, and C signals are decreased by either KMT1F or SUV39H1 transiently knocked down with a higher effect for CENP B and C staining at mitotic chromosomes (p < 0.001; Fig. 3). We then reasoned that the reduced methylation of constitutive heterochromatin after KMT1F knockdown might lead to cell cycle defects, and we followed the progression of the cells throughout the cycle by live cell confocal imaging after transient transfection of siRNA against KMT1F in HeLa cells (Fig. 4, A and B). Prior to these experiments, KMT1F staining was verified in this cell line, and similar results were observed compared with 293T cells (supplemental Fig. S4A). Then we observed that mitotic progression from prophase to anaphase was delayed in HeLa cells transfected with KMT1F siRNA (from 24 to 70 min in control and KMT1F-depleted cells, respectively; Fig. 4B and supplemental movie) and was associated with the presence of lagging chromosomes (at 24 and 27 min; Fig. 4A), suggesting that reduction in the level of KMT1F affects progression into mitosis as previously observed for SUV39H1 (21). In addition, inspection of mitoses in cells with a reduced level of KMT1F revealed a global disorganization of the tubulin pattern (Fig. 4C) and a significant increase in mitotic cells with abnormal spindle (Fig. 4D), suggesting a key role for KMT1F in the regulation of cell cycle progression. However, no overlap was seen between KMT1F and H3K9me3 staining during mitosis (supplemental Fig. S4B).
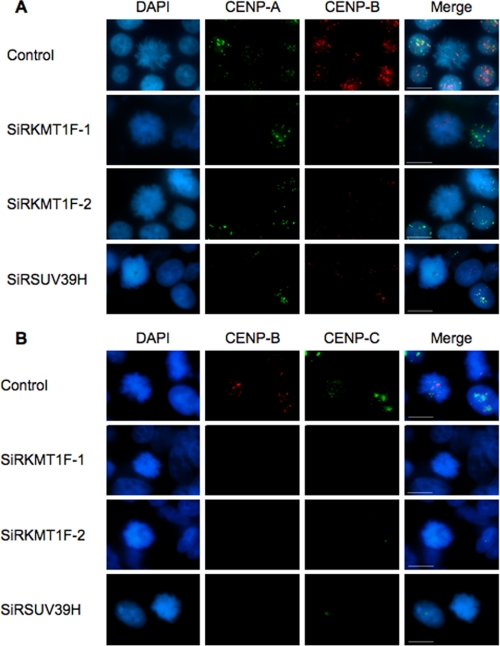
FIGURE 3.
KMT1F depletion correlates with decreased CENP B and C staining at mitotic chromosomes. To further investigate the link between KMT1F and CREST staining, we depleted KMT1F or SUV39H1 in 293T cells and analyzed CENP A, CENP B, or CENP C staining by indirect immunofluorescence in mitotic cells using either mouse anti-CENP A (1/250; Abcam), rabbit anti-CENP B (1/100; Abcam), or mouse anti-CENP C (1/250; Abcam) for 2 h. Secondary anti-mouse and anti-rabbit antibodies were used for detection as previously described. The cells were counterstained with DAPI (Sigma). Imaging of 30–50 mitotic cells was performed using conventional microscopy. A, costaining of CENP A and B. B, costaining of CENP B and C. Scale bars, 10 μm. In KMT1F-depleted cells, CENP A staining is not significantly modified, whereas CENP B (p < 0.001 for SiRKMT1F-1 (58% of cells with loss of CENP B signal) or SiRKMT1F-2 (79% of cells with loss of CENP B signal versus control using a Student's t test)) and CENP C staining are significantly decreased in mitotic cells (p < 0.001 for SiRKMTF-1 (63% of cells with loss of CENP C signal) or SiRKMTF-2 (90% of cells with loss of CENP B signal) versus control using a Student's t test)). Similar results were obtained after depletion in SUV39H (p < 0.001, 86% of cells with loss of CENP B signal or 76% of cells with loss of CENP C signal versus control using a Student's t test), suggesting that both histone methyltransferases contribute to the trimethylation of H3K9me3 residues at centromeres and thereby regulate the assembly of functional centromeres.
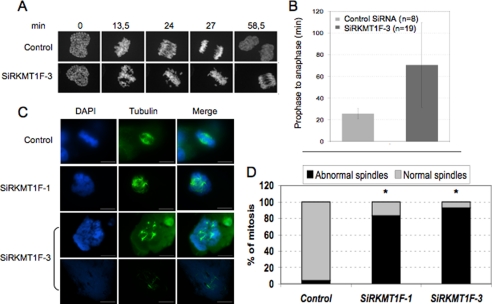
FIGURE 4.
KMT1F depletion delays mitosis. A, live cell confocal imaging of HeLa cells expressing a fusion GFP-H2B vector and transfected either with KMT1F-SiR3 or a control siRNA. B, the duration of mitosis from prophase to anaphase is indicated for each condition. The histograms represent the data collected from 27 cells and 27 different movies. A representative example is shown in the supplemental information. The bars indicate the standard deviations. C, indirect immunofluorescence with anti-β-tubulin antibody in 293T cells, 4 days after transient transfection with two KMT1F siRNA (SiRKMT1F-1 and SiRKMT1F-3) or a control siRNA. KMT1F depletion increases the number of cells containing abnormal spindle and multi-asters. D, histograms represent the rate of mitotic figures with either normal or abnormal spindle in 293T cells transfected with a control siRNA (3250 cells analyzed, n = 100 cells in metaphasis, 1.53% of necrotic cells), SiRKMT1F-1 (1386 cells analyzed, n = 6 cells in metaphases, 58% of necrotic cells), and SiRKMT1F-3 (852 cells analyzed, n = 55 cells in metaphasis, 2% of necrotic cells). The significant differences are indicated with asterisks (*, p < 0.001). The p values were calculated using a t test.
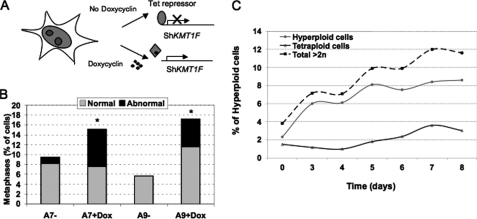
To study the long term consequence of KMT1F inhibition, we stably integrated a Tet-inducible short hairpin RNA against KMT1F into 293T cells (Fig. 5A). Several clones were selected and characterized, and short hairpin RNA production was induced for several days (Fig. 5A and supplemental Fig. S5A). Again, the inhibition of KMT1F confirms the importance of this protein in the regulation of the cell cycle as shown by the increase in cells with abnormal metaphases (Fig. 5B) and multinucleated cells after induction (7.56% versus 1.23% for A7 clone, p < 0.001 and 5.6% versus 0% for A9, p < 0.001 in induced versus noninduced cells; p value was determined by a t test). Moreover, consistent with the delayed anaphase transition and lagging chromosomes detected in transiently transfected cells, the presence in doxycyclin-induced cells of hyperploidy (12% of polyploid cells, 6 days after induction of KMT1F short hairpin RNA compared with 4% in mock-treated cells) and micronuclei, indicative of aberrant segregation, supports the conclusion that the KMT1F defect alters cell cycle progression (Fig. 5C). Collectively, our data show that mitosis is impaired in KMT1F-depleted cells, resulting in delayed chromosome congression and segregation and, consequently, increased polyploidy in daughter cells, suggesting the involvement of this protein in the regulation of the cell cycle, probably through the methylation of histone H3.
FIGURE 5.
Prolonged inhibition of KMT1F is associated with chromosomal abnormalities. A, doxycyclin-inducible KMT1F depletion system. KMT1F expression was assayed in Zeocin-resistant cell populations and isolated clones by reverse transcription-PCR (Fig. S2). Three days after induction, KMT1F expression is significantly decreased in A9 and A7 clones, which were selected for further analysis (60 and 80%, compared with noninduced cells, respectively). B, abnormal metaphase figure count comparison. Between 2500 and 3000 cells were analyzed in A7 and A9 mock treated (−) or doxycyclin-induced (+) cells, and 30, 219, 130, and 242 metaphase figures were respectively analyzed. Upon induction an increased number of cells with aberrant nuclear morphology or nuclei containing multipolar spindles is observed. Significant differences are indicated with asterisks (*, doxycyclin versus mock induced in A7 and A9; p values (<0.001) were determined using a t test). C, the percentages of hyperploid (>2n) and tetraploid (4n) cells were estimated in A9 cells induced or not with doxycyclin for 8 days.
DISCUSSION
The neighbor-joining tree of SET domain proteins present in public databases assigned KMT1F (or SETDB2 or CLLD8) as the closest homolog to ESET/SETDB1 (6, 7). The type and position of domains along both proteins suggest that they belong to the HMTase family. In agreement with previous studies showing that the SET and bifurcated SET can be a signature for lysine methyltransferase, we provide the first evidence that KMT1F is indeed a functional member of the HMTase family and harbors a modest but significant HMTase activity compared with the SET domains of SUV39H1 and G9a (p < 0.005). We also show that KMT1F specifically regulates the levels of H3K9me3 in human cells. KMT1F transiently accumulates at DAPI-intense regions and colocalizes with HP1 foci, suggesting a close connection between this protein and formation of constitutive heterochromatin. Depletion of KMT1F results in the loss of HP1 and H3K9me3 enrichment and alters CENP epitope (CREST) staining by decreasing the association of the α-satellite-bound CENP B and CENP C distribution and CENP A. Moreover, KMT1F-dependent reduction in H3K9me3 delays mitosis likely because of a defect in chromosome segregation. Similar phenotypes have been described for SUV39H1 (21, 22) or after depletion of the yeast homolog, Clr4 (23), suggesting a functional redundancy between SUV39H1 and KMT1F (supplemental Table S1). SUV39H1 is localized at centromeres throughout metaphase and could participate in the recruitment of HP1α, which modulates the targeting of the INCENP complex during mitotic spindle formation (24). The defect in chromosome segregation observed upon KMT1F depletion might also result from an impaired sister chromatin cohesion. Therefore, we propose that together with SUV39H1, the KMT1F-dependent H3K9 methylation participates in heterochromatin formation and that this previously uncharacterized protein contributes to centromere and pericentromere organization and mitotic chromosome functions. However, consistent with the modification of histone methylation pattern at Alu and LINE interspersed repetitive DNA, the resolution of our immunofluorescence analysis does not rule out a genome-wide activity. In agreement with this hypothesis, it has been recently shown that KMT1F associates with SETDB1 and contributes to the methylation of the TAT protein from human immunodeficiency virus (25), suggesting that this protein regulates methylation at different levels.
Interestingly, if SUV39h1 invalidation does not completely abrogate H3K9 trimethylation, it results in a more diffuse and modified pattern in mouse cells (26, 27), and evidence from the literature favors the existence of different pathways for the trimethylation of H3K9 residues, especially at pericentromeres (28, 29). Indeed, SETDB1/KMT1E, which is mainly involved in the regulation of euchromatic sequence and associates with corepressors at silenced promoters (29), was also found to modulate H3K9 trimethylation at pericentric heterochromatin and to interact with HP1 (30, 31), suggesting that it may also contribute to heterochromatin formation. Thus, the mechanism by which KMT1F modulates histone methylation is currently unknown, but despite the fact that SUV39H, G9a, and KMT1F modify the same residue, they may also play unique roles in the distribution of H3K9me3 and/or involve different protein complexes or noncoding RNAs (28, 31).
These last 10 years, a direct link between DNA methylation and organization of the chromatin fiber has been clearly demonstrated by the identification of large complexes containing proteins with affinity for methylated DNA, histones deacetylases, and HMTases. Thus, it became apparent that histone modification has an important role in regulating methylated DNA. The fact that KMT1F, like ESET/SETDB1, contains a MBD raises the possibility that histone methylation functions in concert with DNA methylation. Enzymes like SUV39H1 might be involved in the writing of the epigenetic mark by methylating lysine 9 residues, whereas enzymes containing MBD motifs are involved in the reading of the methylation marks at CpG sites. One can speculate that the MBD of KMT1F would serve as a platform for the subsequent methylation of H3K9 and then HP1 binding. Such a mechanism has been proposed for the mouse pericentric regions (32, 33) or the complex formed by SETDB1 and MBD1 (34), suggesting that KMT1F might also link DNA to histone methylation at heterochromatin and thereby strengthening the intimate link existing between these marks.
Supplementary Material
Acknowledgments
We thank T. Jenuwein (IMP Vienna), R. Losson (IGBMC, Strasbourg), I. Stancheva (Wellcome Trust Center for Cell Biology), S. Lowe and M. Narita, W. Herr and E. Julien (CSHL), H. Clevers (the Hubrecht laboratory), Y. Zhang (Lineberger Comprehensive Cancer Center, Chapel Hill), S. Kochbin and C. Caron (Institut Albert Bonniot, Grenoble, France), V. Ogrysko (IGR, Paris), and Y. Shinkai (Kyoto University) for providing reagents. We are grateful to J. Seeler, V. Doye, A. Echard, K. Monier, and D. Anguelov for advice. We acknowledge the facilities of the IFR 128 for microscopy (PLATIM) and Flow Cytometry.
This work was supported by grants from La Ligue Nationale contre le Cancer (Comité de la Drôme and Comité du Rhône, to G. F.; and Equipe Labelisée, to E. G.), l'Association pour la Recherche sur le Cancer (Programme National et Programme Alliance des Recherches sur le Cancer), Cancéropole Rhône-Alpes, “RISCRAD, an FP6 program from European Economic Community (to E. G.), and an Action Concertée Incitative grant from the French Minister of Research (to G. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Movie S1, Table S1, and Figs. S1–S5.
- HP1
- heterochromatin protein 1
- HMTase
- histone methyltransferase
- MBD
- methyl-binding domain
- DAPI
- 4′,6-diamidino-2-phenylindole
- siRNA
- small interfering RNA.
REFERENCES
- 1.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 2.Stewart M. D., Li J., Wong J. (2005) Mol. Cell. Biol. 25, 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reik W. (2007) Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 4.Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D. (2005) Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 5.Chen E. S., Zhang K., Nicolas E., Cam H. P., Zofall M., Grewal S. I. (2008) Nature 451, 734–737 [DOI] [PubMed] [Google Scholar]
- 6.Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. (2004) Genes Dev. 18, 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allis C. D., Berger S. L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., Zhang Y. (2007) Cell 131, 633–636 [DOI] [PubMed] [Google Scholar]
- 8.Yang L., Xia L., Wu D. Y., Wang H., Chansky H. A., Schubach W. H., Hickstein D. D., Zhang Y. (2002) Oncogene 21, 148–152 [DOI] [PubMed] [Google Scholar]
- 9.Sun X. J., Wei J., Wu X. Y., Hu M., Wang L., Wang H. H., Zhang Q. H., Chen S. J., Huang Q. H., Chen Z. (2005) J. Biol. Chem. 280, 35261–35271 [DOI] [PubMed] [Google Scholar]
- 10.Mabuchi H., Fujii H., Calin G., Alder H., Negrini M., Rassenti L., Kipps T. J., Bullrich F., Croce C. M. (2001) Cancer Res. 61, 2870–2877 [PubMed] [Google Scholar]
- 11.Harte P. J., Wu W., Carrasquillo M. M., Matera A. G. (1999) Cytogenet. Cell Genet. 84, 83–86 [DOI] [PubMed] [Google Scholar]
- 12.Huang N., vom Baur E., Garnier J. M., Lerouge T., Vonesch J. L., Lutz Y., Chambon P., Losson R. (1998) EMBO J. 17, 3398–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waring P. M., Cleary M. L. (1997) Curr. Top. Microbiol. Immunol. 220, 1–23 [DOI] [PubMed] [Google Scholar]
- 14.Stec I., Wright T. J., van Ommen G. J., de Boer P. A., van Haeringen A., Moorman A. F., Altherr M. R., den Dunnen J. T. (1998) Hum. Mol. Genet. 7, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 15.Ryu H., Lee J., Hagerty S. W., Soh B. Y., McAlpin S. E., Cormier K. A., Smith K. M., Ferrante R. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19176–19181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheatley S. P., Wang Y. (1996) J. Cell Biol. 135, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Small J. V. (1981) J. Cell Biol. 91, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magdinier F., Yusufzai T. M., Felsenfeld G. (2004) J. Biol. Chem. 279, 25381–25389 [DOI] [PubMed] [Google Scholar]
- 19.Bannister A. J., Kouzarides T. (2004) Methods Enzymol. 376, 269–288 [DOI] [PubMed] [Google Scholar]
- 20.Maison C., Bailly D., Peters A. H., Quivy J. P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G. (2002) Nat. Genet. 30, 329–334 [DOI] [PubMed] [Google Scholar]
- 21.Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 22.Melcher M., Schmid M., Aagaard L., Selenko P., Laible G., Jenuwein T. (2000) Mol. Cell. Biol. 20, 3728–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekwall K., Nimmo E. R., Javerzat J. P., Borgstrøm B., Egel R., Cranston G., Allshire R. (1996) J. Cell Sci. 109, 2637–2648 [DOI] [PubMed] [Google Scholar]
- 24.Ainsztein A. M., Kandels-Lewis S. E., Mackay A. M., Earnshaw W. C. (1998) J. Cell Biol. 143, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Duyne R., Easley R., Wu W., Berro R., Pedati C., Klase Z., Kehn-Hall K., Flynn E. K., Symer D. E., Kashanchi F. (2008) Retrovirology 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters A. H., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Martens J. H., Jenuwein T. (2003) Mol. Cell 12, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 27.Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y., Allis C. D. (2003) Mol. Cell 12, 1591–1598 [DOI] [PubMed] [Google Scholar]
- 28.Wu R., Terry A. V., Singh P. B., Gilbert D. M. (2005) Mol. Biol. Cell 16, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Rauch T., Chen Z. X., Szabó P. E., Riggs A. D., Pfeifer G. P. (2006) J. Biol. Chem. 281, 19489–19500 [DOI] [PubMed] [Google Scholar]
- 30.Lee J., Hagerty S., Cormier K. A., Kim J., Kung A. L., Ferrante R. J., Ryu H. (2008) Hum. Mol. Genet. 17, 1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kourmouli N., Sun Y. M., van der Sar S., Singh P. B., Brown J. P. (2005) Biochem. Biophys. Res. Commun. 337, 901–907 [DOI] [PubMed] [Google Scholar]
- 32.Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. (2003) Curr. Biol. 13, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 33.Fuks F., Hurd P. J., Deplus R., Kouzarides T. (2003) Nucleic Acids Res. 31, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarraf S. A., Stancheva I. (2004) Mol. Cell 15, 595–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.