Abstract
Tandem gene duplication is one of the major gene duplication mechanisms in eukaryotes, as illustrated by the prevalence of gene family clusters. Tandem duplicated paralogs usually share the same regulatory element, and as a consequence, they are likely to perform similar biological functions. Here, we provide an example of a newly evolved tandem duplicate acquiring novel functions, which were driven by positive selection. CG32708, CG32706, and CG6999 are 3 clustered genes residing in the X chromosome of Drosophila melanogaster. CG6999 and CG32708 have been examined for their molecular population genetic properties (Thornton and Long 2005). We further investigated the evolutionary forces acting on these genes with greater sample sizes and a broader approach that incorporate between-species divergence, using more variety of statistical methods. We explored the possible functional implications by characterizing the tissue-specific and developmental expression patterns of these genes. Sequence comparison of species within D. melanogaster subgroup reveals that this 3-gene cluster was created by 2 rounds of tandem gene duplication in the last 5 Myr. Based on phylogenetic analysis, CG32708 is clearly the parental copy that is shared by all species. CG32706 appears to have originated in the ancestor of Drosophila simulans and D. melanogaster about 5 Mya, and CG6999 is the newest duplicate that is unique to D. melanogaster. All 3 genes have different expression profiles, and CG6999 has in addition acquired a novel transcript. Biased polymorphism frequency spectrum, linkage disequilibrium, nucleotide substitution, and McDonald–Kreitman analyses suggested that the evolution of CG6999 and CG32706 were driven by positive Darwinian selection.
Keywords: Drosophila, positive selection, tandem duplication, young gene
Introduction
It is well recognized that gene duplication is prevalent in eukaryotes. Genomic analyses of model organisms have shown that over one-third of all protein-coding genes belong to multigene families (Rubin et al. 2000; Kent et al. 2003). The mechanisms of gene duplication can be classified based on its scale (e.g., whole-genome duplication, segmental duplication, and tandem gene duplication) or whether it is RNA mediated (retroposition and transposition). Comparative genomic analysis between closely related species has revealed that tandem duplication is one of major mechanisms creating new genes, particularly genes clustered into a gene family, which have been documented in many organisms (e.g., Anderson and Roth 1977; Stark 1993; Brown et al. 1998; Eichler and Sankoff 2003; Leister 2004; Cardoso et al. 2006; Ponce and Hartl 2006; Shoja and Zhang 2006; Tuskan et al. 2006; Hazkani-Covo and Graur 2007). It is believed that tandem gene duplication could arise by unequal crossing over, which results from homologous recombination between paralogous sequences or nonhomologous recombination by replication-dependent chromosome breakages (Arguello et al. 2007).
A newly duplicated gene must overcome substantial hurdles before fixation. Once fixed, duplicated genes may face different fates: 1 of the 2 copies could lose its function and become pseudogenized due to the accumulation of degenerative mutations or both copies can maintain the same function. It is also possible that the 2 copies can accumulate different mutations leading to the duplicated genes taking on different roles that had previously been performed by the original gene, a process known as subfunctionalization. The most remarkable fate of gene duplication is neofunctionalization, whereby the new copy evolves a novel function driven and maintained by selection, whereas the old copy still retains the original function.
The location of duplicated copy can be adjacent to the original (tandem) or somewhere else in the genome (dispersed), for example, the duplicate generated by RNA-mediated retrotransposition. Separated from their regulatory elements, the dispersed duplicated copies will likely evolve novel functions by recruiting new regulatory elements (e.g., Wang et al. 2002). In contrast, the tandemly duplicated gene would tend to maintain a similar function to their parental copy due to their sharing the same regulatory elements and this has been demonstrated in many examples (e.g., Li et al. 2006; Ponce and Hartl 2006; Arisue et al. 2007). Given the apparent importance of tandem gene duplication for gene expansion in the eukaryotes, it is of great interest to know whether the tandem gene duplication can also generate novel functions. It has been recognized that gene duplication followed by divergence is one of the most important mechanism for generating new genes with novel functions, and such genetic novelty could involve in increasing organismic complexity, speciation, and adaptation processes (Long et al. 2004; Roth et al. 2007).
Here, we provide an example of tandemly duplicated genes acquiring novel transcription patterns, which could potentially lead to novel biological functions. CG32708, CG32706, and CG6999 are a 3-gene cluster on the X chromosome of Drosophila melanogaster. By investigating their homologous counterparts in the D. melanogaster subgroup species, we have found that this 3-gene cluster was created by 2 rounds of tandem gene duplication in the last 5 Myr. Though the newly duplicated copies (CG32706 and CG6999) have diverged biological functions from their parental copy (CG32708), they share great similarity both in their DNA and protein sequences with only few substitutions in D. melanogaster. All the 3 genes have different expression patterns, which can potentially lead to diverged biological functions. Particularly, we have found that the newest duplicate, CG6999, has a novel transcript with shorter sequence compared with its major transcript. We further observed that the homologous copy of CG32706 in species Drosophila simulans, Drosophila mauritiana, and Drosophila sechellia has undergone extensive sequence divergence compared with D. melanogaster CG32706. Sequence divergence and population genetic tests strongly suggested that CG6999 and CG32706 evolved under positive selection.
Materials and Methods
Stocks, Sampling, and DNA Extraction
We used isofemale stocks of D. melanogaster (Oregon-R), D. mauritiana, D. sechellia, Drosophila yakuba, Drosophila teissieri, Drosophila santomea, and Drosophila erecta, which have been kept in our laboratory for over 50 generations. We sequenced a collection of Drosophila lines to generate the polymorphism for the 3 genes in D. melanogaster and D. simulans. The 26 isofemale D. melanogaster strains sampled include 10 from North America (NA), 7 from Zimbabwe (ZS), 5 from Taiwan, and 4 from Israel (FS). The polymorphism of D. simulans was generated from 22 population samples, of which 6 are from Africa, 6 from NA, 5 from France, 2 from FS, 2 from South America, and 1 from Southern Pacific Cook Island. To avoid potential problem with population structure within McDonald–Kreitman (MK) test (McDonald and Kreitman 1991), we restricted our analysis to the 6 D. simulans African ancestral strains to test the evolution of CG32706 using a MK test.
Genomic DNA of D. melanogaster, D. simulans, D. mauritiana, D. sechellia, D. yakuba, D. teissieri, D. santomea, and D. erecta was extracted using Puregene DNA isolation kits (Gentra Systems, Minneapolis, MN) from 25–30 flies (for microarray hybridization, Southern blotting, and genomic DNA polymerase chain reaction [PCR] amplification) or single male fly (for the D. melanogaster and D. simulans population genetic analysis). We did not observe any potential heterozygous sites in the sequence traces, so the sequence from each individual was considered to be a haplotype.
Duplication Identification, DNA Amplification, and Sequencing
We first identified the potential duplicated candidates in the D. melanogaster subgroup species using microarray-based comparative genomic hybridization (CGH) methods. Genomic DNA was digested using DNaseI, and 3′ termini of the fragmentation products were labeled with biotin-dideoxyuridine triphosphate (ddUTP). The target DNA fragments (∼100–150 bp) were hybridized onto The GeneChip Drosophila melanogaster Genome Array following the standard Affymetrix protocol (Affymetrix, Santa Clara, CA). The ratio of pairwise comparisons for each probe was calculated using hybridization intensity among 8 species, and the median value of intensity fold-change in all probes for each feature was taken as threshold for gene duplication criterion. The detailed methodology for the duplication identification was described in Fan and Long (2007).
PCR were performed in the standard thermal cycler using Invitrogen Taq polymerase following the manufacturer's protocol, with annealing temperature adjusted based on the length of fragments with 1 kb/min. The double-stranded PCR products were purified using a Qiagen PCR purification kit or a Qiagen miniprep Gel purification system. Purified PCR products were sequenced using Applied Biosystems 3730XL 96-capillary automated DNA sequencer. The entire fragments of the blocks were sequenced using the sequence walk procedure. Sequences were edited and assembled. ClustalX was used to align sequences for further analyses (Thompson et al. 1997). Manual adjustments were made where necessary.
Expression Analysis
Retrotranscription (RT)–PCR was used to analyze the expression profile in different developmental stages and tissues. Total RNA was extracted from D. melanogaster adult virgin females, males, 2-hour-old eggs, second- and third-instar larvae, and pupae using a Qiagen total RNA extraction kit. We examined tissue differential expression pattern by RT-PCR with RNA extractions from the head, body without ovary/testis, accession gland, and ovary/testis. Testis and ovary were obtained by dissecting mature male and female flies in saline solution, and removed testis and ovary were preserved in RNA later solution. Total RNA was extracted from flies or tissues following the Qiagen protocol.
Population Genetic Analysis
Basic population genetic analyses were performed in DnaSP (Rozas et al. 2003). The sequence diversity was quantified as nucleotide diversity (π) (Nei 1987) and Watterson's θ (1975). Tests of deviation from neutrality were conducted using tests from Tajima (1989), Fu & Li (1993), and Fay & Wu (2000), and significance was assessed using coalescent simulations. The neutral coalescent process was simulated using 2,000 replicates with the number of segregating sites set to that observed in the data. However, these approaches, based on the polymorphic spectrum, are of limited utility in testing for neutrality in young genes because a reduced level of diversity and skew toward rare are expected (Thornton 2007). Therefore, we also used MK test as implemented in DnaSP. In the MK test for CG32706, we compared the polymorphism generated from the D. simulans lines and fixed mutations between D. simulans and D. mauritiana. In addition, we also investigated linkage disequilibrium (LD).
Phylogenetic Analysis and Sequence Divergence Calculation
The phylogenetic analysis for both DNA and protein sequence was performed using the Neighbor-Joining and maximum likelihood methods implemented in PAUP* 4.0b10 (Swofford 2002), with 10,000 bootstrap replicates to assess support. We further calculated the number and rate of nonsynonymous and synonymous substitution for 3 genes in D. melanogaster using codon model (Codeml) implemented in PAML (Yang 1997, 2007) under a model in which all branches were allowed their own Ka/Ks (ω) value. To generate the data using free ratio for each branch, we aligned the coding sequences of CG32708, CG32706, and CG6999. Because we have determined CG32708 as the parental copy of the other 2 genes, the tree for Codeml analysis used CG32708 as outgroup. The numbers of synonymous and nonsynonymous substitution along each branch were calculated under a model in which the Ka/Ks ratio (ω) was free to vary along each branch. (Goldman and Yang 1994; Yang 1997).
We calculated the Ka/Ks ratio using maximum likelihood algorithm using Perl script incorporated PAML for CG32706 homologues between species D. simulans, D. mauritiana, and D. sechellia. The significance of Ka/Ks that deviated from neutrality (=1) was tested using likelihood ratio test (Yang 1998).
Results
Tandem Duplication in D. simulans, D. mauritiana, and D. melanogaster
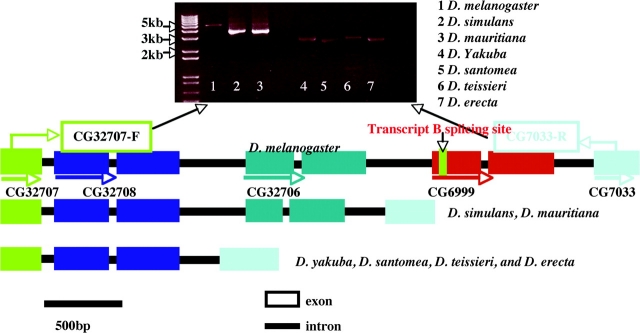
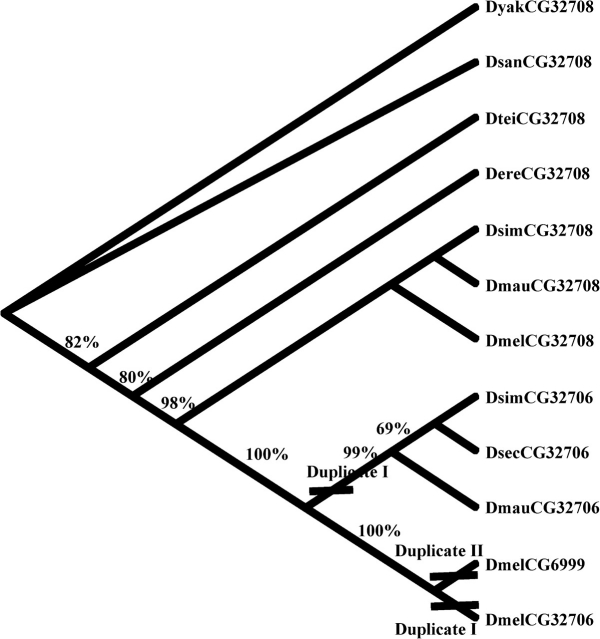
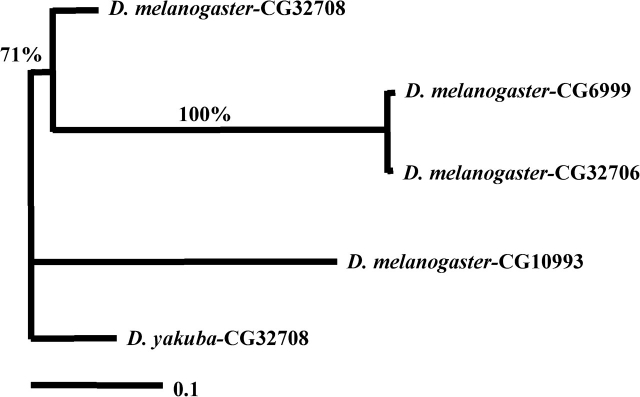
Our initial microarray CGH suggested that there are multiple homologous copies of CG6999 in the species of D. melanogaster. By blasting the candidate sequences that we identified from CGH against the genomic sequence of D. melanogaster, we found 3 copies, CG32708, CG32706, and CG6999, closely adjacent with only 500 bp separating the protein-coding sequences on the X chromosome near 8C5 (fig. 1). To characterize the gene content and structure in all D. melanogaster subgroup species, we designed a pair of primers that were located in the flanking sequences of CG32708 and CG6999 (fig. 1) to amplify and obtain the homologous sequences in all D. melanogaster subgroup species. The PCR and sequencing results indicated that a single homologue is present in D. yakuba, D. erecta, D. santomea, and D. teissieri, and 2 homologous copies in D. simulans and D. mauritiana (fig. 1). Phylogenetic analysis clearly showed that 2 duplication events occurred in the last 5 Myr. The first duplication event occurred before the divergence of ancestor of D. melanogaster and D. simulans approximately 5 Mya, and a more recent duplication happened in the branch of D. melanogaster in the last 1–2 Myr (fig. 2).
FIG. 1.—
The PCR products and schematic graphs of the genomic fragment between CG32707-F and CG7033-R in 7 Drosophila melanogaster subgroup species. The homologous copies in the species are shown as same color. The splicing site for the CG6999 transcript “B” is marked as green bar. CG32707-F and CG7033-R are 2 primers used for PCR amplification.
FIG. 2.—
Neighbor-Joining tree of CG32708, CG32706, and CG6999 in Drosophila melanogaster subgroups. Bootstrap support and duplication events are shown.
Expression Analysis by RT-PCR
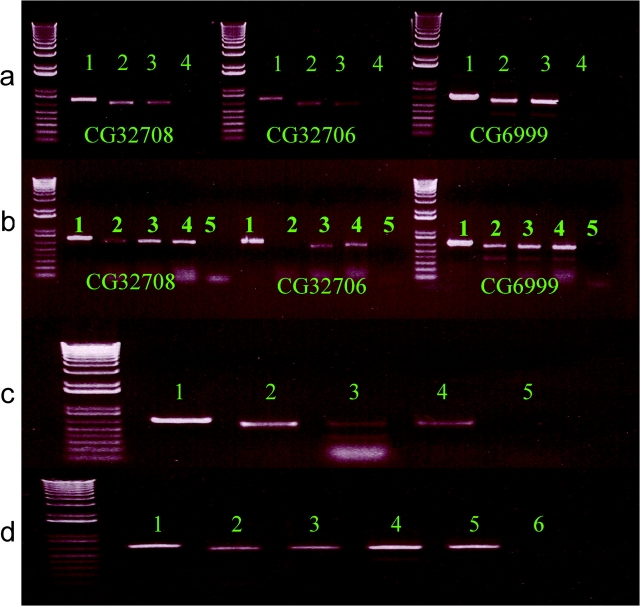
We performed RT-PCR to investigate the pattern of gene expression across tissues and developmental stages in D. melanogaster for all 3 genes. Overall, the expression levels were different among the 3 genes, with CG6999 having the highest expression and CG32706, the lowest expression (fig. 3). Interestingly, we found 2 transcripts of CG6999 in both female and male flies (fig. 3a). Transcript “B” is a novel shorter transcript that has a 5′ splicing site located in the first exon of transcript “A.” To dissect the differential expressions of the novel transcript of CG6999, we conduct RT-PCR to examine the expression profile using different tissues and found that only reproductive organs (testis and ovary) show the expression of the novel CG6999 transcript “B” (fig. 3c and d). The differential expression profiles of the 3 genes also appear to be consistent across different developmental stages. CG32708 and CG32706 tend to have lower expression in second-instar larva than in third-instar larva and pupa. CG6999, however, has an equal expression level in both transcripts (A and B) (fig. 3b).
FIG. 3.—
The expression profile of CG32708, CG32706, and CG6999. Two transcripts of CG6999 are marked as “A” and “B”. Panel (a), in adult flies—1: genomic DNA, 2: female fly cDNA, 3: male fly cDNA, and 4: negative control. Panel (b), in developmental stages—1: genomic DNA, 2: second larva cDNA, 3: third larva cDNA, 4: pupa cDNA, and 5: negative control. Panel (c), CG6999 expression in female fly tissues—1: genomic DNA 2: head cDNA 3: ovary cDNA, 4: body without ovary cDNA, and 5: negative control. Panel (d), CG6999 expression in male fly tissues—1: genomic DNA 2: head cDNA 3: accessory gland cDNA, 4: testis cDNA, 5: body without testis cDNA, and 6: negative control.
Sequence Divergence of the 3 Genes
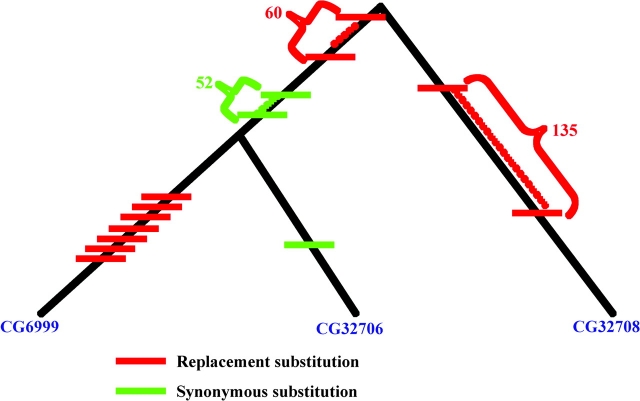
We estimated the Ka/Ks ratio of CG32708 and CG32706 across species. The average Ka/Ks ratio for CG32708 in all D. melanogaster subgroup species is equal to 0.23, which indicates that CG32708 are under strong functional constraints. However, the sequence of CG32706 is highly diverged between the clade of D. simulans–D. mauritiana–D. sechellia and the clade of D. melanogaster, and there are extensive deletions in the 5′ and 3′ ends of the sequences (supplementary fig. 1, Supplementary Material online), which is even higher than the sequence divergence in the intron regions (data not shown). Because CG6999 is a novel gene in the species of D. melanogaster, we calculated its Ka/Ks values against D. melanogaster CG32706. Overall, 7 nonsynonymous substitutions, 1 synonymous substitution, and a 6-base insertion are observed (fig. 4). The ratio of Ka/Ks (1.6) along the CG6999 lineage indicates that CG6999 has undergone accelerated divergence after the gene duplication event, and because the ratio is largely greater than 1, it seems likely that this was driven by positive selection. The distribution of the substitutions and insertions are primarily located at near 5′ or 3′ end of the gene. This biased distribution of substitutions is also seen in D. simulans CG32706.
FIG. 4.—
The tree of CG6999, CG32706, and Cg32708 in Drosophila melanogaster showing the number of substitutions in each branch.
Positive Selection of CG6999 in D. melanogaster
To further investigate whether adaptive evolution had affected these 3 genes, we collected polymorphism data from 26 D. melanogaster lines. Because the local population structure can lead to a departure from neutrality under certain tests, we tested for gene flow and population subdivision using Fst. The Fst values clearly show the high gene flow and low population subdivision among 4 local populations (table 1). Among the 3 genes, only CG6999 shows a significant bias in the site-frequency spectrum away from neutral expectations (table 2). The negative values of these tests suggest that either positive selection or demographic process (e.g., older bottleneck, population expansion, recently fixed duplication, and hidden population structure) drove the evolution of CG6999.
Table 1.
Gene Flow and Population Subdivision Test of Drosophila melanogaster Local Populations (Fst)
| Pop1 | Pop2 | 3-Gene Cluster | CG32708 | CG32706 | CG6999 |
| NA | ZS | 0.45109 | 0.16667 | 0.46336 | 0.15781 |
| NA | FS | 0.06476 | 0.07407 | 0.0331 | 0.05761 |
| NA | TWN | −0.05289 | 0.04762 | −0.10624 | −0.06702 |
| ZS | FS | 0.44062 | 0 | 0.608 | 0 |
| ZS | TWN | 0.49308 | 0 | 0.5858 | 0 |
| FS | TWN | 0.03468 | 0 | −0.06061 | 0 |
NOTE.—TWN, Taiwan.
Table 2.
Levels of Polymorphism in Drosophila melanogaster and Neutrality Tests on the Site-Frequency Spectrum
| Summary Statistic | CG32708 | CG32706 | CG6999 |
| N | 26 | 26 | 26 |
| L | 798 | 766 | 786 |
| S | 4 | 11 | 14 |
| Π | 0.00064 | 0.0043 | 0.0017 |
| Θ | 0.00131 | 0.0038 | 0.0053 |
| Tajima's D | −1.36, P = 0.08 | 0.47, P = 0.73 | −2.30a, P = 0.001 |
| Fu & Li's D* | −0.90, P = 0.32 | −0.48, P = 0.37 | −3.64a, P = 0.001 |
| Fay & Wu's H | 0.39, P = 0.55 | −2.043b, P = 0.019 | −10.38a, P = 0.001 |
NOTE.—The Fay & Wu's H of CG32708 and CG32706 was calculated using homologous sequences of Drosophila simulans as outgroup and that of CG6999 was estimated using Drosophila melanogaster CG32706 sequence as outgroup. N, population size; L, gene length (bp); S, the number of segregation sites.
The significance as P < 0.01.
The significance as P < 0.05.
We further investigated the above possibilities by performing LD analysis. An LD test covering the entire of the 3 genes regions was conducted, and the significant associations were estimated using Chi-square tests. To dissect the association within genes and between genes, we partitioned the region into 3 fragments, with each fragment corresponding to 1 gene. The partitions were based on the breaking point of gene duplication by alignment of 3 genes including flanking regions. The 47 polymorphic sites were pairwise combined into 1,081 comparisons. Among 1,081 comparisons, 165 comparisons show the significant association and 56 of them remain significant after Bonferroni correction: 32 of these 56 comparisons are within genes and 24 are between genes. Among 3 genes, CG32708 has the least number of significant associations (2 SNPs; single nucleotide polymorphism) and CG6999 has the highest number of significant associations (19 SNPs) (table 3). This suggests that CG6999 may be currently undergoing a selective sweep.
Table 3.
The Number of Significant Associations between Pairs of SNPs
| Region | CG32708 | CG32706 | CG6999 | CG32708∼CG32706 | CG32708∼CG6999 | CG32706∼CG6999 | Total |
| Distance (bp) | 1,106 | 1,227 | 1,236 | 2,333 | 3,569 | 2,463 | 3,569 |
| Significant associationsa | 2 | 11 | 19 | 7 | 8 | 9 | 56 |
Pairwise comparisons show a significant association after applying the Bonferroni procedure.
Positive Selection of CG32706 Orthologs in D. simulans, D. sechellia, and D. mauritiana
We calculated the Ka/Ks ratio of CG32706 in the species D. simulans, D. sechellia, and D. mauritiana (table 4). The average Ka/Ks is equal to 2.067. The Ka/Ks ratio between D. sechellia and D. mauritiana is significantly greater than 1 (3.406, P = 0.05), suggesting positive selection (table 4). The result of the MK test revealed a significant excess of replacement substitutions between species, indicating strong positive selection acting on CG32706 after the species diverged within the D. simulans clade (table 5). Moreover, the polymorphism analysis revealed that CG32706 is not a pseudogene, as shown by the excess of synonymous substitution in D. simulans polymorphism spectrum (table 6).
Table 4.
Ka/Ks of CG32706 in Drosophila simulans(Dsim), Drosophila sechellia(Dsec), and Drosophila mauritiana(Dmau)
| Ks | Ka | Ka/Ks | Likelihood Ratio Test | |
| Dsim/Dsec | 0.063 | 0.072 | 1.143 | P = 0.81 |
| Dsim/Dmau | 0.040 | 0.097 | 2.425 | P = 0.42 |
| Dmau/Dsec | 0.032 | 0.109 | 3.406 | P = 0.05a |
| Average | 0.045 | 0.093 | 2.067 | P = 0.19 |
The significance as P < 0.05.
Table 5.
MK Test of CG32706 in Drosophila simulans
| Substitution | Divergence | Polymorphism |
| Nonsynonymous | 30 | 3 |
| Synonymous | 6 | 5 |
| Fisher exact test | P = 0.015a |
The significance as P < 0.05.
Table 6.
Neutrality Test of Polymorphism Substitutions in Drosophila simulans CG32706
| Expected value | Observed value | Chi-square test | |
| Synonymous | 2.7 | 8 | χ2 = 4.46a |
| Nonsynonymous | 10.3 | 5 | P = 0.03 |
The significance as P < 0.05.
Discussion
Positive Selection Drive the Evolution of CG32706 and CG6999
The sequence and phylogenetic analyses from the D. melanogaster subgroup species clearly suggest the 3-gene cluster in D. melanogaster is a product of 2 rounds of gene duplication, with CG6999 originating 1–2 Mya and CG32706 derived from 5 Mya. Thornton and Long (2005) previously generated sequence polymorphism data for CG6999 and CG32708 (synonymns to CG6997 in Thornton and Long 2005) from 10 ZS D. melanogaster lines. They compared the parologs of CG32708 and CG6999 in D. melanogaster using population genetics and MK analyses and found no evidence for selection for new protein functions after gene duplication. In this study, we further pursued this question by using a combination of polymorphism and divergence analyses using comparative sequences from all D. melanogaster subgroup species. Several complement lines of evidence suggest that the evolution of CG6999 was likely to have been driven by positive Darwinian selection. First, the significant skew toward rare alleles in the site-frequency spectrum suggests an excess of rare allele in the D. melanogaster population particularly in the gene CG6999. Second, the LD test indicates that CG6999 has a remarkably high number of the significantly associated sites, consistent with the notion that CG6999 is linked to a site that is under selection within or immediately outside the gene region. Third, the excess of nonsynonymous substitutions occurs after gene duplication between CG32706 and CG6999 in D. melanogaster (fig. 4).
We further noticed that the sequences of CG32706 in D. simulans and D. mauritiana are highly diverged with a significant Ka/Ks ratio deviating from neutrality, which apparently shows the accelerated rate of evolution after it diverged from its ancestor CG32708. Moreover, the significant Fay & Wu's test for CG32706 indicated that selection drove the excess of high-frequency alleles in the D. melanogaster population, and the MK test of CG32706 orthologues in D. simulans and D. mauritiana strongly suggests positive selection after species divergence in the D. simulans sister group.
Functional Divergence, Speciation, and Novel Transcript of Tandemly Duplicated Genes
CG32708, CG32706, and CG6999 are believed to play a role in the RNA-binding and alternative-spicing pathway (Park et al. 2004), though they act at different stages. CG32706 is an RNA-binding protein, which interacts selectively with premessenger RNA or messenger RNA (mRNA). CG6999 plays a role in the regulation of alternative nuclear mRNA splicing via the spliceosome (Dimova et al. 2003; Park et al. 2004). The cellular and biological function of CG32708 remains unclear. Based on the homologous sequences of its paralog in D. melanogaster, we believe it also plays a role in the RNA-splicing pathway. However, our evidence from both the expression data and the evolutionary analysis of sequences demonstrated that the new genes are likely to have replaced the major functions of their parental copies with the expansion of molecular and biological functionality. Interestingly, we have found a nontandem duplicated homologous copy, CG10993, in this gene family in D. melanogaster. CG10993 is located in 12C5 of X chromosome. To determine the relationship and function of CG10993 with other 3 homologous members, we performed a phylogenetic analysis and further calculated Ka and Ks values. The phylogenetic tree shows CG10993 resides in the basal lineage and has a closer relationship with D. yakuba CG32708 (fig. 5). Therefore, CG10993 is likely to be the ancestor of the 3 tandem genes diverged over 10 Mya. The Ka/Ks (0.3363/0.5614 = 0.599) test between CG10993 and CG32708 suggested that the 2 duplicated genes were subject to purifying selection.
FIG. 5.—
Maximum likelihood tree of Drosophila melanogaster CG6999 gene family. Drosophila yakuba CG32708 serves as outgroup. Bootstrap supports shown above the branches.
It has been observed that X-linked genes evolved rapidly after gene duplication and the X chromosome appears to be a fertile ground for gene duplication and evolution. For example, studies from Drosophila and mammals found that the significant excess of retroposed genes originated from X chromosome, likely under adaptive evolution (Betran et al. 2002; Emerson et al. 2004). A previous genome analysis has predicted an excess of new genes on the X that are tandem duplicates, based on the fact that Ks seems to be low on the X between duplicates compared with the autosomes (Thornton and Long 2002). A recent investigation further demonstrated that the Drosophila X chromosome not only shows rapid origination and evolution of retroposed genes but also imposes noncoding RNA genes under positive selection through DNA-level duplication (Levine et al. 2006). Notably, our 3 gene cluster resides closely (less 1 cM distance) between CG32712 and CG32690, the 2 fast-evolved noncoding RNA genes found by Levine et al. (2006). Such coincidence may indicate a hot spot in the X chromosome for the novel gene origination under adaptive evolution. Furthermore, a whole-genome polymorphism and divergence analysis in D. simulans similarly found significantly less polymorphism and faster divergence on the X chromosome, indicating that X-linked genes are influenced by adaptive evolution (Begun et al. 2007).
It is well known that transcription is driven by regulatory elements, the interaction between the sequence-specific binding transcriptional factors (the trans-elements) and their DNA recognition sites (the cis-elements). In addition to the cis–trans interaction, the spatial and temporal expression of genes coding transcription factor genes can also regulate expression patterns, in which transcription factors with similar DNA-binding properties can control distinct biological processes (Duarte et al. 2006). Our RT-PCR expression experiments indicate that the 3 genes have differential expression profiles. Particularly, we have found an additional novel transcript in CG6999. We therefore applied the Neutral Network Promoter Prediction (NNPP) program for the flanking region of all 3 genes to detect their putative transcription regulatory element. We found high scores for putative sequences that appear to be cis-regulatory elements. The sequences of cis-element for 3 genes are highly similar with CG32708 and CG6999 sharing identical sequences (supplementary fig. 2, Supplementary Material online). We further aligned the entire franking region of 3 genes and observed very highly conserved sequences (99% identity). Therefore, we claim that the novel transcription of CG6999 might be caused by trans-regulatory factors that expressed differentially.
Supplementary Materials
Supplementary figures 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors thank a number of people: Chung-I Wu, Jerry Coyne, Peter Andolfatto, and Eviatar Nevo for providing fly strains; Xinming Li for performing the microarray hybridization; members in the Long laboratory for the valuable discussions and inputs; Adam Eyre-Walker and Stuart Wigby for critical reading the manuscript; and 3 anonymous reviewers for their critical comments and valuable suggestions. This work is supported by National Institutes of Health and National Science Foundation grants to M.L.
References
- Anderson RP, Roth JR. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Arguello JR, Fan C, Wang W, Long M. Origination of chimeric genes through DNA-level recombination. Genome dynamics: protein and gene evolution, Volff J-N eds, 2007;Vol. 3 doi: 10.1159/000107608. Basel (Switzerland): Karger. p. 131–146. [DOI] [PubMed] [Google Scholar]
- Arisue N, Hirai M, Arai M, Matsuoka H, Horii T. Phylogeny and evolution of the SERA multigene family in the Genus Plasmodium. J Mol Evol. 2007;65:82–91. doi: 10.1007/s00239-006-0253-1. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (13 authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;6:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Todd K, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Cardoso JC, Pinto VC, Vieira FA, Clark MS, Power DM. Evolution of secretin family GPCR members in the metazoa. BMC Evol Biol. 2006;6:108. doi: 10.1186/1471-2148-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, DePamphilis CW. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- Eichler E, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Fan C, Long M. A new retroposed gene in Drosophila heterochromatin detected by microarray-based comparative genomic hybridization. J Mol Evol. 2007;64:272–283. doi: 10.1007/s00239-006-0169-9. [DOI] [PubMed] [Google Scholar]
- Fay J, Wu C-I. Hitchhiking under positive Darwinian Selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Hazkani Covo E, Graur D. A comparative analysis of numt evolution in human and chimpanzee. Mol Biol Evol. 2007;24:13–18. doi: 10.1093/molbev/msl149. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci USA. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, et al. (13 coauthors) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2004;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce R, Hartl DL. The evolution of the novel Sdic gene cluster in Drosophila melanogaster. Gene. 2006;376:174–183. doi: 10.1016/j.gene.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles DA. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J Exp Zoolog B Mol Dev Evol. 2007;308:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rubin G, Yandell MD, Wortman JR, et al. (50 co-authors) Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoja V, Zhang L. A roadmap of tandemly arrayed genes in the genomes of human, mouse, and rat. Mol Biol Evol. 2006;23:2134–2141. doi: 10.1093/molbev/msl085. [DOI] [PubMed] [Google Scholar]
- Stark GR. Regulation and mechanisms of mammalian gene amplification. Adv Cancer Res. 1993;61:87–113. doi: 10.1016/s0065-230x(08)60956-2. [DOI] [PubMed] [Google Scholar]
- Swofford D. Sunderland (MA): Sinauer Associates; 2002. PAUP: phylogenetic analysis using parsimony. Version 4.0b10. [Google Scholar]
- Tajima F. Statistical methods for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton KR. The neutral coalescent process for recent gene duplications and copy-number variants. Genetics. 2007;177:987–1000. doi: 10.1534/genetics.107.074948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton KR, Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X Chromosome. Mol Biol Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- Thornton KR, Long M. Excess of amino acid substitutions relative to polymorphism between X-linked duplications in Drosophila melanogaster. Mol Biol Evol. 2005;22:273–284. doi: 10.1093/molbev/msi015. [DOI] [PubMed] [Google Scholar]
- Tuskan G, DiFazio S, Jansson S, et al. (110 co-authors) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Wang W, Brunet FG, Nevo E, Long M. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.