Abstract
High-throughput technology has facilitated genome-scale analyses of transcriptomic adjustments in response to environmental perturbations with an oxygen deprivation component, such as transient hypoxia or anoxia, root waterlogging, or complete submergence. We showed previously that Arabidopsis (Arabidopsis thaliana) seedlings elevate the levels of hundreds of transcripts, including a core group of 49 genes that are prioritized for translation across cell types of both shoots and roots. To recognize low-oxygen responses that are evolutionarily conserved versus species specific, we compared the transcriptomic reconfiguration in 21 organisms from four kingdoms (Plantae, Animalia, Fungi, and Bacteria). Sorting of organism proteomes into clusters of putative orthologs identified broadly conserved responses associated with glycolysis, fermentation, alternative respiration, metabolite transport, reactive oxygen species amelioration, chaperone activity, and ribosome biogenesis. Differentially regulated genes involved in signaling and transcriptional regulation were poorly conserved across kingdoms. Strikingly, nearly half of the induced mRNAs of Arabidopsis seedlings encode proteins of unknown function, of which over 40% had up-regulated orthologs in poplar (Populus trichocarpa), rice (Oryza sativa), or Chlamydomonas reinhardtii. Sixteen HYPOXIA-RESPONSIVE UNKNOWN PROTEIN (HUP) genes, including four that are Arabidopsis specific, were ectopically overexpressed and evaluated for their effect on seedling tolerance to oxygen deprivation. This allowed the identification of HUPs coregulated with genes associated with anaerobic metabolism and other processes that significantly enhance or reduce stress survival when ectopically overexpressed. These findings illuminate both broadly conserved and plant-specific low-oxygen stress responses and confirm that plant-specific HUPs with limited phylogenetic distribution influence low-oxygen stress endurance.
Oxygen is required for the efficient production of ATP by plants and other aerobes. Despite the extremely high affinities for oxygen of the oxidases involved in aerobic respiration (Km of 0.08–1 μm; Hoshi et al., 1993), obligate and facultative aerobes regularly experience oxygen deprivation for myriad reasons. Although oxygen is a by-product of photosynthesis, plants lack a circulatory system to mobilize oxygen to heterotrophic roots, tubers, meristems, germinating pollen, and developing seeds. These and flooded organs are vulnerable to oxygen deficiency. In mammals, intermittent tissue or cellular hypoxia can occur due to inhibition of pulmonary respiration (i.e. sleep apnea; Azad et al., 2009) and blood flow (i.e. stroke; Mense et al., 2006), a low-oxygen environment (i.e. high altitude), or high cellular density and metabolic activity (i.e. tumor cells [Chen et al., 2008] and ischemia [Fang et al., 2009]). In the case of microbes, oxygen availability is influenced by the density and identity of surrounding organisms and can be modulated over the course of a day or season, as observed in the microbial mats found in thermal springs (Steunou et al., 2006). It is not surprising that organisms can be predisposed to endure or avoid a low-oxygen environment. For example, submergence-tolerant rice (Oryza sativa) varieties adopt a quiescence strategy that limits the consumption of energy reserves and growth when covered by deep floodwaters, whereas deepwater rice varieties that experience a progressive deep flood during establishment can accelerate elongation growth to extend leaves above water (Fukao et al., 2006; Hattori et al., 2009). Other constitutive or induced adaptations of plants to oxygen deficiency include the formation of aerenchyma connecting aerial and underwater organs and alterations in organ or subcellular architecture of roots and leaves to oxygen status (Bailey-Serres and Voesenek, 2008). Animals also display adaptations that improve oxygenation, ranging from changes in vascular physiology and development (Bertout et al., 2008; Bishop-Bailey, 2009) to the formation of gills (van der Meer et al., 2005).
At the cellular level, a common eukaryotic response to oxygen deficiency is the Pasteur effect, whereby glycolysis and fermentation are promoted and the tricarboxylic acid (TCA) cycle and mitochondrial respiration are repressed (animals [Hochachka et al., 1996; Papandreou et al., 2006], yeast [Kwast et al., 2002], and plants [Bailey-Serres and Voesenek, 2008]). However, some organisms greatly curtail energy consumption to conserve precious reserves and extend survival during prolonged oxygen deficiency (i.e. turtles [Bickler and Buck 2007] and rice [Bailey-Serres and Voesenek, 2008]). Low-oxygen-induced metabolic adjustments involve sensing and signaling mechanisms that precipitate complex reconfiguration in gene expression that are far better understood in animals, fungi, and bacteria than in plants (Bruick, 2003; Bailey-Serres and Chang, 2005). The production and detoxification of reactive oxygen species (ROS) is likely to play a widespread role in signaling and damage at the onset and release from low-oxygen stress (Bailey-Serres and Chang, 2005; Rhoads and Subbaiah, 2007; Semenza, 2007).
Genome-scale technologies facilitate the appraisal of responses to environmental stimuli in a wide variety of organisms but are generally used to decipher responses within a single species. The interrogation of multispecies data sets within and across kingdoms can recognize evolutionarily conserved and species-specific responses to environmental perturbation. A meta-analysis of transcriptomic data from five species exposed to hydrogen peroxide stress identified common transcriptional adjustments, including one protein family globally induced in prokaryotes and eukaryotes (Vandenbroucke et al., 2008). Despite the growing number of independent evaluations of plant (Arabidopsis [Arabidopsis thaliana], poplar [Populus trichocarpa], rice, and Chlamydomonas reinhardtii), animal, fungal, and bacterial responses to low oxygen, there has been no systematic comparison of responses across plant species, kingdoms, or experimental systems. Here, we took advantage of 34 published microarray data sets that record the responses to varying degrees of oxygen deficiency of 21 organisms, representing plants, animals, fungi, and bacteria. The coupling of gene expression data with protein ortholog assignments exposed conserved and species-specific responses. The regulation of mRNAs associated with core cellular responses, including reconfiguration of central carbon metabolism and induction of heat shock proteins (HSPs), was generally preserved across organisms. By contrast, signaling protein and transcription factor (TF) genes responding to the stress were largely plant or species specific. We also considered genes encoding hypoxia-responsive proteins of unknown function that were highly coexpressed with genes associated with the core metabolic adjustments but varied in species distribution (Horan et al., 2008). We confirmed that transgenic Arabidopsis seedlings that ectopically express HYPOXIA-RESPONSIVE UNKNOWN PROTEIN (HUP) genes exhibit altered endurance of low-oxygen stress.
RESULTS AND DISCUSSION
Data Collection and Identification of Differentially Expressed Genes
Our initial objective was to perform a meta-analysis of alterations in mRNA abundance in response to transient oxygen deprivation in evolutionarily distant species, including model monocot and eudicot, animal, fungal, and bacterial species. Publicly available gene expression data were obtained for 21 species, in which stress conditions varied from anoxia to partial oxygen deprivation (i.e. hypoxia, 0.1%–10% oxygen; Supplemental Table S1). The plant survey included experiments that monitored the responses of Arabidopsis seedlings to varying degrees of hypoxia, poplar roots to flooding, rice seeds and coleoptiles to anoxia, rice seedling leaves to submergence, and Chlamydomonas cells to anoxia. Gene transcripts that were significantly up- or down-regulated in response to the experimental treatments were identified as differentially expressed genes (DEGs) by uniform processing of the data sets, using 1.8- or 2-fold change in abundance and a false discovery rate of 0.1 to 0.01 as criteria for differential expression (Supplemental Tables S2 and S3). In cases where raw microarray data were unavailable, published confidence values were used.
The overall number of DEGs in each experiment and organism varied as expected for a data set comprising diverse experimental parameters and species (Fig. 1A; Supplemental Table S1). Notably, the highest numbers of DEGs prevailed in plant samples, with up to 2,996 induced and 3,042 reduced mRNAs. In most cases, the number of induced DEGs exceeded the number of reduced DEGs; however, the number of reduced DEGs was greater when treatment duration was extended. Most animal data sets identified fewer than 100 DEGs, with the exception of stagnant human cell cultures (more than 1,000 DEGs; Guimbellot et al., 2009). DEG numbers in microorganisms were similar to those of animals, despite their lower gene contents. Notably, the facultative anaerobes Saccharomyces cerevisiae and Escherichia coli, which reconfigure metabolism and sustain growth under low oxygen, displayed the largest number of DEGs of the microorganisms sampled. Two other trends were observed: DEG number increased with the severity of stress in plants and bacteria, and it was influenced by the duration of the treatment in plants and to a lesser degree in yeast (Fig. 1, B and C; Supplemental Table S1). This overview confirms that low-oxygen stress induces a programmed transcriptomic response in organisms of four kingdoms, with the most dramatic reconfiguration in plants. This marked adjustment of the plant transcriptome most likely reflects complex strategies of acclimation and adaptation.
Figure 1.
Number of induced and repressed genes following oxygen deprivation in diverse organisms. A, Bars indicate the minimum and maximum number of DEGs recognized when multiple studies were evaluated (Supplemental Table S1). Induced DEGs are plotted as positive numbers; reduced DEGs are plotted as negative numbers. Species investigated are as follows: Arabidopsis thaliana, Populus trichocarpa, Oryza sativa, Chlamydomonas reinhardtii, Homo sapiens, Mus musculus, Danio rerio, Drosophila melanogaster, Caenorhabditis elegans, Trichoderma reesei, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, Cryptococcus neoformans, Escherichia coli, Mycobacterium tuberculosis, Pseudomonas aeruginosa, Pseudomonas stutzeri, Synechocystis species PCC 6803, Sinorhizobium meliloti, and Zymomonas mobilis. B, Responses of organisms over a range of oxygen concentrations and time. C, Responses of organisms over time. gen, Generation.
Recognition of Orthologous Induced and Reduced Genes
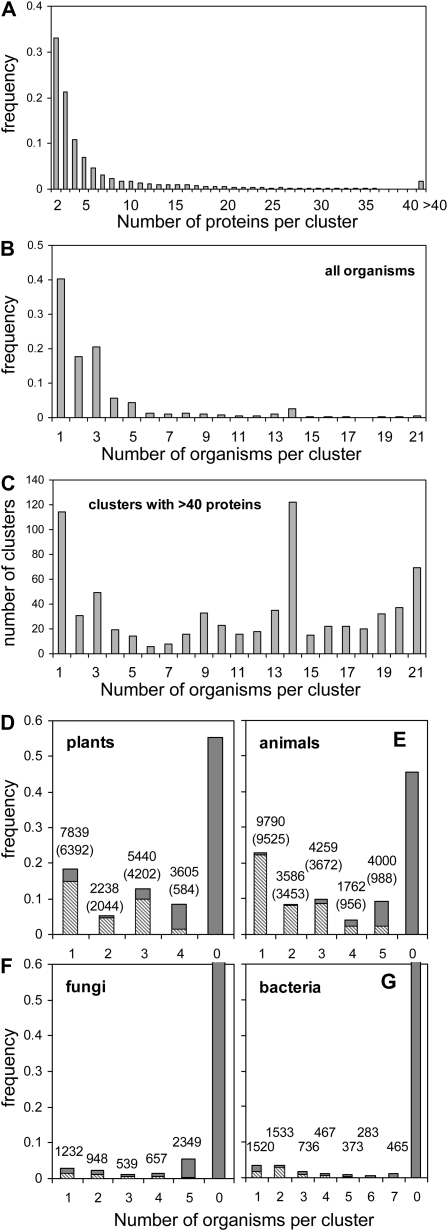
To compare the transcriptomic response across these diverse species, it was necessary to identify genes encoding evolutionarily related proteins. To accomplish this, the proteomes of the 21 organisms were coclustered into protein families using the OrthoMCL method (OMCL), which performs an all-against-all BLAST comparison of protein sequences with subsequent Tribe-Markov clustering (Li et al., 2003). The analysis of 367,643 proteins in total resolved 42,830 clusters of related proteins with two to 2,185 proteins per OMCL cluster (Supplemental Tables S4 and S5). For simplicity, the proteins within a single cluster (family) are termed orthologous, although conservation in function was not tested. We found that clusters with only two or three proteins were the most frequent (33% and 21% of all clusters, respectively), with the frequency decreasing with the number of proteins per cluster (Fig. 2, A and B). Clusters with more than 40 proteins included highly expanded gene families in one or a few organisms, such as the transposable element proteins of rice and known evolutionarily conserved proteins including glycolytic enzymes (glyceraldehyde 3-phosphate dehydrogenase, pyruvate kinase, enolase), ribosomal proteins, tRNA synthetases, RNA polymerases, and ATP-binding cassette transporters (Supplemental Table S4). Protein clusters with representatives from a few species were the most frequent, as recognized previously (Li et al., 2003; Vandenbroucke et al., 2008). For example, 40% of the clusters contained genes from one organism, whereas only 0.4% possessed entries from all 21 organisms (Fig. 2C). Over 1,000 clusters (2.4%) included entries from the 14 eukaryotes in the data set, and 38 clusters (0.1%) included exclusively entries from the seven prokaryotes evaluated. The analysis recognized 19,122 clusters with plant genes (Supplemental Table S4). Of these, only 8.4% (3,605) had entries in all four species (Fig. 2D), and most of these clusters (84%) also included proteins from nonplant species. By contrast, clusters limited to one to three higher plants were predominantly kingdom specific (Fig. 2D, lighter gray sections of bars). For example, Arabidopsis proteins were present in 11,806 clusters, from which 8,951 (75%) were conserved across higher plants and 1,066 (9%) were Arabidopsis specific. Similar trends were evident for animal clusters (Fig. 2E).
Figure 2.
OMCL clustering of 367,643 proteins from 21 organisms resolved 42,830 clusters/protein families. A, Cluster size frequency. B, Frequency of number of organisms per cluster. C, Distribution of clusters with more than 40 genes across organisms. D to G, Frequency of organisms per cluster in individual kingdoms. The 0 bar indicates clusters not identified in the kingdom. Values above bars are numbers of protein clusters. Lighter gray regions indicate frequency of clusters limited to kingdom, with numerical values in parentheses.
The small clusters of evolutionarily distinct proteins are likely to reflect the ongoing ontogeny and evolution of proteins. Indeed, proteins of unknown function (PUFs) and proteins of no defined motif tend to be species specific (Gollery et al., 2006; Horan et al., 2008). Reflective of this is the observation that 30% (3,595) of the low-oxygen-induced DEGs and 25% (3,353) of the reduced DEGs found in one of the four plants evaluated encode a PUF (Supplemental Fig. S1; Supplemental Table S6). Although species-specific stress-induced PUFs can play a role in abiotic stress tolerance (Luhua et al., 2008), the approximately 450 cross-species-conserved hypoxia-responsive unknown proteins (e.g. HUPs) that are induced in multiple plant species are attractive targets for reverse genetic analysis.
Broadly Conserved Induced and Reduced Genes
To recognize evolutionarily conserved versus species-specific responses to oxygen deprivation, 12,685 induced and 15,966 reduced DEGs were sorted to an OMCL cluster, and enrichment of specific clusters was tested by hypergeometric distribution analyses (Supplemental Table S6). This identified 2,409 clusters that contained two or more induced DEGs. Twenty percent (466 clusters) included members from three or more organisms. The cluster with induced DEGs from the greatest number of organisms (11 species, four kingdoms) encoded sugar transporters; enzymes of primary carbon metabolism, secondary metabolism (i.e. coproporphyrinogen III oxidase, C-5 sterol desaturase, squalene monooxygenase), HSPs, and enzymes that ameliorate ROS were also induced in the majority of the species (Supplemental Fig. S2; Supplemental Table S6b). Mitogen-activated protein kinases (MAPKs) were the only signal transduction pathway component induced in diverse eukaryotes (plants, human, and yeast). The broadly reduced DEGs resolved into 3,136 clusters with two or more proteins, of which 719 clusters contained sequences from three or more organisms (Supplemental Fig. S2; Supplemental Table S6d). The most frequently represented clusters encoded ribosomal proteins, TCA cycle enzymes, and aerobic respiratory complex subunits (i.e. isocitrate dehydrogenase, F0-ATPase δ -subunit; Supplemental Figs. S2 and S3). Gene Ontology (GO) analysis confirmed that many species reduced mRNAs encoding proteins associated with aerobic respiration (2.77E-12) and biogenesis of ribosomes (7.59E-34), organelles (1.95E-09), and cell walls (1.06E-11; Supplemental Table S7d). The decline in mRNAs associated with biosynthetic processes strongly suggests that restriction of ATP-consuming processes is an evolutionarily conserved coping mechanism. Interestingly, a reduction in mRNA abundance is not a prerequisite for down-regulation of gene expression, since the transient selective repression of translation occurs in both animals and plants during low-oxygen stress (Thomas and Johannes, 2007; Branco-Price et al., 2008).
Diversity in Anaerobic Energy Production
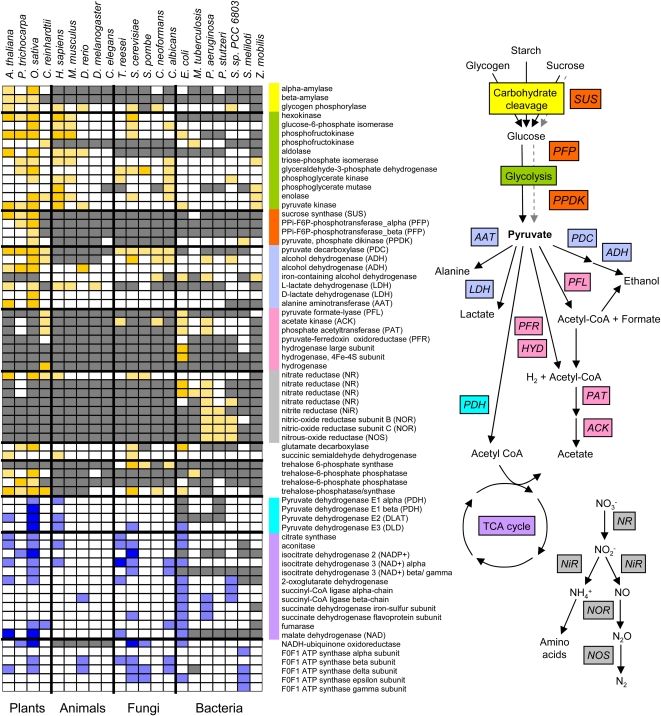
In general, oxygen deprivation reconfigures central carbon metabolism to promote substrate-level ATP production (Fig. 3; Supplemental Fig. S4). Frequently, glycolysis is activated and the excess NADH is recycled through fermentative metabolism of pyruvate, regenerating the NAD+ needed to maintain glycolytic flux. However, there is remarkable diversity in anaerobic metabolism. In animals, anaerobic metabolism of pyruvate is limited to the formation of lactate (lactate dehydrogenase in OMCL2360; Supplemental Table S6). In plants, lactate is not the major fermentation end product; rather, pyruvate is predominantly converted to acetaldehyde by pyruvate decarboxylase (PDC; OMCL1074), which is further metabolized to ethanol by alcohol dehydrogenase (ADH; OMCL198 and OMCL286; Branco-Price et al., 2008; van Dongen et al., 2009). Consistently, PDC and ADH were induced in rice, Arabidopsis, and poplar. Plants also produce Ala (Ala aminotransferase [OMCL240] and Asp aminotransferase [OMCL347]) and γ -aminobutyric acid (Glu decarboxylase [OMCL595]) and accumulate succinate under low-oxygen stress (Bailey-Serres and Voesenek, 2008). PDC and ADH were also induced in the five fungal species evaluated, which produce ethanol under anaerobiosis (Fig. 3; Supplemental Table S6). In bacteria, pyruvate metabolism is considerably more complex. It can be converted into acetyl-CoA and formate by pyruvate-formate lyase (OMCL17942) or to acetyl-CoA and hydrogen by pyruvate-ferredoxin oxidoreductase and hydrogenases (OMCL12153, OMCL22614, and OMCL40976). Acetyl-CoA can be further metabolized into acetate by the sequential action of phosphate acetyltransferase (OMCL6584) and acetate kinase (OMCL4953) or into ethanol by ADH-E (OMCL1642). The algae Chlamydomonas presents a special case, in which anaerobic metabolism of pyruvate can be metabolized to lactate or ethanol via the pathways characteristic of higher plants as well as to formate, hydrogen, and acetate by the pathways characteristic of bacteria (Mus et al., 2007; Fig. 3).
Figure 3.
OMCL clusters of genes encoding proteins involved in glycolysis, fermentation, and the TCA cycle. Colors of bars correspond to proteins in the metabolic pathway at right. A single gene can be represented by multiple OMCL groups because of multisubunit enzyme complexes or large evolutionary differences in genes encoding the protein. Bright yellow, cluster has more than one induced DEG in that organism; light yellow, cluster has one induced DEG in that organism; bright blue, cluster includes more than one reduced DEG in that organism; light blue, cluster includes one reduced DEG in that organism; white, no DEG ortholog identified in that organism; gray, orthologous cluster/protein not identified in that organism. Extended data are shown in Supplemental Figure S4.
Microorganisms can additionally produce nitrogen/nitric oxide to facilitate anaerobic survival (nitrate reductase [NR], OMCL10813, OMCL10728, and OMCL16045; nitrite reductase, OMCL34060; nitric oxide reductase, OMCL23279 and OMCL15967; nitrous oxide reductase, OMCL23273 [Sherman et al., 2001; Alvarez-Ortega and Harwood, 2007]). Interestingly, plants induce NRs under low-oxygen stress (OMCL130), although the enzyme was distinguished from the bacterial NRs by OMCL analysis. A function of plant NR could be, as for bacterial NRs, the removal of reduced NADH. Accordingly, plants survive low-oxygen stress for longer periods if nitrate is provided rather than ammonia (Fan et al., 1997; Allègre et al., 2004). NR mutants of tobacco (Nicotiana tabacum) are also more sensitive to hypoxia than the wild type (Stoimenova et al., 2003).
Substrate-level ATP production requires the catabolism of soluble or stored carbohydrates (i.e. Suc and starch, or glycogen in animals). To circumvent the exhaustion of soluble carbohydrate reserves, enzymes responsible for the cleavage of stored carbohydrates such as glycogen phosphorylases and related proteins activate the catabolism of stored carbohydrates (OMCL873). Genes encoding these enzymes are induced in representatives of all four kingdoms. Plants notably have alternative catabolic pathways that allow hydrolysis of starch to Glc by amylases (OMCL1389 and OMCL18208; Loreti et al., 2003) and ATP-independent catabolism of Suc by Suc synthase (OMCL855; Ricard et al., 1998). A complete set of starch-degrading enzymes, including α -amylase, β -amylase, debranching enzyme, and α -glucosidase, were induced in coleoptiles and germinating seeds of rice (Supplemental Fig. S5; Lasanthi-Kudahettige et al., 2007; Narsai et al., 2009). However, amylases were not up-regulated strongly in all plants and organs (i.e. Arabidopsis seedlings, poplar roots), indicating that starch reserves may be unavailable or inaccessible. It was found previously that low-oxygen-sensitive seeds of cereal species, such as wheat (Triticum aestivum) and barley (Hordeum vulgare), do not induce amylases under anoxia, in contrast to seeds of anaerobic germination-competent rice seeds (Guglielminetti et al., 1995). Neither Arabidopsis seedlings nor poplar roots induced amylases under oxygen deprivation; however, we cannot exclude amylase induction in seeds or other organs in these species.
Accompanying the induction of genes associated with the mobilization of sugars into glycolysis is the elevation of glycolytic enzyme and the decline in TCA cycle enzyme mRNAs in species of all four kingdoms (Fig. 3; Supplemental Figs. S4 and S5; Supplemental Table S6). Glycolytic enzymes are often encoded by gene families in eukaryotes with differentially regulated individual members. The reduced DEGs encoding TCA cycle enzymes and mitochondrial respiratory complex subunits were broadly conserved, although less evident in animals. mRNAs encoding pyruvate dehydrogenase kinase, which inhibits pyruvate dehydrogenase, thereby limiting pyruvate entry into the TCA cycle, were induced in studies of humans and plants (OMCL985; Papandreou et al., 2006; Supplemental Fig. S6). Translational control of gene expression reduces TCA cycle enzyme synthesis in Arabidopsis, as demonstrated by the rapidly reversible shift of these mRNAs off of polysomes during low-oxygen stress (Branco-Price et al., 2008).
The cross-species analysis supports the hypothesis that plants enhance overall ATP production during an energy crisis by utilizing alternative glycolytic enzymes that consume pyrophosphate (PPi) instead of ATP. These include PPi-Fru-6-P phosphotransferase and pyruvate-orthophosphate dikinase (Bailey-Serres and Voesenek, 2008). This strategy was observed at all developmental stages evaluated for rice, which induced PPi-Fru-6-P phosphotransferase subunits, pyruvate-orthophosphate dikinase (OMCL946), and isoforms of the phosphoenolpyruvate carboxykinase (OMCL2547; Huang et al., 2008), thereby improving energy usage in pyruvate metabolism under anoxia (Fig. 3; Supplemental Fig. S5). Genes encoding these PPi-dependent enzymes were more strongly induced in leaves of submergence-tolerant rice [compare M202(Sub1) with M202 in Supplemental Table S8]. Furthermore, a vacuolar PPi-dependent proton pump (OMCL1960), an alternative to ATP-dependent proton pumps, was preferentially induced in submergence-tolerant rice during submergence (Supplemental Fig. S5; Supplemental Table S8; Liu et al., 2010). As confirmed by mutational analyses of Suc synthase genes (Ricard et al., 1998; Bieniawska et al., 2007) and by decreasing PPi content of potatoes (Solanum tuberosum; Mustroph et al., 2005), these observations indicate a benefit of ATP-independent enzymes during low-oxygen stress in plants.
The meta-analysis also illuminated the evolutionarily conserved importance of trehalose-6-phosphate (T6P) metabolism (Fig. 3; Supplemental Fig. S4). T6P synthase (T6PS) transcripts (OMCL252) were induced in seven of 21 organisms under hypoxia, particularly in plants, fungi, and E. coli. Additionally, a T6P phosphatase was induced in plants and fungi (OMCL1954 and OMCL5544). T6P regulates glycolysis in yeast by inhibition of hexokinase (Blázquez et al., 1993). In plants, elevated T6P activates the starch-synthesizing enzyme ADP-Glc pyrophosphorylase, which subsequently limits glycolysis (Paul, 2007). In Arabidopsis, elevation of T6P increased growth by limiting the activity of the energy-sensing SNF1-related protein kinase 1 (SnRK1/AtKIN10/AtKIN11; Zhang et al., 2009). We found that submergence-tolerant rice induced three T6PS mRNAs to a greater extent than intolerant rice, raising the possibility that modulation of T6P may be relevant to the regulation of growth during submergence (Supplemental Table S8).
Conserved Induction of Stress Response Protein Genes
The transition to hypoxia and return to oxygenation promote the formation of ROS. The meta-analysis of ROS-regulated mRNAs found that HSPs were induced in both prokaryotes and eukaryotes (Vandenbroucke et al., 2008). Similarly, this study confirmed the induction of HSP and ROS network mRNAs in 16 and 10 organisms, respectively, including all the plants evaluated (Supplemental Fig. S7A). In addition to HSP mRNAs, heat shock TFs were DEGs in plants and yeast. Consistent with a functional role of HSPs during the stress, pre-heat-stressed Arabidopsis seedlings were more tolerant to anoxia (Banti et al., 2008, 2010). The prevalence of induced DEGs associated with the amelioration of ROS suggests that their detoxification is important for low-oxygen stress survival in diverse organisms (Supplemental Fig. S7B). Consistent with this, a correlation with the ability to ameliorate ROS and survival was reported for Iris pseudacorus (Monk et al., 1987) and rice (Ella et al., 2003). Along with ROS amelioration enzymes, plants uniformly induced mRNAs encoding the mitochondrial alternative oxidase (OMCL4174), which limits ROS formed by the mitochondrial electron transport chain upon reoxygenation (Rhoads and Subbaiah, 2007). Along with alternative oxidase, a conserved mitochondrial uncoupling protein (OMCL215) was an induced DEG in plants and animals (Supplemental Fig. S2). Finally, plasma membrane-localized respiratory burst oxidases that are implicated in signaling during hypoxia in plants and animals (Baxter-Burrell et al., 2002; Schroeder et al., 2009) were induced DEGs in Arabidopsis and Caenorhabditis elegans (OMCL290; Supplemental Table S6). The induction of HSPs and ROS network proteins in diverse organisms provides convincing evidence that the low-oxygen response includes components of a universal stress response (Feder and Hofmann, 1999; Swindell et al., 2007; Timperio et al., 2008).
Limited Conservation of Regulatory Proteins
Despite the conserved reconfiguration of primary metabolic enzymes and stress-associated protein mRNAs, there was limited conservation in the regulation of protein kinase (PK) and TF mRNAs (Supplemental Fig. S6). Exceptions included conservation in low-oxygen-induced mRNAs encoding MAPKs and MAPKKs (OMCL224 and OMCL302; Supplemental Table S6) as well as the CAMK-type kinases associated with energy sensing (i.e. OMCL212 and OMCL1019, including mammalian AMP-activated kinases [AMPKs; Fisslthaler and Fleming, 2009] and yeast SNF1 kinase [Hedbacker and Carlson, 2008]). Orthologs of the noncatalytic β -subunit of AMPK/SNF1 were induced in Arabidopsis, rice, and yeast (OMCL1419) but not in animals. By contrast, orthologs of the catalytic α -subunit were induced in rice (Os08g37800; SnRK1B) and the fungus Trichoderma reesei (OMCL1019). Although the Arabidopsis SNF1s AtKIN10 and AtKIN11 were not induced by hypoxia, the posttranslational regulation of these kinases mediates the sensing of energy status and gene regulation in leaf protoplasts (Baena-González et al., 2007). Interestingly, rice SnRK1A (Os05g45420 and OMCL1019) is regulated by a calcineurin B-like-interacting protein kinase (CIPK15; Os11g02240 and OMCL39391) and is necessary to promote starch degradation during germination under low-oxygen conditions (Lee et al., 2009). The absence of close orthologs of CIPK15 in other plants may explain the unusual capacity of rice to germinate under anaerobic conditions. The AGC/RPS6K kinase family (OMCL318) also had DEGs in multiple organisms, including induced ribosomal S6-kinase members in humans (NP_037389), yeast (YMR104C), and higher plants (Supplemental Fig. S6; Supplemental Table S6). Although S6-kinase mRNAs are elevated in Arabidopsis seedlings by oxygen deprivation, they are not engaged with polysomes until reoxygenation (Branco-Price et al., 2008). A number of plant-specific PK mRNAs were differentially regulated by hypoxia in a species- or genotype-specific manner, including SHR5 receptor-like (OMCL123), Ser/Thr (OMCL139), and APK1A-like (OMCL5149) PKs (Supplemental Table S6). For example, some SHR5 receptor-like PKs had significantly stronger induction in leaves of submergence-tolerant rice, although the significance of the differential regulation remains unknown (Supplemental Table S8). The receptor-like PKs induced in Chlamydomonas were also largely distinct from those in higher plants (i.e. cluster OMCL39).
The meta-analysis also resolved differential expression of TF mRNAs in plants and other organisms (Supplemental Fig. S6). The regulation of mRNAs encoding factors involved in low-oxygen sensing was consistent with published reports (i.e. induction of Sinorhizobium meliloti and Pseudomonas aeruginosa FixJ/FixK TFs [Green et al., 2009; OMCL8812], Mycobacterium tuberculosis DosR TF [Sherman et al., 2001; OMCL2741], fungal Hap1/Rox1 TFs that measure cellular heme [Hon et al., 2003; OMCL4393 and OMCL1263], and MGA2 that regulates fatty acid desaturase [Jiang et al., 2001; OMCL6345] and no induction in posttranscriptionally regulated TFs, including E. coli redox-sensing ArcA/ArcB system and FNR TFs [Shalel-Levanon et al., 2005; Malpica et al., 2006] and mammalian hypoxia-inducible factor-1 α -subunit [Bruick, 2003]; Supplemental Table S6). Plants lack orthologs of bacterial, yeast, and animal TFs associated with direct or indirect low-oxygen sensing (Bailey-Serres and Chang, 2005). However, orthologs of the oxygen-dependent prolyl hydroxylases, which sense physiological decline in oxygen in animals (EGL9 homolog [OMCL2339]; Supplemental Table S6; Bruick, 2003), were induced DEGs in multiple plants (OMCL700; Vlad et al., 2007), although their function in low-oxygen responses in plants is unknown. The meta-analysis of plant DEGs recognized the prevalent induction of TFs encoding the ethylene-responsive factor (ERF; MCL50), MYB (MCL46), CCCH-type zinc finger (MCL767), WRKY (MCL92), and bZIP (MCL196) family members (Supplemental Fig. S6; Supplemental Table S6). Several of these TFs affect survival in low-oxygen or flooding stress (i.e. MYBs [Hoeren et al., 1998; Lee et al., 2007; Mattana et al., 2007], ERFs [Fukao et al., 2006; Xu et al., 2006; Hattori et al., 2009], and NAC [Christianson et al., 2009]). Despite conservation of some TF families between higher plants and Chlamydomonas (Supplemental Table S4), there was only limited induction of closely related TFs in the plant and algae species evaluated. These included a family of MADS TFs, which were induced in Arabidopsis, rice, and Chlamydomonas and also in humans and yeast (OMCL171), and an orphan TF family (OMCL7327) induced in Arabidopsis, rice, and Chlamydomonas (Supplemental Table S6). Overall, this survey indicates that the TFs that mediate responses to oxygen deprivation are evolutionarily new relative to the conserved cellular acclimations they regulate.
Evolutionarily Divergent Adaptive Responses That Mediate Oxygen Delivery
Multicellular animals and plants can alter development in response to low-oxygen stress to facilitate oxygen delivery. For example, mammals can adjust pulmonary respiration rate, increase development of capillaries in hypoxic tissues, and stimulate red blood cell formation, as evidenced by enrichment in DEGs encoding proteins associated with these processes (Supplemental Tables S6 and S7a; i.e. vascular endothelial growth factor [OMCL4003; Ferrara et al., 2003] and adrenomedullin [OMCL16253; Guidolin et al., 2008]). Plant adaptation to a semiaquatic environment includes the formation of gas-filled aerenchyma that connect aerial leaves with submerged stems and roots, elongation of underwater stems and leaf petioles, and the initiation of adventitious roots (Bailey-Serres and Voesenek, 2008). Both the development of aerenchyma and underwater elongation growth involve ethylene. Plant-specific DEGs included several genes associated with ethylene biosynthesis (i.e.1-amino-cyclopropane-1-carboxylate synthase [OMCL714] and 1-amino-cyclopropane-1-carboxylate oxidase [OMCL28112]) and perception (OMCL4190). The induction of members of the expansin (OMCL4264 and OMCL6686) and xyloglucan endotransglucosylase (OMCL1722 and OMCL2239) gene families, which are involved in cell elongation growth, was also observed in all higher plants examined (Supplemental Table S5).
Conserved Versus Species-Specific Low-Oxygen-Responsive Genes of Plants
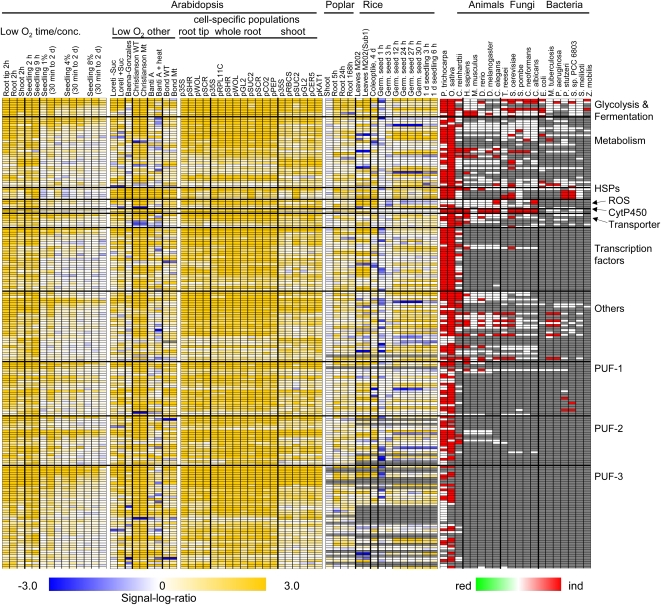
Our secondary objective was to recognize the core of plant-specific genes that respond to low-oxygen stress using Arabidopsis as a reference plant and model for reverse genetic analyses. Toward this goal, we examined data from seedling experiments that varied in duration or degree of oxygen deprivation, cell type, or genotype (Fig. 4; Supplemental Table S9, a and b). The analysis exposed strong similarity in the most highly induced DEGs across Arabidopsis experiments, despite the variation in experimental parameters. The most highly induced DEGs of Arabidopsis were also induced in multiple cell-specific mRNA populations obtained by immunopurification of polysomes (Mustroph et al., 2009). The elevation of transcripts was less dramatic under less severe conditions and in shoots, as evidenced by lower induction of HSPs, cytochrome P450s, and a number of TF mRNAs. A similar evaluation was performed with human data sets (Supplemental Fig. S8; Supplemental Table S10). mRNAs associated with anaerobic metabolism and signaling were induced in most root and shoot cell types. However, in contrast to plants, the survey of human cell types from different organs revealed considerable variation in induced DEGs of cells with distinct identity (Chi et al., 2006). The analysis also considered the reduced DEGs. As might be expected due to distinctions between cell types, the reduced mRNAs showed greater variation between cell-specific populations in both Arabidopsis and humans (Supplemental Figs. S9 and S10). In conclusion, the extent to which metabolism is reconfigured depends not only upon environmental parameters (Fig. 1) but also on genotype, organ, and cell type in plants and animals (Ellis et al., 1999; Chi et al., 2006; Mustroph et al., 2009).
Figure 4.
Low-oxygen-induced genes of Arabidopsis: expression across experiments, cell types, and orthologs in other organisms. The top 200 induced DEGs were selected from Supplemental Table S9. Each row represents one gene. Genes were manually grouped into functional categories. Left, Signal-log ratio values for genes across different plant experiments (Arabidopsis experiments, Loreti et al., 2005; Baena-González et al., 2007; Branco-Price et al., 2008; Bond et al., 2009; Christianson et al., 2009; Mustroph et al., 2009; van Dongen et al., 2009; Banti et al., 2010; Arabidopsis cell-specific populations, Mustroph et al., 2009, root tip, whole root, whole shoot; poplar experiments, Kreuzwieser et al., 2009; rice experiments, Lasanthi-Kudahettige et al., 2007; Narsai et al., 2009; this study). Poplar and rice orthologs were identified by selecting the most highly expressed probe set either belonging to the same OMCL group as the Arabidopsis proteins or having a BLASTP similarity E value of 10−6 or less. conc., Concentration. Right, Orthologs in different organisms from plants (P. trichocarpa, O. sativa, Chlamydomonas), animals (H. sapiens, M. musculus, D. rerio, D. melanogaster, C. elegans), fungi (T. reesei, S. cerevisiae, S. pombe, C. neoformans, C. albicans), and bacteria (E. coli, M. tuberculosis, P. aeruginosa, P. stutzeri, S. meliloti, Synechocystis species PCC 6803, Z. mobilis). Orthologs were identified by a BLASTP similarity cutoff of E ≤ 10−6. Red, induced orthologs present in that organism; white, ortholog(s) not DEG(s); gray, no ortholog in that organism.
To more accurately identify evolutionarily related genes with similar regulation in diverse species, we performed a BLASTP comparison of Arabidopsis proteins with whole proteomes of other organisms (Supplemental Table S9e). This provided a similarity score (E value) between proteins, allowing identification of proteins with strong to limited similarity (cutoff E ≤ 10−6). Of the 200 most highly induced DEGs of Arabidopsis, 75% had induced orthologs in rice leaves, coleoptiles, and/or germinating seeds (Fig. 4; Supplemental Table S9e). In poplar, where the response of root waterlogging was assessed, 50% of the Arabidopsis genes had induced orthologs. This value was only 15% for oxygen-deprived Chlamydomonas. Consistent with the OMCL analysis, the highly induced genes were associated with fermentation, Suc transport, ROS regulation, and heat shock response. Nonplant species possessed induced putative orthologs of 48 of the 200 most induced Arabidopsis genes, primarily in the metabolic and HSP categories (Supplemental Table S9e). The 30 most highly induced TFs were plant specific, with the exception of a C3H zinc finger and MADS domain family protein with modest similarity to TFs induced in other eukaryotes. The most conserved induced TF mRNAs of higher plants were zinc finger and LOB domain proteins. Highly induced TFs included group VII ERFs associated with the regulation of submergence responses (i.e. rice Sub1A, Sub1C [Os09g11460], and ERF67 [Os07g47790]; poplar ERF [grail3_0003056501]; and Arabidopsis ERF71 [At2g47520] and ERF73 [At1g72360]). The conserved induction of group VII ERFs provides strong evidence for an ancestral role of this ERF TF subfamily in the survival of conditions with a low-oxygen component.
The analysis also identified a number of plant-specific induced DEGs encoding PUFs as annotated by the GO and/or SwissProt databases or based on the absence of a defined protein sequence motif in the PFAM database (Horan et al., 2008). Remarkably, nearly half (739) of the 1,860 low-oxygen-induced genes of Arabidopsis encode PUFs (e.g. HUP genes). Of these, 89% (659) have one or more putative ortholog in other plant species, including 58% (432) that are induced by one or more conditions with a low-oxygen component (Supplemental Fig. S11; Supplemental Table S9e). It is notable that 89 of the 200 most highly induced genes of Arabidopsis encode PUFs, most of which have orthologs limited to higher plants (Fig. 4). Examples of plant-specific hypoxia-responsive PUF families are universal stress proteins (OMCL2826 and OMCL3100), phloem-associated proteins (OMCL6676), and wound-responsive proteins (OMCL7410; Fig. 4; Supplemental Table S9c).
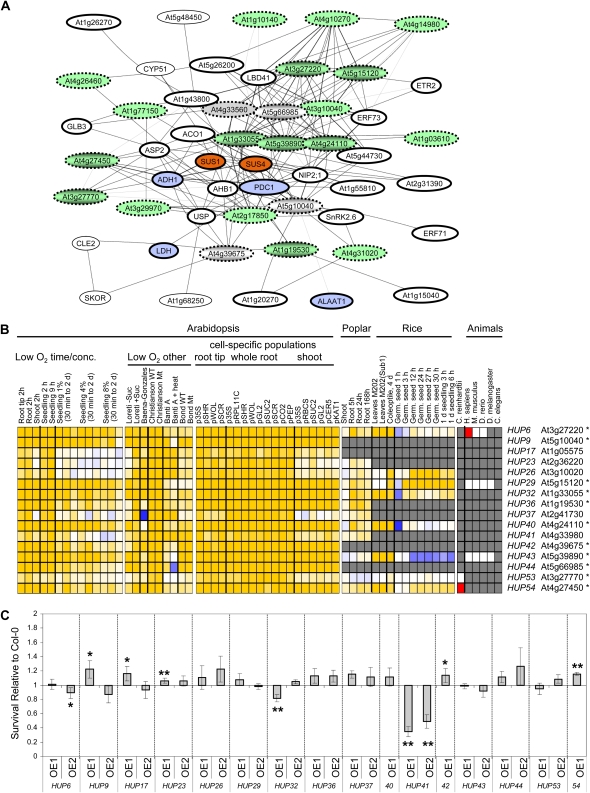
To investigate the role of proteins that lack a functionally defined protein domain in stress survival, mutants were established for 11 PUFs coexpressed with enzymes essential to anaerobic metabolism such as ADH1 (Fig. 5A) and five genes coexpressed with other stress-induced proteins (Table I). These genes, designated as HUPs, varied in number of gene family members, predicted subcellular location, presence versus absence in other plants, and cell-specific pattern of expression (Fig. 5B; Supplemental Table S9). Comparison of the survival of homozygous mutant seedlings and wild-type (ecotype Columbia [Col-0]) seedlings confirmed that ectopic overexpression of HUP cDNAs, by use of the nearly constitutive 35S promoter, significantly modified the tolerance of prolonged oxygen deprivation in 12 of the 16 genes evaluated, including four Arabidopsis-specific genes. Overexpression of HUP6, HUP9, HUP17, HUP23, HUP32, HUP41, HUP42, and HUP54 altered tolerance after 8 h of stress (Fig. 5C; Supplemental Fig. S12), whereas HUP6, HUP9, HUP17, HUP29, HUP36, HUP37, HUP41, and HUP44 overexpressors showed altered tolerance after 12 h of stress (Supplemental Fig. S12). This confirmed that genes in several coexpression networks influence low-oxygen endurance. However, only some of the HUP overexpression lines were affected in low-oxygen survival. This could be due to a number of reasons. For example, a complex network may regulate the stress response, with products of most genes acting as modifiers because of redundancy in gene function and pathways. Alternatively, the use of the nearly constitutive 35S promoter may be ineffective if the level, timing, location, and conditional nature of gene expression are important parameters in determining the response to a transient stress. Interestingly, overexpression of HUP6, HUP32, and HUP41 markedly reduced seedling tolerance of low oxygen, with the most dramatic decline observed in HUP41 overexpressors (Supplemental Fig. S12K). The reduction in tolerance as a result of ectopic expression may indicate that these genes are critical in counterbalancing aspects of the stress response, such as carbohydrate consumption or ROS production (Fukao and Bailey-Serres, 2004). By contrast, overexpression of HUP9 and HUP17 was correlated with enhanced tolerance in independent transgenic lines, consistent with constitutive accumulation of the protein (Supplemental Table S11). T-DNA insertion homozygotes evaluated for six of the HUPs were altered in stress survival (Supplemental Fig. S12), but these mutants had limited impact, possibly due to gene redundancy or incomplete gene disruption. Notably, the distribution and induction of HUP orthologs in the species evaluated was variable. HUP9, HUP17, HUP42, and HUP44 were Arabidopsis or Brassicaceae specific; HUP23, HUP36, HUP37, and HUP41 orthologs were present and induced in poplar; HUP29 was present and induced in Arabidopsis, poplar, and rice; and HUP54 was present and induced in higher plants and Chlamydomonas. These analyses confirm that PUFs with both broad and limited distribution within the plant kingdom can contribute to the survival of low-oxygen stress.
Figure 5.
HUP genes coregulated with genes associated with anaerobic metabolism modify stress survival. A, Gene coexpression network for key enzymes associated with Suc catabolism and fermentation from the analysis of Ma et al. (2007). ACO1, 1-Aminocyclopropane-1-carboxylate oxidase (At2g19590); ADH1, alcohol dehydrogenase (At1g77120); AHB1, Arabidopsis hemoglobin 1 (At2g16060); ALAAT1, Ala aminotransferase (At1g17290); ASP2, Asp aminotransferase (At5g19550); CLE2, clavata 3/ESR related (At4g18510); CYP51, cytochrome P450 (At1g11680); ERF71 and ERF73, ERF group VII family TFs (At2g47520 and At1g72360); ETR2, ethylene response receptor (At3g23150); GLB3, Arabidopsis hemoglobin 3 (At4g32690); LBD41, LOB domain family protein (At3g02550); LDH, lactate dehydrogenase (At4g17260); NIP2;1, NOD26-like intrinsic protein (At2g34390); PDC1, pyruvate decarboxylase 1 (At4g33070); SKOR, Shaker family potassium ion channel (At3g02850); SnRK2.6, At4g33950; SUS1 and SUS4, Suc synthases (At5g20830 and At3g43190); USP, universal stress protein (At3g03270). Induced genes are indicated by thick ellipses; fill colors correspond to Figure 3. HUPs are indicated by dashed ellipses in gray (limited to Arabidopsis and near relatives) or green (present Arabidopsis, poplar, and rice) ellipses with genes targeted in mutant analyses shaded. B, Low-oxygen-induced HUPs of Arabidopsis: expression across experiments, cell types, and orthologs in other plants. conc., Concentration. Data are from Figure 4. Genes indicated with asterisks are present in the coexpression network shown in A. C, HUP mutant seedling survival of 8-h oxygen deprivation in dim light. Seedling viability was compared with Col-0 seedlings grown on the same plate. Error bars represent sd (n = 3). Asterisks indicate significant differences in survival of mutant relative to Col-0 (* P ≤ 0.1, ** P ≤ 0.05).
Table I. HUP genes analyzed for function in low-oxygen survival.
| Gene Name | Arabidopsis Genome Initiative Locus | Ortholog(s) in Plants | Family Membersa | Molecular Mass | Coexpressed Genesb |
| kD | |||||
| HUP6 | At3g27220 | Rice, poplar | 1_1_1 | 48.5 | Anaerobic metabolism |
| HUP9 | At5g10040 | None | 2_1_1 | 9.8 | Anaerobic metabolism |
| HUP17 | At1g05575 | None | 2_2_1 | 9.5 | ERFs, WRKYs |
| HUP23 | At2g36220 | Poplarc | 2_2_1 | 29.0 | PP2C, ERF4 |
| HUP26 | At3g10020 | Rice, poplarc | 1_1_1 | 16.7 | HSPs, SnRK2, T6PS |
| HUP29 | At5g15120 | Rice, poplar | 5_2_1 | 33.0 | Anaerobic metabolism |
| HUP32 | At1g33055 | Rice, poplar | 1_1_1 | 7.5 | Anaerobic metabolism, ETR2, ZF |
| HUP36 | At1g19530 | Poplar | 1_1_1 | 13.7 | Anaerobic metabolism |
| HUP37 | At2g41730 | Poplarc | 2_2_2 | 13.3 | ACS2, NAC, ROS response |
| HUP40 | At4g24110 | Rice, poplarc | 1_1_1 | 28.6 | Anaerobic metabolism |
| HUP41 | At4g33980 | Poplarc | 1_1_1 | 24.6 | PDC2, ABF1, APRR5, ZF |
| HUP42 | At4g39675 | None | 1_1_1 | 8.2 | Anaerobic metabolism |
| HUP43 | At5g39890 | Rice, poplar | 19_7_1 | 30.8 | Anaerobic metabolism |
| HUP44 | At5g66985 | None | 1_1_1 | 9.4 | Anaerobic metabolism |
| HUP53 | At3g27770 | Ricec, poplarc | 8_1_1 | 36.7 | Anaerobic metabolism, KCO1 |
| HUP54 | At4g27450 | Rice, poplar | 5_2_2 | 27.6 | Anaerobic metabolism |
Number of Arabidopsis genes with 35% or greater, 50% or greater, or 70% or greater amino acid sequence identity from Horan et al. (2008).
Coexpression network based on data of Ma et al. (2007) and Horan et al. (2008). Anaerobic metabolism coexpressed genes include one or more of the following: ADH1, PDC1, SUS1, SUS4, and HB1. ABF1, Abscisic acid-responsive element-binding factor 1; ACS2, 1-aminocyclopropane-1-carboxylate synthase 2; APRR5, pseudoresponse regulator 5; ERF, ethylene responsive factor; ETR2, ethylene triple response 2; HSPs, heat shock proteins; KCO1, potassium channel 1; NAC, NAC domain TF family; PP2C, protein phosphatase 2C; SnRK2, SNF1-related protein kinase 2; T6PS, trehalose-6-phosphate synthase; WRKY, WRKY TF family; ZF, zinc finger protein.
Putative ortholog(s) identified but not induced by low oxygen (Supplemental Table S9e).
CONCLUSION
Here, the transcriptomic reconfiguration by low-oxygen stress in 21 organisms from four kingdoms was systematically evaluated to identify conservation and specificity in stress responses in plants. The grouping of proteins into OMCL clusters and recognition of protein families enriched in DEGs confirmed both broadly conserved and kingdom-specific characteristics in carbohydrate catabolism and fermentation. The carbohydrate catabolic pathways characteristic of higher plants contrasted with alternative metabolic pathways present in Chlamydomonas and some microorganisms. In addition to core metabolic reconfigurations that enhance substrate-level ATP production and NAD+ regeneration, many species displayed increases in mRNAs encoding HSPs and enzymes associated with ROS networks as well as decreases in mRNAs encoding proteins associated with ribosome biogenesis and cell wall synthesis. Despite these conserved responses, the components of signal transduction pathways and transcriptional regulators were markedly distinct across organisms, indicating independent evolution of the regulatory networks at the kingdom and phylum levels. The comparison of the low-oxygen transcriptomes obtained with diverse experimental systems for three land plants (Arabidopsis, poplar, and rice) and across cell types of the root and the shoot of Arabidopsis exposed a large degree of conservation in response, extending to regulatory proteins. This study also found that the low-oxygen-induced genes encoding PUFs (e.g. HUP genes) of Arabidopsis were primarily limited to higher plants, in contrast to genes associated with reconfiguration of metabolism and general stress tolerance. Phenotypic evaluation of mutants overexpressing HUPs confirmed a role for these poorly characterized proteins in low-oxygen survival. We propose that these phylogenetically limited proteins act as modifiers to fine-tune responses to low-oxygen stress, although further investigation is necessary to determine their precise function. This study demonstrates both uniformity and distinction in the response to low-oxygen stress across the biological kingdoms, highlighting the evolution of regulatory networks and acclimation responses unique to higher plants.
MATERIALS AND METHODS
Data Set Collection and Analysis
Low-oxygen response expression data were obtained for 21 organisms from databases and publications: Arabidopsis (Arabidopsis thaliana; Branco-Price et al., 2008; van Dongen et al., 2009; Mustroph et al., 2009), rice (Oryza sativa; Lasanthi-Kudahettige et al., 2007; Narsai et al., 2009), poplar (Populus trichocarpa; Kreuzwieser et al., 2009), Chlamydomonas reinhardtii (Mus et al., 2007), Homo sapiens (Kim et al., 2006; Chen et al., 2008; Guimbellot et al., 2009), Mus musculus (Greijer et al., 2005; Allen et al., 2006; Hu et al., 2006), Danio rerio (van der Meer et al., 2005; Marques et al., 2008), Drosophila melanogaster (Liu et al., 2006; Azad et al., 2009), Caenorhabditis elegans (Shen et al., 2005), Trichoderma reesei (Bonaccorsi et al., 2006), Saccharomyces cerevisiae (Linde et al., 1999; Lai et al., 2005; Protchenko et al., 2008), Schizosaccharomyces pombe (Todd et al., 2006), Candida albicans (Setiadi et al., 2006), Cryptococcus neoformans (Chun et al., 2007), Escherichia coli (Salmon et al., 2003; Kang et al., 2005; Covert et al., 2008), Mycobacterium tuberculosis (Park et al., 2003), Pseudomonas aeruginosa (Alvarez-Ortega and Harwood, 2007), Pseudomonas stutzeri (Dou et al., 2008), Sinorhizobium meliloti (Becker et al., 2004), Synechocystis species PCC 6803 (Summerfield et al., 2008), and Zymomonas mobilis (Yang et al., 2009). Additional Arabidopsis and human data sets evaluated responses across experimental conditions and cell types (Arabidopsis, Loreti et al., 2005; Baena-González et al., 2007; Bond et al., 2009; Christianson et al., 2009; Banti et al., 2010; human, Chi et al., 2006; Mense et al., 2006; Irigoyen et al., 2007; Ricciardi et al., 2008; Calzado et al., 2009; Fang et al., 2009; E-MIMR-644 [http://www.ebi.ac.uk/microarray-as/ae/], GSE15583 [http://www.ncbi.nlm.nih.gov/geo]). For the rice leaf samples, cv M202 and the Sub1 introgression line M202(Sub1) were grown for 14 d and submerged for 1 d in a greenhouse exactly as described by Fukao et al. (2006). Total RNA was extracted from aerial tissue (Fukao and Bailey-Serres, 2008), and two independent biological replicate hybridizations to Rice Genome Arrays (GeneChip; Affymetrix) were performed using the manufacturer's protocol. These data are available from the Gene Expression Omnibus database as accession GSE18930.
Genes with a significant change in steady-state mRNA abundance in response to treatments were identified as follows. If Affymetrix CEL files were available, the data were reanalyzed by use of the R program (http://cran.at.r-project.org/) and Bioconductor package (http://www.bioconductor.org/; Gentleman et al., 2004). GC-corrected robust multichip average normalization was employed for the rice data sets, and the robust multichip average normalization was used for all other organisms. MAS5 present calls were obtained to remove probe sets that did not contain a P call in all replicates of at least one sample set. The LIMMA package was used for pairwise comparisons of stress treatment versus control and to obtain the signal-log ratio (Smyth, 2004). The Benjamini and Hochberg (1995) method was selected to adjust P values for multiple testing and to determine false discovery rates. In other cases, data were obtained from publications (Supplemental Table S1). To identify DEGs, a 2-fold change value was used for the initial selection. If fewer than 50 DEGs were identified for one organism, the change criterion was relaxed to 1.8-fold. Gene lists were further refined based on the robustness of data using a false discovery rate of 0.01. In some cases, other criteria were used due to the array setup or data availability (Supplemental Table S1). If multiple data sets were available, up to three from one organism were analyzed. Gene annotations were transformed into RefSeq accession numbers from the National Center for Biotechnology Information or another basic annotation system. Affymetrix (https://www.affymetrix.com/support) and PubMed (www.ncbi.nlm.nih.gov) data were used for annotation conversions.
BLAST Analysis of DEGs
FASTA protein sequences of whole proteomes were obtained: C. albicans, http://www.candidagenome.org/download/sequence/Assembly21/current/; S. pombe, http://www.sanger.ac.uk/Projects/S_pombe/protein_download.shtml; S. cerevisiae, http://downloads.yeastgenome.org/sequence/genomic_sequence/; T. reesei, http://genome.jgi-psf.org/Trire2/Trire2.download.ftp.html (Martinez et al., 2008); Chlamydomonas, http://genome.jgi-psf.org/Chlre3/Chlre3.download.ftp.html (Merchant et al., 2007); Arabidopsis, ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR8_blastsets/; rice, http://rice.plantbiology.msu.edu/data_download.shtml; poplar, http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.download.ftp.html (Tuskan et al., 2006); and all other organisms, ftp://ftp.ncbi.nih.gov/genomes/.
An OMCL analysis of protein sequences was performed to identify homologous proteins as described by Li et al. (2003). First, an all-against-all BLASTP comparison was performed (Altschul et al., 1990). A P value of 10−5 or less was used for similarity cutoff. A similarity matrix was created based on P values, which was normalized for within- and between-species comparisons. Based on this similarity matrix, proteins were grouped with their homologs/orthologs by use of the Tribe-Markov clustering algorithm (Enright et al., 2002). Clusters of related proteins were formed with an inflation factor of 1.2 (clusters labeled with OMCL). Since this analysis resulted in a large number of small clusters, we additionally performed an MCL analysis of unnormalized P values according to Vandenbroucke et al. (2008) using an inflation factor of 1.5 to generate larger clusters, particularly to resolve regulatory proteins (clusters labeled with MCL). The resulting clusters are presented in Supplemental Table S5.
GO and PUF Assignment of Genes
GO annotations were obtained from public databases. First, all protein identifiers were transformed into UniprotKB identifiers by use of the ID Mapping feature of the Web site http://www.uniprot.org/. Then, all associated GO terms were obtained for the Uniprot identifiers using GOA files for each organism (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/). For some organisms, GO annotations were directly obtained from organism-specific Web sites (http://genome.jgi-psf.org [Chlamydomonas, poplar, T. reesei], www.arabidopsis.org [Arabidopsis], and http://rice.plantbiology.msu.edu/ [rice]). GO enrichment analysis was performed with the GOHyperGall function according to Horan et al. (2008). Arabidopsis genes encoding PUFs were recognized based on Horan et al. (2008). Additionally, similarity to proteins by SwissProt (ftp://ftp.ebi.ac.uk/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz) was used to recognize PUFs in Arabidopsis and other organisms. Proteins were defined as “unknown” if the similarity E value to any protein of the SwissProt collection was lower than 10−6.
BLAST Search for Related Arabidopsis Genes
For the stress-induced and -repressed genes, we performed BLAST analyses of Arabidopsis protein sequences against whole proteomes of the other 20 organisms (see above). The “blastall” function and a similarity E value cutoff of 10−6 or less were used to identify putative homologs/orthologs, with stress-induced or -repressed genes preferred over noninduced or nonrepressed genes, respectively, even if a noninduced or nonrepressed gene was more similar. The best of any hit for each Arabidopsis protein was selected.
Production and Stress Treatment of Arabidopsis Transgenics
p35S:HF, created from the binary T-DNA vector pPZP111 that contains a 1,343-bp cauliflower mosaic virus 35S promoter followed by the tobacco mosaic virus Ω 5 ′ untranslated leader, an N-terminal His6-FLAG tag, and a 3 ′ NOS terminator (Zanetti et al., 2005), was modified to create two Gateway-compatible vectors: p35S:HF-GATA and p35S:GATA-HF for N- and C-terminal epitope tagging, respectively. The N-terminal tag is M(H)6(G)3DYKDDDDK(G)7, the C-terminal tag is (G)7DYKDDDDK(G)3(H)6, and GATA is the att1-cmR-ccdb-att2 Gateway cassette in reading frame A (Invitrogen). Full-length cDNAs were obtained from the Arabidopsis Biological Research Collection or by amplification from reverse-transcribed mRNA from low-oxygen-stressed seedlings using gene-specific primers for At1g05575, At1g19530, At1g33055, At2g36220, At2g41730, At3g10020, At3g27220, At3g27770, At4g24110, At4g27450, At4g33980, At4g39675, At5g10040, At5g15120, At5g39890, and At5g66985 (Supplemental Table S11). Full-length open reading frames were amplified by PCR and cloned into the pENTR/D-TOPO vector (Invitrogen). Epitope tagging was at the C terminus, unless the predicted protein had a hydrophobic C terminus. Following recombination, the sequences of the tag junction and coding region were verified by sequencing (Institute for Integrated Genome Biology Core Facility, University of California, Riverside). Col-0 ecotype transformation, transgenic plant selection, and establishment of single-copy insertion lines were as described previously (Zanetti et al., 2005; Mustroph et al., 2009). Protein expression was confirmed as described in Supplemental Materials and Methods S1. At least two independent transgenic lines were established for each HUP gene.
Seeds were propagated on Murashige and Skoog medium (0.43% [w/v] Murashige and Skoog salts, 1% [w/v] Suc, and 0.4% [w/v] phytagel, pH 5.75) in square petri dishes in a vertical orientation at 23°C with a 16-h-day (50 μ mol photons m−2 s−1) and 8-h-night cycle for 7 d (Branco-Price et al., 2008), and then seedlings were deprived of oxygen for 8 or 12 h or mock treated under dim light (0.15 μ mol photons m−2 s−1) as detailed previously (Branco-Price et al., 2008). Treatments commenced at the end of the light period. After treatment, samples were returned to the growth chamber for 3 d. Viability of overexpressor lines, knockout mutants (Supplemental Table S11), and Col-0 seedlings grown on the same plate was scored as follows: the numbers of nondamaged, damaged, and dead plants (scores 5, 3, and 1, respectively) were summed for each genotype. Nondamaged plants had green aerial tissue and no evident tissue damage, damaged plants had anthocyanin-pigmented or bleached cotyledons or leaves, and dead plants had dead aerial meristems. Root growth was attenuated in all genotypes after 8 h of stress. Viability of overexpressor and knockout lines was compared with that of Col-0, using Student's t test to identify significant differences between means of replicates (n = 3) of different genotypes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. OMCL clusters for hypoxia-induced and hypoxia-reduced DEGs.
Supplemental Figure S2. OMCL clusters with DEGs in at least six organisms of the 21 organisms analyzed.
Supplemental Figure S3. OMCL clusters associated with ribosomes.
Supplemental Figure S4. OMCL clusters with genes involved in glycolysis, fermentation, and the TCA cycle.
Supplemental Figure S5. Genes involved in primary metabolism: comparison across different treatment conditions, stages, and genotypes in higher plants.
Supplemental Figure S6. OMCL clusters of protein kinases and transcription factors.
Supplemental Figure S7. OMCL clusters with genes associated with heat shock ROS networks.
Supplemental Figure S8. Induced DEGs in low-oxygen-stressed human cells: expression patterns across experiments and cell types.
Supplemental Figure S9. Low-oxygen-reduced DEGs of Arabidopsis: expression patterns across experiments, cell types, and orthologs in other organisms.
Supplemental Figure S10. Reduced DEGs in low-oxygen-stressed human cells: expression patterns across experiments and cell types.
Supplemental Figure S11. Species distribution of low-oxygen-induced PUFs.
Supplemental Figure S12. Phenotype data for transgenic Arabidopsis plants expressing low-oxygen-induced HUPs.
Supplemental Table S1. Selected organisms and microarray experiments evaluating low-oxygen responses used in this paper.
Supplemental Table S2. Compilation of low-oxygen-induced genes in 21 organisms from four kingdoms.
Supplemental Table S3. Compilation of low-oxygen-reduced genes in 21 organisms from four kingdoms.
Supplemental Table S4. OMCL clustering of proteomes.
Supplemental Table S5. Assignment of proteomes of 21 organisms to OMCL clusters.
Supplemental Table S6. OMCL clustering of hypoxia-induced and hypoxia-reduced genes.
Supplemental Table S7. GO enrichment analysis of hypoxia-induced and -reduced genes in multiple organisms.
Supplemental Table S8. DEGs between two genotypes of the rice variety M202.
Supplemental Table S9. Observation of experimental and cell type variations in gene expression of hypoxia-induced and -reduced genes from Arabidopsis.
Supplemental Table S10. Observation of experimental and cell type variations in gene expression of hypoxia-induced and -reduced genes from human.
Supplemental Table S11. Summary of PUFs that were studied by mutant analysis.
Supplemental Materials and Methods S1.
Acknowledgments
We thank Thomas Girke for guidance in bioinformatics.
References
- Allègre A, Silvestre J, Morard P, Kallerhoff J, Pinelli E. (2004) Nitrate reductase regulation in tomato roots by exogenous nitrate: a possible role in tolerance to long-term root anoxia. J Exp Bot 55: 2625–2634 [DOI] [PubMed] [Google Scholar]
- Allen JW, Khetani SR, Johnson RS, Bhatia SN. (2006) In vitro liver tissue model established from transgenic mice: role of HIF-1alpha on hypoxic gene expression. Tissue Eng 12: 3135–3147 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alvarez-Ortega C, Harwood CS. (2007) Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65: 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad P, Zhou D, Russo E, Haddad GG. (2009) Distinct mechanisms underlying tolerance to intermittent and constant hypoxia in Drosophila melanogaster. PLoS One 4: e5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signaling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R. (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot (Lond) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LA. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Banti V, Mafessoni F, Loreti E, Alpi A, Perata P. (2010) The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 152: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banti V, Loreti E, Novi G, Santaniello A, Alpi A, Perata P. (2008) Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ 31: 1029–1037 [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. (2002) ROPGAP4-dependent Rop GTPase rheostat controls of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Becker A, Bergès H, Krol E, Bruand C, Rüberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, et al. (2004) Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant Microbe Interact 17: 292–303 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bertout JA, Patel SA, Simon MC. (2008) The impact of O2 availability on human cancer. Nat Rev Cancer 8: 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickler PE, Buck LT. (2007) Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu Rev Physiol 69: 145–170 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Bishop-Bailey D. (2009) Tumour vascularisation: a druggable target. Curr Opin Pharmacol 9: 96–101 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Lagunas R, Gancedo C, Gancedo JM. (1993) Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett 329: 51–54 [DOI] [PubMed] [Google Scholar]
- Bonaccorsi ED, Ferreira AJ, Chambergo FS, Ramos AS, Mantovani MC, Farah JP, Sorio CS, Gombert AK, Tonso A, El-Dorry H. (2006) Transcriptional response of the obligatory aerobe Trichoderma reesei to hypoxia and transient anoxia: implications for energy production and survival in the absence of oxygen. Biochemistry 45: 3912–3924 [DOI] [PubMed] [Google Scholar]
- Bond DM, Wilson IW, Dennis ES, Pogson BJ, Jean Finnegan E. (2009) VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J 59: 576–587 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Bruick RK. (2003) Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev 17: 2614–2623 [DOI] [PubMed] [Google Scholar]
- Calzado MA, de la Vega L, Möller A, Bowtell DD, Schmitz ML. (2009) An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat Cell Biol 11: 85–91 [DOI] [PubMed] [Google Scholar]
- Chen JL, Lucas JE, Schroeder T, Mori S, Wu J, Nevins J, Dewhirst M, West M, Chi JT. (2008) The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet 4: e1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Børresen-Dale AL, et al. (2006) Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med 3: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CD, Liu OW, Madhani HD. (2007) A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog 3: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Xiao N, Chen TJ, Karr JR. (2008) Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics 24: 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou YT, Yan YL, Ping SZ, Lu W, Chen M, Zhang W, Wang YP, Jin Q, Lin M. (2008) Expression profile analysis of the oxygen response in the nitrogen-fixing Pseudomonas stutzeri A1501 by genome-wide DNA microarray. Chin Sci Bull 53: 1197–1204 [Google Scholar]
- Ella ES, Kawano N, Ito O. (2003) Importance of active oxygen-scavenging system in the recovery of rice seedlings after submergence. Plant Sci 165: 85–93 [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ. (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW, Higashi RM, Frenkiel TA, Lane AN. (1997) Anaerobic nitrate and ammonium metabolism in flood-tolerant rice coleoptiles. J Exp Bot 48: 1655–1666 [Google Scholar]
- Fang HY, Hughes R, Murdoch C, Coffelt SB, Biswas SK, Harris AL, Johnson RS, Imityaz HZ, Simon MC, Fredlund E, et al. (2009) Hypoxia-inducible factors 1 and 2 are important transcriptional effectors in primary macrophages experiencing hypoxia. Blood 114: 844–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61: 243–282 [DOI] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, LeCouter J. (2003) The biology of VEGF and its receptors. Nat Med 9: 669–676 [DOI] [PubMed] [Google Scholar]
- Fisslthaler B, Fleming I. (2009) Activation and signaling by the AMP-activated protein kinase in endothelial cells. Circ Res 105: 114–127 [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. (2004) Plant responses to hypoxia: is survival a balancing act?. Trends Plant Sci 9: 449–456 [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollery M, Harper J, Cushman J, Mittler T, Girke T, Zhu JK, Bailey-Serres J, Mittler R. (2006) What makes species unique? The contribution of proteins with obscure features. Genome Biol 7: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Crack JC, Thomson AJ, LeBrun NE. (2009) Bacterial sensors of oxygen. Curr Opin Microbiol 12: 145–151 [DOI] [PubMed] [Google Scholar]
- Greijer AE, van der Groep P, Kemming D, Shvarts A, Semenza GL, Meijer GA, van de Wiel MA, Belien JA, van Diest PJ, van der Wall E. (2005) Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J Pathol 206: 291–304 [DOI] [PubMed] [Google Scholar]
- Guglielminetti L, Yamaguchi J, Perata P, Alpi A. (1995) Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiol 109: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin D, Albertin G, Spinazzi R, Sorato E, Mascarin A, Cavallo D, Antonello M, Ribatti D. (2008) Adrenomedullin stimulates angiogenic response in cultured human vascular endothelial cells: involvement of the vascular endothelial growth factor receptor 2. Peptides 29: 2013–2023 [DOI] [PubMed] [Google Scholar]
- Guimbellot JS, Erickson SW, Mehta T, Wen H, Page GP, Sorscher EJ, Hong JS. (2009) Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis. BMC Med Genomics 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. (2008) SNF1/AMPK pathways in yeast. Front Biosci 13: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Buck LT, Doll CJ, Land SC. (1996) Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA 93: 9493–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. (1998) Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon T, Dodd A, Dirmeier R, Gorman N, Sinclair PR, Zhang L, Poyton RO. (2003) A mechanism of oxygen sensing in yeast: multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem 278: 50771–50780 [DOI] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T. (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi Y, Hazeki O, Tamura M. (1993) Oxygen dependence of redox state of copper in cytochrome oxidase in vitro. J Appl Physiol 74: 1622–1627 [DOI] [PubMed] [Google Scholar]
- Hu CJ, Iyer S, Sataur A, Covello KL, Chodosh LA, Simon MC. (2006) Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol Cell Biol 26: 3514–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi?. Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Irigoyen M, Ansó E, Martínez E, Garayoa M, Martínez-Irujo JJ, Rouzaut A. (2007) Hypoxia alters the adhesive properties of lymphatic endothelial cells: a transcriptional and functional study. Biochim Biophys Acta 1773: 880–890 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Vasconcelles MJ, Wretzel S, Light A, Martin CE, Goldberg MA. (2001) MGA2 is involved in the low-oxygen response element-dependent hypoxic induction of genes in Saccharomyces cerevisiae. Mol Cell Biol 21: 6161–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. (2005) Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187: 1135–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, Whelan J. (2009) Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol 149: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast KE, Lai LC, Menda N, James DT, III, Aref S, Burke PV. (2002) Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol 184: 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LC, Kosorukoff AL, Burke PV, Kwast KE. (2005) Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol Cell Biol 25: 4075–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P. (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, Seo YW. (2007) A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol Plant 129: 375–385 [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Roy J, Johnson EA. (2006) Identification and function of hypoxia-response genes in Drosophila melanogaster. Physiol Genomics 25: 134–141 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang Q, Burton RA, Shirley NJ, Atwell BJ. (2010) Expression of vacuolar H(+)-pyrophosphatase (OVP3) is under control of an anoxia-inducible promoter in rice. Plant Mol Biol 72: 47–60 [DOI] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Yamaguchi J, Alpi A, Perata P. (2003) Sugar modulation of alpha-amylase genes under anoxia. Ann Bot (Lond) 91: 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R. (2008) Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol 148: 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. (2007) An Arabidopsis gene network based on the graphical Gaussian model. Genome Res 17: 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Sandoval GR, Rodriguez C, Franco B, Georgellis D. (2006) Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal 8: 781–795 [DOI] [PubMed] [Google Scholar]
- Marques IJ, Leito JT, Spaink HP, Testerink J, Jaspers RT, Witte F, van den Berg S, Bagowski CP. (2008) Transcriptome analysis of the response to chronic constant hypoxia in zebrafish hearts. J Comp Physiol [B] 178: 77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al. (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei. Nat Biotechnol 26: 553–560 [DOI] [PubMed] [Google Scholar]
- Mattana M, Vannini C, Espen L, Bracale M, Genga A, Marsoni M, Iriti M, Bonazza V, Romagnoli F, Baldoni E, et al. (2007) The rice Mybleu transcription factor increases tolerance to oxygen deprivation in Arabidopsis plants. Physiol Plant 131: 106–121 [DOI] [PubMed] [Google Scholar]
- Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ, Zhang L. (2006) Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics 25: 435–449 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk LS, Braendle R, Crawford RMM. (1987) Catalase activity and post-anoxic injury in monocotyledonous species. J Exp Bot 38: 233–246 [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Albrecht G, Hajirezaei M, Grimm B, Biemelt S. (2005) Low levels of pyrophosphate in transgenic potato plants expressing E. coli pyrophosphatase lead to decreased vitality under oxygen deficiency. Ann Bot (Lond) 96: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J. (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. (2006) HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3: 187–197 [DOI] [PubMed] [Google Scholar]
- Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. (2003) Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol 48: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. (2007) Trehalose 6-phosphate. Curr Opin Plant Biol 10: 303–309 [DOI] [PubMed] [Google Scholar]
- Protchenko O, Shakoury-Elizeh M, Keane P, Storey J, Androphy R, Philpott CC. (2008) Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot Cell 7: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC. (2007) Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194 [DOI] [PubMed] [Google Scholar]
- Ricard B, Toai TV, Chourey P, Saglio P. (1998) Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiol 116: 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi A, Elia AR, Cappello P, Puppo M, Vanni C, Fardin P, Eva A, Munroe D, Wu X, Giovarelli M, et al. (2008) Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Mol Cancer Res 6: 175–185 [DOI] [PubMed] [Google Scholar]
- Salmon K, Hung SP, Mekjian K, Baldi P, Hatfield GW, Gunsalus RP. (2003) Global gene expression profiling in Escherichia coli K12: the effects of oxygen availability and FNR. J Biol Chem 278: 29837–29855 [DOI] [PubMed] [Google Scholar]
- Schroeder K, Kohnen A, Aicher A, Liehn EA, Büchse T, Stein S, Weber C, Dimmeler S, Brandes RP. (2009) NADPH oxidase Nox2 is required for hypoxia-induced mobilization of endothelial progenitor cells. Circ Res 105: 537–544 [DOI] [PubMed] [Google Scholar]
- Semenza GL. (2007) Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J 405: 1–9 [DOI] [PubMed] [Google Scholar]
- Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. (2006) Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J Mol Biol 361: 399–411 [DOI] [PubMed] [Google Scholar]
- Shalel-Levanon S, San KY, Bennett GN. (2005) Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol Bioeng 92: 147–159 [DOI] [PubMed] [Google Scholar]
- Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. (2005) Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem 280: 20580–20588 [DOI] [PubMed] [Google Scholar]
- Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. (2001) Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc Natl Acad Sci USA 98: 7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: article 3 [DOI] [PubMed] [Google Scholar]
- Steunou AS, Bhaya D, Bateson MM, Melendrez MC, Ward DM, Brecht E, Peters JW, Kühl M, Grossman AR. (2006) In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA 103: 2398–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoimenova M, Libourel IGL, Ratcliffe RG, Kaiser WM. (2003) The role of nitrate reduction in the anoxic metabolism of roots. II. Anoxic metabolism of tobacco roots with or without nitrate reductase activity. Plant Soil 253: 155–167 [Google Scholar]
- Summerfield TC, Toepel J, Sherman LA. (2008) Low-oxygen induction of normally cryptic psbA genes in cyanobacteria. Biochemistry 47: 12939–12941 [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Linde JJ, Liang H, Davis RW, Steensma HY, van Dijken JP, Pronk JT. (1999) Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J Bacteriol 181: 7409–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Johannes GJ. (2007) Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA 13: 1116–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timperio AM, Egidi MG, Zolla L. (2008) Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J Proteomics 71: 391–411 [DOI] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. (2006) Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol 26: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Vandenbroucke K, Robbens S, Vandepoele K, Inzé D, Van de Peer Y, Van Breusegem F. (2008) Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol 25: 507–516 [DOI] [PubMed] [Google Scholar]
- van der Meer DL, van den Thillart GE, Witte F, de Bakker MA, Besser J, Richardson MK, Spaink HP, Leito JT, Bagowski CP. (2005) Gene expression profiling of the long-term adaptive response to hypoxia in the gills of adult zebrafish. Am J Physiol Regul Integr Comp Physiol 289: R1512–R1519 [DOI] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Spano T, Vlad D, Daher FB, Ouelhadj A, Fragkostefanakis S, Kalaitzis P. (2007) Involvement of Arabidopsis prolyl 4 hydroxylases in hypoxia, anoxia and mechanical wounding. Plant Signal Behav 2: 368–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yang S, Tschaplinski TJ, Engle NL, Carroll SL, Martin SL, Davison BH, Palumbo AV, Rodriguez M, Jr, Brown SD. (2009) Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics 10: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]