Abstract
The two-step oxidation of proline in all eukaryotes is performed at the inner mitochondrial membrane by the consecutive action of proline dehydrogenase (ProDH) that produces Δ1-pyrroline-5-carboxylate (P5C) and P5C dehydrogenase (P5CDH) that oxidizes P5C to glutamate. This catabolic route is down-regulated in plants during osmotic stress, allowing free Pro accumulation. We show here that overexpression of MsProDH in tobacco and Arabidopsis or impairment of P5C oxidation in the Arabidopsis p5cdh mutant did not change the cellular Pro to P5C ratio under ambient and osmotic stress conditions, indicating that P5C excess was reduced to Pro in a mitochondrial-cytosolic cycle. This cycle, involving ProDH and P5C reductase, exists in animal cells and now demonstrated in plants. As a part of the cycle, Pro oxidation by the ProDH-FAD complex delivers electrons to the electron transport chain. Hyperactivity of the cycle, e.g. when an excess of exogenous l-Pro is provided, generates mitochondrial reactive oxygen species (ROS) by delivering electrons to O2, as demonstrated by the mitochondria-specific MitoSox staining of superoxide ions. Lack of P5CDH activity led to higher ROS production under dark and light conditions in the presence of Pro excess, as well as rendered plants hypersensitive to heat stress. Balancing mitochondrial ROS production during increased Pro oxidation is therefore critical for avoiding Pro-related toxic effects. Hence, normal oxidation of P5C to Glu by P5CDH is key to prevent P5C-Pro intensive cycling and avoid ROS production from electron run-off.
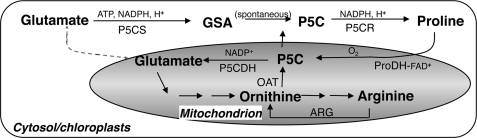
Stress-related changes in Pro biosynthesis and degradation are conserved in different organisms and used to induce essential signaling pathways, such as p53-mediated apoptosis in mammalian cells (1) or osmo-protecting mechanisms in plants (2) and bacteria (3). Pro is one of the most common osmolytes that, together with glycine-betaine and soluble sugars, accumulates in plants in response to a wide array of abiotic and biotic stresses (2, 4, 5). Although the role of Pro as an osmo-protecting molecule in prokaryotes is well established (3, 6), the overall function of stress-accumulated free Pro in plants is still under debate (2, 7–9). In most plants, stress-induced free Pro accumulation results from enhanced Pro synthesis and concomitant silencing of Pro degradation (10–13). Pro is produced in the cytosol and chloroplasts (14, 15) and degraded in the mitochondria by a short, strictly regulated cyclic pathway (Fig. 1). In the anabolic part of the pathway, glutamate is converted to Δ1-pyrroline-5-carboxylate (P5C)3 by P5C synthetase (P5CS), and P5C-reductase (P5CR) reduces P5C to Pro. In an alternative pathway, P5C is generated from ornithine by mitochondrial ornithine-aminotransferase (16, 17) and is reduced to Pro by P5CR in the cytosol. Thus, a possible movement of P5C among organelles and cytosol is assumed. P5CS is considered the key enzyme in the synthesis route, and its expression is increased by osmotic stress in many plants (11, 16–18). A single gene encodes P5CR in Arabidopsis (19) and a post-transcriptional regulation mechanism maintains constant levels of the enzyme during heat and salt stresses (20). In the catabolic route, proline dehydrogenase (ProDH) catalyzes Pro oxidation to P5C, which is maintained in a pH-dependent nonenzymatic equilibrium with glutamate semialdehyde (GSA) (21). ProDH is an FAD-dependent enzyme localized to the inner mitochondrial membrane and transfers electrons directly to the mitochondrial electron chain during Pro oxidation (21). In all plants studied to date, transcription and enzymatic activity of ProDH decrease during osmotic stress and rapidly increase during the consecutive recovery period. These changes are inversely correlated with cellular free Pro levels (22–25). In Arabidopsis, most of the studies have described AtProDH1 (At3g30775) silencing during osmotic stress. Microarray expression data indicated that AtProDH2 (At5g38710) is transcribed at low levels during different stresses (Genevestigator) (26) and is regulated by sucrose in a similar way to AtProDH1 (27). Although little is known about the function of AtProDH2, many studies identified AtProDH1 as the major Pro oxidizing enzyme (7, 28, 29). P5C, generated in mitochondria by ProDH activity, is further oxidized to Glu by P5CDH, encoded by a single-copy gene in Arabidopsis (30). An antisense overlapping of AtP5CDH and AtSRO5, whose transcription is induced by salinity (150 mm NaCl), enables salt-induced production of nat-small interfering RNA that initiates RNA interference-based AtP5CDH mRNA degradation (31). However, during 100 mm NaCl treatment, in both Arabidopsis and alfalfa, P5CDH transcript levels remained unchanged or even increased at late stages of the stress period (12, 32).
FIGURE 1.
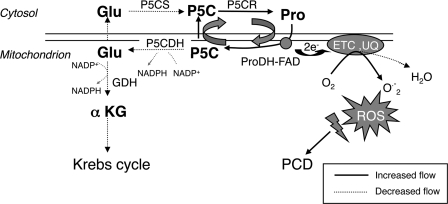
Summary of Pro synthesis and Pro catabolism pathways in plants. Pro is synthesized from Glu in the cytosol and chloroplasts. The bifunctional enzyme P5CS catalyzes the ATP-dependent phosphorylation of Glu to γ-glutamyl phosphate and then the NADPH-dependent reduction to GSA. P5C and GSA exist in a nonenzymatic equilibrium. GSA/P5C is also generated from ornithine by ornithine aminotransferase (OAT) in the mitochondria and is suggested to serve as a Pro precursor in the cytosol. Following P5C reduction to Pro by P5CR, Pro is transported to the mitochondria and subjected to two-step oxidation by ProDH-FAD complex and P5CDH, both of which are linked to the inner mitochondrial membrane. ARG, arginase.
In contrast to endogenous stress-accumulated Pro, exogenously provided Pro is toxic at millimolar concentrations, leading to programmed cell death in yeast, human cancer cells, and plant cells (33–36). Several studies demonstrated that mutants impaired in their ability to oxidize Pro are more sensitive to its external application (7, 28, 37). Likewise, enhanced Pro oxidation in transgenic and mutant plants decreased plant sensitivity to exogenous Pro (7, 28, 31, 37). In yeast, overexpression of PUT1 (the yeast ProDH) induces apoptosis (35, 38). Furthermore, high levels of P5C in the yeast put2 mutant, which lacks P5CDH expression, were associated with increased ROS production. It was therefore suggested that Pro toxicity in yeast results from increased P5C levels (36). The toxic effect of exogenously applied Pro in plants was also attributed to increased P5C levels (34, 37).
We describe here a novel plant P5C-Pro cycling mechanism, whereby P5C excess, resulting from Pro oxidation by mitochondrial ProDH, is reduced by cytosolic P5CR to Pro and transported back to the organelle. This cycle is highly similar to the “proline cycle” in mammalian cells (1, 39). We demonstrate that neither enhanced production nor reduced oxidation of P5C changes the cellular Pro to P5C ratio, which is maintained by a rapid P5C reduction to Pro. When the intake of externally provided Pro exceeds its physiological levels accumulated under stress conditions, P5C oxidation by P5CDH is essential for limiting P5C-Pro cycle activity and avoiding high levels of mitochondrial ROS production.
EXPERIMENTAL PROCEDURES
Reagents
MS salts were obtained from Duchefa (Haarlem, Netherlands) (12). l-Thiazolidine-4-carboxylic acid (T4C), O-amino-benzaldehyde, 2,4-dinitrophenylhydrazine, dl-P5C-2,4-dinitrophenylhydrazine-hydrochloride double salt, and PD10 desalting columns were purchased from Sigma-Aldrich. Purified P5C was obtained by removing the 2,4-dinitrophenylhydrazine salt with acetophenone (supplemental data).
Plant Material and Growth Conditions
Nicotiana tabacum cv. Samsun (NN) and Arabidopsis thaliana cv. Columbia (Col-0) were grown at 25 or 22 °C, respectively, in a 16-h light/8-h dark regime, at light intensity of 100 μmol cm−2 s−1. The p5cdh T-DNA knock-out mutant was obtained from ABRC, and homozygous plants were identified using the SIGnAL (Salk Institute Genomic Analysis Laboratory) recommended primers. This mutant (Salk_018453) had already been described by Deuschle et al. (37) as p5cdh2 with a T-DNA insert in the ninth exon and exhibits similar characteristics as other p5cdh mutants described previously (31, 37).
Stress Treatments
For salt stress, which lasted 5 weeks, 6-week-old WT and ProDH-OE tobacco plants were irrigated with 300 mm NaCl (a total of 2 liters was applied gradually during the stress period to 2 liters of soil). Drought (dehydration) stress was imposed for 72 h on 3-week-old Arabidopsis plants grown on MS-soaked vermiculite (supplemental data). These plants were then rehydrated for 3 days by adding the lost water volume. Water loss of vermiculite containers and values of relative water content were similar for WT and p5cdh plants during the drought and recovery periods (supplemental Fig. S6). l-Pro was applied to the seedlings by saturating the vermiculite in bottom-pierced cups with 100 mm l-Pro solution. Root elongation in response to heat stress (38 °C) was measured in seedlings grown on MS plates as described previously (40). Heat stress survival assay was carried out at 41 °C for 2 h according to Suzuki et al. (41).
T4C Survival Assay
T4C competes with Pro in the translation process (42, 43) and can be degraded by Pro dehydrogenase in plants and bacteria (44, 45). One-week-old WT and ProDH-OE tobacco seedlings (T2) were transferred to MS medium containing 50 mm NaCl, 3 mm T4C, or 50 mm NaCl + 3 mm T4C. Pictures were taken 3 weeks later.
Toxicity Assays
Toxicity assays of 1 mm P5C, 100 mm Pro, or 1 mm 2,4-dinitrophenyl hydrazine hydrochloride were conducted on 2-week-old Arabidopsis plants in 6-well plates containing liquid MS.
Molecular and Biochemical Analyses
Transgenic tobacco and Arabidopsis plants, constitutively expressing MsProDH1 cDNA (12), driven by the cauliflower mosaic virus 35 S promoter, were generated by Agrobacterium-mediated transformation as described previously (12, 46) (supplemental data). Total RNA isolation and Northern blot analyses were performed according to Miller et al. (12). Pro oxidation was measured in vitro, using leaf proteins as the enzyme source, Pro and NAD+ as substrates. NAD+ reduction was measured at 340 nm (supplemental data).
cDNA Synthesis and Quantitative Reverse Transcription- PCR
Total RNA was extracted from frozen Arabidopsis tissues and treated with RNase-free DNaseI using the SV total RNA isolation kit (Promega) according to the manufacturer's instructions. cDNA was synthesized using 3 μg of total RNA as a template, an oligo(dT) primer, and Moloney murine leukemia virus reverse transcriptase (Promega), according to the manufacturer's instructions. Quantitative reverse transcription-PCR was performed using 1/100 volume of the cDNA reaction as the template (supplemental data).
Proline and P5C Measurements
Similarly to Pro, P5C reacts with ninhydrin (47) in the commonly used Pro colorimetric determination of Miller et al. (12) and Bates et al. (48) (supplemental Fig. S1A). By quantifying purified P5C and l-Pro, using separate ninhydrin reactions, we could show that the absorbance of the P5C-ninhydrin complex is ∼65% of that of the Pro-ninhydrin complex in a range of known concentrations (supplemental Fig. S1B). To the best of our knowledge all previous studies that have used the ninhydrin assay for Pro quantification have not taken into account the absorbance of the P5C-ninhydrin complex.
To estimate the net levels of Pro in vivo, P5C values determined by the O-amino-benzaldehyde assay (see below) were subtracted from the Pro+P5C values measured by the ninhydrin reaction according to the following procedure: leaf samples were homogenized in 3% (w/v) sulfosalicylic acid and centrifuged for 10 min. Half of the supernatant was used for Pro determination, and half was used for P5C determination by the O-amino-benzaldehyde assay at 440 nm (49, 50). Known concentrations of l-Pro and dl-P5C as 2,4-dinitrophenylhydrazine-hydrochloride-double salt (detailed in the supplemental data) were used for estimating Pro and P5C concentrations in plant extracts.
Metabolic Profiling
Primary metabolites of WT and p5cdh plants were analyzed by gas chromatography coupled to mass spectrometry according to Ref. 51. Plants were grown on vermiculite and exposed to dehydration as described above. Five biological replicates, each comprising a mixture of leaves collected from 15–20 plants (100 mg of fresh weight), were analyzed in each treatment.
Mitochondrial ROS Evaluation
MitoSox-Red (5 μm; Molecular Probes-Invitrogen) was used for imaging superoxide accumulation in mitochondria of root cells, as described in the supplemental data.
Accession Numbers
The sequence data used in this study are found in the NCBI GenBankTM data base under the following accession numbers: AY556386 (MsProDH1), At5g62530 (AtP5CDH), At5g14800 (AtP5CR), At2g39800 (AtP5CS1), At3g55610 (AtP5CS2), At3g30775 (AtProDH1), and At5g38710 (AtProDH2).
RESULTS
Overexpression of MsProDH in Tobacco and Arabidopsis
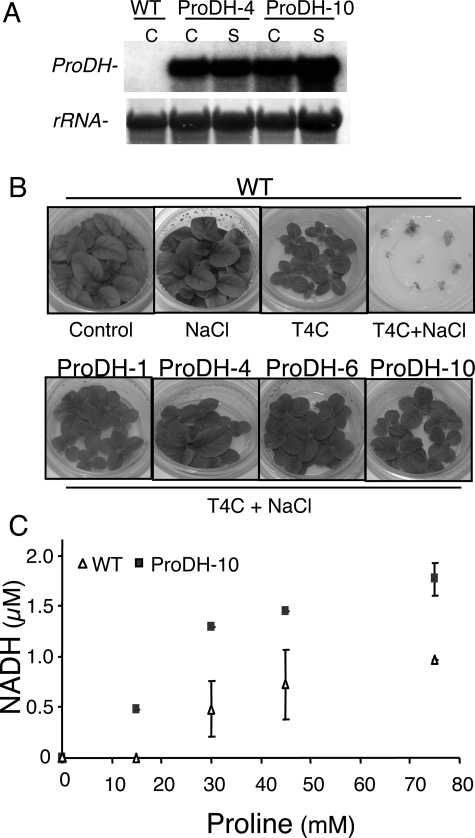
The most prominent changes in cellular Pro levels occur in plants during the imposition of different abiotic stresses and subsequent recovery processes, when high levels of Pro are being synthesized and catabolized, respectively, within a short time (7, 12). To estimate the linkage between down-regulated Pro catabolism and Pro accumulation during stress, we generated transgenic tobacco plants that constitutively overexpress alfalfa (Medicago sativa) MsProDH1 (12), assuming that these plants (ProDH-OE) will show reduced levels of Pro during exposure to stress. Northern blot analysis of leaf RNA extracted from ProDH-OE plants (T2 generation) confirmed the presence of high levels of MsProDH1 transcripts under normal and salt stress conditions (Fig. 2A). Pro oxidizing capacity in planta was evaluated in ProDH-OE lines (T3 generation) by examining their ability to detoxify T4C. This Pro analogue can be oxidized by plant or prokaryotic Pro-dehydrogenases (30, 45). WT tobacco plants showed retarded growth in the presence of T4C (Fig. 2B, upper panel). The addition of a low NaCl concentration (50 mm), which represses endogenous ProDH expression (12), greatly increased the sensitivity of WT plants to T4C (Fig. 2B, upper panel, T4C+NaCl treatment). In contrast, transgenic plants that constitutively overexpressed MsProDH were much more tolerant to T4C, even under salt stress conditions (Fig. 2B, lower panel). Furthermore, the Pro oxidation assay, performed in vitro using increasing l-Pro concentrations as the substrate, showed higher ProDH activity in leaves of drought-stressed ProDH-OE plants in comparison with WT plants (Fig. 2C). These results confirmed that in the transgenic tobacco lines the ectopic MsProDH is not only synthesized but is also enzymatically active and capable of degrading Pro and T4C, thereby eliminating T4C toxicity when the two endogenous NtProDHs (52) are less active (Fig. 2C).
FIGURE 2.
Transgenic tobacco plants overexpressing MsProDH show an increased Pro oxidation activity. A, RNA blot showing the constitutive high MsProDH expression in leaves of transgenic plants grown under normal conditions (Control, C) or irrigated with 150 mm NaCl for 48 h (S). MsProDH cDNA was used as a probe. B, ProDH-dependent tolerance to the Pro toxic analogue T4C. One-week-old tobacco seedlings were transferred to MS, MS+ 50 mm NaCl, MS + 3 mm T4C, or MS+50 mm NaCl + 3 mm T4C medium and photographed 3 weeks later. Upper panel, WT plants highly sensitive to T4C + NaCl combination (right photo). Lower panel, ProDH-OE plants grown on MS + NaCl + T4C with no toxic effects. C, Pro oxidation assay in WT and ProDH-10 plants. Leaf proteins were extracted from 10-day water-stressed plants. The activity was measured in the presence of increasing Pro concentrations, using NAD+ as the electron acceptor. Standard deviation represents three replicates. ProDH-1, -4, -6, and -10 are independent transgenic lines harboring the MsProDH expression cassette.
We next exposed WT and ProDH-OE plants to salt stress to evaluate the effects of increased Pro oxidation on Pro accumulation and stress tolerance. Six-week-old WT and ProDH-OE plants were irrigated with 300 mm NaCl, and the free Pro content of leaves was measured after 5 weeks of stress. No significant decrease in free Pro accumulation was detected in ProDH-OE plants. Both WT and ProDH-OE plants increased their Pro levels by more than 230-fold (Fig. 3A). Furthermore, no significant effect on growth rate or osmotolerance of the ProDH-OE compared with WT plants was observed (supplemental Fig. S3). Shorter exposures of 48 and 72 h to 150 mm NaCl also showed similar levels of Pro accumulation in both plant types (data not shown).
FIGURE 3.
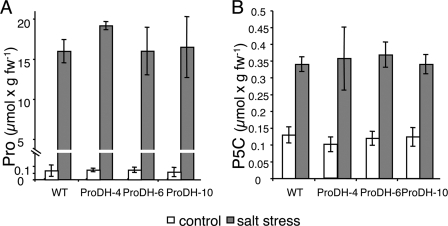
Comparison of Pro and P5C levels in leaves of salt-stressed WT and MsProDH-overexpressing tobacco plants. Six-week-old plants, grown on soil, were irrigated with 300 mm NaCl. Pro and P5C levels in leaves were measured after 5 weeks of salt irrigation. A, net Pro concentration calculated by subtracting the corresponding P5C amount from the Pro + P5C value obtained by the ninhydrin reaction. B, P5C content determined by O-amino-benzaldehyde assay. The standard deviation represents samples from three different plants of the same line.
Increased Pro oxidation in ProDH-OE plants was expected to result in elevated levels of P5C, the first breakdown product (Fig. 1). However, comparable levels of P5C were observed in both WT and ProDH-OE plants, with a 2.5–3-fold increase in P5C content during the salt stress treatment (Fig. 3B). Compared with the 230-fold elevation in Pro content (Fig. 3A), this 3-fold P5C increase is very minor (Fig. 3), indicating that P5C did not accumulate to high levels during salinity stress, not even when the introduced MsProDH remained active. Thus, in ProDH- OE plants, as in WT, the equilibrium between Pro and P5C is maintained.
To examine whether a general tendency to maintain a balanced Pro to P5C ratio in plants exposed to abiotic stresses exists, A. thaliana (Col-0) plants overexpressing MsProDH1 were generated and exposed to dehydration stress. In these plants, the transcript levels of the ectopic MsProDH were 4–7-fold higher than the levels of the endogenous ProDH1 and ProDH2 transcripts after 48 h of dehydration (supplemental Fig. S4A). However, despite the high expression of MsProDH, comparable levels of accumulated Pro and P5C were observed in WT, ProDH-4, and ProDH-5 plants (supplemental Fig. S4, B and C). These results indicated that P5C produced by the foreign MsProDH is converted to its WT levels in tobacco and Arabidopsis plants grown under ambient and stress conditions.
Pro and P5C Levels in the p5cdh Mutant of A. thaliana
The comparable high Pro to P5C ratio observed in WT and ProDH-OE plants during exposure to salinity or dehydration stress, despite increased Pro oxidation in ProDH-OE plants, suggested that Pro and P5C are constantly cycled between the cytosol and mitochondria and are reversibly oxidized and reduced, respectively. Alternatively, the mitochondrially generated P5C might be further oxidized by P5CDH to glutamate, which in turn serves again as a substrate for P5CS and P5CR to rapidly reproduce Pro (Fig. 1). In both cases, there is no net gain of P5C or Pro levels. Therefore, only blocking of the P5C oxidation step could indicate which one is active.
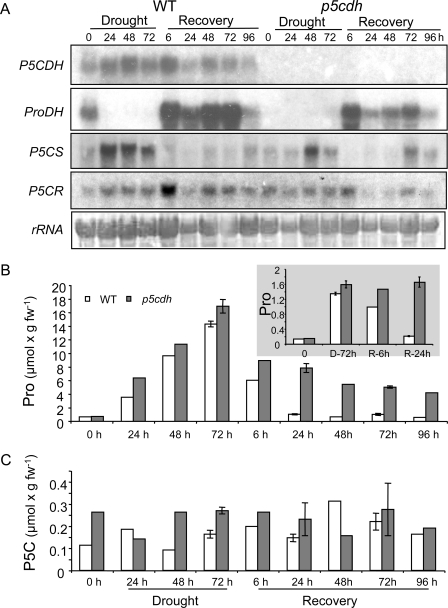
To achieve a complete inactivation of P5C oxidation to Glu, the A. thaliana p5cdh-T-DNA knock-out mutant (Salk_018453) was used. This mutant was described previously as p5cdh2 (37), possessing a T-DNA insert in AtP5CDH-exon 9, upstream to the dehydrogenase domain. The levels of Pro and P5C were measured in WT and p5cdh plants exposed to 3-day dehydration and 4-day recovery. RNA blot analysis showed that the absence of active P5CDH in the mutant did not affect the expression pattern of AtP5CSs, AtP5CR, and AtProDHs during drought stress and subsequent recovery (Fig. 4A). Although the extent of drought-induced Pro accumulation was slightly higher in p5cdh plants (Fig. 4B), the expression of P5CSs was lower throughout the drought period and during the first 48 h of recovery. P5CR transcript levels were also lower in the mutant compared with WT plants during the recovery period. It should be noted that the higher levels of the P5CR transcript in WT plants might not reflect an increased P5CR activity, because it was reported that in Arabidopsis plants P5CR protein levels remain essentially unchanged regardless of changes in its transcript levels induced by salinity stress (20). Although the observed ProDH transcript levels were lower in the p5cdh mutant compared with WT plants (Fig. 4A), the general pattern of stress-induced silencing of ProDH and recovery-up-regulated expression of ProDH was maintained in the mutant (Fig. 4A). This pattern is very typical to stress responses and appears in many plant species studied to date (12, 23, 53). However, compared with WT plants, higher Pro levels were recorded in p5cdh plants during the recovery period (Fig. 4B). The initial apparent decrease in Pro content observed in the p5cdh mutant after 6 h of rehydration (Fig. 4B) may be attributed to the rapid uptake of water during the first stage of rehydration as can be deduced from the level of Pro normalized to dry weight (Fig. 4B, inset). Furthermore, P5C levels were only 1.2–2.5-fold higher in the mutant compared with WT plants throughout the drought and recovery periods. No P5C increase was observed even after 24–96 h of recovery (Fig. 4C), regardless of the increase in ProDH expression (Fig. 4A). These results indicated that throughout the recovery period P5C was most likely reduced back to Pro by P5CR via the P5C-Pro cycle, and thereby steady-state P5C levels were maintained (Fig. 4C). Because further P5C oxidation to Glu does not exist in the p5cdh mutant, the observed slower and gradual Pro decrease during the recovery period (Fig. 4B) may be attributed to Pro incorporation into proteins and a concomitant decrease in Pro synthesis resulting from rehydration-induced down-regulation of P5CS transcription (Fig. 4A and Ref. 54).
FIGURE 4.
Comparison of Pro and P5C content in p5cdh and WT Arabidopsis plants during drought stress and recovery and correlation with the transcript levels of genes involved in Pro biosynthesis and oxidation. Three-week-old WT and p5cdh plants were dehydrated for 72 h and thereafter recovered by adding the water volume lost by evaporation. A, RNA blot analysis of total RNA (10 μg/lane) probed with P5CDH, ProDH, P5CS, and P5CR cDNA fragments. B and C, leaf Pro and P5C content during drought and recovery. The inset in B displays Pro levels normalized to dry weight, showing that during 24 h of water absorption in the recovery period, no loss of Pro is observed in the p5cdh plants. The standard deviation represents three leaf samples, each pooled from 15–20 plants.
Leaf Metabolic Profiling; Comparison of WT and p5cdh Mutant
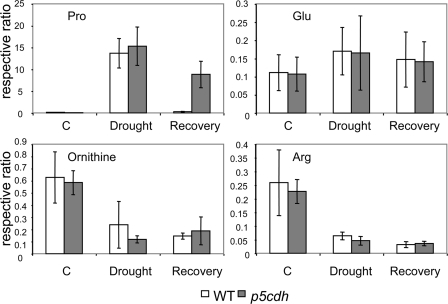
To further validate the existence of the P5C-Pro cycle, we compared the metabolic profiles of WT and p5cdh plants. Three-week-old plants grown under normal conditions were dehydrated for 72 h and then rehydrated for 24 h. Throughout these dehydration and rehydration periods, comparable relative water content values were measured in WT and p5cdh plants (supplemental Fig. S6). Leaf primary metabolites from samples, collected at the end of the stress or after the recovery period, were analyzed by gas chromatography-mass spectrometry. supplemental Table S1 summarizes the identified changes in amino acid and sugar levels. An approximately 1.5-fold increase in glucose, fructose, and trehalose levels was observed in the p5cdh mutant compared with WT plants in response to drought. No significant differences in Pro levels were observed in response to dehydration. In both WT and p5cdh plants, Pro content increased by more than 2 orders of magnitude during the drought stress. In contrast, after 24 h rehydration, Pro levels still remained 30-fold higher in the mutant compared with WT plants (Fig. 5 and supplemental Table S1). These results are in agreement with the ninhydrin evaluation of Pro content (Fig. 4B). The levels of Glu, ornithine, and Arg that might serve as P5C precursors or products (Fig. 1) remained essentially the same in both WT and p5cdh plants during all treatments (Fig. 5), whereas a higher level of Pro during the 24-h recovery period was recorded in p5cdh plants. Because Glu and P5C levels remained similar in both plant types (Figs. 5 and 4C, respectively), it can be concluded that P5C, generated by ProDH activity during the recovery period, was reduced back to Pro by the P5C-Pro cycle, in the p5cdh mutant.
FIGURE 5.
Comparison of the levels of Pro and P5C potential precursors in WT and p5cdh plants during drought stress and following recovery. Gas chromatography-mass spectrometry analysis was performed on leaves of WT and p5cdh plants after 72 h of drought or 24 h of recovery as detailed in Fig. 4. The values are calculated relatively to the ribitol internal standard and normalized to the average dry weight value at each time point, as presented in supplemental Table S1. C (control), no stress. The standard deviation represents five leaf samples, each pooled from 15 to 20 plants.
Enhanced Sensitivity of the p5cdh Mutant to Heat Stress
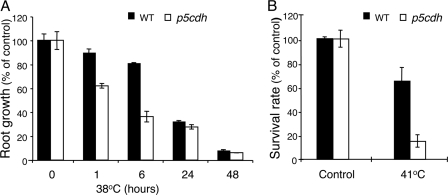
In contrast to osmotic stress, Pro accumulation at high temperatures is toxic to plants (55), whereas the ability to oxidize Pro is important for acquiring thermotolerance in Arabidopsis (29). Therefore, the response of p5cdh and WT plants to heat stress was compared. Five-day-old p5cdh seedlings showed reduced root growth compared with WT when exposed to 38 °C for 6 h (Fig. 6). Such seedlings also displayed dramatically reduced thermotolerance in response to 2 h of exposure to 41 °C, showing 15% survival compared with 65% of WT plants (Fig. 6). These results and the previous reports (29, 55) highlight the importance of regulating Pro catabolism during exposure to high temperatures and possibly during other yet undiscovered physiological conditions.
FIGURE 6.
Deficiency in P5C oxidation to Glu increases thermosensitivity. A, the effect of a continuous heat stress on root elongation of young Arabidopsis seedlings of WT and p5cdh plants. The percentage of root elongation during heat stress (38 °C) is expressed relative to nonstressed seedlings (control). The standard deviation represents an average of three repeats of 10 seedlings each. B, basal thermotolerance of WT and p5cdh seedlings to heat stress. Five-day-old seedlings were exposed to 41 °C for 2 h, and survival was estimated 2 days later. The standard deviation represents three repeats, each comprising 100 seedlings.
P5C-Pro Cycle Hyperactivity Leads to Mitochondrial ROS Formation
According to our hypothesis, mitochondrial P5C excess is reduced back to Pro in the cytosol. Pro is then imported back to the mitochondria, providing a constant substrate for ProDH-linked FAD reduction. Thus, this cycle is coupled to a concomitant FADH2-mediated transfer of electrons to the electron transport chain (mtETC) prior to complex III (21). We speculated that the absence of P5C oxidation to Glu would increase the extent of P5C-Pro cycling and the rate of electron transfer to the mtETC and to O2, resulting in elevated production of superoxide ions (O2˙̄) (56). Paraquat (PQ, methyl viologen) was used as an enhancer for detecting mitochondrial O2˙̄ levels. PQ generates toxic superoxide ions (O2˙̄) when reduced by electron donors. It is frequently used as an oxidative stress-inducing agent (12, 57) and a mediator of increased O2˙̄ production in mitochondria both in vivo and in vitro (58). MitoSox-Red fluorescent staining (58, 59) was used to specifically detect O2˙̄ formed by e− flow through the mtETC to O2.
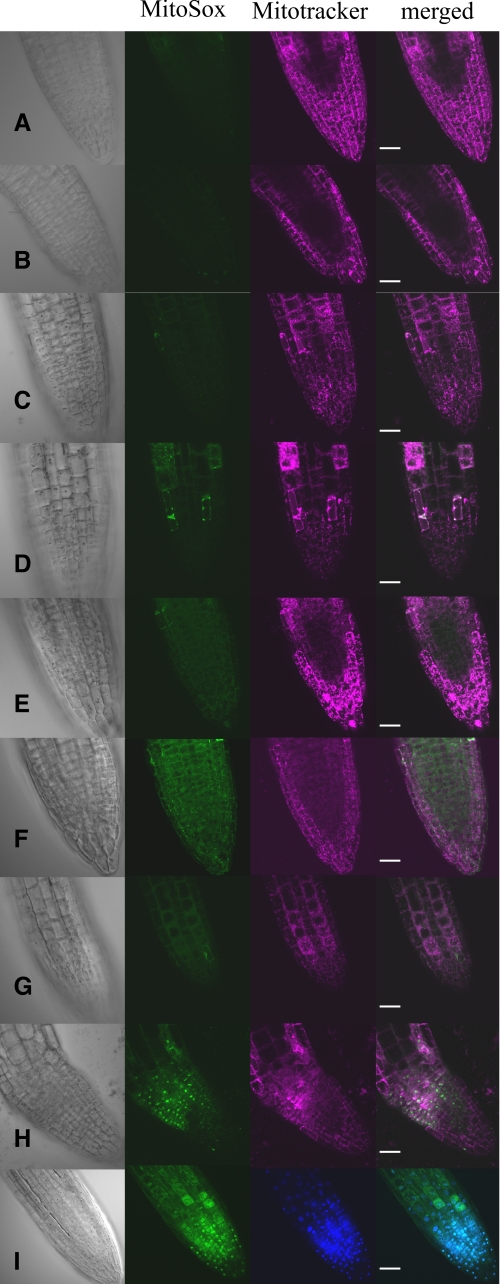
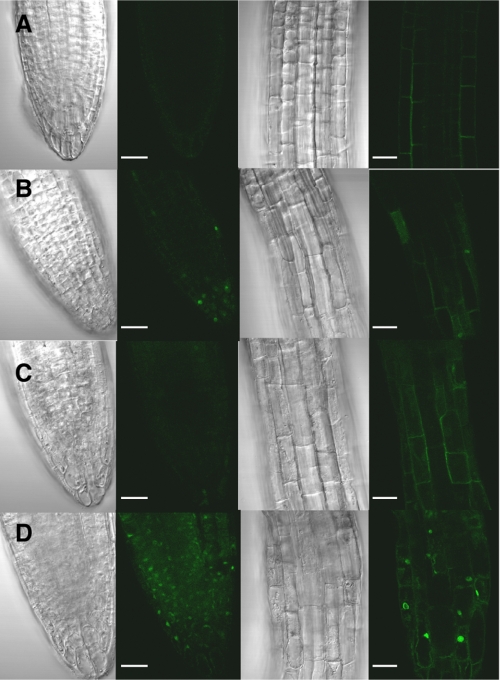
The roots of 5-day-old WT and p5cdh seedlings, exposed for 24 h in the dark to 100 mm Pro, 100 μm PQ, 100 mm Pro + 100 μm PQ, or untreated (control), were stained with MitoSox and Mitotracker for detecting O2˙̄ generation and mitochondrial localization, respectively (supplemental data). Growth on MS as a control treatment resulted in a very weak MitoSox-Red fluorescent signal in both WT and p5cdh roots (Fig. 7, A and B). The addition of Pro (100 mm) generated a relatively higher O2˙̄ level only in the p5cdh mutant, as was visualized by the increased MitoSox fluorescence (Fig. 7D), compared with the WT control (Fig. 7C), indicating increased P5C-Pro cycling in the mutant. PQ (100 μm) application enhanced O2˙̄ detection in mitochondria of WT roots, although the O2˙̄ level in p5cdh roots was much higher (Fig. 7, E and F, respectively), indicating that the extent of electron transfer to O2 in mitochondria of p5cdh exceeded that of WT plants. These effects were prominent in root tips whose Pro levels and Pro metabolism are normally very high (60, 61). The combined application of Pro and PQ (Fig. 7, G–I) generated even higher levels of O2˙̄ in the mutant compared with the WT, but only in the mutant did additional stained dots appear that did not colocalize with MitoTracker staining (Fig. 7, H and I). 4′,6-diamidino-2-phenylindole staining identified the dots as stained nuclei, indicating that in the Pro + PQ treatment MitoSox was able to penetrate the nuclei of root cells in the p5cdh mutant (Fig. 7I). Thus, increased leakage of ROS from mitochondria might have allowed the entrance of MitoSox into nuclei and the staining of nuclear DNA. These data confirmed that the increased P5C-Pro cycle-linked mtETC activity in p5cdh plants leads to higher mitochondrial O2˙̄ formation.
FIGURE 7.
Exogenous Pro application induces mitochondrial ROS generation in WT and p5cdh roots under dark conditions. Confocal root tip images of 5-day-old WT (A, C, E, and G) and p5cdh (B, D, F, H, and I) seedlings exposed to the following treatments for 24 h in the dark: A and B, control, no treatment; C and D, 100 mm Pro; E and F, 100 μm PQ; G–I, 100 mm Pro + 100 μm PQ. Left column, Numarski images. MitoSox fluorescence images are in green pseudo-color (second from the left). MitoTracker Deep-Red images are in magenta pseudo-color (third from the left), and the merged fluorescence is in the right column. Nuclear localization of MitoSox (I) is shown by costaining with 4′,6-diamidino-2-phenylindole (blue). Bar, 25 mm.
Pro Toxicity and Hypersensitivity of p5cdh Plants
Exogenously applied Pro was shown to be toxic to Arabidopsis plants, especially those lacking either ProDH or P5CDH activity (7, 28, 31, 32, 37). The hypersensitivity of p5cdh plants to Pro was attributed to the overaccumulation of P5C, suggested to cause ROS formation by an undefined mechanism (31, 34, 37).
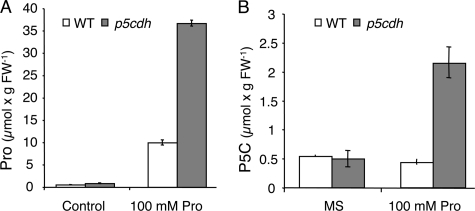
To verify the effect of exogenously provided Pro on the endogenous equilibrium that exists between P5C and Pro, Pro (100 mm) was provided for 48 h to 3-week-old p5cdh and WT plants. The p5cdh plants accumulated 3.7-fold more free Pro in comparison with WT plants (Fig. 8A). Under these conditions, Pro levels in p5cdh plants exceeded the endogenous Pro levels induced by drought stress (Fig. 4B) by more than 2-fold, indicating the need for P5CDH activity for overcoming unnaturally high Pro levels.
FIGURE 8.
The effect of exogenous Pro application on Pro and P5C content in leaves of p5cdh and WT plants. Three-week-old plants, grown on vermiculite, were treated with MS (Control) or MS containing 100 mm Pro. Free Pro (A) and P5C (B) levels of leaf samples were determined 48 h later. The standard deviation of three replicates is presented.
P5C levels were similar in both WT and p5cdh plants under normal conditions and remained unchanged in WT plants following Pro treatment (Fig. 8B). However, compared with WT, p5cdh plants had a 4.35-fold increase in P5C content in response to Pro treatment (Fig. 8B), reaching a maximal value of 2.3 μmol/gram of fresh weight (Fig. 8B). Thus, the increased P5C and Pro content in the p5cdh mutant above WT levels (Fig. 8) indicated that P5C oxidation to Glu is required for overcoming the high uptake of exogenous Pro and preventing the nonphysiological P5C accumulation. We next tested the proposed toxic effect of P5C by using the only commercially available P5C (Sigma-Aldrich) in the form of 2,4-dinitrophenylhydrazine hydrochloride salt (P5C-DNPH-Cl2), which stabilizes P5C under mild acidic conditions (supplemental Fig. S5 and Ref. 49). P5C-DNPH-Cl2 was found to be highly toxic to Arabidopsis seedlings, whereas the purified P5C did not cause a significant damage (supplemental Fig. S5). Hence, it is very likely that in the p5cdh mutant, the extensive activity of the P5C-Pro cycle coupled to elevated mitochondrial ROS production (Fig. 7, G–I) is responsible for the toxicity of the exogenously provided Pro excess rather than P5C (31, 37). O2˙̄ generation was more prominent in roots of light grown p5cdh seedlings (Fig. 9) that showed MitoSox fluorescence in root tips even in the control treatment (no Pro) (Fig. 9, A and B). Pro addition further increased MitoSox fluorescence and penetration to the nuclei in p5cdh roots (Fig. 9, C and D), indicating that ROS production in the light is higher than in dark and does not require PQ amplification for visualization. These results further emphasize the role of P5C oxidation to Glu (Fig. 9C compared with Fig. 9D) in limiting P5C-Pro cycle activity, especially during extreme cases when the cells are overwhelmed with Pro.
FIGURE 9.
Pro-induced ROS generation in roots of light grown seedlings. Confocal images of roots of 5-day-old WT (A and C) and p5cdh (B and D) light grown seedlings, untreated (A and B) or treated with 100 mm l-Pro (C and D) for 24 h. Root tips (left, first two columns) and elongation zones (third and fourth columns) are shown. Numarski images are in gray tone (first and third columns) and MitoSox fluorescence in green pseudo-color (second and fourth columns). Bar, 25 mm.
DISCUSSION
Pro catabolism in plants is subjected to developmental, metabolic, and stress control. ProDH and P5CDH are highly expressed in floral organs and meristematic zones, as well as in vegetative parts in the dark upon sugar depletion. The most prominent Pro catabolic activity occurs during recovery from abiotic stresses (7, 12, 25, 37, 62), when the stress-accumulated Pro (Fig. 1) serves as an energy source for the recovery processes (2). Thus, free Pro, whose synthesis is photosynthetically dependent (63), is not only acting as a compatible solute during osmotic stress but is also used as a means of shuttling photosynthetic energy to the mitochondria.
In this study we revealed a novel aspect of Pro metabolism in plants by demonstrating the existence of a P5C-Pro cycle (Fig. 10). This cytosolic-mitochondrial cycle is responsible for the preferential Pro accumulation, keeping a high cellular Pro to P5C ratio and tunneling electrons to the mitochondrial electron transport chain.
FIGURE 10.
The hypothesized P5C-Pro cycle and its hyperactivity during uncoupled P5C oxidation. In response to abiotic stresses, Pro is gradually accumulated by increased synthesis and suppressed oxidation. Once the stress is relieved, ProDH-FAD complex oxidizes Pro to P5C while transferring electrons to the mtETC. P5C further oxidation to Glu and then to α ketoglutarate (αKG) is coupled to NAD(P)H formation. When Pro is supplied in excess and ProDH transcription and activity are induced, P5CDH activity does not keep up with ProDH activity, and Pro oxidation becomes uncoupled. Under such conditions, the rate of P5C generation in the mitochondria exceeds its further oxidation to Glu. P5C excess is transported to the cytosol and reduced by P5CR to Pro that is transported to the mitochondria. This P5C-Pro intensive cycling elevates the flow of electrons through ProDH-FAD to the mtETC and to O2, leading to concomitant generation of ROS. In the p5cdh mutant, the intensive operation of this cycle enhances ROS generation and their spreading to the nuclei and very likely induces programmed cell death (PCD). ETC, electron transfer chain; UQ, ubiquinone.
P5C-Pro Cycle
The cytosolic reduction of P5C, generated by mitochondrial Pro oxidation (P5C-Pro cytoplasmic-mitochondrial cycle; Fig. 10), was first identified in ProDH-overexpressing plants and then further confirmed by examining the P5CDH-deficient mutant (p5cdh2). Constitutive overexpression of alfalfa ProDH in tobacco and Arabidopsis increased Pro oxidizing capacity but did not reduce Pro content, compared with WT plants, under normal, salinity, or drought conditions. Furthermore, no increase in P5C levels or hypersensitivity to drought or salinity stresses were recorded (Fig. 3A and supplemental Fig. S3). The maintenance of comparable Pro and P5C levels in WT and ProDH-OE plants indicates that compensation of P5C overproduction is achieved by its immediate reduction to Pro and thereby WT-like balance between the two compounds is kept. Interestingly, AtProDH-overexpressing plants accumulated Pro to a lesser extent than WT plants after 24 h of exposure to polyethylene glycol 6000 (7). However, in both cases no changes in sensitivity to osmotic stress were recorded.
P5C-Pro cycling was observed in mammalian cells and termed the “proline cycle” (39). Our results suggest that this seemingly futile cycle that constantly recycles P5C and Pro between mitochondria and cytosol with no net energy gain exists in plants and is responsible for preventing P5C accumulation as well as shuttling redox equivalents from the cytosol to the mtETC (Fig. 10).
To further support the existence of such a P5C-Pro cycle in Arabidopsis, Pro and P5C levels were measured in the p5cdh mutant that cannot oxidize P5C to Glu. Dehydration-induced Pro accumulation was similar in both WT and p5cdh plants; however, only the mutant retained high Pro levels during the post-stress rehydration period, when the endogenous ProDH was highly up-regulated (Fig. 4). Yet no increase in P5C level accompanied this high ProDH activity. Furthermore, there was no increase in the in the p5cdh mutant of other potential P5C precursors, such as Glu, Arg, and ornithine (Fig. 5). These data strongly support the operation of a Pro-P5C cycle that prevents P5C excess caused by the absence of P5CDH in the p5cdh mutant by reducing it back to Pro.
Linkage to ROS Generation
High activity of the P5C-Pro cycle in the ProDH-OE and p5cdh plants provides an excess of electrons to the mitochondrial electron transport chain via FAD-dependent-ProDH (Fig. 10). The increased electron flow is partially diverted to O2, resulting in O2˙̄ formation, as was evident in the roots of the p5cdh mutant by specific MitoSox staining of mitochondrial ROS (Figs. 7 and 9). This ROS generation occurs in the mitochondria under dark and light conditions (Figs. 7 and 9, respectively).
Although the existence of such a cycle has not been reported previously in plants, a similar cycle operates in mammalian cells causing ROS formation by p53-induced proline oxidase (HsProDH1, PIG6), leading to the induction of programmed cell death in normal cells (1, 39, 64). When P5CDH (ALDH4 in mammals) is normally operating and oxidizing P5C, it exerts a negative control on p53-induced apoptosis, by oxidizing P5C and reducing P5C-Pro pool (39, 65).
Our model suggests that the operation of the P5C-Pro cycle controls the cellular excess of P5C that is not immediately oxidized to Glu by the mitochondrial P5CDH and exported to the cytosol (Fig. 10). Therefore, its further reduction to Pro depends on P5CR availability and the possible existence of yet undefined P5C transporter(s) in the mitochondrial outer membrane. Until now only a Pro-specific inward transporter and Pro (inward)/Glu (outward) antiporter have been identified in the mitochondrial membrane of durum wheat, responsible for Pro entrance (66). However, these transporters have not been tested for their ability to transfer P5C. Even in mammalian cells with a functional P5C-Pro cycle, no information exists regarding mitochondrial P5C transporters.
In mammalian cells, P5C and Pro constitute a redox couple whose interconversion can affect NADP+/NADPH ratio and thereby control metabolic pathways and gene expression (67–71). In plants, the operation of the cycle is likely P5C-dependent and probably relies on the balance between P5CR and P5CDH activities, responsible for maintaining a constant low level of cellular P5C. In plants with higher P5C generation, such as ProDH-OE and p5cdh plants, prevention of P5C accumulation is probably controlled by P5CR activity, regardless of changes in its transcript levels, as was observed in the p5cdh mutant (Fig. 4). This immediate reduction of P5C to Pro accounted for the maintenance of significantly higher Pro levels in the mutant, compared with WT, during the recovery period that followed 72 h of drought stress (Fig. 4).
Down-regulation of P5CDH expression, caused by a nat-siRNA, was shown to occur only during salt stress (31). Mutants unable to produce this nat-small interfering RNA could not suppress P5CDH expression and showed less Pro accumulation in response to a 6-h treatment with 150 mm NaCl (31). This natural suppression of P5CDH during salt stress (31), in addition to transcriptional silencing of ProDH (12), further suggests that the regulation of P5C-Pro levels plays an important role in plants.
The Role of P5CDH
Pro accumulation is believed to support salinity, cold, and drought tolerance (2, 18) but is probably toxic to plants exposed to high temperatures (55). The absence of P5CDH activity renders Arabidopsis seedlings more sensitive to heat shock (Fig. 6), suggesting that P5C oxidation, which reduces the pool of P5C and eventually the activity of the P5C-Pro cycle, is essential for survival during heat stresses. By contrast, this cycle is significantly less active during drought and salinity because of the stress-induced down-regulation of ProDH transcription (Fig. 4 and Ref. 12). Still, under salinity (in tobacco) and drought stresses (Arabidopsis) the normal function of the P5C-Pro cycle and Pro to P5C balance are maintained, as was evident in ProDH-OE plants where the ectopic MsProDH was active during stress (Fig. 3 and supplemental Fig. S4). This indirect control of the Pro-P5C cycle raises the question of how the hyperactivity of this cycle is being regulated, for example, when an excess of Pro is provided?
Initially it was suggested that in response to exogenous Pro treatment, the induced expression of ProDH precedes that of P5CDH (32), leading to P5C accumulation and ROS formation. However, no P5C-derived mechanism of ROS generation has been proposed. Here we demonstrate that exogenously provided Pro led to an increase in Pro but not in P5C content in WT plants, whereas in p5cdh plants, Pro and P5C levels showed 3.7- and 4.35-fold increases, respectively (Fig. 8). These findings indicate that when an excess of Pro is provided to Arabidopsis plants, the normal Pro to P5C balance is maintained by the P5C-Pro cycle despite the abnormal high flux of Pro through the cycle. Under such circumstances, the involvement of P5CDH that reduces P5C levels by oxidation to Glu is essential for limiting Pro-P5C cycle operation and ROS generation through the mtETC (Fig. 10). Hence P5CDH controls P5C flow between the P5C-Pro cycle and P5C oxidation route. Accordingly, in p5cdh plants, the extent of mitochondrial ROS formation was by far higher than that generated in WT plants and spread to other cellular compartments as was evident by the staining of nuclei by MitoSox (Figs. 7, H and I, and 9D). Furthermore, the inability to oxidize P5C to Glu and then further to α-ketoglutarate in p5cdh plants might significantly decrease mitochondrial NAD(P)H generation, which in turn can affect the regeneration of reduced glutathione by NADPH-dependent glutathione reductase (72) and slow down the mitochondrial ascorbate/glutathione cycle leading to enhanced ROS levels in the mitochondria. Taken together, P5CDH can be considered as an important regulator in plants, involved in controlling the activity of the P5C-Pro cycle and contributing to mitochondrial redox control and to prevention of ROS overproduction.
Supplementary Material
Acknowledgments
We thank Prof. Amram Eshel (Tel Aviv University) for help in the water retention measurements. The contribution of Samuel Cohen-Salmon to the initial development of the Pro oxidation assay is greatly appreciated. We also thank Israel Science Foundation 214/08, COST Action FA0605, and EU FP7-Marie Curie 447.
This work was supported by EU-FP5 Grant EU QLK5-CT-2002-00841 (to A. Z.) and Israel Science Foundation Grant 1432/08.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S6.
- P5C
- Δ1-pyrroline-5-carboxylate
- GSA
- glutamate semialdehyde
- P5CDH
- P5C dehydrogenase
- P5CR
- P5C reductase
- P5CS
- P5C synthetase
- PQ
- paraquat
- ProDH
- Proline dehydrogenase
- ProDH-OE
- ProDH overexpressing plants
- T4C
- l-thiazolidine-4-carboxylic acid
- WT
- wild type plants
- ROS
- reactive oxygen species
- mtETC
- mitochondrial electron transport chain
- nat-siRNA
- natural sense and antisense small interfering RNA.
REFERENCES
- 1.Hu C. A., Donald S. P., Yu J., Lin W. W., Liu Z., Steel G., Obie C., Valle D., Phang J. M. (2007) Mol. Cell Biochem. 295, 85–92 [DOI] [PubMed] [Google Scholar]
- 2.Hare P. D., Cress W. A. (1997) Plant Growth Regul. 21, 79–102 [Google Scholar]
- 3.Csonka L. N. (1989) Microbiol. Rev. 53, 121–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delauney A. J., Verma D. P. S. (1993) Plant J. 4, 215–223 [Google Scholar]
- 5.Chen T. H., Murata N. (2002) Curr. Opin. Plant Biol. 5, 250–257 [DOI] [PubMed] [Google Scholar]
- 6.Takagi H., Iwamoto F., Nakamori S. (1997) Appl. Microbiol. Biotechnol. 47, 405–411 [DOI] [PubMed] [Google Scholar]
- 7.Mani S., Van De Cotte B., Van Montagu M., Verbruggen N. (2002) Plant Physiol. 128, 73–83 [PMC free article] [PubMed] [Google Scholar]
- 8.Kishor P., Hong Z., Miao G. H., Hu C., Verma D. (1995) Plant Physiol. 108, 1387–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanjo T., Kobayashi M., Yoshiba Y., Sanada Y., Wada K., Tsukaya H., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. (1999) Plant J. 18, 185–193 [DOI] [PubMed] [Google Scholar]
- 10.Yoshiba Y., Kiyosue T., Nakashima K., Yamaguchi-Shinozaki K., Shinozaki K. (1997) Plant Cell Physiol. 38, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 11.Roosens N. H., Willem R., Li Y., Verbruggen I. I., Biesemans M., Jacobs M. (1999) Plant Physiol. 121, 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller G., Stein H., Honig A., Kapulnik Y., Zilberstein A. (2005) Planta 222, 70–79 [DOI] [PubMed] [Google Scholar]
- 13.Xue X., Liu A., Hua X. (2009) BMB Rep. 42, 28–34 [DOI] [PubMed] [Google Scholar]
- 14.Verbruggen N., Hermans C. (2008) Amino Acids 35, 753–759 [DOI] [PubMed] [Google Scholar]
- 15.Székely G., Abrahám E., Cséplo A., Rigó G., Zsigmond L., Csiszár J., Ayaydin F., Strizhov N., Jásik J., Schmelzer E., Koncz C., Szabados L. (2008) Plant J. 53, 11–28 [DOI] [PubMed] [Google Scholar]
- 16.Delauney A. J., Hu C. A., Kishor P. B., Verma D. P. (1993) J. Biol. Chem. 268, 18673–18678 [PubMed] [Google Scholar]
- 17.Roosens N. H., Thu T. T., Iskandar H. M., Jacobs M. (1998) Plant Physiol. 117, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J. K. (2002) Annu. Rev. Plant Biol. 53, 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X. J., van de Cotte B., Van Montagu M., Verbruggen N. (1997) Plant Physiol. 114, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua X. J., Van de Cotte B., Van Montagu M., Verbruggen N. (2001) Plant J. 26, 157–169 [DOI] [PubMed] [Google Scholar]
- 21.Tanner J. J. (2008) Amino Acids 34, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayapati P. J., Stewart C. R. (1991) Plant Physiol. 95, 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Z., Lu Q., Verma D. P. S. (1996) Mol. Gen. Genet. 253, 334–341 [DOI] [PubMed] [Google Scholar]
- 24.Mattioni C., Lacerenza N. G., Troccoli A., De L. A., Di F. N. (1997) Physiol. Plant. 101, 787–792 [Google Scholar]
- 25.Nakashima K., Satoh R., Kiyosue T., Yamaguchi-Shinozaki K., Shinozaki K. (1998) Plant Physiol. 118, 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004) Plant Physiol. 136, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson J., Hanssen M., Wiese A., Hendriks M. M., Smeekens S. (2008) Plant J. 53, 935–949 [DOI] [PubMed] [Google Scholar]
- 28.Nanjo T., Fujita M., Seki M., Kato T., Tabata S., Shinozaki K. (2003) Plant Cell Physiol. 44, 541–548 [DOI] [PubMed] [Google Scholar]
- 29.Larkindale J., Vierling E. (2008) Plant Physiol. 146, 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elthon T. E., Stewart C. R. (1982) Plant Physiol. 70, 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borsani O., Zhu J., Verslues P. E., Sunkar R., Zhu J. K. (2005) Cell 123, 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deuschle K., Funck D., Hellmann H., Däschner K., Binder S., Frommer W. B. (2001) Plant J. 27, 345–356 [DOI] [PubMed] [Google Scholar]
- 33.Bonner C. A., Williams D. S., Aldrich H. C., Jensen R. A. (1996) Plant Sci. 113, 43–58 [Google Scholar]
- 34.Hellmann H., Funck D., Rentsch D., Frommer W. B. (2000) Plant Physiol. 123, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxwell S. A., Davis G. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13009–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura M., Takagi H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12616–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deuschle K., Funck D., Forlani G., Stransky H., Biehl A., Leister D., van der Graaff E., Kunze R., Frommer W. B. (2004) Plant Cell 16, 3413–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Borchert G. L., Surazynski A., Hu C. A., Phang J. M. (2006) Oncogene 25, 5640–5647 [DOI] [PubMed] [Google Scholar]
- 39.Yoon K. A., Nakamura Y., Arakawa H. (2004) J. Hum. Genet. 49, 134–140 [DOI] [PubMed] [Google Scholar]
- 40.Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005) Plant Cell 17, 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki N., Bajad S., Shuman J., Shulaev V., Mittler R. (2008) J. Biol. Chem. 283, 9269–9275 [DOI] [PubMed] [Google Scholar]
- 42.Fowden L., Lewis D., Tristram H. (1967) Adv. Enzymol. Relat. Areas Mol. Biol. 29, 89–163 [DOI] [PubMed] [Google Scholar]
- 43.Budisa N., Minks C., Medrano F. J., Lutz J., Huber R., Moroder L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elthon T. E., Stewart C. R., Bonner W. D. (1984) Plant Physiol. 75, 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deutch C. E. (1992) J. Gen. Microbiol. 138, 1593–1598 [DOI] [PubMed] [Google Scholar]
- 46.Koncz C., Martini N., Mayerhofer R., Koncz-Kalman Z., Körber H., Redei G. P., Schell J. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H. R., Rho H. W., Park J. W., Park B. H., Kim J. S., Lee M. W. (1994) Anal. Biochem. 223, 205–207 [DOI] [PubMed] [Google Scholar]
- 48.Bates L. S., Waldren I. P., Teare I. D. (1973) Plant Soil 39, 205–207 [Google Scholar]
- 49.Mezl V. A., Knox W. E. (1976) Anal. Biochem. 74, 430–440 [DOI] [PubMed] [Google Scholar]
- 50.Yang C. W., Kao C. H. (1999) Plant Growth Regul. 27, 191–194 [Google Scholar]
- 51.Brosché M., Vinocur B., Alatalo E. R., Lamminmäki A., Teichmann T., Ottow E. A., Djilianov D., Afif D., Bogeat-Triboulot M. B., Altman A., Polle A., Dreyer E., Rudd S., Paulin L., Auvinen P., Kangasjärvi J. (2005) Genome Biol. 6, R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribarits A., Abdullaev A., Tashpulatov A., Richter A., Heberle-Bors E., Touraev A. (2007) Planta 225, 1313–1324 [DOI] [PubMed] [Google Scholar]
- 53.Kiyosue T., Yoshiba Y., Yamaguchi-Shinozaki K., Shinozaki K. (1996) Plant Cell 8, 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oono Y., Seki M., Nanjo T., Narusaka M., Fujita M., Satoh R., Satou M., Sakurai T., Ishida J., Akiyama K., Iida K., Maruyama K., Satoh S., Yamaguchi-Shinozaki K., Shinozaki K. (2003) Plant J. 34, 868–887 [DOI] [PubMed] [Google Scholar]
- 55.Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., Mittler R. (2004) Plant Physiol. 134, 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhoads D. M., Umbach A. L., Subbaiah C. C., Siedow J. N. (2006) Plant Physiol. 141, 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizhsky L., Davletova S., Liang H., Mittler R. (2004) J. Biol. Chem. 279, 11736–11743 [DOI] [PubMed] [Google Scholar]
- 58.Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., Pacher P. (2007) Biochem. Biophys. Res. Commun. 358, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson K. M., Janes M. S., Pehar M., Monette J. S., Ross M. F., Hagen T. M., Murphy M. P., Beckman J. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15038–15043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verslues P. E., Sharp R. E. (1999) Plant Physiol. 119, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raymond M. J., Smirnoff N. (2002) Ann. Bot. 89, 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savouré A., Jaoua S., Hua X. J., Ardiles W., Van Montagu M., Verbruggen N. (1995) FEBS Lett. 372, 13–19 [DOI] [PubMed] [Google Scholar]
- 63.Abrahám E., Rigó G., Székely G., Nagy R., Koncz C., Szabados L. (2003) Plant Mol. Biol. 51, 363–372 [DOI] [PubMed] [Google Scholar]
- 64.Rivera A., Maxwell S. A. (2005) J. Biol. Chem. 280, 29346–29354 [DOI] [PubMed] [Google Scholar]
- 65.Liu Y., Borchert G. L., Surazynski A., Phang J. M. (2008) Oncogene 27, 6729–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Martino C., Pizzuto R., Pallotta M. L., De Santis A., Passarella S. (2006) Planta 223, 1123–1133 [DOI] [PubMed] [Google Scholar]
- 67.Hagedorn C. H., Phang J. M. (1983) Arch. Biochem. Biophys. 225, 95–101 [DOI] [PubMed] [Google Scholar]
- 68.Phang J. M. (1985) Curr. Top. Cell Regul. 25, 91–132 [DOI] [PubMed] [Google Scholar]
- 69.Mixson A. J., Phang J. M. (1988) J. Biol. Chem. 263, 10720–10724 [PubMed] [Google Scholar]
- 70.Yeh G. C., Phang J. M. (1988) J. Biol. Chem. 263, 13083–13089 [PubMed] [Google Scholar]
- 71.Nemoto N., Sakurai J. (1991) Jpn. J. Cancer Res. 82, 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moller I. M. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.