Abstract
Recombination and pairing of homologous chromosomes are critical for bivalent formation in meiotic prophase. In many organisms, including yeast, mammals, and plants, pairing and recombination are intimately interconnected. The POOR HOMOLOGOUS SYNAPSIS1 (PHS1) gene acts in coordination of chromosome pairing and early recombination steps in plants, ensuring pairing fidelity and proper repair of meiotic DNA double-strand-breaks. In phs1 mutants, chromosomes exhibit early recombination defects and frequently associate with non-homologous partners, instead of pairing with their proper homologs. Here, we show that the product of the PHS1 gene is a cytoplasmic protein that functions by controlling transport of RAD50 from cytoplasm to the nucleus. RAD50 is a component of the MRN protein complex that processes meiotic double-strand-breaks to produce single-stranded DNA ends, which act in the homology search and recombination. We demonstrate that PHS1 plays the same role in homologous pairing in both Arabidopsis and maize, whose genomes differ dramatically in size and repetitive element content. This suggests that PHS1 affects pairing of the gene-rich fraction of the genome rather than preventing pairing between repetitive DNA elements. We propose that PHS1 is part of a system that regulates the progression of meiotic prophase by controlling entry of meiotic proteins into the nucleus. We also document that in phs1 mutants in Arabidopsis, centromeres interact before pairing commences along chromosome arms. Centromere coupling was previously observed in yeast and polyploid wheat while our data suggest that it may be a more common feature of meiosis.
Keywords: maize, meiosis, recombination, chromosome pairing
Meiotic prophase I encompasses a large number of tightly regulated and coordinated processes (1). Although many structural proteins involved in these processes are now known, mechanisms regulating meiotic prophase progression remain largely unexplored. In this study, we describe the function of the POOR HOMOLOGOUS SYNAPSIS (PHS1) gene in higher plants that regulates prophase I by coordinating recombination and homologous chromosome pairing (2). Meiotic recombination is initiated by SPO11-mediated formation of double-strand breaks (DSBs) in chromosomal DNA (3). In plants, this occurs in or before early leptotene (2, 4). The DSBs are subsequently resected by the MRN protein complex in plants and animals, and a similar MRX complex in S. cerevisiae, to generate single-stranded DNA (ssDNA) overhangs. The MRN complex contains MRE11, RAD50, and NBS1 (5, 6). MRE11 is a DNA-binding protein with endonuclease, exonuclease, and helicase activities that directly facilitate ssDNA overhang formation (6, 7). RAD50 most likely plays a structural role in the complex (6, 8). NBS1 regulates the activity of MRE11 and signals DSB presence (5, 6). ssDNA tails created by DSB resection are coated by two DNA strand-exchange proteins, RAD51 and DMC1 (9). The nucleoprotein filaments formed in this way find and invade double-stranded DNA regions on the homologous chromosome in the process of single-end invasion (SEI).
In plants, mammals, and fungi, although not in C. elegans or Drosophila, the initiation and progression of meiotic recombination are closely linked to homologous chromosome pairing (10). Pairing is an interaction between chromosomes that includes recognition of homology (the homology search) and juxtaposition of the homologs, which is closely followed by synapsis, a process of installing a proteinaceous structure of the synaptonemal complex (SC) (3). Plant, mammalian, and fungal mutants defective in meiotic recombination frequently show pairing defects. In Arabidopsis, mutations in the two meiosis-specific SPO11 homologs, SPO11–1 and SPO11–2, prevent chromosome pairing and synapsis, leading to the presence of univalents (unsynapsed chromosomes) at pachytene (11, 12). Similar defects are observed in Arabidopsis RAD50 and MRE11 knockouts (13, 14). A well-studied link between pairing and recombination is the RAD51/DMC1-mediated SEI step of meiotic recombination. Both RAD51 and DMC1 have been shown to facilitate homologous interactions between kilobase-long DNA substrates in vitro (15). Moreover, severe pairing defects have been observed in Arabidopsis rad51 and dmc1 mutants, the maize rad51 mutant, as well as in rad51 and dmc1 mutants in yeast, and the dmc1 mutant in mouse (16–20).
In contrast to the lack of synapsis observed in the spo11, mre11, and rad50 mutants, the phs1-O mutant in maize showed an unusual phenotype wherein synapsis took place almost exclusively (95% of the time) between nonhomologous chromosome segments (2). The mutant also exhibited a severe recombination defect, showing less than 1% of the normal number of RAD51 foci at mid-zygotene, even though the formation of meiotic DSBs and accumulation of the RAD51 protein in anthers were not affected. Here, we demonstrate that PHS1 is a cytoplasmic protein that regulates the progression of meiotic prophase I by controlling the entry of MRN complex protein RAD50 to the nucleus.
Results
The Arabidopsis PHS1 Homolog Exhibits a Similar Role in Chromosome Pairing as the Maize Phs1 Gene, Despite the Large Difference in Genome Size and Complexity Between the Two Species.
The maize genome is about 2.4 Gb in size and contains about 70% repetitive DNA (21). In addition, it shows extensive internal duplications as a consequence of its tetraploid origin (22). This genome complexity suggests that the nonhomologous chromosome associations observed in the maize phs1 mutant may be the result of ectopic pairing between repetitive DNA elements and/or duplicated (homoeologous) chromosome segments. To examine this possibility, we investigated chromosome pairing in mutants in the Arabidopsis homolog of Phs1. The Arabidopsis genome is about 20-fold smaller than the genome of maize, contains about 100-fold less repetitive DNA, and does not show large duplicated collinear chromosome segments (23). Consequently, in Arabidopsis, a mutation in a gene that prevents pairing between homoeologous genome regions or repetitive elements would not lead to extensive ectopic chromosome associations.
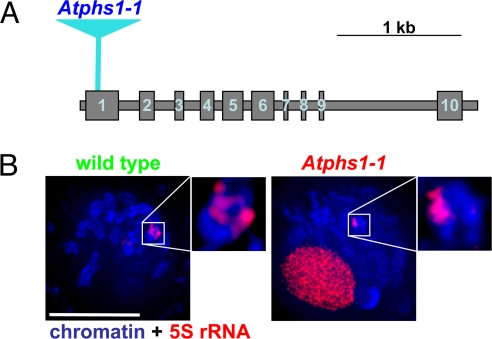
Only a single Arabidopsis gene (At1g10710, AtPHS1) shows significant homology to maize Phs1 (Fig. S1). To explore the function of AtPHS1, we generated RNAi knockdown lines using constructs with the constitutive CaMV 35S promoter and the meiosis-specific AtDMC1 promoter (24). Analyses of seven lines carrying the 35S:AtPHS1 construct and ten lines containing the pDMC1:AtPHS1 construct indicated that the presence of the transgene was associated with strong meiotic sterility (Fig. S2). In addition, we identified a line in the RIKEN collection (25) that carried an insertion of a modified maize Ds transposon in the first exon of AtPHS1 (Fig. 1A) resulting in severe sterility (Fig. S3). The sterility phenotypes corresponded to defects in meiotic chromosome behavior. Analyses of male meiocytes in the Ds-induced mutant (named Atphs1–1), and two RNAi lines, each carrying a different silencing construct, revealed that although chromosomes in pachytene showed complete or nearly complete bivalent formation, pairing defects, such as misalignment of chromosome ends in bivalents, were visible in many cells (Fig. S4). In diakinesis, mostly univalents were observed.
Fig. 1.
The Arabidopsis thaliana PHS1 homolog. (A) The AtPHS1 gene. Blue triangle = position of the Ds insertion in the Atphs1–1 mutant. (B) Paring of 5S rRNA loci in a wild-type and Atphs1–1 mutant meiocytes. Only one 5S locus associated with a nonhomologous chromosome segment is shown in the mutant. Closeups are shown in insets. Images are flat projection of several consecutive optical sections but do not represent entire nuclei. (Scale bar, 5 μm.)
To examine homologous chromosome pairing in the mutant meiocytes, we used 3-D microscopy coupled with fluorescent in situ hybridization (FISH) with probes recognizing the 5S and 25S rRNA loci, which we found to be the most robustly working probes in our previous experiments (Fig. 1B and Table S1). In wild-type meiocytes, we always observed distinct pairs of homologously associated 5S and 25S rRNA loci in pachytene. In contrast, in mutant meiocytes we found homologous pairing at the 5S and 25S rRNA loci only about 45% of the time, while in the other 55% of cases, we observed associations of the rRNA loci with nonhomologous chromosome regions. Overall, the Arabidopsis phs1 mutants showed significant homologous pairing defects that were similar, albeit less severe, to the defects observed in the phs1 mutant in maize (2). These data suggest that the nonhomologous chromosome associations in phs1 mutants do not result from ectopic pairing between repetitive genome regions. Instead, the target for the PHS1-mediated regulation of chromosome pairing is most likely the gene-rich fraction of the genome.
Chromosome Dynamics: Arabidopsis phs1 Mutants Show Coupling of Centromeres.
To further understand the pairing defects in the Atpsh1 mutants, we investigated the telomere behavior during prophase I in the Atphs1–1 mutant and in a pDMC1:AtPHS1 RNAi line using a telomere-specific FISH probe. Telomere dynamics in mutant meiocytes, including prezygotene telomere clustering around the nucleolus (26) was indistinguishable from the wild-type (Fig. S5).
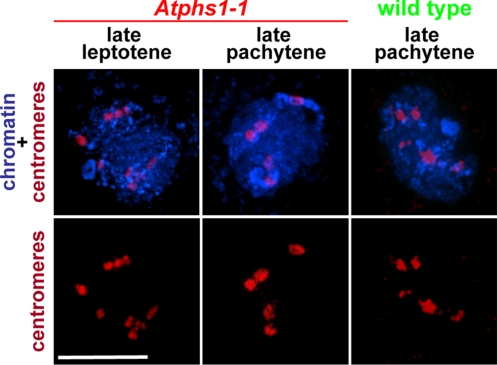
We also used a centromere-specific probe to examine the behavior of centromeres during prophase I. We found, surprisingly, that pachytene meiocytes from both mutant sources showed always the normal number of five centromere FISH signals (Fig. 2 and Fig. S5) (for Atphs1–1 n = 14; for the pDMC1:AtPHS1 RNAi line n = 11). This observation indicated that even though loci located on chromosome arms mispaired frequently, centromeric regions always associated with other centromeric regions, although we do not know whether these associations were between centromeres of homologous or nonhomologous chromosomes. This centromere coupling was first seen in zygotene, while in leptotene the 10 centromeres were always separate. Centromere coupling was preceded by a transient clustering of most, or sometimes all, Arabidopsis centromeres in early zygotene. The early zygotene clustering of all centromeres has been previously observed in wild-type Arabidopsis meiosis (26). However, in wild-type meiocytes, following the resolution of the cluster, the centromeres pair homologously concomitantly with pairing along the chromosome arms, and centromere coupling has not been observed as a separate chromosome pairing step.
Fig. 2.
Centromere coupling in the Arabidopsis Atphs1 mutant. Each image is a flat projection across the entire nucleus. (Scale bar, 5 μm.)
Synaptonemal Complex and Chromosome Axis Components Are Installed on Chromosomes in phs1 Mutants.
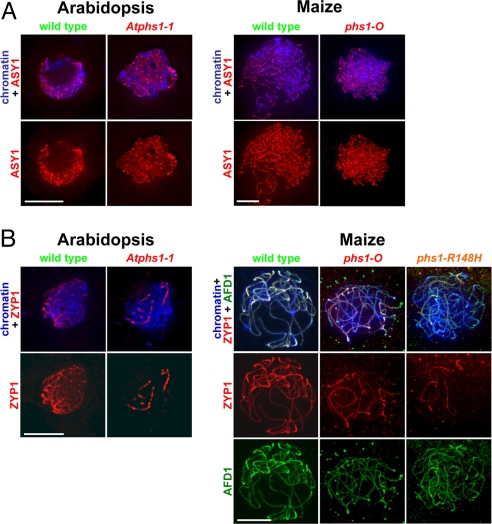
To elucidate whether the nonhomologous chromosome associations in phs1 mutants are caused by SC defects, we examined installation of chromosome axis and SC proteins. The chromosome axis forms in early leptotene and becomes the lateral element of the SC after chromosomes synapse in zygotene (3). To investigate chromosome axis formation, we analyzed installation of ASY1 (Fig. 3A), a protein associated with the chromosome axis in plants (27, 28). In maize, we also examined the installation of AFD1 (Fig. 3B), a homolog of the meiosis-specific alpha-kleisin Rec8 required for meiotic sister-chromatid cohesion and chromosome axis formation (28). However, in both cases, we found that mutant meiocytes were very similar to wild-type meiocytes.
Fig. 3.
Installation of chromosome axis and SC proteins on meiotic chromosomes in phs1 mutants in Arabidopsis and maize. (A) Installation of ASY1, a chromosome-axis associated protein in zygotene meiocytes. (B) Installation of ZYP1, the central element protein of the SC, and AFD1, a meiosis-specific cohesin, in pachytene. (Scale bar, 5 μm.)
To investigate installation of the central element of the SC, we followed localization of the central element protein ZYP1 (29). In wild-type meiocytes in both species, the ZYP1 antibody showed staining from zygotene to diplotene and was localized along each bivalent, between the two homologous chromosomes (Fig. 3B). In phs1 mutants, ZYP1 installation was delayed as some bivalent segments, and occasionally entire bivalents, still lacked ZYP1 at the end of zygotene (Fig. 3B). However, the ZYP1 protein was only installed in bivalents and the ZYP1 stretches were contiguous, suggesting that the basic pattern of the SC central element installation was similar to the wild-type.
Overall, these data showed that the patterns of installation of chromosome axis and SC proteins in phs1 mutant meiocytes were not significantly different from those in the wild-type, which suggests the pairing defects in phs1 mutants are unlikely to be results of abnormal chromosome axis or SC installation.
A Maize Point Mutation Allele, phs1-R148H, Identifies an Essential Functional Domain in PHS1.
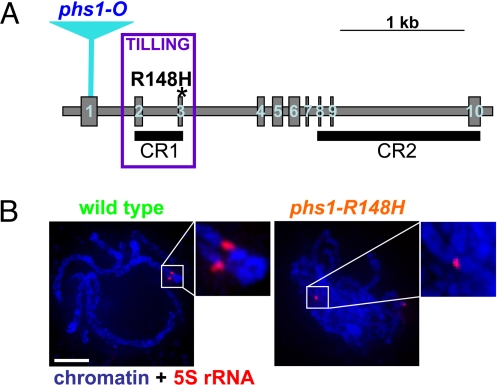
Sequence alignment of PHS1 homologs from several species identified two relatively short conserved regions, CR1 (Conserved Region 1) and CR2 (Fig. S1). The C-terminal CR2 encompasses a putative SUMOylation site (30). Modifications by SUMO are associated with various cellular processes, including transport from cytosol to the nucleus (31). In contrast, no known functional sequence motifs were apparent in CR1, which is located in the central portion of PHS1. To investigate whether CR1 corresponds to a functional domain, we generated a point mutation in this region using a reverse genetics TILLING approach (32). The mutation, named phs1-R148H, changes a conserved arginine at position 148 to a histidine (Fig. 4A and Fig. S1).
Fig. 4.
The maize phs1-R148H mutation. (A) The maize Phs1 gene. Blue triangle = position of Mutator transposon insertion resulting in the original phs1-O mutation. Violet box = region used for the TILLING screen. Asterisk = positions of the phs1-R148H mutation. (B) Paring of 5S rRNA loci in a wild-type and phs1-R148H mutant meiocytes. Both loci in the mutant are associated with nonhomologous chromosome segments. Closeups of homologously paired 5S rRNA loci in the wild-type and one of the loci in the mutant are shown in insets. The other 5S locus in the mutant is visible in the lower right corner of the cell. Images are flat projection of several consecutive optical sections but do not represent entire nuclei. (Scale bar, 5 μm.)
Plants homozygous for the phs1-R148H allele exhibited univalents at diakinesis and showed meiotic sterility (Fig. S6). Using FISH with the 5S rRNA locus probe we found that they showed pairing defects (Fig. 4B) similar to those observed in the maize phs1-O mutant (2). The 5S loci were homologously paired in only 8% of mutant meiocytes at pachytene while in 92% of cells, they were associated with nonhomologous chromosome segments. These data suggest that CR1 defines a functional domain required for the role of PHS1 in homologous chromosome pairing.
In contrast to phs1-R148H, none of several point mutations outside of the conserved CR1 and CR2 regions that we examined in Arabidopsis (Fig. S1) when searching for phs1 mutant alleles, showed meiotic defects, suggesting that the nonconserved PHS1 regions are not critical for the protein function.
PHS1 Is Required for Nuclear Localization of RAD50 in Meiosis.
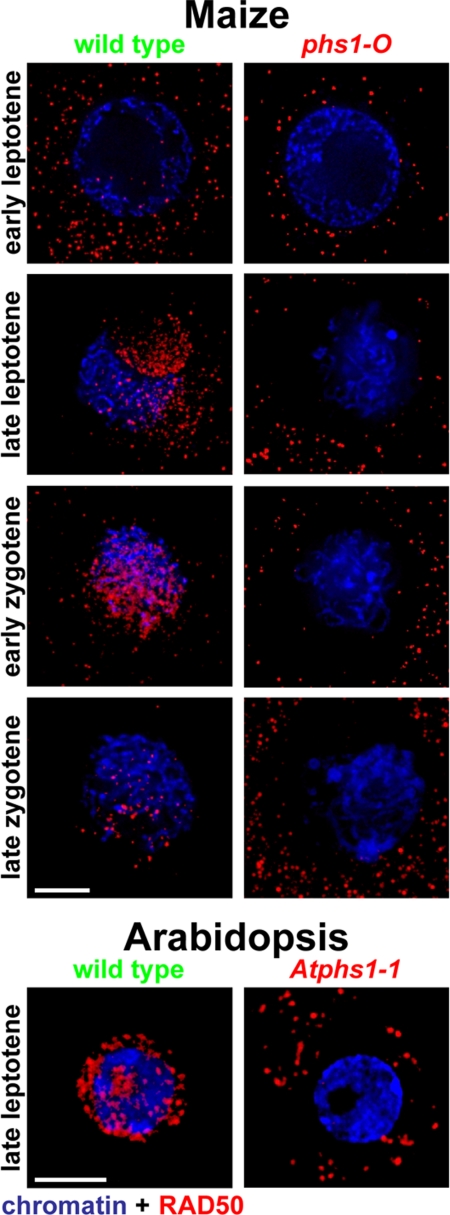
Analyses of the maize phs1-O mutant showed that meiotic DSBs were generated in chromosomal DNA but the number of RAD51 foci on chromosomes was greatly reduced (2). To identify the first step of the recombination pathway that is defective in phs1 mutants, we studied the localization patterns of RAD50, a well-conserved component of the MRN complex involved in an early step of DSB processing that precedes RAD51 loading onto the DSB sites. Absence of RAD50 leads to defective DSB repair (13). In wild-type maize and Arabidopsis meiocytes, the anti-AtRAD50 antibody showed staining from leptotene to pachytene, both in the cytoplasm and inside the nucleus, with some foci located on chromatin and some present in the nucleoplasm but not associated with chromatin (Fig. 5). In maize, the average number of foci inside the nucleus in leptotene was 364 (±71, n = 19). The foci number peaked at 548 per nucleus (±98, n = 17) in early zygotene and then decreased to 275 (±33, n = 25) in mid-zygotene. Only about 20 foci were still present in mid to late pachytene. No RAD50 signal was observed in diplotene. In Arabidopsis, the dynamics of RAD50 foci was similar to maize (Fig. 5) but the foci numbers were smaller: 171 foci per nucleus (±60, n = 16) in early zygotene and 8 (±4, n = 9) in pachytene. The anti-RAD50 antibody did not detect any signal in the Arabidopsis rad50–1 mutant (13), confirming that it was specific to RAD50.
Fig. 5.
Immunolocalization of RAD50 in wild-type and phs1 mutant meiocytes. Each image is a single optical section. (Scale bar, 5 μm.)
In contrast to the wild-type, in the phs1 mutants in both maize and Arabidopsis at all meiosis stages, very few RAD50 foci were observed inside the nucleus (Fig. 5) even though the protein was still present in the cytoplasm. For example, in the maize phs1-O mutant, only 26 foci (±4, n = 38) were found at mid zygotene, which represents less than 5% of the foci number observed in the wild-type at the same stage. Overall, our findings suggest that RAD50 fails to enter the nucleus from the cytoplasm in phs1 mutants.
The PHS1 Protein Localizes to the Cytoplasm During Leptotene and Zygotene.
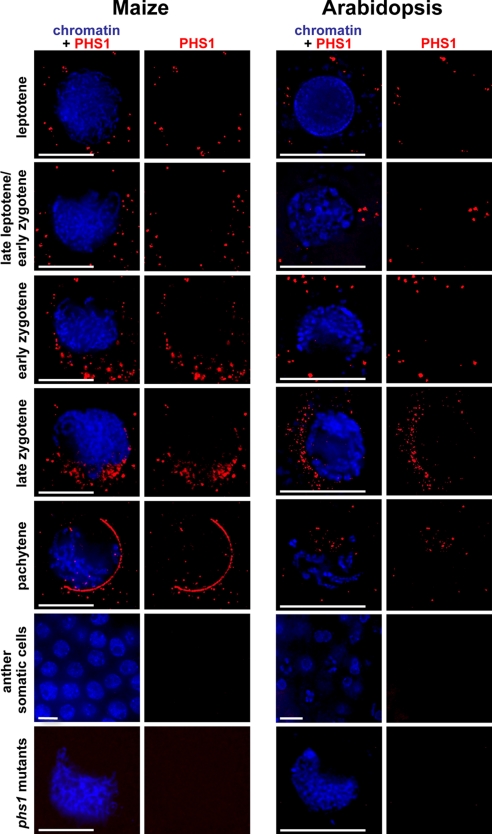
To investigate the subcellular localization of PHS1, we developed two polyclonal antibodies, one against the maize PHS1 and another one against the Arabidopsis homolog. In wild-type meiocytes in both species in leptotene and zygotene, PHS1 showed diffuse staining throughout the meiocyte but mostly formed numerous granules in the cytoplasm and not in the nucleus (Fig. 6). The protein was first visible in mid-leptotene but the peak of PHS1 accumulation was during zygotene. In zygotene, some maize meiocytes (fewer than 10%) showed PHS1 foci clusters specifically located along the nuclear envelope. However, such pattern was never observed in Arabidopsis. In late zygotene in both maize and Arabidopsis, PHS1 staining in the cytoplasm decreased and a few PHS1 foci were found inside the nucleus. After pachytene, the protein was no longer detectable. PHS1 was also absent from somatic cells in maize and Arabidopsis anthers and Arabidopsis petals (Fig. 6). No detectable PHS1 signal was also seen in the maize phs1-O and the Arabidopsis Atphs1–1 mutants (Fig. 6), indicating that the antibodies were PHS1-specific and confirming that phs1-O and Atphs1–1 are both null mutants.
Fig. 6.
Immunolocalization of the PHS1 protein. The phs1 mutants pictured are Atphs1–1 in Arabidopsis and phs1-O in maize. Each image is a single optical section. (Scale bar, 10 μm.)
Subcellular Localization of PHS1 Does Not Depend on Meiotic Recombination.
To elucidate whether the localization pattern of PHS1 is dependent on meiotic recombination, we examined the protein localization in male meiocytes of the Arabidopsis spo11–1-1 mutant. We found that the PHS1 localization pattern in the mutant meiocytes was similar to the wild-type (Fig. S7). These data suggest that PHS1 localization does not depend on SPO11 or the presence of DSBs on meiotic chromosomes.
Discussion
PHS1 Controls Nuclear Import of RAD50 in Meiosis.
Our observations indicate that the earliest defect in phs1 mutants is the failure of RAD50 to enter the nucleus. In plants, the MRN complex proteins are not required for the formation of meiotic DSBs, in contrast to budding yeast or C. elegans (6, 13, 14). However, their nuclear import is not a response to DSB formation because normal numbers of nuclear foci of MRE11 are present in Arabidopsis mutants lacking meiotic DSBs (33). As neither RAD50 nor MRE11 contain nuclear localization signals, these data suggest that the import of the MRN protein complex into the nucleus is a regulated process independent from recombination initiation.
Our experiments in maize and Arabidopsis using two different species-specific antibodies showed that the PHS1 protein accumulates in the cytoplasm during leptotene and most of zygotene. During the same time, PHS1 affects nuclear localization of RAD50. These results indicate that PHS1 regulates the import of RAD50 into the nucleus by acting in the cytoplasm. PHS1 does not have a broad role in shuttling meiotic proteins into the nucleus since several other meiotic proteins, including ASY1, AFD1, and ZYP1, do enter the nucleus in phs1 mutants. In mammalian cells, NBS1 is known to be required for nuclear localization of the MRN complex (34). Interestingly, nbs1 mutants in Arabidopsis do not show a meiotic defect on their own (5), suggesting that MRN proteins in plants may use a different mechanism to enter the nucleus. Although it is possible that PHS1 is directly involved in physically shuttling RAD50 into the nucleus, we do not favor this possibility because if this was the case, we would expect to see larger amounts of the PHS1 protein inside the nucleus during leptotene and zygotene. Instead, we propose that PHS1 plays a regulatory role in this process.
We previously showed that in the maize phs1-O mutant meiotic DSBs were formed but RAD51 foci were absent from the sites of DSBs (2). As RAD51 is recruited to single-stranded DNA ends created through resection of meiotic DSBs by MRN (9), the absence of RAD51 foci in the phs1-O mutant is a direct consequence of the absence of RAD50 from the nucleus. However, the presence of RAD50 in the nucleus is not completely eliminated in phs1 mutants, suggesting that an alternative mechanism, albeit a less efficient one, exists for nuclear import of the two proteins. The roughly 5% remaining RAD50 foci likely lead to proper repair of some DSBs and could account for the observation of a small number of RAD51 foci in the maize phs1-O mutant (2). These foci may also be responsible for the fact that the chromosome fragmentation observed in rad50 knockout mutants (13, 14) is not seen in phs1 mutants, although even in the rad50 knockouts most DSBs are repaired by MRN-independent mechanisms since the numbers of observed broken chromosome fragments in these mutants is minuscule compared to the number of SPO11-generated DSBs.
Why Do Homologous Chromosomes Fail to Pair Properly in phs1 Mutants?
Maize and Arabidopsis phs1 mutants show severe defects in chromosome homology recognition and meiotic recombination. In contrast, other prophase I processes are not defective, including telomere clustering, sister-chromatid cohesion, and chromosome axis formation. Chromatin structure appears normal in light microscopy examination. Installation of the SC central element protein ZYP1 is also largely unaffected, although it is delayed and may be incomplete. Based on these data, we conclude that the failure of RAD50 to enter the nucleus and resect meiotic DSB is the primary defect in phs1 mutants.
Arabidopsis mutants lacking MRN complex proteins RAD50 and MRE11 show chromosome pairing defects, although these defects are mostly lack of synapsis (13, 14) rather than the extensive nonhomologous synapsis that we observe in the phs1 mutants. However, in phs1 mutants, a small number of DSBs are properly processed and repaired, as evidenced by the few RAD50 and RAD51 foci present on chromosomes. This hypomorphic phenotype could result in a different chromosome pairing defect than the complete absence of RAD50 in a knockout mutant. A hypomorphic mre11S mutation in budding yeast, which blocks DBS processing but not their formation, has been shown to lead to nonhomologous synapsis (35). Conceivably, a small number of ssDNA ends that are available for the RAD51/DMC1-facilitated process of homology search could lead to ectopic pairing interactions between nonhomologous loci.
Centromere Coupling.
The observation of centromere coupling in phs1 mutants suggests that interactions between centromeric regions precede pairing along chromosome arms. Presynaptic centromere coupling has been reported in yeast and polyploid wheat (36, 37). Our observations suggest that it may be a more universal phenomenon. Although centromere coupling is not distinguishable as a separate chromosome pairing step in wild-type Arabidopsis, it may be because in wild-type meiosis centromere coupling is ephemeral and quickly followed by chromosome arm pairing. Centromere coupling in wheat precedes meiosis and is also present in somatic cells that do not become meiocytes (37). In contrast to wheat, but similarly to Arabidopsis, centromere coupling in yeast is transient and occurs at the onset of meiotic chromosome pairing (36). However, centromere coupling in yeast is mediated by the installation of the SC central element protein at the centromeres (36, 38), while our observations showed that in some cases the ZYP1 protein appeared to be absent from entire bivalents in the Atphs1 mutant, which suggests that the mechanism of centromere coupling in Arabidopsis may be different from the one in yeast. Centromere coupling in the Arabidopsis phs1 mutant implies that PHS1 is required for homologous pairing along chromosome arms, but chromosome interactions at the centromeres are governed by a different mechanism.
PHS1 Is a Part of a Regulatory System That Controls Nuclear Localization of Meiotic Proteins.
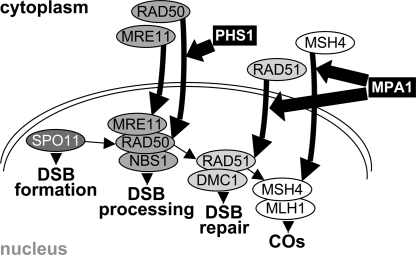
An intriguing question is whether the action of PHS1 is a unique way of regulating meiosis or a part of a broader system that directs meiosis progression by controlling entry of meiotic proteins into the nucleus. Meiotic prophase I events require coordination between seemingly independent processes (1). It is conceivable that for this complex coordination to work, meiotic proteins must be delivered to their place of action at a specific time and in the correct order. The Arabidopsis MPA1 gene is be required for nuclear localization of the RAD51 and MSH4 proteins (39) and it is possible that it plays a direct role in regulating the entry of RAD51 and MSH4 to the nucleus. We propose that PHS1 and MPA1 are both components of a regulatory network that controls the progression of meiotic prophase by regulating nuclear import of specific meiotic proteins (Fig. 7).
Fig. 7.
A model for meiotic prophase regulation by controlling transport of meiotic proteins into the nucleus. PHS1 regulates DSB resection by controlling nuclear import of RAD50. MPA1 regulates DSB repair by controlling nuclear import of RAD51 and MSH4.
We found sequence homologs of PHS1 only in Angiosperms. This recent evolutionary origin may be linked to the function of this protein. In contrast to other taxa, plants show little temporal regulation of meiotic gene expression: plant homologs of some meiotic genes that show tight transcriptional regulation in other species, such as SPO11 or HOP2, are ubiquitously expressed in plants (11, 40). Regulating the entry of meiotic proteins into the nucleus may be a way of compensating for the lack of transcriptional control.
Materials and Methods
AtPHS1 RNAi Lines.
To silence the AtPHS1 gene with RNAi, we used a 524-bp fragment of AtPHS1 (nt 26–549), and followed the method described by Siaud et al. (24).
Screening for TILLING Alleles.
In maize, EMS-mutagenized maize lines were screened with primers TGATTAGATTTCCTCGCACTCGCAGAA and TGCATAAGGGCAACACTGAAAGACATG. In Arabidopsis, primers TCTTCGACGGTTTCTCTCCACATTGTC and GCTTCACAGTTGTTTGGCCTGCATCTA were used.
PHS1 Antibody.
Full-length maize and Arabidopsis PHS1 cDNAs were cloned in to the pDONR221 vector (Invitrogen). The constructs were then recombined into the pET160-DEST vector (Invitrogen) and transformed into E. coli Arctic express (DE3) cells (Stratagen). The recombinant proteins were purified using Ni-NTA columns (Qiagen) under native conditions and the antigen was used to immunize rats (Rockland Immunochemicals). The antibodies were affinity-purified using their respective antigens.
Fluorescent in Situ Hybridization (FISH).
Arabidopsis 5S rRNA and 25S rRNA and maize 5S RNA repeats were labeled with fluorescein isothiocyanate (FITC) using nick translation (Roche). A cyanine 3-labeled oligonucleotide (CCCTAAA)4 was used to detect telomeres. A cyanine 5-labeled oligonucleotide YGGTTGCGGTTTAAGTTCTTATACTCAATC (Proligo) was used to detect the AL1 repeat present in Arabidopsis centromeres (41). FISH and 3-D microscopic imaging were carried out as previously described (42, 43).
Immunolocalization.
Immunolocalization experiments were performed as previously described (28, 43) using the following antibodies: rabbit anti-AtASY1 (26) diluted 1:50, rabbit anti-AtZYP1 (29) diluted 1:100, rat anti-AFD1 (28) diluted 1:100, guinea pig anti-ZmZYP1, diluted 1:50, and rabbit anti-AtRAD50 (44) diluted 1:200. The anti-PHS1 antibodies were used at 1:100.
Supplementary Material
Acknowledgments.
We thank Chris Franklin and Susan Armstrong (University of Birmingham, Birmingham, United Kingdom) for the Arabidopsis ASY1 and ZYP1 antibodies, Sylviane Dadoual-Coterell and Charles White (Clermont Université, France) for the RAD50 antibody and the Atrad50 mutant, Zac Cande and Rachel Wang (University of California, Berkeley, CA) for the maize ZYP1 and AFD1 antibodies, Matilde Grelon (Institut National de la Recherche Agronomique, Versailles, France) for the spo11-1-1 mutant, and Rita-Ann Monde and Cliff Weil (Purdue University, West Lafayette, IN) for the maize TILLING allele; Gagan Sidhu for help with FISH; and Chris Bozza, Lorrie Anderson, and Graham Moore for comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 19751.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906273106/DCSupplemental.
References
- 1.Pawlowski WP, Cande WZ. Coordinating the events of the meiotic prophase. Trends Cell Biol. 2005;15:674–681. doi: 10.1016/j.tcb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Pawlowski WP, et al. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science. 2004;303:89–92. doi: 10.1126/science.1091110. [DOI] [PubMed] [Google Scholar]
- 3.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterworth WM, et al. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007;52:41–52. doi: 10.1111/j.1365-313X.2007.03220.x. [DOI] [PubMed] [Google Scholar]
- 6.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 7.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 9.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey NJ, et al. Arabidopsis SPO11–2 functions with SPO11–1 in meiotic recombination. Plant J. 2006;48:206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- 13.Bleuyard JY, Gallego ME, White CI. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma. 2004;113:197–203. doi: 10.1007/s00412-004-0309-1. [DOI] [PubMed] [Google Scholar]
- 14.Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16:1968–1978. doi: 10.1105/tpc.104.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggler AL, Inman RB, Cox MM. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J Biol Chem. 2002;277:39280–39288. doi: 10.1074/jbc.M204328200. [DOI] [PubMed] [Google Scholar]
- 16.Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 17.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida K, et al. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 19.Li W, et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA. 2004;101:10596–10601. doi: 10.1073/pnas.0404110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, et al. Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics. 2007;176:1469–1482. doi: 10.1534/genetics.106.062604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haberer G, et al. Structure and architecture of the maize genome. Plant Physiol. 2005;139:1612–1624. doi: 10.1104/pp.105.068718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siaud N, et al. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 2004;23:1392–1401. doi: 10.1038/sj.emboj.7600146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuromori T, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong SJ, Franklin FC, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong SJ, Caryl AP, Jones GH, Franklin FC. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- 28.Golubovskaya IN, et al. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci. 2006;119:3306–3315. doi: 10.1242/jcs.03054. [DOI] [PubMed] [Google Scholar]
- 29.Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: A web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Carvalho CE, Colaiacovo MP. SUMO-mediated regulation of synaptonemal complex formation during meiosis. Genes Dev. 2006;20:1986–1992. doi: 10.1101/gad.1457806. [DOI] [PubMed] [Google Scholar]
- 32.Weil CF, Monde RA. Induced mutations in maize. Israel J Plant Sci. 2007;55:183–190. [Google Scholar]
- 33.Lohmiller LD, et al. Cytological analysis of MRE11 protein during early meiotic prophase I in Arabidopsis and tomato. Chromosoma. 2008;117:277–288. doi: 10.1007/s00412-007-0147-z. [DOI] [PubMed] [Google Scholar]
- 34.Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nairz K, Klein F. mre11S–a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Perez E, Shaw P, Moore G. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature. 2001;411:204–207. doi: 10.1038/35075597. [DOI] [PubMed] [Google Scholar]
- 38.Tsubouchi T, Macqueen AJ, Roeder GS. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008;22:3217–3226. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Moran E, Jones GH, Franklin FC, Santos JL. A puromycin-sensitive aminopeptidase is essential for meiosis in Arabidopsis thaliana. Plant Cell. 2004;16:2895–2909. doi: 10.1105/tpc.104.024992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 2003;36:1–11. doi: 10.1046/j.1365-313x.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- 41.Maluszynska J, Heslop-Harrison JS. Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1991;1:159–166. [Google Scholar]
- 42.Golubovskaya IN, Harper LC, Pawlowski WP, Schichnes D, Cande WZ. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays, L. ) Genetics. 2002;162:1979–1993. doi: 10.1093/genetics/162.4.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlowski WP, Golubovskaya IN, Cande WZ. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests involvement of RAD51 in the meiotic homology recognition. Plant Cell. 2003;8:1807–1816. doi: 10.1105/tpc.012898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daoudal-Cotterell S, Gallego ME, White CI. The plant Rad50-Mre11 protein complex. FEBS Lett. 2002;516:164–166. doi: 10.1016/s0014-5793(02)02536-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.