Abstract
Luteinizing Hormone Receptor (LHR) mRNA is post-transcriptionally regulated during ligand-induced downregulation. This process involves interaction of LHR mRNA with a specific mRNA binding protein (LRBP), identified as Mevalonate Kinase (MVK), resulting in inhibition of translation followed by targeting the ribonucleoprotein complex to accelerated degradation. The present study investigated the endogenous association of LRBP with the translational machinery and its interaction with LHR mRNA during LH/hCG-induced downregulation. Ovaries were collected from rats that were injected with the ligand, hCG, to induce downregulation of LHR mRNA expression. Western blot analysis showed significantly higher levels of LRBP in polysomes from downregulated ovaries compared to controls. Western blot analysis of ribosome-rich fractions from FPLC-assisted gel filtration of post-mitochondrial supernatants confirmed the presence of LRBP in translating ribosomes isolated from the downregulated state but not from controls. The association of LRBP with LHR mRNA in the downregulated polysomes was demonstrated by immunoprecipitation with LRBP antibody followed by qPCR analysis of the associated RNA. Increased association of LHR mRNA with LRBP during downregulation was also demonstrated by subjecting the polysome-associated RNAs to oligo(dT) cellulose chromatography followed by immunoprecipitation and qPCR analysis. Additionally, analysis of in vitro translation of LHR mRNA showed increased inhibition of translation by polysomes from downregulated ovaries compared to control. This study provides strong in vivo and in vitro evidence to show that during ligand-induced downregulation, LRBP translocates to ribosomes and associates with LHR mRNA to form an untranslatable ribonucleoprotein complex and inhibits LHR mRNA translation, paving the way to its degradation.
Keywords: Luteinizing hormone receptor, mRNA decay, mRNA binding protein, ribosomes, translational inhibition
1. Introduction
Luteinizing Hormone (LH) Receptor, a member of the rhodopsin-like family of G protein-coupled receptors, is post transcriptionally regulated by an RNA binding protein during downregulation [1, 2]. The RNA-binding protein in rat ovary as well as in human granulosa cells binds to a cytidine-rich region of the LH receptor mRNA [1–4]. RNA gel electrophoretic mobility shift assay (REMSA) and hydroxyl-radical RNA footprinting of the partially purified LH receptor mRNA-binding protein (LRBP), showed that the LRBP binding site is located in the LH receptor open reading frame and binds to a polypyrimidine sequence within nucleotides 203 and 220 of the LH receptor mRNA with a high degree of specificity [2]. The identity of this LH receptor mRNA-binding protein was then established as mevalonate kinase (MVK) [5]. LRBP, purified to homogeneity, was able to bind LHR mRNA directly. Furthermore, purified LRBP was recognized by rat mevalonate kinase antibody in Western blots performed with one- and two-dimensional SDS-polyacrylamide gels [5]. Recombinant mevalonate kinase produced in human embryonic kidney cells (293 cells) showed all of the characteristics of LRBP with respect to specificity of LH receptor mRNA binding sequence [5]. The functional role of LRBP in LH receptor mRNA downregulation has been established in our laboratory [6] and, independently, by Ikeda et al [7]. Furthermore, the binding of the protein to LH receptor mRNA was shown to be enhanced during ligand-induced downregulation [8]. We have also demonstrated that in a cell-free translation system, partially purified LRBP prevents the translation of LH receptor mRNA and that the intact catalytically important residues of MVK are necessary for translational suppression [9, 10]. The ability of LRBP to block translation of LH receptor mRNA was specific, since translation of non-relevant mRNA such as human β-actin was not affected [10]. While these studies established a role of LRBP in suppressing LH receptor mRNA translation, the association of LRBP with ribosomes has not been demonstrated during ligand-induced downregulation.
In the present study, we hypothesize that LRBP, a cytosolic protein, translocates to the ribosomes during downregulation and interacts with LH receptor mRNA to cause translational suppression. Our results show that during down regulation, there is an increased association of LRBP with the translating LH receptor mRNA to form an untranslatable mRNP complex, thus inhibiting translation and leading to its degradation.
2. Materials and Methods
2.1. Materials
Pregnant mare serum gonadotrophin (PMSG) was purchased from Calbiochem (San Diego, CA). Highly purified human chorionic gonadotrophin (hCG; CR 127) was purchased from Dr. A. F. Parlow (National Hormone and Peptide Program, Torrance, CA). Enlightening reagent, [α-32P] UTP and Redivue L-[35S] methionine were obtained from Perkin Elmer Life Sciences (Waltham, MA). mMessage mMachine T7 ultra and Maxiscript T7 were products of Ambion (Austin, TX). EDTA-free protease inhibitor mixture tablets and Quickspin (G-50 Sephadex) columns for radiolabeled RNA purification were purchased from Roche Applied Science (Indianapolis, IN) and Anti-FLAG M2-agarose affinity gel was purchased from Sigma (St. Louis, MO). RNAse inhibitor (rRNasin) and Flexi Rabbit Reticulocyte Lysate System were from Promega (Madison, WI). Primers specific for LH receptor, β-actin, GAPDH, and RPS6 mRNA and 18S rRNA (TaqMan Assay-on-Demand Gene Expression Product) and Multiscribe reverse transcriptase were from Applied Biosystems (Foster City, CA). Since LRBP was identified as MVK, anti-N-terminal mevalonate kinase IgG was raised against the first 15 N-terminal amino acids of MVK (MLSEVLLVSAPGKVI) and this antibody is referred to as the LRBP antibody in the text. Purified antibodies against ribosomal protein S6 (RPS6; Cell Signaling, Beverly, MA) and heat shock protein 90β (HSP90β; Assay Designs, Ann Arbor, MI) were commercial products. The Super Signal West Femto chemiluminescence kit and anti-rabbit/anti mouse IgG conjugated to horseradish peroxidase were obtained from Pierce (Rockford, IL). BCA reagent and Oligo(dT) cellulose were products of GE Healthcare Life Sciences (Piscataway, NJ).
2.2. Animals and Tissues
Pseudopregnancy was induced in 23 day old Sprague-Dawley rats by subcutaneous injection of 50 IU of PMSG followed by 25 IU of hCG 56 h later. The day of hCG injection was taken as 0. LH receptor downregulation was induced by the injection of 50 IU of hCG on the fifth day of pseudopregnancy. Ovaries were collected 0, 2, 4, 6 and 12 h after hCG injection and were frozen in liquid nitrogen until further use.
2.3. Preparation of tissue extracts, polysomes and ribosomal salt wash
Ovaries from control and hCG-injected rats were homogenized in solution A (1 mM potassium acetate, 2 mM Mg(Ac)2, 2 mM dithiothreitol, and 10 mM Tris acetate, pH 7.6) at 4°C. After centrifugation at 10,000× g for 10 min, the supernatants were layered over a cushion of solution B (solution A containing 30% sucrose) and centrifuged at 130,000 × g for 2.5 h. The polyribosome pellets were resuspended in solution A and stored at −80 °C. Quantitation of polysomes was performed by measuring the absorbance at 260 nm. To obtain ribosomal salt wash (RSW), equal amounts of polysomes from control and downregulated conditions were resuspended in solution A and were supplemented with 4M KCl to a final concentration of 0.5 M and incubated at 4°C for 15 min. The high salt treated ribosomes were layered on top of solution B and centrifuged at 130,000× g for 2.5 h at 4 °C to separate ribosomes and ribosome associated proteins. The proteins were removed from the interface between the two layers, buffer exchanged and concentrated. Total protein content of the RSW was measured using BCA assay.
2.4. Western Blot analysis
Polysomes and other protein samples were incubated with SDS sample loading buffer and subjected to 10% SDS-PAGE under reducing conditions followed by Western blot analysis as previously described [6]. The presence of immune complexes was detected by chemiluminescence.
2.5. Fast Protein Liquid Chromatography (FPLC) assisted gel-filtration
Post-mitochondrial supernatants (S10) were prepared by homogenizing the rat ovaries in 0.4ml of lysis buffer containing 20mM Tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5% NP-40 and 100 U/ml RNasin followed by centrifugation of the lysate at 10,000×g for 10min at 4 °C. S10 was then applied to a Superose 6 HR 10/30 column (GE Healthcare Life Sciences) equilibrated in lysis buffer without NP-40. Gel filtration was run on BioLogic DuoFlow FPLC device (Bio-Rad) with a flow rate of 0.5ml/min and 24 fractions of 1 ml were collected. Total RNA was extracted from 0.5ml of each fraction with phenol/chloroform according to standard procedure, precipitated with ethanol and dissolved in 1×TBE. Samples were resolved on a 1.2% agarose gel and visualized by ethidium bromide. The remaining 0.5ml of each fraction was concentrated and used for Western blot analysis.
2.6. Immunoprecipitation of the RNP complex from ribosomes
Polysomes were homogenized in NET-2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.05% Nonidet P-40) containing RNasin (100 units/0.5 ml of buffer) and protease inhibitors. The homogenates were centrifuged at 10,000 × g for 10 min at 4°C. The supernatants were collected and 1–1.5 mg of protein/ml of the extracts were used for immunoprecipitation of RNP complex as described before [10], using the procedure developed by Lerner and Steitz [11].
2.7. Real-Time PCR (qPCR) analysis
Total RNAs were reverse-transcribed and subjected to real time PCR quantitation as described before [6]. The fold change in gene expression was calculated using the ΔΔCt method [12] with 18S rRNA as the internal control.
2.8. Oligo(dT) cellulose chromatography of the ribosomes
To isolate poly (A) mRNA-mRNP complexes, the polysomes were first suspended in a solution containing 30 mM EDTA, 0.5% NP-40, 20 mM Tris-HCl, pH 7.5 (Pellet Buffer), kept on ice for 10 min and centrifuged at 100,000 × g for 2 h. The mRNPs, free 40 and 60 S ribosomal subunits and a small subfraction of monosomes were pelleted. The pellet was then resuspended in the pellet buffer and spun at 14,000 × g for 2 min. The supernatant was saved, KCl concentration adjusted to 200 mM and then incubated with 40 μl of pre-washed oligo(dT) cellulose (100 mg/ml) with constant rotation overnight at 4°C. Poly (A) mRNAs and associated proteins were eluted using 10 mM Tris-HCL, pH 7.5 at 65°C or using SDS sample buffer. The sample buffer eluate was used for Western blot analysis and the Tris buffer eluate used for immunoprecipitation with LRBP antibody followed by qPCR analysis using RNA isolated from the immune complex. qPCR analysis was also done using RNA isolated directly from Tris buffer eluate.
2.9. In vitro transcription
Rat LH receptor cDNA containing T7 promoter at the 5′ end and FLAG tag at the 3′ end and β-actin cDNA with T7 RNA polymerase promoter at the 5′ end and T3 RNA polymerase promoter at the 3′ end were synthesized as previously described [10]. The full-length capped and FLAG-tagged rat LH receptor mRNA and human β-actin mRNA were synthesized using mMessage mMachine T7 Ultra kit. The cDNA for generating the LRBP binding site (5′-GGCCUCGCCAGACUAUCUCUCACCUAUCUCCCU GUCAAAGU-3′; LBS) of LH receptor mRNA was chemically synthesized with T7 RNA polymerase promoter at the 5′ end [2]. For gel shift analysis, [α-32P]-labeled RNA was in vitro transcribed from this cDNA template using Ambion in vitro transcription kit (Maxiscript T7).
2.10. In vitro translation
In vitro translation reactions (25-μl reaction volume) were performed using a Flexi rabbit reticulocyte lysate system as per the manufacturer’s instructions (Promega). FLAG-tagged in vitro translated rat LH receptor was immunoprecipitated using anti-FLAG M2-agarose affinity gel according to manufacturer’s instructions (Sigma) and subjected to SDS-PAGE and autoradiography.
2.11. RNA Electrophoretic Mobility Shift Analysis
RNA electrophoretic mobility shift analysis (REMSA) was performed by incubating RSW from control and downregulated ovaries with a fixed concentration of [α-32P]UTP-labeled LBS, as described previously [5]. The RNA-protein complexes were resolved by 5% native polyacrylamide (70:1) gel electrophoresis and analyzed by autoradiography.
2.12. Statistical Analysis
Statistical analysis was carried out using one-way ANOVA followed by the Tukey multiple comparison test. Values were considered statistically significant for p<0.05. Each experiment was repeated at least three times with similar results. Blots and autoradiograms shown are representative of a minimum of three experiments.
3. Results
3.1. Downregulation of LH receptor mRNA in the post-mitochondrial supernatants and ribosomes of rat ovaries in response to hCG treatment
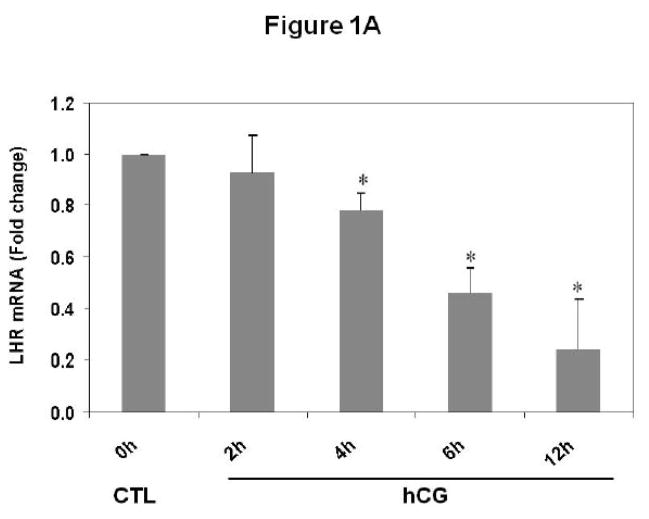
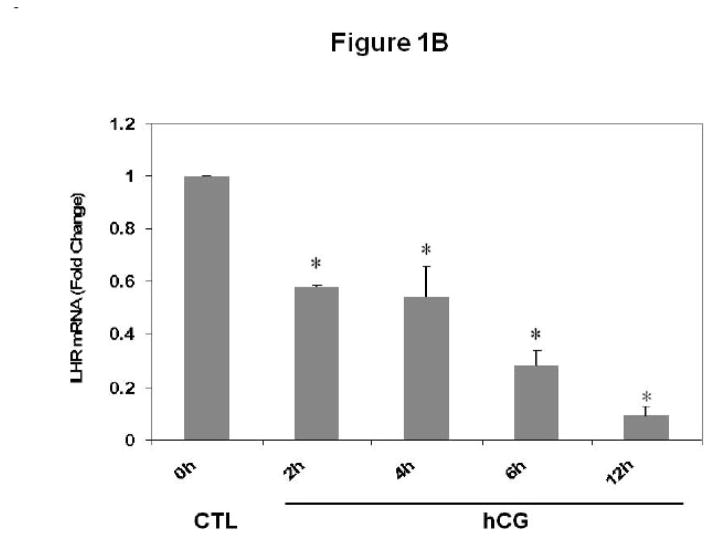
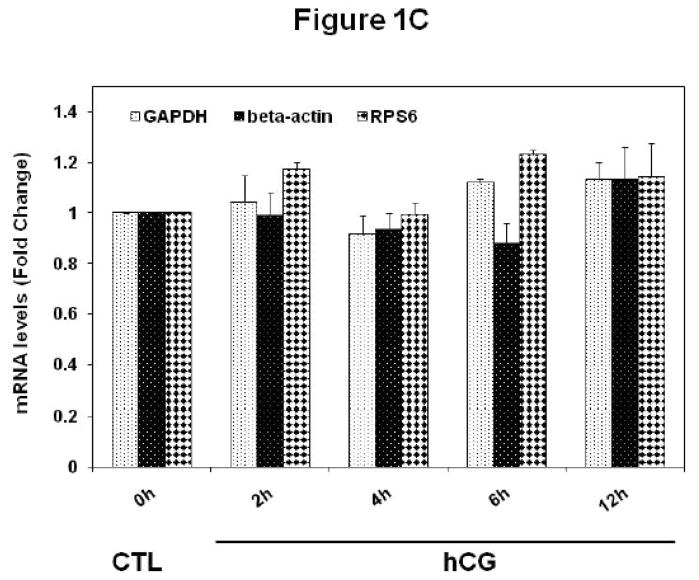
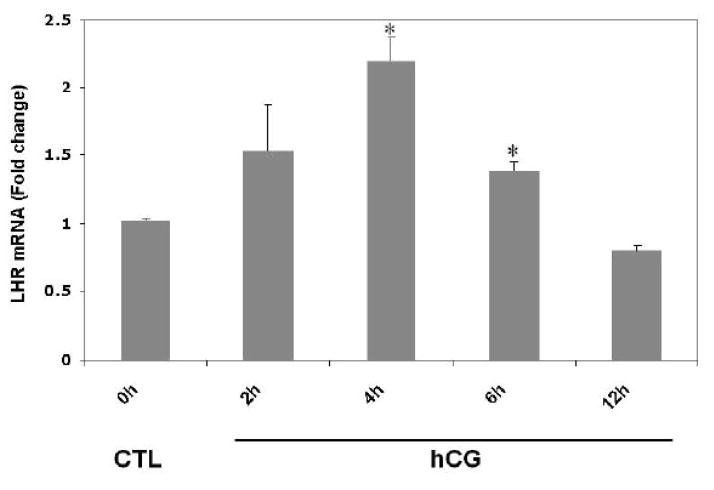
Experiments were conducted to examine whether downregulation of LH receptor mRNA in response to hCG treatment occurs in polysome preparations. Pseudopregnant rats were injected with hCG and ovaries were collected 0, 2, 4, 6 and 12 h post-injection. Post-mitochondrial supernatants and polysomes were isolated from the ovaries. LH receptor mRNA expression was analyzed in the post-mitochondrial supernatants (S10) and polysomes using real-time PCR, as described in Materials and Methods. The results are shown in Fig 1. Consistent with our previous results [8], LH receptor mRNA levels in the S10 fractions were found to progressively decrease after hCG treatment, reaching a minimum level at 12 h (Fig 1A). LH receptor mRNA levels were significantly lower at 4, 6 and 12 h (p<0.05 vs. Control). The polysome fractions showed a similar trend (Fig 1B). There was a 40% reduction in the mRNA levels as early as 2 hours after hCG treatment. The levels continued to decline in a similar manner as the S10 fractions and reached a maximum decline at 12 h. There was no significant difference in the levels of LH receptor mRNA between different control samples at these time points (data not shown). In addition, our earlier work has shown that no significant differences exist in the levels of LRBP between control ovaries collected after 0, 2, 4, 6 or 12 h of saline treatment [13]. Therefore only one control, isolated at 0 h, was used for all the experiments. There was no significant change in the levels of non-specific mRNAs like β-actin, GAPDH or Ribosomal Protein S6 in the polysome fractions following hCG treatment when compared to control, thus providing further proof to the specificity of LH receptor downregulation (Fig 1C).
Fig 1. Downregulation of LH receptor mRNA in the post-mitochondrial supernatants and ribosomes of pseudopregnant rats injected with hCG.
Rats were injected with hCG on the fifth day of pseudo-pregnancy, ovaries were collected 0, 2, 4, 6 and 12 h later and polysomes were isolated. Total RNA from the control (CTL) or hCG-treated post-mitochondrial supernatant (S10) were reverse transcribed, and the resulting cDNAs were subjected to real-time PCR using predesigned primers and probes for rat LH receptor mRNA (A). Total RNA from the control (CTL) or hCG-treated polysomes were reverse transcribed, and the resulting cDNAs were subjected to real-time PCR using predesigned primers and probes for rat LH receptor mRNA (B), β–actin, GAPDH and RPS6 (C), as described in Materials and Methods. The graphs represent changes in mRNA levels normalized to 18S rRNA and shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=3.
3.2. Mobilization of LRBP to the ribosomes in downregulated rat ovaries
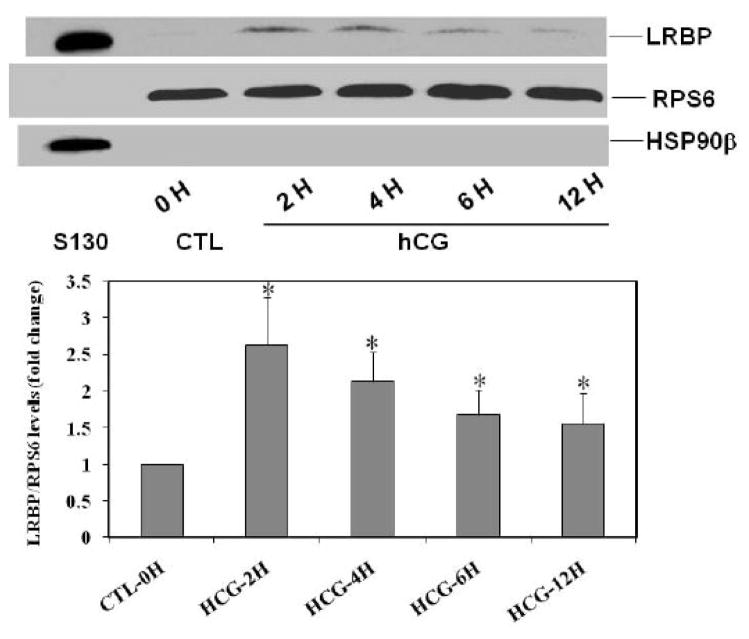
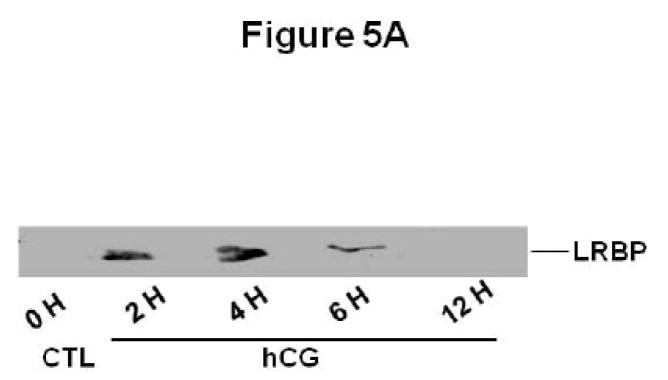
To demonstrate that LRBP is mobilized to the ribosomes following hCG treatment to downregulate the receptor, we examined the levels of LRBP in the polysomes isolated from rat ovaries using Western blot analysis with anti-LRBP antibody. As shown in Fig 2, there was an increase (2.62±0.6-fold vs. CTL) in the levels of LRBP in the polysomes isolated from downregulated ovaries, observed after 2 h, remaining high until 4 h (2.12±0.4-fold vs. CTL) and decreasing slowly thereafter. To confirm the purity of the isolated polysome fractions, the blots were stripped and reprobed with antibody against ribosomal protein S6, RPS6. The lanes containing the polysomes gave intense signals for RPS6. A cytosolic fraction (S130) from downregulated rat ovary, which was used as a positive control for LRBP, did not give any signal for RPS6, but cross reacted with the antibody against the cytosolic protein, Heat Shock Protein 90β (HSP90β). The isolated polysome fractions did not give any signal for HSP90β, which is not known to associate with ribosomes [14].
Fig 2. Mobilization of LRBP to the ribosomes in downregulated ovaries.
Polysomes were isolated from 0 h control (CTL) and 2, 4, 6 and 12 h hCG treated ovaries, solubilized in SDS sample buffer and subjected to Western Blot analysis to detect LRBP using LRBP antibody. The membranes were stripped and reprobed for RPS6 and then for HSP90β. The blot shown is a representative of five independent experiments. The lower panel shows the quantitative expression of LRBP normalized to RPS6. The graph represents the mean of five experiments and error bars represent mean ±SE. *p<0.05, n=5.
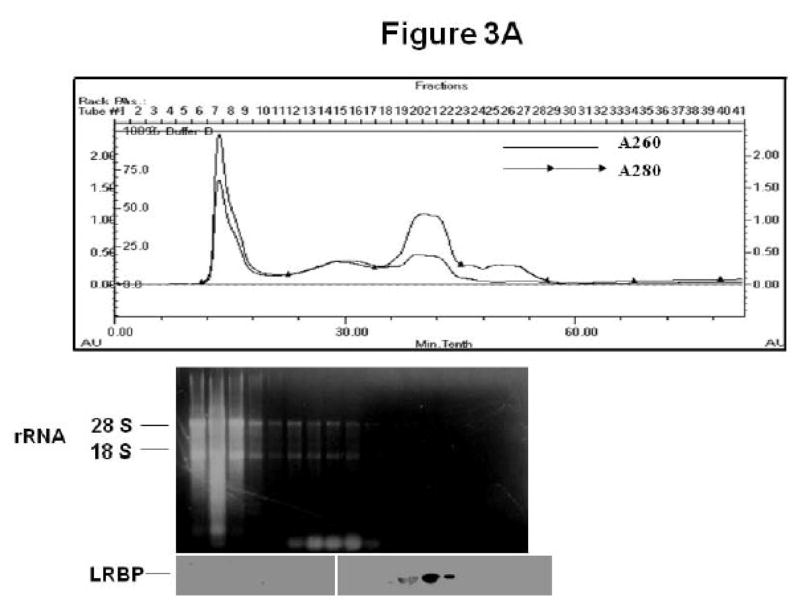
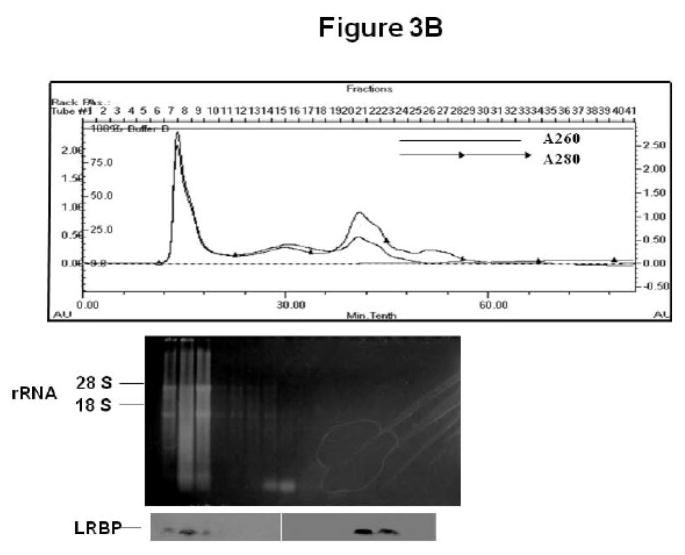
To further confirm the translocation of LRBP to the ribosomes, we performed an FPLC-assisted gel filtration analysis of the post-mitochondrial supernatants from the control and 4 h downregulated ovaries. In the absorbance profiles in Figs 3A & B (top panels), ribosomes formed the initial peak since the specific fractions from the gel filtration column corresponding to that peak (fractions 7–9) showed the presence of 18S and 28S rRNA in agarose gel electrophoresis (middle panels; Figs 3A & B). Western blot analysis of the same fractions (ribosomal fractions 7–9) showed intense bands for LRBP in the downregulated samples (Fig 3B; left lower panel) but not in control samples (Fig 3A; left lower panel), whereas in both conditions the same fractions (fractions 7–9) showed the presence of RPS6 (data not shown). It should be noted that the elution profiles of both samples showed positive signals in Western blot analysis for LRBP (Figs 3A & B; right lower panels) in the fractions containing free unbound cytosolic proteins (fractions 20–22).
Fig 3. Gel filtration chromatography of post-mitochondrial supernatants from control (CTL; A) and downregulated (hCG; B) ovaries.
Rats were injected with hCG or saline on the fifth day of pseudopregnancy, ovaries were collected after 4 h and the S10 fractions were subjected to FPLC-assisted gel filtration. The top panels in A and B show the absorbance profile at 280 and 260 nm. The middle panels show the agarose gel electrophoresis profile of RNA extracted from fractions 7–24 from the gel filtration column, placed under the corresponding fractions in the absorbance profile. The bottom panels show Western blot analysis to detect LRBP using LRBP antibody in fractions 7–24, with the LRBP lanes placed directly under the corresponding fractions of the absorbance profile.
3.3. Association of LRBP with LH receptor mRNA in the downregulated rat ovaries
In order to demonstrate a physical connection between LRBP and LH receptor mRNA during downregulation, we performed an immunoprecipitation-coupled real- time PCR analysis. Briefly, ribosomes from control and downregulated ovaries were solubilized in NET-2 buffer and were immunoprecipitated using LRBP antibody. The RNA isolated from this immune complex was then analyzed for the presence of LH receptor mRNA by performing real time RT-PCR using LH receptor cDNA-specific primers for amplification. As shown in Fig 4, the levels of LH receptor mRNA bound to LRBP in the RNP complex were significantly higher in the downregulated ovaries when compared to control ovaries. There was a steady increase in the levels of LH receptor mRNA with downregulation, reaching a 2.5-fold difference by 4 h and declining afterwards. A significant (1.5-fold vs. control) increase in the levels was observed even at 6 h, which however, declined to below control levels by 12 h. However, amplification of actin or GAPDH using specific primers showed undetectable CT values in real time PCR analysis (data not shown), thus confirming that there was no significant association of non-specific mRNAs with the immune complex isolated from the ribosomes. This experiment provides proof for a direct and specific association between LRBP and LH receptor mRNA in the ribosomes of LH receptor downregulated ovaries.
Fig 4. LH receptor mRNA co-precipitates with LRBP in the ribosomes from downregulated ovaries.
Polysomes prepared from 0 h control (CTL) and downregulated (hCG) pseudopregnant rat ovaries collected 2, 4, 6 and 12 h after hCG treatment were subjected to immunoprecipitation using LRBP antibody. Total RNA extracted from the immune complex was reverse transcribed and the cDNAs were used for real-time PCR using pre-designed primers and probes for rat LH receptor mRNA. The graph represents changes in LH receptor mRNA levels normalized to 18S rRNA and shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=3.
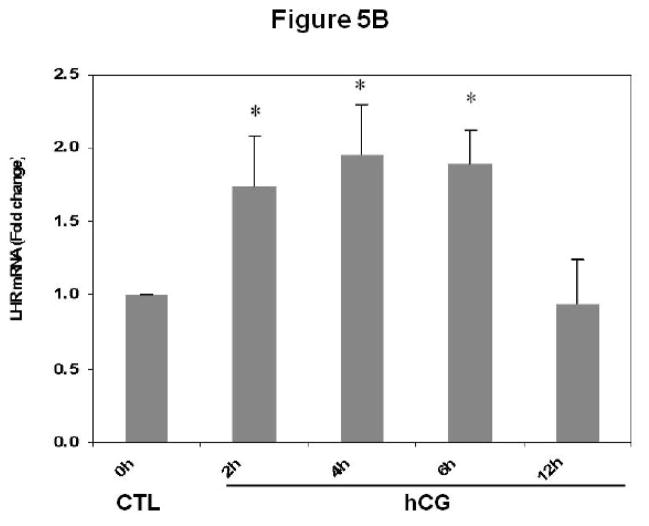
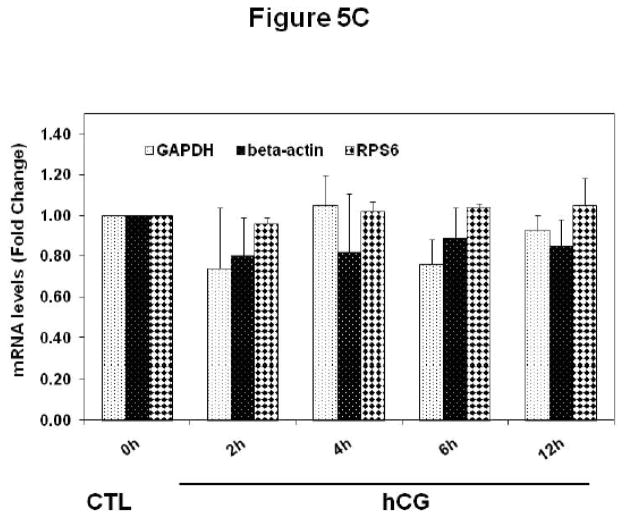
This observation was further confirmed by a different approach, by first performing oligo(dT) cellulose chromatography to isolate mRNA with associated proteins, followed by Western blot analysis to detect LRBP. The mRNA-protein complex was then immunoprecipitated with LRBP antibody followed by real-time PCR to identify LH receptor mRNA in the mRNP complex. To start, polysomes were resuspended in Tris/HCl buffer containing 30 mM EDTA to disrupt the polyribosomes and to release poly (A) mRNA-mRNP complexes. The fractions containing mRNPs, free 40s, and 60s subunits and a small subfraction of monosomes were then subjected to oligo(dT) cellulose chromatography, as described in the Materials and Methods section. Poly (A) mRNA-binding proteins eluted with SDS sample buffer were subjected to Western blot analysis using LRBP antibody. The results are presented in Fig 5A. LRBP was found to be associated with the mRNP complexes at 2, 4 and 6 h after treatment with hCG to downregulate LH receptor mRNA expression. Additionally, elution of poly(A) mRNA-mRNP complexes using Tris/HCl buffer without KCl followed by immunoprecipitation with LRBP antibody, RNA isolation and real-time PCR analysis were done to confirm that LRBP is bound specifically to LH receptor mRNA. Consistent with the earlier results, there was a significant increase in the levels of LH receptor mRNA associated with LRBP in the ribosomes following 2, 4 and 6 h of hCG treatment, approximately 1.6 to 2- fold increase when compared to control (Fig 5B). Importantly, the association was found to decrease to below control level at 12 h, in a similar manner as observed in the previous experiment (Fig 4). To confirm that equal amounts of poly(A) mRNA-mRNP complexes were eluted from the column, the eluates were subjected to real time PCR analysis using primers for non specific genes like β-actin, GAPDH, and RPS6. There was no change in the levels of β-actin, GAPDH, or RPS6 mRNA between control or downregulated samples (Fig 5C).
Fig 5. Co-precipitation of LRBP and LH receptor mRNA in the oligo(dT) eluted mRNP fraction.
mRNA-binding proteins in the polysomes from 0 h control (CTL) and downregulated (hCG) pseudopregnant rat ovaries collected 2, 4, 6 and 12 h after hCG treatment were separated using oligo(dT) cellulose chromatography as described in detail in Materials and Methods. Proteins from the oligo(dT) cellulose bound mRNP complexes were eluted using SDS sample buffer and subjected to Western blot analysis using LRBP antibody (A). The blot shown is a representative of three independent experiments. Oligo(dT) cellulose bound mRNPs were eluted using 10 mM Tris/HCl, pH 7.5 and subjected to immunoprecipitation using LRBP antibody. The immune complex was reverse transcribed and the cDNAs were used for real time PCR using pre-designed primers and probes for rat LH receptor mRNA (B). Total RNA was also extracted from the mRNP eluate directly and was reverse transcribed and subjected to real time PCR analysis using pre-designed primers and probes for rat β–actin, GAPDH and RPS6 (C). The graphs represents changes in mRNA levels normalized to 18S rRNA, and are shown as fold changes vs. control. Error bars represent mean ±SE. *p<0.05, n=3.
3.4. Evidence for the LH receptor mRNA binding activity in the polysomes
To examine the binding ability of ribosome-associated LRBP to bind LH receptor mRNA, gel shift analysis was performed by incubating RSW with a fixed concentration of radiolabeled LRBP binding site (LBS) of LH receptor mRNA [2]. Polysomes were isolated from control and downregulated ovaries after 4 h of hCG injection and used for ribosomal salt wash preparation. This time frame was chosen based on our earlier experiments which showed maximum RNA binding activity occurring at the onset of accelerated receptor-mRNA degradation [8] and the strong association of LRBP with ribosomes observed at 4 h (fig 2). Samples were processed as described in Materials and Methods and were resolved by 5% native-polyacrylamide gel electrophoresis. As shown in Fig 6, an appreciable amount of mRNA binding activity was exhibited by LRBP present in the RSW. A distinct band representing LH receptor mRNP complex was observed in the samples containing RSWs but not in the negative control containing only labeled LBS. Samples containing RSW prepared from downregulated polysomes showed increased mRNA binding activity when compared to the control. This experiment clearly shows that LRBP present in the polysomes retains its ability to bind to LH receptor mRNA and further supports the notion that mobilization of LRBP to the ribosomes occurs following ligand-induced downregulation.
Fig 6. RNA mobility shift analysis.
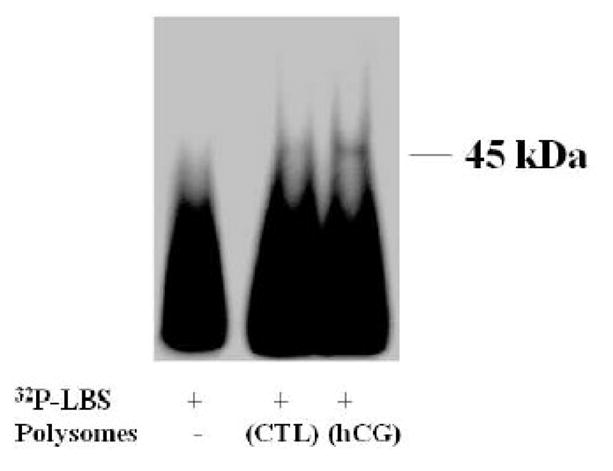
Gel mobility shift analysis was performed with [32p] labeled rat LBS (1.5 × 105 c.p.m) using no protein (LBS only), or equal amounts of protein from the RSW extracted from control (CTL) and 4 h hCG-treated ovaries (hCG), as described in Materials and Methods. The autoradiogram shown is representative of three independent experiments.
3.5. Inhibition of translation of LH receptor mRNA by polysomes from downregulated ovaries
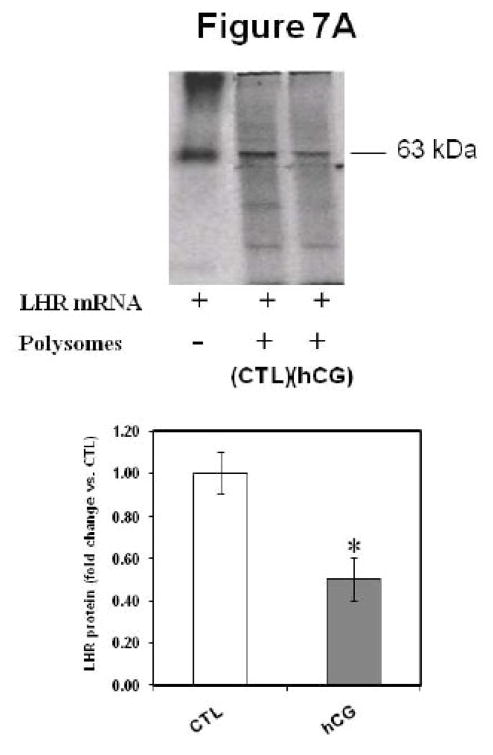
To demonstrate the role of ribosome-associated LRBP in the inhibition of translation, in vitro translation of FLAG-tagged LH receptor mRNA was performed in the presence of polysomes from control and 4 h downregulated ovaries, and the resulting translated proteins were subjected to immunoprecipitation with anti FLAG M2 followed by SDS-PAGE and autoradiography. The results (Fig 7A) show that there is a decrease (0.5±0.03-fold vs. CTL, p<0.05) in the amount of translated protein in the sample containing polysomes from downregulated ovaries, when compared to the control. This supports our hypothesis that interaction of ribosome-associated LRBP with LH receptor mRNA results in the inhibition of LH receptor mRNA translation. This is not due to general impairment of the translation machinery since there is no change in the translation of the pool of mRNAs generally present in the ribosomes or that of a house-keeping gene like β-actin (Fig 7B). In the absence of any exogenously added mRNA, the translation rate was unaffected by the addition of polysomes from control or downregulated ovaries (lanes 1 and 2). Similarly, when β-actin mRNA was in vitro translated in the presence of control and downregulated polysomes (lanes 3 & 4), there was no change in the protein synthesis, thus indicating that the inhibitory effect on LH receptor mRNA translation by polysomes from downregulated ovaries was specific to LH receptor mRNA.
Fig 7. in vitro translation of LH receptor mRNA; Effect of Polysome-associated LRBP.
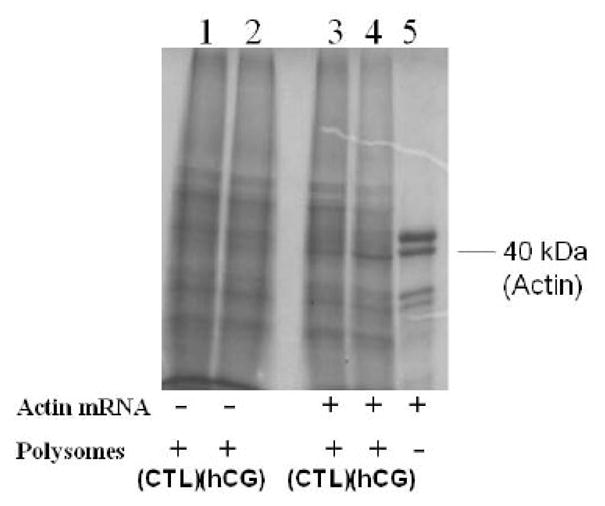
200 ng of FLAG-tagged rat LH receptor mRNA (A) was in vitro translated using 15 μCi of [35S] methionine in the presence of ribosomes from control (CTL) and 4 h hCG-injected (hCG) ovaries. The translated LH receptor protein was immunoprecipitated and subjected to SDS-PAGE. The gel was dried and exposed to X-ray film for autoradiography. The experiment was repeated 4 times with the same results. The lower panel shows the quantitative expression of LHR. The graph represents the mean of four experiments and error bars represent mean ±SE. *p<0.05, n=4. B. in vitro translation was conducted in the presence of ribosomes from control (CTL) and 4 h hCG-treated (hCG) ovaries in the absence of any exogenously added mRNA (Lanes 1 & 2) or by using human β-actin mRNA (100 ng; Lanes 3 & 4). Lane 5 was β-actin mRNA (200 ng) without polysomes. Translated proteins were directly subjected to SDS-PAGE and autoradiography.
4. Discussion
The results presented in this study show that LRBP mobilizes to the polysomes and associates with LH receptor mRNA during hCG-induced downregulation, providing strong evidence for the notion that LRBP interacts with the translational apparatus via its association with LH receptor mRNA. The data from Western blot analysis of the polysomes and FPLC-assisted gel filtration of the post-mitochondrial supernatant from the ovarian homogenates substantiate the association of LRBP with the polysomes during ligand-induced downregulation. The association of specific mRNA binding proteins with ribosomes appears to be a general mechanism of post-transcriptional regulation. Other examples are ribosomal association of Fragile X Mental Retardation Protein, more commonly referred to as FMRP [15, 16], association of the RNA-binding protein ELAV/HuR with HSP70 mRNA in the ribosomes following H2O2-mediated oxidative stress [17], and the ribosomal association of polysome associated mRNA endonuclease, PMR1, that initiates the destabilization of albumin mRNA causing the endonuclease mediated mRNA decay [18]. However, there are other examples where proteins directly bind to the ribosomal subunits. For example, Rbm3, a glycine-rich RNA-binding protein which is enhanced under conditions of mild hypothermia, has been demonstrated to bind directly to the 60s ribosomal subunit and regulate global levels of protein synthesis under normal and cold-stress conditions [19]. Our present results demonstrate that the association of LRBP with polyribosomes occurs through its interaction with LH receptor mRNA.
The association of LH receptor mRNA with LRBP in the ribosomes was seen as early as 2 h of downregulation and remains high even after 6 h (Fig 4). There was no observable association after 12 h of hCG treatment; in fact it was even below the control levels. This is not surprising, considering the fact that there is hardly any mRNA left in the ribosomes at this time period (Fig 1B). This supports the notion that during downregulation, LRBP mobilizes to the ribosomes, associates with LH receptor mRNA and then the LH receptor mRNA-LRBP complex is subsequently routed for degradation. Control of mRNA stability by translational regulation has been well documented [20]. Examples include c-fos and granulocyte-macrophage colony-stimulating factor ARE subsets that mediate selective mRNA degradation through polysome-associated mechanisms coupled with ongoing translation [21]. Another example is SgrS, a bacterial small RNA that has been shown to form a specific ribonucleoprotein complex with RNase E through RNA chaperone Hfq and inhibits the translation and RNase E-dependent degradation of ptsG mRNA [22].
The loss of cell surface receptors can be due to several mechanisms including the sequestration of the receptor to the intracellular compartments, the loss of ligand binding activity or decreased receptor expression. Our previous studies have shown that LH receptor levels closely correlate with the LH receptor mRNA levels and the transcription rate is unaffected during downregulation [4]. These observations suggest that the loss of LH receptor seen during hCG-induced downregulation is attributed mainly due to the loss of mRNA. The demonstration of LH receptor mRNA binding activity and inhibition of translation by LRBP-rich polysomes from downregulated ovaries in vitro supports the notion that LRBP associates with mRNA leading to mRNA decay in vivo. Several mechanisms have been proposed for the degradation of mRNAs involving interaction with trans-acting factors, including, but not limited to, stress granules and p bodies [20, 23–26]. Stress granules are transient structures, assembled as a consequence of interrupted RNA translation, and contain mRNA still associated with some of the translational machinery [23]. P-bodies contain 5′→3′ exonucleolytic activity as well as some of the RNA silencing machinery [27] and hence can serve both as sites of mRNA silencing and decay [28, 29]. Since LH receptor mRNA is almost totally depleted in the ovary by treatment with hCG [30, 31], It will be interesting to see if LH receptor mRNAs are compartmentalized into P bodies or stress granules.
The present study provides mechanistic insights into the inhibition of translation of LH receptor mRNA by LRBP. Here we show that LRBP mobilizes to the ribosomes during ligand-induced downregulation, prior to the observed decrease in the levels of LH receptor mRNA. Using different biochemical approaches, we also show that the mobilized LRBP binds directly to the LH receptor mRNA associated with the ribosomes and inhibits its translation. The ability to regulate the LH receptor expression at the post-transcriptional level could be a rapid and efficient way to control the receptor expression, especially in response to the constantly changing hormonal milieu during the ovarian cycle. The present study reveals a novel mechanism for the regulation of expression of LH receptor, a crucial molecule required for mammalian reproduction.
Acknowledgments
This work was supported by National Institutes of Health Grant R37 HD06656. We thank Dr. Anil K Nair and members of the laboratory for many helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kash JC, Menon KMJ. Identification of a hormonally regulated luteinizing hormone/human chorionic gonadotropin receptor mRNA binding protein. Increased mrna binding during receptor down-regulation. J Biol Chem. 1998;273:10658–10664. doi: 10.1074/jbc.273.17.10658. [DOI] [PubMed] [Google Scholar]
- 2.Kash JC, Menon KMJ. Sequence-specific binding of a hormonally regulated mRNA binding protein to cytidine-rich sequences in the lutropin receptor open reading frame. Biochemistry. 1999;38:16889–16897. doi: 10.1021/bi9915770. [DOI] [PubMed] [Google Scholar]
- 3.Nair AK, Peegel H, Menon KMJ. The role of luteinizing hormone/human chorionic gonadotropin receptor-specific mRNA binding protein in regulating receptor expression in human ovarian granulosa cells. J Clin Endocrinol Metab. 2006;91:2239–2243. doi: 10.1210/jc.2005-2739. [DOI] [PubMed] [Google Scholar]
- 4.Menon KMJ, Nair AK, Wang L. A novel post-transcriptional mechanism of regulation of luteinizing hormone receptor expression by an RNA binding protein from the ovary. Mol Cell Endocrinol. 2006;246:135–141. doi: 10.1016/j.mce.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 5.Nair AK, Menon KMJ. Isolation and characterization of a novel trans-factor for luteinizing hormone receptor mRNA from ovary. J Biol Chem. 2004;279:14937–14944. doi: 10.1074/jbc.M309484200. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Nair AK, Menon KMJ. Ribonucleic acid binding protein-mediated regulation of luteinizing hormone receptor expression in granulosa cells: relationship to sterol metabolism. Mol Endocrinol. 2007;21:2233–2241. doi: 10.1210/me.2007-0102. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda S, Nakamura K, Kogure K, Omori Y, Yamashita S, Kubota K, Mizutani T, Miyamoto K, Minegishi T. Effect of estrogen on the expression of luteinizing hormone-human chorionic gonadotropin receptor messenger ribonucleic acid in cultured rat granulosa cells. Endocrinology. 2008;149:1524–1533. doi: 10.1210/en.2007-1163. [DOI] [PubMed] [Google Scholar]
- 8.Nair AK, Kash JC, Peegel H, Menon KMJ. Post-transcriptional regulation of luteinizing hormone receptor mRNA in the ovary by a novel mRNA-binding protein. J Biol Chem. 2002;277:21468–21473. doi: 10.1074/jbc.M111653200. [DOI] [PubMed] [Google Scholar]
- 9.Nair AK, Young MA, Menon KMJ. Regulation of luteinizing hormone receptor mRNA expression by mevalonate kinase--role of the catalytic center in mRNA recognition. FEBS J. 2008;275:3397–3407. doi: 10.1111/j.1742-4658.2008.06490.x. [DOI] [PubMed] [Google Scholar]
- 10.Nair AK, Menon KMJ. Regulation of luteinizing hormone receptor expression: evidence of translational suppression in vitro by a hormonally regulated mRNA-binding protein and its endogenous association with luteinizing hormone receptor mRNA in the ovary. J Biol Chem. 2005;280:42809–42816. doi: 10.1074/jbc.M503154200. [DOI] [PubMed] [Google Scholar]
- 11.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Menon KMJ. Regulation of luteinizing hormone/chorionic gonadotropin receptor messenger ribonucleic acid expression in the rat ovary: relationship to cholesterol metabolism. Endocrinology. 2005;146:423–431. doi: 10.1210/en.2004-0805. [DOI] [PubMed] [Google Scholar]
- 14.Niksic M, Slight J, Sanford JR, Caceres JF, Hastie ND. The Wilms’ tumour protein (WT1) shuttles between nucleus and cytoplasm and is present in functional polysomes. Hum Mol Genet. 2004;13:463–471. doi: 10.1093/hmg/ddh040. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 16.Tamanini F, Meijer N, Verheij C, Willems PJ, Galjaard H, Oostra BA, Hoogeveen AT. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 17.Amadio M, Scapagnini G, Laforenza U, Intrieri M, Romeo L, Govoni S, Pascale A. Post-transcriptional regulation of HSP70 expression following oxidative stress in SH-SY5Y cells: the potential involvement of the RNA-binding protein HuR. Curr Pharm Des. 2008;14:2651–2658. doi: 10.2174/138161208786264052. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Schoenberg DR. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci U S A. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 21.Winstall E, Gamache M, Raymond V. Rapid mRNA degradation mediated by the c-fos 3′ AU-rich element and that mediated by the granulocyte-macrophage colony-stimulating factor 3′ AU-rich element occur through similar polysome-associated mechanisms. Mol Cell Biol. 1995;15:3796–3804. doi: 10.1128/mcb.15.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita T, Maki K, Yagi M, Aiba H. Analyses of mRNA destabilization and translational inhibition mediated by Hfq-binding small RNAs. Methods Enzymol. 2008;447:359–378. doi: 10.1016/S0076-6879(08)02218-0. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha N, Tisdale S, Hickman T, Anderson P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 2008;448:521–552. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- 24.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 26.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Stoecklin G, Anderson P. In a tight spot: ARE-mRNAs at processing bodies. Genes Dev. 2007;21:627–631. doi: 10.1101/gad.1538807. [DOI] [PubMed] [Google Scholar]
- 28.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman YM, Peegel H, Sprock MJ, Zhang QY, Menon KMJ. Evidence that human chorionic gonadotropin/luteinizing hormone receptor down-regulation involves decreased levels of receptor messenger ribonucleic acid. Endocrinology. 1991;128:388–393. doi: 10.1210/endo-128-1-388. [DOI] [PubMed] [Google Scholar]
- 31.Peegel H, Towns R, Nair A, Menon KMJ. A novel mechanism for the modulation of luteinizing hormone receptor mRNA expression in the rat ovary. Mol Cell Endocrinol. 2005;233:65–72. doi: 10.1016/j.mce.2004.12.009. [DOI] [PubMed] [Google Scholar]