SPN2 represses katanin translation to prevent mitotic defects (independently of ubiquitin-mediated katanin degradation).
Abstract
In Caenorhabditis elegans, the MEI-1–katanin microtubule-severing complex is required for meiosis, but must be down-regulated during the transition to embryogenesis to prevent defects in mitosis. A cullin-dependent degradation pathway for MEI-1 protein has been well documented. In this paper, we report that translational repression may also play a role in MEI-1 down-regulation. Reduction of spn-2 function results in spindle orientation defects due to ectopic MEI-1 expression during embryonic mitosis. MEL-26, which is both required for MEI-1 degradation and is itself a target of the cullin degradation pathway, is present at normal levels in spn-2 mutant embryos, suggesting that the degradation pathway is functional. Cloning of spn-2 reveals that it encodes an eIF4E-binding protein that localizes to the cytoplasm and to ribonucleoprotein particles called P granules. SPN-2 binds to the RNA-binding protein OMA-1, which in turn binds to the mei-1 3′ untranslated region. Thus, our results suggest that SPN-2 functions as an eIF4E-binding protein to negatively regulate translation of mei-1.
Introduction
The levels of protein products must be tightly controlled during development for the normal execution of stage-specific processes. For example, in many organisms, the transition from developing to mature oocyte and then to embryo depends on maternally provided mRNAs and proteins. Some proteins are needed at specific times during oocyte development and maturation, and the synthesis of these proteins must be precisely repressed and activated. Proteins that are needed in oogenesis or meiosis can be harmful for embryogenesis, and their accumulation after fertilization must be prevented. Ubiquitin-mediated proteolysis often serves to degrade oocyte proteins after fertilization. In Caenorhabditis elegans, the CUL-3–based ubiquitin ligase regulates the meiosis-to-mitosis transition of fertilized embryos by triggering the degradation of MEI-1 (Bowerman and Kurz, 2006). MEI-1 is a subunit of a katanin microtubule-severing enzyme essential for the assembly of the meiotic spindle, but its activity has to be eliminated before mitosis to form a robust mitotic spindle that orients properly (Clark-Maguire and Mains, 1994a,b; Srayko et al., 2000). Interestingly, however, mei-1 mRNA persists in embryos long after meiosis (Clark-Maguire and Mains, 1994b). This observation suggests that additional mechanisms could be required to prevent the synthesis of new MEI-1 protein.
Translational control also plays an important role in regulating protein levels during oogenesis and the egg-to-embryo transition (for reviews see Evans and Hunter, 2005; Stitzel and Seydoux, 2007; Vardy and Orr-Weaver, 2007). For example, Xenopus laevis Maskin represses the translation of cyclin until oocyte maturation, and Drosophila Cup represses the translation of oscar and nanos mRNAs so that only those mRNAs that are targeted to the posterior cytoplasm of the egg and embryo are activated for translation. Maskin and Cup act at the level of translational initiation as eIF4E-binding proteins (4E-BPs). eIF4E binds to the 5′ cap of mRNAs and in conjunction with eIF4G mediates the recruitment of the 40S ribosomal subunit (Gingras et al., 1999). 4E-BPs compete with eIF4G for binding to eIF4E, thus preventing translation initiation. 4E-BPs also play important roles in a variety of processes such as cell cycle progression, oncogenic transformation, and modulation of neuronal activity (Richter and Sonenberg, 2005). Although some 4E-BPs generally repress translation, others like Maskin and Cup are targeted to a small number of mRNAs through interactions with RNA-binding proteins (Richter and Sonenberg, 2005). Here, we report the identification of the first 4E-BP orthologue in C. elegans, SPN-2. SPN-2 is required for down-regulation of MEI-1 levels during embryonic mitosis, thereby identifying translational regulation as another mechanism for controlling MEI-1 levels during the egg-to-embryo transition.
Results and discussion
SPN-2 is required for normal spindle orientation in early embryos
A single allele of the spn-2 gene (spindle orientation defective) was identified in a screen for maternal-effect lethal mutations that disrupt asymmetric division in the C. elegans embryo. Homozygous spn-2 worms also show oogenesis defects and reduced embryo production. The spn-2(it149) allele behaves as a recessive, loss-of-function mutation, and all phenotypes are more severe at higher temperatures (Tables I and S1).
Table I.
Mutations in spn-2 cause defects in nuclear and spindle positioning
| Genotype and conditions | P0 spindle transversea | Ectopic cleavage furrowsb | Mispositioned AB or P1 nucleic | Abnormal AB or P1 spindle positiond |
| Wild type filmed at 23–24C° | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) |
| spn-2(it149) filmed at 23–24°C | 1/23 (4%) | 14/23 (61%) | 13/22 (59%) | 10/22 (45%) |
| spn-2(it149) filmed > 25°C | 3/13 (23%) | 12/12 (100%) | 11/11 (100%) | 7/10 (70%) |
| spn-2(it149) shifted to 25°C; filmed at 23–24°C | 11/31 (35%) | 24/27 (89%) | 13/22 (59%) | 10/20 (50%) |
| spn-2(it149);mei-1(RNAi) shifted to 25°C; filmed at 23–24°C | 0/17 (0%) | 6/19 (32%) | 3/20 (15%) | 5/20 (25%) |
| F56F3.1(RNAi) filmed at 23–24°C | 5/19 (26%) | 12/19 (63%) | 11/18 (61%) | 12/17 (71%) |
Hermaphrodites were raised at 20°C and shifted to 25°C for 1–2 h if indicated. Embryos were scored using DIC microscopy during the first two divisions, but those with a transverse P0 spindle, cytokinesis defects, or osmosensitivity were excluded from the analysis of AB and P1 defects.
The spindle was positioned at a >45° angle relative to the A-P axis of 0° at either metaphase or anaphase. Of the 20 embryos with this phenotype, 17 failed to center and rotate, but then the spindle became normally aligned by anaphase in 11 of those. The remaining three centered and rotated but the spindle moved to a posterior transverse position during metaphase/anaphase.
More than two ingressing cleavage furrows during and just after cytokinesis.
Nuclei were not centrally positioned after cytokinesis.
The AB spindle aligned within 45° of the A-P axis and/or the P1 spindle was transverse to the A-P axis; in wild type, AB spindles are transverse and P1 spindles are aligned.
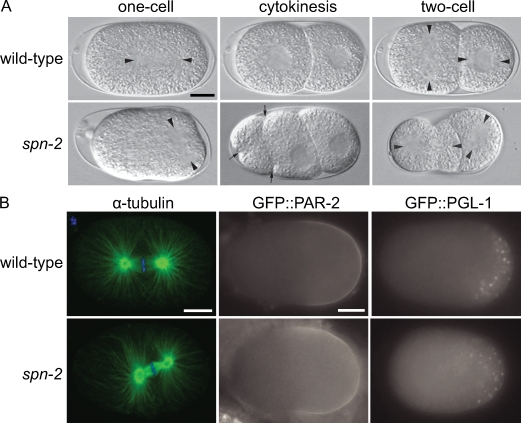
Wild-type C. elegans embryos undergo a series of asymmetric divisions that exhibit precise spindle orientations. After fertilization, an anterior-posterior (A-P) polarity axis is established in the one-cell embryo by the PAR proteins (Galli and van den Heuvel, 2008). The male and female pronuclei meet in the posterior, and the pronuclear–centrosome complex moves to the center and rotates onto the A-P axis (Fig. 1 A). Spindle displacement toward the posterior results in unequal cell division. Similar spindle movements are repeated in the smaller posterior daughter cell, P1, but not the anterior AB cell (Fig. 1 A). In most embryos from spn-2(it149) mutant mothers grown at 20°C and analyzed at a temperature of 23–24°C, nuclear and spindle positioning in the one-cell stage appeared normal, but alterations in second division spindle orientations were observed. When shifted to higher temperature just before or during filming, some embryos exhibited a posteriorly positioned spindle that was transverse to the A-P axis of the embryo (Fig. 1 A and Table I). Astral microtubules were robust in spn-2 embryos (Fig. 1 B); however, metaphase spindles were shorter (9.8 ± 0.2 µm; n = 8) than in wild type (13.1 ± 0.9 µm; n = 6). Most mutant embryos also exhibited ectopic furrows during cytokinesis, and mispositioned nuclei at the two-cell stage before spindle orientation (Fig. 1 A and Table I). For simplicity, embryos from spn-2 mutant mothers will be referred to hereafter as spn-2 mutants or embryos. All further analyses were performed using spn-2(it149), except for the use of RNAi as evidence for the identity of the spn-2 gene (Table I).
Figure 1.
SPN-2 is required for proper spindle positioning. (A) DIC images of live mitotic embryos. Arrowheads mark the centrosomes and arrows point to ectopic cleavage furrows. (B) Confocal micrographs of α-tubulin (green) and DAPI (blue) staining of one-cell metaphase embryos; epifluorescence images of embryos expressing GFP::PAR-2 or GFP::PGL-1. Bars, 10 µm.
To determine if the spn-2 spindle positioning phenotypes are caused by polarity defects, we examined the localization of PAR-2 and P granules (Galli and van den Heuvel, 2008). As in the wild type, PAR-2::GFP was restricted to the posterior periphery of single-cell spn-2 embryos (Fig. 1 B; 9/10, including two embryos with transverse spindles; PAR-2 was lateral in one embryo). Similarly, PGL-1::GFP, a constitutive component of P granules (Kawasaki et al., 1998), was localized to the posterior half of spn-2 embryos (Fig. 1 B; 11/12; one had lateral P granules). Finally, the AB cell divided before P1, as in wild type (n = 17). These results suggest that SPN-2 affects spindle positioning downstream of the PAR proteins or in an independent pathway.
MEI-1 ectopically localizes to the mitotic spindle in spn-2 embryos
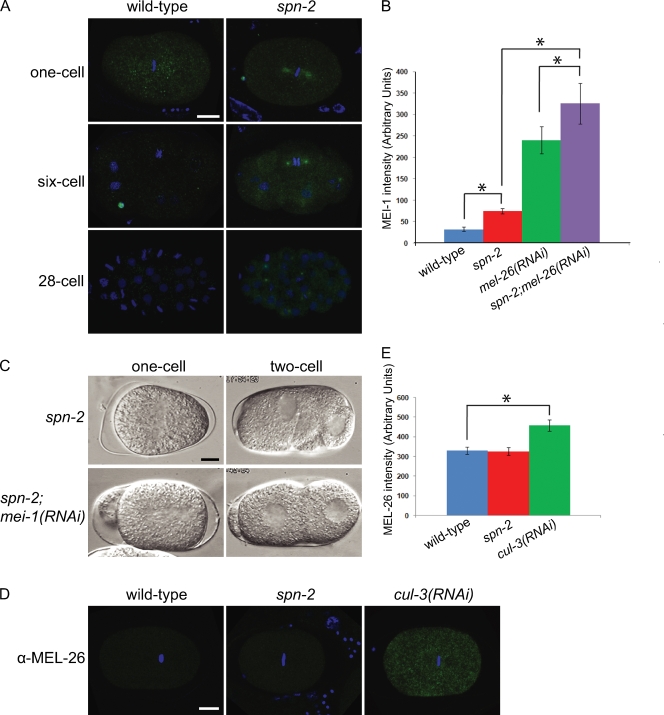
Posteriorly mispositioned spindles, ectopic furrows, and mispositioned nuclei are also observed in mutants defective in MEI-1 degradation (Kurz et al., 2002; Pellettieri et al., 2003). This led us to examine spn-2 embryos for ectopic MEI-1 during mitosis. α-MEI-1 antibodies stained the mitotic spindle in all one- and two-cell spn-2 embryos (n = 65) but not in wild type (n = 88; Fig. 2, A and B; and Table S2); MEI-1 staining persisted in spn-2 embryos to at least the 28-cell stage (n = 10; Fig. 2 A). Depletion of MEI-1 by RNAi in spn-2 mutants suppressed the spindle positioning defects in one-cell stage embryos, and suppressed the ectopic furrows and mispositioned nuclei in many spn-2 embryos; two-cell stage spindle positioning defects were also reduced (Fig. 2 C and Table I). These results indicate that many of the phenotypes observed in early spn-2 embryos are caused by ectopic MEI-1 localization.
Figure 2.
MEI-1 ectopically localizes to mitotic spindles in spn-2 embryos. (A) Confocal micrographs of α-MEI-1 (green) and DAPI (blue) staining. (B) Quantification of spindle MEI-1 levels in one-cell metaphase embryos. (C) DIC images of one-cell anaphase (left) and two-cell interphase (right) embryos. (D) Confocal micrographs of α-MEL-26 (green) and DAPI (blue) staining. (E) Quantification of MEL-26 levels in one-cell embryos. Error bars represent SEM; asterisks indicate statistical significance (P < 0.05). Bar, 10 µm.
MEL-26 is the substrate-specific adaptor that targets MEI-1 to the CUL-3 complex for degradation after meiosis, and thus reduction of MEL-26 leads to ectopic MEI-1 (Kurz et al., 2002; Pintard et al., 2003a,b). At the same time, MEL-26 is ubiquitinated by CUL-3, and thus defects in the CUL-3 pathway, including the upstream neddylation pathway, lead to higher levels of MEL-26 in addition to failure of MEI-1 degradation (Luke-Glaser et al., 2007). We therefore examined MEL-26 by indirect immunofluorescence as a readout for the function of the CUL-3 pathway. MEL-26 levels were normal in spn-2 embryos, in contrast to what was observed after cul-3(RNAi) (Fig. 2, D and E; and Table S2). Thus, the degradation pathway for MEI-1 does not appear to be perturbed in spn-2 embryos. In addition, MEI-1 levels in spn-2; mel-26(RNAi) were higher than those in spn-2 or mel-26(RNAi) embryos, which is consistent with SPN-2 acting in parallel to the MEL-26 degradation pathway (Fig. 2 B and Table S2).
SPN-2 is an eIF4E-binding protein
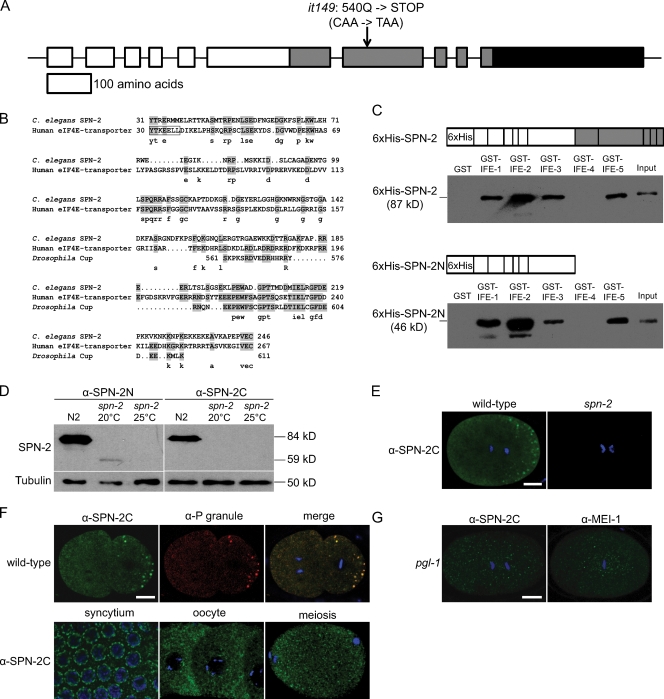
To address how SPN-2 regulates MEI-1 protein levels, we cloned the spn-2 gene. Mapping and germ-line transformation placed spn-2 on the cosmid F56F3 (see Materials and methods). Two lines of evidence indicate that spn-2 corresponds to predicted gene F56F3.1: (1) RNAi of the F56F3.1 gene phenocopied the spn-2 mutant (Table I), and (2) sequencing of DNA from spn-2(it149) mutants revealed a nonsense mutation in F56F3.1 (Fig. 3 A). Basic Local Alignment Search Tool (BLAST) searches (Altschul et al., 1997) and alignment by Clustal W (Larkin et al., 2007) revealed that the predicted F56F3.1/SPN-2 protein shares limited sequence similarity to two 4E-BPs, human eukaryotic translation initiation factor 4E transporter (4E-T), and Drosophila Cup (Fig. 3 B). 4E-T is required for the nucleocytoplasmic shuttling of eIF4E (Dostie et al., 2000) and promotes RNA decay in processing bodies (Ferraiuolo et al., 2005). Cup prevents precocious translation of oskar and nanos mRNA (Piccioni et al., 2005). SPN-2 and Cup both contain clusters of glutamine residues in their C-terminal regions, but otherwise the C termini do not show sequence similarity (Nakamura et al., 2004).
Figure 3.
spn-2 encodes an eIF4E-binding protein that localizes to the cytoplasm and P granules. (A) Diagram of spn-2 intron/exon structure and the it149 mutation. White, protein coding sequence; gray, Q-rich domain; black, 3′ UTR. (B) Alignments of the N-terminal region of C. elegans SPN-2 with human 4E-T and an internal region of Drosophila Cup; these regions are 27% and 37% identical, respectively, to SPN-2 (identities shaded). The eIF4E-binding motif (YxxxxLϕ, where ϕ is a hydrophobic residue) is boxed. (C) 6xHis-tagged SPN-2 or SPN-2N were incubated with GST-eIF4E isoforms; eluates were analyzed by Western blotting with α-6xHis. (D) Western blots of wild-type (N2) and spn-2 worm extracts probed with α-SPN-2N or α-SPN-2C and reprobed with α-tubulin as a loading control. (E–G) Confocal images of wild type or mutants stained with the indicated antibodies and DAPI (blue). (E) One-cell embryos. (F, top) Two-cell embryos. (F, bottom) Wild-type germ-line syncytium, oocytes, and a meiotic embryo. (G) One-cell embryos. Bars, 10 µm.
SPN-2 lacks the consensus eIF4E-binding motif found in several 4E-BPs including 4E-T and Cup; however, binding through noncanonical sites has been observed (Stebbins-Boaz et al., 1999; Amiri et al., 2001; Nelson et al., 2004). Therefore, we performed binding assays with GST-tagged eIF4E isoforms and 6xHis-tagged SPN-2 protein expressed and purified from Escherichia coli. C. elegans has five eIF4E isoforms (Jankowska-Anyszka et al., 1998; Keiper et al., 2000). 6xHis-SPN-2 copurified with GST–IFE-1, -2, -3, and -5 but not with GST–IFE-4 or GST alone (Fig. 3 C). A SPN-2 N-terminal fragment containing the region similar to other 4E-BPs was sufficient to bind the same eIF4E isoforms (Fig. 3 C). The specificity of the eIF4E–SPN-2 interaction was confirmed by quantitative assays using IFE-1 and full-length 6xHis-SPN-2, which showed saturable binding with a Kd of 0.033 µM (Fig. S1). Thus, we conclude that SPN-2 is a bona fide 4E-BP.
SPN-2 localizes to the cytoplasm and to P granules
To determine the cellular localization of SPN-2, affinity purified antibodies against the SPN-2 N terminus (SPN-2N) and C terminus (SPN-2C) were generated. Western blot assays of embryo extracts using α-SPN-2N revealed an 84-kD protein in wild type and a 59 kD protein in spn-2 worms (Fig. 3 D). The levels of truncated product in spn-2 worms were greatly reduced compared with wild type, especially at 25°C, which is consistent with the more severe phenotype observed at higher temperatures. The α-SPN-2C antibody also recognized an 84-kD protein in wild-type but not the truncated SPN-2 protein.
In situ immunolocalization using both antibodies gave similar staining patterns in wild type, and staining was absent in spn-2 mutants when α-SPN-2C was used (Fig. 3 E), or greatly reduced when α-SPN-2N was used. In embryos, SPN-2 staining was observed in the cytoplasm and on large punctae that were partitioned to the posterior of the one-cell embryo (n = 17; Fig. 3 E), as well as to the germ-line P blastomeres in older embryos (n = 91). Double-labeling confirmed that the SPN-2 punctae colocalize with P granules (Fig. 3 F), ribonucleoprotein particles important for germ-line development that contain several translational regulators (Strome, 2005). The localization of SPN-2 on P granules thus supports the model that SPN-2 is a 4E-BP that functions in translational control. Cytoplasmic staining and perinuclear punctae were also observed in the meiotic germ line, but only cytoplasmic staining was seen in late oocytes and meiotic embryos, when P granules are dispersed (Fig. 3 F).
MEI-1 is synthesized during oogenesis and is essential for meiosis. Thus, the presence of SPN-2 at these stages raised the question of when SPN-2 regulates MEI-1. Quantification showed that MEI-1 levels in oocytes and meiotic embryos were not affected in spn-2 mutants (Table S2), which suggests that SPN-2 down-regulates MEI-1 after meiosis. To determine if P granule localization of SPN-2 is required for MEI-1 regulation, we examined pgl-1 mutants. In pgl-1 germ lines and embryos, SPN-2 failed to localize to P granule–like structures but was still present in the cytoplasm (Fig. 3 G). However, pgl-1 embryos did not have ectopic MEI-1 staining (n = 11; Fig. 3 G). Together, these results suggest that the cytoplasmic pool of SPN-2 is sufficient to prevent ectopic MEI-1 expression but that some additional regulation must restrict SPN-2's effect to mitosis.
OMA-1 interacts with SPN-2 and the mei-1 3′ untranslated region (UTR)
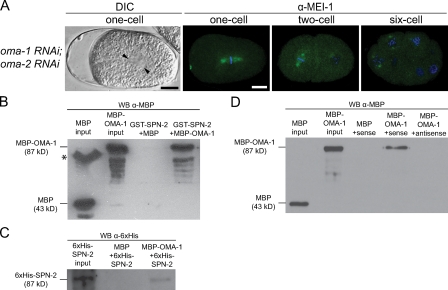
The identification of SPN-2 as a 4E-BP suggests it controls MEI-1 expression via translational repression. 4E-BPs do not bind target mRNAs directly; rather, specificity is provided by interacting with RNA-binding proteins (Piccioni et al., 2005). We investigated whether SPN-2 represses mei-1 translation via OMA-1 and OMA-2, two known RNA-binding proteins and translational repressors (Jadhav et al., 2008). OMA-1 and OMA-2 function redundantly in oocyte maturation (Detwiler et al., 2001). Partial depletion of oma-1 and oma-2 by RNAi allows maturation and results in embryos with division defects (Nishi and Lin, 2005). We examined early division in oma-1(RNAi); oma-2(RNAi) embryos and observed transverse spindles at first division (7/13; Fig. 4 A), ectopic cleavage furrows during cytokinesis (5/5), and mispositioned nuclei in AB or P1 cells (4/5). All embryos showed ectopic MEI-1 staining during first mitosis (n = 9; Fig. 4 A), and MEI-1 persisted in later stage embryos (n = 24; Fig. 4 A). In addition, MEL-26 levels were normal (Table S2), as seen in spn-2. These results suggest that OMA-1 and OMA-2 could be involved in the same pathway as SPN-2 to regulate MEI-1. As OMA-1 and OMA-2 share a high degree (64%) of identity throughout the entire protein, only OMA-1 was tested in further assays.
Figure 4.
OMA-1 interacts with SPN-2 and the mei-1 3′ UTR. (A) DIC image of a metaphase embryo with skewed spindle (arrowheads mark the centrosomes), and confocal images of embryos stained for MEI-1 (green) and DAPI (blue). Bar, 10 µm. (B and C) In vitro pull-down assays using the proteins shown; eluates were analyzed by Western blotting (WB) with the indicated antibodies. The asterisk in B indicates a band of aggregated MBP in the MBP input lane. (D) MBP–OMA-1 or MBP was incubated with biotinylated sense or antisense RNA from the mei-1 3′ UTR. RNA-bound proteins were analyzed by Western blotting.
To determine if OMA-1 and SPN-2 interact, in vitro pull-down assays were performed. Maltose-binding protein (MBP)-tagged OMA-1 copurified with GST–SPN-2 bound to glutathione beads, but MBP alone did not; similarly, 6xHis–SPN-2 copurified with MBP–OMA-1 immobilized on amylose resin (Fig. 4, B and C). 3′ UTRs are often the target for translational control (de Moor et al., 2005; Merritt et al., 2008). Thus, to test whether OMA-1 binds directly to mei-1 mRNA, we performed RNA pull-down assays using biotinylated RNA corresponding to the sense or antisense strand of the mei-1 3′ UTR. RNAs were incubated with purified MBP–OMA-1 or MBP, and then bound to streptavidin beads. Western blotting of bound proteins showed that the sense strand of mei-1 3′ UTR associated with MBP–OMA-1 but not MBP alone (Fig. 4 E). These results suggest that OMA-1 recruits SPN-2 to the mei-1 mRNA.
All of our results are consistent with the model that SPN-2 acts with OMA-1 to translationally repress mei-1 mRNA and thus down-regulate MEI-1 protein levels during mitosis. The activities of severing enzymes for both microfilaments and microtubules must be tightly regulated for normal cytoskeletal function (Yu et al., 1992; Sun et al., 1999). In C. elegans, multiple pathways have been described for MEI-1 regulation during the egg-to-embryo transition. The best characterized is the CUL-3–MEL-26 ubiquitin pathway (Luke-Glaser et al., 2005). Although SPN-2 could regulate components of this pathway, our data argue that SPN-2 acts in parallel to MEL-26. The anaphase-promoting complex (APC) and the MBK-2 kinase have also been shown to promote MEI-1 degradation after meiosis in a MEL-26–independent pathway (Lu and Mains, 2007), and MBK-2 can target OMA-1 for degradation in the early embryo (Nishi and Lin, 2005; Stitzel et al., 2006). However, the depletion of APC and MBK-2 only result in ectopic MEI-1 during the first division. In contrast, MEI-1 persists in later embryos in both spn-2 and oma-1(RNAi); oma-2(RNAi) embryos (Figs. 2 A and 4 A). Further, the levels of OMA-1::GFP expressed under the control of oma-1 regulatory sequences were comparable in spn-2 and wild type (Table S2), which indicates that SPN-2 does not act by controlling OMA-1 levels. Recently, phosphorylation of MEI-1 has also been implicated in regulating MEI-1 activity, but this appears to act independently of MEI-1 protein levels, in contrast to what is observed for SPN-2 (Han et al., 2009). Thus, we propose that translational repression of mei-1 mRNA by SPN-2 via its interaction with OMA-1 is yet another mechanism to regulate MEI-1 and prevent the deleterious effects of ectopic MEI-1 during mitosis. Northern blot analysis revealed that wild-type mitotic embryos have high levels of mei-1 mRNA for at least 3 h after meiosis (Clark-Maguire and Mains, 1994b), and in situ hybridization supports the persistence of mei-1 mRNA in embryos (Y. Kohara, personal communication). Thus, translational repression by SPN-2 would prevent the synthesis of new MEI-1 protein from this maternally loaded mRNA.
SPN-2 is predicted to act similarly to the Cup and Maskin 4E-BPs, which mediate repression in conjunction with specific RNA-binding proteins. In this case, OMA-1 would bind the mei-1 3′ UTR, and recruit SPN-2, which would then bind to eIF4E and prevent translation initiation. Although OMA-1 levels are greatly reduced by the end of the one-cell stage, roles for OMA-1 and -2 in the first two divisions have been reported (Nishi and Lin, 2005; Guven-Ozkan et al., 2008). After the two-cell stage, repression of MEI-1 could be mediated either by very low levels of OMA-1 and -2, or, alternatively, by other RNA-binding proteins; a precedent for sequential action by different repressors has been described for nos-2 in C. elegans (Jadhav et al., 2008). Both SPN-2 and OMA-1 are also abundant in oocytes and meiotic embryos, but SPN-2 does not appear to repress mei-1 expression at these stages. 4E-BPs in other systems are regulated by phosphorylation, and thus one possibility is that SPN-2 activity is regulated by the meiosis-to-mitosis transition (Richter and Sonenberg, 2005). Alternatively, other proteins may interact with mei-1 mRNA, SPN-2, or OMA-1 to regulate the timing of repression.
In conclusion, we have identified SPN-2 as a 4E-BP protein in C. elegans that was not previously identified through homology searches. Although some 4E-BPs have a general effect on translation, others such as Cup and Maskin regulate a subset of messages. We speculate that SPN-2 likewise has a small number of mRNA targets. Although spn-2/deletion animals have defects in the late stages of oogenesis, overall larval and adult development appeared normal (Table S1). Further, embryonic polarity and levels of MEL-26 and OMA-1 were normal in spn-2 embryos, and mei-1(RNAi) suppressed most spn-2 phenotypes. The future study of SPN-2 mRNA targets and interacting proteins will provide new insights into mechanisms of translational regulation during the egg-to-embryo transition.
Materials and methods
Strains and genetics
C. elegans were cultured on MYOB plates (Church et al., 1995) using standard conditions (Brenner, 1974). The following strains were used in this study. BC4637: dpy-17 (e164) sDf130 (s2427) unc-32(e189)III; sDp (III, f). BC4638: dpy-17 (e164) sDf127 (s2428) unc-32(e189)III; sDp (III, f). BC4697: dpy-17 (e164) sDf121 (s2098) unc-32(e189)III; sDp (III, f). BW1092a: pgl-1 (ct131)IV (Kawasaki et al., 1998). CB4856: wild type, Hawaiian variant. JH1623: unc-119 (ed3); axIs1182[pie-1:gfp::par-2 + unc-119(+)] (Hao et al., 2006). KK781: spn-2(it149) unc-36 (e251)/qC1 III. MT8388: dpy-17 (e164) mpk-1 (oz140)/dpy-17 (e164) unc-79 (e1068). N2: wild type, Bristol variant. RL104: spn-2(it149) dpy-17(e164)/qC1 III. SS747: bnIs1[pie-1::gfp::pgl-1 + unc-119(+)] (Spike et al., 2008). TX189: unc-119 (ed3) III; teIs[oma-1::gfp+ unc-119(+)] (Lin, 2003). BC4637, BC4638, and BC4697 were provided by D. Baillie (Simon Fraser University, Burnaby, British Columbia, Canada); BW1092a and SS747 were provided by S. Strome (University of California, Santa Cruz, Santa Cruz, CA); JH1623 was provided by G. Seydoux (Johns Hopkins University, Baltimore, MD); and the remaining strains were obtained from the Caenorhabditis Genetics Center at the University of Minnesota or created in the lab.
Molecular identification of spn-2
A single allele of spn-2, it149, was identified in a screen of 10,000 haploid genomes for maternal-effect lethal mutations (Rose and Kemphues, 1998). Standard linkage and recombination analysis placed spn-2 on chromosome III, between daf-2 and unc-36. The maternal effect lethality of spn-2 was complemented by deletion sDf121 but not sDf130, placing spn-2 between dpy-27 and mab-21 (Stewart et al., 1998). Single nucleotide polymorphism (SNP) mapping further defined an ∼100-kb region between mpk-1 and mab-21 that contains spn-2. For SNP mapping (Wicks et al., 2001), spn-2 (it149) dpy-17/qC1 hermaphrodites (Bristol variant) were mated to CB4856 (Hawaiian variant) males to generate heterozygotes, and the dpy-17 nonspn-2 recombinants were analyzed for the presence of SNP-F25F2[1] and SNP-C28A5[2] (http://www.wormbase.org/). Of 36 recombinants, 24 recombined between dyp-17 and SNP-F25F2[1], and 12 recombined between SNP-F25F2[1] and SNP-C28A5[2], which suggests that spn-2 was to the right or very close to SNP-C28A5[2]. To identify the spn-2 gene within this region, DNA from spn-2(it149) homozygous adults was used for PCR amplification and sequencing of the predicted coding regions of H38K22.2, R07E5.3, R07E5.10, R07E5.17, C28A5.6, C28A5.3, F56F3.1, F56F3.5, and C07G2.3, which are predicted to be involved in microtubule function or were reported to show sterility or cell division phenotypes after RNA-mediated interference. The F56F3 cosmid (from the C. elegans Sequence Consortium) was coinjected (Mello and Fire, 1995) with a plasmid containing pY110A7A.20::GFP (which expresses GFP in cilated neurons; provided by G. Ou and J. Scholey, University of California, Davis, Davis, CA) into spn-2(it149) dpy-17(e164)/qC1 hermaphrodites. Heritable GFP-positive lines were obtained, and GFP spn-2 segregants that gave rise to >20 adult progeny were scored as rescued.
RNAi
For spn-2 and cul-3 RNAi, antisense and sense RNAs were transcribed in vitro (MEGAscript; Applied Biosystems) from linear DNA templates made by the polymerase chain reaction, using T3 (5′-GAAAGCCTTGTTACGCATGGATGC-3′) and T7 (5′-CGTCATCAACTGGTCTCAAGAC-3′) to amplify partial cDNAs from the RB1 library (R. Barstead, Oklahoma Medical Research Foundation, Oklahoma City, OK). Double-stranded RNAs (dsRNAs) were annealed as described previously (Fire et al., 1998). L4 worms were soaked (Tabara et al., 1998) in a 1.5-mg/ml solution of spn-2 dsRNA for 16–24 h at 20°C, or injected with cul-3 dsRNA and analyzed 24 h later. RNAi was performed by feeding (Kamath and Ahringer, 2003) wild-type or spn-2 L4 larvae on freshly induced plates at 20°C for 24 h, 30 h, and 20 h for mei-1, mel-26, oma-1, and oma-2, respectively; the L4440 plasmid was used as a control.
Antibodies and immunolocalization
Fragments of the spn-2 gene encoding the N-terminal (1–399 aa, SPN-2N) and C-terminal portions of SPN-2 (542–761 aa, SPN-2C) were amplified from the yk635e3 cDNA (provided by Y. Kohara, National Institute of Genetics, Yata, Mishima, Japan) and cloned into pDEST 17 and pDEST 15 vectors (Gateway; Invitrogen) to generate 6×His-tagged and GST-tagged SPN-2 fusions. Fusion proteins were expressed in bacteria and column purified on Ni2+-NTA agarose (QIAGEN) or glutathione Sepharose beads (GE Healthcare) using standard procedures. GST–C-terminal SPN-2 and 6×His–N-terminal SPN-2 were injected into rabbits (Covance). Polyclonal antisera were affinity purified against GST:SPN-2N and GST:SPN-2C cross-linked to glutathione beads (http://www.flemingtonlab.com/Protocols/AbAffinityPurificationProt.pdf). Antibodies were eluted with 0.2 M glycine-HCl, pH 2.5, immediately neutralized to 1 M K2HPO4, and dialyzed against 1:1 PBS/glycerol overnight at 4°C.
Whole-worm extracts from ∼50 gravid adult worms were resolved on 10% SDS-polyacrylamide gels, and subjected to Western blotting using standard procedures (Harlow and Lane, 1988). Blots were incubated with α-SPN-2N (1:300,000 in phosphate buffered saline with Tween 20), α-SPN-2C (1:600,000), and α-tubulin DM1a (1:2,000; Sigma-Aldrich) and visualized using the ECL detection system (GE Healthcare).
For in situ immunolocalization, worms were raised at 20°C and processed immediately. For SPN-2 and P granules, worms were cut in egg buffer on poly-lysine–coated slides, freeze-cracked, and fixed in −20°C methanol (Miller and Shakes, 1995). Slides were preblocked with 30% goat serum for 30 min at room temperature. Primary antibodies used were as follows: rabbit α-SPN-2N (1:3,000), rabbit α-SPN-2C (1:6,000), α-P granules (mAbOIC1D4, 1:2; Developmental Studies Hybridoma Bank, developed by Strome and Wood [1983]). For MEI-1 and MEL-26, embryo fixation and primary antibody incubations were performed as described for MEI-1 (1:100; Clark-Maguire and Mains, 1994a) and MEL-26 (1:30; Johnson et al., 2009). For α-tubulin, slides were fixed in room temperature methanol for 5 min and incubated with DM1a (1:400; Sigma-Aldrich) for 45 min at room temperature, followed by secondary antibody incubation. FITC goat α-rabbit and rhodamine goat α-mouse antibodies (1:200 in PBS; Jackson ImmunoResearch Laboratories) were used as secondary antibodies after preabsorption with acetone powder made from wild-type worms. DAPI (4, 6-diamidino-2-phenylindole dihydrochloride) was used to label nuclei for determination of cell cycle stages.
Microscopy and quantification of staining intensities
For live analysis, embryos were cut from hermaphrodites in 80% egg buffer and mounted on poly-lysine–coated slides as described previously (Rose and Kemphues, 1998) to prevent flattening. Embryos were filmed under differential interference contrast (DIC) Nomarski optics using a time-lapse video recorder and charge-coupled device (CCD) camera (Panasonic), or examined by epifluorescence using a 100× UPLan-Fl NA 1.30 oil immersion lens (Olympus) on a photomicroscope (BX60; Olympus) equipped with a 12-bit digital CCD camera (Orca; Hamamatsu) and Openlab software (PerkinElmer). For DIC analysis, worms were raised and filmed at the temperatures listed in Table I. GFP::PGL-1 worms were raised at 20°C and shifted to 25°C for 1 h, and GFP::PAR-2 worms were raised at 24°C to ensure GFP reporter expression, and then filmed at 23–24°C.
For immunolocalization, z series (1 µm optical sections) through the entire embryo were obtained using a laser scanning confocal microscope (FV1000 Fluoview) equipped with a 60× Plan-Apochromat NA 1.42 oil immersion lens (both from Olympus). All images for a particular antibody were taken with identical settings, with the gain adjusted below saturation, using FV10-ASW software (version 1.5.0.14; Olympus). Images were cropped in Photoshop and assembled in Illustrator (both from Adobe). ImageJ (http://rsb.info.nih.gov/ij/) was used to measure the mean pixel intensities of areas as follows. For quantification of MEI-1 and MEL-26, four confocal sections at mid-spindle plane or mid-cell plane were merged using maximum-intensity mode. Spindle MEI-1 levels in one-cell embryos were measured within a box of fixed size (16 µm × 8 µm) around the metaphase spindle as determined by DAPI labeling. Spindle MEI-1 levels in meiotic embryos were measured in a circle of 10-µm diameter. Spindle intensities were corrected by subtracting the cytoplasmic contribution measured from circles/boxes of the same sizes. Cytoplasmic MEI-1 values in oocytes were measured from circles of 5-µm diameter. For MEL-26, measurements were from 15-µm-diameter circles in the one-cell cytoplasm. To quantify OMA-1, spn-2 (it149) dpy17/qC1; OMA-1::GFP was created by crossing RL104 and TX189. Cytoplasmic intensities were measured in one-cell metaphase embryos from 15-µm-diameter circles in the anterior and posterior and averaged for each embryo. For all measurements, cytoplasmic intensities were corrected by subtracting the intensity of the slide background measured from circles/boxes of the same sizes.
Binding assays
To test the interaction between SPN-2 and eIF4E, full-length cDNAs for the five eIF4E isoforms of C. elegans were subcloned into pDEST 15. GST-tagged eIF4Es or GST expressed in E. coli were bound to glutathione Sepharose beads (GE Healthcare), then incubated with purified 6×His-SPN-2 or 6×His-SPN-2N in binding buffer (30 mM Hepes, pH 7.5, 100 mM KCl, 10% glycerol, 1% Triton X-100, 1 mM MgCl2, 1 mM DTT, and protease inhibitor cocktail) at 4°C for 2 h. The beads were washed with binding buffer four times and boiled in SDS sample buffer. Eluates were separated by SDS-PAGE and analyzed by Western blotting with α-6xHis monoclonal antibody (Abcam). To generate a saturation binding curve, increasing concentrations of 6xHis-SPN-2 were added to glutathione beads bound with a fixed concentration of GST–IFE-1, or a fixed concentration of 6xHis-SPN-2 was added to glutathione beads bound with increasing concentrations of GST–IFE-1. The purified 6×His-SPN-2 and GST–IFE-1 were detected by Coomassie blue staining and quantified using Image J.
Binding between GST–SPN-2 and MBP–OMA-1 was performed as described for SPN-2 and eIF4E, except that blots were probed with an α-MBP monoclonal antibody (New England Biolabs, Inc.). MBP–OMA-1 or MBP bound to amylose resin (New England Biolabs, Inc.) were incubated with purified 6×His-SPN-2 in binding buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10 mM β-mercaptoethanol, and protease inhibitor cocktail). Resin was washed with binding buffer four times and boiled in SDS sample buffer. The eluates were analyzed by Western blotting with α-6xHis.
Biotinylated RNA pull-down assay
PCR-amplified products containing the T7 promoter served as templates for the in vitro synthesis of biotinylated mei-1 sense 3′ UTR and antisense 3′ UTR using the AmpliScribe T7-Flash transcription kit (EPICENTRE Biotechnologies). 0.5–1 µg of purified MBP–OMA-1 or MBP was incubated with 1 µg of biotinylated RNA for 30 min at room temperature. RNA–protein complexes were isolated using prewashed streptavidin-conjugated Dynabeads (Dynabeads M-280 streptavidin; Invitrogen). Proteins were eluted with SDS sample buffer and analyze by Western blotting with α-MBP.
Online supplemental material
Fig. S1 shows that the interaction between eIF4E and SPN-2 is saturable by a series of GST pull-down assays. Table S1 shows that mutations in spn-2 result in embryo lethality and reduced embryo production by quantifying the brood size and hatch rate. Table S2 displays quantification of MEI-1, OMA-1::GFP, and MEL-26 levels in different genotypes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903003/DC1.
Acknowledgments
We are grateful to P. Mains and K. Kemphues for strains, antibodies, and valuable discussions. We thank Y. Kohara for cDNAs, and S. Strome and G. Seydoux for strains. Other strains were obtained from the Caenorhabditis Genetics Center, supported by the National Institutes of Health National Center for Research Resources. We also thank F. McNally, D. Starr, J. Scholey, and G. Ou for helpful comments, and A. Hayashi for technical assistance.
This work was supported by National Institutes of Health grants R01GM068744 to L. Rose and R01HD037933 to R. Lin.
Footnotes
Abbreviations used in this paper:
- 4E-BP
- eIF4E-binding protein
- A-P
- anterior-posterior
- DIC
- differential interference contrast
- MBP
- maltose-binding protein
- UTR
- untranslated region
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Nucleic Acids Res. 25:3389–3402 doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A., Keiper B.D., Kawasaki I., Fan Y., Kohara Y., Rhoads R.E., Strome S. 2001. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans.Development. 128:3899–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Kurz T. 2006. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis.Development. 133:773–784 doi:10.1242/dev.02276 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans.Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D.L., Guan K.L., Lambie E.J. 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans.Development. 121:2525–2535 [DOI] [PubMed] [Google Scholar]
- Clark-Maguire S., Mains P.E. 1994a. Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component.J. Cell Biol. 126:199–209 doi:10.1083/jcb.126.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., Mains P.E. 1994b. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases.Genetics. 136:533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor C.H., Meijer H., Lissenden S. 2005. Mechanisms of translational control by the 3′ UTR in development and differentiation.Semin. Cell Dev. Biol. 16:49–58 doi:10.1016/j.semcdb.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Detwiler M.R., Reuben M., Li X., Rogers E., Lin R. 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans.Dev. Cell. 1:187–199 doi:10.1016/S1534-5807(01)00026-0 [DOI] [PubMed] [Google Scholar]
- Dostie J., Ferraiuolo M., Pause A., Adam S.A., Sonenberg N. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E.EMBO J. 19:3142–3156 doi:10.1093/emboj/19.12.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T.C., Hunter C.P. 2005. Translational control of maternal RNAs. WormBook, editor. The C. elegans Research Community, Wormbook. doi/10.1895/wormbook.1.34.1. http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo M.A., Basak S., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay.J. Cell Biol. 170:913–924 doi:10.1083/jcb.200504039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans.Nature. 391:806–811 [DOI] [PubMed] [Google Scholar]
- Galli M., van den Heuvel S. 2008. Determination of the cleavage plane in early C. elegans embryos.Annu. Rev. Genet. 42:389–411 doi:10.1146/annurev.genet.40.110405.090523 [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation.Annu. Rev. Biochem. 68:913–963 doi:10.1146/annurev.biochem.68.1.913 [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T., Nishi Y., Robertson S.M., Lin R. 2008. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4.Cell. 135:149–160 doi:10.1016/j.cell.2008.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Gomes J.E., Birmingham C.L., Pintard L., Sugimoto A., Mains P.E. 2009. The role of protein phosphatase 4 in regulating microtubule severing in the Caenorhabditis elegans embryo.Genetics. 181:933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Boyd L., Seydoux G. 2006. Stabilization of cell polarity by the C. elegans RING protein PAR-2.Dev. Cell. 10:199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D.P. 1988. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Jadhav S., Rana M., Subramaniam K. 2008. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells.Development. 135:1803–1812 doi:10.1242/dev.013656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska-Anyszka M., Lamphear B.J., Aamodt E.J., Harrington T., Darzynkiewicz E., Stolarski R., Rhoads R.E. 1998. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures.J. Biol. Chem. 273:10538–10542 doi:10.1074/jbc.273.17.10538 [DOI] [PubMed] [Google Scholar]
- Johnson J.L., Lu C., Raharjo E., McNally K., McNally F.J., Mains P.E. 2009. Levels of the ubiquitin ligase substrate adaptor MEL-26 are inversely correlated with MEI-1/katanin microtubule-severing activity during both meiosis and mitosis.Dev. Biol. 330:349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Ahringer J. 2003. Genome-wide RNAi screening in Caenorhabditis elegans.Methods. 30:313–321 [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S. 1998. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans.Cell. 94:635–645 doi:10.1016/S0092-8674(00)81605-0 [DOI] [PubMed] [Google Scholar]
- Keiper B.D., Lamphear B.J., Deshpande A.M., Jankowska-Anyszka M., Aamodt E.J., Blumenthal T., Rhoads R.E. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans.J. Biol. Chem. 275:10590–10596 doi:10.1074/jbc.275.14.10590 [DOI] [PubMed] [Google Scholar]
- Kurz T., Pintard L., Willis J.H., Hamill D.R., Gönczy P., Peter M., Bowerman B. 2002. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway.Science. 295:1294–1298 doi:10.1126/science.1067765 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0.Bioinformatics. 23:2947–2948 doi:10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lin R. 2003. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality.Dev. Biol. 258:226–239 [DOI] [PubMed] [Google Scholar]
- Lu C., Mains P.E. 2007. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis.Dev. Biol. 302:438–447 doi:10.1016/j.ydbio.2006.09.053 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Lu C., Mains P.E., Peter M. 2005. The BTB protein MEL-26 promotes cytokinesis in C. elegans by a CUL-3-independent mechanism.Curr. Biol. 15:1605–1615 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Roy M., Larsen B., Le Bihan T., Metalnikov P., Tyers M., Peter M., Pintard L. 2007. CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo.Mol. Cell. Biol. 27:4526–4540 doi:10.1128/MCB.01724-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C., Fire A. 1995. DNA transformation.Methods Cell Biol. 48:451–482 [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G. 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline.Curr. Biol. 18:1476–1482 doi:10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M., Shakes D.C. 1995. Immunofluorescence microscopy.Methods Cell Biol. 48:365–394 [PubMed] [Google Scholar]
- Nakamura A., Sato K., Hanyu-Nakamura K. 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis.Dev. Cell. 6:69–78 doi:10.1016/S1534-5807(03)00400-3 [DOI] [PubMed] [Google Scholar]
- Nelson M.R., Leidal A.M., Smibert C.A. 2004. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression.EMBO J. 23:150–159 doi:10.1038/sj.emboj.7600026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi Y., Lin R. 2005. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans.Dev. Biol. 288:139–149 doi:10.1016/j.ydbio.2005.09.053 [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S.K., Seydoux G. 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans.Dev. Cell. 5:451–462 doi:10.1016/S1534-5807(03)00231-4 [DOI] [PubMed] [Google Scholar]
- Piccioni F., Zappavigna V., Verrotti A.C. 2005. A cup full of functions.RNA Biol. 2:125–128 [DOI] [PubMed] [Google Scholar]
- Pintard L., Kurz T., Glaser S., Willis J.H., Peter M., Bowerman B. 2003a. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans.Curr. Biol. 13:911–921 doi:10.1016/S0960-9822(03)00336-1 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J.H., Willems A., Johnson J.L., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., Peter M. 2003b. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase.Nature. 425:311–316 doi:10.1038/nature01959 [DOI] [PubMed] [Google Scholar]
- Richter J.D., Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins.Nature. 433:477–480 doi:10.1038/nature03205 [DOI] [PubMed] [Google Scholar]
- Rose L.S., Kemphues K. 1998. The let-99 gene is required for proper spindle orientation during cleavage of the C. elegans embryo.Development. 125:1337–1346 [DOI] [PubMed] [Google Scholar]
- Spike C.A., Bader J., Reinke V., Strome S. 2008. DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells.Development. 135:983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M., Buster D.W., Bazirgan O.A., McNally F.J., Mains P.E. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis.Genes Dev. 14:1072–1084 [PMC free article] [PubMed] [Google Scholar]
- Stebbins-Boaz B., Cao Q., de Moor C.H., Mendez R., Richter J.D. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E.Mol. Cell. 4:1017–1027 doi:10.1016/S1097-2765(00)80230-0 [DOI] [PubMed] [Google Scholar]
- Stewart H.I., O'Neil N.J., Janke D.L., Franz N.W., Chamberlin H.M., Howell A.M., Gilchrist E.J., Ha T.T., Kuervers L.M., Vatcher G.P., et al. 1998. Lethal mutations defining 112 complementation groups in a 4.5 Mb sequenced region of Caenorhabditis elegans chromosome III.Mol. Gen. Genet. 260:280–288 [DOI] [PubMed] [Google Scholar]
- Stitzel M.L., Seydoux G. 2007. Regulation of the oocyte-to-zygote transition.Science. 316:407–408 doi:10.1126/science.1138236 [DOI] [PubMed] [Google Scholar]
- Stitzel M.L., Pellettieri J., Seydoux G. 2006. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation.Curr. Biol. 16:56–62 doi:10.1016/j.cub.2005.11.063 [DOI] [PubMed] [Google Scholar]
- Strome S. 2005. Specification of the germ line. WormBook, editor. The C. elegans Research Community, Wormbook. doi/10.1895/wormbook.1.9.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Strome S., Wood W.B. 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos.Cell. 35:15–25 [DOI] [PubMed] [Google Scholar]
- Sun H.Q., Yamamoto M., Mejillano M., Yin H.L. 1999. Gelsolin, a multifunctional actin regulatory protein.J. Biol. Chem. 274:33179–33182 doi:10.1074/jbc.274.47.33179 [DOI] [PubMed] [Google Scholar]
- Tabara H., Grishok A., Mello C.C. 1998. RNAi in C. elegans: soaking in the genome sequence.Science. 282:430–431 [DOI] [PubMed] [Google Scholar]
- Vardy L., Orr-Weaver T.L. 2007. Regulating translation of maternal messages: multiple repression mechanisms.Trends Cell Biol. 17:547–554 doi:10.1016/j.tcb.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Wicks S.R., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map.Nat. Genet. 28:160–164 [DOI] [PubMed] [Google Scholar]
- Yu F.X., Sun H.Q., Janmey P.A., Yin H.L. 1992. Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin.J. Biol. Chem. 267:14616–14621 [PubMed] [Google Scholar]