Abstract
The organizer anchors the primary embryonic axis, and balance between dorsal (organizer) and ventral domains is fundamental to body patterning. Ligand of Numb protein-X (LNX) is a RING finger and four PDZ domain containing E3 ubiquitin ligase1,2. LNX serves as a binding platform and may have a role in cell fate determination, but its in vivo functions are unknown1–5. Here we show that Lnx-l (Lnx-like) acts as a critical regulator of dorso-ventral (D-V) axis formation in zebrafish. Depletion of Lnx-l using specific antisense morpholinos (MO), caused strong embryonic dorsalization. We identified Bozozok (Boz; also called Dharma or Nieuwkoid) as a binding partner and substrate of Lnx-l. Boz is a homeodomain-containing transcriptional repressor induced by canonical Wnt signaling that is critical for dorsal organizer formation6–12. Lnx-l induced K48-linked polyubiquitination of Boz, leading to its proteasomal degradation in human 293T cells and in zebrafish embryos. Dorsalization induced by Boz overexpression was suppressed by raising the level of Lnx-l, but Lnx-l failed to counteract dorsalization caused by mutant Boz lacking a critical motif for Lnx-l binding. Further, dorsalization induced by depletion of Lnx-l was alleviated by attenuation of Boz expression. We conclude that Lnx-l modulates Boz activity to prevent the invasion of ventral regions of the embryo by organizer tissue. These studies introduce a ubiquitin ligase, Lnx-l, as a balancing modulator of axial patterning in the zebrafish embryo.
A role for proteolysis in early embryogenesis was first suggested by the finding that degradation of β-catenin is selectively inhibited on the future dorsal side of the Xenopus embryo, marking the initial step in D-V axis specification13–15. Components similar to the SCF-β-TrCP complex which mediates β-catenin destruction, also limit Xom/Vent2/Vox stability16. Further, the Smurf1 E3 ubiquitin ligase is involved in D-V patterning by mediating degradation of Smad1 and reducing BMP signaling17,18. While tight regulation of the levels of multiple proteins is undoubtedly required during early embryonic development, the involvement of E3 ubiquitin ligases besides the SCF-β-TrCP complex and Smurf1 have not been reported to date.
Three closely related Lnx homologues are expressed during zebrafish development (Lnx-1: BC124171, Lnx-2: FJ156085, Lnx-like: FJ156084, Supplementary Fig. 1, Supplementary Fig. 2). Among the Lnx homologues, Lnx-like is strongly expressed maternally, with zygotic expression starting around the onset of gastrulation in marginal blastomeres, exhibiting a ventral-to-dorsal gradient (Supplementary Fig. 2). To explore the embryonic role of Lnx-l, we inhibited its expression with antisense MOs which are effective in blocking lnx-l translation (Fig. 1a). Injection of UTR or ATG targeted MOs caused similar embryonic defects, displaying oval shape at early somitogenesis and degenerated tail and trunk at 24 hours post fertilization (hpf), both hallmarks of dorsalization (Fig. 1b–d, Supplementary Fig. 3). The dorsalized morphant phenotype was rescued by the co-injection of MO-resistant lnx-l mRNA (Supplementary Fig. 3). Since a splice blocking MO, expected to inhibit zygotic expression of Lnx-l, failed to induce dorsalization (data not shown), we conclude that maternal transcripts are responsible for the effect of Lnx-l on D-V patterning. Dorsalization caused by Lnx-l depletion was confirmed by analyzing several D-V markers. In the morphants, the organizer markers goosecoid (gsc) and chordin (chd) were expanded ventro-laterally at the expense of the expression domain of the ventral markers vox and vent (Fig. 1e–l). Likewise, the expression domains of neuroectodermal markers (otx2 and hoxb1b) extended into ventral regions in the morphants (Fig. 1m and n). Overexpression of Lnx-l caused axial mesoderm defects including anterior notochord and forebrain malformations and a shortened body axis, reminiscent of the strongest boz mutant phenotype8 (Supplementary Fig. 4). These results indicate that maternal lnx-l transcripts are critical for D-V patterning of the embryo.
Figure 1. Depletion of Lnx-l causes dorsalization.
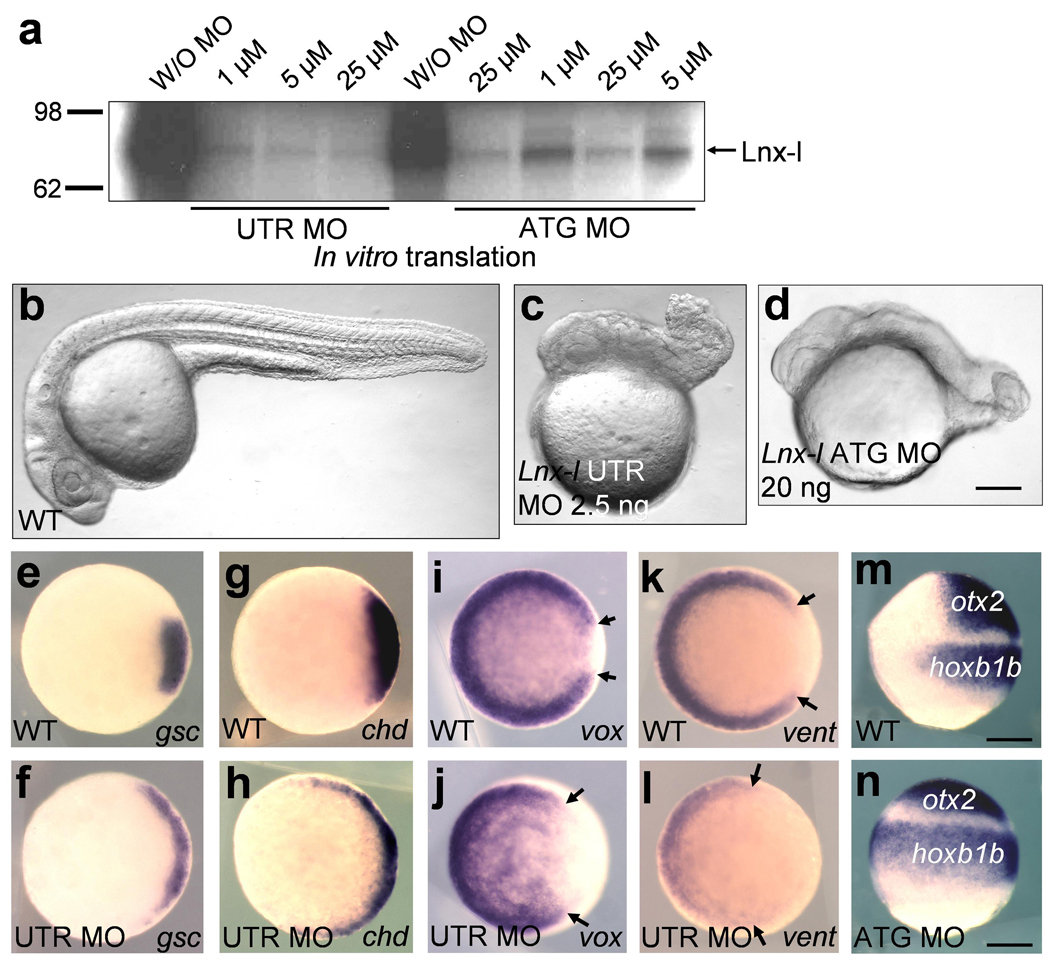
a, Both MOs inhibit in vitro translation of lnx-l mRNA; lnx-l UTR MO is more efficient than ATG MO. b–d, lnx-l UTR MO (2.5 ng) and ATG MO (20 ng) both caused similar embryonic defects displaying degenerated tail and trunk; 26 hpf embryos, lateral views. e–l, lnx-l morphants (UTR MO, 2.5 ng) show ventro-laterally expanded expression of gsc (f) and chd (h), and reduced expression of vox (j) and vent (l), with arrows indicating enlarged dorsal gap in the vox and vent domains; germ ring stage, animal pole views. m and n, lnx-l ATG MO (20 ng) injected embryos showed extended expression of otx2 and hoxb1b; 80% epiboly stage, lateral view, dorsal is to the right. Scale bar, 200 µm.
Since mammalian LNX acts as an E3 ubiquitin ligase1,2,19, we searched for potential target proteins by carrying out a yeast two-hybrid screen, and isolated a clone encoding a partial homeobox domain of vent. As full length Vent failed to bind to Lnx-l (data not shown), we tested other homeobox genes expressed in the early embryo, and found strong interaction between Lnx-l and Boz in the yeast two-hybrid system (Fig. 2a). Boz is a key regulatory factor in organizer formation in the embryo6–8,20. The physical interaction between Lnx-l and Boz was verified by showing that epitope-tagged Boz and wild type Lnx-l, Lnx-l Mu (conserved histidine and cysteine residues in the RING domain were replaced by alanine; H63A, C66A), and Lnx-l ΔN (carrying a RING domain deletion) could be co-immunoprecipitated using purified proteins in vitro (Fig. 2b) and in transfected cell lysates (Fig. 3a). Incubation of Boz and Lnx-l isolated from cultured cells in an in vitro ubiquitination system (see Methods) produced ubiquitinated derivatives of Boz (Fig. 2c). No ubiquitination was seen when a RING domain mutant of Lnx-l was used, or when the substrate was Boz All K/R, a mutant in which all five lysines in Boz were changed to arginines (Fig. 2c); lysine residues are the most common target sites, resulting in an isopeptide bond between ubiquitin and the substrate21.
Figure 2. Lnx-l binds and ubiquitinates Boz but not Gsc.
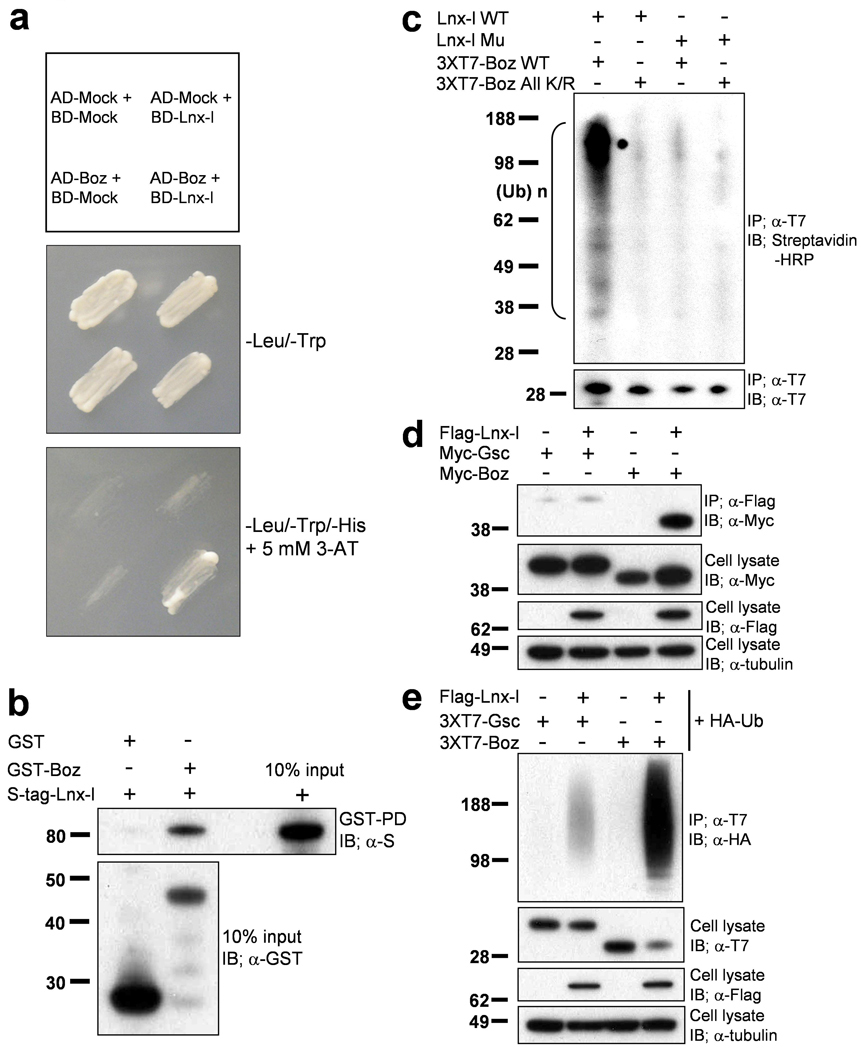
a, Lnx-l-Boz interaction in yeast. Yeast cells were cotransformed with the indicated fusion constructs (upper panel; see Methods). The transformants were plated on –Leu-Trp drop-out plates (middle panel) for 3 days. Individual transformants were transferred to –Leu-Trp-His drop-out plates containing 5 mM 3-AT, and incubated for 4 days (bottom panel). Only Boz-Lnx-l co-transformants showed substantial growth, indicating interaction between the two proteins. b, Direct interaction between Lnx-l and Boz was confirmed by the GST-pull down assay with proteins purified from bacteria (see Methods); GST alone could not pull-down S-tagged Lnx-l. c, In vitro ubiquitination assay was performed as described in Methods. Wt Boz mixed with extracts containing Wt Lnx-l yielded abundant ubiquitinated products, while RING mutant Lnx-l did not ubiquitinate Wt Boz, and Wt Lnx-l did not ubiquitinate Boz All K/R. d, Lnx-l interacts with Boz but not with Gsc. Flag-tagged Lnx-l was contransfected into 293T cells with 6xMyc-tagged Gsc or Boz. After 48 h, IP and IB were performed with the indicated antibodies. e, Lnx-l ubiquitinates Boz but not Gsc. Flag-tagged Lnx-l was contransfected with 3xT7 tagged Gsc or Boz. The yield of polyubiquitinated Gsc is very low compared to that of Boz (see also Fig. 3, below).. α-tubulin was used as loading control.
Figure 3. Lnx-l-mediated polyubiquitination destabilizes Boz.
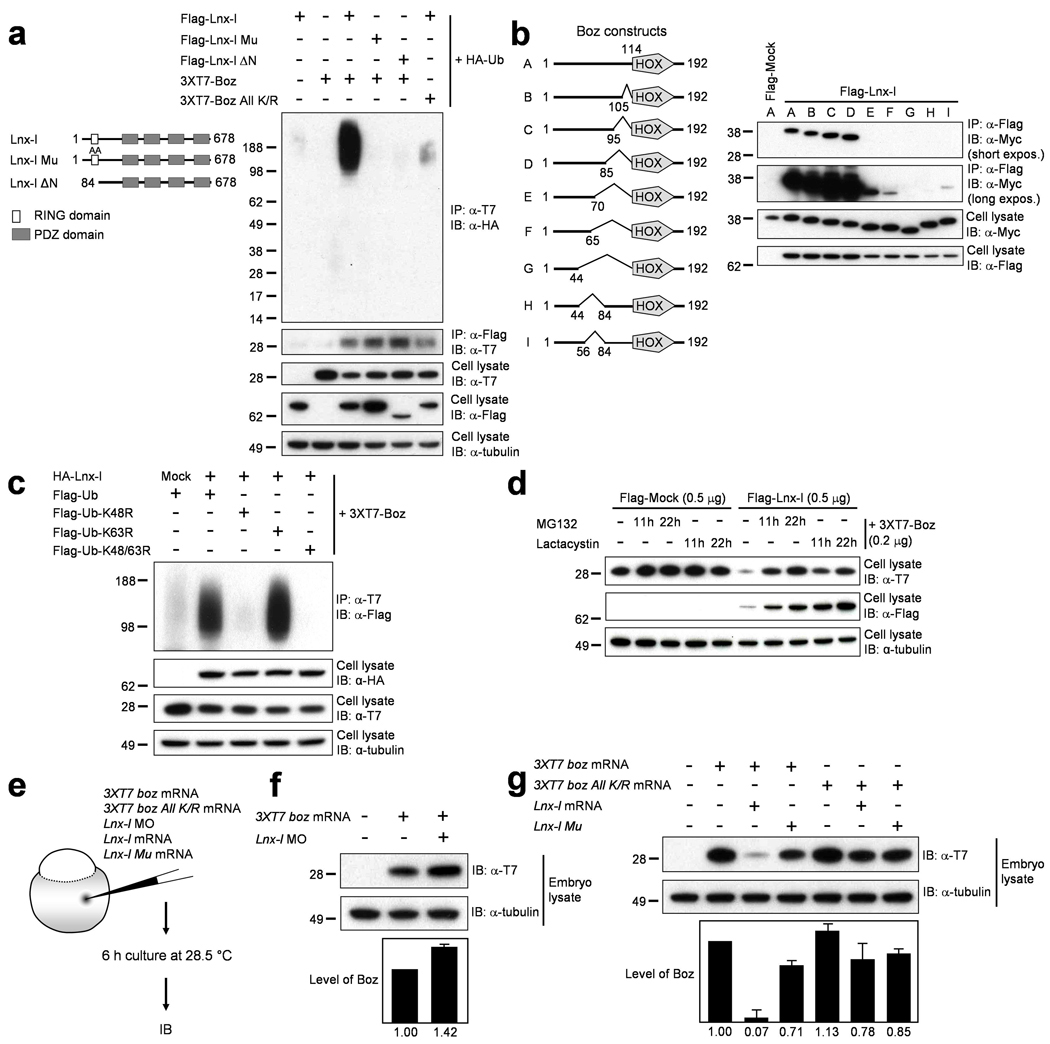
a,Flag-tagged Lnx-l, 3xT7-Boz, and HA-ubiquitin were cotransfected into 293T cells as indicated. Lnx-l constructs are shown on the left. Only Wt Boz is significantly polyubiquitinated by Wt Lnx-l. Molecular size markers are indicated at the left. b, Schematic diagrams of Boz deletion constructs are shown at the left. 6xMyc epitope-tagged Boz constructs were cotransfected with Flag-Lnx-l. Upon IP with anti-Flag antibody, the coprecipitates were subjected to IB with anti-Myc antibody. Short and long exposures are shown to illustrate the range of interaction strength between Lnx-l and Boz. c, HA-tagged Lnx-l and 3xT7-tagged Boz were cotransfected with Flag-tagged Wt or mutant ubiquitin. The K48R ubiquitin mutant could not support Boz polyubiquitination. d, Transfected cells were incubated with MG132 or lactacystin, each at 10 µmolar, for the indicated times. Destabilization of Boz by Lnx-l cotransfection was inhibited by MG132 or lactacystin. Lnx-l itself was stabilized in a time-dependent manner by the proteasome inhibitors. e, Schematic diagram of experimental design in (f) and (g). f, 20 pg of 3xT7 boz mRNA was injected with or without 2.5 ng of lnx-l UTR MO. Depletion of endogenous Lnx-l increased Boz levels. g, 20 pg 3xT7 boz or 3xT7 boz All K/R mRNA was injected with 50 pg lnx-l or 50 pg lnx-l Mu mRNA. Wt Lnx-l strongly destabilizes Wt Boz. Error bars in f and g were obtained from two repeats each of two independent experiments. α-tubulin was used as loading control.
As in the in vitro reaction, polyubiquitinated Boz was detected after cotransfection of Boz and Lnx-l into cultured cells, and the expression level of Boz was reduced (Fig. 3a). Even though Boz All K/R did coprecipitate with Lnx-l, this mutant was not ubiquitinated in cultured cells (Fig. 3a), confirming the in vitro results (Fig. 2c). Additionally, Boz All K/R was not destabilized by Lnx-l when injected into zebrafish embryos (Fig. 3g). To check the specificity of the Boz-Lnx-l interaction we tested whether Gsc, an organizer homeodomain protein and the closest homolog of Boz, could act as a substrate of Lnx-l. Binding between Gsc and Lnx-l and polyubiquitination of Gsc by Lnx-l were very weak and probably not significant as expression levels of Gsc were not substantially affected by co-transfection with Lnx-l (Fig. 2d and e). These data indicate that Boz is a specific target of Lnx-l, and that the RING domain is important for ubiquitination of Boz by Lnx-l but is dispensable for their physical interaction (Fig. 3a).
The Lnx-l-binding domains in Boz were mapped by testing deletion constructs, pointing to a critical region upstream of the homeodomain (Supplementary Fig. 5a and b). More detailed mapping of the binding domain in Boz identified amino acid (aa) residues 44–84 as essential (Fig. 3b). The aa 44–84 deletion mutant, Boz ΔBD, will be used in biological experiments below. The aa 44–84 domain was necessary but not sufficient to interact with Lnx-l (Fig. 3b; Supplementary Fig. 5c and d), indicating that additional domain(s) are required. Indeed, the C-terminal 15 amino acids of Boz are essential for efficient binding to Lnx-l (Supplementary Fig. 5a and b).
Polyubiquitin chains have diverse structures due to the presence of seven lysine residues in ubiquitin21–23. Different chain conformations have different biological consequences, with K48-linked polyubiquitinated proteins usually undergoing 26S proteasome-dependent proteolysis. We examined the nature of Boz polyubiquitination by Lnx-l by co-transfection of Boz with wild type or mutant ubiquitin constructs (Ub-K48R, Ub-K63R, Ub-K48/63R), all of which allow formation of a monoubiquitinated substrate but limit the type of polyubiquitin chain outgrowth. As shown in Figure 3c, K63R mutant ubiquitin was as effective as wild type in modification of Boz, while Ub-K48R or Ub-K48/63R were largely inactive. These results indicate that Boz ubiquitination by Lnx-l leads to K48-linked chains. This fact is consistent with the observation that Boz degradation following co-transfection with Lnx-l was blocked by the proteasome inhibitors MG132 and lactacystin (Fig. 3d). In zebrafish embryos, Boz levels increased after depleting endogenous Lnx-l, whereas lnx-l mRNA-injected embryos showed a dramatic reduction in the level of Boz protein (Fig. 3e–g, Supplementary Fig. 5e). This reduction depends on Lnx-l E3 ubiquitin ligase activity as Lnx-l Mu or Lnx-l ΔN failed to induce a similar lowering of Boz protein (Fig. 3g, Supplementary Fig. 5e). In addition, Boz All K/R levels were stable in the presence of Lnx-l, suggesting that Boz destabilization depends on polyubiquitin chain formation (Fig. 3g). These results indicate that Lnx-l negatively regulates Boz levels by post-translational regulation, providing an explanation for the dorsalization observed after depletion of Lnx-l from the embryo.
In our experiments we used the 6xMyc tag in binding studies because of its high sensitivity. However, we noted that Myc-tagged Boz became stabilized rather than undergoing proteasomal degradation after cotransfection with Lnx-l (Fig. 2d). Further, 6xMyc-tagged Boz showed K63-linked polyubiquitin chain elongation, and Lnx-l was able to polyubiquitinate 6xMyc-tagged Boz All K/R (data not shown). These observations suggest that lysine residues within the Myc tag served as targets for Lnx-l-mediated ubiquitination. Therefore we used the lysine-free T7 epitope (MASMTGGQQMG) in all ubiquitination experiments; as expected, T7-tagged Boz All K/R was resistant to Lnx-l dependent ubiquitination (Fig. 2c and Fig. 3a). These observations show that the lysine(s) in an epitope tag can serve as targets for E3 ubiquitin ligases, potentially endangering the conclusions derived from constructs with lysine-containing tags.
In the zebrafish embryo, boz activation by a Wnt/β-catenin signal11,12 mediates organizer formation by suppressing the expression of ventralizing genes such as bmp, vox (vega1), vent (vega2) and ved 20,24–29. The similarity of defects caused by Boz overexpression and Lnx-l depletion, and of boz null mutation8,10 and Lnx-l gain-of-function, suggested to us that Lnx-l counteracts Boz in vivo by ubiquitination-dependent destabilization. Therefore we tested functional interactions between these two factors by injecting boz mRNA, with or without lnx-l mRNA, into zebrafish embryos. Embryos injected with 1 pg boz mRNA were strongly dorsalized as previously reported7, and this phenotype was reversed by co-injection of lnx-l in a dosage-dependent manner (Fig. 4a). Dorsalization was also tested by expansion of the gsc expression domain at gastrula. Ectopic gsc expression induced by boz (100% = G2-G4) was significantly suppressed by lnx-l (45% = G1), but not by lnx-l ΔN (Fig. 4b). Functional interaction between Lnx-l and Boz is also indicated by the synergy between low levels of Lnx-l suppression and Boz overexpression (1 ng lnx-l MO: 13% = G2; 1 pg boz mRNA: 22% = G4; 1 ng lnx-l MO + 1 pg boz mRNA: 80% = G4) (Fig. 4b). We predicted that the Lnx-l binding domain mutant, Boz ΔBD, should retain dorsalizing activity, as it contains an intact Goosecoid-Engrailed homology domain at its N-terminus (Fig. 4c)6. We found that Boz ΔBD induced dorsalization more effectively than the wild type, and that this phenotype was resistant to co-injection with lnx-l or lnx-l ΔN mRNA (Fig. 4d). As shown in Fig. 4e, Boz ΔBD was only very weakly polyubiquitinated by Lnx-l and showed a stable expression level in the presence of Lnx-l.
Figure 4. Lnx-l counteracts Boz-mediated dorsalization.
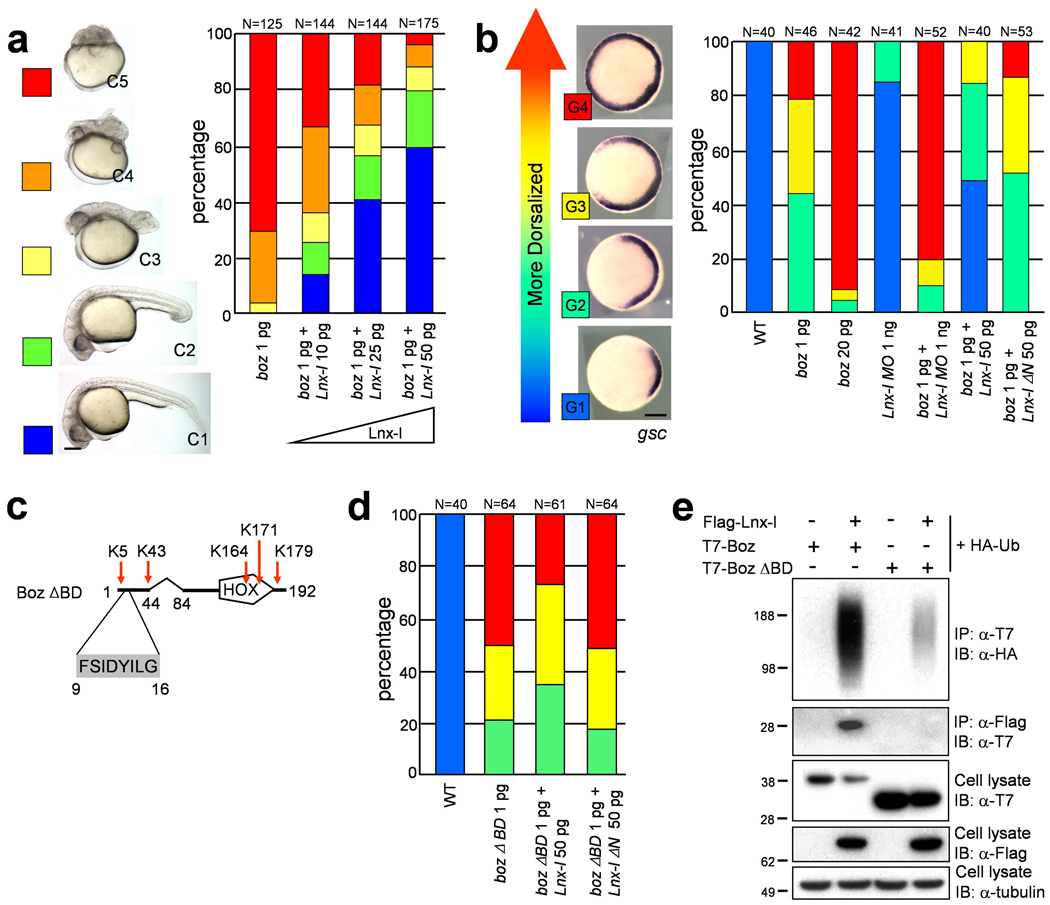
a, b, d, The amounts of injected RNA or MO are shown beneath each bar, and number of embryos above each bar. a, Coinjection of lnx-l mRNA rescues boz-induced dorsalization. The embryonic morphology was classified as C1-C5 at 26 hpf, as illustrated at the left. b, Dorsalization was analyzed by observing the domain of gsc expression at the germ ring stage. Boz and lnx-l MO show synergism, while lnx-l mRNA counteracts dorsalization. c, Schematic drawing of Boz lacking the Lnx-l binding domain (Boz ΔBD), but retaining the Goosecoid-Engrailed homology domain (amino acid 9–16) and all lysine residues. d The gsc expression level, analyzed as in b, shows that Boz ΔBD is resistant to Lnx-l (compare to b). e, T7 epitope-tagged Wt Boz or Boz ΔBD was cotransfected into 293T cells with Flag-Lnx-l and HA-Ub. Boz ΔBD ubiquitination is almost absent. Scale bar, 200 µm.
Next we investigated the genetic interaction between boz and lnx-l. Since the expression levels of two boz target genes (vent and vox)25–27 decreased through development in lnx-l morphants (Supplementary Fig. 6a–s), we tested whether this gradual reduction is a consequence of a reciprocal increase in boz expression. The initial level and range of boz expression was not affected in lnx-l morphants (Fig. 5a and b), but the boz expression domain was expanded by late blastula stage (Fig. 5c and d). Likewise, gradual expansion of gsc and chd was seen in the morphants (Supplementary Fig. 6t–e’). To explore whether dorsalization of lnx-l morphants could be restored by depletion of endogenous boz, we designed a splicing MO against boz. This MO specifically and efficiently blocked normal splicing of boz, strongly suppressed noggin1 expression30, and affected shield formation (Supplementary Fig. 6f’-m’). Knockdown of boz rescued the embryonic dorsalized phenotype obtained by Lnx-l depletion (Fig. 5e–g). In addition, ectopic gsc expression in the lnx-l morphants was alleviated in the boz-lnx-l double morphants (Fig. 5h–k). These data provide genetic evidence that lnx-l MO-dependent embryonic dorsalization can be suppressed by the attenuation of Boz expression.
Figure 5. Boz depletion mitigates dorsalization in lnx-l morphants.
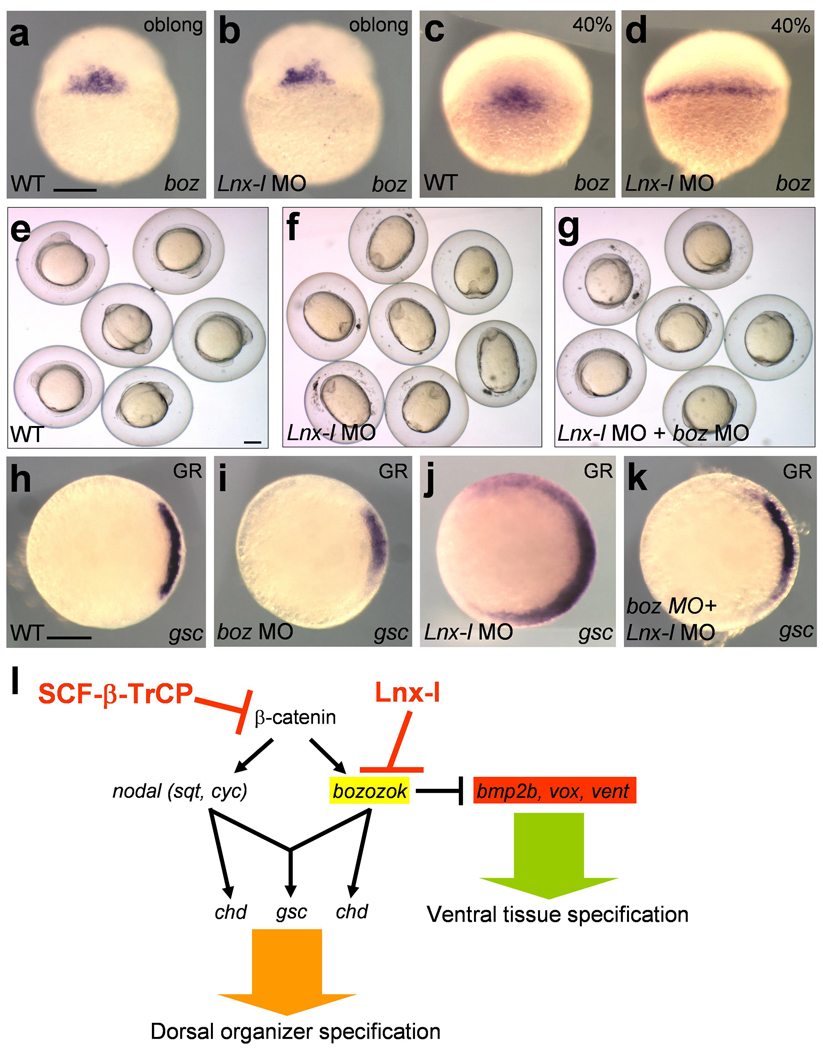
a–d, lnx-l morphants showed normal expression of boz at the oblong stage (30/30), but in some morphants, boz expression was expanded laterally at 40% epiboly (12/31); 2.5 ng lnx-l UTR MO was injected into each embryo in b and d. e–g, 5-somite stage. e, Uninjected control. f, Embryos injected with 20 ng lnx-l ATG MO; the oval shape indicates dorsalization (92%, N=100). g, boz MO (5 ng), injected together with 20 ng lnx-l ATG MO, alleviated the morphological defects of lnx-l depletion (37% dorsalized, N=116). h–k, gsc expression in embryos injected with indicated MOs. h, uninjected. i, 5 ng of boz MO. j, 2.5 ng lnx-l UTR MO (89% expanded gsc expression, N=28). k, 2.5 ng lnx-l UTR MO plus 5 ng boz MO (32% expanded gsc expression, N=28). l, A schematic model of the role of Lnx-l during establishment of the D–V body axis. Scale bar, 200 µm.
Boz is a strong transcriptional repressor that is expressed transiently during early embryonic development6,7,20,25–27. This temporally restricted expression suggests that boz should be tightly regulated at the transcriptional and post-transcriptional level. Here we provide evidence for post-translational regulation of Boz. We propose that Lnx-l regulates the output of early Wnt signaling by limiting the duration and range of Boz activity (Fig. 5l). It is possible that Lnx-l modulates early body patterning by affecting other molecules in addition to Boz because the lnx-l morphant phenotype could not be rescued completely by boz depletion, and the gain-of-function of lnx-l caused stronger embryonic malformations than the boz null mutation. However, regulation of Gsc does not appear to be involved as Lnx-l neither binds nor ubiquitinates Gsc to a substantial extent (Fig. 2d and e). Our data indicate that the Lnx-l/Boz interaction constitutes the major mechanism by which Lnx-l limits the size of the organizer domain in the zebrafish embryo. The data presented here represent evidence that not only β-catenin but a direct target molecule of β-catenin signaling is down-regulated by the action of a specific E3 ubiquitin ligase to achieve patterning of the D-V body axis during early embryonic development.
METHODS
Yeast two-hybrid screening
The pAS2-1 and pGAD10 plasmids (Clontech) encode the GAL-4 DNA binding domain and the transcriptional activation domain. Lnx-l open reading frame (ORF) fused to the GAL-4 DNA binding domain was generated by PCR cloning. cDNA libraries from zebrafish and Xenopus embryos were purchased from Clontech, and screening was performed according to the manufacturer’s instructions. pAS2-1/Lnx-l ORF and each cDNA library were cotransformed into the CG1945 yeast strain. Approximately 1.4 × 108 Xenopus and 4.5 × 107 zebrafish independent clones were screened. In interaction experiments (Fig. 2a), the same yeast strain was used, and mock refers to transfection with the corresponding empty vectors.
Cell culture and Transfection
Cos7 and 293T cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented 10% fetal bovine serum (FBS). Cells in plates were transfected with various plasmid constructs by FuGene6 (Roche). After 24–48 hr, cells were harvested and the cell lysates were used for further assay.
Immunoprecipitation and Western blotting
Cells were lysed by M-PER Mammalian Protein Extraction Reagent (Pierce) containing Complete protease inhibitor cocktail (Roche), and mixed with 15 µl of Protein G Sepharose 4 Fast Flow (GE Healthcare). The total volume of lysate was adjusted to 800 µl with NP-40 buffer (150 mM NaCl, 0.02% NaN3, Complete protease inhibitor cocktail (Roche), 10 mM Tris-Cl, pH 7.2, 0.5% NP-40, 1 mM DTT). Precipitates were washed at least three times with ice-cold NP-40 buffer (1 ml), and were analyzed by Western blotting. In order to analyze polyubiquitination, immunoprecipitation used high stringency conditions (150 mM NaCl, 0.02% NaN3, Complete protease inhibitor cocktail (Roche), 10 mM Tris-Cl, pH 7.2, 0.5% NP-40, 0.1% SDS, 1% sodium deoxycholate, 1 mM DTT). After separation in 12% Nu-PAGE (Invitrogen) and transfer to PVDF membranes, membranes were incubated for 1 hr in blocking solution (4% skim milk in Tris-buffered saline with 0.05% Tween-20) at room temperature, incubated with primary antibodies for 1 hr, followed by incubation with HRP-conjugated secondary antibody (Jackson, 1:1000 – 1:10000) for 1 hr. Detection used the ECL detection system (Pierce). Embryonic lysates were obtained by adding 10 µl CompLysis Protein Extraction Reagent (SignaGen) per embryo. Antibodies were obtained from Roche (rat HA), Sigma (mouse Flag), Calbiochem (mouse T7, mouse T7-HRP conjugate, α-tubulin), Clontech (mouse Myc) and Millipore (rabbit Myc).
Whole mount in situ hybridization
Antisense riboprobes were generated using the appropriate RNA polymerase following manufacturers’ instructions (Roche). Hybridization signal was detected using pre-absorbed anti-digoxigenin-AP Fab fragment (Roche) diluted (1:2000) in blocking solution.
In vitro translation
In vitro translation reactions were performed using TNT T7 Coupled Reticulocyte Lysate (Promega). We subcloned the Lnx-l ORF including the 5’ untranslated region (UTR) into the modified pcGlobin231 vector to construct the template.
Site directed mutagenesis
For amino acid substitution in Lnx-l H63A/C66A, a mega-primer was generated by PCR using TLA polymerase (Bioneer Inc.) (forward primer; 5’- GGGATCCGCCATGACGGAGTCTAAGACGTCTTCACTGCCG-3’, reverse primer; 5’-GAGGCACTGGAAGGCATAAGTGGCGCCGCACAATGTG-3’, mutated sequences underlined). The resulting mega-primer together with another primer (5’-GCTCGAGTTAAACCAGACTGCCAGGCCAGGAGACCACGG −3’) were used to PCR-amplify the full length mutant lnx-l, which was subcloned into T-vector (pGEM-T Easy Vector Systems; Promega). Lysine free Boz mutant was generated by substitution of amino acids 5, 43, 164, 171 and 179 to arginine using the aforementioned mega-priming method.
Boz deletion constructs
Internal deletion constructs of Boz were generated using a PCR-based method. For example, Boz ΔBD was generated using four primers. P1: 5’-GATGAATTCGAACATGGCAACTCAGAAGTTTTC3’; P2: 5’-CCCTGCATAGTAAGTCGACTTTCTCAAGTGCCCCTG-3’; P3: 5-CAGGGGCACTTGAGAAAGTCGACTTACTATGCAGGG-3’ and P4: 5’-GATCTCTAGACTAATCTGATTCCTGATGATC-3’. Primary PCR reactions were carried out with two different sets of primers (P1 + P2 and P3 + P4) using PfuUltra DNA polymerase (Stratagene). Each PCR product was purified, mixed with the complementary product, and used as template in the second PCR reaction with P1 and P4 primers.
mRNA and MO Microinjection
For microinjection, cDNA for lnx-l, lnx-l ΔN, boz and boz ΔBD were individually subcloned into the pCS2+ or modified pCS2+ vector. mRNAs were synthesized from linearized constructs using the mMESSAGE mMACHINE kit (Ambion Inc.) according to the manufacturer’s instruction. After purification following the manufacturer’s recommendation, mRNAs were dissolved in DEPC treated 0.1 M KCl. mRNAs and MOs were pressure injected into the yolk of 1–4 cell stage embryos. In order to reduce the p53 dependent off-targeting effect of MOs (lnx-l ATG MO and lnx-l splicing MO), we coinjected the MOs with a p53 MO32. Three different MOs against lnx-l and one boz splice-blocking MO were obtained from Gene Tools, LLC.
Lnx-l UTR MO: CCTACGCCTCTTTCACAGCTCACAA.
Lnx-l ATG MO: AGACTCCGTCATGGCCTGGAGAAGT.
Lnx-l splicing MO: GTAAGTGATGCAATACCATCTTCGC.
Boz splicing MO: AAATTGAAAATGCATACCGGCTACG.
In vitro binding assay
BL21 (DE3) E. coli cells (Invitrogen) were used for expression of GST, GST-tagged Boz and S-tagged Lnx-l. The boz ORF was subcloned into the pGEX4T-1 (GE Healthcare Life Science), and lnx-l or lnx-l Mu was subcloned into pET29a (+) (Novagen). After adding IPTG (final concentration 1 mM), the bacteria were cultured for 2 h at 30° C, and lysed with B per solution (Pierce). The fusion proteins were purified as inclusion bodies, and were denatured and renatured using Rapid GST Inclusion Body Solubilization and Renaturation Kit (Cell Biolabs, Inc.) following the manufacturer’s instruction.
Purified GST or GST-Boz was incubated with S-tagged Lnx-l and Glutathione Sepharose 4 Fast Flow (GE Healthcare Life Science) for 2 h at 4°C with gentle agitation. Mixtures were then centrifuged and pellets were washed four times with ice-cold NP-40 buffer (150 mM NaCl, 0.02% NaN3, Complete protease inhibitor cocktail (Roche), 10 mM Tris-Cl, pH 7.2, 0.5% NP-40, 1 mM DTT). Bound proteins were eluted by boiling after adding 30 µl of gel loading buffer. Eluted proteins were separated by SDS-PAGE (12%), transferred to PVDF membrane, and S-tagged Lnx-l was detected by immunoblotting using anti-S·tag antibody (Novagen) and chemiluminescent detection system (Pierce).
In vitro ubiquitination assay
Expression constructs for 3xT7-Boz, 3xT7-Boz All K/R, Flag-Lnx-l and Flag-Lnx-l Mu were transfected into separate plates of 293T cells. Each culture was lysed, and nuclear fractions were isolated from Boz transfectants, while cytoplasmic fractions were isolated from Lnx-l transfectants. Extracts were combined in various combinations (see Fig. 2c), and subjected to IP with anti-T7 antibody. After washing several times with IP buffer, the protein-bound beads were directly used for the in vitro ubiquitination assay. The beads were incubated with the components of the ubiquitination machinery which includes biotinylated ubiquitin, following the manufacturer’s instructions (Ubiquitinylation Kit, BIOMOL). As E2 we used UbcH5b, which proved the most effective among those tested (UbcH1, UbcH2, UbcH5a, UbcH5b, UbcH5c, UbcH8 and Ubc13). After 1 h incubation at 37°C, the reaction was terminated by adding IP buffer containing SDS (0.5%; final concentration). The reacted mixtures were heated for 5 min at 80°C in order to reliably dissociate Boz from Lnx-l or any other potential interacting protein. After adding 1 ml of ice-cold IP buffer, 15 µl of fresh protein G sepharose beads and anti-T7 antibody, T7-tagged Boz or Boz All K/R proteins were recovered after 4 h incubation at 4°C with gentle agitation. After gel electrophoresis, biotin-labeled ubiquitinated Boz was detected using the VECTASTAIN ABC Kit (Vector Laboratories, Inc.) and SuperSignal West Dura Extended Duration Substrate (Pierce).
Supplementary Material
Acknowledgments
We thank Mark Rath and John Gonzales for help with fish maintenance, and Dr. Seok-Yong Choi, Chonnam National University Medical School, Republic of Korea, for helpful comments. This work was supported by the Intramural Research Program of the NICHD, NIH.
REFERENCES
- 1.Dho SE, et al. The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein LNX. J. Biol. Chem. 1998;273:9179–9187. doi: 10.1074/jbc.273.15.9179. [DOI] [PubMed] [Google Scholar]
- 2.Nie J, et al. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002;21:93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sollerbrant K, et al. The Coxsackievirus and adenovirus receptor (CAR) forms a complex with the PDZ domain-containing protein ligand-of-numb protein-X (LNX) J. Biol. Chem. 2003;278:7439–7444. doi: 10.1074/jbc.M205927200. [DOI] [PubMed] [Google Scholar]
- 4.Kansaku A, et al. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene. 2006;25:5071–5084. doi: 10.1038/sj.onc.1209468. [DOI] [PubMed] [Google Scholar]
- 5.Rice DS, Northcutt GM, Kurschner C. The Lnx family proteins function as molecular scaffolds for Numb family proteins. Mol. Cell Neurosci. 2001;18:525–540. doi: 10.1006/mcne.2001.1024. [DOI] [PubMed] [Google Scholar]
- 6.Koos DS, Ho RK. The nieuwkoid gene characterizes and mediates a Nieuwkoop-center-like activity in the zebrafish. Curr. Biol. 1998;8:1199–1206. doi: 10.1016/s0960-9822(07)00509-x. [DOI] [PubMed] [Google Scholar]
- 7.Yamanaka Y, et al. A novel homeobox gene, dharma, can induce the organizer in a non-cell-autonomous manner. Genes Dev. 1998;12:2345–2353. doi: 10.1101/gad.12.15.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fekany K, et al. The zebrafish bozozok locus encodes Dharma, a homeodomain protein essential for induction of gastrula organizer and dorsoanterior embryonic structures. Development. 1999;126:1427–1438. doi: 10.1242/dev.126.7.1427. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu T, et al. Cooperative roles of Bozozok/Dharma and Nodal-related proteins in the formation of the dorsal organizer in zebrafish. Mech. Dev. 2000;91:293–303. doi: 10.1016/s0925-4773(99)00319-6. [DOI] [PubMed] [Google Scholar]
- 10.Solnica-Krezel L, Driever W. The role of the homeodomain protein Bozozok in zebrafish axis formation. Int. J. Dev. Biol. 2001;45:299–310. [PubMed] [Google Scholar]
- 11.Ryu SL, et al. Regulation of dharma/bozozok by the Wnt pathway. Dev. Biol. 2001;231:397–409. doi: 10.1006/dbio.2000.0150. [DOI] [PubMed] [Google Scholar]
- 12.Leung T, Söll I, Arnold SJ, Kemler R, Driever W. Direct binding of Lef1 to sites in the boz promoter may mediate pre-midblastula-transition activation of boz expression. Dev. Dyn. 2003;228:424–432. doi: 10.1002/dvdy.10408. [DOI] [PubMed] [Google Scholar]
- 13.Larabell CA, et al. Establishmengt of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J. Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JR, et al. Establishment of the Dorsal–Ventral Axis in Xenopus Embryos Coincides with the Dorsal Enrichment of Dishevelled That Is Dependent on Cortical Rotation. J. Cell Biol. 1999;146:427–437. doi: 10.1083/jcb.146.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, et al. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Z, Kirschner M. Regulated proteolysis of Xom mediates dorsoventral pattern formation during early Xenopus development. Dev. Cell. 2002;3:557–568. doi: 10.1016/s1534-5807(02)00270-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 18.Podos SD, Hanson KK, Wang YC, Ferguson EL. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell. 2001;1:567–578. doi: 10.1016/s1534-5807(01)00057-0. [DOI] [PubMed] [Google Scholar]
- 19.Weiss A, Baumgartner M, Radziwill G, Dennler J, Moelling K. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase Ligand-of-Numb protein X1. FEBS Lett. 2007;581:5131–5136. doi: 10.1016/j.febslet.2007.09.062. [DOI] [PubMed] [Google Scholar]
- 20.Leung T, et al. bozozok directly represses bmp2b transcription and mediates the earliest dorsoventral asymmetry of bmp2b expression in zebrafish. Development. 2003;130:3639–3649. doi: 10.1242/dev.00558. [DOI] [PubMed] [Google Scholar]
- 21.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 22.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 23.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Fekany-Lee K, Gonzalez E, Miller-Bertoglio V, Solnica-Krezel L. The homeobox gene bozozok promotes anterior neuroectoderm formation in zebrafish through negative regulation of BMP2/4 and Wnt pathways. Development. 2000;127:2333–2345. doi: 10.1242/dev.127.11.2333. [DOI] [PubMed] [Google Scholar]
- 25.Melby AE, Beach C, Mullins M, Kimelman D. Patterning the early zebrafish by the opposing actions of bozozok and vox/vent. Dev. Biol. 2000;224:275–285. doi: 10.1006/dbio.2000.9780. [DOI] [PubMed] [Google Scholar]
- 26.Kawahara A, Wilm T, Solnica-Krezel L, Dawid IB. Antagonistic role of vega1 and bozozok/dharma homeobox genes in organizer formation. Proc. Natl. Acad. Sci. USA. 2000;97:12121–12126. doi: 10.1073/pnas.97.22.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawahara A, Wilm T, Solnica-Krezel L, Dawid IB. Functional interaction of vega2 and goosecoid homeobox genes in zebrafish. Genesis. 2000;28:58–67. doi: 10.1002/1526-968x(200010)28:2<58::aid-gene30>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez EM, et al. Head and trunk in zebrafish arise via coinhibition of BMP signaling by bozozok and chordino. Genes Dev. 2000;14:3087–3092. doi: 10.1101/gad.852400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu T, Yamanaka Y, Nojima H, Yabe T, Hibi M, Hirano T. A novel repressor-type homeobox gene, ved, is involved in dharma/bozozok-mediated dorsal organizer formation in zebrafish. Mech. Dev. 2002;118:125–138. doi: 10.1016/s0925-4773(02)00243-5. [DOI] [PubMed] [Google Scholar]
- 30.Sirotkin HI, Dougan ST, Schier AF, Talbot WS. bozozok and squint act in parallel to specify dorsal mesoderm and anterior neuroectoderm in zebrafish. Development. 2000;127:2583–2592. doi: 10.1242/dev.127.12.2583. [DOI] [PubMed] [Google Scholar]
- 31.Ro H, Soun K, Kim EJ, Rhee M. Novel vector systems optimized for injecting in vitro-synthesized mRNA into zebrafish embryos. Mol. Cells. 2004;17:373–376. [PubMed] [Google Scholar]
- 32.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onichtchouk D, et al. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- 34.Melby AE, Clements WK, Kimelman D. Regulation of dorsal gene expression in Xenopus by the ventralizing homeodomain gene vox. Dev. Biol. 1999;221:293–305. doi: 10.1006/dbio.1999.9296. [DOI] [PubMed] [Google Scholar]
- 35.Trindade M, Tada M, Smith JC. DNA-binding specificity and embryological function of Xom (Xvent-2) Dev. Biol. 1999;216:442–456. doi: 10.1006/dbio.1999.9507. [DOI] [PubMed] [Google Scholar]
- 36.Imai Y, et al. The homeobox genes vox and vent are redundant repressors of dorsal fates in zebrafish. Development. 2001;128:2407–2420. doi: 10.1242/dev.128.12.2407. [DOI] [PubMed] [Google Scholar]
- 37.Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat. Genet. 2000;2:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


