Abstract
Lipoic acid is a sulfur-containing cofactor required for the function of several multienzyme complexes involved in the oxidative decarboxylation of α-keto acids and glycine. Mechanistic details of lipoic acid metabolism are unclear in eukaryotes, despite two well defined pathways for synthesis and covalent attachment of lipoic acid in prokaryotes. We report here the involvement of four genes in the synthesis and attachment of lipoic acid in Saccharomyces cerevisiae. LIP2 and LIP5 are required for lipoylation of all three mitochondrial target proteins: Lat1 and Kgd2, the respective E2 subunits of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, and Gcv3, the H protein of the glycine cleavage enzyme. LIP3, which encodes a lipoate-protein ligase homolog, is necessary for lipoylation of Lat1 and Kgd2, and the enzymatic activity of Lip3 is essential for this function. Finally, GCV3, encoding the H protein target of lipoylation, is itself absolutely required for lipoylation of Lat1 and Kgd2. We show that lipoylated Gcv3, and not glycine cleavage activity per se, is responsible for this function. Demonstration that a target of lipoylation is required for lipoylation is a novel result. Through analysis of the role of these genes in protein lipoylation, we conclude that only one pathway for de novo synthesis and attachment of lipoic acid exists in yeast. We propose a model for protein lipoylation in which Lip2, Lip3, Lip5, and Gcv3 function in a complex, which may be regulated by the availability of acetyl-CoA, and which in turn may regulate mitochondrial gene expression.
Several oxidative decarboxylation reactions are carried out in prokaryotes and eukaryotes by multienzyme complexes. The function of these complexes requires the action of a sulfur-containing cofactor, lipoic acid (6,8-thioctic acid) (1, 2). Lipoic acid is covalently attached via an amide linkage to a specific lysine residue on the surface of the conserved lipoyl domain of the E2 subunits of pyruvate dehydrogenase (PDH),3 α-ketoglutarate dehydrogenase (α-KDH), the branched chain α-keto acid dehydrogenase complexes, and the H protein of the glycine cleavage (GC) enzyme (3). The lipoyl moiety serves as a swinging arm that shuttles reaction intermediates between active sites within the complexes (1). Despite the well characterized function of lipoic acid as a prosthetic group, the mechanisms of its synthesis and attachment to proteins are the subject of ongoing investigations (4–7).
These reactions are best understood in Escherichia coli, which has two well defined pathways for lipoic acid synthesis and attachment: a de novo pathway and a salvage pathway (8). Octanoic acid, synthesized on the acyl carrier protein (ACP) (9), is the substrate for the de novo pathway. Lipoyl synthase (LipA) catalyzes the addition of two sulfur atoms to form lipoic acid from octanoic acid either before or after transfer to the target protein (10) by lipoyl(octanoyl)-ACP:protein transferase (LipB) (11, 12). The preferred order of these two reactions is attachment of octanoic acid by LipB, followed by addition of sulfur by LipA (13). By contrast, in the salvage pathway, lipoate-protein ligase (LplA) attaches free lipoic acid to proteins in a two-step reaction. Lipoic acid, which can be scavenged from the medium, is first activated to lipoyl-AMP and then the lipoyl group is transferred to the proteins (14).
Lipoic acid synthesis and attachment to target proteins are less well understood in eukaryotes. Homologs of the E. coli enzymes have been found in fungi, plants, protists, and mammals, but many mechanistic details are unclear (15–17). In Saccharomyces cerevisiae, the mitochondrial type II fatty acid biosynthetic pathway (FAS II) synthesizes octanoyl-ACP, which is the substrate for de novo lipoic acid synthesis (18). Lip2 and Lip5, the respective yeast homologs of E. coli LipB and LipA, were shown to be required for respiratory growth on glycerol medium, PDH activity (19), and lipoic acid synthesis (20), indicating functional roles in de novo lipoic acid synthesis and attachment. However, there has been no previous report of an LplA-like lipoate-protein ligase homolog in yeast. Furthermore, lip2 and lip5 mutant strains cannot grow on medium containing lipoic acid (19, 20), suggesting that yeast either cannot use exogenously supplied lipoic acid or there is no yeast equivalent of the E. coli LplA-driven salvage pathway.
Here we report the involvement of two additional enzymes in protein lipoylation in yeast mitochondria. The first, Lip3, is a lipoate-protein ligase homolog and is required with Lip2 and Lip5 for lipoylation of the E2 subunits of PDH (Lat1) and α-KDH (Kgd2). The second enzyme, Gcv3, the H protein of the GC enzyme, is absolutely required for lipoylation of all proteins in yeast.
EXPERIMENTAL PROCEDURES
Strains and Media
S. cerevisiae strains and their genotypes are listed in Table 1. The deletions in the strains of interest in the EUROSCARF collection were verified by PCR using a gene-specific 5′ upstream primer and a kanamycin resistance gene 3′ primer. Cells were grown in various media as follows: YEPD (1% (w/v) yeast extract, 2% (w/v) peptone, and 2% (w/v) glucose); YEPG (1% (w/v) yeast extract, 2% (w/v) peptone, and 3% (w/v) glycerol), WO −ura (0.67% (w/v) yeast nitrogen base without amino acids or ammonium sulfate, 0.5% (w/v) ammonium sulfate, 2% (w/v) glucose, 20 μg ml−1 methionine, 100 μg ml−1 leucine, and 20 μg ml−1 histidine), synthetic complete (SCD) ((Sigma) containing 2% (w/v) glucose), and SCD lacking uracil (SC −ura). After plasmid transformation using the lithium acetate method (21), uracil was omitted as a supplement in WO −ura medium to select for URA+ transformants. WO −ura glycerol plates contained 3% (w/v) glycerol and 0.01% (w/v) glucose as carbon sources. Solid media contained 2% (w/v) agar.
TABLE 1.
Strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| S. cerevisiae | ||
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | EUROSCARF |
| cbp2Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, cbp2::kanMX4 | 18 |
| lip3Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, lip3::kanMX4 | This study |
| lip2Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, lip2::kanMX4 | This study |
| lip5Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, lip5::kanMX4 | This study |
| lat1Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, lat1::kanMX4 | EUROSCARF |
| kgd2Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, kgd2::kanMX4 | EUROSCARF |
| gcv3Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, gcv3::kanMX4 | This study |
| lpd1Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, lpd1::kanMX4 | EUROSCARF |
| gcv1Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, gcv1::kanMX4 | EUROSCARF |
| gcv2Δ | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, gcv2::kanMX4 | EUROSCARF |
| Lip3-GFP wild-type | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, LIP3-GFP::HIS3MX6 | This study |
| Lip3(K249L)-GFP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, lip3(K249L)-GFP::HIS3MX6 | This study |
| Gcv3-GFP wild-type | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, GCV3-GFP::HIS3MX6 | 50 |
| Gcv3-GFP Δkgd2 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, kgd2::kanMX4, GCV3-GFP::HIS3MX6 | This study |
| Gcv3(K102L)-GFP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, gcv3(K102L)-GFP::HIS3MX6 | This study |
| Gcv3(K102R)-GFP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, gcv3(K102R)-GFP::HIS3MX6 | This study |
| E. coli | ||
| JRG33-lip9 | F−, Δ(gpt-proA)62, lacY1, lipA9supE44?, galK2, purB15, hisG4, rpsL35, xyl-5, mtl-1, thi-1, λ− | 51 |
Genomic Screen
Details of the genomic screen have been reported previously (18).
Disruption of Open Reading Frames
The LIP3, LIP2, LIP5, and GCV3 genes were deleted in BY4741 to confirm the phenotypes of the EUROSCARF strains. Each open reading frame was deleted by homologous recombination with the kanamycin resistance (kanMX4) cassette. lip3Δ::kanMX4, lip2Δ::kanMX4, lip5Δ::kanMX, and gcv3Δ::kanMX4 constructs were generated by amplification of the kanMX4 cassette using primers specific to the 5′ upstream and 3′ downstream regions of the genes. The PCR products were gel-purified and transformed into the wild-type strain using the lithium acetate method followed by selection for geneticin resistance. Each of the remade deletion strains had the same growth and RNA processing phenotypes as the original EUROSCARF strains.
Construction of Plasmids
Teasy-LIP3-GFP was generated by ligation of 293 bp of the LIP3 5′-untranslated region sequence, the LIP3 open reading frame without the stop codon, the GFP-HIS3MX6 cassette from plasmid pFA6a-GFP(S65T)-HIS3MX6 (containing the Saccharomyces kluyveri HIS3 module) (22), and 40 bp of LIP3 3′-untranslated region into the pGEM-T Easy vector (Promega, Madison, WI). The Lys-249 to Leu mutation in Lip3-GFP was generated by site-directed mutagenesis (23) in pGEM-T Easy containing the LIP3 open reading frame to produce Teasylip3(K249L)-GFP. To generate the Teasygcv3(K102L)-GFP and Teasygcv3(K102R)-GFP plasmids, the GCV3 open reading frame was amplified by PCR and ligated to pGEM-T Easy. The K102L and K102R mutations were made by site-directed mutagenesis. The GFP-containing constructs were excised from the pGEM-T Easy vector, gel-purified, and transformed into the wild-type strain or the Gcv3-GFP strain using the lithium acetate method followed by selection for histidine prototrophy. The sequences of all PCR products were verified.
Isolation and Fractionation of Mitochondria
For Western blot analysis, cells were grown to stationary phase in YEPD or WO −ura medium at 30 °C. Mitochondrial fractions were isolated and prepared as described (18). Post-mitochondrial supernatant (PMS) fractions were collected immediately following centrifugation of mitochondria. Laemmli sample buffer was added to the mitochondrial pellets resuspended in TE buffer plus protease inhibitors, and to the PMS fractions for analysis by gel electrophoresis and Western blot. For enzyme activity assays, cells were grown to A600 nm 6.0–6.5 in YEPD at 30 °C. Mitochondrial fractions were prepared in the same manner as above. Mitochondrial pellets were suspended in breaking buffer (0.1 m KPO4 buffer, pH 7.4, 1 mm EDTA, 10 μm thiamine pyrophosphate, 0.2 mm phenylmethanesulfonyl fluoride, 0.5 mg liter−1 leupeptin, 0.68 mg liter−1 pepstatin A) and broken by the addition of Triton X-100 to a final concentration of 1%. Particulate matter was removed by centrifugation for 20 min at 47,800 × g.
Western Blot Analysis
Approximately 40 μg of mitochondrial and PMS protein was resolved on SDS-polyacrylamide gels following the protocol described by Laemmli (24). Generally, protein transfer and antibody incubations were performed as described (18). For visualization of Lip3-GFP and Gcv3-GFP, however, the blots were reacted with anti-GFP monoclonal antibody (Covance, Berkeley, CA) diluted 1:4000, and proteins were visualized using ECL chemiluminescent substrate (Pierce). Anti-IDH antiserum was a gift from Lee McAlister-Henn, University of Texas Southwestern Medical Center, San Antonio.
Enzyme Assays
Protein concentrations in mitochondrial extracts were estimated by the Bradford assay (Bio-Rad). Bovine serum albumin (Sigma) was used as a standard. A modification of the method described by Kresze and Ronft (25) was used to assay PDH and α-KDH activities. The activities were measured by following the formation of NADH at 340 nm. The reaction mixture contained 0.1 m KPO4 buffer, pH 8.0, 0.05% Triton X-100, 1 mm MgCl2, 0.5 mm CaCl2, 2 mm dithiothreitol, 0.5 mm EDTA, 0.2 mm thiamine pyrophosphate, 5 mm sodium pyruvate or 5 mm α-ketoglutarate, 2.5 mm NAD+, and enzyme in a total volume of 0.99 ml. The reactions were initiated by the addition of 10 μl of 15 mm CoASH. Measurement of fumarase activity was described previously (26). The activities were measured at 25 °C and are based on the initial rates.
Light Microscopy
Localization of Lip3-GFP was attempted by fluorescence microscopy as described previously (18).
Lipoic Acid Analysis
Lipoic acid content of yeast strains was monitored by a biological assay described previously (27, 28) using the lipoic acid-deficient JRG33-lip9 E. coli strain, with minor modifications to the protocol. Yeast strains were grown in 50 ml of SCD media instead of YEPD, and acid hydrolysis was carried out in 0.5 ml of 9 NH2SO4. JRG33-lip9 cultures were inoculated at an initial A600 nm of 0.015 in 2 ml of 1× basal growth medium (29) containing 50 mm sodium succinate and grown for 36–48 h. The growth response of the strain was linear between 0.05 and 0.5 ng ml−1 lipoic acid in the cultures. Each result represents the mean lipoic acid content of five different yeast cultures ± S.D.
RESULTS
Lip3 Is Homologous to Lipoate-Protein Ligases
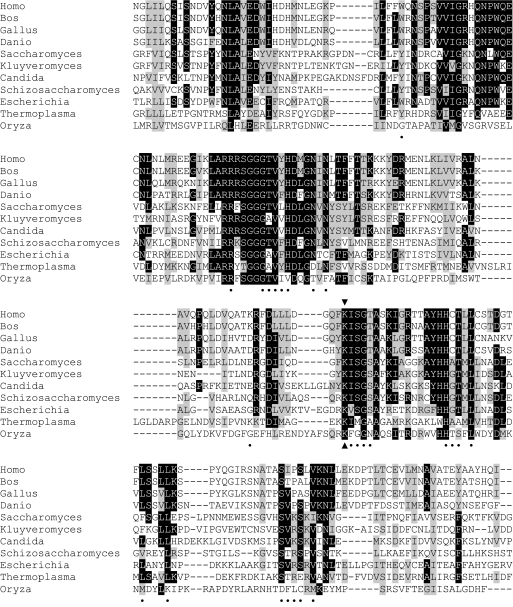
We identified a candidate gene for a lipoate-protein ligase during the course of studies on RNA processing. The EUROSCARF collection of haploid S. cerevisiae strains carrying deletions in nonessential nuclear genes was screened for novel nucleus-encoded factors that affect mitochondrial RNA processing. The respiration-deficient yjl046wΔ strain was found to be defective in processing 5′ leader sequences of mitochondrial tRNAs (data not shown). The defect was in the same step as, but less severe than, in mutants defective in the mitochondrial fatty acid synthesis pathway, which provides octanoic acid for the synthesis of lipoic acid (18). Accordingly, we looked for homology with enzymes known to be involved in lipoic acid metabolism in other organisms. Alignment of the predicted amino acid sequence of YJL046w using ClustalW2 (30) revealed homology to a diverse set of lipoate-protein ligases and lipoyltransferases (Fig. 1).
FIGURE 1.
Amino acid sequence alignment of lipoate-protein ligases and lipoyltransferases. The predicted amino acid sequence of S. cerevisiae Lip3 starting with residue 115 and ending with residue 330 is aligned with enzymes from Homo sapiens, Bos taurus, Gallus gallus, Danio rerio, Kluyveromyces lactis, Candida albicans, Schizosaccharomyces pombe, E. coli, Thermoplasma acidophilum, and Oryza sativa. The aligned residues define the N-terminal domain of B. taurus lipoyltransferase, which contains the lipoyl-AMP binding pocket (37). Black boxes denote identical amino acid residues, and gray boxes denote similar amino acid residues. Residues that interact with lipoyl-AMP (within 4.0 Å) are marked by a dot. The arrowheads denote the highly conserved lysine residue. The sequence alignment was carried out using ClustalW2 (30).
Lipoate-protein ligases are a family of proteins that attach the cofactor lipoic acid to target enzymes (31). We wondered whether the YJL046w gene product functions in lipoic acid attachment in yeast. Lipoic acid-deficient (lip) mutants defining four genes found previously in a large collection of respiration-deficient yeast strains (32) were crossed to the yjl046wΔ strain, and the diploids were tested for complementation of respiratory deficiency (data not shown). The yjl046wΔ strain did not complement the lip3 mutation in the G49 lipoic acid-deficient mutant strain. Sequencing of YJL046w in the G49 strain revealed a premature stop codon at position 75. We conclude that the YJL046w gene product, Lip3, is required for lipoic acid biosynthesis and/or, more likely, attachment.
Lip3 Is Required for Lipoic Acid-dependent Enzyme Activities
To examine the role of Lip3 in protein lipoylation, the total lipoic acid content of the lip3Δ strain was measured after release of the cofactor from mitochondrial enzymes by acid hydrolysis (27, 28). The mutant strain contained less than 10% of the level of lipoic acid in the wild-type strain (Table 2). This phenotype is equivalent to that observed for the lip5 yeast mutant strains (20) and suggests that Lip3 may also be required for lipoic acid synthesis and/or attachment. In the absence of attachment, the cofactor may be degraded. The respiration-deficient cbp2Δ strain, which is defective in COB mRNA intron processing (33), was also analyzed as a control for possible general effects of respiratory deficiency on lipoylation. The cbp2Δ strain contained only slightly less lipoic acid than wild type, as reported previously for another respiration-deficient strain (20).
TABLE 2.
Lipoic acid content in the cbp2Δ and lip3Δ strains
Lipoic acid assays were performed as described under “Experimental Procedures.” The yeast strains were grown in WO medium. The growth response of the JRG33-lip9 strain was followed by measuring turbidity at A600 nm. Each result represents the mean lipoic acid content of five different yeast cultures ± S.D.
| Strain | Lipoic acid content ng/g cells, WO medium |
|---|---|
| Wild type | 147 ± 28.3 |
| cbp2Δ | 127 ± 30.6 |
| lip3Δ | 10.2 ± 3.1 |
Because lipoic acid is required for activity of PDH and α-KDH, the lip3Δ strain was assayed for these two lipoic acid-dependent enzyme activities. PDH and α-KDH activities were completely absent in the lip3Δ strain (Table 3), confirming that Lip3 is essential for the activities of both multienzyme complexes. Fumarase activity, assayed as a representative of all non-lipoic acid-dependent enzymes in mitochondria, decreased by a third in the respiration-deficient lip3Δ mutant strain relative to wild type.
TABLE 3.
PDH and α-KDH activity in the lip3Δ strain
Mitochondrial extracts were prepared as described under “Experimental Procedures.” Specific enzyme activities for PDH and α-KDH are expressed as nanomoles of NADH formed × min−1 mg−1 of mitochondrial proteins. Specific enzyme activity for fumarase is expressed as micromoles of fumarate formed × min−1 mg−1 of mitochondrial protein. Fumarase activity was measured as a control to ensure the mitochondria were functional. The experiment was performed in triplicate, and the data shown are the average of the measurements ± S.E.
| Strain | Specific activity |
||
|---|---|---|---|
| PDH | α-KDH | Fumarase | |
| nmol of NADH min−1mg−1 | μmol of fumarate min−1mg−1 | ||
| Wild type | 86 ± 2 | 95 ± 0 | 1.44 ± 0.06 |
| lip3Δ | 0 | 0 | 0.96 ± 0.11 |
Lip3 Is Required with Lip2 and Lip5 for Synthesis and Attachment of Lipoic Acid to Lat1 and Kgd2
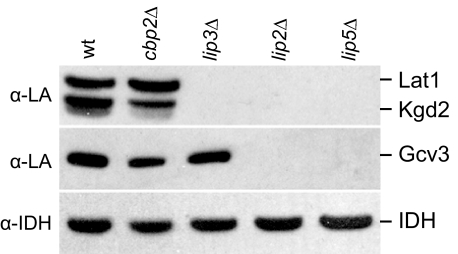
In E. coli, LipB (Lip2) plus LipA (Lip5) synthesize and attach lipoic acid to the H protein of GC and to the E2 subunits of PDH and α-KDH, or free lipoic acid is attached by LplA (Lip3). Two hints suggested that, by contrast, there were not two redundant pathways in Saccharomyces. First, each of the lip mutants is respiration-deficient. Second, lip2 and lip5 mutants are not rescued by growth on lipoic acid (19, 20), which would be expected if lipoic acid is imported and Lip3 could substitute for a Lip2 plus Lip5 pathway. To investigate the effects of individual lip mutations in yeast on the lipoylation of Lat1 (the E2 subunit of PDH), Kgd2 (the E2 subunit of α-KDH), and Gcv3 (the H protein of GC), mitochondrial extracts were analyzed on Western blots challenged with anti-lipoic acid antiserum (34) (Fig. 2). The antiserum specifically detected all three lipoylated proteins in the wild-type strain and in the respiration-deficient cbp2Δ control strain. The verification of the identity of the three target proteins is shown later in this work. Lipoylated Lat1 or Kdg2 was not detected in any of the three mutant strains. However, lipoylated Gcv3 was detected in the lip3Δ strain. These data suggest that all three enzymes, Lip3, Lip2, and Lip5, are required for lipoylation of Lat1 and Kgd2, but only Lip2 and Lip5 are required for lipoylation of Gcv3.
FIGURE 2.
Analysis of lipoylated proteins in lipoylation mutant strains. Mitochondrial extracts from wild-type (wt), cbp2Δ, a respiration-deficient control strain, and the lipoic acid synthesis-attachment mutant strains lip3Δ, lip2Δ, and lip5Δ strains were analyzed by Western blot using an antiserum directed against lipoic acid (α-LA). Lat1 and Kgd2 are the lipoylated E2 subunits of PDH and α-KDH, respectively, and Gcv3 is the lipoylated H protein of GC. Mitochondrial isocitrate dehydrogenase (antiserum α-IDH) was used as a loading control.
Lip3-GFP Is Localized to Mitochondria
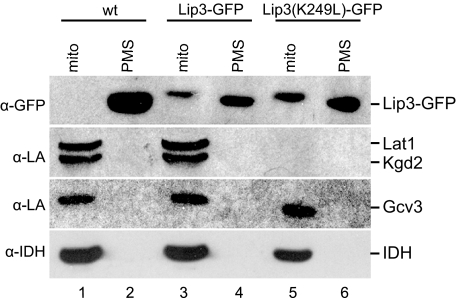
The MitoProt program (35) predicted with 95% probability that the YJL046w gene product is localized to mitochondria. To test this prediction, we made a Lip3-GFP fusion protein and expressed it at the LIP3 genomic locus. The strain was respiration-competent, indicating functional expression of the Lip3-GFP fusion protein. Mitochondrial and post-mitochondrial supernatant (PMS) fractions were analyzed by Western blot using an anti-GFP antibody (Fig. 3, top panel). A protein of ∼105 kDa, which corresponds to the predicted molecular mass of the Lip3-GFP polypeptide, was detected in the Lip3-GFP strain but not in the wild-type strain(Fig. 3, lanes 1–4). Moreover, the polypeptide was recognized only in the mitochondrial fraction and not in the PMS fraction. A protein of similar mass was recognized nonspecifically by the antibody in all PMS fractions regardless of Lip3 fusion to GFP. These data show that the Lip3-GFP protein is expressed and is targeted to mitochondria.
FIGURE 3.
Functional analysis of Lip3. Mitochondrial (mito) and PMS fractions from wild-type (wt), Lip3-GFP, and Lip3(K249L)-GFP strains were analyzed by Western blot using the lipoic acid (α-LA) antiserum. An anti-GFP antibody (α-GFP) was used to detect Lip3-GFP (top panel). A protein of similar mass was recognized nonspecifically in the PMS fractions by the α-GFP antibody regardless of Lip3 fusion to GFP. The α-IDH antiserum was used as a loading control.
To verify that Lip3-GFP is a functional substitute for Lip3, we analyzed the lipoylation state of the target proteins from the mitochondrial and PMS fractions in the same experiment shown in Fig. 3. All three lipoic acid-modified proteins, Lat1, Kgd2, and Gcv3, were detected at wild-type levels in the mitochondrial fractions of the wild-type and Lip3-GFP strains (Fig. 3, lanes 1 and 3), suggesting that Lip3-GFP is fully functional in lipoic acid synthesis and attachment.
Conserved Lysine in the Lipoate-Protein Ligase Active Site of Lip3 Is Required for Lipoylation
Lip3 contains a lysine residue that is strictly conserved in lipoyl ligase, lipoyltransferase, and biotinyl ligase families (see sequence alignment in Fig. 1) (31). The yeast Lip3 lysine residue (Lys-249) is in the lipoyl-AMP binding pocket and forms a hydrogen bond with the carbonyl oxygen of lipoyl-AMP, facilitating transfer of the lipoyl group to the ϵ-amino group of the specific lysine acceptor residue in target apoproteins (36–38). To test whether the predicted lipoyl-protein ligase activity of Lip3 is necessary for its role in protein lipoylation, we mutated Lys-249 to Leu in the Lip3-GFP fusion protein expressed at the LIP3 locus. Mitochondrial localization and stability of the K249L mutant form of Lip3-GFP was confirmed by Western blot using the anti-GFP antibody (Fig. 3, top panel, lanes 5 and 6). The anti-lipoic acid antiserum did not detect any lipoylated Lat1 or Kgd2 protein in the mitochondrial fraction of the strain expressing the Lip3(K249L)-GFP protein (Fig. 3, lane 5), as compared with strains expressing wild-type Lip3 (lane 1) or Lip3-GFP (lane 3). However, lipoylated Gcv3 was detected (Fig. 3, lane 5). This result suggests that the Lys-249 to Leu mutation in Lip3-GFP affects the activity of the enzyme and that the activity of Lip3-GFP is required for lipoylation of Lat1 and Kgd2, but not of Gcv3. This phenotype is equivalent to that of the lip3 deletion mutant strain.
Gcv3, a Target of Lipoylation, Is Required for Lipoylation of Lat1 and Kgd2
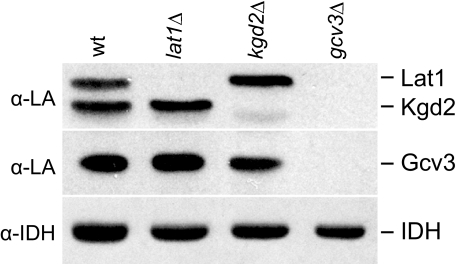
We assumed that our assignment of the target proteins visualized on Western blots with anti-lipoic acid antiserum matched the predicted masses for Lat1 (52 kDa), Kgd2 (50 kDa), and Gcv3 (19 kDa) (Saccharomyces Genome Data base). Because some proteins run anomalously on gels, and mitochondrially targeted proteins have different length N-terminal sequences that are proteolytically removed upon entry into the organelle, we wanted to verify the identity of the bands by Western blot analysis of mitochondrial proteins from the lat1Δ, kgd2Δ, and gcv3Δ strains with the anti-lipoic acid antiserum (Fig. 4). As expected, the largest of the three proteins was the only band not detected in the lat1Δ strain as compared with wild type. Likewise, the slightly smaller protein was the only band not detected in the kgd2Δ strain. Strikingly, however, lipoylated proteins were not detected at all in the gcv3Δ strain (Fig. 4). Even prolonged exposure of the Western blot did not reveal any bands (data not shown). A faint band visible below Kgd2 in Fig. 4, 1st to 3rd lanes, may be an as yet unidentified fourth protein previously detected by the anti-lipoic acid antiserum (39). We conclude that Gcv3, a target of lipoylation, is absolutely required for lipoylation of Lat1 and Kgd2. That a target of lipoylation is required for lipoylation is a novel result that suggests a level of regulation in yeast not previously recognized in the well studied prokaryotic system.
FIGURE 4.
Identification of lipoylated proteins. Mitochondrial extracts from wild type (wt), lat1Δ, kgd2Δ, and gcv3Δ were analyzed by Western blot using an antiserum directed against lipoic acid (α-LA). Antiserum α-IDH was used as a loading control.
Glycine Cleavage Is Not Required for Lipoylation
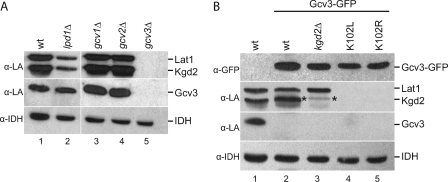
Because loss of Gcv3 knocks out lipoylation of mitochondrial target proteins completely, we wondered whether this is because of a loss of GC activity per se or is specific to the Gcv3 protein itself. To test the necessity of GC activity for lipoylation, mitochondrial extracts from the gcv1Δ and gcv2Δ strains, which are deficient in the T and P proteins of the GC enzyme (40), respectively, and the lpd1Δ strain, which lacks the L protein, were analyzed by Western blot using the anti-lipoic acid antiserum (Fig. 5A). The LPD1 gene encodes dihydrolipoamide dehydrogenase, a subunit common to all three lipoic acid-dependent multienzyme complexes (3). All three lipoylated proteins were recognized in the lpd1Δ, gcv1Δ, and gcv2Δ strains, as compared with the gcv3Δ strain. However, there was an approximate 2-fold decrease in the levels of lipoylated proteins in the lpd1Δ strain as compared with wild type, whereas gcv1Δ and gcv2Δ had wild-type levels of lipoylation. These data show that GC activity per se is not required for protein lipoylation.
FIGURE 5.
Analysis of glycine cleavage and Gcv3-GFP mutant strains. A, mitochondrial extracts from wild-type (wt), lpd1Δ, gcv1Δ, gcv2Δ, and gcv3Δ strains were analyzed by Western blot using the lipoic acid (α-LA) antiserum. B, mitochondrial extracts from wild-type, Gcv3-GFP, Gcv3-GFP kgd2Δ, Gcv3(K102L)-GFP, and Gcv3(K102R)-GFP strains were analyzed by Western blot using the lipoic acid antiserum. The α-GFP antibody was used to detect Gcv3-GFP (top panel). The asterisk in the Gcv3-GFP kgd2Δ lane marks lipoylated Gcv3-GFP. The α-IDH antiserum was used to detect IDH as a loading control.
Lipoylated Gcv3 Is Required for Lipoylation of Lat1 and Kgd2
Because Gcv3, a target of lipoylation, is also required for lipoylation (Fig. 4), we explored whether the lipoylated form of Gcv3, in contrast to the apoprotein form, is required for this activity. To test this hypothesis, the specific lysine residue that is covalently modified by lipoic acid within the lipoylation domain of Gcv3, Lys-102, was mutated to leucine and to arginine. The K102R mutation preserved the positive charge while rendering the residue inactive as a lipoic acid acceptor. Both mutations were made in a strain that expresses a GFP-tagged version of Gcv3 at the GCV3 locus. The strain that expresses wild-type Gcv3-GFP is respiration-competent and has wild-type levels of lipoylated Lat1 and Kgd2 (Fig. 5B, lane 2). Interestingly, the Gcv3 K102L and K102R mutations eliminated lipoylation of Lat1 and Kgd2 (Fig. 5B, 4th and 5th lanes), suggesting that the lipoylated form of Gcv3 is required for lipoylation of the E2 subunits, Lat1 and Kgd2. The molecular mass of the Gcv3-GFP polypeptide is similar to that of the Kgd2 polypeptide, making lipoylated Gcv3-GFP (Fig. 5B, lane 2, marked with an asterisk) difficult to distinguish from lipoylated Kgd2. Therefore, the KGD2 gene was deleted from the Gcv3-GFP strain, and lipoylated Gcv3-GFP (Fig. 5B, lane 3, marked with as asterisk) was visible.
DISCUSSION
Here we show that four enzymes are involved in protein lipoylation in yeast mitochondria; one is a target of lipoic acid modification itself. Lip2 and Lip5, the homologs of E. coli LipB and LipA, are required for lipoylation of three lipoic acid-modified proteins as follows: Lat1, Kgd2, and Gcv3. Lip3, the homolog of lipoate-protein ligase (LplA) in E. coli, is required for lipoylation of two of the three lipoate-modified proteins, Lat1 and Kgd2. The invariant Lys-249 residue in the active site of Lip3 is essential for this function. Finally, lipoylated Gcv3 is also required for lipoylation of Lat1 and Kgd2. We conclude that protein lipoylation in yeast is different from E. coli in three ways as follows: 1) Lip2 plus Lip5 are not sufficient to lipoylate all target proteins; 2) Lip3 is not sufficient to lipoylate any target protein alone; and 3) Gcv3, a target of lipoylation, is required for lipoylation.
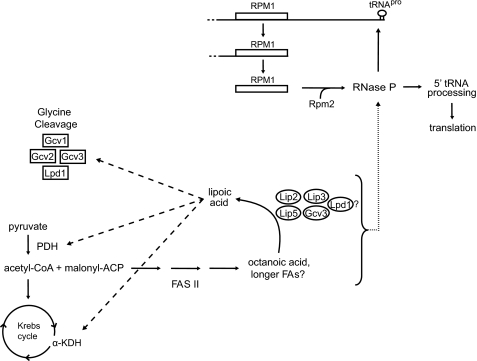
Lip2, Lip5, Lip3, and Gcv3 were all first identified in studies of other cellular processes (19, 20, 41). Our data showing that all four proteins are required for protein lipoylation, and thus for respiratory competence, lead us to suggest a model in which the four proteins function in a lipoylation complex (Fig. 6). As deletion of LPD1, which codes for the dihydrolipoamide dehydrogenase subunit common to all three lipoic acid-dependent enzyme complexes, causes a reduction in protein lipoylation (Fig. 5A), some molecules of Lpd1 may function also in the lipoylation complex. An alternative hypothesis is that loss of Lpd1 may lead to destabilization of the complexes. The model proposes that some molecules of Gcv3 function in the glycine cleavage enzyme, whereas others participate in the lipoylation complex. In independent support of the complex model, Lip2 purified with Gcv3 in a global tandem affinity purification tag study for protein interactions (42). Alternatively, the enzymes may function separately as part of one lipoylation pathway in which Lip2 and Lip3 are both required for the transfer of lipoic acid, and lipoylated Gcv3 is required, in some novel way, for the lipoylation of Lat1 and Kgd2.
FIGURE 6.
Model of protein lipoylation in yeast. The mitochondrial FAS II pathway produces octanoic acid, the precursor for lipoic acid synthesis. Lip2, Lip3, Lip5, Gcv3, and Lpd1 may function in a lipoylation complex. We previously reported that FAS II and tRNA processing intersect (18), and we hypothesize that this intersection exists as a point of regulation for mitochondrial gene expression. A product of FAS II is required for assembly or activity of mitochondrial RNase P, which processes the 5′ leader sequences of tRNAs. Because RNase P activity is required for the maturation of its own mitochondrially encoded RNA subunit, a positive feedback loop exists for RNase P maturation and activity in response to the FAS II pathway. Pyruvate dehydrogenase, which requires lipoic acid for its function, produces acetyl-CoA, which is fed into the FAS II pathway. The dashed arrows represent the requirement of lipoic acid for PDH, α-KDH, and GC enzymatic activity. The dotted arrow represents the requirement of a product of the FAS II pathway for RNase P activity or assembly.
Regardless of the existence of a complex, the organization of the yeast enzymes in a single pathway differs from the two alternative lipoylation pathways in E. coli. In E. coli, attachment of lipoic acid to apoproteins can be catalyzed by one of two different enzymes, LipB or LplA. LipB transfers the lipoyl (or octanoyl) group endogenously synthesized on ACP to the acceptor lysine residue of target apoproteins (11, 12). Alternatively, LplA catalyzes the attachment of exogenously supplied lipoic acid in a two-step reaction. ATP and lipoic acid are first joined in formation of a lipoyl-AMP intermediate, and then the lipoyl group is transferred to the conserved lysine residue within the acceptor lipoyl domain of the targets. Although E. coli LplA can independently lipoylate target proteins using lipoic acid from the medium, lipoylation of Lat1 and Kgd2 in yeast requires Lip2, Lip5, and Gcv3 in addition to the homolog of LplA, Lip3. Hence, the role of yeast Lip3 is not the same as that of E. coli LplA.
Our most detailed view of the function of LplA-like enzymes comes from analyses of crystal structures of bacterial, archaeal, and bovine enzymes (36–38). The crystal structure of E. coli LplA revealed N-terminal and C-terminal folded domains (38). The crystal structure of LplA from the archaeon Thermoplasma acidophilum (Ta LplA) with no substrate bound (38, 43) or complexed with ATP or lipoyl-AMP has also been solved (38). Similar to E. coli LplA, Ta LplA catalyzes both the activation and transfer of lipoic acid in vivo, however, the protein expressed for crystallization is homologous to only the N-terminal domain of E. coli LplA. This form of the enzyme is inactive and needs the noncovalent association of the product of an adjacent gene that has homology to the E. coli C-terminal domain (43).4 However, lipoyl-AMP binds to the crystallized Ta LplA domain at a site structurally equivalent to that of the bovine enzyme (see below) (37). Lipoyl-AMP adopts a U-shaped structure within the bifurcated binding pocket. The lipoyl group of the intermediate is buried within a hydrophobic channel, and the AMP moiety forms hydrogen bonds with residues in the other tunnel of the two-lobed pocket. The bent arrangement orients the carbon atom subject to nucleophilic attack at the surface of Ta LplA. The carbonyl oxygen atom of the lipoyl group interacts with Lys-145 (the invariant lysine residue analogous to Lys-133 in E. coli and Lys-249 in S. cerevisiae) in the binding pocket. This interaction likely aids nucleophilic attack of the exposed carbon atom of lipoyl-AMP by the ϵ-amino group of the acceptor lysine, resulting in an amide linkage.
Two separate enzymes are required in cows for catalysis of the two reactions carried out by lipoate-protein ligases in prokaryotes. Lipoate-activating enzyme, which is a medium-chain acyl-CoA synthetase, activates lipoic acid with GTP (44), whereas lipoyltransferase catalyzes the transfer of the lipoyl group to the acceptor lysine residue of target proteins (45). The crystal structure of bovine lipoyltransferase, similar to E. coli LplA, contains both N-terminal and C-terminal domains (37). Interestingly, the N-terminal domain of the recombinant bovine protein produced in E. coli had lipoyl-AMP bound in the active site, indicating a high affinity of bovine lipoyltransferase for lipoyl-AMP. The invariant lysine residue in bovine lipoyltransferase (Lys-135) forms similar bonds with lipoyl-AMP as does Lys-145 of Ta LplA and, most likely, Lys-133 of E. coli LplA. The gene encoding the human lipoyltransferase has been isolated (16), and amino acid sequence alignment reveals regions of significant similarity with other LplA-like enzymes and includes the invariant lysine residue (Fig. 1). It seems likely that the mammalian lipoyltransferase transfers lipoic acid obtained from the diet or from intestinal bacteria to target proteins, but recent reports of a soluble mitochondrial ACP (46) and of lipoic acid synthase (a Lip5-like enzyme) (47, 48) indicate the existence of a de novo lipoic acid biosynthetic pathway, which may be important for development or for certain tissues.
A third major difference between lipoylation in yeast and that in E. coli is the requirement for Gcv3, the lipoylated subunit of the GC complex. How is Gcv3, a target of lipoylation, required for lipoylation? Because lipoylated Gcv3 is required for lipoylation of Lat1 and Kgd2, Gcv3 may act as a sensor for the metabolic state of the cell. When acetyl-CoA is abundant, the lipoylation complex model proposes that octanoic acid produced by the FAS II pathway is available for lipoylation of Gcv3 by Lip2 and Lip5. Lipoylated Gcv3 in turn may be required either for assembly of the active lipoylation complex or for a conformational change in one or more enzymes in the complex. The assembled, active complex then lipoylates Lat1 and Kgd2, leading to cellular respiration. Under conditions of low availability of acetyl-CoA, low levels of lipoylated Gcv3 may act as a switch to turn off PDH and α-KDH activities by blocking lipoylation of Lat1 and Kgd2. Because PDH requires lipoic acid for the production of acetyl-CoA that is fed into the FAS II pathway, lower levels of octanoic acid and therefore lipoic acid are produced. However, Kgd2 and Gcv3 are lipoylated in the lat1Δ strain, indicating that, when the cells are grown in rich glucose medium, acetyl-CoA from another source, such as amino acid breakdown, β-oxidation of fatty acids, or the pyruvate dehydrogenase bypass pathway, can be fed into the FAS II pathway. Inactive PDH and α-KDH lead to a decrease in or loss of cellular respiration, and mitochondrial gene expression may also be affected by inhibition of tRNA processing (18).
Together, our results suggest that proteins are lipoylated in yeast mitochondria via one pathway or lipoylation complex, unlike in E. coli, which has two redundant lipoylation pathways. Future investigations will reveal whether protein lipoylation in mammalian mitochondria is similar to that in yeast. Of particular interest will be discovering whether the eukaryotic pathway/complex is regulated in response to the availability of acetyl-CoA, and then in turn serves to regulate mitochondrial gene expression via tRNA processing (18). One piece of evidence reported by Autio et al. (49) suggests that mitochondrial gene expression may be regulated by the availability of acetyl-CoA in human cells. A bicistronic cDNA was found to encode an FAS II enzyme and an RNase P protein subunit. This gene arrangement has been conserved throughout vertebrate evolution from fish to humans. The link between mitochondrial fatty acid synthesis and RNA processing in yeast (18) and humans (49) along with the finding that protein lipoylation in yeast occurs via one pathway/complex suggest that eukaryotic mitochondrial function is highly regulated in response to cellular metabolism.
Acknowledgments
We thank Telsa Mittelmeier, John Little, Alex Tzagoloff, John E. Cronan, and Joseph Boyd for critical reading of the manuscript. We thank Mike Rice for help in the laboratory. We also thank Lee McAlister-Henn for the generous gift of anti-IDH antiserum.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34893 (to C. L. D.).
Christensen, Q. H., and Cronan, J. E. (2009) J. Biol. Chem. 284, 21317–21326
- PDH
- pyruvate dehydrogenase
- α-KDH
- α-ketoglutarate dehydrogenase
- GC
- glycine cleavage
- ACP
- acyl carrier protein
- Lip
- lipoyl synthase
- Lpl
- lipoate-protein ligase
- IDH
- isocitrate dehydrogenase
- FAS II
- type II fatty acid synthesis pathway
- GFP
- green fluorescent protein
- PMS
- post-mitochondrial supernatant.
REFERENCES
- 1.Perham R. N. (2000) Annu. Rev. Biochem. 69,961–1004 [DOI] [PubMed] [Google Scholar]
- 2.Herbert A. A., Guest J. R. (1975) Arch. Microbiol. 106,259–266 [DOI] [PubMed] [Google Scholar]
- 3.Reed L. J., Hackert M. L. (1990) J. Biol. Chem. 265,8971–8974 [PubMed] [Google Scholar]
- 4.Cronan J. E., Zhao X., Jiang Y. (2005) Adv. Microb. Physiol. 50,103–146 [DOI] [PubMed] [Google Scholar]
- 5.Zhao X., Miller J. R., Cronan J. E. (2005) Biochemistry 44,16737–16746 [DOI] [PubMed] [Google Scholar]
- 6.Morris T. W., Reed K. E., Cronan J. E., Jr. (1995) J. Bacteriol. 177,1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cicchillo R. M., Booker S. J. (2005) J. Am. Chem. Soc. 127,2860–2861 [DOI] [PubMed] [Google Scholar]
- 8.Booker S. J. (2004) Chem. Biol. 11,10–12 [DOI] [PubMed] [Google Scholar]
- 9.Jordan S. W., Cronan J. E., Jr. (1997) J. Biol. Chem. 272,17903–17906 [DOI] [PubMed] [Google Scholar]
- 10.Miller J. R., Busby R. W., Jordan S. W., Cheek J., Henshaw T. F., Ashley G. W., Broderick J. B., Cronan J. E., Jr., Marletta M. A. (2000) Biochemistry 39,15166–15178 [DOI] [PubMed] [Google Scholar]
- 11.Jordan S. W., Cronan J. E., Jr. (2003) J. Bacteriol. 185,1582–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesbitt N. M., Baleanu-Gogonea C., Cicchillo R. M., Goodson K., Iwig D. F., Broadwater J. A., Haas J. A., Fox B. G., Booker S. J. (2005) Protein Expr. Purif. 39,269–282 [DOI] [PubMed] [Google Scholar]
- 13.Zhao X., Miller J. R., Jiang Y., Marletta M. A., Cronan J. E. (2003) Chem. Biol. 10,1293–1302 [DOI] [PubMed] [Google Scholar]
- 14.Morris T. W., Reed K. E., Cronan J. E., Jr. (1994) J. Biol. Chem. 269,16091–16100 [PubMed] [Google Scholar]
- 15.Kang S. G., Jeong H. K., Lee E., Natarajan S. (2007) Gene 393,53–61 [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara K., Suzuki M., Okumachi Y., Okamura-Ikeda K., Fujiwara T., Takahashi E., Motokawa Y. (1999) Eur. J. Biochem. 260,761–767 [DOI] [PubMed] [Google Scholar]
- 17.Ma Q., Zhao X., Nasser, Eddine A., Geerlof A., Li X., Cronan J. E., Kaufmann S. H., Wilmanns M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schonauer M. S., Kastaniotis A. J., Hiltunen J. K., Dieckmann C. L. (2008) Mol. Cell. Biol. 28,6646–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marvin M. E., Williams P. H., Cashmore A. M. (2001) FEMS Microbiol. Lett. 199,131–136 [DOI] [PubMed] [Google Scholar]
- 20.Sulo P., Martin N. C. (1993) J. Biol. Chem. 268,17634–17639 [PubMed] [Google Scholar]
- 21.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350,87–96 [DOI] [PubMed] [Google Scholar]
- 22.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14,953–961 [DOI] [PubMed] [Google Scholar]
- 23.Zheng L., Baumann U., Reymond J. L. (2004) Nucleic Acids Res. 32,e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U. K. (1970) Nature 227,680–685 [DOI] [PubMed] [Google Scholar]
- 25.Kresze G. B., Ronft H. (1981) Eur. J. Biochem. 119,573–579 [DOI] [PubMed] [Google Scholar]
- 26.Remes A. M., Rantala H., Hiltunen J. K., Leisti J., Ruokonen A. (1992) Pediatrics 89,730–734 [PubMed] [Google Scholar]
- 27.Hayden M. A., Huang I. Y., Iliopoulos G., Orozco M., Ashley G. W. (1993) Biochemistry 32,3778–3782 [DOI] [PubMed] [Google Scholar]
- 28.Brody S., Oh C., Hoja U., Schweizer E. (1997) FEBS Lett. 408,217–220 [DOI] [PubMed] [Google Scholar]
- 29.Herbert A. A., Guest J. R. (1970) Methods Enzymol. 18,269–272 [Google Scholar]
- 30.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics. 23,2947–2948 [DOI] [PubMed] [Google Scholar]
- 31.Reche P. A. (2000) Protein Sci. 9,1922–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzagoloff A., Dieckmann C. L. (1990) Microbiol. Rev. 54,211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGraw P., Tzagoloff A. (1983) J. Biol. Chem. 258,9459–9468 [PubMed] [Google Scholar]
- 34.Humphries K. M., Szweda L. I. (1998) Biochemistry 37,15835–15841 [DOI] [PubMed] [Google Scholar]
- 35.Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241,779–786 [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara K., Toma S., Okamura-Ikeda K., Motokawa Y., Nakagawa A., Taniguchi H. (2005) J. Biol. Chem. 280,33645–33651 [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara K., Hosaka H., Matsuda M., Okamura-Ikeda K., Motokawa Y., Suzuki M., Nakagawa A., Taniguchi H. (2007) J. Mol. Biol. 371,222–234 [DOI] [PubMed] [Google Scholar]
- 38.Kim D. J., Kim K. H., Lee H. H., Lee S. J., Ha J. Y., Yoon H. J., Suh S. W. (2005) J. Biol. Chem. 280,38081–38089 [DOI] [PubMed] [Google Scholar]
- 39.Onder O., Yoon H., Naumann B., Hippler M., Dancis A., Daldal F. (2006) Mol. Cell. Proteomics 5,1426–1436 [DOI] [PubMed] [Google Scholar]
- 40.Piper M. D., Hong S. P., Ball G. E., Dawes I. W. (2000) J. Biol. Chem. 275,30987–30995 [DOI] [PubMed] [Google Scholar]
- 41.Nagarajan L., Storms R. K. (1997) J. Biol. Chem. 272,4444–4450 [DOI] [PubMed] [Google Scholar]
- 42.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Nature 440,637–643 [DOI] [PubMed] [Google Scholar]
- 43.McManus E., Luisi B. F., Perham R. N. (2006) J. Mol. Biol. 356,625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujiwara K., Takeuchi S., Okamura-Ikeda K., Motokawa Y. (2001) J. Biol. Chem. 276,28819–28823 [DOI] [PubMed] [Google Scholar]
- 45.Fujiwara K., Okamura-Ikeda K., Motokawa Y. (1994) J. Biol. Chem. 269,16605–16609 [PubMed] [Google Scholar]
- 46.Cronan J. E., Fearnley I. M., Walker J. E. (2005) FEBS Lett. 579,4892–4896 [DOI] [PubMed] [Google Scholar]
- 47.Morikawa T., Yasuno R., Wada H. (2001) FEBS Lett. 498,16–21 [DOI] [PubMed] [Google Scholar]
- 48.Yi X., Maeda N. (2005) Mol. Cell. Biol. 25,8387–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Autio K. J., Kastaniotis A. J., Pospiech H., Miinalainen I. J., Schonauer M. S., Dieckmann C. L., Hiltunen J. K. (2008) FASEB J. 22,569–578 [DOI] [PubMed] [Google Scholar]
- 50.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425,686–691 [DOI] [PubMed] [Google Scholar]
- 51.Herbert A. A., Guest J. R. (1968) J. Gen. Microbiol. 53,363–381 [DOI] [PubMed] [Google Scholar]