Abstract
We developed a novel approach for de novo genome assembly using only sequence data from high-throughput short read sequencing technologies. By combining data generated from 454 Life Sciences (Roche) and Illumina (formerly known as Solexa sequencing) sequencing platforms, we reliably assembled genomes into large scaffolds at a fraction of the traditional cost and without use of a reference sequence. We applied this method to two isolates of the phytopathogenic bacteria Pseudomonas syringae. Sequencing and reassembly of the well-studied tomato and Arabidopsis pathogen, PtoDC3000, facilitated development and testing of our method. Sequencing of a distantly related rice pathogen, Por1_6, demonstrated our method's efficacy for de novo assembly of novel genomes. Our assembly of Por1_6 yielded an N50 scaffold size of 531,821 bp with >75% of the predicted genome covered by scaffolds over 100,000 bp. One of the critical phenotypic differences between strains of P. syringae is the range of plant hosts they infect. This is largely determined by their complement of type III effector proteins. The genome of Por1_6 is the first sequenced for a P. syringae isolate that is a pathogen of monocots, and, as might be predicted, its complement of type III effectors differs substantially from the previously sequenced isolates of this species. The genome of Por1_6 helps to define an expansion of the P. syringae pan-genome, a corresponding contraction of the core genome, and a further diversification of the type III effector complement for this important plant pathogen species.
New technologies have _rapidly reduced the time and cost of whole-genome sequencing. Illumina (formerly known as Solexa sequencing) and 454 Life Sciences (Roche) (hereafter 454; Margulies et al. 2005) sequencers provide shorter and more error-prone reads than Sanger sequencing (Sanger and Coulson 1975), but cost orders of magnitude less per base sequenced (Mardis 2008). Both technologies have been developed largely for “resequencing” of closely related individuals in cases where high-quality reference genome sequences exist; thus, identification of new polymorphisms in eukaryotes (Hillier et al. 2008;Ossowski et al. 2008), including some linked to human disease (Chen et al. 2008; Morin et al. 2008), is feasible. These technologies are expanding to previously unsequenced organisms (Hogg et al. 2007; McCutcheon and Moran 2007), but assembly of these genomes requires either a reference guided approach (Salzberg et al. 2008) or significant oversampling of the genome sequence (depth of coverage), thus detracting significantly from the cost savings.
The Illumina and 454 sequencing methods each have drawbacks that make de novo assembly difficult. 454 reads are currently shorter than Sanger reads (250 bp vs. >650 bp) but are less than one-tenth the cost per base (Mardis 2008). 454 reads are also prone to high rates of small insertions and deletions (indels). Such frameshifts inhibit gene prediction in de novo assemblies more than single nucleotide errors. Illumina sequencing is less than one-hundredth the cost of Sanger sequencing, but reads are currently 36 bp in length, preventing significant assembly of even short repeats. For example, one estimate suggests that the 4.65-Mb Yersinia pestis genome has a theoretical maximum N50 size of ∼26 kbp if only 30-bp reads are used in a de novo assembly (Pop and Salzberg 2008). Illumina reads also have a relatively high error rate and an apparent bias against sequencing of AT-rich regions (Hillier et al. 2008).
De novo sequencing and assembly of bacterial pathogen genomes is essential for understanding ecological and evolutionary relationships between strains and species, as well as for investigating virulence mechanisms. Members of a given eukaryotic species generally have few gene content differences. However, due largely to horizontal gene transfer and gene loss, bacterial isolates of the same species that are highly related at housekeeping loci often share a surprisingly low fraction of overall gene content (Ochman and Moran 2001). For example, ∼75% of genes are shared between any two of the three fully sequenced pathovars (strains isolated from a particular plant species) of Pseudomonas syringae (Feil et al. 2005; Joardar et al. 2005). Such divergence in gene content limits our ability to obtain complete genome sequences for bacteria using resequencing or reference-assisted assembly (Salzberg et al. 2008), because horizontally transferred fragments of the genome may assemble poorly even if there is a previously sequenced closely related isolate. Often these horizontally transferred fragments provide genes essential for adaptive phenotypes that allow the bacteria to persist and exploit a variety of environments and hosts (Lindeberg et al. 2008); thus, it is essential that these fragments be assembled well.
We focus on P. syringae because it is a plant pathogen that infects crop plants worldwide and is related to the human pathogen, P. aeroginosa. Although other genes can contribute to the success of infections, growth and virulence within the plant host is primarily determined by a type III secretion system that delivers strain-specific suites of “type III effectors” into eukaryotic host cells (He et al. 2004) and, in many strains, provides the regulation for production of specific toxins (Bender et al. 1987) that can mimic plant hormones (Katsir et al. 2008). Type III effector proteins are virulence factors and can directly disrupt plant immune function and signaling within host tissues, allowing the extracellular bacteria to survive, multiply, and disperse (Mudgett 2005; Grant et al. 2006). The type III effector complement can be highly variable between pathovars of P. syringae, likely due in part to their over-representation within the horizontally transferred gene pool and to strong selective pressures for both their presence (as virulence factors) and their absence (since, as virulence factors, they can be recognized by the plant immune system; Jones and Dangl 2006; Ma et al. 2006). Genes encoding both the type III secretion system and type III effector proteins described to date are coregulated in P. syringae (Huynh et al. 1989; Xiao and Hutcheson 1994). Type III effector proteins often share homologous modules over part of their length and may often be truncated within a particular strain (Chang et al. 2005). These gene fragments can be used as is or reused evolutionarily via recombination that places them downstream of proper promoter and amino-terminal coding contexts (Stavrinides et al. 2006, 2008). These traits, combined with the fact that many type III effector genes are associated with putative horizontally acquired DNA (Hacker and Kaper 2000), make whole-genome de novo sequencing and assembly the only way to capture type III effector variation, toxin variation, and whole-genome evolution in full.
Here we sequence and de novo assemble two isolates of P. syringae using relatively low-coverage, and hence economical, short read sequencing. Previous work demonstrated that combining new sequencing technologies with traditional Sanger sequencing can improve assembly and reduce the cost of de novo sequencing (Goldberg et al. 2006; McCutcheon and Moran 2007). Our approach takes this concept a step further by assembling only Illumina and low-coverage 454 reads. Because our assembly approach is novel, reassembly of a previously sequenced reference genome, P. syringae pv. tomato DC3000 (PtoDC3000) (Buell et al. 2003), was essential for method validation as well as for understanding the limitations of our approach. We demonstrate the efficacy of our method by sequencing a novel genome, P. syringae pv. oryzae 1_6 (Por1_6), a rice (monocot) pathogen. This isolate also represents a fourth phylogenetic clade of P. syringae (Hwang et al. 2005) for which a genome sequence is now available. It is significantly diverged from the three previously sequenced genomes, making this new sequence a valuable comparative resource.
Results
Minimal sequencing from two short read technologies results in ∼29× coverage, high-quality whole-genome sequence
Sequencing and filtering of a single lane from the University of North Carolina-Chapel Hill (UNC-CH) high-throughput sequencing facility's Illumina GA1 yielded 4,905,077 reads 35 bp in length, or 26.3× coverage for PtoDC3000 (Table 1; published chromosome of 6.4 Mb; Buell et al. 2003). Alignment of these reads to the reference genome (see Methods) showed that the Illumina reads were of high quality: ∼88% of reads had one or fewer errors, and the overall error rate was 1.5% (Table 1). One-quarter plate of 454 runs yielded 77,466 reads at 240-bp average length or ∼2.85× coverage. These reads had 0.0018 syntenic indels per base pair (0.43 errors per read, Table 1). Additionally, the 454 reads did not cover a total of ∼50 kb of the larger of two PtoDC3000 plasmids (GenBank; pDC3000A: NC_004633.1 size 73.6 kb). We believe that the single colony we picked for 454 sequencing may have been a deletion mutant for part or all of this plasmid, as recently documented in PtoDC3000 (Landgraf et al. 2006). The same plasmid polymorphism was not detected in our Illumina data, and no large deletions were seen in the genome of PtoDC3000. One-quarter plate of 454 paired ends yielded 94,262 pairs where both right- and left-end sequences were present. The mean span length for these pairs was 2487 bp, with a standard deviation of 689 bp (Table 1). The distribution of the size of the fragments between the paired ends was significantly skewed to the right (Supplemental Fig. 1).
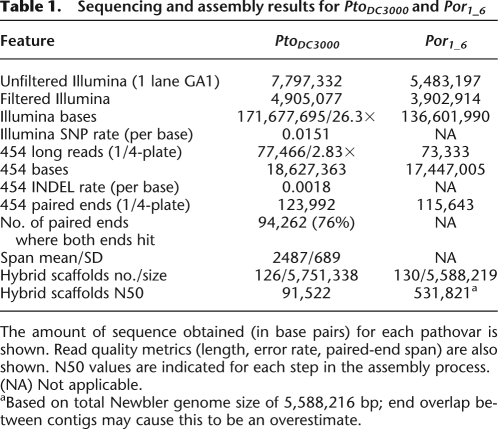
Table 1.
Sequencing and assembly results for PtoDC3000 and Por1_6
The amount of sequence obtained (in base pairs) for each pathovar is shown. Read quality metrics (length, error rate, paired-end span) are also shown. N50 values are indicated for each step in the assembly process.
(NA) Not applicable.
aBased on total Newbler genome size of 5,588,216 bp; end overlap between contigs may cause this to be an overestimate.
Short read sequencing covers the entire PtoDC3000 genome
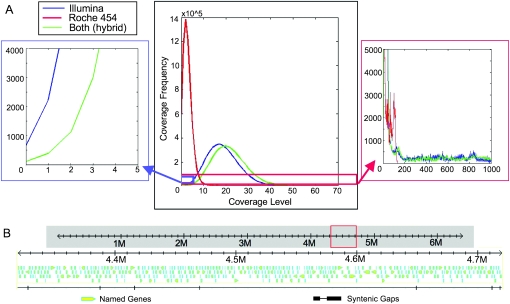
Simple syntenic alignment of the high-quality Illumina GA1 reads to the reference PtoDC3000 sequence revealed that only 656 bp of the reference had zero coverage; alignment of the 454 reads showed 535,820 bp of the reference with no coverage. However, syntenic alignment of the two sequence types together showed that only 107 bp remained unsequenced (Fig. 1A, left inset). These 107 missed bases were broken into 14 short unsequenced regions randomly distributed across the genome (e.g., Fig. 1B). Additionally, there were hundreds of positions in the PtoDC3000 reference genome where far more reads match than would be expected by chance (Fig. 1A, right inset). These likely represent regions of repetitive sequences larger than 35 bp. In these large repeated regions, a single Illumina read cannot simultaneously span the repeated sequence and overlap unique sequence.
Figure 1.
Syntenic reassembly of PtoDC3000 demonstrates that few bases were not sequenced and that the PtoDC3000 genome has multiple stretches of repetitive sequence. (A) Histogram (center) showing the number of bases (×105) of the PtoDC3000 reference genome that are covered by sequence data at a given coverage level (x-axis). One lane of Illumina GA1 sequence, 1/4-plate of 454 sequence, and the combination of both sequence sets are shown. Only 656 bp remain unsequenced from the single lane of Illumina sequence (blue line), whereas 535,820 bp remain unsequenced from the plate of 454 long reads (red line). Combination of both sequence types (green) reduces this to 107 bp (blue box, center, expanded at left). PtoDC3000 has many repetitive regions over 35 bp in length (red box, center, expanded at right). This will cause difficulty in de novo assembly of only Illumina reads, as repeats will break assembly by most methods. However, the longer 454 reads and 454 paired-end reads partially ameliorate this problem. (B) Example of a typical 400-kbp genomic region with ORFs derived from the reference sequence as yellow arrows and unsequenced bases noted as ticks in the syntenic gaps line. The missing bases are scattered randomly and tend to be only one or a few bases in length.
Hybrid assembly of PtoDC3000 improves scaffold size and quality
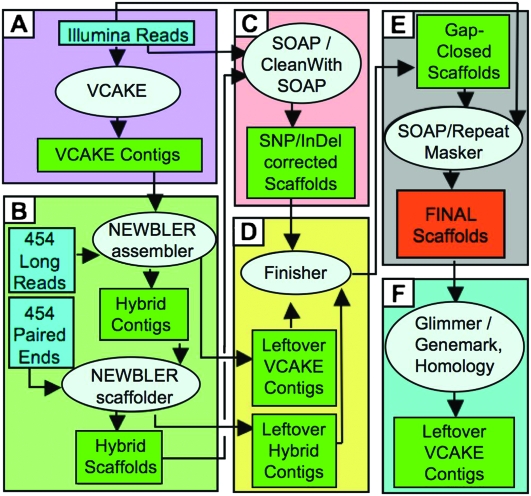
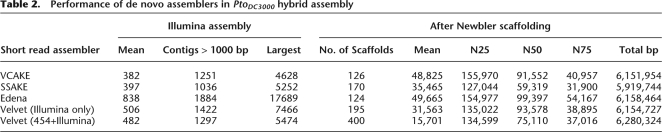
Illumina reads were combined with 454 long reads and paired ends in the hybrid assembly process outlined in Figure 2 (see Methods; Supplemental Fig. 2). First, Illumina reads were extended into longer contigs using the assembler VCAKE v1.5 (Jeck et al. 2007; see Methods; Supplemental Figs. 4, 5). A total of 99.4% of the PtoDC3000 genome was assembled into thousands of small (70–7000 bp) Illumina contigs. VCAKE contigs are of high quality: From 10,095 PtoDC3000 VCAKE contigs over 200 bp, only 3% had errors (determined post hoc by synteny to the reference). This equates to an overall error rate, or false single nucleotide polymorphism (SNP) rate, of 1/9000 bp, similar to Sanger assemblies (Sanger and Coulson 1975). Roche's 454 assembler, Newbler, assembled the 454 long reads and VCAKE Illumina contigs into 1864 hybrid contigs (Fig. 2; see Methods). The Newbler scaffolder (Margulies et al. 2005) then used 454 paired ends to order the PtoDC3000 hybrid contigs into 126 scaffolds (Fig. 2B). Any VCAKE or Newbler-derived contigs not included in the final scaffolds (unscaffolded contigs) were reserved for later use (Fig. 2D). For comparison, we assembled the same data through the same pipeline using three other short read assemblers: Velvet (Zerbino and Birney 2008); Edena (Hernandez et al. 2008); and SSAKE 3.2.1 (R. Warren, G. Sutton, S. Jones, and R. Holt, http://www.bcgsc.ca/platform/bioinfo/software/ssake) (Table 2; see Methods). VCAKE and Edena produced comparable results that were marginally superior to those from Velvet and SSAKE (Table 2). We were therefore satisfied that VCAKE was a suitable tool for initial, de novo assembly.
Figure 2.
Hybrid genome assembly pipeline. (A) Illumina reads are de novo assembled with VCAKE into VCAKE contigs. (B) Newbler then assembles VCAKE contigs and 454 long reads into hybrid contigs. The Newbler scaffolder orients the hybrid contigs into larger hybrid scaffolds using 454 paired-end data. (C) Hybrid scaffolds are cleaned of 1- to 2-bp indels using Illumina read depth; (D) longer gaps within scaffolds are filled with unused VCAKE and hybrid contigs in Finisher. (E) Polymorphism and coverage in the scaffolds are used to identify any putative repeat regions (see also Supplemental Figs. 2, 3). (F) Final scaffolds are used as templates for gene prediction using both homology and gene prediction software (Glimmer and GeneMark). See Methods for a complete description.
Table 2.
Performance of de novo assemblers in PtoDC3000 hybrid assembly
We used BLAST to align our hybrid PtoDC3000 scaffolds to the PtoDC3000 reference genome and to calculate assembly quality metrics. The scaffolds totaled 5,751,338 bp with an N50 size of 91,552 bp and a largest scaffold of 388,624 bp (Table 1; Fig. 3). The PtoDC3000 scaffolds represent a total of 90.6% of the reference genome. Addition of 2816 nonredundant unscaffolded contigs (Fig. 2D and Fig. 3, green line) brought the genome size to 6,924,419 bp. The previously determined genome size of PtoDC3000 including its two plasmids is 6,538,260 bp (Buell et al. 2003). We conclude that there is redundancy in the unscaffolded contigs because the majority (52%) have at least one repeat over 35 bp, and many of them (12%) contain apparent errors relative to the reference. This could lead to redundancy in cases where the hybrid assembler fails to reconcile errors and repeats, and instead outputs several versions of the same sequence. Additionally, short (<18 bp) end overlap between contigs can accumulate and contribute to redundant sequencing data. As Illumina sequencing is known to be poor in AT-rich regions (Hillier et al. 2008), we also calculated the GC content of our final assembly of PtoDC3000 and compared it with the published reference genome. We found that the GC content for our assembly (58.48%) was similar to that of the reference genome (58.39%).
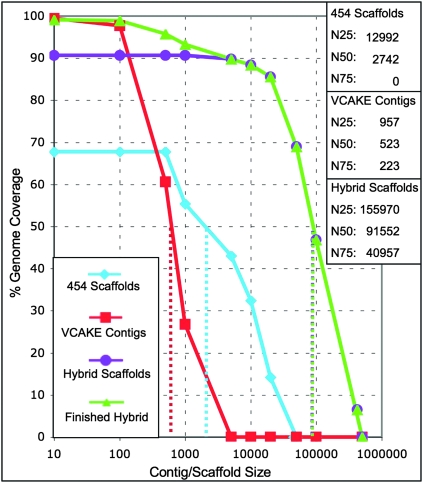
Figure 3.
Hybrid assembly outperforms assembly of either Illumina or 454 data sets alone. We reassembled the PtoDC3000 genome de novo to validate our assembly approach. For a given reassembly, the percentage of the reference PtoDC3000 genome (y-axis) covered by contigs of a given size (x-axis) is shown. The ∼3× coverage 454-only assembly (blue diamonds) reassembles only ∼70% of the PtoDC3000 genome, with most scaffolds in the 1- to 10-kb range. De novo assembly of the ∼26.3× Illumina read coverage only using the VCAKE assembler (red squares) leads to essentially complete genomic coverage, but in small contigs of less than 100 kbp. Hybrid assembly combining both technologies (purple circles) leads to 90% genomic coverage with hybrid contigs over 500 kbp. The subsequent finishing step incorporating unscaffolded contigs (Supplemental Fig. 1) leads to final coverage of 99.1% of the PtoDC3000 reference genome (green triangles). N50 values (dotted lines) increase at each step.
Although some regions of the genome will be over-represented in the final assembly, retaining as much of the potentially coding genome as possible is important for downstream analyses of gene content. Thus, including all unscaffolded contigs, we cover 99.1% of the genome; a total of 54,817 bp of PtoDC3000 remain unassembled. Syntenic comparison showed that these unassembled bases consist of 1102 gaps ranging in size from 1 to 940 bp, with a median size of 33 bp (Fig. 4A, Supplemental data). As demonstrated by synteny (above), these gaps were covered by the raw reads and are thus products of imperfect hybrid assembly. They do not appear in any genomic region preferentially (e.g., Fig. 4B) but do disrupt a few open reading frames (ORFs) because of the high gene density of the PtoDC3000 genome. There were also 20 rearrangements in our PtoDC3000 scaffolds relative to the reference sequence (Table 3; Supplemental data), as a result of imperfect assembly.
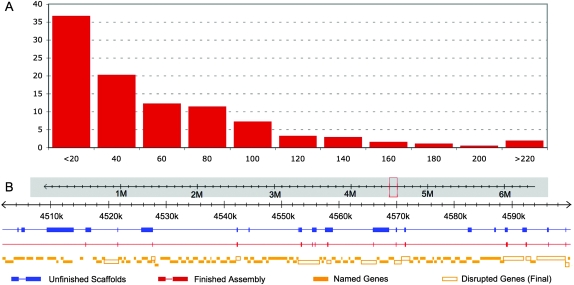
Figure 4.
The final de novo PtoDC3000 hybrid assembly has few, mostly small gaps. (A) Histogram of gaps remaining in the PtoDC3000 hybrid assembly. Less than 1% of the reference genome (55,164 base pairs) remains unassembled into contigs or scaffolds. These bases are from a total of 1102 syntenic gaps with a median size of 55 bp. The largest gap is 987 bp (see also Supplemental data). (B) A typical 100-kbp region of the PtoDC3000 reference genome with ORFs shown (“Named Genes,” solid orange box) exhibits gaps remaining in the reassembled scaffold. “Unfinished Scaffolds” (blue) display the size and distribution of gaps before scaffold gaps are filled using unscaffolded contigs (Fig. 2D). Several large gaps are present. After finishing, most large gaps and many small gaps are eliminated (“Finished Scaffolds,” red). As a result, few (11/115) PtoDC3000 ORFs are disrupted (“Disrupted Genes,” open orange box) in this region.
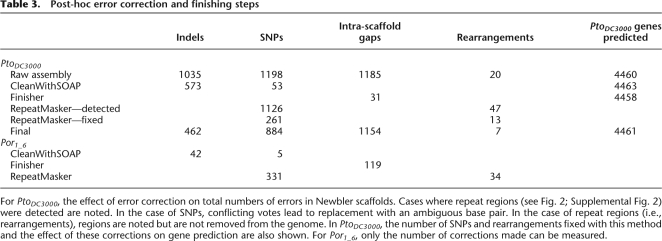
Table 3.
Post-hoc error correction and finishing steps
For PtoDC3000, the effect of error correction on total numbers of errors in Newbler scaffolds. Cases where repeat regions (see Fig. 2; Supplemental Fig. 2) were detected are noted. In the case of SNPs, conflicting votes lead to replacement with an ambiguous base pair. In the case of repeat regions (i.e., rearrangements), regions are noted but are not removed from the genome. In PtoDC3000, the number of SNPs and rearrangements fixed with this method and the effect of these corrections on gene prediction are also shown. For Por1_6, only the number of corrections made can be measured.
Illumina depth-corrects errors in PtoDC3000 scaffolds
We aligned all of our high-quality Illumina sequences to our final scaffolds (modified from McCutcheon and Moran 2007) and used the consensus of aligned reads to correct any false SNPs and small indels that were present in the hybrid assembly (Supplemental Fig. 2C; see Methods). Indel correction is essential, as 454 sequencing is known to add or remove bases around homopolymer stretches (Margulies et al. 2005), resulting in small indels. Using this “depth-of-coverage” approach, we corrected 53 false SNPs and 573 indels in our sequenced and reassembled PtoDC3000. Subsequent realignment of this PtoDC3000 reassembly to the reference genome showed a total of 1145 SNPs and 462 small indels still present in our PtoDC3000 scaffolds, for an overall error rate of 1/3500 (Table 3).
Inspection of our short oligonucleotide alignment program (SOAP) alignment (Li et al. 2008) revealed that many of the remaining SNPs and indels were clustered in small regions. We determined that these regions are duplicated in the PtoDC3000 reference genome and that these duplicate regions had been improperly reassembled by our pipeline. To detect all potential single-nucleotide assembly errors, we developed an automated approach (SOAP/RepeatMasker; Fig. 2E) to note all regions where there was both high polymorphism among the Illumina reads and high coverage (Supplemental Fig. 3). A total of 1126 highly polymorphic bases in these regions were changed to the ambiguous nucleotide code (Cornish-Bowden 1985). This reduced our apparent SNP rate to 1/7000. Additionally, we successfully identified 70 potential repeat regions, including 13 of the 20 known rearrangements. The remaining seven rearrangements appear to be caused by perfect repeats and thus would not be detected using this algorithm. The resulting cleaned and verified 126 final hybrid scaffolds (Table 1) were then ready for ORF prediction and comparison to the ORF predictions from the reference assembly.
The PtoDC3000 de novo hybrid assembly contains all previously known genes and is a suitable substrate for ab initio gene prediction
Using BLAST, we aligned the reference PtoDC3000 ORFs to our hybrid assembly and found that 5476/5476 (100%) of the genes were present. However, ∼20% of ORFs (1133/5476) matched less than 90% of the reference gene length. In many cases, this was due to a single gene “bridging” two or more hybrid scaffolds or contigs. However, 326 of these 1133 partially matched genes were assembled to over 90% of their length after allowing for bridging, leading to a total of 4669/5476 (85%) of the genes identified to over 90% of their length and 5026/5476 (92%) identified to over 80% of their length.
Our ultimate goal is to identify with high fidelity ORFs in genomes for which no reference sequence exists. We used ab initio consensus ORF predictions from Glimmer and GeneMark (see Methods). To determine whether assembly quality would affect gene prediction, we applied our gene prediction pipeline to both the PtoDC3000 reference genome and our PtoDC3000 hybrid assembly. We then compared predicted ORFs to the 5476 curated ORFs in the PtoDC3000 reference genome (Buell et al. 2003) (see also http://pseudomonas-syringae.org/) using BLAST. A total of 5045 ORFs were predicted from the reference genome, and BLAST found 84% (4622/5476) of the curated ORFs among the ab initio ORFs. Our pipeline defined 6100 ORFs from our hybrid assembly of PtoDC3000. Of these, 2076 were partial predictions containing a sequence linking contigs or scaffolds at either the 5′ or 3′ end (see Methods). We found that 84% (4602/5476) of the PtoDC3000 curated ORFs were found among the 6100 ab initio ORFs predicted from our hybrid assembly. If only full-length (no linker) ab initio ORFs were compared with the curated ORFs, then 71% of the curated ORFs (3888/5476) were found. This percentage changed only slightly when gene prediction was performed after each finishing step (Table 3). Hence, the effectiveness of our ORF calling pipeline was not substantially different if we used our PtoDC3000 hybrid assembly or the corresponding reference assembly.
Significantly more of the ORFs predicted in our PtoDC3000 hybrid assembly were truncated on their 3′ end (750) compared with those ORFs predicted from the reference genome (46). This is partly due to cases where the 3′ end of a gene is represented in a second contig or scaffold as described above. Given these data, our conservative Glimmer and GeneMark pipeline is expected to predict ab initio ∼84% of novel genes in newly sequenced strains, like Por1_6 (see below).
Partial assembly of PtoDC3000 plasmids
In addition to the ∼6.4-Mb circular chromosome, PtoDC3000 has two plasmids (pDC3000A: NC_004633.1, size = 73.6 kb; pDC3000B: NC_004632.1, size = 67.5 kb). Both were assembled to near completion (98.9% for pDC3000A and 98.5% for pDC3000B). However, pDC3000A and pDC3000B were more fragmentary and had far more putative errors (1.5% and 4.0%, respectively) than the chromosome. This is due to large regions of homology between the two plasmids and portions of the chromosome that confound hybrid assembly. For example, Scaffold_00010 of the PtoDC3000 hybrid assembly is a “mosaic” assembly of large portions of both plasmids, hitting neither plasmid perfectly over its full length. This is unsurprising, as we anticipated that assembly of large repeat regions using only short reads would be a problem. Although the plasmids were assembled poorly compared with the main chromosome of PtoDC3000, the genes that are unique to the plasmids were assembled reasonably well; all 135 plasmid-borne genes were found in our PtoDC3000 hybrid assembly, and 103 were assembled over 90% or more of their full length. The error rate in these genes (0.8%) was lower than the remainder of the plasmid sequence, suggesting that most of the error was limited to nongenic repeated regions.
Extensive structural divergence between the Por1_6 genome and those of other P. syringae pathovars means that the Por1_6 genome must be assembled de novo
Sequence similarity, gene content, and gene order differ dramatically among P. syringae pathovar strains. The three sequenced reference genomes of P. syringae share less than 75% of their genes (Feil et al. 2005; Joardar et al. 2005). Further, multilocus sequence typing (MLST) analysis (Hwang et al. 2005) suggests that Por1_6 is likely to be highly diverged from any of the three sequenced strains, as it is a member of a fourth phylogenetic clade. We estimated the divergence between our Por1_6 Illumina reads and the PtoDC3000 reference genome using SOAP (Li et al. 2008). SOAP requires that the read match across its entire length and with a minimum number of errors (we allowed up to five SNPs or one indel in 35 bp). From 3,902,914 Por1_6 Illumina reads (Table 1), only 1,615,055 (41%) matched the PtoDC3000 reference genome under these conditions. Moreover, the error rate among even these 1,615,055 matching reads is 7.7%.
In bacteria, assembly solely based on synteny, or reference guided assembly (Pop et al. 2004), will not effectively assemble regions that have undergone significant recombination (Yan et al. 2008) or complex rearrangements, will miss novel horizontally transferred regions, and will struggle to assemble deletions and duplications properly. As these genomic changes are likely to be associated with host specialization and virulence in the pathovars of P. syringae, it is essential that they be assembled effectively de novo.
The Por1_6 genome was de novo assembled into lower-error, longer scaffolds than PtoDC3000
For the genome of Por1_6, we generated 3,902,914 Illumina reads of 36 bp (trimmed to 35 bp) in length, 73,333 long 454 reads averaging 237 bp in length, and 115,643 454 paired ends (each from 1/4-plate runs; Table 1). Hybrid assembly was as described for PtoDC3000. The 130 resulting scaffolds totaled 5,588,219 bp. We used PCR and Sanger sequencing to confirm the orientation of 13 contigs (Supplemental Table 1, “gaps”), lending considerable confidence to the hybrid assembly. After addition of 2002 unscaffolded contigs, the total length was increased to 6,718,951 bp. Based on the PtoDC3000 results, we estimate that the true Por1_6 genome size lies between these values. To estimate N50 values, we considered the conservative total scaffold size of 5,588,219 bp to be the apparent genome size and simply summed the length of the scaffolds from largest to smallest until 50% of apparent genome size was reached (Fig. 5). By these estimates, the Por1_6 assembly has a largest scaffold of 794,194 bp, with 87% of the genome assembled into just 14 contigs all over 100,000 bp and an N50 of 531,821 bp (Fig. 5, pink; Table 1). As a control, we performed the same analysis on PtoDC3000 (Fig. 5, violet) and obtained similar results to those noted above (Fig. 3). The GC content of the Por1_6 assembly is 57.76%, about 0.5% less than PtoDC3000.
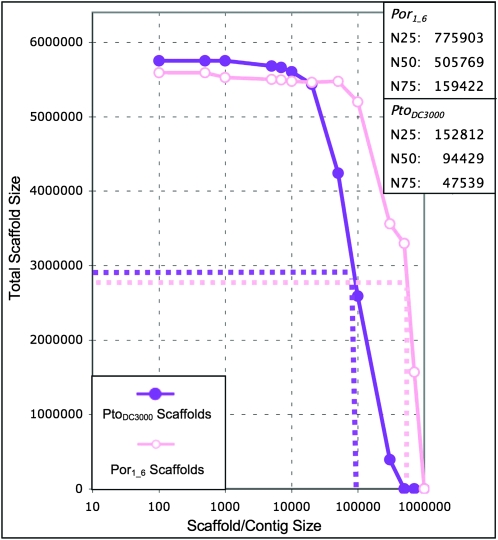
Figure 5.
The Por1_6 genome assembled into contigs larger than 700,000 bp. Our hybrid assembly of Por1_6 is better than our hybrid assembly of PtoDC3000. The Por1_6 scaffolds (open pink circles) are significantly larger on average than the PtoDC3000 scaffolds (solid purple circles). The estimated N50 for Por1_6 is over 500,000 bp, with some scaffolds longer than 700,000 bp. N25, N50, and N75 values are estimated for Por1_6 based on total scaffold size (for Por1_6, 5,588,216 bp) rather than percentage of the genome covered as in Figure 3. This method is accurate: The PtoDC3000 N25, N50, and N75 values using this method are comparable to those derived from our syntenic approach (see Fig. 3).
We applied our “depth-of-coverage” approach for error correction to Por1_6 (Methods). A total of 5 false SNPs and 42 indels were corrected in the Por1_6 scaffolds using Illumina depth, and 119 intra-scaffold contig gaps were closed (Table 2). Additionally, 34 problematic repeat regions in our hybrid assembly of Por1_6 were identified, and 331 polymorphic bases were replaced with the ambiguous nucleotide code (Cornish-Bowden 1985) (see Methods). Because there is no reference genome for Por1_6, we cannot calculate an overall error rate. However, because there were fewer errors to correct in the scaffolds and fewer repeat regions, our Por1_6 hybrid assembly likely has an error rate less than or equal to our hybrid assembly of PtoDC3000.
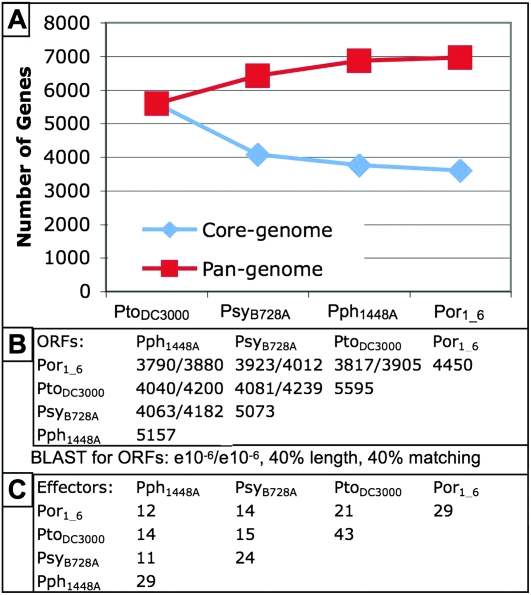
The Por1_6 genome redefines the core genome and pan-genome of P. syringae
We defined a core genome from the three reference P. syringae sequences and aligned the 3755 ORFs shared by them to our Por1_6 hybrid assembly (see Methods). A total of 3594/3755 of the core genes were present in Por1_6 hybrid assembly, indicating that up to 161 of the genes previously thought to be part of the P. syringae core genome are not (Fig. 6A). We developed an algorithm to confirm these gene losses by verifying the presence of flanking genes from PtoDC3000 (Methods). For 102/161 of the missing genes, both flanking genes (defined as the nearest neighbors to the missing gene in PtoDC3000) were on the same scaffold. Of these, only 39 contained one or more “linker” sequences, indicating that the missing genes could be present in Por1_6 but not assembled. We checked the assembly accuracy of 13 sites with flanking genes on the same scaffold using PCR and Sanger sequencing. We obtained a product for 12/13 sequences and all 12 closely matched the Por1_6 assembly, including four cases where a linker was bridged (Supplemental Table 1). We therefore concluded that our core-genome estimates are sound and that the core genome of P. syringae, as we define it (see Methods) currently contains 3594 genes (Fig. 6B). This number should continue to decrease with sequencing of additional P. syringae isolates.
Figure 6.
Por1_6 genome redefines the P. syringae pan-genome and type III effectorome. (A) The Por1_6 genome adds 97 unique genes to the pan-(distributed) genome of P. syringae (red boxes) and reduces the core (overlapping) genome by 161 genes (blue diamonds). (B) Por1_6 shares about 4000 of its 4450 predicted ORFs with each of the other three fully sequenced P. syringae strains. Shown are predictions based on a BLAST homology (E < 10−6). (C) Por1_6 contains a unique combination of type III effector proteins compared with the three previously sequenced strains. Type III effector proteins in Por1_6 were predicted using both homology with the known type III effectors in the other three fully sequenced P. syringae strains and gene prediction software (Methods). Note that the presence of a given type III effector gene, or homolog, does not necessarily imply that it is translocated (Chang et al. 2005; Schechter et al. 2006).
We annotated Por1_6 using a combination of homology and ab initio gene prediction. First, we used TBLASTX to align all known genes from the three previously sequenced P. syringae strains to the Por1_6 scaffolds. Matching sequences were defined as putative ORFs. Then we used Glimmer and GeneMark to define ORFs ab initio. After removing redundant predictions, we found that Por1_6 has 4450 predicted full-length ORFs. Another 2895 ORFs are only partial in length because a contig ends in the middle of the gene. Some of these ORFs may be artifacts, while others may be true genes. This compares to the 5,157, 5073 and 5476 total ORFs predicted from the reference sequences of Pph1448A, PsyB728A, and PtoDC3000 genomes, respectively (Buell et al. 2003; Feil et al. 2005; Joardar et al. 2005).
To determine the total gene content (the pan-genome) of the four sequenced P. syringae pathovars, we used TBLASTX to iteratively compare the ORFs of each pathovar (Methods). We chose to use only the 4450 full-length predicted Por1_6 ORFs for this analysis. 97 genes were found to be unique to Por1_6 among the sequenced P. syringae strains. Sixty-six of these were truly novel hypothetical genes (e.g., no BLAST hit in GenBank at E = 10−5), and 31 others matched either phage or cellular proteins from other microbial species. Addition of these 97 unique Por1_6 ORFs to the other three sequenced P. syringae strains brings the size of the pan-genome to 6965 (Fig. 6). We anticipate that the pan-genome will continue to grow with the sequencing of additional genomes of P. syringae.
Por1_6 has a unique type III effector complement
Type III effector proteins are a major determinant of P. syringae virulence. These proteins are delivered through the type III pilus into plant host cells and generally work to disrupt plant immunity. Hence, they are collectively candidate virulence factors (Mudgett 2005; Grant et al. 2006; Stavrinides et al. 2008). Unsurprisingly, the plant immune system has evolved to monitor the plant cell for the presence of these type III effectors, in many cases by monitoring the integrity of their host cellular target(s) (Chisholm et al. 2006; Jones and Dangl 2006). We used BLAST (E = 10−6) to discover which of the validated type III effector proteins from the three sequenced P. syringae strains and from literature-based curation (list from http://pseudomonas-syringae.org/) are present in Por1_6. We found 34 sequences with homology with known type III effectors (Supplemental Table 2). Five of these appear to be pseudogenes (Ma et al. 2006). The HopI1 and HopAF1 homologs contain frameshifts leading to premature stop codons, while premature stop codons in HopZ1, AvrE1, and HopH1 homologs are due to point mutations. Of the remaining 29, 26 matched a predicted Por1_6 ORF at essentially full length. Three others matched an ORF but appeared truncated. Alignment of Por1_6 scaffold sequences to type III effector homologs revealed that these genes were present at full length without frameshifts but hit across a “linker” sequence. Hence, Por1_6 shares an overlapping, but unique, complement of at least 29 apparently functional type III effector genes with the other sequenced P. syringae strains (Fig. 6C). It is of course likely that there are additional, novel, type III effector genes present in this strain that await discovery by other means.
Por1_6 likely harbors an integrated plasmid
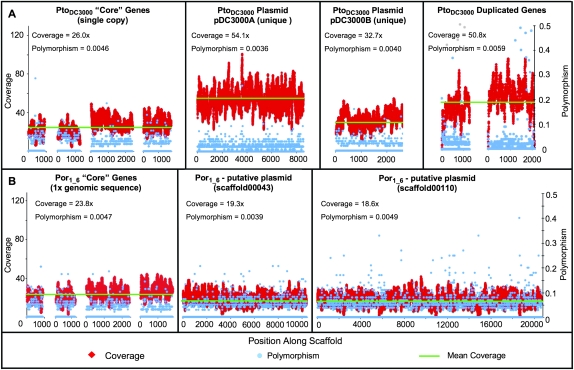
To determine whether Por1_6 had one or more plasmids, we used BLAST to search the genome for genes found within P. syringae plasmids. Multiple plasmid genes were present in our Por1_6 assembly, and these were found mainly within scaffolds 43 and 100 (Fig. 7A). Upon further inspection, these two scaffolds contain significant sequence similarity to the known P. syringae plasmid pPSR1 (Sundin et al. 2004). If these two scaffolds were part of one or more plasmids, we would expect coverage levels within the assembly to be significantly higher than the rest of the genome. For instance, in our PtoDC3000 hybrid assembly unique portions of each of the two plasmids are found at higher levels of coverage than that of four single-copy genes commonly used in MLST studies but at similar levels of coverage as two duplicated genes in PtoDC3000 (Fig. 7A). However, the coverage of scaffolds 43 and 100 within our Por1_6 assembly is no different than that of the MLST genes in its own genome. Although we cannot rule out the presence of a low copy number plasmid, it is likely that Por1_6 contains a plasmid integrated into the main genome, as noted for other P. syringae plasmids (Jackson et al. 2000; Rohmer et al. 2003; Pitman et al. 2005).
Figure 7.
Por1_6 likely has an integrated plasmid. We surveyed all Por1_6 scaffolds for sequences similar to known P. syringae plasmid genes. Por1_6 scaffolds 34 and 110 showed significant levels of similarity to a known plasmid, pPSR1 (Sundin et al. 2004). Coverage levels within the raw reads were determined for these scaffolds as well as for unique portions of each PtoDC3000 plasmid and unique genomic regions from both Por1_6 and PtoDC3000. (red diamonds). Four single-copy chromosomal genes commonly used in MLST studies (rpoD, fruK, gyrB, gltA) were chosen from PtoDC3000 and Por1_6 as controls for chromosome-level coverage, and two duplicated genes (ilvA2 and PSPTO_0034) were randomly chosen as positive controls for duplicated regions. Coverage levels for sequences from both PtoDC3000 plasmids were consistently higher than those for the single-copy PtoDC3000 chromosomal sequences (A). Hence, there are multiple copies of each plasmid present within a cell relative to the chromosome. In contrast, the coverage levels of Por1_6 scaffolds 43 and 110 were very similar to that of the four control genes (B), implying that these regions are present on a very low copy number plasmid or have been integrated into the Por1_6 chromosome.
Discussion
We have shown that de novo hybrid assembly of an ∼6.5-Mb bacterial genome is possible using low-coverage, high-throughput sequencing, without the use of a reference. We validated that our de novo hybrid assembly method produces high-quality assemblies by sequencing and reassembling a standard research strain PtoDC3000 (Fig. 4; Table 2). We then measured error rates and genomic coverage of our hybrid assembly compared with the reference sequence of PtoDC3000. By these standards, our hybrid assembly pipeline is effective and is as useful as other currently available methods for de novo assembly of short read DNA sequence (Table 2).
We used the hybrid assembly method on short read sequences from a strain of P. syringae, Por1_6, originally isolated as a causal agent of disease on rice. Our assembly of Por1_6 contains many scaffolds longer than 500,000 bp (Fig. 5; Table 1) and contained 4450 predicted genes (Fig. 6). This is the first sequenced P. syringae strain isolated from monocots, and it is highly diverged from any of the three previously sequenced strains (estimated 10% nucleotide divergence and ∼85% of predicted ORFs shared with PtoDC3000). The Por1_6 genome contains unique genes (97; Fig. 6) with respect to the other sequenced P. syringae strains and a unique combination of type III effector virulence proteins. The low cost of this method (<$6000 per bacterial genome) makes whole-genome de novo sequencing and assembly economical and will open the door for sequencing and analyses of many more strains of this, and other, bacterial pathogens of plants and animals.
De novo bacterial genome assembly
Previous attempts to de novo assemble bacterial genomes with short read data have been of two kinds: (1) either extremely high levels of coverage were used (Dohm et al. 2007; Hogg et al. 2007) or (2) lower Illumina coverage was used, but assembly of bacterial genomes resulted in thousands of short contigs (Jeck et al. 2007; Warren et al. 2007; Chaisson and Pevzner 2008; Hernandez et al. 2008; Zerbino and Birney 2008). The high levels of coverage required for efficient assemblies using these algorithms significantly reduce the per base cost savings of either 454 or Illumina sequencing technologies. For example, Dohm et al. (2007) used the Illumina GA1 instrument to sequence a Helicobacter acinonychis genome to a depth of ∼285× before de novo assembly. This greatly reduces the 1:100 per base pair cost-advantage of Illumina over Sanger sequencing (Mardis 2008). Likewise, high-coverage 454 sequencing (30×) can produce useful assemblies (Hogg et al. 2007) but also reduces the cost savings of pyrosequencing compared with Sanger. Even with such high coverage, these assemblies are still more fragmentary and error-rich than traditional Sanger sequences.
Low-coverage Illumina assemblies requiring less sequence data have been attempted; however, the resulting assemblies are fragmentary and have limited application. For example, Hernandez et al. (2008) sequenced Staphylococcus aureus strain MW2 to ∼48× coverage using the Illumina GA1 instrument and produced about 1000 contigs with an N50 of 6 kb using their assembler Edena (as well as several others).
We resolve these problems by combining light coverage of both types of short read data with assembly algorithms suited to handling such data. Our hybrid assembly method is successful because the two sequencing technologies overcome each other's shortcomings. Short indels present in 454 data are problematic for accurate assembly. Because these errors are rare in Illumina sequencing, we used Illumina data during and after assembly to correct them. 454 reads (in this case of ∼250 bp, but soon to be >400 bp) are much longer than current Illumina reads (36 bp), so the addition of light coverage of 454 reads bridges short repeats without difficulty. Finally, longer repeats are bridged with the use of 454 paired ends, condensing several thousand contigs into roughly one hundred scaffolds. Final assembly resulted in 126 scaffolds with an N50 of 91 kb for PtoDC3000 and 130 scaffolds with an estimated N50 of 523 kb for Por1_6. By these estimates, our method assembles the largest contigs yet reported for minimal cost (∼$634/Mbp in the final assembly).
Using de novo hybrid assembly of PtoDC3000 as a metric, we demonstrate that our error rates are low and reasonable (1/7000; Table 2) and that the genome is nearly complete (99.1% genome assembled, 5476 of 5476 genes present). The less than 1% of unsequenced bases are in small gaps, all under 1 kb in length (Fig. 4). Based on the observation that our Por1_6 hybrid assembly is, by several metrics, of higher quality than our PtoDC3000 hybrid assembly (Fig. 5) with fewer errors corrected (Table 2), we are confident that the Por1_6 genome is at least as well assembled as our PtoDC3000.
We believe the higher quality of the Por1_6 assembly results from better quality 454 sequence data or the presence of fewer repeats in this strain. For instance, PtoDC3000 has two plasmids whereas Por1_6 appears to have none (Fig. 7).
The genome of P. syringae pv. oryzae 1_6
The sequencing and assembly of the first three pathovars of P. syringae has led to greater understanding of plant bacterial pathogenicity than would otherwise be possible without whole sequenced genomes. Additionally, because P. syringae infects the tractable model plant Arabidopsis thaliana, it is an attractive model for bacterial pathogenesis in mammals. Its close relative P. aeruginosa mostly causes opportunistic infections in humans, though some strains are broad host and also infect plants. Not surprisingly, these strains share many genes (Joardar et al. 2005).
Addition of the Por1_6 genome brings the number of P. syringae clades with a member sequenced to four out of five (Hwang et al. 2005). We found that Por1_6 contains homologs for 29 of the previously identified type III effector proteins and that some but not all of these are present in one or more or the previously sequenced P. syringae genomes (Fig. 6). A total of 4450 full-length proteins and an additional 2895 truncated genes were predicted in the Por1_6 genome using both homology and gene prediction software. We used the predicted ORFs to curate a new core genome for the four sequenced strains of P. syringae, reducing the number of genes shared between them, the core genome, to 3594 (Fig. 6). Deeper comparative analysis of the Por1_6 genome with the other three sequenced strains (to be published elsewhere) will potentially give insight into what allows this clade to infect monocots.
Conclusions
We developed and implemented a de novo hybrid assembly pipeline that can generate nearly complete genome sequence for diverse bacterial isolates. To show the feasibility of this method, we sequenced and reassembled an isolate of P. syringae with a reference genome sequence. Metrics for this analysis proved useful as benchmarks for de novo hybrid assembly of a high-quality draft genome sequence of another, distantly related P. syringae isolate. Our approach greatly reduces the cost of bacterial genome sequencing and provides high-quality sequence that can be used for phylogenetic and evolutionary comparisons.
Methods
DNA preparation and sequencing
Frozen cultures of PtoDC3000 and Por1_6 were received from the Dangl laboratory and Ministry of Agriculture, Fisheries and Forestry, Japan (MAFF) (no. 311107), respectively. For each of the sequencing runs, bacteria underwent a small number of passages on agar plates, and then a single colony was picked and used to produce a 250-mL culture. Genomic PtoDC3000 DNA was prepared at two separate times so that sequences from 454 and Illumina sequencing runs were the product of two separate colonies. DNA was isolated and prepared for 454 long read and paired-end sequencing following standard protocols (Roche). DNA was prepared from a separate culture for sequencing on the Illumina GA1 sequencer using standard Illumina protocols.
A single lane of the UNC-CH Illumina GA1 sequencer was loaded with the DNA from each of the two strains. The Illumina GA1 sequencer was run with 36 cycles using the standard flow cell. Raw Illumina GA1 sequence image data were phased and filtered for quality using default GERALD parameters for unaligned reads (analysis: NONE, Use_Bases: 35). For 454 sequencing, a 1/4-plate of long reads and a 1/4-plate of paired ends was loaded and sequenced for each pathovar. 454 sequencing was performed by Roche/454.
Sequence quality control
PtoDC3000 Illumina sequences were aligned to the reference genome using SOAP. The SOAP results were restricted to the best hit for a given sequence (−r 1) and allowing up to five SNPs or one indel (−v 5, −g 1, −e 3). We used these alignments with custom Perl scripts to estimate error rates in the sequencing data (Table 1). The Perl scripts estimate error rates by counting the number of mismatches within a hit. We used BLAST to estimate the span of the PtoDC3000 454 Paired ends (Supplemental Fig. 1) and error rates for PtoDC3000 454 long reads (Table 1). For paired ends, BLAST was run with basic parameters and then parsed to only include pairs that had a span shorter than the upper limit of 7000 bp and only had unambiguous partners. For long reads, results were restricted to the best hit for a given sequence (−b 1), and errors were counted using custom Perl scripts.
Por1_6 raw Illumina sequences were also aligned with SOAP to the PtoDC3000 genome to estimate the divergence of the Por1_6 genome. The same “error analysis” was run as described above, although in this case errors were presumed to be bases that had diverged from PtoDC3000, rather than sequencing errors.
Hybrid assembly overview
Filtered Illumina GA1 data were input into the de novo short read assembler VCAKE v. 1.5. VCAKE is designed and optimized for low-coverage assemblies of Illumina data because it makes use of reads that include errors and will assemble at local coverage as low as 2×, provided both reads agree. We first did an analysis to determine whether our 26.3× coverage would be sufficient for proper assembly (Supplemental Fig. 5A). We found that the genomic coverage by VCAKE contigs had plateaued by 25×, although the size of the contigs appeared to still be increasing.
We next reran the assembly of the reference PtoDC3000 genome under several different parameters to determine the set that maximized error-free assembly (Supplemental Fig. 6). After this analysis we ran VCAKE v. 1.5 (http://sourceforge.net/projects/vcake) (Jeck et al. 2007) using the following parameters: –k 35 –n 20 –m 18 –v 3 –x 35 –q T –u T –i T. A second VCAKE run was done using the following parameters: –k 35 –n 20 –m 18 –v 10 –y 10 –q T –u T –i T (Fig. 2A). The first parameter set is designed to build Illumina contigs from nonrepetitive sequences by preventing assembly of regions where Illumina read depth is above 35× (-x 35). The second run is designed to only build Illumina contigs from repetitive sequences, by preventing assembly of regions where Illumina read depth is below 10× (-y 10). Appropriate –x and –y parameters were determined empirically by rerunning assembly of the known genome of PtoDC3000 (data not shown). Both runs together capture and assemble most repeat classes appropriately (see Supplemental Fig. 2A) (Jeck et al. 2007).
VCAKE sequence and quality output results from both parameter sets were combined, and contigs shorter than 70 bp in length were removed. The VCAKE contigs were input into Roche's 454 assembler (Newbler) along with 454 long reads and 454 paired-end reads (Fig. 2B). Newbler was run using standard parameters (40-bp overlap, 90% identity, 2500-bp paired-end span; Supplemental Fig. 1). The scaffolds formed by Newbler in this step form the bulk of our assembly and are termed “hybrid contigs.” Hybrid contig assembly is a two-step process—first, 454 reads and VCAKE contigs are assembled by sequence overlap into longer contigs, then these are ordered into scaffolds using 454 paired ends. After Newbler assembly, any remaining VCAKE or hybrid contigs that were not included in the final scaffolding step (Fig. 2B) were identified (by virtue of failing to align completely with any of the final scaffolds) and set aside. Redundant contigs were removed from this contig set using a custom program and the alignment program BLAT (Kent 2002). These unscaffolded contigs are then used in subsequent clean-up steps (Fig. 2D) and are also included as part of the final assembly.
For comparison, we ran our assembly pipeline, substituting three other short read assemblers (Velvet, Edena, and SSAKE v3.2.1) for the initial Illumina assembly step. We ran under standard parameters for all three programs. We also attempted de novo assembly loading both Illumina and 454 long reads into Velvet, but the results were poor compared to Velvet's assembly using Illumina sequences alone. Each de novo Illumina assembly was then assembled further using 454 reads and 454 paired ends as above for VCAKE. Although initial assembly for the three other assemblers yielded longer and fewer contigs than did VCAKE, the number and length of final scaffolds were comparable between VCAKE, Velvet, and Edena (Table 2). We are therefore confident that VCAKE is an appropriate tool to use in our assembly pipeline.
Assembly quality control in PtoDC3000
After each step of the PtoDC3000 assembly process, we were able to perform quality control using BLASTN (E = 10−8, and post-hoc removal of redundant hits) to align sequences to the reference genome. We calculated errors (Table 2) and estimated the quality of that assembly (contig/scaffold length, N50, and missing sequences; see Figs. 3, 4). This process was essential for assessing the initial performance and optimizing our assembly approach. Obviously, we cannot use these metrics for de novo genome assembly of an unknown genome. However, careful monitoring of the assembly process was essential for our method validation using PtoDC3000.
De novo error correction using Illumina reads
We piled our 4,905,077 Illumina GA1 reads onto our assembled scaffolds to correct any residual false SNPs or small indels in our assembly (Fig. 2C; Supplemental Fig. 2C). We combined the alignment program SOAP (Li et al. 2008) (parameters –g 2 –e 3 –v 3 –r 2) with custom Perl scripts to identify and correct small indels and SNPs in the final scaffolds. In this process, putative errors were verified and corrected by consensus of all overlapping reads in the region, based on the SOAP alignment data. We also determined that 25× coverage was enough to correct as many indels as were likely to be corrected using this method (Supplemental Fig. 4B).
Next, we used Illumina sequence depth to identify potentially duplicated (or triplicated) regions. We used the SOAP alignment feature (Li et al. 2008) to identify regions in our assembled scaffolds with consistently two- or threefold more reads than the genome average and an excess of high-frequency polymorphic bases—bases with more or less equal numbers of reads “voting” for two or more different bases at that position (Figs. 2E, 7; Supplemental Fig. 3). These regions likely define short stretches of truly duplicated sequence. We used coverage to determine where the boundaries of a repeat region were (Supplemental Fig. 3). We then marked these regions as potential repeats (repeat masking) and replaced potentially polymorphic bases with the degenerate genetic code (Cornish-Bowden 1985).
Scaffold finishing
After error correction, we closed gaps between contigs within the final scaffolds using a combination of the alignment program BLAT and a custom program (Supplemental Fig. 2D; Table 3, intra-scaffold gaps). Newbler represents such gaps as strings of N's. We determined whether any of these gaps could be filled in using the unscaffolded (and hence, as yet unassigned) contigs. If an unscaffolded contig overlapped with the sequences directly before and after the gap, we replaced the string of N's with the sequence from the matching contig. We also allowed the 3′ and 5′ end of a gap to simply match one another—in these cases, the two ends were fused to remove the gap.
Genome annotation
Sequence homology can only find genes similar to those already known in other organisms and thus misses all novel or heretofore unknown genes. A variety of tools for ab initio gene prediction are available. We used Glimmer (Delcher et al. 2007) and GeneMark (Borodovsky et al. 2003; Besemer and Borodovsky 2005), the tools first used to annotate the previous P. syringae genomes. Glimmer and GeneMark were trained and parameterized on the published PtoDC3000 genome before use on our draft PtoDC3000 and Por1_6 assemblies. Following Dohm et al. (2007) to predict ORFs with Glimmer and GeneMark in our assemblies, we substituted a sequence with a start and stop codon in each frame (NNNNNCATTCCATTCATTAATTAATTAATGAATGAATGNNNNN) everywhere there was a break between contigs. This allows the programs to predict partial ORFs even in cases where a contig begins or ends in the middle of a gene. We used the most conservative (shortest) ORF predicted between Glimmer and GeneMark as these programs are known to overpredict gene number and length (Guigo et al. 2000; Reese et al. 2000; Brent 2007).
For Por1_6, we also used TBLASTX (E = 10−6) to align all proteins from the three sequenced strains of P. syringae to the Por1_6 scaffolds. Complete hits were annotated as putative ORFs. Cases where genes were truncated due to hitting the end of a contig were annotated as partial, putative ORFs. We then removed redundant hits between the homology and ab initio predictions to form our final gene annotation. The homology-based and ab initio predictions were pooled, and redundant genes were removed using a custom program and the alignment program BLAT (Kent 2002).
Defining the core genome and pan-genome
We redefined the core genome of the four sequenced strains of P. syringae using an iterative TBLASTX (E = 10−6, 40% homology, 40% length hit) strategy. Beginning with the ORFs of PtoDC3000 (Buell et al. 2003), we performed TBLASTX versus the PsyB728A genome to determine which genes were not present in PsyB728A. We took the shared genes and performed TBLASTX versus the Pph1448A (Joardar et al. 2005) genome and again removed any genes that were not present in Pph1448A. Finally, we took the remaining (core) genes and performed TBLASTX versus our Por1_6 scaffolds and contigs. Because of small gaps in our assembly, we called genes as present if they hit multiple, adjacent sequences for a total of 40% or more of the gene's original length. To determine whether poor assembly could be causing genes to be called as missing when they were actually present, we located the nearest neighbors of each gene (flanking genes) from the PtoDC3000 genome and found their location in our Por1_6 assembly using TBLASTX. The location of the flanking genes was characterized as (1) on separate scaffolds, (2) on same scaffold separated by a linker, or (3) on same scaffold and not separated by a linker. We chose 13 from cases 2 and 3 to perform PCR and Sanger sequencing to validate the quality of the assembly between flanking sequences.
To determine the total gene content (the pan-genome) of the four sequenced P. syringae strains, we again used an iterative TBLASTX (E = 10−6, 40% homology, 40% length hit) strategy. Beginning with the predicted ORFs in PtoDC3000 (Buell et al. 2003), we used TBLASTX to determine which genes in PsyB728A (Feil et al. 2005) were not present in PtoDC3000. These genes were combined with the PtoDC3000 genes, and this file was compared with all the ORFs in Pph1448A (Joardar et al. 2005) with TBLASTX. The genes from Pph1448A that were not present were added to the previous file to generate the pan-genome, and this was compared with the ORFs in Por1_6. Additional, complete ORFs predicted by the Glimmer/GeneMark pipeline in Por1_6 were added to the new pan-genome as new genes.
Acknowledgments
Funding was provided by NIH grant GM066025 (J.L.D.), NIH NRSA Ruth Kirchstein Award GM082279-01 (D.B.), and a UNC Junior Faculty Development Award and funds from the Carolina Center for Genome Sciences (C.D.J.). The University of North Carolina at Chapel Hill's Office of the Vice Chancellor for Research and Economic Development provided support for open-access publication. We thank Dr. Piotr Miecskowski, Hemant Kelkar, and Jesse Walsh of the UNC-CH HTS core for technical assistance, and Chris Willet and Chuck Perou of UNC-CH for use of equipment. We thank Nassib Nassar and Charles Schmidt for the prerelease version of VCAKE 1.5. We also thank Steven Salzberg and Mihai Pop (University of Maryland), Vincent Magrini and Elaine R. Mardis (Genome Sequencing Center, Washington University of St. Louis), and Jim Carrington and Chris Sullivan (Oregon State University) for useful discussions.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data for the Por1_6 genome have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession no. ABZR00000000.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083311.108.
References
- Bender C.L., Stone H.E., Cooksley D.A. Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol. Mol. Plant Pathol. 1987;30:273–283. [Google Scholar]
- Besemer J., Borodovsky M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M., Mills R., Besemer J., Lomsadze A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr. Protoc. Bioinformatics. 2003 doi: 10.1002/0471250953.bi0405s01. [DOI] [PubMed] [Google Scholar]
- Brent M.R. How does eukaryotic gene prediction work? Nat. Biotechnol. 2007;25:883–885. doi: 10.1038/nbt0807-883. [DOI] [PubMed] [Google Scholar]
- Buell C.R., Joardar V., Lindeberg M., Selengut J., Paulsen I.T., Gwinn M.L., Dodson R.J., Deboy R.T., Durkin A.S., Kolonay J.F., et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J., Pevzner P.A. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.H., Urbach J.M., Law T.F., Arnold L.W., Hu A., Gombar S., Grant S.R., Ausubel F.M., Dangl J.L. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. 2005;102:2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Kim Y.C., Jung Y.C., Xuan Z., Dworkin G., Zhang Y., Zhang M.Q., Wang S.M. Scanning the human genome at kilobase resolution. Genome Res. 2008;18:751–762. doi: 10.1101/gr.068304.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: Recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J.C., Lottaz C., Borodina T., Himmelbauer H. SHARCGS, a fast and highly accurate short-read assembly algorithm for de novo genomic sequencing. Genome Res. 2007;17:1697–1706. doi: 10.1101/gr.6435207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil H., Feil W.S., Chain P., Larimer F., DiBartolo G., Copeland A., Lykidis A., Trong S., Nolan M., Goltsman E., et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg S.M., Johnson J., Busam D., Feldblyum T., Ferriera S., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.R., Fisher E.J., Chang J.H., Mole B.M., Dangl J.L. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- Guigo R., Agarwal P., Abril J.F., Burset M., Fickett J.W. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 2000;10:1631–1642. doi: 10.1101/gr.122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J., Kaper J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- He S.Y., Nomura K., Whittam T.S. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Hernandez D., Francois P., Farinelli L., Osteras M., Schrenzel J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L.W., Marth G.T., Quinlan A.R., Dooling D., Fewell G., Barnett D., Fox P., Glasscock J.I., Hickenbotham M., Huang W., et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- Hogg J.S., Hu F.Z., Janto B., Boissy R., Hayes J., Keefe R., Post J.C., Ehrlich G.D. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 2007;8:R103. doi: 10.1186/gb-2007-8-6-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T.V., Dahlbeck D., Staskawicz B.J. Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- Hwang M.S., Morgan R.L., Sarkar S.F., Wang P.W., Guttman D.S. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 2005;71:5182–5191. doi: 10.1128/AEM.71.9.5182-5191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.W., Mansfield J.W., Arnold D.L., Sesma A., Paynter C.D., Murillo J., Taylor J.D., Vivian A. Excision from tRNA genes of a large chromosomal region, carrying avrPphB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola. Mol. Microbiol. 2000;38:186–197. doi: 10.1046/j.1365-2958.2000.02133.x. [DOI] [PubMed] [Google Scholar]
- Jeck W.R., Reinhardt J.A., Baltrus D.A., Hickenbotham M.T., Magrini V., Mardis E.R., Dangl J.L., Jones C.D. Extending assembly of short DNA sequences to handle error. Bioinformatics. 2007;23:2942–2944. doi: 10.1093/bioinformatics/btm451. [DOI] [PubMed] [Google Scholar]
- Joardar V., Lindeberg M., Jackson R.W., Selengut J., Dodson R., Brinkac L.M., Daugherty S.C., Deboy R., Durkin A.S., Giglio M.G., et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 2005;187:6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf A., Weingardt H., Tsiamis G., Boch J. Different versions of Pseudomonas syringae pv. Tomato DC3000 exist due to the activity of an effector transposon. Mol. Plant Pathol. 2006;7:355–364. doi: 10.1111/j.1364-3703.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Lindeberg M., Myers C.R., Collmer A., Schneider D.J. Roadmap to new virulence determinants in Pseudomonas syringae: Insights from comparative genomics and genome organization. Mol. Plant Microbe Interact. 2008;21:685–700. doi: 10.1094/MPMI-21-6-0685. [DOI] [PubMed] [Google Scholar]
- Ma W., Dong F.F., Stavrinides J., Guttman D.S. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2:e209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J.P., Moran N.A. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett M.B. New insights to the function of phytopathogenic bacterial Type III effectors in plants. Annu. Rev. Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- Ochman H., Moran N.A. Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman A.R., Jackson R.W., Mansfield J.W., Kaitell V., Thwaites R., Arnold D.L. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr. Biol. 2005;15:2230–2235. doi: 10.1016/j.cub.2005.10.074. [DOI] [PubMed] [Google Scholar]
- Pop M., Salzberg S.L. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24:142–149. doi: 10.1016/j.tig.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M., Phillippy A., Delcher A.L., Salzberg S.L. Comparative genome assembly. Brief. Bioinform. 2004;5:237–248. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- Reese M.G., Hartzell G., Harris N.L., Ohler U., Abril J.F., Lewis S.E. Genome annotation assessment in Drosophila melanogaster. Genome Res. 2000;10:483–501. doi: 10.1101/gr.10.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Kjemtrup S., Marchesini P., Dangl J.L. Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol. Microbiol. 2003;47:1545–1562. doi: 10.1046/j.1365-2958.2003.03402.x. [DOI] [PubMed] [Google Scholar]
- Salzberg S.L., Sommer D.D., Puiu D., Lee V.T. Gene-boosted assembly of a novel bacterial genome from very short reads. PLoS Comput. Biol. 2008;4:e1000186. doi: 10.1371/journal.pcbi.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Schechter L.M., Vencato M., Jordan K.L., Schneider S.E., Schneider D.J., Collmer A. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 2006;19:1180–1192. doi: 10.1094/MPMI-19-1180. [DOI] [PubMed] [Google Scholar]
- Stavrinides J., Ma W., Guttman D.S. Terminal reassortment drives the quantum evolution of Type III effectors in bacterial pathogens. PLoS Pathogens. 2006;2:e104. doi: 10.1371/journal.ppat.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides J., McCann H.C., Guttman D.S. Host-pathogen interplay and the evolution of bacterial effectors. Cell. Microbiol. 2008;10:285–292. doi: 10.1111/j.1462-5822.2007.01078.x. [DOI] [PubMed] [Google Scholar]
- Sundin G.W., Mayfield C.T., Zhao Y., Gunasekera T.S., Foster G.L., Ullrich M.S. Complete nucleotide sequence and analysis of pPSR1 (72,601 bp), a pPT23A-family plasmid from Pseudomonas syringae pv. syringae A2. Mol. Genet. Genomics. 2004;270:462–476. doi: 10.1007/s00438-003-0945-9. [DOI] [PubMed] [Google Scholar]
- Warren R.L., Sutton G.G., Jones S.J., Holt R.A. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Hutcheson S.W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Liu H., Mohr T.J., Jenrette J., Chiodini R., Zaccardelli M., Setubal J.C., Vinatzer B.A. Role of recombination in the evolution of the model plantpathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Appl. Environ. Microbiol. 2008;74:3171–3181. doi: 10.1128/AEM.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]