Abstract
Ran GTPase plays essential roles in multiple cellular processes, including nucleocytoplasmic transport, spindle formation, and postmitotic nuclear envelope (NE) reassembly. The cytoplasmic Ran GTPase activating protein RanGAP is critical to establish a functional RanGTP/RanGDP gradient across the NE and is associated with the outer surface of the NE in metazoan and higher plant cells. Arabidopsis thaliana RanGAP association with the root tip NE requires a family of likely plant-specific nucleoporins combining coiled-coil and transmembrane domains (CC-TMD) and WPP domain–interacting proteins (WIPs). We have now identified, by tandem affinity purification coupled with mass spectrometry, a second family of CC-TMD proteins, structurally similar, yet clearly distinct from the WIP family, that is required for RanGAP NE association in root tip cells. A combination of loss-of-function mutant analysis and protein interaction data indicates that at least one member of each NE-associated CC-TMD protein family is required for RanGAP targeting in root tip cells, while both families are dispensable in other plant tissues. This suggests an unanticipated complexity of RanGAP NE targeting in higher plant cells, contrasting both the single nucleoporin anchor in metazoans and the lack of targeting in fungi and proposes an early evolutionary divergence of the underlying plant and animal mechanisms.
INTRODUCTION
The small GTPase Ran is a Ras-related nuclear protein that acts in diverse cellular processes, including nuclear transport, mitotic spindle assembly, and postmitotic nuclear envelope (NE) reassembly (Dasso, 2002; Hetzer et al., 2002; Arnaoutov and Dasso, 2003, 2005; Quimby and Dasso, 2003). Ran hydrolyzes GTP to GDP with the aid of a cytosolic Ran GTPase-activating protein (RanGAP) and its coactivator RanBP1 (Bischoff et al., 1994, 1995). Cytoplasmic Ran-GDP is shuttled back to the nucleus by employing the Ran-specific nuclear transport factor NTF2 as a carrier (Quimby et al., 2000). Replacement of GDP with GTP is achieved by the action of a nuclear-localized guanine nucleotide exchange factor, RCC1 (Bischoff and Ponstingl, 1991). Thus, a RanGDP versus RanGTP gradient across the NE is established by the spatial distribution of RanGAP and RCC1. This gradient is critical for maintaining directionality of nucleocytoplasmic transport during interphase (Izaurralde et al., 1997; Kalab et al., 2002).
During vertebrate mitosis, the action of RanGTP can counteract the inhibitory effect of bound importins on specific proteins, such as TPX2, which is required for spindle formation (Gruss et al., 2001). In addition, Ran also plays roles at mitotic kinetochores. It is essential for regulation of the spindle assembly checkpoint and for assembly of microtubule fibers that attach kinetochores to spindle poles (Arnaoutov and Dasso, 2003, 2005). Furthermore, nuclear reassembly during telophase was shown to depend on the local concentration of RanGTP and its hydrolysis around the daughter chromosomes (Hetzer et al., 2000; Zhang and Clarke, 2000; Zhang et al., 2002).
RanGAP, which is critical for the Ran cycle, has different subcellular locations in different species. In yeast and some fungi that undergo closed mitosis (mitosis occurring inside an intact nuclear membrane), RanGAP is predominantly cytoplasmic (Hopper et al., 1990; De Souza et al., 2004). However, in vertebrates and higher plants, RanGAP is cytoplasmic during interphase, with a clear concentration at the outer NE (Mahajan et al., 1997; Rose and Meier, 2001). The targeting of RanGAP to the NE is through protein–protein interaction. The domain structure of mammalian and plant RanGAPs and their distinct targeting mechanisms indicate that their anchor proteins at the NE might differ (Mahajan et al., 1998; Rose and Meier, 2001). Mammalian RanGAP has a unique C-terminal domain required for its targeting to the nuclear pore and kinetochore. Upon sumoylation of a Lys residue in the C-terminal domain, the sumoylated C terminus binds to RanBP2/Nup358, and the two proteins colocalize at the NE during interphase and in the vicinity of the spindle and at kinetochores during mitosis (Matunis et al., 1998; Joseph et al., 2002, 2004). By contrast, all known plant RanGAPs possess a unique N-terminal domain, which in turn is not conserved in mammalian or yeast RanGAPs (Meier, 2000). This domain, called the WPP domain after a highly conserved Trp-Pro-Pro motif, is necessary and sufficient for NE targeting of Arabidopsis thaliana RanGAP1 during interphase and to the cell plate during cytokinesis (Rose and Meier, 2001; Jeong et al., 2005).
In addition to RanGAP1 and RanGAP2, the Arabidopsis genome codes for three small proteins with high similarity to the WPP domain: WPP1, WPP2, and WPP3 (Patel et al., 2004). WPP1 and WPP2, but not WPP3, are concentrated at the NE during interphase in undifferentiated root cells. The NE targeting mechanism of WPP1, WPP2, and RanGAP1 share the requirement for the conserved WPP motif (Patel et al., 2004).
There is no homolog of RanBP2/Nup358 in plants. Instead, targeting of RanGAP1 to the NE in Arabidopsis root tips depends on a family of three related proteins: WPP domain–interacting protein 1 (WIP1), WIP2a, and WIP3 (Xu et al., 2007). The WIP family is dispensable for targeting of RanGAP1 to the NE in differentiated cells and to the cell plate during mitosis. We have now identified by tandem affinity purification coupled with mass spectrometry a structurally similar, yet clearly distinct, second family of NE-associated proteins required for RanGAP NE association in root tip cells. Our data suggest that at least one member of each protein family is required for RanGAP targeting in root tip cells, while both families are dispensable in other plant tissues.
RESULTS
Identification by Tandem Affinity Purification of Novel WPP2-Interacting Proteins
To identify proteins associated with WPP2 in Arabidopsis, transgenic plants containing WPP2 fused to a tandem affinity purification tag (Rohila et al., 2004) were created. After two purification steps using protein A and the calmodulin binding domain of the fusion protein, eluted proteins were resolved by SDS-PAGE. Silver-stained bands were excised and subjected to in-gel trypsin digestion and mass spectrometry. Obtained peptide sequences were used for NIH nonredundant database search and protein identification.
At5g11390 was identified in a 90-kD band based on the alignment of four peptides corresponding to its deduced amino acid sequence (Figures 1A and 1B). At1g68910 matched a single peptide identified in two different excised slices: 90 and 85 kD. The two proteins have a high percentage of amino acid sequence identity (34%) and similarity (56%).
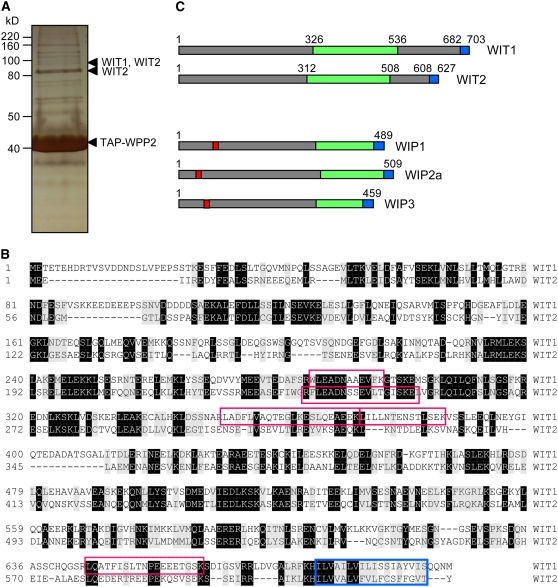
Figure 1.
Two Novel Coiled-Coil Transmembrane Domain Proteins Interact with Arabidopsis WPP2.
(A) Silver-stained gel of tandem affinity purification from TAP-WPP2–expressing 14-d-old Arabidopsis seedlings. Peptides corresponding to WIT1 and WIT2 were identified using tandem mass spectrometry in-gel slices excised from the regions marked with arrowheads. The position of TAP-WPP2 is also marked.
(B) Amino acid sequence alignment of WIT1 and WIT2. Identical and similar amino acids are shaded in black and gray, respectively. The four peptides corresponding to the WIT1 protein sequence that were identified by tandem mass spectrometry are boxed in red. The single peptide corresponding to WIT2 protein sequence that was identified in two gel slices by tandem mass spectrometry from TAP-WPP2 pull down is also represented by a red box. Predicted transmembrane domains are marked with a blue box.
(C) Domain structures of WIT and WIP protein families are similar. The domain structure is characterized by a presence of extended coiled-coil domain (green) and a single C-terminal transmembrane domain (blue). The position of the transmembrane domain allows classification of both protein families as tail-anchored proteins. WIP protein family members have a bipartite nuclear localization signal motif (red), which has not been identified in the WIT protein sequences.
The two proteins are characterized by a presence of a long predicted coiled-coil domain (Figure 1C; Rose et al., 2004) and a C-terminal putative transmembrane domain (Figure 1B). Based on the position of the predicted transmembrane domain, they can be further classified as tail-anchored proteins (Borgese et al., 2003) and were named WIT1 (At5g11390) and WIT2 (At1g68910) for WPP domain–interacting tail-anchored proteins. No significant similarity of the WIT amino acid sequence outside the coiled-coil region was detected with any sequence present in GenBank. Based on domain organization, WIT1 and WIT2 resemble the recently described WIP protein family (Figure 1C; Xu et al., 2007). However, the two protein families have no amino acid similarity outside the coiled-coil domain. mRNA expression patterns based on microarray data obtained from the Genevestigator database (Zimmermann et al., 2004) show that WIT1 and WIT2 are expressed ubiquitously in all Arabidopsis organs and developmental stages (see Supplemental Table 1 online). The expression level is consistently higher for WIT1 than for WIT2. WIT1 protein was detected in all organs tested, with a high level in roots and a lower level in rosette leaves, in agreement with the mRNA data (see Supplemental Figure 1 online).
The Interaction of WIT1 with WPP, RanGAP, and WIP Protein Families
To address the specificity of the WIT1–WPP2 interaction, we tested binding of WIT1 to all WPP domain protein family members and RanGAP1/RanGAP2 by yeast two-hybrid and coimmunoprecipitation assays. In yeast two-hybrid assays, WIT1 interacted with WPP1 and WPP2 but not with WPP3 (Figure 2A). WIT1 also bound to full-length RanGAP1 and RanGAP2 and to the WPP domain of RanGAP1 (RanGAP1ΔC, amino acids 1 to 119) but not RanGAP1ΔN (amino acids 120 to 535, RanGAP1 without the WPP domain).
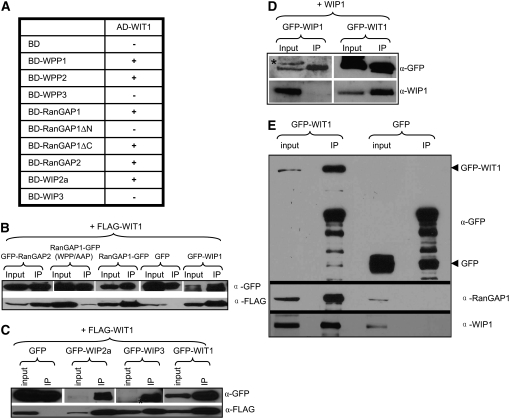
Figure 2.
WIT1 Interacts with WPP1, WPP2, RanGAP, and WIP Family Members and with Itself.
(A) Interaction of WIT1 with WIP family or WPP domain proteins in yeast two-hybrid assays. RanGAP1ΔC, WPP domain of RanGAP1; RanGAP1ΔN, RanGAP1 without WPP domain; AD, GAL4 activation domain; BD, GAL4 DNA binding domain. Plus (+), positive interaction; minus (−), no interaction. Both WIP1 and WIT1 self-activate as BD fusions, which were therefore not included in this assay.
(B) WIT1 interacts with RanGAP1 and 2 and with WIP1 in planta. Interaction with RanGAP1 is abolished by the WPP/AAP mutation in the WPP domain of RanGAP1. FLAG-WIT1 was coexpressed with GFP, RanGAP1-GFP, GFP-RanGAP2, GFP-WIP1, or RanGAP1 (WPP/AAP)-GFP in N. benthamiana. Immunoprecipitation was performed using the anti-GFP antibody, and coimmunoprecipitated protein was detected with the anti-FLAG antibody.
(C) WIT1 interacts with WIP proteins and has homodimerization ability in planta. FLAG-WIT1 was coexpressed with GFP, GFP-WIP2a, GFP-WIP3, or GFP-WIT1 in N. benthamiana. Immunoprecipitation was performed using the anti-GFP antibody, and coimmunoprecipitated protein was detected with the anti-FLAG antibody. A nonspecific band detected with the anti-GFP antibody is indicated with an asterisk.
(D) WIP1 does not homodimerize in planta. WIP1 was coexpressed with GFP-WIP1 or GFP-WIT1 constructs in N. benthamiana. Immunoprecipitation was performed using the anti-GFP antibody, and coimmunoprecipitated protein was detected with the anti-WIP1 antibody. A nonspecific band detected with the anti-GFP antibody is indicated with an asterisk.
(E) WIT1 interacts with endogenous RanGAP1 and WIP1 in Arabidopsis. Samples immunoprecipitated from GFP and GFP-WIT1 transgenic lines using a monoclonal anti-GFP antibody were probed with the anti-RanGAP1 and the anti-WIP1 antibody.
Since both the WIT and the WIP protein families contain predicted coiled-coil domains with possible dimerization ability, the interaction of WIT1 with WIP proteins was also tested. WIT1 interacted with WIP2a but not with WIP3 (Figure 2A). Since both WIT1 and WIP1 alone activate the reporter genes when fused to the GAL4 DNA binding domain (data not shown), their interaction could not be tested using this assay. These data show that WIT1 binds to the same WPP proteins that interact with the WIP family members (Xu et al., 2007) and are targeted to the NE and that, as for WIP family members, the WPP domain of RanGAP1 is necessary and sufficient for binding.
To test protein–protein interactions in planta, we employed coimmunoprecipitation (co-IP) assays using agroinfiltrated Nicotiana benthamiana (Figures 2B to 2D) and green fluorescent protein (GFP)-WIT1–expressing transgenic Arabidopsis plants (Figure 2E). Co-IP from N. benthamiana showed that WIT1 interacted with both RanGAP1 and RanGAP2 as well as with WIP protein family members and with itself (Figures 2B and 2C). While capable of interacting with WIT1, WIP1 did not show homodimerization ability in the same assay (Figure 2D). Affinity of RanGAP1 binding for WIT1 was much reduced by the WPP/AAP mutation in the WPP domain, which was shown to disrupt NE association (Jeong et al., 2005; Figure 2B). Co-IP from Arabidopsis confirmed the interaction between GFP-WIT1 and endogenous RanGAP1 and WIP1 (Figure 2E).
Subcellular Localization of WIT1 Protein and Domain Requirement for Subcellular Targeting
To explore the localization of WIT1 in Arabidopsis, an anti-WIT1 antibody was generated using a partial cDNA encoding amino acids 1 to 317, fused with a His tag and expressed in Escherichia coli. The anti-WIT1 antibody recognized a protein of ∼80 kD in wild-type Arabidopsis, while no protein of the same size was detected in the wit1-1 mutant (see below). We used both live cell imaging and immunofluorescence with anti-WIT1 or anti-GFP to characterize the subcellular localization of endogenous WIT1 and of the cauliflower mosaic virus 35S promoter (35S promoter)–driven GFP-WIT1 in root tip cells of Arabidopsis seedlings. Figure 3A shows that endogenous WIT1 associated with the NE in both undifferentiated and differentiated cells in roots. Similarly, GFP-WIT1 was clearly located at the NE (Figure 3B). During cytokinesis, it appeared that GFP-WIT1 was concentrated around the cell plate. Monoclonal antitubulin was used to mark the position of the phragmoplast (Figure 3B). Live imaging of GFP-WIT1 transgenic roots suggested that GFP-WIT1 decorated NEs in a dotted pattern (Figure 3C), indicative of an association with nuclear pores.
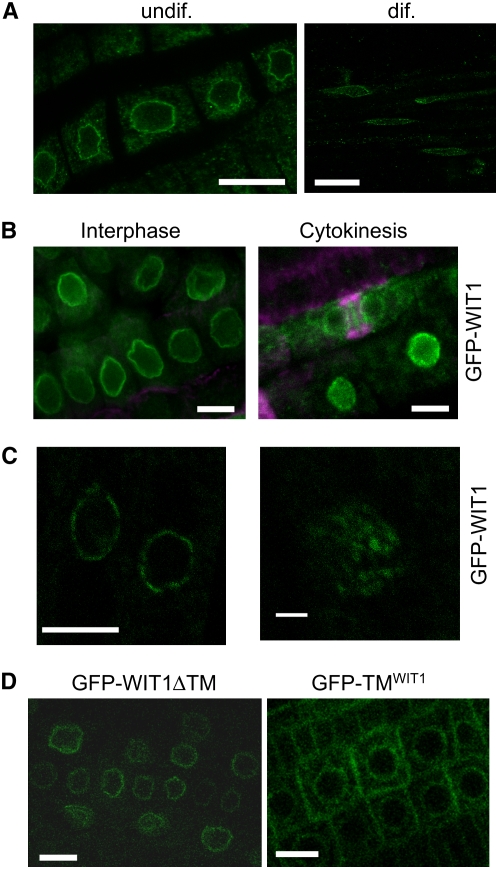
Figure 3.
WIT1 Is Targeted to the NE in Arabidopsis Root Tips.
(A) Endogenous WIT1 is targeted to the NE in both differentiated cells and undifferentiated cells in Arabidopsis roots. WIT1 is detected by the anti-WIT1 antibody in immunofluorescence analysis. Bars = 10 μm.
(B) GFP-WIT1 is targeted to the NE during interphase and cell plate during cytokinesis. Double immunofluorescence of GFP-WIT1 (green) and tubulin (magenta) in Arabidopsis root tip cells. The gain setting of the green channel is increased in the cytokinesis image to highlight the cell plate. Wild-type Arabidopsis plants were used as a negative control, and no fluorescence was detected under the same gain settings (data not shown). Bars = 10 μm.
(C) GFP-WIT1 localizes to the NE in a punctate pattern. Confocal images were taken directly from the root tip cells of Arabidopsis expressing GFP-WIT1. The right panel shows three-dimensional maximal projection of confocal images spanning half of the nucleus from Arabidopsis root tip cells. Bar = 10 μm for the left panel and 1 μm for the right panel.
(D) The transmembrane domain is dispensable for NE targeting. Root tip cells of a transgenic Arabidopsis line expressing GFP-WIT1 lacking its C-terminal transmembrane domain (GFP-WIT1ΔTM) (left panel) and of a line expressing GFP fused to the WIT1 transmembrane domain alone (GFP-TMWIT1) (right panel). Bars = 10 μm.
To investigate domain requirement for NE targeting, we fused different WIT1 deletion constructs to GFP and visualized them in root cells of transgenic Arabidopsis. The construct GFP-WIT1ΔTM represents the fusion of GFP with N-terminal part of WIT1 corresponding to amino acids 1 to 660, and GFP-TMWIT1 has GFP fused to the C-terminal 43 amino acids of WIT1. The data showed that a fraction of GFP-TMWIT1 was still associated with the nuclear periphery (Figure 3D). However, more signal was now detected in the cytoplasm and possibly the plasma membrane. When the putative transmembrane domain was deleted, the remaining protein GFP-WIT1ΔTM still associated with the NE (Figure 3D). We concluded that the C-terminal TMD of WIT1 was partially sufficient for targeting GFP to the nuclear periphery but was not necessary for NE association of WIT1. One possible explanation is that WIT1 is held at the NE via additional protein–protein interaction (see below).
Role of the WIT Family in RanGAP Anchoring
Our previous study showed that in root tip cells of a wip1-1 wip2-1 wip3-1 triple mutant, RanGAP1 is absent from the NE, while its association with the NE in differentiated root cells or with the cell plate during cytokinesis is not changed (Xu et al., 2007). These data indicate that, although RanGAP NE anchoring at the root tip depends on WIP family members, additional activities must exist for NE association in differentiated cells. Therefore, we tested whether WIT family members play roles in anchoring RanGAP1 at the NE during interphase or at other RanGAP1 locations during mitosis. T-DNA insertion lines for WIT1 and WIT2 were identified from the SALK and GABI-Kat collection, respectively (Alonso et al., 2003; Rosso et al., 2003). The T-DNA in line 470E06 (wit1-1) is inserted within the first coding exon of WIT1, while the T-DNA in line SALK_127765 (wit2-1) is in the second coding exon of WIT2 (Figure 4A). In wit1-1, no WIT1 mRNA across the T-DNA insertion could be detected (Figure 4B, top left panel). In an immunoblot with the anti-WIT1 antibody, a band of 80 kD corresponding to WIT1 was detected in wild-type Arabidopsis but not in the wit1-1 mutant (Figure 4B, right panel). Therefore, we conclude that wit1-1 represents a null mutation. In wit2-1, no WIT2 mRNA could be detected, either across the insertion or downstream of the insertion, again consistent with a knockout mutation (Figure 4B, bottom left panel). At the same time in wit2-1, WIT1 was expressed at the wild-type level, additionally confirming the specificity of the anti-WIT1 antibody to WIT1 (Figure 4B, right panel).
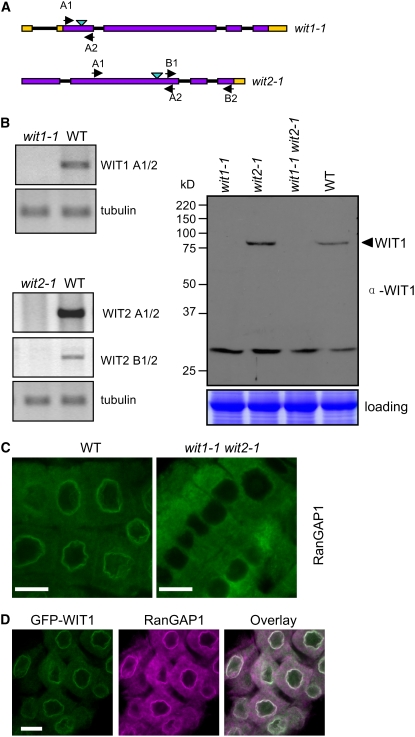
Figure 4.
In the wit1-1 wit2-1 Double Mutant, RanGAP1 Is Dislocated from the Root Tip NE.
(A) Schematic representation of T-DNA insertions in wit1-1 and wit2-1. Open reading frames, introns, and untranslated regions are indicated by purple boxes, black lines, and yellow boxes, respectively. Light-blue arrowheads are marking T-DNA insertion sites. Positions of primers used for RT-PCR are labeled with black arrowheads.
(B) WIT mRNA and protein expression analysis in wit1-1, wit2-1, and wit1-1 wit2-1. The top left panel represents wit1-1 RT-PCR analysis; the bottom left panel represents wit2-1 RT-PCR analysis with two pairs of primers, spanning the T-DNA insertion site (A1/2) or 3′ to the T-DNA insertion site (B1/2). Tubulin, β-tubulin primers used as RT-PCR control. The right panel shows an immunoblot analysis of wit1-1, wit2-1, and wit1-1 wit2-1 using the anti-WIT1 antibody. The position of WIT1 protein is marked with an arrowhead. A section of a Coomassie blue–stained replica gel is shown at the bottom as loading control.
(C) Immunofluorescence localization of RanGAP1 in the wild type and wit1-1 wit2-1 double mutant in Arabidopsis root tip cells. Bars = 10 μm.
(D) Double immunofluorescence localization of GFP-WIT1 (green) and RanGAP1 (magenta) in the root tip cells of the wit1-1 wit2-1 double mutant transformed with GFP-WIT1. Bar = 10 μm.
RanGAP1 localization in individual homozygous insertion lines was tested by immunofluorescence, but no significant difference was observed in either undifferentiated or differentiated root cells compared with wild-type plants (data not shown). Therefore, two homozygous lines were crossed to obtain a wit1-1 wit2-1 double mutant. Surprisingly, endogenous RanGAP1 did not accumulate at the NE in root tip cells of the wit1-1 wit2-1 double mutant line (Figure 4C). By contrast, the localization of endogenous RanGAP1 was not changed in differentiated root cells, and RanGAP1 association with the cell plate was not lost in the mitotic root cells of the wit1-1 wit2-1 double mutant (see Supplemental Figure 2 online). To confirm that the lack of RanGAP1 NE targeting in the root tip cells was due to the absence of WIT1 and WIT2 proteins, we transformed the wit1-1 wit2-1 double mutant with a construct expressing GFP-WIT1 under the control of the 35S promoter. Figure 4D shows that in the wit1-1 wit2-1 double mutant expressing GFP-WIT1, RanGAP1 NE association was reestablished in root tip cells. This clearly demonstrates that GFP-WIT1 is functional and that WIT1 is necessary for RanGAP1 NE targeting in root tip cells.
Since wit1-1 wit2-1 phenocopies wip1-1 wip2-1 wip3-1 in terms of mislocalization of RanGAP1 in root tip cells (Xu et al., 2007), we tested whether in a wit1-1 wit2-1 double mutant WIP1 protein level is affected, whether the interaction between WIP1 and RanGAP1 is lost, and whether WIP1 is dislocated from the NE. Immunoblot data showed that the endogenous WIP1 protein level was not changed in wit1-1 wit2-1 (see Supplemental Figure 3A online). Co-IP from the wild type and wit1-1 wit2-1 indicated that the interaction between endogenous RanGAP1 and WIP1 was not affected (see Supplemental Figure 3B online). To analyze the localization of WIP1 in wit1-1 wit2-1, we transformed wit1-1 wit2-1 with a construct expressing GFP-WIP1 under the control of the 35S promoter. Supplemental Figure 3C online shows that GFP-WIP1 was properly targeted to the NE in wit1-1 wit2-1. However, although 35S promoter–driven expression of GFP-WIP1 complements the wip1-1 wip2-1 wip3-1 phenotype (Xu et al., 2007), it could not compensate for the loss of WIT1 and WIT2, and RanGAP1 NE association was not reestablished in root tip cells (see Supplemental Figure 3C online). Therefore, we concluded that WIT1 and WIT2 are required for RanGAP1 NE targeting in root tip cells in a mechanism independent of WIP1 protein abundance and NE targeting.
WIT1 Is Downregulated in the wip1-1 wip2-1 wip3-1 Triple Mutant and Cannot Functionally Replace the WIP Family
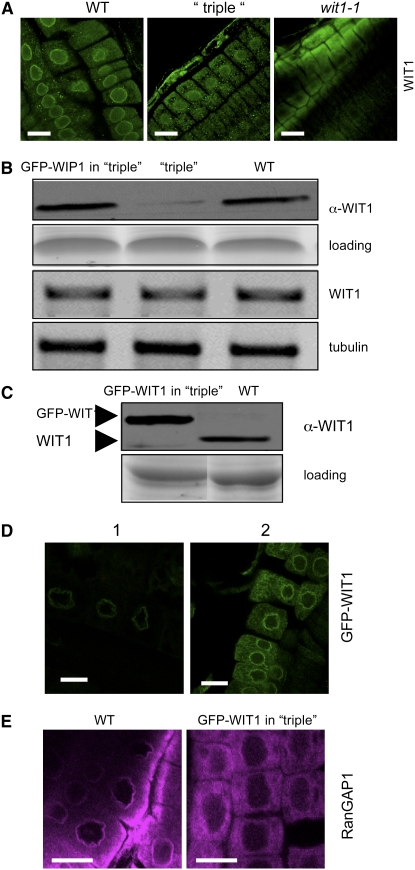
To explore the functional relationship between members of the WIP and WIT protein families, we investigated the localization and expression level of endogenous WIT1 in the wip1-1 wip2-1 wip3-1 mutant. Interestingly, WIT1 accumulation at the NE was greatly reduced in root tip cells of wip1-1 wip2-1 wip3-1 (Figure 5A). Immunoblot data showed that endogenous WIT1 expression level was significantly decreased in wip1-1 wip2-1 wip3-1 compared with the wild type. The corresponding WIT1 mRNA was detected in wip1-1 wip2-1 wip3-1 (Figure 5B).
Figure 5.
WIT1 Cannot Functionally Replace the WIP Family.
(A) Immunofluorescence localization of WIT1 in the wild-type, the wip1-1 wip2-1 wip3-1 triple mutant (“triple”), and the wit1-1 mutant. Bars = 10 μm.
(B) WIT1 protein level is reduced in the wip1-1 wip2-1 wip3-1 triple mutant (“triple”). Protein extracts from the wild type, the wip1-1 wip2-1 wip3-1 triple mutant, and GFP-WIP1 in the wip1-1 wip2-1 wip3-1 triple mutant were probed with anti-WIT1 antibody. A section of a Coomassie blue–stained replica gel is shown as loading control. WIT1 mRNA level is not changed in the wip1-1 wip2-1 wip3-1 triple mutant. The third panel from the top shows RT-PCR analysis with WIT1-specific primers. The bottom panel shows tubulin as RT-PCR control.
(C) Protein extract from the wild type and GFP-WIT1 in the wip1-1 wip2-1 wip3-1 triple mutant (“triple”) were probed with anti-WIT1 antibody. A Coomassie blue–stained replica gel shown at the bottom represents loading control.
(D) Overexpressed GFP-WIT1 is targeted to the NE in the wip1-1 wip2-1 wip3-1 triple mutant. GFP-WIT1 in the wip1-1 wip2-1 wip3-1 triple mutant was detected by live confocal imaging (1) or by immunofluorescence with a polyclonal anti-GFP antibody (ab290) (2). Bars = 10 μm.
(E) Immunofluorescence localization of RanGAP1 in root tip cells in the wild-type background (left panel) and in wip1-1 wip2-1 wip3-1/GFP-WIT1 (right panel). Bars = 10 μm.
Based on these findings, it is formally possible that the delocalization of RanGAP1 in wip1-1 wip2-1 wip3-1 is caused by the reduction of WIT1 and that WIT1 and not a WIP family member is the actual anchor of RanGAP at the NE. To test this hypothesis, we expressed 35S promoter–driven GFP-WIT1 in wip1-1 wip2-1 wip3-1. If reduction of WIT1 is the only cause of RanGAP delocalization in wip1-1 wip2-1 wip3-1, we reasoned that overexpression of GFP-WIT1 should restore RanGAP NE targeting in the root tip. First, we tested the level and localization of GFP-WIT1. A line was identified in which GFP-WIT1 accumulated in the wip1-1 wip2-1 wip3-1 background to a level comparable to WIT1 in wild-type plants (Figure 5C). Next, we analyzed the localization of GFP-WIT1 in wip1-1 wip2-1 wip3-1. Figure 5D shows that GFP-WIT1 was properly targeted to the NE in the root tips, suggesting that reduction of NE-associated WIT1 in wip1-1 wip2-1 wip3-1 was largely due to reduced protein abundance and not reduced NE targeting. We also tested the interaction of GFP-WIT1 with RanGAP1 in the wild type and wip1-1 wip2-1 wip3-1 triple mutant background. RanGAP1 was coimmunoprecipitated by GFP-WIT1 from both the wild type and wip1-1 wip2-1 wip3-1, indicating that GFP-WIT1 could bind RanGAP1 independent of WIP family members (see Supplemental Figure 4A online). Finally, we analyzed the localization of RanGAP1 in wip1-1 wip2-1 wip3-1/35S-GFP-WIT1 by immunofluorescence. Clearly, RanGAP1 was absent from the NE of root tip cells despite the presence of GFP-WIT1 at the NE (Figure 5E). This indicates that WIT1 is not sufficient to compensate for the loss of WIP1/WIP2/WIP3 and suggests that RanGAP NE targeting in root tip cells requires the activity of at least one member of each protein family.
An additional indication that the reduced WIT1 level was not sufficient to cause RanGAP1 delocalization in wip1-1 wip2-1 wip3-1 came from the analysis of WIT1 protein level in wip double mutants and the wip1-1 wip2-1 wip3-1 triple mutant expressing 35S promoter–driven GFP-WIP1, GFP-WIP2a, or GFP-WIP3 (see Supplemental Figure 4B online). Although WIT1 protein level was reduced to a comparable level in wip1-1 wip2-1, wip1-1 wip2-1 wip3-1, wip1-1 wip2-1 wip3-1/GFP-WIP2a, and wip1-1 wip2-1 wip3-1/GFP-WIP3, RanGAP1 delocalization can be observed only in wip1-1 wip2-1 wip3-1 (Xu et al., 2007), suggesting that WIT1 reduction is not its sole cause. The simplest model consistent with all presented data is therefore that a critical step of RanGAP anchoring to the NE in the root tips of Arabidopsis requires a complex consisting of at least one WIP and one WIT family member.
The Binding Affinity of WIT1 for RanGAP1 Is Increased in the Presence of WIP1
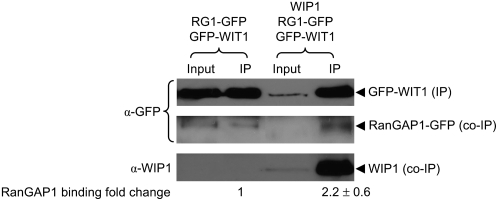
The hypothesis that RanGAP anchoring in the specific region of the root of Arabidopsis requires at least one of both WIP and WIT family members was further tested in the heterologous system of N. benthamiana agroinfiltration. We reasoned that although WIT1 itself was capable of binding RanGAP1 in this system (see Figure 2B), coexpression of WIP1 with WIT1 might increase the affinity of WIT1 for RanGAP1. Figure 6 shows that, indeed, in the presence of WIP1, WIT1 immunoprecipitated about two times more RanGAP1 than when only WIT1 and RanGAP1 were coexpressed, suggesting that WIP1 increases the binding affinity of WIT1 and RanGAP1.
Figure 6.
The Binding Affinity of WIT1 for RanGAP1 Is Increased in the Presence of WIP1.
GFP-WIT1 was coexpressed with RanGAP1-GFP in N. benthamiana either in the presence (right two lanes) or absence (left two lanes) of coexpressed WIP1. Immunoprecipitation was performed using the anti-WIT1 antibody, and immunoprecipitated and coimmunoprecipitated proteins were detected with the anti-GFP antibody (top and middle panels) and the anti-WIP1 antibody (bottom panel). The “fold change” of RanGAP1 binding indicated below the lanes was calculated by quantifying the ratio of coimmunoprecipitated RanGAP1-GFP signal to GFP-WIT1 IP signal intensity. The value for GFP-WIT1 binding of RanGAP1-GFP in the absence of WIP1 was set to 1. The “fold change” value is a mean value (±sd) obtained from five independent repetitions of the experiment (three biological replicates).
DISCUSSION
Association of RanGAP1 with the Arabidopsis Root Tip NE Requires Two Distinct Nuclear Pore–Associated Coiled-Coil Protein Families
The Ran cycle is evolutionarily conserved among different kingdoms. RanGAP is the only known GTPase-activating protein for Ran (Bischoff et al., 1994). Therefore, its subcellular localization is an important indicator of where RanGTP hydrolysis is happening and required. Despite its functional conservation (Pay et al., 2002), RanGAP localization patterns differ among different kingdoms. Yeast RanGAP is predominantly cytoplasmic throughout the cell cycle (Hopper et al., 1990). However, both vertebrate and plant RanGAPs contain kingdom-specific targeting domains for additional NE association (Matunis et al., 1996; Mahajan et al., 1998; Rose and Meier 2001; Jeong et al., 2005). In vertebrates, the C-terminal domain of RanGAP binds to the nucleoporin RanBP2/Nup358 after sumoylation, thereby anchoring RanGAP at the NE (Matunis et al., 1996; Mahajan et al., 1998). In plants, there is no evidence that RanGAP is sumoylated, and there is no homolog of RanBP2 in the known plant genomes. Instead, plant RanGAP binds with its N-terminal WPP domain to WIP family members, which are necessary in Arabidopsis root tips to target RanGAP to the NE (Xu et al., 2007).
Here, we have identified by tandem affinity purification coupled with mass spectrometry a second protein family (WIT1 and WIT2) involved in RanGAP anchoring in Arabidopsis. WIT1 interacts in yeast two-hybrid assays with all WPP domain–containing proteins that accumulate at the NE (Rose and Meier, 2001; Patel et al., 2004) and interacts with RanGAP1 and RanGAP2 in planta. Binding is significantly decreased if a mutation (WPP to AAP) is introduced that disrupts NE targeting (Jeong et al., 2005; Xu et al., 2007). Like RanGAP1, WIT1 is associated with the NE during interphase in cells of all developmental stages and with the cell plate during cytokinesis (Rose and Meier, 2001; Jeong et al., 2005). In a wit1-1 wit2-1 double mutant, RanGAP1 is delocalized from the NE in the Arabidopsis root tip region, and NE targeting is restored upon expression of GFP-WIT1 in wit1-1 wit2-1.
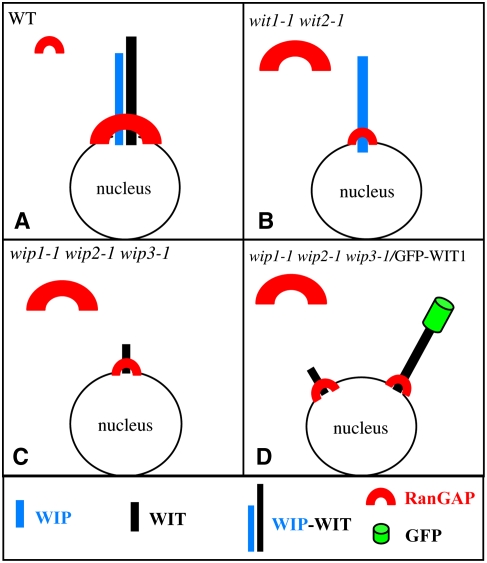
The same molecular phenotype of RanGAP1 mistargeting specifically in the root tip has been reported for a wip1-1 wip2-1 wip3-1 triple mutant (Xu et al., 2007). We have shown here that in this mutant the WIT1 protein level is significantly reduced. By contrast, the level of WIP1 is not affected in wit1-1 wit2-1, suggesting that WIP1 is not sufficient to anchor RanGAP1 in the absence of WIT family members. When GFP-WIT1 was expressed in wip1-1 wip2-1 wip3-1 at a level similar to that of WIT1 in the wild type, RanGAP1 NE targeting to the root tip was not restored, indicating that WIT1 is also not sufficient to anchor RanGAP1 in the absence of WIP family members. Together, the data presented here indicate that in root tip cells, at least one member of each protein family, WIP and WIT, has to be expressed for the mechanism that targets RanGAP to the NE to be functional. The simplest model consistent with all data is that a heterocomplex containing at least one WIT and one WIP family member is the critical molecule required for RanGAP1 targeting at the root tip NE (Figure 7).
Figure 7.
Requirement of Both WIP and WIT Family Members for RanGAP NE Targeting in Root Tip Cells.
(A) In the wild type, a complex containing both a WIP and a WIT family member is present at the NE and RanGAP is bound to this complex.
(B) In wit1-1 wit2-1, WIP1 abundance and NE association are unchanged. However, in the absence of WIT family members, only a small amount of RanGAP is associated with the NE.
(C) In wip1-1 wip2-1 wip3-1, WIT1 abundance is drastically reduced. Remaining WIT1 is likely associated at least in part with the NE but is not sufficient for efficient RanGAP targeting.
(D) Overexpressing GFP-WIT1 in wip1-1 wip2-1 wip3-1 does not alter RanGAP localization, indicating that abundant GFP-WIT1 at the NE (which complements wit1-1 wit2-1) is not sufficient for RanGAP targeting in the absence of WIP family members.
The size of the symbols in (A) to (D) represents the abundance of the molecule in the respective cellular location.
WIT1 and WIP1 Are Both Present in a Protein Complex That Binds RanGAP1 with High Affinity
The hypothesis that a protein complex containing both a WIP and WIT family member is the bona fide RanGAP1 anchor in Arabidopsis root tips is further supported by yeast two-hybrid data and by immunoprecipitation data obtained from transient expression in N. benthamiana leaves. In yeast, WIT1 interacts with each WIP family member. In N. benthamiana, WIT1 was shown to form homomultimers and to interact with all three WIP proteins. By contrast, WIP1 does not interact with itself (Figure 2D) or with either WIP2a or WIP3 in N. benthamiana leaves (data not shown). Moreover, the affinity of WIT1 for RanGAP1 binding increased in the presence of WIP1 (Figure 6). Combined with the wit1-1 wit2-1 double and wip1-1 wip2-1 wip3-1 triple mutant analysis with respect to RanGAP1 delocalization, the existence of a WIP-WIT heterocomplex required for anchoring RanGAP1 to the root tip NE is strongly supported (Figure 7A). In wit1-1 wit2-1, neither WIT1 nor WIT2 is present, and the WIP1 level is not decreased (Figure 7B). Based on the absence of detectable WIP1–WIP1 interactions in N. benthamiana, we propose that the WIP1 dimer concentration is low and that under wild-type concentrations, WIP1 is not sufficient for RanGAP1 binding. This notion is confirmed by weak interaction between WIP1 and RanGAP1 in N. benthamiana (data not shown; Figure 7B). In the wip1-1 wip2-1 wip3-1 triple mutant, WIP1, WIP2a, and WIP3 are not present and the WIT1 level is significantly decreased. Based on the interaction data described above, remaining WIT1 presumably forms a homomultimer, not sufficient to anchor RanGAP1 to the NE (Figure 7C). However, the same low level of WIT1 in the wip1-1 wip2-1 double mutant (see Supplemental Figure 4B) is sufficient to target RanGAP1 to the NE (Xu et al., 2007). The most likely explanation is that the remaining low amount of both WIP3 and WIT1 form a complex sufficient for RanGAP1 NE binding. We indeed show that the affinity of the hetero WIP1-WIT1 complex for RanGAP1 binding is higher than RanGAP1 binding by WIT1 alone (Figure 6). A similar scenario for WIP3 and WIT1 is conceivable. When a 35S promoter–driven GFP-WIT1 construct is introduced into wip1-1 wip2-1 wip3-1, a majority of RanGAP1 remains detached from the NE (Figure 7D). Although the GFP-WIT1 level in the wip1-1 wip2-1 wip3-1 triple mutant background is comparable with endogenous WIT1 level in the wild type (Figure 5C), it is not sufficient to target RanGAP1 to the nuclear rim.
Does RanGAP1 Association with the NE Require a Whole Set of Different Anchors?
Contrary to the RanGAP anchoring mechanism described in mammalian cells, where Nup358/RanBP2 has this specific function both during interphase and mitosis, the situation in plants seems to be significantly more complex. Plants use at least two different strategies to anchor RanGAP1, depending on developmental and cell cycle stage. The proposed model is restricted to the root tip zone, while RanGAP NE targeting appears undisrupted in differentiated root cells and during cytokinesis of both wit1-1 wit2-1 (shown here; see Supplemental Figure 2A) and wip1-1 wip2-1 wip3-1 (Xu et al., 2007). Based on promoter-β-glucuronidase fusions, tissue-specific expression patterns with high expression in the root tip, gradually decreasing throughout the differentiation zone, have been reported for both RanGAP1 and WIP family members (Xu et al., 2007; Meier et al., 2008). Here, we show by immunocytochemistry that WIT1 has a similar expression pattern with respect to developmental stage (Figure 3A). Therefore, all three protein families are present in differentiated cells, albeit likely at reduced levels. Therefore, the simplest working hypothesis is that in differentiated cells, the level of redundancy of nuclear pore–associated coiled-coil proteins involved in RanGAP anchoring is even higher, and our current mutant combinations are therefore not sufficient to disrupt the process. Alternatively, an unrelated, unknown protein could act as RanGAP anchor in these cell types.
Do Plants Have a Unique Class of Coiled-Coil Transmembrane Domain Nucleoporins?
Yeast and vertebrate nuclear pore complexes (NPCs) are similar with respect to general morphology and the number of proteins (Rout et al., 2000; Cronshaw et al., 2002; Alber et al., 2007a). However, it appears that the membrane-associated anchoring nucleoporins, which differ significantly among animals and fungi, have a more recent evolutionary origin and show a larger degree of kingdom specificity (Bapteste et al., 2005). Three integral membrane nucleoporins each have been described in yeast (Pom 34, Pom152, and Ndc1) and in mammalian cells (gp210, POM121, and NDC1) (Rout et al., 2000; Stavru et al., 2006a). While the plant NPC composition has not been studied in detail, proteins with convincing similarity to NDC1 and gp210, but not the other membrane-associated Nups, have been identified in Arabidopsis (Cohen et al., 2001; Stavru et al., 2006a). Based on several lines of evidence, we have suggested recently that WIP family members represent novel, plant-specific transmembrane nucleoporins (Xu et al., 2007; Meier et al., 2008). While demonstration of nuclear pore association of WIT1 awaits high-resolution electron microscopy localization data, several lines of evidence shown here suggest that WIT1 and WIP1 are in close proximity in the cell. The GFP-WIT1 signal also appears in a punctate pattern in the nuclei of Arabidopsis root cells, resembling the one described for WIP1 (Figure 3B; Xu et al., 2007). WIT1 and WIT2 show a domain organization similar to WIP family members (Figure 1C) and bind to WIP1 and RanGAP1, thus together functionally replacing the mammalian nucleoporin Nup358/RanBP2. The coiled-coil domain, present in both WIP and WIT proteins, is one of the eight fold types assigned to the NPC constituents (Devos et al., 2006). It is tempting to speculate that the coiled-coil transmembrane domain proteins discovered in this study and by Xu et al. (2007) have functions at the plant nuclear pore beyond providing a docking mechanism for RanGAP.
The yeast NPC has six coiled-coil–containing nucleoporins (Devos et al., 2006; Alber et al., 2007a; Schrader et al., 2008). Among those, Nic96 and Nup82 represent linker nucleoporins with an important structural role in bridging the inner and outer rings, as well as in recruiting functionally relevant FG repeat-containing subcomplexes (Alber et al., 2007a, 2007b; Schrader et al., 2008). By means of their coiled-coil domains, WIP and WIT proteins could similarly act as keystone nucleoporins, connecting several subcomplexes within the plant NPC. Interestingly, it was shown that depletion of mammalian Nup107 causes codepletion of several nucleoporins via proteolysis (Boehmer et al., 2003), remarkably analogous to the WIP1 depletion–caused codepletion of WIT1 reported here.
The absence of more than subtle molecular phenotypes in the mutant combinations discussed does not argue against a structural role of these novel proteins in the plant nuclear pore. It is not without precedence that structurally diverse nucleoporins can functionally replace one another. Single and double deletion mutants of yeast transmembrane nucleoporins Pom34 and Pom152, for example, are not affected in growth, NPC assembly, or nucleocytoplasmic protein transport (Madrid et al., 2006). Mammalian gp210 and POM121 are also dispensable for NPC assembly (Stavru et al., 2006b). Although NDC1 was shown to be essential for yeast viability (Winey et al., 1993), the loss of NDC1 function in Caenorhabditis elegans is not lethal (Stavru et al., 2006a). Considering the high level of tolerance of NPC biogenesis to the loss of one or several of its constituents, it will be interesting to next obtain a quintuple wit1-1 wit2-1 wip1-1 wip2-1 wip3-1 mutant and to further combine it with mutations in putative Arabidopsis gp210 and NDC1. The combination of this genetic analysis with a biochemical analysis of the protein interaction network of WIP1 and WIT1 will further clarify whether they indeed represent a novel class of plant-specific nucleoporins and shed much-needed light on the currently poorly understood plant NPC composition.
METHODS
Computational Analysis
The related sequences of WIT1 (At5g11390) and WIT2 (At1g68910) were identified based on BLAST and WU-BLAST searches. No significant similarity outside the coiled-coil region was noticed with any other protein sequences in GenBank, except for putative WIT orthologs from other plant species, which were identified using WU-BLAST search performed with The Institute for Genomic Research Plant Transcript Assemblies (http://tigrblast.tigr.org/euk-blast/plantta_blast.cgi). Sequence alignments were performed using MEGALIGN (DNASTAR) using the Clustal algorithm as previously described (Rose and Meier, 2001). The amino acid sequence identity and similarity was determined using bl2seq (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). The coiled-coil prediction was performed using Multicoil (http://groups.csail.mit.edu/cb/multicoil/cgi-bin/multicoil.cgi), and the transmembrane domain prediction was accessed through the Aramemnon database (http://aramemnon.botanik.uni-koeln.de/). Quantity One 4.1.1 (Bio-Rad) was used for the quantification of the co-IP signals in Figure 6.
Constructs
The WIT1 cDNA was cloned by RT-PCR from Arabidopsis thaliana (Columbia ecotype) root RNA into the pENTR/D-TOPO vector (Invitrogen). The insert was confirmed by sequencing. To construct N-terminal GFP fusion proteins, we moved each cDNA into pK7WGF2 (Karimi et al., 2002) by LR recombination cloning (Invitrogen). All GFP fusions with WIT1 fragments were generated in the same way. GFP-WIP1, GFP-WIP2a, and GFP-WIP3 were described previously (Xu et al., 2007). RanGAP1-GFP cloning and mutagenesis was also described previously (Jeong et al., 2005). WIT1 cDNA was moved into ProQuest Y2H vectors pDEST22 and pDEST32 (Invitrogen) by LR recombination for expression of GAL4 AD and BD fusion proteins. BD-RanGAP1, BD-RanGAP1ΔN, BD-N of RanGAP1, BD-RanGAP2, BD-WIP2a, BD-WIP3, BD-WPP1, BD-WPP2, and BD-WPP3 were previously described (Xu et al., 2007). The WIT1 cDNA was moved into pEarleyGate202 vector (Earley et al., 2006). The WPP2 gene was subcloned from pBD-GAL4 vector (Patel et al., 2004) to pENTR3C vector (Invitrogen), using the EcoRI restriction site. The N-terminal TAP-WPP2 fusion construct was obtained by LR recombination (Invitrogen) with the NTAPi binary vector (Rohila et al., 2004). See Supplemental Table 2 online for detailed description of all constructs used.
Yeast Two-Hybrid Assays
All plasmid pairs were transformed into the yeast strain PJ694A (James et al., 1996) in accordance with published protocols (Dohmen et al., 1991). Handling of yeast cultures and plate growth assays were as described in the Clontech Yeast Protocols Handbook (1996).
Antibody Development
For WIT1 antibody production, a partial protein consisting of the N-terminal 317 amino acids was expressed as a His tag fusion protein from pDEST17 (Invitrogen). The pellet from a 500+mL culture expressing His-WIT1 was used for purification with Ni-NTA resin under denaturing conditions following the protocol of the QIAexpressionist handbook (Qiagen). After purification and preparative SDS-PAGE, a rabbit antiserum (OSU 206) was produced by Cocalico Biologicals.
Plant Material and Generation of Transgenic Plants
The T-DNA GABI-Kat line 470E06 (wit1-1) was acquired from the Max Plank Institute for Plant Breeding Research, and the T-DNA SALK_line 127765 (wit2-1) was acquired from the ABRC. The double mutant line wit1-1 wit2-1 was obtained as F2 progeny of a cross between wit1-1 and wit2-1. The triple-mutant line wip1-1 wip2-1 wip3-1 was described previously (Xu et al., 2007). Plasmids expressing TAP-WPP2, GFP-WIT1, GFP-WIT1ΔTM, and GFP-TMWIT1 were mobilized into the Agrobacterium tumefaciens strain ABI. Transconjugants were selected on LB plates containing 50 μg/mL spectinomycin, 34 μg/mL chloramphenicol, and 50 μg/mL kanamycin. Arabidopsis Columbia wild-type, wit1-1 wit2-1, and wip1-1 wip2-1 wip3-1 were transformed by floral dipping (Clough and Bent, 1998) and selected by kanamycin or BASTA resistance.
Agroinfiltration of Nicotiana benthamiana
WIT1 in pEarlyGate202 or WIP1 in pH2GW7,0 were transformed into the Agrobacterium strain ABI. Transconjugants were selected on LB plates containing 50 μg/mL spectinomycin, 34 μg/mL chloramphenicol, and 50 μg/mL kanamycin. The Agrobacterium strains transformed with GFP-WIP1, GFP-WIP2a GFP-WIP3, RanGAP1-GFP, RanGAP1(WPP/AAP), and GFP-RanGAP2 were described previously (Jeong et al., 2005; Xu et al., 2007). Agrobacterium cultures containing different plasmids were either infiltrated or coinfiltrated transiently into N. benthamiana leaves as described previously (Zhao et al., 2006).
RNA Isolation and RT-PCR
Total RNA was extracted with the RNeasy plant mini kit (Qiagen) from rosette leaves of 30-d-old Arabidopsis plants grown on soil or from roots of 14-d-old Arabidopsis seedlings. After digestion with DNase I (amplification grade; Invitrogen), cDNA synthesis was performed with oligo(dT) primer and the Thermoscript RT-PCR system (Invitrogen). cDNA templates were amplified with gene-specific primers: 5′-CACCATGGAAACAGAAACGGAACATGATAGA-3′ and 5′-CTGACCCAAAGATTGTTCAGTAGCATTC-3′ were used for WIT1 spanning the insertion site; 5′-GAATATGTGAAGTCAGCTGAACAAAAGCT-3′ and 5′-GTTGAGTTCAGAGTTTGTGGTAGA-3′ were used for WIT2 spanning the insertion site; 5′-TCTACCACAAACTCTGAACTCAAC-3′ and 5′-TTAATAAGTCACACCAAAGAATGAACAAAACAGC-3′ were used for WIT2 3′ to the insertion site; 5′-CTCAAGAGGTTCTCAGCAGTA-3′ and 5′-TCACCTTCTTCATCCGCAGTT-3′ were used for tubulin product.
Tandem Affinity Purification
Twenty five grams (fresh weight) of TAP-WPP2 Arabidopsis seedlings grown in liquid Murashige and Skoog medium for 14 d under continuous white light was ground in liquid nitrogen to a fine powder. Total proteins were extracted using 2.5 volumes of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.25% dodecyl maltoside [Sigma-Aldrich], protease inhibitor cocktail [1:100; Sigma-Aldrich], 1 mM phenylmethylsulphonyl fluoride, 2 μg/μL antipain, 2 μg/μL leupeptin, 2 μg/μL aprotinin, 10 μM chymostatin, and 1 μM E-64 [Sigma-Aldrich]) by occasional vortexing for 30 min at 4°C. After filtration through Miracloth, the extract was centrifuged at 1450g for 10 min at 4°C. The supernatant was incubated with 1.25 mL IgG-agarose beads (Sigma-Aldrich) for 2 h at 4°C with gentle rotation. IgG-agarose beads were recovered by centrifugation at 150g for 3 min at 4°C, washed three times with 30 mL washing buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, and 0.1% Nonidet P-40), and once with 30 mL TEV cleavage buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, 0.5 mM EDTA, pH 8.0, 1 mM DTT, and 1 μM E-64). TAP-tagged proteins were released from IgG beads by cleavage with 800 units of AcTEV protease (Invitrogen) in 12 mL of TEV cleavage buffer for 3 h at 4°C with gentle rotation. After centrifugation at 150g for 3 min at 4°C, the supernatant was diluted with 3 volumes of Calmodulin binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, 1 mM Mg-acetate, 1 mM imidazole, 2 mM CaCl2, and 10 mM β-mercaptoethanol) and 1.2 μL of 2.5 M CaCl2 per mL supernatant was added to titrate EDTA from the TEV cleavage buffer. The mixture was incubated with 1.25 mL Calmodulin affinity resin (Stratagene) for 1 h at 4°C with gentle rotation, after which it was packed into a Poly-prep chromatography column (Bio-Rad). The column was washed two times with 50 mL Calmodulin binding buffer. Bound proteins were eluted three times with 4 mL Calmodulin elution buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, 1 mM Mg-acetate, 1 mM imidazole, 2 mM EGTA, and 10 mM β-mercaptoethanol). The eluate was concentrated using an iCON Concentrator 7 mL/9 K (Pierce). Proteins were separated using preparative SDS-PAGE gel and silver stained with Silver Stain Plus (Bio-Rad).
Mass Spectrometry Analysis
In-gel digests were performed by incubation of the gel slices at 37°C overnight with sequencing grade modified trypsin (Promega). The resulting peptide mixture was extracted from the gel, concentrated using Captrap peptide trap (Michrom Bioresources), and separated on a BioBasic C18 PicoFrit microcapillary column (New Objective) by a 60-min gradient of 7 to 93% acetonitrile in water and 0.1% formic acid. Eluting peptides were electrosprayed directly into a DECA-XP Plus ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (Thermo Fisher Scientific). Full mass spectrometry as well as tandem mass spectrometry spectra were recorded. BioWorks 3.2 software (Thermo Fisher Scientific) based on the SEQUEST algorithm was used for data analysis. The search was performed using the NIH nonredundant database. Peptide/spectra matches were retained if the following criteria were met: XCor scores higher than 1.5 for peptides charged +1, higher than 2.0 for peptides charged +2, higher than 2.5 for peptides charged +3, Delta Cn scores higher than 0.08, and peptide probability lower than 0.05.
Immunoblot and Protein Interaction Analysis
Arabidopsis Columbia wild-type expressing GFP-WIT1 or GFP and GFP-WIT1–transformed wip1-1 wip2-1 wip3-1 triple mutant plants were grown in constant light for 14 d. N. benthamiana plants were grown for 3 d after agroinfiltration was performed. Whole Arabidopsis seedling or infiltrated leaves of N. benthamiana were collected and ground into fine powders. Extracts for co-IP were prepared at 4°C in a buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 3 mM DTT, 1 mM phenylmethylsulphonyl fluoride, and protease inhibitor cocktail (1:100; Sigma-Aldrich). Immunoprecipitates from both Arabidopsis and N. benthamiana were prepared either with monoclonal anti-GFP antibody (A11120; Molecular Probes), polyclonal anti-GFP antibody (ab290; Abcam Cambridge), or anti-WIT1 or anti-WIP1 antibody bound to protein A-sepharose beads (GE Healthcare) with 1 to 3 h binding. The immunoprecipitates were then analyzed by SDS-PAGE, transferred to nitrocellulose or PVDF membranes, and probed with anti-WIT1 (1:2000), anti-RanGAP1 (1:3000), anti-WIP1 (1:2000), or anti-FLAG M2-HRP (1:1000; Sigma-Aldrich) antibody. Ponceau staining was performed with Ponceau S solution (Sigma-Aldrich) for 15 min at room temperature. Two types of anti-GFP antibodies were used for detection of immunoprecipitates: polyclonal (A11122, 1:4000; Molecular Probes) and monoclonal (MO48-3, 1:1000; Medical and Biological Laboratories). A dilution of 1:25,000 was used for the horseradish peroxidase (HRP)–conjugated anti-rabbit secondary antibody (GE Healthcare). HRP-conjugated anti-mouse secondary antibody (Sigma-Aldrich) was used at a dilution of 1:10,000. SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) was used as HRP detection system.
Immunolabeling and Confocal Microscopy
Whole-mount immunolocalization in Arabidopsis roots was performed as described (Sauer et al., 2006). Polyclonal anti-RanGAP1 (1:200), polyclonal anti-WIT1 (1:400), monoclonal anti-tubulin (DM1A, 1:250; Sigma-Aldrich), polyclonal anti-GFP (A11122, 1:200; Molecular Probes), and monoclonal anti-GFP (A11120, 1:100; Molecular Probes) primary antibodies and anti-rabbit or anti-mouse secondary antibodies conjugated with Alexa Fluor 488 or 568 (1:200; Molecular Probes) were used. Images were collected from a PCM 2000/Nikon Eclipse E600 confocal laser scanning microscope as described (Rose and Meier, 2001).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or the GenBank/EMBL data libraries under accession numbers NP_196700 (At5g11390, WIT1), NP_177057 (At1g68910, WIT2), BT003145 (WIP1), AY735734 (WIP2a), AY045697 (WIP3), NP_191872.1 (RanGAP1), NP_197433.1 (RanGAP2), NP_568959.1 (tubulin), NP_199121.1 (WPP1), NP_564498.1 (WPP2), and NP_568504.1 (WPP3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. WIT1 Is Expressed Ubiquitously in Arabidopsis.
Supplemental Figure 2. RanGAP1 Is Targeted to the NE in the Differentiation Zone and during Cytokinesis in the Roots of wit1-1 wit2-1.
Supplemental Figure 3. The Fate of WIP1 in wit1-1 wit2-1.
Supplemental Figure 4. WIT1-RanGAP Interaction in wip1-1 wip2-1 wip3-1 and WIT1 Abundance in Different WIP Double Mutant Combinations.
Supplemental Table 1. Selected Organ-Specific Expression Profiles of WIT1 and WIT2 from Genevestigator Expression Data.
Supplemental Table 2. List of Plasmids Used in This Study.
Supplementary Material
Acknowledgments
We thank the ABRC and MPIZ for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, Irina Sorokina for useful suggestions related to the mass spectrometry service, Thushani Rodrigo-Peiris for the help with initial cloning of WIT1, Xianfeng Morgan Xu for the help with three-dimensional projection confocal imaging, Biao Ding for the use of his confocal microscope, and Sumire Fujiwara and Xiao Zhou for critical reading of the manuscript. Financial support by the National Science Foundation is greatly acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Iris Meier (meier.56@osu.edu).
Online version contains Web-only data.
References
- Alber, F., Dodukovskaya, S., Veenhoff, L.M., Zhang, W., Kipper, J., Devos, D., Suprapto, A., Karni-Schmidt, O., Williams, R., Chait, B.T., Rout, M.P., and Sali, A. (2007. b). Determining the architectures of macromolecular assemblies. Nature 450 683–694. [DOI] [PubMed] [Google Scholar]
- Alber, F., Dodukovskaya, S., Veenhoff, L.M., Zhang, W., Kipper, J., Devos, D., Suprapto, A., Karni-Schmidt, O., Williams, R., Chait, B.T., Sali, A., and Rout, M.P. (2007. a). The molecular architecture of the nuclear pore complex. Nature 450 695–701. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arnaoutov, A., and Dasso, M. (2003). The Ran GTPase regulates kinetochore function. Dev. Cell 5 99–111. [DOI] [PubMed] [Google Scholar]
- Arnaoutov, A., and Dasso, M. (2005). Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 4 1161–1165. [DOI] [PubMed] [Google Scholar]
- Bapteste, E., Charlebois, R.L., MacLeod, D., and Brochier, C. (2005). The two tempos of nuclear pore complex evolution: Highly adapting proteins in an ancient frozen structure. Genome Biol. 6 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., Klebe, C., Kretschmer, J., Wittinghofer, A., and Ponstingl, H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., Krebber, H., Kempf, T., Hermes, I., and Ponstingl, H. (1995). Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc. Natl. Acad. Sci. USA 92 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., and Ponstingl, H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354 80–82. [DOI] [PubMed] [Google Scholar]
- Boehmer, T., Enninga, J., Dales, S., Blobel, G., and Zhong, H. (2003). Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 100 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese, N., Colombo, S., and Pedrazzini, E. (2003). The tale of tail-anchored proteins: Coming from the cytosol and looking for a membrane. J. Cell Biol. 161 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, M., Wilson, K.L., and Gruenbaum, Y. (2001). Membrane proteins of the nuclear pore complex: Gp210 is conserved in Drosophila, C. elegans and A. thaliana. Gene Ther. Mol. Biol. 6 47–55. [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso, M. (2002). The Ran GTPase: Theme and variations. Curr. Biol. 12 502–508. [DOI] [PubMed] [Google Scholar]
- De Souza, C.P., Osmani, A.H., Hashmi, S.B., and Osmani, S.A. (2004). Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14 1973–1984. [DOI] [PubMed] [Google Scholar]
- Devos, D., Dodukovskaya, S., Williams, R., Alber, F., Eswar, N., Chait, B.T., Rout, M.P., and Sali, A. (2006). Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA 103 2172–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., Strasser, A.W., Honer, C.B., and Hollenberg, C.P. (1991). An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7 691–692. [DOI] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., Carazo-Salas, R.E., Schatz, C.A., Guarguaglini, G., Kast, J., Wilm, M., Le Bot, N., Vernos, I., Karsenti, E., and Mattaj, I.W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104 83–93. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., Bilbao-Cortes, D., Walther, T.C., Gruss, O.J., and Mattaj, I.W. (2000). GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell 5 1013–1024. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., Gruss, O.J., and Mattaj, I.W. (2002). The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4 177–184. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K., Traglia, H.M., and Dunst, R.W. (1990). The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J. Cell Biol. 111 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I.W., and Gorlich, D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S., Rose, A., Joseph, J., Dasso, M., and Meier, I. (2005). Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 42 270–282. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Liu, S.-T., Jablonski, S.A., Yen, T.J., and Dasso, M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14 1–20. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Tan, S.H., Karpova, T.S., McNally, J.G., and Dasso, M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P., Weis, K., and Heald, R. (2002). Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus cell extracts. Science 295 2452–2456. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Madrid, A.S., Mancuso, J., Cande, W.Z., and Weis, K. (2006). The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88 97–107. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., Gerace, L., and Melchior, F. (1998). Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Wu, J., and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, I. (2000). A novel link between ran signal transduction and nuclear envelope proteins in plants. Plant Physiol. 124 1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, I., Xu, X.M., Brkljacic, J., Zhao, Q., and Wang, H.-J. (2008). Going green: Plants' alternative way to position the Ran gradient. J. Microsc., in press. [DOI] [PubMed]
- Patel, S., Rose, A., Meulia, T., Dixit, R., Cyr, R.J., and Meier, I. (2004). Arabidopsis WPP-domain proteins are developmentally associated with the nuclear envelope and promote cell division. Plant Cell 16 3260–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pay, A., Resch, K., Frohnmeyer, H., Fejes, E., Nagy, F., and Nick, P. (2002). Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 30 699–709. [DOI] [PubMed] [Google Scholar]
- Quimby, B.B., and Dasso, M. (2003). The small GTPase Ran: Interpreting the signs. Curr. Opin. Cell Biol. 15 338–344. [DOI] [PubMed] [Google Scholar]
- Quimby, B.B., Lamitina, T., L'Hernault, S.W., and Corbett, A.H. (2000). The mechanism of ran import into the nucleus by nuclear transport factor 2. J. Biol. Chem. 275 28575–28582. [DOI] [PubMed] [Google Scholar]
- Rohila, J.S., Chen, M., Cerny, R., and Fromm, M.E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38 172–181. [DOI] [PubMed] [Google Scholar]
- Rose, A., Manikantan, S., Schraegle, S.J., Maloy, M.A., Stahlberg, E.A., and Meier, I. (2004). Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 134 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A., and Meier, I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, M., Paciorek, T., Benkova, E., and Friml, J. (2006). Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protocols 1 98–103. [DOI] [PubMed] [Google Scholar]
- Schrader, N., Stelter, P., Flemming, D., Kunze, R., Hurt, E., and Vetter, I.R. (2008). Structural basis of the Nic96 subcomplex organization in the nuclear pore channel. Mol. Cell 29 46–55. [DOI] [PubMed] [Google Scholar]
- Stavru, F., Hülsmann, B.B., Spang, A., Hartmann, E., Cordes, V.C., and Görlich, D. (2006. a). NDC1: A crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru, F., Nautrup-Pedersen, G., Cordes, V.C., and Görlich, D. (2006. b). Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J. Cell Biol. 173 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Hoyt, M.A., Chan, C., Goetsch, L., Botstein, D., Byers, B., and Thomas, J.H. (1993). NDC1: A nuclear periphery component required for yeast spindle body duplication. J. Cell Biol. 122 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.M., Meulia, T., and Meier, I. (2007). Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 17 1157–1163. [DOI] [PubMed] [Google Scholar]
- Zhang, C., and Clarke, P.R. (2000). Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288 1429–1432. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Hutchins, J.R., Muhlhausser, P., Kutay, U., and Clarke, P.R. (2002). Role of importin-beta in the control of nuclear envelope assembly by Ran. Curr. Biol. 12 498–502. [DOI] [PubMed] [Google Scholar]
- Zhao, Q., Leung, S., Corbett, A.H., and Meier, I. (2006). Identification and characterization of the Arabidopsis orthologs of Nuclear Transport Factor 2, the nuclear import factor of Ran. Plant Physiol. 140 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.