Abstract
Adipocyte differentiation requires a well-defined programme of gene expression in which the transcription factor C/EBPα (CCAAT/enhancer-binding protein) has a central function. Here, we show that Hzf (haematopoietic zinc-finger), a previously identified p53 transcriptional target, regulates C/EBPα expression. Hzf is induced during differentiation of preadipocyte cell lines, and its suppression by short hairpin RNA disrupts adipogenesis. In Hzf's absence, expression of C/EBPα is severely impaired because of reduced translation of its mRNA. Hzf physically interacts with the 3′ untranslated region of C/EBPα mRNA to enhance its translation. Taken together, these findings underscore a critical role of Hzf in the adipogenesis regulatory cascade.
Keywords: adipocyte, C/EBPα, Hzf, translation
Introduction
Adipose tissue has an important function in energy homoeostasis. It serves as an endocrine organ as well as a major site of energy storage, and dysfunction of adipocyte is strongly associated with metabolic disorders including diabetes. The cellular and molecular mechanisms of adipocyte differentiation have been extensively studied using preadipocyte cell lines such as 3T3-L1 cells. A number of transcription factors that regulate the complex transcriptional network during adipogenesis have been identified, and these factors coordinate the sequential expression of genes required for the establishment of the mature fat-cell phenotype (Rosen et al, 2000). The CCAAT/enhancer-binding proteins (C/EBPα, C/EBPβ, and C/EBPδ) and peroxisome proliferator-activated receptor γ (PPARγ) have central functions in this process. Upon the exposure to the adipogenic inducers, confluent preadipocyte cells reenter the cell cycle and progress through a few round of cell division called clonal expansion. During this period, C/EBPβ and C/EBPδ are activated, which lead to the subsequent induction of relatively low levels of PPARγ and C/EBPα (Cao et al, 1991; Yeh et al, 1995). PPARγ further activates C/EBPα, which drives more PPARγ expression in a positive feedback loop. Thus, C/EBPα and PPARγ have mutually re-enforcing functions in fostering later steps of adipogenesis. These transcription factors, together with ADD1/SREBP1c, work synergistically to transactivate the expression of genes required for adipocyte differentiation and insulin sensitivity (Wu et al, 1999).
Mice nullizygous for C/EBPα exhibit neonatal lethality due to defective gluconeogenesis in the liver (Wang et al, 1995), and cells derived from these animals are defective in undergoing adipogenesis in vitro. The lethal phenotype can be rescued by liver-specific expression of a C/EBPα transgene or by ‘knocking in' the C/EBPβ gene into the C/EBPα locus (Chen et al, 2000; Linhart et al, 2001). In turn, ectopic expression of C/EBPα in fibroblast cell lines triggers adipocytic differentiation (Freytag et al, 1994; Lin and Lane, 1994). However, the enforced expression of PPARγ in C/EBPα-null cells can induce adipogenic differentiation, and C/EBPα is incapable of inducing adipogenesis in the absence of PPARγ (Rosen et al, 2002). Adipocytes lacking C/EBPα activity are entirely devoid of insulin-stimulated glucose transport (El Jack et al, 1999; Wu et al, 1999). Therefore, in addition to maintaining PPARγ activity during adipogenesis, C/EBPα is also required for the control of insulin action in mature fat cells.
Hzf (haematopoietic zinc finger) was originally identified as a gene whose expression is induced in haematopoietic progenitor cells derived from differentiating embryonic stem cells in vitro (Hidaka et al, 2000). Consistent with its expression in haematopoietic precursors, mice lacking the Hzf gene exhibited subtle defects in the maturation of megakaryocyte α-granules (Kimura et al, 2002). We independently identified Hzf as a direct transcriptional target of the p53 tumour suppressor (Sugimoto et al, 2006). Hzf is induced by DNA damage or via the Arf tumour suppressor pathway in a p53-dependent manner, and it appears to be required for the maintenance of prolonged G2 cell cycle arrest and p21Cip1 stability following DNA damage. It has been recently reported that Hzf is directly involved in the transcriptional regulation by p53 (Das et al, 2007). The Hzf protein contains three typical C2H2-type zinc-finger domains that can potentially serve as nucleic acid-binding motifs. Iijima et al (2005) reported that Hzf associates through its zinc-finger motifs with the type 1 inositol 1,4,5-triphosphate receptor (IP3R) mRNA in cerebellar Purkinje cells where it regulates the intracellular localization and translation of the mRNA. Presumably, the role of Hzf in megakaryocyte differentiation and cell cycle checkpoint control might also be attributed to its mRNA-binding function.
In further exploring the biological function of Hzf, we found that it is highly expressed in mouse adipose tissue and is induced in preadipocyte cell lines during their differentiation to mature fat cells. We now show that Hzf directly associates with the 3′ untranslated region (UTR) of C/EBPα mRNA to enhance its efficient translation and to thereby reinforce adipogenesis. Although Hzf is dispensable for adipose tissue development per se, Hzf-null mice exhibit glucose intolerance and reduced insulin sensitivity. Taken together, these findings provide new insights about C/EBPα regulation during adipocyte differentiation, and point to an unexpected role of p53 in controlling glucose metabolism.
Results
HZF is required for adipocytic differentiation of mouse fibroblasts
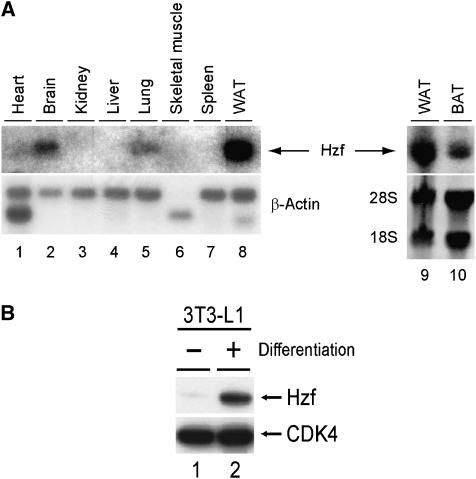
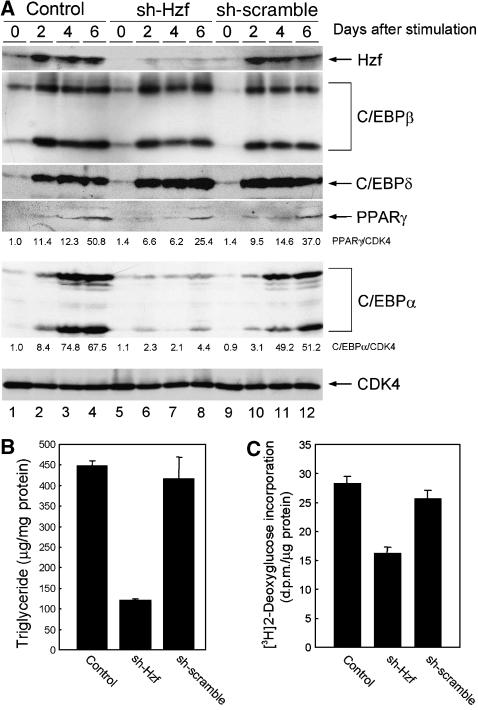
We analysed Hzf mRNA expression in mouse tissues by northern blotting and detected its expression in brain (Iijima et al, 2005), lung, and most prominently in white adipose tissue (Figure 1A, lanes 2, 5, 8–10). Because the Hzf mRNA level in adipose tissue was much higher than that observed in other tissues, we examined Hzf expression in the 3T3-L1 preadipocyte cell line. Hzf protein levels were low in undifferentiated 3T3-L1 cells, but were markedly induced after hormonal stimulation of adipocyte differentiation (Figure 1B). To test the possibility that Hzf contributed directly to this process, 3T3-L1 cells were infected with either a control retrovirus encoding puromycin resistance or with a vector also encoding a short hairpin (sh) RNA that targets the 3′UTR of Hzf mRNA (Sugimoto et al, 2006), or sh-RNA that does not target any known sequence on mouse genome (Maehara et al, 2005). Infected cells were selected with puromycin, after which differentiation was induced with hormones. No significant differences were observed in basal triglyceride levels between uninduced control, sh-Hzf, and sh-scramble (SCR)-transduced cells in which Hzf protein was undetectable (data not shown). In contrast, Hzf protein was readily detected within 48 h after hormonal stimulation of 3T3-L1 cells infected with the control or sh-SCR vector, and its expression was sustained for up to 6 days when more than 60% of the cells had differentiated (Figure 2A top, lanes 1–4, and 9–12). In the presence of inhibitory sh-RNA, Hzf expression was efficiently suppressed (lanes 5–8). Under these conditions, we quantified cytoplasmic triglyceride and insulin-mediated glucose uptake. As expected, triglyceride levels and insulin uptakes were increased upon differentiation in control or sh-SCR-infected 3T3-L1 cells but were inhibited in sh-Hzf-infected cells (Figure 2B and C, data not shown). The specificity of sh-Hzf was further confirmed in Figure 7 that shows re-introduction of Hzf ORF fully restored the defective adipogenesis in sh-Hzf cells.
Figure 1.
Hzf is highly expressed in adipose tissue. (A) Expression of Hzf mRNA was analysed in normal mouse tissues using Hzf cDNA as a probe. WAT (white adipose tissue) and BAT (brown adipose tissue) were obtained from epididymal and interscapular adipose, respectively. (B) Expression of Hzf protein in 3T3-L1 cells (−: undifferentiated, +: differentiated for 2 days) was analysed by immunoblotting. CDK4 was used as a loading control.
Figure 2.
Hzf is required for efficient C/EBPα expression and adipogenesis in fibroblasts. (A) Control, sh-Hzf, and sh-SCR 3T3-L1 cells were stimulated to differentiate for indicated periods, and expression of the indicated proteins was analysed by immunoblotting with CDK4 as a loading control. Values represent the ratios of PPARγ and C/EBPα to CDK4 protein. (B) 3T3-L1 cells infected with indicated retroviruses were induced to differentiate for 6 days, and triglycerides in cell lysates were measured. (C) Differentiated 3T3-L1 cells infected with indicated retroviruses were assayed for 2-deoxyglucose uptake in the presence of 5 μg/ml of insulin.
Figure 7.
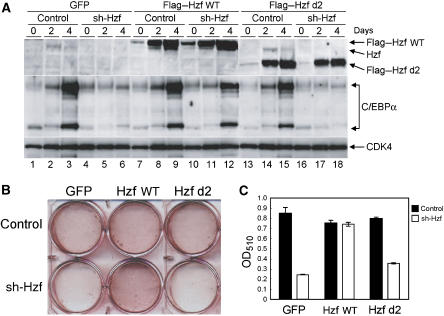
RNA-binding domain of Hzf protein is required for the induction of C/EBPα protein and for adipocyte differentiation. (A) 3T3-L1 cells infected with the indicated retroviruses were cultured in differentiation-inducing medium for various periods. Lysates were prepared and protein expression was analysed by immunoblotting. (B) Cells were stained with Oil Red O after 4 days. (C) Quantification of lipid incorporation by measuring the intensity of Oil Red O staining in (B).
Because our previous findings indicated that Hzf is a direct target of p53 transcriptional activation (Sugimoto et al, 2006), we examined whether the function of Hzf in adipocyte differentiation depends upon p53 or on p21Cip1, a p53-inducible inhibitor of cyclin-dependent kinases that enforces p53-dependent cell cycle arrest. Mouse embryonic fibroblasts (MEFs) prepared from wild-type, p53−/−, or p21Cip1−/− mice were infected with control, sh-Hzf, or sh-SCR retrovirus and subjected to differentiation. Oil Red O staining revealed that Hzf is required for the efficient differentiation of primary MEFs into adipocytes (Supplementary Figure S1). Like MEFs expressing Hzf shRNA, Hzf−/− MEFs also failed to differentiate to adipocytes (Supplementary Figure S2). However, neither p53 nor p21Cip1 were required for adipocyte differentiation (Supplementary Figure S1). Similarly, although other cell cycle regulators including E2Fs, RB family, CDK4, p27Kip1, and p18Ink4c participate in adipogenesis (Morrison and Farmer, 1999; Fajas et al, 2002; Abella et al, 2005; Scime et al, 2005), we have no evidence that Hzf directly affects the activity of these proteins or the clonal expansion of preadipocytes (data not shown and Supplementary Figure S3).
Expression of C/EBPα is impaired in the absence of Hzf
Transcription factors C/EBPα, C/EBPβ, C/EBPδ, and PPARγ are necessary for adipogenesis. Because our results indicated that Hzf-knockdown cells failed to undergo efficient adipogenic differentiation, we compared the expression of these transcription factors in control and Hzf-knockdown 3T3-L1 cells. C/EBPβ, C/EBPδ, PPARγ, and C/EBPα proteins were each induced in control or sh-SCR virus-infected 3T3-L1 cells upon hormonal stimulation (Figure 2A, lanes 1 to 4, and 9–12). C/EBPβ and C/EBPδ proteins were similarly induced in sh-Hzf cells (lanes 5–8), whereas PPARγ accumulated to somewhat lower levels than that observed in control cells. In sharp contrast, the induction of C/EBPα protein was significantly attenuated in Hzf-knockdown cells as well as Hzf−/− MEFs (Supplementary Figure S2). Another transcription factor that leads to PPARγ activation, ADD1/SREBP1c, was induced equally well in both control and Hzf-knockdown cells (Supplementary Figure S4).
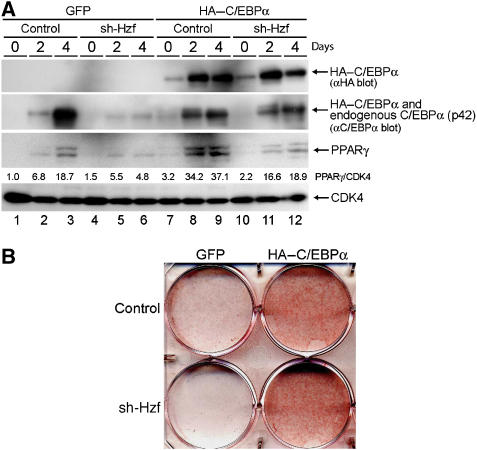
Because the expression of PPARγ during adipogenesis is partly dependent on the activity of C/EBPα (Wu et al, 1999), we reasoned that the attenuated induction of PPARγ protein in sh-Hzf cell may well be attributed to impaired C/EBPα expression in these cells. To directly test this possibility, we retrovirally expressed HA-tagged human C/EBPα together with green fluorescent protein (GFP) or GFP protein alone in control or Hzf-knockdown 3T3-L1 cells. Infected cells were selected and then cultured under differentiation-inducing conditions. For unknown reasons, the exogenous HA–C/EBPα protein levels were low in undifferentiated 3T3-L1 cells, but were increased in response to differentiating stimulation (Figure 3A top, lanes 7–12). The achieved level of the exogenous HA–C/EBPα protein was lower than the endogenous protein, as judged by immunoblotting with an antibody to C/EBPα itself. Induction of PPARγ was restored under these conditions (lanes 11 and 12), suggesting that its weakened induction reflects the loss of C/EBPα expression in sh-Hzf-treated cells. Exogenous C/EBPα protein greatly enhanced adipocytic differentiation as judged by Oil Red O staining, and sh-Hzf did not noticeably affect differentiation under these circumstances (Figure 3B). These results suggest that the defect in undergoing efficient adipogenesis in sh-Hzf cells is attributed to attenuated expression of C/EBPα protein.
Figure 3.
Exogenous C/EBPα restores adipocytic differentiation of sh-Hzf-treated 3T3-L1 cells. Cells expressing the indicated shRNAs and co-infected with retroviral vectors encoding C/EBPα+GFP or GFP alone were stimulated to differentiate for indicated periods. Protein expression was analysed by immunoblotting (A), and cells cultured in differentiation-inducing media (4 days) were stained with Oil Red O (B). Values represent the ratios of PPARγ protein to CDK4 protein in each sample.
Translation of C/EBPα mRNA is impaired in sh-Hzf-treated 3T3-L1 cells
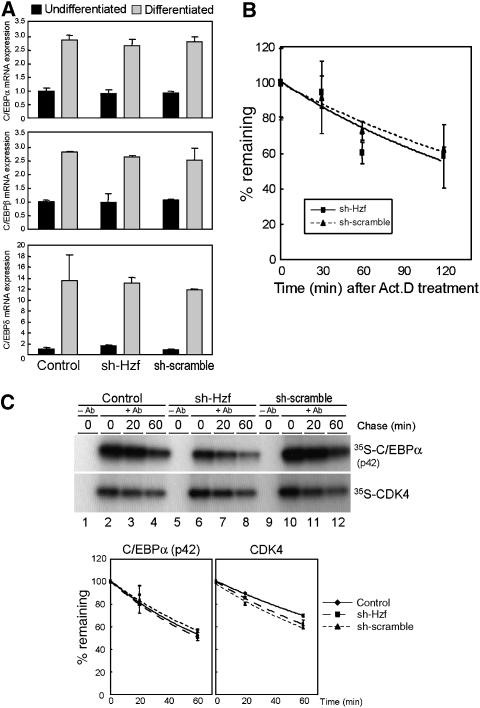
Because Hzf is required for the induction of C/EBPα protein during adipogenesis, we next sought the molecular mechanism that links Hzf to C/EBPα expression. We first employed real-time PCR analysis to compare C/EBP family mRNA expression in control, sh-SCR, and sh-Hzf virus-infected cells. C/EBPβ and δ mRNA levels were low in undifferentiated 3T3-L1 cells, but were dramatically increased after stimulating fat-cell differentiation (Figure 4A). Similarly, C/EBPα mRNA was also induced irrespective of Hzf status (Figure 4A and Supplementary Figure S2). We subsequently checked if loss of Hzf affects the stability of C/EBPα mRNA. Cells were cultured in the presence of actinomycin D, and mRNA decay was analysed by real-time PCR. As shown in Figure 4B, there was no significant difference in the stability of C/EBPα mRNA among these cells. These results imply that Hzf does not affect the expression of the C/EBPα mRNA during adipogenesis.
Figure 4.
Loss of Hzf does not affect the transcription or protein stability of C/EBPα. (A) 3T3-L1 cells infected with the indicated retroviruses were cultured in normal or differentiation-inducing media for 4 days. Expression of C/EBPα, C/EBPβ, and C/EBPδ mRNA was analysed by real-time quantitative PCR. Values were normalized to the expression of GAPDH mRNA. (B) 3T3-L1 cells differentiated for 4 days were treated with 5 μg/ml of actinomycin D for indicated periods, and expression of C/EBPα mRNA was analysed by real-time PCR. Values were normalized to the amount of 18S rRNA in each sample. (C) Cells differentiated (4 days) were pulse-labelled with 35S-Met/Cys for 1 h and chased for the indicated periods. C/EBPα or CDK4 protein was immunoprecipitated, separated on denaturing gels, and detected by autoradiography. Radioactivity in each band was measured (lower panel).
Our previous report suggested that Hzf can affect ubiquitin–proteasome-dependent p21Cip1 stability in cells undergoing radiation-induced DNA damage (Sugimoto et al, 2006). Hence, we next determined whether inhibition of proteasome activity could restore the C/EBPα level in sh-Hzf cells. Control or sh-Hzf virus-infected 3T3-L1 cells were cultured under differentiating conditions for 4 days and treated with the proteasome inhibitor, MG-132 (Supplementary Figure S5). In both transduced cell types, MG-132 treatment had no impact on C/EBPα protein levels; accumulation of C/EBPα was reduced in sh-Hzf-infected cells irrespective of proteasomal activity. To more directly evaluate C/EBPα protein stability, cells were metabolically exposed to radiolabelled amino acids, and C/EBPα protein was immunoprecipitated following a ‘chase' with radioisotope-free medium. Recovered C/EBPα was electrophoretically separated on denaturing polyacrylamide gels and detected by autoradiography. Despite the significant differences in the total C/EBPα protein levels between these cells, there was no significant change in the half-life of C/EBPα protein (Figure 4C).
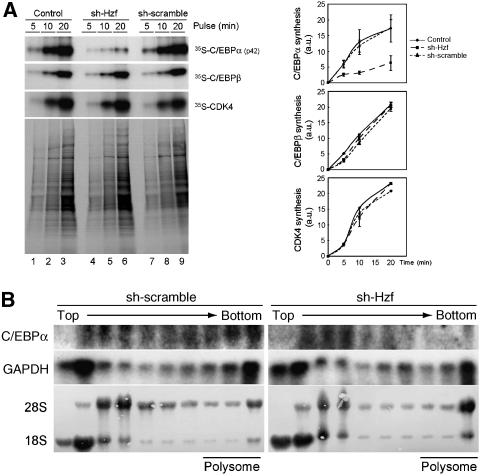
Iijima et al (2005) reported that Hzf regulates the translation of the IP3R mRNA in cerebellar Purkinje cells through direct interaction with a cis-element residing within the 3′UTR of the mRNA. We therefore wished to determine if Hzf also regulates the translation of the C/EBPα mRNA during adipogenesis. Control, sh-Hzf-, or sh-SCR-expressing 3T3-L1 cells were metabolically labelled, and the rate of incorporation of radiolabelled amino-acid precursors into the C/EBPα protein was evaluated. Although there was no significant difference in overall protein synthesis, the de novo synthesis of C/EBPα protein was significantly decreased in cells expressing sh-Hzf (Figure 5A top). We estimate that the rate of C/EBPα synthesis declined to approximately one-third of that of cells expressing the control vector. Under these conditions, loss of Hzf had no impact on the rate of C/EBPβ or CDK4 protein synthesis. To further examine if the translation of Hzf mRNA is regulated by Hzf, cytoplasmic lysates prepared from these cells were fractionated by sucrose gradient centrifugation. RNAs were isolated from each fraction, and C/EBPα mRNA was analysed by northern blotting. The distribution of GAPDH mRNA was virtually identical in these cells irrespective of Hzf status. C/EBPα mRNA was mainly observed around bottom fractions that include polysomal fractions in sh-SCR cells, suggesting that C/EBPα mRNA is actively translated in these cells (Figure 5B, data not shown). However, C/EBPα mRNA was re-distributed into lighter fractions in the absence of Hzf, in which mRNA translation is diminished (Figure 5B). Taken together, these results strongly suggest that Hzf is required for the efficient translation of C/EBPα mRNA in differentiating 3T3-L1 cells.
Figure 5.
Translation of C/EBPα mRNA is impaired in Hzf-knockdown cells. (A) Cells were pulse-labelled with 35S-Met/Cys for the indicated periods, and immunoprecipitated C/EBPα, C/EBPβ, or CDK4 protein was separated by SDS–PAGE and detected by autoradiography. Portions of the total radiolabelled cell extract were analysed in parallel (lower panel). Radioactivity of each protein was measured (right). (B) Cytoplasmic lysates prepared from cells differentiated for 4 days were fractionated using sucrose density gradient centrifugation. rRNA (28S and 18S) was visualized by methylene blue staining. northern blot analysis was performed using C/EBPα or GAPDH cDNA as a probe.
Hzf binds to C/EBPα mRNA and regulates its translation
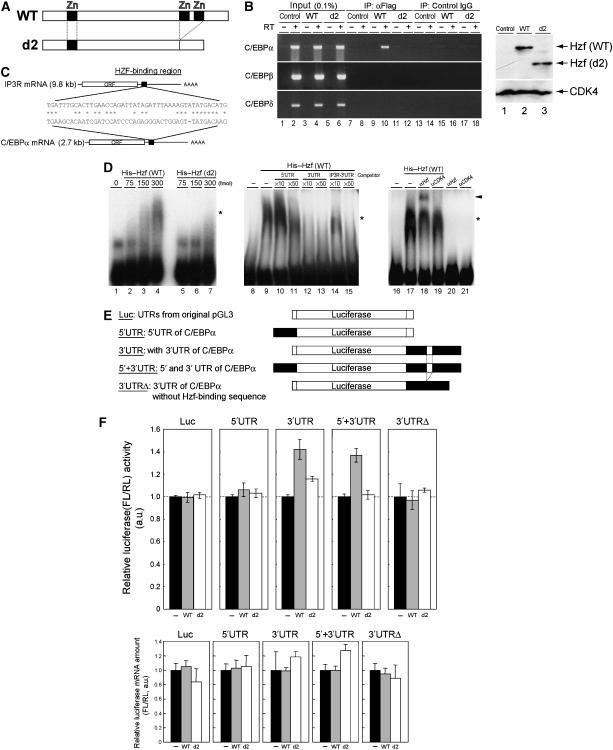
Hzf protein contains three typical C2H2-type zinc-finger motifs and can serve as a sequence-specific RNA-binding domain (Iijima et al, 2005). We therefore asked whether Hzf associates with C/EBPα mRNA in differentiating 3T3-L1 cells. 3T3-L1 cells were infected with retrovirus encoding GFP, Flag-tagged Hzf (together with GFP) or with a Flag-tagged Hzf mutant (d2) that lacks two of the zinc-finger domains near its C terminus (Figure 6A). These cells express comparable amount of exogenous Hzf proteins as judged by immunoblotting using anti-Flag antibody (Figure 6B right). Following induction of differentiation for 4 days, cells were crosslinked using formaldehyde, and cell lysates were prepared and immunoprecipitated using anti-Flag. RNA was eluted from immunoprecipitates after reversal of crosslinking and was subjected to RT–PCR using specific primers for C/EBPα, C/EBPβ, or C/EBPδ mRNA. C/EBPα mRNA was specifically enriched in immune complexes containing wild-type Hzf (Figure 6B, lane 10), but not in control immunoprecipitates performed with an irrelevant antibody (Figure 6B, lane 8). Consistent with the previous report by Iijima et al (2005) and with our own data indicating that the Hzf d2 mutant is defective (Sugimoto et al, 2006), the d2 mutant failed to associate with C/EBPα mRNA in differentiating 3T3-L1 cells (Figure 6B, lane 12). Under the same conditions, we were unable to detect the C/EBPβ or δ mRNA in the Hzf immune complex.
Figure 6.
Hzf associates with C/EBPα mRNA to enhance its translation. (A) Schematic representation of Hzf proteins. (B) 3T3-L1 cells infected with indicated retroviruses were induced to differentiate for 4 days. Following formaldehyde crosslinking, cell lysates were precipitated with antibodies to the Flag epitope or control IgG. RNA eluted from each IP complex was subjected to RT–PCR using C/EBPα, C/EBPβ, or C/EBPδ primers (left panel). The same RT–PCR reaction was performed using 0.1% of supernatant before IP (input). Expression of exogenous Hzf protein was confirmed by immunoblotting with CDK4 as a loading control (right panel). (C) A potential Hzf-binding region in the 3′UTR of C/EBPα mRNA is illustrated. (D) R-EMSA was performed using a radiolabelled RNA probe representing the C/EBPα 3′UTR. The probe was mixed with increasing quantities of bacterially produced wild-type (lanes 2–4) or d2 mutant (lanes 5–7) Hzf proteins. Competition assay was performed on wild-type Hzf proteins with increasing amount of non-radiolabelled C/EBPα 5′UTR (lanes 10 and 11), C/EBPα 3′UTR (lanes 12 and 13), or IP3R 3′UTR (lanes 14 and 15) fragment. Hzf antibody was used for supershift, and CDK4 antibody was used as a control. Complexes were separated by native PAGE and detected by autoradiography. The asterisk and arrowhead indicate Hzf protein–RNA complexes and supershifted complex, respectively. (E) Schematic representation of luciferase reporter mRNAs. Firefly luciferase mRNA in conjunction with 5′ and/or 3′UTRs (either wild-type or mutant sequence that lacks Hzf-binding segment illustrated in (C)) of C/EBPα mRNA was expressed under the regulation of an SV40 promoter and enhancer. (F) Luciferase reporters illustrated in (E) were co-transfected into undifferentiated 3T3-L1 cells together with an SV40-driven Renilla luciferase plasmid used as an internal standard. Upper panel: luciferase activities in each sample were measured, and relative luciferase activities (firefly/Renilla) in each sample were calculated. The relative luciferase activity (FL/RL) of the sample transfected with the control plasmid was normalized to 100% in each group. Lower panel: ratios of firefly and Renilla luciferase mRNA level in each sample were analysed by real-time quantitative PCR analysis.
Because Hzf directly binds to a specific RNA sequence in the 3′UTR of IP3R mRNA (Iijima et al, 2005), we searched for potential Hzf-binding sequences in the C/EBPα mRNA and found one candidate segment (Figure 6C). To test whether Hzf binds to these sequences, we performed an RNA-electrophoretic mobility shift assay (R-EMSA). A radiolabelled RNA probe that contains the complete sequence of the 5′UTR or a portion of 3′UTR of C/EBPα mRNA that consists of the region homologous to the Hzf-binding sequence of IP3R (Figure 6C) and 40 bases of its flanking sequence was mixed with increasing amounts of bacterially produced wild-type or d2 mutant Hzf protein, and RNA–protein complexes were electrophoretically separated on polyacrylamide gels. We detected a significant mobility shift of the 3′UTR probe in the presence of wild-type Hzf protein but not with the same amount of the d2 mutant (Figure 6D). To more directly compare the specificities of interaction of these molecules, R-EMSA was performed in the presence of excess amount of non-radiolabelled competitors, including 5′UTR or 3′UTR of C/EBPα mRNA, or 3′UTR sequence of IP3R mRNA (Figure 6C). As expected, the signal disappeared by adding the excess cold 3′UTR of C/EBPα mRNA (Figure 6D, lanes 12 and 13). 3′UTR of IP3R mRNA also competed with the radiolabelled probe with less efficiency, suggesting that Hzf prefers 3′UTR of C/EBPα to IP3R mRNA under these conditions (lanes 14 and 15). On the other hand, the signal hardly disappeared by 5′UTR of C/EBPα (lanes 10 and 11). We then performed supershift experiment using Hzf or CDK4 antibody. We observed a supershifted signal in the presence of Hzf antibody, but not with control CDK4 antibody (lanes 18 and 19), suggesting that these signals include Hzf–RNA complexes.
Next, we checked if binding of Hzf to the 3′UTR of C/EBPα mRNA could enhance its translation. For this purpose, we constructed luciferase reporter plasmids containing whole 5′UTR and/or 3′UTR (either wild type or mutant lacking Hzf-binding region) derived from C/EBPα mRNA (Figure 6E). The reporter plasmids were co-transfected with wild-type or d2 mutant Hzf expression plasmids together with SV40 promoter-driven Renilla luciferase. Co-expression of Hzf proteins (either wild type or d2 mutant) did not affect the expression of luciferase protein produced from the mRNA that contains vector-derived UTRs or the 5′UTR of C/EBPα mRNA (Figure 6F upper panel). In the presence of the 3′UTR of C/EBPα mRNA, however, we detected a slight but reproducible increase in luciferase activity when cells were co-transfected with wild-type Hzf expression plasmid. Under the same condition, Hzf d2 mutant failed to increase the luciferase activity. Expression of Hzf (wild type or d2) protein did not affect the firefly/Renilla mRNA expression ratios in all samples (lower panel). Consistent with the observations that loss of Hzf did not affect the expression of other C/EBPs, Hzf did not affect the translation of luciferase mRNA conjugated to UTR of other C/EBP (data not shown). Taken together, these results strongly suggest that Hzf associates with the 3′UTR of C/EBPα mRNA to enhance its translation.
The mRNA-binding domain of Hzf protein is required for the induction of C/EBPα and adipogenesis
As Hzf is induced by adipogenic stimulation and associates with C/EBPα mRNA to enhance its translation, we next examined whether the mRNA-binding activity of the Hzf protein is required for adipocyte differentiation. 3T3-L1 cells were infected with the control or sh-Hzf-expressing retrovirus together with wild-type or d2 mutant Hzf, and the cells were cultured under differentiating conditions. We noted increases in both wild-type and d2 mutant Hzf protein levels after hormonal stimulation, which suggests that Hzf might itself be regulated through a post-translational mechanism during adipogenesis. Nonetheless, only wild-type Hzf protein was able to restore the induction of C/EBPα protein in sh-Hzf 3T3-L1 cells after stimulation (Figure 7A). Expression of wild-type Hzf also re-established differentiation in sh-Hzf-treated cells, whereas the d2 mutant failed to induce differentiation under the same conditions (Figure 7B and C). Notably, the overexpression of wild-type Hzf protein itself neither induced nor enhanced C/EBPα expression or differentiation of 3T3-L1 cells treated with the control shRNA vector, implying that endogenous levels of Hzf protein are sufficient to drive this process. Taken together, these results strongly suggest that the mRNA-binding domain of Hzf protein is required for induction of the C/EBPα protein and adipogenesis.
Reduced insulin sensitivity and glucose metabolism in Hzf knockout mice
All of the in vitro studies indicated that Hzf is required for efficient adipocyte differentiation through the translational regulation of C/EBPα mRNA. Therefore, we next wished to know whether Hzf-null animals exhibit defects in adipose tissue development. However, epididymal and inguinal fat pads were virtually normal in adult Hzf-null animals (Supplementary Figure S6, data not shown). Therefore Hzf appears to be dispensable for adipose tissue development in vivo. Consistent with studies using 3T3-L1 or MEFs, the C/EBPα protein level, but not mRNA level, was lower than that of wild type, suggesting that C/EBPα is translationally regulated by Hzf in vivo. Wu et al (1999) reported that C/EBPα expression is not an absolute prerequisite for adipogenic differentiation when PPARγ is activated. However, adipocytes derived from C/EBPα-null cells show a complete absence of insulin-stimulated glucose transport. This prompted us to examine whether Hzf-null mice have defects in glucose homoeostasis. Indeed, we observed impaired glucose tolerance in Hzf-null animals compared with their wild-type counterparts (Supplementary Figure S7A), which was accompanied by resistance to the blood glucose-lowering effects of insulin (Supplementary Figure S7B). Consistent with these observations, plasma insulin levels of Hzf-null animals were higher, and homoeostasis model assessment index, which strongly correlates with insulin resistance (Matthews et al, 1985), was significantly increased in Hzf-null animals (Supplementary Figure S7C), suggesting that lack of Hzf expression leads to insulin resistance. To further investigate functional defects in Hzf-null adipose tissue, we measured plasma adiponectin, which is secreted from adipose tissue and enhances insulin-mediated glucose transport in several tissues (Maeda et al, 2002). Plasma adiponectin levels were significantly lower in Hzf knockout animals than in wild-type animals (Supplementary Figure S7D). Taken together, these results strongly suggest that loss of Hzf causes functional deregulation of adipose tissues, although it may not affect the total fat mass in the body.
Discussion
Although Hzf was first discovered in an attempt to identify genes expressed in haematopoietic precursor cells (Hidaka et al, 2000), its elimination in the mouse germ line contributes only to subtle phenotypic anomalies (Kimura et al, 2002). Our studies now reveal that Hzf is highly expressed in adult adipose tissue and is induced during adipocyte differentiation of cultured cells where it governs C/EBPα expression. Although the inactivation of Hzf significantly slows the efficient adipogenic differentiation of 3T3-L1 cells, adult Hzf-null mice show no defects in fat mass, implying that Hzf activity is not strictly essential for normal fat accumulation and that other mechanisms likely compensate for Hzf loss.
Differentiation into adipocytes requires the sequential expression of the transcription factors, C/EBPβ, C/EBPδ, PPARγ, and C/EBPα. These proteins coordinate the expression of genes responsible for the establishment of the fat-cell phenotype. Of these, sh-Hzf significantly repressed C/EBPα expression. Loss of Hzf also affected accumulation of PPARγ but to a much lesser extent, presumably reflecting its reduced positive feedback regulation by C/EBPα. Although expression of short interference RNA could provoke nonspecific or off-target effects, it is unlikely that the phenotypes of sh-Hzf we observed are attributed to such effects because of the following reasons. First, introduction of relevant double-stranded RNA that does not have any homologous sequence with mouse genome sequence did not elicit any effect on adipocyte differentiation or expression of the transcription factors. Second, re-introduction of full-length Hzf ORF in Hzf-knockdown cells fully restored the C/EBPα expression as well as the ability to differentiate into adipocyte. Moreover, MEFs prepared from Hzf-null animals were also defective in undergoing efficient adipogenesis.
Exogenous C/EBPα protein completely restored adipogenic differentiation and PPARγ expression in Hzf knockdown cells, thereby pinpointing the level at which Hzf functions. However, the enforced overexpression of Hzf did not further increase the level of C/EBPα protein induced by hormonal stimulation, implying that Hzf is not limiting under normal inductive conditions. Ectopic expression of C/EBPα protein confers an ability to differentiate into adipocytes in non-adipogenic cell lines (Freytag et al, 1994). Hzf overexpression, in contrast, did not provoke an adipogenic response in NIH-3T3 cells (data not shown), indicating that Hzf is itself insufficient to drive C/EBPα levels high enough to induce adipogenesis. Presumably, the ‘upstream' transcription factors that regulate C/EBPα expression are required to ensure differentiation.
PPARγ activity may be attenuated in the absence of Hzf, but the C/EBPα gene is transcribed normally irrespective of Hzf expression, suggesting that the residual activity of PPARγ is sufficient at least for the transcriptional activation of C/EBPα. Loss of Hzf does not affect C/EBPα protein turnover, but instead, the de novo synthesis of C/EBPα is significantly decreased in Hzf's absence. Following on the report of Iijima et al (2005) that Hzf directly associates with IP3R mRNA to regulate its translation and intracellular localization, our experiments similarly indicate that Hzf directly interacts with C/EBPα mRNA in living cells. As for IP3R mRNA, Hzf directly binds in vitro through its zinc-finger domains to a specific sequence identified within the 3′UTR of C/EBPα mRNA. We reason that this binding mediates translational control of C/EBPα mRNA, because Hzf can enhance the translation of luciferase mRNA conjugated with the 3′UTR, but not the 5′UTR, of C/EBPα mRNA. However, Hzf was unable to increase the luciferase activity when the luciferase mRNA is conjugated to minimal Hzf-binding sequence illustrated (data not shown); therefore, it is conceivable that Hzf associates with other proteins on C/EBPα mRNA to facilitate the access or activity of ribosomes. It has been reported that C/EBPα is translationally regulated by RNA-binding proteins such as calreticulin and hnRNP E2 (Perrotti et al, 2002; Timchenko et al, 2002). These proteins bind to 5′UTR of C/EBPα mRNA and repress its translation, which could cause acute or chronic myeloid leukaemia (Perrotti et al, 2002; Helbling et al, 2004). So far we have no evidence suggesting that Hzf functionally interacts with calreticulin, or calreticulin itself is involved in adipogenic process.
In addition to the translational regulation, C/EBPα may also be under post-translational regulation during adipogenesis, as the exogenous HA–C/EBPα protein, of which transcript was driven by retroviral promoter and does not retain Hzf-binding UTR sequence, was increased in response to adipogenic stimulation (Figure 3A). However, the exogenous C/EBPα protein was increased regardless of Hzf status, therefore indicating that Hzf is not involved in such regulation.
By sharp contrast to in vitro studies demonstrating Hzf's contribution to adipocyte differentiation, we did not observe obvious morphological abnormalities in the epididymal and inguinal fat pads of Hzf-null animals. Hzf-null mice are much smaller than wild-type or Hzf+/− littermates at birth, but these differences in size are no longer apparent by the time animals reach 1 month of age. This implies that, although Hzf may affect adipose development during embryonic development or at an early stage after birth, the gene is dispensable for the normal development of adult adipose tissue. Presumably, other proteins compensate for the loss of Hzf soon after birth. Similar compensation mechanisms are exemplified by observations that although mice nullizygous for C/EBPβ and C/EBPδ have reduced adipose mass, PPARγ and C/EBPα expression are normal in the remaining adipose tissue (Tanaka et al, 1997). Similarly, ADD1/SREBP1c, which also essentially contributes to adipogenesis in vitro through the activation of PPARγ, is dispensable for adipose tissue development in vivo (Shimano et al, 1997).
In vitro studies suggest that C/EBPα is dispensable for adipogenesis when PPARγ is ectopically activated. However, these adipocytes are entirely devoid of insulin-dependent glucose transport (Wu et al, 1999). Although mice in which C/EBPα genes are postnatally deleted suffer from abnormal glucose metabolism due to defects in gluconeogenesis in the liver (Inoue et al, 2004; Yang et al, 2005), defects within adipose tissue might also contribute. Likewise, animals with adipocyte-specific PPARγ deficiency have insulin resistance in both fat and liver (He et al, 2003). As there is no detectable Hzf expression in the liver, it is unlikely that Hzf is directly involved in liver function. Importantly, however, Hzf-null animals clearly show impaired glucose tolerance and insulin sensitivity. Therefore, we favor the idea that the latter phenotypes are attributed, at least in part, to deregulation of C/EBPα activity in adipose tissue. This is further supported by the observation that insulin-mediated glucose uptake was also impaired in sh-Hzf 3T3-L1 cells.
Besides the direct effect of Hzf on adipose tissue, Hzf may also regulate the insulin sensitivity in other tissues. We observed that Hzf-null animals have lower plasma adiponectin levels. Adiponectin is a hormone that is exclusively expressed in, and secreted from, adipose tissue. Lower adiponectin levels could trigger insulin resistance in other tissues including liver and muscle, and mice nullizygous for adiponectin or its receptor genes may possess normal fat mass in the body, but are defective in insulin-mediated glucose transport (Kubota et al, 2002; Maeda et al, 2002; Yamauchi et al, 2007). Thus, it is possible that loss of Hzf confers insulin resistance also to other tissues through the repression of plasma adiponectin levels, which could contribute to the whole body phenotype observed in Hzf-null mice. Indeed, the phenotype of Hzf-null animals is reminiscent of those of adiponectin-deficient mice or their receptor-deficient counterparts. Therefore, Hzf's function in vivo might be more important for maintenance of adiponectin expression than for adipose tissue development per se. Consistent with these observations, C/EBPα directly regulates adiponectin transcription (Qiao et al, 2005). However, the precise mechanism of adiponectin transcription is still controversial, as others have reported that PPARγ, C/EBPβ, or ADD1 also regulate adiponectin expression during adipogenesis (Iwaki et al, 2003; Park et al, 2004; Seo et al, 2004). DNA microarray analysis indicates that ectopic expression of Hzf results in an increase of adiponectin mRNA in NIH-3T3 cells without an accompanying induction of these other transcription factors (M Sugimoto, unpublished results).
Altered glucose metabolism is a hallmark of cancer cells, and drugs that affect gluconeogenesis in type 2 diabetes are now being tested for their effects as antitumour agents (Garber, 2004; Shaw and Cantley, 2006). Intriguingly, the fact that Hzf is a direct transcriptional target of p53, which regulates the expression of p53 target proteins including p21Cip1 (Sugimoto et al, 2006; Das et al, 2007), points to a potential mechanistic link between stress-induced p53 activity and altered glucose metabolism. Very recently completed genomic surveys for human genes that affect susceptibility to type 2 diabetes have implicated the CDKN2A–CDKN2B gene cluster as possibly contributory (Saxena et al, 2007; Scott et al, 2007; Zeggini et al, 2007). This locus encodes two INK4 cyclin-dependent kinase inhibitors as well as the p14ARF tumour suppressor protein, an oncogene-induced activator of p53. In turn, the human HZF gene resides on chromosome 12q13, a locus involved in susceptibility to type 2 diabetes (Lakka et al, 2003; Wiltshire et al, 2004). Reduced adiponectin levels have similarly been implicated in this disease (Hotta et al, 2000). Taken together, these findings conceptually link two canonical, frequently inactivated tumour suppressors (ARF and p53) through Hzf to adiponectin and to downstream genes regulating glucose metabolism.
Materials and methods
Cells and culture conditions
3T3-L1 and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 2 mM glutamine, and 100 U/ml penicillin and streptomycin (P/S). Primary MEFs were cultured in medium supplemented with 0.1 mM non-essential amino acids, 55 μM 2-mercaptoethanol, and 10 μg/ml gentamycin instead of P/S (Zindy et al, 1998). To induce adipocytic differentiation, cells were kept confluent for 2 days and switched to differentiation medium (DMEM containing 5 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM MIX (Sigma Chemicals, St Louis, MO)). The medium was replaced every 2 days until cells were harvested.
Retrovirus production and infection
Human C/EBPα cDNA was obtained by PCR using the following primers: sense 5′-AAAGAATTCATGGAGTCGGCCGACTTCTA-3′, antisense 5′-AAACTCGAGTCACGCGCAGTTGCCCATGG-3′. The PCR product digested with EcoRI and XhoI was cloned into a murine stem cell retroviral vector containing a 5′ HA tag. Retrovirus production and infection were performed as described previously (Zindy et al, 1998).
Immunoblotting, immunoprecipitation and metabolic labelling
Cell lysates prepared using RIPA buffer (10 mM Na-phosphate, pH 7.2, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Na-deoxycholate, 1% NP-40 and PIs) were separated by SDS–PAGE and transferred to PVDF membranes (Millipore, Bedford, MA). Proteins were detected using antibodies to the Hzf C terminus (Sugimoto et al, 2006), C/EBPα (14AA; Santa Cruz Biotechnology, Santa Cruz, CA), C/EBPβ (C-19; Santa Cruz), C/EBPδ (C-22; Santa Cruz), PPARγ (E-8; Santa Cruz), ADD1/SREBP1c (2A4; Santa Cruz), CDK4 (C-22; Santa Cruz), Flag tag (M2; Sigma Chemicals), or an HA tag (3F10; Roche Diagnostics).
For metabolic labelling, cells were pre-incubated with Met/Cys-free DMEM containing 10% dialysed FCS for 30 min, and labelled in the presence of 200 μCi/ml 35S-Met/Cys mix (Perkin Elmer, Boston, MA) for indicated periods. Cells were then washed with PBS and incubated with radioisotope-free medium containing excess Met/Cys. Lysates were prepared with Tween-20 lysis buffer containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 2.5 mM EGTA, 10 mM β-glycerophosphate, 1 mM DTT, and PIs, pre-cleared with protein-A beads, and immunoprecipitated using antibody (6 μg per IP) to C/EBPα, C/EBPβ or CDK4. Recovered radiolabelled proteins were separated by SDS–PAGE and detected by autoradiography.
Supplementary Material
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Figures S6
Supplementary Figures S7
Supplementary Material
Acknowledgments
We thank Drs Adam Gromley, Hirotaka J Okano, Takatoshi Iijima, and Daisuke Nakayama for valuable discussions and comments on the paper; Dr Martine F Roussel for providing MEFs and plasmids; Drs Azusa Asai and Natsuko Uekawa for technical assistance; and Dr Alan Bernstein for providing Hzf knockout mice. This study was supported by a grant from the Japanese Ministry of Education, Culture, Science and Technology, and Suzuken Memorial Foundation to MS and, initially, by funds from the Howard Hughes Medical Institute to the laboratory of CJS. CJS is an investigator of the Howard Hughes Medical Institute.
References
- Abella A, Dubus P, Malumbres M, Rane SG, Kiyokawa H, Sicard A, Vignon F, Langin D, Barbacid M, Fajas L (2005) Cdk4 promotes adipogenesis through PPARgamma activation. Cell Metab 2: 239–249 [DOI] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL (1991) Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5: 1538–1552 [DOI] [PubMed] [Google Scholar]
- Chen SS, Chen JF, Johnson PF, Muppala V, Lee YH (2000) C/EBPbeta, when expressed from the C/ebpalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol Cell Biol 20: 7292–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW (2007) Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 130: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Jack AK, Hamm JK, Pilch PF, Farmer SR (1999) Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARgamma and C/EBPalpha. J Biol Chem 274: 7946–7951 [DOI] [PubMed] [Google Scholar]
- Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J (2002) E2Fs regulate adipocyte differentiation. Dev Cell 3: 39–49 [DOI] [PubMed] [Google Scholar]
- Freytag SO, Paielli DL, Gilbert JD (1994) Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev 8: 1654–1663 [DOI] [PubMed] [Google Scholar]
- Garber K (2004) Energy boost: the Warburg effect returns in a new theory of cancer. J Natl Cancer Inst 96: 1805–1806 [DOI] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM (2003) Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 100: 15712–15717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbling D, Mueller BU, Timchenko NA, Hagemeijer A, Jotterand M, Meyer-Monard S, Lister A, Rowley JD, Huegli B, Fey MF, Pabst T (2004) The leukemic fusion gene AML1–MDS1–EVI1 suppresses CEBPA in acute myeloid leukemia by activation of calreticulin. Proc Natl Acad Sci USA 101: 13312–13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka M, Caruana G, Stanford WL, Sam M, Correll PH, Bernstein A (2000) Gene trapping of two novel genes, Hzf and Hhl, expressed in hematopoietic cells. Mech Dev 90: 3–15 [DOI] [PubMed] [Google Scholar]
- Hotta K, Funahashi T, Arita Y, Takahashi M, Matsuda M, Okamoto Y, Iwahashi H, Kuriyama H, Ouchi N, Maeda K, Nishida M, Kihara S, Sakai N, Nakajima T, Hasegawa K, Muraguchi M, Ohmoto Y, Nakamura T, Yamashita S, Hanafusa T et al. (2000) Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol 20: 1595–1599 [DOI] [PubMed] [Google Scholar]
- Iijima T, Imai T, Kimura Y, Bernstein A, Okano HJ, Yuzaki M, Okano H (2005) Hzf protein regulates dendritic localization and BDNF-induced translation of type 1 inositol 1, 4, 5-trisphosphate receptor mRNA. Proc Natl Acad Sci USA 102: 17190–17195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ (2004) Disruption of hepatic C/EBPalpha results in impaired glucose tolerance and age-dependent hepatosteatosis. J Biol Chem 279: 44740–44748 [DOI] [PubMed] [Google Scholar]
- Iwaki M, Matsuda M, Maeda N, Funahashi T, Matsuzawa Y, Makishima M, Shimomura I (2003) Induction of adiponectin, a fat-derived antidiabetic and antiatherogenic factor, by nuclear receptors. Diabetes 52: 1655–1663 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hart A, Hirashima M, Wang C, Holmyard D, Pittman J, Pang XL, Jackson CW, Bernstein A (2002) Zinc finger protein, Hzf, is required for megakaryocyte development and hemostasis. J Exp Med 195: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Yamauchi T, Kubota T, Moroi M, Matsui J, Eto K, Yamashita T, Kamon J, Satoh H, Yano W, Froguel P, Nagai R, Kimura S, Kadowaki T, Noda T (2002) Disruption of adiponectin causes insulin resistance and neointimal formation. J Biol Chem 277: 25863–25866 [DOI] [PubMed] [Google Scholar]
- Lakka TA, Rankinen T, Weisnagel SJ, Chagnon YC, Rice T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C (2003) A quantitative trait locus on 7q31 for the changes in plasma insulin in response to exercise training: the HERITAGE Family Study. Diabetes 52: 1583–1587 [DOI] [PubMed] [Google Scholar]
- Lin FT, Lane MD (1994) CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci USA 91: 8757–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ (2001) C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc Natl Acad Sci USA 98: 12532–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, Komuro R, Ouchi N, Kihara S, Tochino Y, Okutomi K, Horie M, Takeda S, Aoyama T, Funahashi T, Matsuzawa Y (2002) Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med 8: 731–737 [DOI] [PubMed] [Google Scholar]
- Maehara K, Yamakoshi K, Ohtani N, Kubo Y, Takahashi A, Arase S, Jones N, Hara E (2005) Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional pRB and p53. J Cell Biol 168: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–419 [DOI] [PubMed] [Google Scholar]
- Morrison RF, Farmer SR (1999) Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem 274: 17088–17097 [DOI] [PubMed] [Google Scholar]
- Park BH, Qiang L, Farmer SR (2004) Phosphorylation of C/EBPbeta at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol Cell Biol 24: 8671–8680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, Iervolino A, Condorelli F, Gambacorti-Passerini C, Caligiuri MA, Calabretta B (2002) BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet 30: 48–58 [DOI] [PubMed] [Google Scholar]
- Qiao L, Maclean PS, Schaack J, Orlicky DJ, Darimont C, Pagliassotti M, Friedman JE, Shao J (2005) C/EBPalpha regulates human adiponectin gene transcription through an intronic enhancer. Diabetes 54: 1744–1754 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM (2002) C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 16: 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM (2000) Transcriptional regulation of adipogenesis. Genes Dev 14: 1293–1307 [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson K, Isomaa B, Lettre G et al. (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336 [DOI] [PubMed] [Google Scholar]
- Scime A, Grenier G, Huh MS, Gillespie MA, Bevilacqua L, Harper ME, Rudnicki MA (2005) Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab 2: 283–295 [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JB, Moon HM, Noh MJ, Lee YS, Jeong HW, Yoo EJ, Kim WS, Park J, Youn BS, Kim JW, Park SD, Kim JB (2004) Adipocyte determination- and differentiation-dependent factor 1/sterol regulatory element-binding protein 1c regulates mouse adiponectin expression. J Biol Chem 279: 22108–22117 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC (2006) Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441: 424–430 [DOI] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD (1997) Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Gromley A, Sherr CJ (2006) Hzf, a p53-responsive gene, regulates maintenance of the G2 phase checkpoint induced by DNA damage. Mol Cell Biol 26: 502–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S (1997) Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 16: 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Iakova P, Welm AL, Cai ZJ, Timchenko NA (2002) Calreticulin interacts with C/EBPalpha and C/EBPbeta mRNAs and represses translation of C/EBP proteins. Mol Cell Biol 22: 7242–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ (1995) Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269: 1108–1112 [DOI] [PubMed] [Google Scholar]
- Wiltshire S, Frayling TM, Groves CJ, Levy JC, Hitman GA, Sampson M, Walker M, Menzel S, Hattersley AT, Cardon LR, McCarthy MI (2004) Evidence from a large UK family collection that genes influencing age of onset of type 2 diabetes map to chromosome 12p and to the MODY3/NIDDM2 locus on 12q24. Diabetes 53: 855–860 [DOI] [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM (1999) Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3: 151–158 [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, Iwabu M, Okada-Iwabu M, Kawamoto S, Kubota N, Kubota T, Ito Y, Kamon J, Tsuchida A, Kumagai K, Kozono H, Hada Y, Ogata H, Tokuyama K, Tsunoda M, Ide T et al. (2007) Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 13: 332–339 [DOI] [PubMed] [Google Scholar]
- Yang J, Croniger CM, Lekstrom-Himes J, Zhang P, Fenyus M, Tenen DG, Darlington GJ, Hanson RW (2005) Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein alpha. J Biol Chem 280: 38689–38699 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL (1995) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9: 168–181 [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR et al. (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF (1998) Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Figures S6
Supplementary Figures S7
Supplementary Material