Abstract
We report that the Arabidopsis thaliana mutant sensitive to ABA and drought2 (sad2), which harbors a T-DNA insertion in an importin β-like gene, is more tolerant to UV-B radiation than the wild type. Analysis of cyclobutane pyrimidine dimer accumulation revealed that less DNA damage occurred in sad2 than in the wild type during UV-B treatment. No significant growth difference was observed between sad2 and the wild type when treated with the genotoxic drug methyl methanesulfonate, suggesting that SAD2 functions in UV-B protection rather than in DNA damage repair. Whereas the R2R3-type transcription repressor MYB4 has previously been shown to negatively regulate the transcription of cinnamate 4-hydroxylase (C4H) and thus to regulate the synthesis of sinapate esters, expression of both MYB4 and C4H and accumulation of UV-absorbing compounds were significantly higher in sad2 than in the wild type. MYB4 did not localize to the nucleus in the sad2 mutant, suggesting that SAD2 is required for MYB4 nuclear trafficking. SAD2 and MYB4 coimmunoprecipitated, indicating that these proteins localize in the same complex in vivo. MYB4 protein specifically bound to its own promoter in gel shift assays and repressed its own expression, demonstrating that MYB4 protein and mRNA are part of a negative autoregulatory loop. This feedback loop is altered in the sad2 mutant due to the absence of MYB4 protein in the nucleus, leading to the constitutive expression of MYB4 and C4H and resulting in accumulation of UV-absorbing pigments that shield the plant from UV-B radiation.
INTRODUCTION
UV radiation has received increasing attention due to the thinning of the earth's stratospheric ozone layer. While UV-C radiation (200 to 280 nm) is completely absorbed by atmospheric gases, a significant amount of UV-B (280 to 320 nm) reaches the earth's surface. Exposure of plants to UV-B radiation results in induction of stress responses, inhibition of photosynthesis, and damage to DNA and other molecules (Jansen et al., 1998).
Plants have developed numerous biochemical mechanisms to cope with environmental change. In response to exposure to UV light, DNA forms cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidinone dimers that in turn trigger DNA repair mechanisms (Frohnmeyer and Staiger, 2003). Genetic studies in Arabidopsis thaliana have identified several mutants defective in DNA repair that are hypersensitive to UV-B light, indicating that DNA protection plays an essential role in the response of the plant to UV-B (Jiang et al., 1997; Nakajima et al., 1998; Liu et al., 2000). Like other abiotic and biotic stresses, UV-B also induces the production of reactive oxygen species and triggers formation of reactive oxygen species scavengers (Mackerness et al., 2001).
Under low-dosage UV-B radiation, plants accumulate more phenylpropanoids like flavonoids and hydroxycinnamic acid derivatives (sinapate esters) as part of an initial defense (production of chemical sunscreens) to block UV-B from reaching and damaging targets. Genetic analysis of Arabidopsis mutants with altered UV-B responses further supports the importance of chemical sunscreens; constitutive accumulation of flavonoids and other phenolics results in resistance of mutants to UV-B (Bieza and Lois, 2001), whereas lack of UV-absorbing compounds results in UV-B hypersensitivity (Lois and Buchanan, 1994).
Recently, a group of MYB transcription factors have been reported to regulate the biosynthesis of secondary metabolites (Stracke et al., 2001). In Arabidopsis, MYB4 acts as a transcription repressor inhibiting expression of the cinnamate 4-hydroxylase (C4H) gene, which encodes a key enzyme in hydroxycinnamate ester biosynthesis. In a myb4 knockout mutant, hydroxycinnamate esters accumulate, resulting in tolerance of the mutant to UV-B, whereas overexpression of MYB4 decreases the level of UV sunscreens and causes UV-B hypersensitivity in the transgenic plants (Jin et al., 2000). Although overexpression of MYB4 repressed chalcone synthase (CHS) expression, the myb4 knockout mutant was less affected in terms of regulation of CHS expression and flavonoid accumulation (Jin et al., 2000). By contrast, a mutation in UVR8, a Regulator of Chromatin Condensation1–like protein, reduced expression of CHS and specifically repressed UV-B–induced flavonoid accumulation but had no effect on accumulation of sinapate esters and resulted in hypersensitivity of the mutant plants to UV-B radiation (Kliebenstein et al., 2002). These results indicate that both flavonoids and sinapate esters are important for plant UV-B tolerance and that diverse regulatory pathways exist in plants to regulate phenylpropanoid biosynthesis.
The precise cellular localization of a protein is essential for its function, and nuclear translocation is the key to the function of transcription factors. In plants, nucleocytoplasmic partitioning of regulatory proteins plays a critical role in various biological processes (Merkle, 2004), including light responses (Stacey et al., 1999; Yang et al., 2000), hormone signaling (Hutchison and Kieber, 2002; Itoh et al., 2002), abiotic stress responses (Lee et al., 2001), and disease resistance (Kinkema et al., 2000). Nuclear transporters have been shown to be involved in regulating different signal transduction pathways during plant development (Bollman et al., 2003) and in plant responses to biotic (Palma et al., 2005) and abiotic stresses (Verslues et al., 2006). However, to date, few targets of known nuclear transporters have been identified in planta (Jiang et al., 2001; Ziemienowicz et al., 2003). In HeLa cells, an importin α protein accumulates reversibly in the nucleus during UV treatment (Miyamoto et al., 2004), indicating that the nuclear transporter is involved in UV responses. Here, we report that an importin β-like protein, SENSITIVE TO ABA AND DROUGHT2 (SAD2), is involved in UV-B responses by regulating accumulation of UV sunscreens through mediation of MYB4 nuclear transport.
RESULTS
sad2 Is a UV-Resistant Mutant
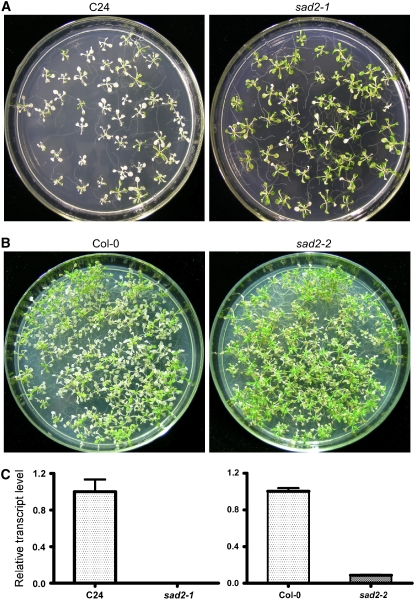
We isolated the mutant sad2-1 from T-DNA–mutagenized pools in the Arabidopsis C24 genetic background harboring a RD29A:LUC reporter gene. Luciferase luminescence and expression of endogenous RD29A in sad2-1 are induced under abscisic acid (ABA), NaCl, cold, and polyethylene glycol treatments, and germination of sad2-1 has previously been shown to be hypersensitive to ABA (Verslues et al., 2006). We found that the mutation in SAD2 increased plant UV-B radiation tolerance. When 12-d-old sad2-1 mutant and wild-type seedlings were exposed to UV-B (5.3 mW/cm2) for 10 min and then incubated in a growth chamber for 3 d, >70% of the wild-type plants were bleached and died. However, the effect of the UV-B treatment on sad2-1 plants was mainly restricted to the cotyledons, and >80% of the plants survived this treatment (Figure 1A). A genomic DNA fragment from 1033 bp upstream of the SAD2 translation start site to 1436 bp downstream of the SAD2 stop codon was isolated from BAC clone T9H9 and cloned into pCAMBIA1200, and the resulting plasmid was transferred into the sad2-1 mutant (Verslues et al., 2006; see Supplemental Figure 1A online). The UV-B–tolerant phenotype was complemented by the SAD2 gene (see Supplemental Figure 1A online). To further confirm that SAD2 is involved in the UV-B response, an Arabidopsis mutant line (SALK_133577) with a T-DNA insertion in the SAD2 gene (sad2-2) was ordered from the ABRC (Alonso et al., 2003). The T-DNA insertion is located in the sixteenth intron of the SAD2 gene and was confirmed by PCR using a gene-specific primer and T-DNA left border primers. Based on analyses using primers upstream of the T-DNA insertions, expression of SAD2 mRNA was abolished in the sad2-1 mutant and was very low in the sad2-2 mutant (Figure 1C). After UV-B treatment, sad2-2 was more tolerant to UV-B than the wild type (Figure 1B; see Supplemental Figure 1C online); however, this tolerance was reduced when compared with that of sad2-1. Overexpression of SAD2 driven by a 35S promoter in the sad2-1 mutant rescued the mutant UV-B–tolerant phenotype (see Supplemental Figure 1B online). When the SAD2 full-length genomic sequence was introduced into sad2-2, it also rescued its UV-B–tolerant phenotype (see Supplemental Figure 1C online). These results demonstrate that the UV-B tolerance of the sad2 mutants is due to the loss of SAD2 function in Arabidopsis.
Figure 1.
sad2-1 Is a UV-B–Tolerant Mutant.
(A) and (B) Twelve-day-old wild-type (left panel) and sad2-1 ([A]; right panel) mutant or sad2-2 ([B]; right panel) seedlings were treated with UV-B (5.3 mW/cm2) for 10 and 12 min, respectively. After treatment, plants were transferred to a continuous light growth chamber for 3 d and then photographed.
(C) Expression of SAD2 in sad2-1 and sad2-2. Total RNA was extracted from 12-d-old seedlings of wild-type and the sad2 mutants. The resulting cDNAs were used for real-time PCR analysis. Error bars indicate sd (n = 3).
The SAD2 Mutation Affects DNA Damage but Not DNA Repair
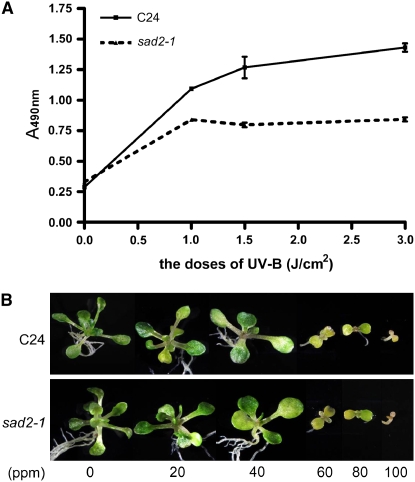
Genomic DNA is one of the major UV-B targets leading to phototransformation and resulting in the accumulation of CPDs and pyrimidine (6-4) pyrimidinone dimers, which turn on DNA damage repair processes. To compare DNA damage in sad2-1 and wild-type plants, genomic DNA was extracted from 15-d-old wild-type and sad2-1 mutants immediately after the plants were exposed to increasing doses of UV-B radiation and subjected to an ELISA assay to measure CPD induction. The results are shown in Figure 2A. When exposed to 1, 1.5, and 3 J/cm2 UV-B (5.3 mW/cm2), the wild type accumulated more CPDs than sad2-1, indicating that the wild type had more UV-B–based DNA damage, and this correlated with the sad2-1 UV-B–tolerant phenotype. To determine if DNA repair is involved in sad2-1 UV-B tolerance, wild-type and sad2-1 plants were treated with methyl methanesulfonate (MMS), a genotoxic chemical, at concentrations of 0, 20, 40, 60, 80, and 100 ppm. The growth of both the wild type and sad2-1 was reduced at 20 ppm and inhibited at 60 ppm (Figure 2B) with no significant growth difference observed between the two, indicating that sad2-1 UV-B tolerance is due to less DNA damage rather than a difference in DNA repair ability.
Figure 2.
Less DNA Damage Occurred in sad2-1.
(A) The sad2-1 mutant accumulated less CPDs than the wild type during UV-B treatment. CPD accumulation in the wild type (solid line) and sad2-1 (dashed line). The number of CPDs in the substrate was measured for each of the indicated treatments. Vertical bars represent the se for three independent experiments.
(B) Four-day-old wild-type and sad2-1 seedlings were treated with the indicated concentrations of the genotoxic drug MMS. Photographs were taken 18 d after treatment.
sad2-1 Accumulates More Soluble Flavonoids and Sinapoyl Esters Than the Wild Type
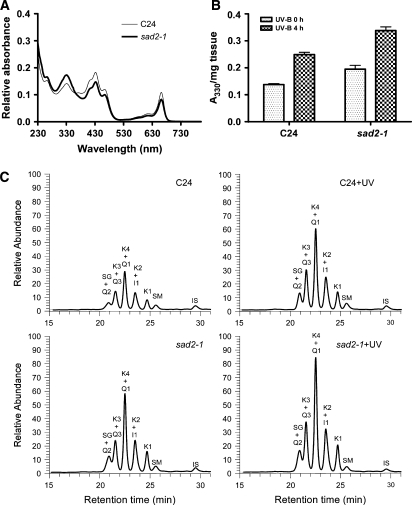
Photoprotective components absorb UV light and protect the plant from damage (Li et al., 1993; Lois, 1994; Lois and Buchanan, 1994; Landry et al., 1995; Mazza et al., 2000). Constitutive accumulation of UV-absorbing pigments at a higher level has been shown to enhance Arabidopsis UV tolerance (Bieza and Lois, 2001). To determine whether induction and accumulation of UV-absorbing pigments play a role in sad2-1 UV tolerance, seedling extracts from the wild type and the sad2-1 mutant were subjected to absorption spectra analysis. UV-absorbing pigments in the region between 260 and 350 nm (Lois and Buchanan, 1994) were significantly higher in sad2-1 than in the wild type (Figure 3A), suggesting a potential mechanism for the increased UV-B resistance measured in sad2-1. Two additional peaks were observed between 400 and 500 nm and between 630 and 680 nm and have been shown to correlate with carotenoids and chlorophyll (Gross, 1991). It appears that sad2-1 accumulates more UV-protective pigments but less carotenoids and chlorophyll under normal conditions when compared with the wild type (Figure 3A). Absorption of phenolic compounds at 330 nm was 30% higher in sad2-1 than in the wild type. Consistent with what has been shown in the wild type (Bieza and Lois, 2001), after UV-B treatment, the levels of these compounds were significantly higher in both the wild type and the sad2-1 mutant, with increases up to 30% (Figure 3B).
Figure 3.
sad2-1 Accumulates More UV-Absorbing Pigments.
(A) Absorption spectra of pigments in the wild type (thin line) and the sad2-1 mutant (bold line) under normal growth conditions.
(B) Content of UV-absorbing pigments at an absorption spectrum value of 330 nm. Pigments were extracted from 12-d-old seedlings with or without UV-B (0.04 mW/cm2) treatment. Vertical bars represent se for three independent experiments.
(C) LC-MS identification of soluble phenolic compounds from sad2-1 and the wild type. Fourteen-day-old wild-type C24 and sad2-1 seedlings with or without UV-B treatment were used for methanol:water (4:1) extraction and liquid chromatography. IS, internal standard; the main flavonol glycosides: K, kaempferol; Q, quercetin; I, isorhamnetin; the main sinapoyl derivatives: E and Z isomers of SG, E and Z isomers of SM.
It has been shown that flavonoids and sinapate esters are the major photoprotective pigments protecting plants from UV radiation (Li et al., 1993; Jin et al., 2000). To further determine the function of SAD2 in phenolic metabolism, methanolic soluble phenolics were extracted from sad2-1 and C24 and analyzed by liquid chromatography–mass spectrometry (LC-MS). The results are shown in Figure 3C and Table 1. Flavonol glycosides like kaempferol (K) and isorhamnetin (I) accumulated at a significantly higher level in the sad2-1 mutant than in the wild type independent of UV-B treatment. Two sinapoyl and coumaroyl derivatives, sinapoyl glucose (SG) and sinapoyl malate (SM), were detected; however, only SG levels increased significantly in the sad2-1 mutant.
Table 1.
LC-MS Determination of Soluble Phenolics
| Compounds | (M-H) | C24 | C24+UV | sad2-1 | sad2-1+UV |
|---|---|---|---|---|---|
| ng/150 mg Fresh Plant Materials | |||||
| Flavonol glycosides | |||||
| K1 | 577 | 26 (4) | 44 (4) | 59 (2) | 91 (19) |
| K2 | 593 | 47 (3) | 84 (6) | 97 (5) | 144 (35) |
| K3 | 739 | 31 (1) | 62 (5) | 71 (2) | 123 (33) |
| K4 | 755 | 50 (4) | 86 (7) | 121 (6) | 185 (47) |
| Q1 | 609 | 57 (4) | 122 (3) | 117 (1) | 207 (47) |
| Q2 | 755 | 8 (1) | 21 (3) | 14 (2) | 27 (6) |
| Q3 | 771 | 22 (5) | 35 (5) | 28 (2) | 44 (6) |
| I1 | 623 | 9 (1) | 20 (2) | 23 (1) | 46 (9) |
| Sinapoyl derivatives | |||||
| SM | 339 | 10 (3) | 17 (2) | 15 (3) | 19 (1) |
| SG | 385 | 18 (2) | 29 (9) | 58 (1) | 85 (22) |
Flavonol glycosides: K, kaempferol; Q, quercetin; I, isorhamnetin. K1, kaempferol 3-O-rhamnoside 7-O-rhamnoside; K2, kaempferol 3-O-glucoside 7-O-rhamnoside; K3, kaempferol 3-O-[6″-O-(rhamnosyl) glucoside] 7-O-rhamnoside; K4, kaempferol 3-O-[6″-O-(glucosyl) glucoside] 7-O-rhamnoside. Q1, quercetin 3-O-glucoside 7-O-rhamnoside; Q2, quercetin 3-O-[6″-O-(rhamnosyl) glucoside] 7-O-rhamnoside; Q3, quercetin 3-O-[6″-O-(glucosyl) glucoside] 7-O-rhamnoside. I1, isorhamnetin 3-O-glucoside 7-O-rhamnoside. Sinapoyl derivatives: E and Z isomers of SG, E and Z isomers of SM. Values represent means (se in parentheses) from three independent experiments with 40 plants each. LC-MS quantitative determination was as described by Abdulrazzak et al. (2006).
Expression of C4H, CHS, and MYB4 mRNA Is Constitutively Higher in sad2-1
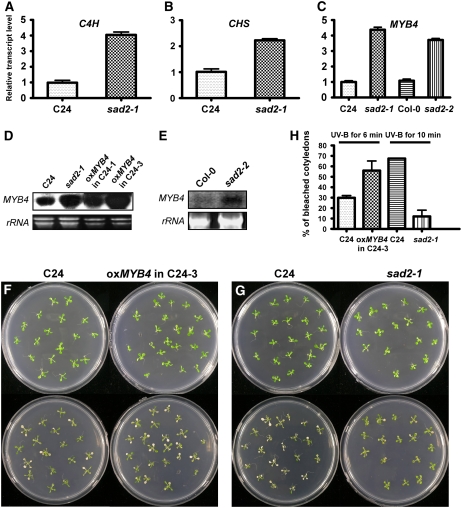
More flavonoids and sinapate esters accumulate in sad2-1 than in the wild type. To determine whether mRNA levels of CHS and C4H are constitutively increased in sad2-1, total RNA was extracted from both 12-d-old wild-type and sad2-1 plants, and real-time PCR analysis was performed with ACTIN2 as an internal control. SAD2 transcript was below the level of detection in the sad2-1 T-DNA line (Figure 1C). The transcriptional level of CHS was twofold higher in sad2-1 than in the wild type, and expression of C4H in sad2-1 was fourfold higher than in the wild type (Figures 4A and 4B). These results are consistent with the constitutive increase in UV-absorbing pigments measured in sad2-1.
Figure 4.
Expression Levels of C4H, CHS, and MYB4 Are Higher in the sad2 Mutant.
(A) to (C) Total RNA was extracted from 12-d-old seedlings of the wild type and the sad2 mutant. The resulting cDNAs were used for real-time PCR analysis. Transcripts of C4H (A), CHS (B), and MYB4 (C) were constitutively higher in sad2-1 than in the wild type. Error bars indicate sd (n = 3).
(C) Real-time PCR analysis shows that MYB4 expression was higher in sad2-2 than in the wild type.
(D) and (E) RNA gel blot analysis of MYB4 expression in the wild type, sad2-1, sad2-2, and two MYB4 overexpression lines. Twenty micrograms of total RNA were probed with MYB4 cDNA. rRNA was used as a loading control.
(F) and (G) Twelve-day-old seedlings of the wild type and an MYB4 overexpression line (F) or the wild type and sad2 (G) were treated with 2 or 3 J/cm2 UV-B, respectively (bottom images). Photographs were taken 3 d after transfer to a continuous light growth chamber.
(H) Percentage of bleached cotyledons from seedlings shown in (F) and (G). Vertical bars represent se for three independent experiments.
It has been shown that MYB4 functions as a transcription repressor and downregulates C4H expression. In the knockout myb4 mutant, C4H mRNA and sinapoyl malate accumulated to high levels when compared with levels in the wild type, while overexpression of MYB4 in the wild type repressed both C4H and CHS expression (Jin et al., 2000). In the sad2-1 mutant, expression of C4H was enhanced to a significantly higher level than CHS (Figures 4A and 4B), suggesting that the expression of MYB4 mRNA might be downregulated in the sad2-1 mutant. To test this, we performed real-time PCR and RNA gel blot analysis. Unexpectedly, the expression of MYB4 mRNA in the sad2 mutants was not reduced but increased significantly in comparison with levels in the wild type (Figures 4C to 4E). This result was surprising because it has been shown that overaccumulation of MYB4 mRNA in Columbia (Col-0) is associated with a decrease in UV-absorbing compounds (Jin et al., 2000). Because the sad2-1 mutant is in the Arabidopsis C24 genetic background, we overexpressed MYB4 under the control of a 35S promoter in C24 (Figure 4D) to investigate whether there is a different effect from overaccumulation of MYB4 in Col-0 and C24; the results are shown in Figure 4F. Similar to MYB4 overexpression in the Col-0 background, overexpression in C24 reduced UV-B tolerance (Figures 4F and 4G). Twelve-day-old seedlings of C24 and the transgenic plants were treated with UV-B (5.3 mW/cm2) for 6 min and incubated in a growth chamber for 3 d, and the bleached cotyledons were counted. More than 50% of the cotyledons in the overexpression plants were bleached compared with <30% in wild-type C24 (Figure 4H). By contrast, the sad2-1 mutant in which MYB4 was also overexpressed was more resistant to UV-B than the wild type (Figures 4G and 4H). RNA gel blot results further confirmed that sad2-1 and two transgenic lines with the UV-B–sensitive phenotype constitutively accumulated more MYB4 transcript than the wild type (Figures 4D and 4E). Our data suggest that the MYB4 gene product may not function in sad2-1 because it is not able to inhibit C4H expression, resulting in the constitutive accumulation of UV-absorbing pigments and, therefore, an increase in sad2-1 UV-B tolerance.
MYB4 Is a Target of SAD2
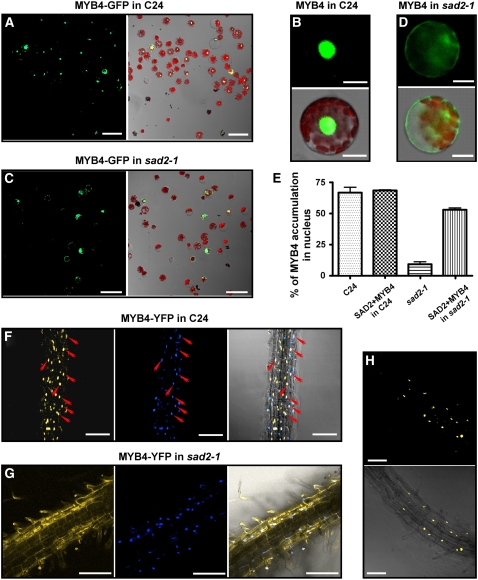
SAD2 encodes an importin β-like protein (Verslues et al., 2006). In nuclear transport complexes, importin β does not directly interact with cargo but recognizes the nuclear pore complex and takes the importin α-cargo complex into the nucleus. Because MYB4 protein does not function in sad2-1, one of the simplest interpretations is that MYB4 is a target of SAD2. In the sad2-1 mutant, MYB4 protein might not be transported into the nucleus to repress C4H expression. This would cause constitutive accumulation of UV-absorbing pigments and result in sad2-1 UV-B tolerance. To test this hypothesis, green fluorescent protein (GFP) was translationally fused to MYB4 at its C terminus under the control of a 35S promoter (35S:MYB4-GFP). The resulting plasmid DNA was then transfected into both wild-type and sad2-1 mesophyll protoplasts. After overnight incubation, at least 300 protoplasts were analyzed with a confocal microscope, and the results are shown in Figure 5. In the wild-type background, >65% of transformed protoplasts exhibited MYB4-GFP localization only in the nucleus (Figures 5A and 5B). However, only ∼15% of the protoplasts from the sad2-1 mutant showed MYB4-GFP signals in the nucleus (Figures 5C and 5D). These results suggest that functional SAD2 is important for the nuclear localization of MYB4. To verify this result, a cotransfection experiment was performed. Cotransfection of 35SP:SAD2 and 35SP:MYB4-GFP in wild-type protoplasts did not increase the percentage of nuclear localization of MYB4-GFP (Figure 5E). Cotransformation of these constructs into sad2-1 mutant protoplasts dramatically increased the percentage (from 15 to 50%) of nuclear-localized MYB4-GFP (Figure 5E). However, MYB4-GFP showed some residual nuclear localization in the sad2-2 mutant (see Supplemental Figure 3 online) consistent with residual SAD2 mRNA expression in this allele (Figure 1C). These results further support SAD2-mediated transport of MYB4 to the nucleus. In Arabidopsis, there are 17 importin β-like proteins, and two of them, At3g59020 and At1g26170, share the highest sequence similarity with SAD2. The T-DNA insertion lines, SALK_052799 and SALK_043918, corresponding to these two genes were obtained from the ABRC, and the T-DNA insertions and the lack of full-length transcripts were confirmed by PCR using the T-DNA left border and gene-specific primers (see Supplemental Figure 2 online). MYB4-GFP plasmids were transfected into protoplasts of these two mutants and the wild type, respectively. There was no difference in MYB4-GFP nuclear localization between the wild type and the two mutants (data not shown).
Figure 5.
SAD2 Is Required for MYB4 Nuclear Localization.
(A) to (E) Transient expression of MYB4-GFP in wild-type and sad2-1 protoplasts.
(A) and (C) MYB4-GFP nuclear localization in wild-type (A) or sad2-1 mutant (C) protoplasts. Left panels, confocal GFP images; right panels, combined bright-field, chloroplast autoflorescence, and GFP images.
(B) A protoplast showing typical nuclear-localized MYB4-GFP in the wild type.
(D) A protoplast showing typical cytoplasmic localized MYB4-GFP in the sad2-1 mutant. In (B) and (D), left panels are confocal GFP images, and right panels show combined bright-field, chloroplast autoflorescence, and GFP images.
(E) Effect of SAD2 on MYB4-GFP nuclear localization. 35SP:MYB4-GFP or 35SP:MYB4-GFP and 35SP:SAD2 were cotransfected into wild-type and sad2-1 protoplasts and the percentage of cells with nuclear-localized MYB4-GFP from each transformation was counted. Vertical bars represent se for three independent experiments.
(F) and (G) MYB4-YFP localized in the nucleus in the wild type (F) and in the cytoplasm in sad2 (G) transgenic plant roots (left panels), 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining (middle panels), and combined MYB4-YFP and DAPI staining (right panels). Arrows point to nuclei and show the colocalization of DAPI staining and the YFP signal (right panel in [F]).
(H) The sad2 MYB4-YFP transgenic plants were crossed to the wild type, and MYB4-YFP localization was determined in F1 heterozygous plants. MYB4 localized to the nucleus. Top panel, confocal GFP image; bottom panel, combined confocal and bright-field images.
Argon laser light (514-nm wavelength, 26% power) was used in (F) and (H); argon laser light (514-nm wavelength, 65.5% power) was used in (G). Bars in (A) and (C) = 50 μm, bars in (F) to (H) = 100 μm, and bars in (B) and (D) = 10 μm.
Transient expression of a gene often causes the gene product to overaccumulate in the cell. To avoid this, we transferred a construct of yellow fluorescent protein (YFP) translationally fused to the MYB4 C terminus, driven by the cauliflower mosaic virus 35S promoter (35SP:MYB4-YFP) into both wild-type and sad2-1 plants. At least eight transgenic plants in the T2 generation were analyzed for MYB4-YFP localization. MYB4-YFP signal was only detected in the nucleus in the wild type (Figure 5F). However, in the sad2-1 mutant, no YFP signal was found in nucleus, but a signal was detected in the cytoplasm (Figure 5G). This result further supports our transient expression data. To avoid any effects associated with transformation, two MYB4-YFP transgenic plants in the sad2-1 background in which the YFP signal was not able to locate to the nucleus were crossed into the wild type to generate SAD2/sad2 heterozygous plants harboring the 35SP:MYB4-YFP construct. F1 plants were then used to investigate the MYB4-YFP localization. In SAD2/sad2 heterozygous plants, MYB4-YFP was located in the nucleus (Figure 5H), indicating that SAD2 is required for MYB4 nuclear localization.
Colocalization of SAD2 and MYB4 in the Plant
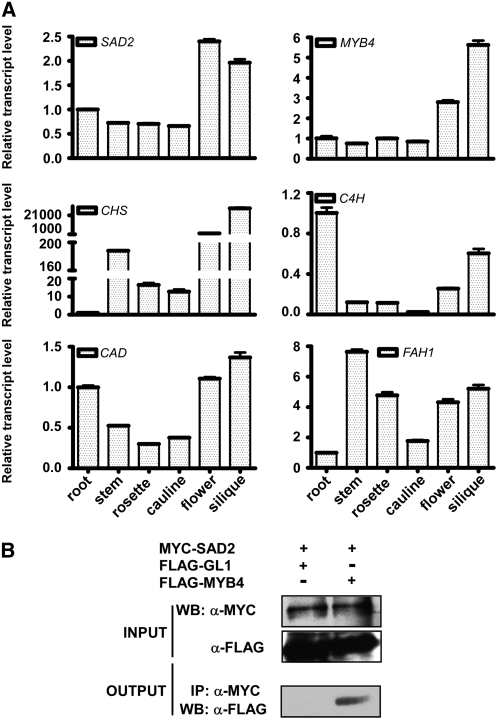
SAD2 is required for MYB4 nuclear localization, and both proteins function in regulating the synthesis of UV-absorbing pigments. To further determine whether the mRNA and protein of MYB4 and SAD2 are colocalized, total RNA was extracted from different organs of 30-d-old plants and subjected to real-time PCR analysis. Transcripts of ferulate-5-hydroxylase (FAH1), cinnamyl alcohol dehydrogenase (CAD), C4H, and CHS were present in the root, leaf, stem, and flower (Figure 6A). To further investigate whether SAD2 and MYB4 colocalize in the same complex, a 6×myc tag was translationally fused to SAD2 and a 3×flag tag was fused to MYB4 and GL1, a homolog of MYB4. All of the tags were fused at the N terminus of their target proteins with MYB4 and GL1 under the control of a 35S promoter and SAD2 driven by a double 35S promoter. The resulting 2×35SP:6×myc-SAD2 plasmid was cotransfected with the 35SP:3×flag-MYB4 or 35SP:3×flag-GL1 constructs into wild-type leaf protoplasts. Protein was extracted from the protoplasts, and 2% of the total extracted protein was subjected to immunoblot analysis with anti-FLAG and anti-MYC antibodies. The input proteins myc-SAD2 and flag-MYB4 or myc-SAD2 and flag-GL1 were detected in each transformation (Figure 6B). MYC-SAD2 protein was then immunoprecipitated with anti-MYC affinity gel. After five washes, the anti-MYC affinity gel with the SAD2 complex was separated on a 12% SDS-PAGE gel, transferred to a membrane, and subjected to immunoblot analysis with anti-FLAG antibody. Only flag-MYB4 could be detected in the SAD2 pull-down product and not flag-GL1 (Figure 6B), suggesting that SAD2 is associated with MYB4 in vivo.
Figure 6.
MYB4 mRNA and Protein Colocalize with SAD2 in Arabidopsis.
(A) Expression of MYB4, SAD2, C4H, CHS, CAD, and FAH1 in Arabidopsis. Total RNA was extracted from roots, leaves, stems, flowers, and siliques of 1-month-old wild-type plants. Real-time PCR analysis (40 cycles) was performed using gene-specific primers. Error bars indicate sd (n = 3).
(B) Coimmunoprecipitation of SAD2 and MYB4 proteins in vivo. The 2×35SP:6×myc-SAD2 plasmid was cotransformed into wild-type protoplasts with 35SP:3×flag-MYB4 or 35SP:3×flag-GL1. Total protein extracts were subjected to protein gel blot (WB) analysis with monoclonal anti-MYC (top panel) and anti-FLAG (middle panel) antibodies to detect the presence of myc-SAD2 and flag-MYB4/flag-GL1. Immunoprecipitation was performed using anti-c-MYC agarose conjugate. Bottom panel, protein gel blot analyzed with anti-FLAG antibody to detect coimmunoprecipitated flag-MYB4 and flag-GL1. Input, crude protein extract. Output, products pulled down by the anti-c-MYC agarose conjugate.
MYB4 Negatively Regulates Its Own Gene Expression
MYB4 transcript is transiently induced by various light treatments and then decreases to a lower level (Jin et al., 2000); under UV-B radiation, this change was more dramatic. The expression of MYB4 was induced by UV-B after 1 h of treatment, reached maximal levels after 6 h, and then reduced to an undetectable level within 24 h (Jin et al., 2000). Overexpression of MYB4 repressed CHS and C4H expression and reduced UV-B resistance (Jin et al., 2000), while the sad2-1 mutation appeared to uncouple the association between overexpression of MYB4 and UV-B resistance. The observation that MYB4 protein was absent from the nucleus and that its mRNA overaccumulated in the sad2-1 mutant suggests that At MYB4 protein may directly repress its own transcription by binding to its promoter and that this autoregulatory feedback loop maintains MYB4 protein and mRNA homeostasis. In the sad2-1 mutant, MYB4 protein is not able to enter the nucleus and this may interrupt this feedback inhibition and result in the constitutive overaccumulation of MYB4 and C4H.
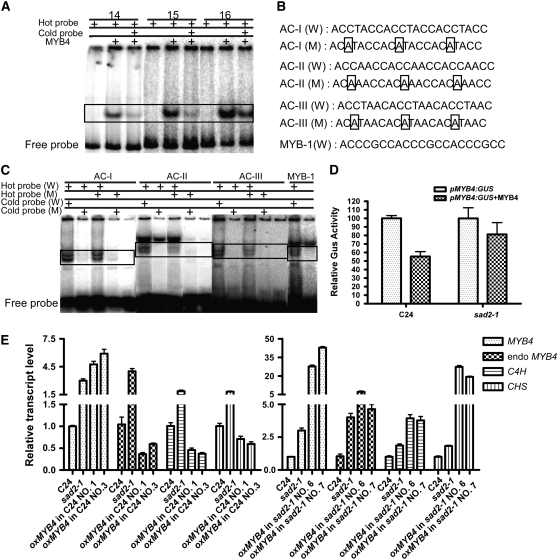
To test this hypothesis, MYB4 was translationally fused to maltose binding protein (MBP), and the fusion protein was purified from Escherichia coli TB1 cells. Seven 300- to 900-bp DNA fragments covering the MYB4 promoter from −3010 bp to −1 bp upstream of the translational start codon ATG were amplified by PCR. The DNA fragments were end-labeled with [γ-32P]ATP by T4 DNA kinase and then incubated with MBP-MYB4 for gel-shift assays. Three fragments, number 14 from −3010 to −2706 bp, number 15 from −2000 to −1701 bp, and number 16 from −1064 to −747 bp were able to bind to MBP-MYB4 (Figure 7A; see Supplemental Figure 4 online). When 100-fold excess unlabeled DNA probe was coincubated with MBP-MYB4 and the [γ-32P]ATP labeled probes, the protein–DNA interaction was reduced, indicating that MYB4 could bind to the MYB4 promoter at different regions. By reducing the length of the DNA fragments and looking for known MYB binding elements, three MYB binding elements, ACCTACC, ACCAACC, and ACCTAAC, which have been described as AC-I, AC-II, and AC-III, respectively (Hatton et al., 1995), were observed at positions −1009 to −1003 bp, −2773 to −2767 bp, −1935 to −1929 bp, and −1766 to −1760 bp (see Supplemental Figure 4 online). These elements could bind to MYB4, but AC-II had less specificity than the other two elements. If the third position C was changed to A in any element, this binding activity was abolished (Figures 7B and 7C). We also found a MYB binding element (named MYB-1) at position −836 to −830 bp. MYB-1 has the sequence ACCCGCC and was also found to bind to AtMYB4 (Figure 7C). The biological function of these putative binding elements remains to be determined.
Figure 7.
MYB4 Binds to Its Own Promoter in EMSAs and Represses Its Own Transcription.
Recombinant MYB4 protein was expressed in bacteria as an MBP fusion and purified by affinity chromatography.
(A) EMSAs were performed with radiolabeled or unlabeled MYB4 promoter fragments and recombinant MYB4 protein; specific combinations are shown above the autoradiograph. Unlabeled fragments were added in 100-fold excess as indicated.
(B) MYB4 binding cis-elements (W) in the MYB4 promoter and their mutant forms (M).
(C) EMSA with MYB4 and the cis-elements identified in (B). Competitors were added in 100-fold excess as indicated. Bands in the boxes show the specific binding.
(D) MYB4 inhibits its own transcription in the wild type but not in sad2. Relative GUS activities were measured after transfection of MYB4:GUS with or without 35SP:MYB4 in wild-type and sad2 protoplasts. The values represent averages of three transfections, and error bars indicate sd.
(E) Overexpression of MYB4 repressed endogenous MYB4, CHS, and C4H expression in the wild type but not in the sad2 mutant. Total RNA was extracted from 12-d-old seedlings of the wild type, the sad2 mutant, and overexpression MYB4 transgenic plants in both the C24 and sad2 background. The resulting cDNAs were used for real-time PCR analysis. Error bars indicate sd (n = 3).
To investigate whether the direct interaction between MYB4 and its own promoter has an effect on MYB4 gene expression, a 2468-bp MYB4 promoter fragment from −3010 to −542 bp upstream of the MYB4 ATG was transcriptionally fused to a 46-bp 35S minimal promoter with a TATA box and the β-glucuronidase (GUS) reporter gene (MYB435smini:GUS) or a fragment from −3010 to −1 bp upstream of the MYB4 ATG was fused to GUS (MYB4:GUS). A 46-bp, 35S minimal promoter:GUS vector (35SP46mini:GUS) was used as a control. The pMYB4:GUS, pMYB435smini:GUS, and 35SP46mini:GUS plasmids were transfected with or without the 35SP:MYB4 plasmid into wild-type and sad2-1 leaf protoplasts (Figure 7D; see Supplemental Figure 4 online). Expression of 35SP46mini:GUS was at a background level and not affected by MYB4 in either the wild type or the sad2-1 mutant (data not shown). The expression of pMYB4:GUS and pMYB435smini:GUS was reduced ∼50% by cotransformation of 35SP:MYB4 in the wild type (Figure 7D). However, this expression was only slightly affected by cotransfection of 35SP:MYB4 in the sad2-1 mutant (Figure 7D), suggesting that MYB4 represses its own transcription and that the sad2-1 mutant disrupts this activity.
Overexpression of MYB4 reduced the UV-B tolerance of transgenic C24 plants (Figure 4) but not transgenic sad2-1 mutant plants (data not shown). To investigate whether these phenotypes are correlated with the expression of CHS and C4H and whether ectopic expression of MYB4 represses endogenous MYB4 expression, real-time PCR analysis was performed. As expected, expression of CHS, C4H, and endogenous MYB4 was reduced in the C24 transgenic plants, supporting the model that MYB4 represses C4H and its own transcription by directly binding their promoters and that it also plays a role in the regulation of CHS expression. However, in the sad2-1 mutant, overexpression of MYB4 could not repress its endogenous expression, and transcription levels of C4H and CHS were not reduced but significantly increased (Figure 7E).
DISCUSSION
Accurate nuclear translocation of signal molecules is essential for plant growth and development. However, little is known about how this regulation takes place in planta. Analysis of the sad2 mutants and the relationship between SAD2 and MYB4 revealed that SAD2, an importin β-like protein, is required to transport MYB4 into the nucleus in Arabidopsis. Studying this nuclear translocation process has allowed us to show that the MYB4 transcript and protein are involved in a negative autoregulatory feedback loop and that they are important for maintaining levels of UV-absorbing pigments in plants.
The sad2 mutants are tolerant to UV-B radiation due to the inability of MYB4 to locate to the nucleus and to repress C4H and likely also CHS expression, resulting in the constitutive accumulation of UV-absorbing pigments. To translocate a protein from the cytoplasm to the nucleus requires a nuclear import complex. In most cases, the nuclear import receptor, importin β, does not directly interact with the cargo. Instead, it needs an adapter protein, such as importin α. The importin α proteins recognize and bind to basic nuclear localization signals in the cargo. Importin β interacts directly with the N-terminal importin β binding domain of importin α, recognizes the nuclear pore complex, and imports cargo to nucleus (Merkle, 2004). SAD2 and MYB4 coimmunoprecipitated, indicating that they can function in the same complex.
Several results demonstrate the unique role of SAD2 in the UV-B response in Arabidopsis. First, no UV-B–resistant phenotype was observed in the T-DNA mutants of seven out of nine Arabidopsis importin α-like genes or in the T-DNA mutants of two Ran GTPase genes (data not shown). Importin α and Ran GTPase family proteins in Arabidopsis share very high sequence similarity, implying that they may overlap functionally in response to UV-B radiation. Second, the UV-B response was unique to SAD2. Importin β proteins share relatively lower sequence similarity among members of this gene family. The closest homologs in Arabidopsis, At3g59020 and At1g26170, show 56 and 20% sequence identity to SAD2, respectively. However, knockout mutants of these two importin β genes were not altered in their UV-B response or MYB4 nuclear translocation (data not shown). Moreover, the nuclear localization of GL1, which functions in trichome initiation and development, was not affected by the SAD2 mutation and its protein did not coimmunoprecipitate with SAD2 (data not shown; Figure 6B). Our data demonstrate that SAD2 specifically mediates MYB4 nuclear trafficking and is consistent with our result that MYB4 was mainly present in the cytoplasm in the sad2 mutants.
MYB4 is a negative regulator in the sinapate ester biosynthesis pathway. Overexpression of MYB4 resulted in the reduction of C4H and CHS expression and hypersensitivity to UV-B, suggesting that regulation of MYB4 transcription is essential for UV-B tolerance in Arabidopsis (Jin et al., 2000). Overexpression of MYB4 in the sad2-1 mutant did not repress C4H and CHS gene expression; by contrast, it increased their expression. MYB4 protein binds to its own promoter and inhibits the level of its transcription. Overexpression of MYB4 represses endogenous MYB4, C4H, and CHS expression in wild-type plants but not in the sad2-1 mutant (Figure 7E). In the sad2 mutants, nuclear translocation of MYB4 was affected; therefore, expression of MYB4, CHS, and C4H was increased. Our data demonstrate that MYB4 protein and mRNA form a negative autoregulative feedback loop. This explains the transcriptional regulation of MYB4 (Jin et al., 2000), which includes transient induction of MYB4 by various wavelengths of light and by wounding. The transcription levels of C4H and CHS were reduced 23-fold and 4-fold, respectively, by cotransfection of MYB4 with the C4H or CHS promoter and the GUS reporter (Jin et al., 2000). However pMYB4:GUS expression was reduced only to approximately one-half by cotransfection with 35SP:MYB4 (Figure 7D), suggesting that the selective repression of target gene expression by MYB4 protein is critical for the plant's response to UV-B. A high level of MYB4 transcript was observed in 35SP:MYB4-YFP transgenic plants, but only a weak MYB4-YFP signal was measured, and MYB4-YFP protein could not be detected by protein gel blots with anti-GFP antibody. It is not known how MYB4 fine-tunes its target gene expression. At the beginning of the UV-B treatment, both of the MYB4 and C4H transcripts were induced, and then MYB4 was reduced to an undetectable level by 24 h, but C4H was continuously induced to a very high level (Jin et al., 2000). It is likely that MYB4 is regulated additionally at the posttranslational level or that other cofactors are required especially during UV-B response to repress MYB4 transcription and release C4H and CHS expression to produce more UV-absorbing pigments. In the myb4 knockout mutant, the expression of CHS was not reduced; however, it was repressed during the overexpression of MYB4 transgenic lines (Jin et al., 2000; Figure 7E).
Our LC-MS results show that the concentration of both flavonol and sinapoyl derivatives are enhanced in the sad2-1 mutant, indicating that MYB4 is only one SAD2 target in response to UV-B and that the sad2 mutants do not work exclusively through SAD2's effect on MYB4. Overexpression of MYB4 in sad2-1 significantly enhanced the expression levels of CHS and C4H (Figure 7E), indicating that other CHS and C4H regulating factors may also be affected by SAD2. The expression of only a few genes was affected in the sad2-1 mutant; among them, MYB4 is the only transcription factor (Verslues et al., 2006). SAD2 might affect transport of other transcription regulators that function in flavonol biosynthesis but that do not regulate in their own expression. Recently, three MYB transcription factors have been shown to regulate flavonol biosynthesis (Stracke et al., 2007); these proteins may be targets of SAD2.
sad2 mutants were hypersensitive to ABA (Verslues et al., 2006; data not shown). Two myb4 T-DNA insertion lines were ordered from the Cold Spring Harbor Laboratory, and homozygous lines were generated. No ABA-sensitive phenotype was detected in these two myb4 T-DNA lines, suggesting that the ABA-sensitive and UV-B–tolerant phenotypes of sad2 are due to effects on different targets.
METHODS
Plant Growth and UV-B Treatment
Arabidopsis thaliana C24 was the wild-type plant used in this study. C24, sad2-1, and sad2-2 complemented transgenic plants (com-1 and com-2) were grown on Murashige and Skoog (MS) medium with 0.3% agar (Sigma-Aldrich) under continuous light (Philips TLD 30W/54 YZ30RR25) for 12 d. Seedlings were treated with UV-B light (UVP 34-0042-01) as indicated in the text. Fifteen-day-old Col-0 and sad2-2 seedlings were treated with UV-B (5.3 mW/cm2) for 12 min and subsequently incubated for 3 d in a continuous-light growth chamber. At the end of this incubation, plants were photographed and the percentage of bleached cotyledons was calculated.
ELISA Assays
Fifteen-day-old seedlings grown on MS medium were irradiated with three different doses of UV-B light. The plants were immediately harvested under green light to avoid CPD repair by photolyases, and the DNA was extracted using a Plant DNA Extraction Mini Kit (Qiagen). CPDs were determined by ELISA using TMD-2 antibody (Tanaka et al., 2002) according to the manufacturer's instructions. Briefly, 50 μL of 0.006% protamine sulfate solution (Sigma-Aldrich) was added to each well of a 96-well microtiter plate (Nunc). The plates were dried at 37°C overnight followed by three washes with 100 μL/well of distilled water. Denatured DNA solution (1 ng/μL; 50 μL/well) was added to the protamine sulfate precoated plates, and the DNA-coated plates were washed five times with 150 μL/well PBS-T (0.05% Tween-20 in PBS). To prevent nonspecific antibody binding, 150 μL 2% fetal bovine serum in PBS was added to each well and incubated for 30 min at 37°C. The plates were washed five times with 150 μL/well of PBS-T. TMD-2 antibodies diluted with PBS (1:2000) were added to each well (100 μL/well) and incubated for 30 min at 37°C followed by five washes with 150 μL/well of PBS-T. Anti-mouse IgG (whole molecule) peroxidase conjugate (Sigma-Aldrich) diluted 1:10,000 with PBS was added to each well (100 μL/well) and incubated for 30 min at 37°C followed by five washes with 150 μL/well of PBS-T. The plates were then washed once with 150 μL/well of citrate-phosphate buffer, pH 5.0. Immediately after removal of the citrate-phosphate buffer, 100 μL/well of the substrate solution (8 mg o-phenylene diamine, 4 μL 35% H2O2, and 20 mL citrate-phosphate buffer, pH 5.0) was added to each well and incubated for 30 min at 37°C. Then, 2 M H2SO4 (50 μL/well) was added to each well to stop the enzyme reaction. After gentle mixing, the absorbance at 490 nm of each well was determined using a spectrophotometer (Bio-Rad).
MMS Treatment
Four-day-old seedlings on MS medium with 0.3% agar (Sigma-Aldrich) were transferred to Petri dishes containing liquid MS medium supplemented with different concentrations of MMS. The plants were incubated in a continuous-light growth chamber. Photographs were taken 18 d after transfer to the growth chamber.
Spectrophotometric Analysis
Pigment extracts were prepared by incubating 0.1 g of 12-d-old seedlings in 500 mL of 80% (v/v) ethanol at 65°C for 15 min. The extracts were centrifuged for 15 min at 12,000g, and the supernatants were used to obtain the absorption spectra with a spectrophotometer (Beckman DU 800).
LC-MS Analyses of Soluble Phenolics
Soluble phenolics were extracted from 14-d-old seedlings treated with or without UV-B (0.04 mW/cm2) for 4 h according to the method described by Abdulrazzak et al. (2006). Forty seedlings (similar size and age) per assay were collected for LC-MS analysis in 8 mL of a methanol:water (4:1; v/v) mixture in the presence of morin hydrate (2.5 μg/μL; Sigma-Aldrich) as an internal standard. The extracts were ultrafiltrated (0.2 mm; Minipore) before injection into an HPLCPDA/electrospray ionization-MS system comprising a Finnigan LCQ DECA XP MAX mass spectrometer (Thermo). HPLC was performed on a ZORBAX Eclipse XDB-C18 column (Agilent; Narrow-Bore 2.1 ×150 mm, particle size 3.5 mm) at a flow rate of 0.165 mL/min. The elution was performed using a linear elution gradient from 5 to 60% solvent B (CH3CN + HCOOH, 0.1%) in solvent A (H2O + HCOOH, 0.1%) within 30 min. The mass spectral data were collected in the negative mode with the ion trap mass spectrometer equipped with an electrospray interface. The source voltage was set to 4 kV, and the capillary was heated to 350°C. Product peaks were identified by their mass spectrometric fragmentation pattern and relative to data reported in the literature (Veit and Pauli, 1999; Bloor and Abrahams, 2002; Tohge et al., 2005; Abdulrazzak et al., 2006). Flavonol glycosides and sinapoyl derivatives were analyzed according to the supplemental data published by Abdulrazzak et al. (2006).
Plasmid Constructs
To overexpress MYB4 in plants, full-length cDNA of MYB4 was amplified with the MYB4-specific primers 5′-AACTGCAGATGGGAAGGTCACCGTGCTGT-3′ and 5′-GACTAGTTTATTTCATCTCCAAGCTTCGAAAGC-3′ and cloned into pCAMBIA1207 between EcoRV and SpeI enzyme sites and driven by a 35S promoter. The constructs were introduced into Arabidopsis plants by Agrobacterium tumefaciens–mediated vacuum infiltration. At least eight independent transgenic lines were analyzed for each construct. To transiently express MYB4 in protoplasts, the MYB4 coding region was amplified with primers 5′-CGGGATCCATGGGAAGGTCACCGTGC-3′ and 5′-GGAATTCTTATTTCATCTCCAAGCTTCGAAAGC-3′, harboring BamHI and EcoRI sites, respectively. The PCR product was cloned into the vector pRT105-GFP (Töpfer et al., 1993). To construct the 35SP:MYB4-YFP plasmid for plant transformation, the MYB4 fragment was inserted into the binary vector pCAMBIA1205-YFP between the BamHI and EcoRI sites. For the 35SP:CFP-SAD2 plasmid, the SAD2 coding region was amplified with primers 5′-GGAATTCATGGATCTGCATAGCCTCGC-3′ and 5′-ACGCGTCGACTCATAGAGCAGTAACAGGAGTAG-3′ and cloned into pCAMBIA1205-CFP between the EcoRI and SalI sites.
To transiently express SAD2, MYB4, and GL1 in Arabidopsis protoplasts for coimmunoprecipitation, three constructs, 2×35SP:6myc-SAD2, 35SP:3×flag-MYB4, and 35SP:3×flag-GL1, were used. For 2×35S:6myc-SAD2, SAD2 was digested with BamHI and SalI from the 35SP:CFP-SAD2 plasmid, inserted into pRT107-6×myc (Töpfer et al., 1993) under the control of a double cauliflower mosaic virus 35S promoter. For 35SP:3×flag-MYB4, MYB4 was excised from the 35SP:MYB4-YFP plasmid with BamHI and EcoRI and cloned into the pRT105-3×flag plasmid. For 35SP:3×flag-GL1, the GL1 coding region was amplified with the primers 5′-CGGGATCCATGAGAATAAGGAGAAGAG-3′ and 5′-GGAATTCCTAAAGGCAGTACTCAACATC-3′. The PCR product was digested with BamHI and EcoRI, and the fragment was cloned into the pRT105-3×flag plasmid.
To express MYB4 protein in Escherichia coli, the MYB4 open reading frame was amplified with the primers 5′-CGGGATCCATGGGAAGGTCACCGTGC-3′ and 5′-ACGCGTCGACTTATTTCATCTCCAAGC-3′, cloned into the plasmid pMAL-c2X (New England Biolabs) between the BamHI and SalI sites, and translationally fused to MBP. All PCR products were sequenced.
To detect the transcriptional level of MYB4, the DNA fragment in the promoter region of MYB4 was amplified with primers 5′-ACGCGTCGACGATAGTGGTTTGTATTTACC-3′ and 5′-GGGGTACCGTAATCAAAGAAATTTCTC-3′, and transcriptionally fused to the GUS reporter gene.
Real-Time PCR and RT-PCR Analysis
Total RNA was extracted with the TRI reagent (Ambion) from 12-d-old seedlings grown on MS plates or 1-month-old plants grown in pots in the greenhouse under an 16-h-light/8-h-dark photoperiod. Ten micrograms of total RNA was treated with RNase-free DNase I (Promega) to remove DNA, and 1 μg of RNA was used for reverse transcription with M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. The cDNAs were used for quantitative real-time PCR amplification with the following primers: Actin as internal control, forward 5′-GTCGTACAACCGGTATTGTG-3′ and reverse 5′-GAGCTGGTCTTTGAGGTTTC-3′; SAD2, forward 5′- CGTCCAATCCTCCCTCAGTTACTTGA-3′ and reverse 5′-TACTTATGGCACGCAAACAACC-3′; C4H, forward 5′-CCGGGATTATATTGGCATTG-3′ and reverse 5′-CTGAATTGTCCACCTTTCTC-3′; CHS, forward 5′-AGAGAAGATGAGGGCGACACG-3′ and reverse 5′-GCCACACCATCCTTAGCTGAC-3′; MYB4, forward 5′-CAAGGGCATGGAAAGTCAAC-3′ and reverse 5′-TTGCTGCTACCTCCGACTAC-3′; endogenous MYB4, forward 5′-CAAGGGCATGGAAAGTCAAC-3′ and reverse 5′-AAGTTTTGTACAGTTACGCCTAA-3′; FAH1, forward 5′-GGATGTTGTCGATACCGATATGG-3′, and reverse 5′-GCTCCGTAATAACTCCGTTAAGG-3′; CAD, forward 5′-ACGGGAAATTGATTCTCATGGG-3′, and reverse 5′-CCTCTCAAACGCAATGTTAAGC-3′.
RNA Gel Blot Analysis
Total RNA was isolated from 19-d-old plants with the TRI reagent as described above. Twenty micrograms of total RNA from each sample was separated on 1% agarose gels and transferred onto nitrocellulose membranes (Amersham Biosciences). The full-length cDNA of MYB4 was labeled with 32P-dCTP by Klenow (Takara). RNA gel blot analysis was performed as described by Sambrook and Russell (2001).
Protoplast Transformation
One-month-old Arabidopsis plants grown in the greenhouse under short-day conditions were used to prepare leaf protoplasts according to the protocol of Sheen (2001). GFP fluorescence was observed after 10 to 15 h of recovery using a Zeiss LSM 510 META confocal microscope.
Coimmunoprecipitation
The 2×35SP:6×myc-SAD2 plasmid was cotransformed with either 35SP:3×flag-MYB4 or 35SP:3×flag-GL1 into wild-type protoplasts. After a 10- to 12-h incubation, the protoplasts were harvested and lysed in 300 μL extraction buffer containing 10 mM Tris, pH 7.5, 0.5% Nonidet P-40, 2 mM EDTA, 150 mM NaCl, 1 mM PMSF, and 1% protease inhibitor cocktail (Sigma-Aldrich). Ten microliters of anti-c-myc agarose conjugate (Sigma-Aldrich) was incubated with the extract supernatant overnight at 4°C. After washing five times in 1 mL of extraction buffer, the immunoprecipitation product was detected by protein gel blots according to the manufacturer's instructions. Both anti-c-myc (Sigma-Aldrich) and anti-flag (Sigma-Aldrich) antibodies were used at 1:5000 dilution.
EMSA
E. coli cells (strain TB1) were used to express recombinant MBP-MYB4, and the protein was purified from the cells with amylase prepacked columns (New England Biolabs) according to the manufacturer's instructions.
DNA fragments with AC cis-elements in the MYB4 promoter were amplified with the following primers: fragment 14, forward 5′-ACGATAGTGGTTTGTATTTAC-3′ and reverse 5′-TCAGAAGAGTAAGATGCTAC-3′; fragment 15, forward 5′-GATTCAGATGTAAATGGACG-3′ and reverse 5′-TGTGTACTGTCCATGTAT-3′; fragment 16, forward 5′-TCATAGGCTATATAGTTGAC-3′ and reverse 5′-CCGATTTAGTACAAACAG-3′. The oligos used for MYB binding elements, including the mutations, were as follows: elements AC-I (W), forward 5′-ACCTACCACCTACCACCTACC-3′ and reverse 5′-GGTAGGTGGTAGGTGGTAGGT-3′; elements AC-I (M), forward 5′-ACATACCACATACCACATACC-3′ and reverse 5′-GGTATGTGGTATGTGGTATGT-3′; elements AC-II (W), forward 5′-ACCAACCACCAACCACCAACC-3′ and reverse 5′-GGTTGGTGGTTGGTGGTTGGT-3′; elements AC-II (M), forward 5′-ACAAACCACAAACCACAAACC-3′ and reverse 5′-GGTTTGTGGTTTGTGGTTTGT-3′; elements AC-III (W), forward 5′-ACCTAACACCTAACACCTAAC-3′ and reverse 5′-GTTAGGTGTTAGGTGTTAGGT-3′; elements AC-III (M), forward 5′-ACATAACACATAACACATAAC-3′ and reverse 5′-GTTATGTGTTATGTGTTATGT-3′; elements MYB-1 (W), forward 5′-ACCCGCCACCCGCCACCCGCC-3′ and reverse 5′-GGCGGGTGGCGGGTGGCGGGT-3′. The DNA probes were labeled with [γ-32P]ATP using T4 polynucleotide kinase and purified using a Sephadex G-25 column (Amersham Biosciences). Three hundred nanograms of DNA fragment and 1 μg of recombinant MYB4 protein were used in gel-shift assays. The DNA binding reactions were performed in a buffer containing 10 mM HEPES/KOH, pH 7.5, 0.1 mM EDTA, 75 mM KCl, 1.25 mM MgCl2, 0.2 mM DTT, 2 μg/mL BSA, and 5% glycerol. The binding solution was preincubated for 30 min on ice and then incubated with the labeled probe (0.02 to 0.08 pmole) for 30 min on ice. For competition experiments, unlabeled competitor (2 to 8 pmole) was added to the reaction. After DNA binding reactions, the samples were loaded onto 5% polyacrylamide native gels, which had been prerun at 180 V for 30 min at 4°C, and run at 180 V for 30 to 50 min at 4°C in 0.5× TGE buffer (12.5 mM Tris, 96 mM glycine, and 0.5 mM EDTA, pH 8.3). After electrophoresis, the gel was dried under vacuum at 80°C for 15 min on filter paper and exposed to a phosphor imager for 2 to 10 h.
Homozygous Mutant Identification
The sad2-1 mutant was identified with the SAD2-specific primers 5′-TTCAAGTCAGATGAGGAGAGGA-3′ and 5′-GCTCAAATGTAATACCTCAAGAAGA-3′ and T-DNA left border primers, AtLB1 5′-ATACGACGGATCGTAATTTGTC-3′ and AtLB2 5′-TAATAACGCTGCGGACATCTAC-3′. The following primers were used to identify the sad2-2 (ABRC SALK_133577) homozygous mutant: 5′-AGTTGACTCCATCACGGTAGC-3′ and 5′-ATACACCAAGCGTCAAACCTG-3′ and T-DNA left border primers LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′ and LBb1 5′-GCGTGGACCGCTTGCTGCAACT-3′.
GUS Activity Measurements
The plasmid pRT105-AtMYB4 was cotransformed with pAtMYB4:GUS or p35Smini:GUS into protoplasts for transient expression as described above. The protoplasts were collected after incubation in W5 solution (Sheen, 2001) for at least 12 h at room temperature. GUS assay buffer (50 mM sodium phosphate, pH 7.0, 5 mM DTT, 1 mM EDTA, and 0.1% Triton X-100) was used to release total protein from the protoplasts. The total protein was quantified by the Bradford method (Bradford, 1976). Eight micrograms of protein was incubated with 0.25 mM 4-Methylumbelliferyl-β-d-glucuronide in a 400-μL reaction for 1 h at 37°C, after which 900 μL of 0.2 M Na2CO3, pH 9.5, was added to stop the reaction. GUS activities were detected using an F-4500 fluorimeter (Hitachi) with the excitation filter at 365 nm and the emission filter at 450 nm.
Accession Numbers
Sequence data for sad2-1 and sad2-2 T-DNA left border flanking sequences can be found in the GenBank/EMBL data libraries under accession numbers EU196514 (sad2-1) and EU196513 (sad2-2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of sad2 Mutants by the SAD2 Gene.
Supplemental Figure 2. Expression of SAD2 Homologs At3g59020 and At1g26170 in the Wild Type and Their Corresponding Mutants.
Supplemental Figure 3. Transient Expression of MYB4-GFP in Wild-Type and sad2-2 Protoplasts.
Supplemental Figure 4. A Schematic Representation of MYB4 Binding cis-Elements in the MYB4 5′ Upstream Sequence.
Supplementary Material
Acknowledgments
We thank Jianmin Zhou and Wanhong Cao for critical reading of the manuscript and stimulating discussions; Liqin Fu, Cheng Zhan, and Jun Zhang for excellent technical assistance and the ABRC (Ohio State University) for the T-DNA insertion lines. We also thank Xiangning Jiang and Weiqi Chen for LC-MS analyses of soluble phenolics. This work was supported by National Basic Research Program of China Grant 2006CB100100 and National High Technology Research and Development Program of China 863 Grant 2003AA210100 to Y.G., by National Natural Science Foundation of China Grant 30771156 to W.H.Z., and by Department of Energy/Energy Biosciences Grant DE-FG02-04ER15616/G to K.S.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy as described in the Instructions for Authors (www.plantcell.org) is: Yan Guo (guoyan@nibs.ac.cn).
Online version contains Web-only data.
References
- Abdulrazzak, N., et al. (2006). A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 140 30–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bieza, K., and Lois, R. (2001). An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol. 126 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor, S.J., and Abrahams, S. (2002). The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59 343–346. [DOI] [PubMed] [Google Scholar]
- Bollman, K.M., Aukerman, M.J., Park, M.-Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130 1493–1504. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Frohnmeyer, H., and Staiger, D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, J. (1991). Pigments in vegetables: Chlorophylls and carotenoids. In AVI Book, J. Gross, ed (New York: Van Nostrand Reinold Publishing), p. 303.
- Hatton, D., Sablowski, R., Yung, M.H., Smith, C., Schuch, W., and Bevan, M. (1995). Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 7 859–876. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.E., and Kieber, J.J. (2002). Cytokinin signaling in Arabidopsis. Plant Cell 14 S47–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, M.A.K., Gaba, V., and Greenberg, B.M. (1998). Higher plants and UV-B radiation: Balancing damage, repair and acclimation. Trends Plant Sci. 3 131–135. [Google Scholar]
- Jiang, C.J., Shoji, K., Matsuki, R., Baba, A., Inagaki, N., Ban, H., Iwasaki, T., Imamoto, N., Yoneda, Y., Deng, X.W., and Yamamoto, N. (2001). Molecular cloning of a novel importin a homologue from rice, by which COP1 NLS-protein is preferentially nuclear imported. J. Biol. Chem. 276 9322–9329. [DOI] [PubMed] [Google Scholar]
- Jiang, C.-Z., Yee, J., Mitchell, D.L., and Britt, A.B. (1997). Photorepair mutants of Arabidopsis. Proc. Natl. Acad. Sci. USA 94 7441–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Lim, J.E., Landry, L.G., and Last, R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human Regulator of Chromatin Condensation 1. Plant Physiol. 130 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, L.G., Chapple, C.C.S., and Last, R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B., and Zhu, J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Ou-Lee, T.M., Raba, R., Amundson, R.G., and Last, R.L. (1993). Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Hossain, G., Showkat, I.-O., Maria, A., Mitchell, D.L., and Mount, D.W. (2000). Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J. 21 519–528. [DOI] [PubMed] [Google Scholar]
- Lois, R. (1994). Accumulation of UV-absorbing flavonoids induced by UV-B radiation in Arabidopsis thaliana. Planta 4 498–503. [Google Scholar]
- Lois, R., and Buchanan, B.B. (1994). Severe sensitivity to ultraviolet radiation in an Arabidopsis mutant deficient in flavonoid accumulation. Planta 4 504–509. [Google Scholar]
- Mackerness, A.H.S., John, C.F., Jordan, B., and Thomas, B. (2001). Early signaling components in ultraviolet-B responses: Distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett. 489 237–242. [DOI] [PubMed] [Google Scholar]
- Mazza, C.A., Boccalandro, H.E., Giordano, C.V., Battista, D., Scopel, A.L., and Ballare, C.L. (2000). Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol. 122 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle, T. (2004). Nucleo-cytoplasmic partitioning of proteins in plants: Implications for the regulation of environmental and developmental signalling for the regulation of environmental and developmental signaling. Curr. Genet. 44 231–260. [DOI] [PubMed] [Google Scholar]
- Miyamoto, Y., Saiwaki, T., Yamashita, J., Yasuda, Y., Kotera, I., Shibata, S., Shigeta, M., Hiraoka, Y., Haraguchi, T., and Yoneda, Y. (2004). Cellular stresses induce the nuclear accumulation of importin α and cause a conventional nuclear import block. J. Cell Biol. 165 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, S., Sugiyama, M., Iwai, S., Hitomi, K., Otoshi, E., Kim, S.T., Jiang, C.Z., Todo, T., Britt, A.B., and Yamamoto, K. (1998). Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 26 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma, K., Zhang, Y., and Li, X. (2005). An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15 1129–1135. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Stacey, M.G., Hicks, S.N., and Von Arnim, A.G. (1999). Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., and Weisshaar, B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Tanaka, A., Sakamoto, A., Ishigaki, Y., Nikaido, O., Sun, G., Hase, Y., Shikazono, N., Tano, S., and Watanabe, H. (2002). An ultraviolet-B-resistant mutant with enhanced DNA repair in Arabidopsis. Plant Physiol. 129 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge, T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Töpfer, R., Maas, C., Höricke-Grandpierre, C., Schell, J., and Steinbiss, H.-H. (1993). Expression vector for high-level gene expression in dicotyledonous and monocotyledonous plants. Methods Enzymol. 217 66–78. [DOI] [PubMed] [Google Scholar]
- Veit, M., and Pauli, G.F. (1999). Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62 1301–1303. [DOI] [PubMed] [Google Scholar]
- Verslues, P.E., Guo, Y., Dong, C.-H., Ma, W., and Zhu, J.-K. (2006). Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. 47 776–787. [DOI] [PubMed] [Google Scholar]
- Yang, H.-Q., Wu, Y.-J., Tang, R.-H., Liu, D., Liu, Y., and Cashmore, A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103 815–827. [DOI] [PubMed] [Google Scholar]
- Ziemienowicz, A., Haasen, D., Staiger, D., and Merkle, T. (2003). Arabidopsis transportin1 is the nuclear import receptor for the circadian clock-regulated RNA-binding protein AtGRP7. Plant Mol. Biol. 53 201–212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.