Abstract
The bacterial ribosome is an extremely complicated macromolecular complex the in vivo biogenesis of which is poorly understood. Although several bona fide assembly factors have been identified, their precise functions and temporal relationships are not clearly defined. Here we describe the involvement of an Escherichia coli GTPase, CgtAE, in late steps of large ribosomal subunit biogenesis. CgtAE belongs to the Obg/CgtA GTPase subfamily, whose highly conserved members are predominantly involved in ribosome function. Mutations in CgtAE cause both polysome and rRNA processing defects; small- and large-subunit precursor rRNAs accumulate in a cgtAE mutant. In this study we apply a new semiquantitative proteomic approach to show that CgtAE is required for optimal incorporation of certain late-assembly ribosomal proteins into the large ribosomal subunit. Moreover, we demonstrate the interaction with the 50S ribosomal subunits of specific nonribosomal proteins (including heretofore uncharacterized proteins) and define possible temporal relationships between these proteins and CgtAE. We also show that purified CgtAE associates with purified ribosomal particles in the GTP-bound form. Finally, CgtAE cofractionates with the mature 50S but not with intermediate particles accumulated in other large ribosome assembly mutants.
Although assembly of prokaryotic ribosomes from purified ribosomal proteins (r-proteins) and rRNAs can occur independently in vitro (51, 52, 75), accumulating evidence suggests that, as in eukaryotes, in vivo prokaryotic ribosome biogenesis depends on the aid of nonribosomal factors. The higher temperature, increased Mg2+ concentration, and longer incubation times necessary for in vitro relative to in vivo conditions (51) imply that the likely role of accessory factors is to expedite ribosome maturation by reducing the activation energies for the rate-limiting reactions. Although not complicated by the involvement of different cellular compartments, the prokaryotic ribosome assembly process is far from simple, requiring coordinated synthesis of 3 rRNAs (5S, 16S, and 23S) and 55 r-proteins, processing and modification of these components, and their appropriate sequential unification to produce mature ribosomes. The details of how this process is controlled temporally, even spatially, in the small bacterial cell are incompletely understood.
More than 170 nonribosomal proteins that transiently associate with different preribosomal particles have been identified in Saccharomyces cerevisiae (19, 22, 38, 62), largely due to progress in combining biochemical affinity purification methods with newly developed proteomic techniques (24, 25, 29, 54, 58, 61). By contrast, only a few such assembly factors have been found in bacteria, and most were identified via conventional genetic methods. These proteins consist of RNA-modifying enzymes such as methyltransferases and pseudouridine synthases, RNA-remodeling proteins such as RNA helicases, chaperones, GTPases, and proteins with unknown functions (1, 5, 7, 10, 11, 18, 26, 32, 33, 48-50, 60, 72, 76). Understanding of the molecular mechanisms by which these factors monitor and influence the ribosomal assembly process and a comprehensive picture of the interactions among these different constituents, however, are still lacking.
Based on phylogenetic analysis, it is hypothesized that all GTPases are derived from an ancestral GTPase with a role in translation (39). The Obg subfamily is a class of highly conserved small monomeric GTPases that appear to be involved primarily in assembly of the large ribosomal subunit. In Saccharomyces cerevisiae, the mitochondrial Obg ortholog Mtg2p is involved in the biogenesis of the large (54S) mitochondrial ribosomal subunit (14), and the nucleolar Obg protein Nog1p is important for pre-60S particle assembly (34, 62). In Bacillus subtilis, Obg associates with the ribosome, and this association can be stabilized by the addition of GTP (82). More specifically, Obg has been shown to bind ribosomal protein L13 by an affinity blot assay (66). The Caulobacter crescentus Obg protein CgtAC cofractionates exclusively with the 50S ribosomal particle (42), and strains expressing a temperature-sensitive allele of cgtAC had a reduced level of 50S subunits compared to the wild type, even at the permissive temperature (15). Likewise, Escherichia coli CgtAE associates with the large ribosome subunit (60, 80), interacts with rRNAs and several r-proteins, and copurifies with the known 50S ribosome assembly factor CsdA (60, 80). In a cgtAE mutant, the ribosome profile is perturbed and a defect in 16S rRNA processing is observed (60). Furthermore, CgtAE has been genetically implicated in the assembly of the 50S subunit based on its ability to suppress an rrmJ mutant. RrmJ is an RNA methyltransferase that is involved in late 50S ribosome assembly. The deletion of rrmJ causes slow growth and a polysome defect, both of which can be suppressed by overexpression of CgtAE (72). All these data are consistent with the role of Obg/CgtA proteins in ribosome assembly and/or 70S coupling.
In this study we further characterize the association between the ribosome and the E. coli CgtAE protein and show that they interact, with the GTP-bound form of CgtAE having a higher affinity for the ribosome. Further, we demonstrate that CgtAE is crucial for the late steps of 50S ribosome subunit assembly. A mutant form of cgtAE displays an altered polysome profile similar to those seen in late ribosome assembly mutants. A new semiquantitative proteomic technique, iTRAQ (isotope tag for relative and absolute quantitation), allows us to describe the proteome of the accumulated 50S particles in the cgtAE mutant. This analysis provides new insights into the precise function of CgtAE in ribosome assembly.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and growth measurements.
E. coli strains used are listed in Table 1. E. coli cells were grown in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter) or on LB agar plates (LB plus 1.5% agar), containing antibiotics as required, at the indicated temperatures. Culture growth was monitored by measuring the absorbance at 600 nm. Antibiotics were used at the following concentrations: 100 μg/ml ampicillin, 30 μg/ml kanamycin, 20 μg/ml chloramphenicol, or 12 μg/ml tetracycline.
TABLE 1.
Escherichia coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference | ||
|---|---|---|---|---|
| Strains | ||||
| MG1655 | rph-1 | 4 | ||
| DH5α | Δ(lab)U169 φ80 Δ(lacZ)M15 hsdR17 endA1 gyrA96 recA1 supE44 thi-1 | 28 | ||
| JM3733 | WJW45 rna-30 ΔcsdA | 10 | ||
| JM3734 | WJW45 ΔsrmB | 11 | ||
| GN5002 | MG1655 ΔcgtAE::kan + pGK14 | 35 | ||
| GN5003 | MG1655 ΔcgtAE::kan + pGK15 | 35 | ||
| JM1138 | BL21+pJM1138 | 80 | ||
| JM3903 | MG1655 ΔcgtAE::kan + pMJ31 | This work | ||
| JM3907 | MG1655 ΔcgtAE::kan + pMJ32 | This work | ||
| JM4711 | JM3903 + pJW3146 | This work | ||
| JM4714 | JM3907 + pJW3146 | This work | ||
| Plasmids | ||||
| pCR2.1TOPO | ori-PUC kan bla | Invitrogen | ||
| pGD103-bla | ori-pSC101 bla | 16 | ||
| pGK14 | ori-pSC101 cat PBADcgtAE+ | 35 | ||
| pGK15 | ori-pSC101 cat PBADcgtAE(G80E D85N) | 35 | ||
| pJW3146 | PT5-lachis-rrmJ | http://ecoli.aist-nara.ac.jp | ||
| pJM1138 | PT7his-cgtAE | 80 | ||
| pMJ25 | ori-PUC kan bla cgtAE+ | This work | ||
| pJM3738 | ori-PUC kan bla cgtAE(G80E D85N) | This work | ||
| pMJ27 | ori-PUC kan bla PCgtAE | This work | ||
| pMJ33 | ori-pUC kan bla PCgtAEcgtAE+ | This work | ||
| pMJ29 | ori-pUC kan bla PCgtAEcgtAE(G80E D85N) | This work | ||
| pMJ31 | ori-pSC101 bla PCgtAEcgtAE+ | This work | ||
| pMJ32 | ori-pSC101 bla PCgtAEcgtAE(G80E D85N) | This work | ||
Strains and plasmid construction.
Full-length cgtAE+ or the cgtAE(G80E D85N) allele was amplified from MG1655 genomic DNA or pGK15 (35), respectively, by PCR using primers YhbZ3-new (5′-GAGAATCATATGAAGTTTGTTGATGAA-3′) and YhbZ-end-HindIII (5′-GATTAAGCTTTTATCATCAGTGATTAA-3′). The 1.17-kb PCR products were cloned into the pCR2.1-TOPO TA cloning vector (Invitrogen Life Technologies), generating pMJ25 and pJM3738, respectively. The cgtAE promoter region was amplified using primers YhbZ-prom-up (5′-GTCTGCAGGGTCGTGACCACACTCTG-3′) and YhbZ-prom-down (5′-GCCATATGGTATTCCCTGCAAAGCGCATT-3′) and cloned into the pCR2.1-TOPO vector. The resulting plasmid was designated pMJ27. To place cgtAE alleles under the control of the cgtAE promoter, the cgtAE alleles were excised from pMJ25 or pJM3738 using NdeI and BamHI and were ligated to pMJ27 digested with the same enzymes. The resulting plasmids, pMJ33 and pMJ29, were digested with PstI and BamHI and were ligated to the low-copy-number plasmid pGD103-bla, a derivative of pGD103 (16), digested with the same enzymes. The subsequent constructs, pMJ31 and pMJ32, were verified by sequencing and transformed into GN5002 and GN5003, respectively. The original plasmids in these two strains, pGK14 and pGK15, were lost by growing the transformants in LB supplemented with ampicillin and kanamycin and screening for chloramphenicol sensitivity, thus generating JM3903 and JM3907 [strains harboring cgtAE+ and cgtAE(G80E D85N), respectively].
Preparation of cell lysates for ribosome profiles.
E. coli strains were grown at the following temperatures: 37°C for MG1655; 30°C for JM3903, JM3907, JM4711, and JM4714; and 20°C for JM3733 and JM3734. Chloramphenicol (FisherBiotech) was added to a final concentration of 200 μg/ml 30 s before harvest. Cells were harvested at an optical density at 600 nm (OD600) of 0.4 to 0.8 by centrifugation at 10,000 × g and 4°C for 10 min in an SLA-1500 rotor (Sorvall). The cell pellet was resuspended in 1 ml lysis buffer (10 mM Tris-Cl [pH 7.5], 10 mM MgCl2, 30 mM NH4Cl, 100 μg/ml chloramphenicol) per 100 ml of culture. The cell lysate was mixed with an equal volume of glass beads (300 μm; Sigma) and vortexed for 5 min at 4°C. The lysate was clarified by a 10-min centrifugation at 32,000 × g and 4°C in an SA-600 rotor (Sorvall). The supernatant was carefully collected and quantified by UV absorbance at 260 nm.
Protein and ribosome purification.
His-CgtAE was purified from JM1138 as previously described (80), with the following modification. After the nickel-nitrilotriacetic acid column, the CgtAE-containing fractions were pooled, dialyzed against core buffer (50 mM Tris [pH 8.0], 10% glycerol, 1 mM dithiothreitol), and loaded onto a 50-ml Toyopearl DEAE-650 M column (TosoHaas), and CgtAE was eluted with a 100-ml linear gradient of core buffer containing 0 to 400 mM NaCl. The relevant fractions were then purified over a 100-ml Sephadex G-75 column as previously described (40). Finally, similarly dialyzed CgtAE fractions were purified over a 1.3-ml UNO Q1 column (Bio-Rad) and eluted with a 30-ml linear gradient of core buffer containing 0 to 1 M NaCl. The concentration of purified CgtAE was determined by a Bradford assay (Bio-Rad). Purified ribosomes were obtained as previously described (13).
Polyribosome fractionation.
Sucrose gradients were formed using a gradient maker (SG15 or SG50; Hoefer) under the indicated buffer conditions. Approximately 13 OD260 units of the cell lysates was loaded gently onto the top of the 10-ml 7 to 47% sucrose gradients, and the gradients were centrifuged in a Beckman SW41 Ti rotor for 3 h at 41,000 rpm (210,000 × g) or in a Beckman SW40 Ti rotor for 4 h at 35,000 rpm (155,000 × g) and 4°C. In work with the ΔsrmB or ΔcsdA strains, in order to separate the 40S from the 50S particles, 35-ml 7 to 47% gradients were used and centrifuged in a Beckman SW28 rotor for 7 h at 28,000 rpm (103,000 × g) and 4°C. The resulting gradients were fractionated as previously described (42). For the experiments in which purified CgtAE and purified ribosomes were used, 2 mM GDP, GTP, or GMP-PNP (5′-guanylyl-imidodiphosphate trisodium salt) was added to the purified protein before the addition of equimolar amounts of ribosomes (8 OD260 units), and the mixture was separated over 10 to 30% sucrose gradients by centrifugation in a Beckman SW40 Ti rotor for 4 h at 35,000 rpm (155,000 × g) and 4°C. Protein samples were precipitated with 15% trichloroacetic acid (TCA) and 0.03% deoxycholic acid, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subjected to immunoblot analysis.
Immunoblot analysis.
Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (NEN Life Science Products) with a Hoefer TE77 semidry transfer apparatus (GE Healthcare). The immunoblot analyses were carried out as previously described (42). The following antibody concentrations were used: for anti-His (Sigma), 1:2,000; for anti-CgtAE, 1:2,000; for anti-L3, 1:4,000; for anti-L33, anti-L34, and anti-L16, 1:10,000.
RNA preparations, RNA electrophoresis, and Northern blot analysis.
Frozen cell pellets (stored at −80°C) from 10 ml JM3903 or JM3907 mid-log-phase cell cultures were resuspended in 800 μl 0.5-mg/ml lysozyme-Tris-EDTA (pH 8.0). SDS and sodium acetate were added to final concentrations of 1% and 0.1 M, respectively. RNA was extracted once at 64°C with an equal volume of water-saturated phenol and once with an equal volume of chloroform at room temperature. RNA was precipitated with 1/10 volume of 3 M sodium acetate, 1 mM EDTA, and 2.5 volumes of 100% ethanol at −80°C, resuspended in water, and quantified by UV absorbance at 260 nm. Equal amounts of RNA (5 μg) from each strain were separated by a 1.2% agarose gel in TBE (90 mM Tris-borate [pH 8.3], 1 mM EDTA) followed by ethidium bromide staining or, for Northern blotting, were mixed with 2 volumes of sample buffer (8% formaldehyde, 1.3× morpholinepropanesulfonic acid [MOPS], 65% formamide), denatured at 65°C for 5 min, and separated via a 1.2% agarose-MOPS gel run at 150 V for 6 h. RNA was transferred overnight to a nylon membrane (GeneScreen Plus; Perkin-Elmer) by capillary action. After transfer, both sides of the membrane were UV cross-linked using an FB-UVXL-1000 cross-linker (FisherBiotech). The membrane was prehybridized in buffer I (1% bovine serum albumin, 0.5 M sodium phosphate [pH 7.2], 1 mM EDTA [pH 8.0], 5% SDS) for 2 h at 65°C and then hybridized in the same buffer with 32P-labeled probes (Fig. 3B) (11) for 1 h at 60°C. Washing conditions and probes were as previously described (11). The blots were exposed to X-ray films for images, and the radioactive signals were quantified by a Molecular Imager FX PhosphorImager and Quantity One software (Bio-Rad). The blot was stripped in boiling buffer II (0.2 × SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% SDS) and reprobed with different probes.
FIG. 4.
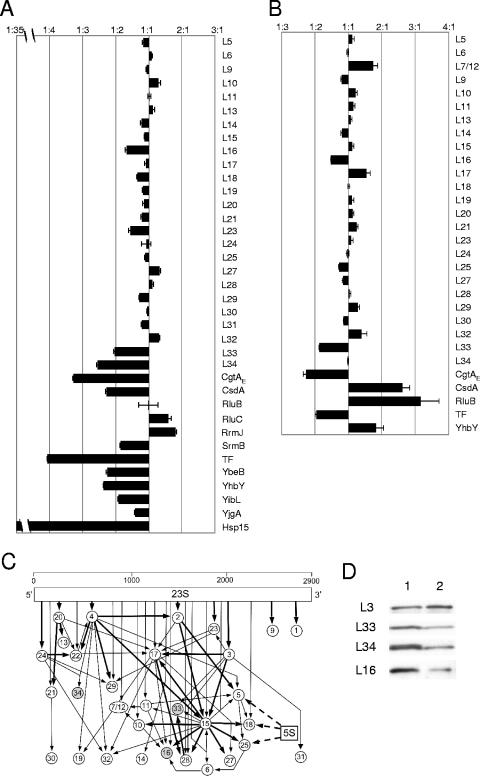
The 50S* particle in the cgtAE(G80E D85N) mutant is not a mature 50S particle. (A and B) Semiquantitative iTRAQ results [shown as the average cgtAE(G80E D85N)/cgtAE+ ratio] for the indicated proteins. Proteins from the relevant ribosomal peaks from cgtAE(G80E D85N) (JM3907) and cgtAE+ (JM3903) cell extracts under high-Mg2+, low-salt (A) (50S* and 50S, respectively) or low-Mg2+, high-salt (B) (40S* and 50S, respectively) conditions (see Fig. 2) were digested with trypsin, labeled with isobaric tags, and mixed, and peptides were quantified as described in Materials and Methods. Shown are the relative levels of key r-proteins and associated proteins, expressed as the ratio of protein in cgtAE(G80E D85N) subunits to that in wild-type subunits. Error bars, standard errors of the means. (C) In vitro assembly map of the 50S ribosomal subunit modified from reference 31. The approximate location of protein binding to the 23S rRNA is indicated by the position of the arrows. Proteins that bind directly to the 5S rRNA are indicated by dashed arrows. Assembly relationships are indicated by arrows (thick arrows for a strong requirement, thin arrows for a weak requirement). Circles representing three proteins that are reduced in the cgtAE(G80E D85N) mutant 50S* are shaded. (D) Equal amounts of proteins from wild-type 50S (lane 1) and mutant 40S* (lane 2) under low-Mg2+, high-salt conditions were subjected to immunoblot analysis using an anti-L3 antibody. The membrane was stripped and reprobed with anti-L33, anti-L34, and anti-L16, as indicated.
Sample preparation for iTRAQ labeling.
Approximately 25 OD260 units of cell lysates was sedimented through 5 to 20% sucrose gradients in a Beckman SW28 rotor for 7 h at 28,000 rpm (103,000 × g) and 4°C and was fractionated as described above. Fractions representing the desired ribosome particles from multiple gradients were pooled and precipitated with TCA as described above. The pellets were washed twice with acetone and resuspended in double-distilled H2O, and the amount of protein was estimated by a Bradford assay (Bio-Rad). Protein samples were lyophilized by vacuum centrifugation, resuspended to a final concentration of 1 or 2.4 mg/ml in 20 μl 0.5 M triethylammonium bicarbonate, and digested with trypsin (porcine modified, 1:16 [wt/wt]; Promega) at 40°C overnight as described previously (12). Labeling with the isobaric tagging iTRAQ reagent was performed as described previously (59). Briefly, 1 U of the iTRAQ reagent was added directly to the peptide mixtures (70% ethanol [final concentration]), and samples were incubated at room temperature for 1 h. The reactions were quenched by addition of 9 volumes 0.1% trifluoroacetic acid (TFA) in water (Optima grade; Fisher Scientific), and reaction products were combined and stored at −20°C. For particles obtained under high-Mg2+, low-salt conditions, iTRAQ reagent 114 was used to label tryptic peptides from cgtAE+ 50S* particles, and iTRAQ reagents 116 and 117 were used to label tryptic peptides from independently isolated cgtAE(G80E D85N) 50S* particles. For particles obtained under low-Mg2+, high-salt conditions, iTRAQ reagents 114 and 115 were used to label cgtAE+ 50S particles, and iTRAQ reagents 116 and 117 were used to label cgtAE(G80E D85N) 40S* particles.
Two-dimensional LC and tandem mass spectrometry (2DLC-MS-MS).
An aliquot of the peptide mixture (200 μg) was fractionated on a strong cation-exchange column (SCX microSpin column; Poly LC, Southborough, MA) equilibrated with 10 mM KH2 phosphate (pH 4.5)-20% CH3CN. For peptide adsorption to the column, washing, and elution, a centrifugal force was applied in approximately 2-s bursts such that 50 μl of solution passed through the column over a 50- to 60-s interval. Excess reagent was washed from the column with 800 μl equilibration buffer. Peptides were eluted using 50-μl volumes of KCl in a stepwise gradient (25, 50, 75, 100, 150, 225, and 350 mM KCl) in equilibration buffer. Fractions were dried in a vacuum centrifuge and reconstituted with 43 μl 0.1% trifluoroacetic acid. Samples (40 μl) from each KCl elution step were separated by C18 nano-liquid chromatography (nano-LC) (Zorbax 300SB; diameter, 5 μm; 5 by 0.3 mm; Agilent Technologies, Palo Alto, CA) using an Agilent 1100 high-performance LC system (Agilent Technologies, Palo Alto, CA) as described previously (12). The column effluent was mixed online with matrix (2 mg/ml α-cyano-4-hydroxycinnamic acid in CH3OH-isopropanol-CH3CN-H2O-acetic acid [12:33.3:52:36:0.7, by volume] containing 10 mM ammonium phosphate) in a mixing Tee (micro-Tee; Agilent Technologies, Palo Alto, CA) and spotted at 30-s intervals onto stainless steel matrix-assisted laser desorption ionization (MALDI) targets (192 wells/plate; Applied Biosystems).
MS-MS spectra were acquired on a model 4700 tandem time-of-flight (TOF/TOF) mass spectrometer (Applied Biosystems/MDX Sciex, Foster City, CA) in positive-ion mode. MS survey spectra were acquired from 800 to 3,500 Da for each fraction. The eight most intense peaks in each MS spectrum above a signal-to-noise ratio threshold of 80 were selected for MS-MS analysis. For peaks observed in consecutive fractions, only the most intense instance was selected for MS-MS analysis. Fragmentation of the labeled peptides was induced by the use of atmosphere as a collision gas with a pressure of ∼6 ×10−7 torr and a collision energy of 1 kV. Samples from the high-Mg2+, low-salt conditions were reapplied to the TOF/TOF mass spectrometer, and an include list was used to determine MS-MS spectrum selection. Peptides were identified using GPS Explorer (version 3.0; Applied Biosystems) as a front end for the Mascot search engine (version 1.9; Matrix Science, London, United Kingdom). The MS-MS spectrum were searched against the SwissProt E. coli database. Trypsin specificity with one missed cleavage was selected. S-Mercaptomethylcysteine and the N-terminal and lysine iTRAQ labels were selected as fixed modifications. Oxidized methionine and iTRAQ-labeled tyrosine were considered as variable modifications. The precursor tolerance and MS-MS fragment tolerances were set to ±0.7 and ±0.3 Da, respectively.
The peak areas at 114.1, 115.1, 116.1, and 117.1 m/z were corrected for overlapping isotope contributions from the iTRAQ tags according to the certificate of analysis provided by Applied Biosystems. Each data set was normalized to the relative ratio of parent ions from large ribosomal proteins, which was set to average 1:1 for each pairwise combination. Duplicate measurements (116, 117 for high-Mg2+, low-salt conditions; 114, 115 and 116, 117 for low-Mg2+, high-salt conditions) were averaged after normalization and the average signal for each parent ion used to determine subsequent ratios. Parent ions for which no measurement for one or more reporter group was obtained were removed from the analysis. Proteins identified in this study are listed in Tables S1 and S2 in the supplemental material.
RESULTS
CgtAE association with ribosomes.
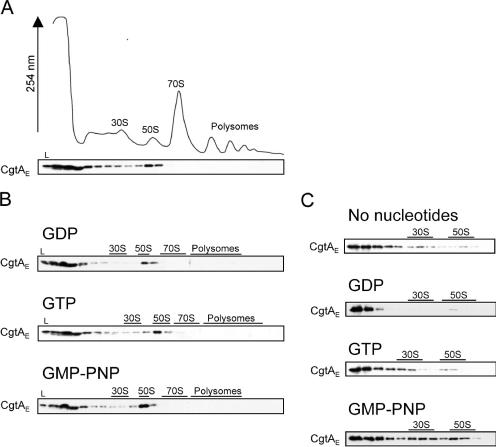
Both His-tagged and endogenous CgtAE proteins associate primarily with the free 50S ribosomal subunit (80). It has been demonstrated, however, that the ribosome associations of a number of bacterial GTPases are affected by their guanine nucleotide occupancy status. For example, the association between the B. subtilis Obg protein and ribosomes can be stabilized by GTP, as judged by velocity centrifugation analysis (82). The addition of exogenous nucleotides can also change the association patterns of several small ribosome-associated GTPases, such as RsgA (YjeQ) and Era (13, 32, 63). CgtAE binds to both GTP and GDP with moderate affinities (80). To determine the effect of guanine nucleotide occupancy on CgtAE-ribosome interactions, we examined the localization of CgtAE in the absence or presence of guanine nucleotides by sucrose density gradient ultracentrifugation of MG1655 cell extracts followed by Western blot analysis with an anti-CgtAE antibody. Consistent with previous observations, in the absence of exogenous nucleotide, a peak of CgtAE was observed cosedimenting with the free 50S subunits, but no CgtAE was associated with the 70S monosomes or the translating polysomes (Fig. 1A). This pattern suggests that CgtAE dissociates from the 50S particle before the 50S is assembled into the functional 70S, consistent with a role for CgtAE as a ribosome assembly factor. Interestingly, in this strain, the majority of CgtAE was found at the top of the gradient, indicating that most CgtAE is not bound to ribosomes in the absence of nucleotide. This suggests either that the in vivo ribosome binding sites for CgtAE were saturated under these experimental conditions or that CgtAE dissociated from the 50S particle during the ultracentrifugation step, consistent with the presence of CgtAE throughout many of the early fractions (Fig. 1A).
FIG. 1.
CgtAE association with ribosomes. Cell lysates from MG1655 (A and B) or the mixture of purified CgtAE and 70S ribosomes (C) in the presence or absence of 2 mM GDP, GTP, or GMP-PNP were sedimented through 7 to 47% sucrose (10 mM MgCl2, 100 mM NH4Cl) (A and B) or 10 to 30% sucrose (1 mM MgCl2, 100 mM NH4Cl) (C) gradients. The resulting UV profile (absorbance at 254 nm) from panel A is shown. The positions of the 30S and 50S subunits, the 70S monosome, and the polysomes are indicated. Fractions were TCA precipitated, and equal volumes of protein from each fraction were analyzed by SDS-PAGE and immunoblotting with an anti-CgtAE antibody. L, 1/100 of the total sample loaded onto the gradient.
To address the form of CgtAE preferentially bound to ribosomes, excess GDP, GTP, or GMP-PNP (a nonhydrolyzable GTP analog) was added to the cell lysates. The addition of these nucleotides did not, however, change the overall polysome profiles (data not shown) or the ribosome association of CgtAE (Fig. 1B), findings similar to those observed for C. crescentus (42). One explanation for the unchanged ribosome association in the presence of excess nucleotides is inhibition of free nucleotide exchange by CgtAE in the whole-cell lysates. To test this possibility, we combined purified CgtAE protein and 70S ribosomes and subjected them to sucrose density centrifugation under low-Mg2+ conditions, in which 70S monosomes dissociate into 30S and 50S particles. In contrast to the observation with whole-cell lysates, there was a dramatic change in the association pattern of CgtAE in the presence of different nucleotides when purified and dissociated 70S particles were used (Fig. 1C). In the absence of nucleotides, a small amount of CgtAE associated with both the 30S and 50S subunits, while the majority of protein was at the top of the gradient. A similar profile was observed with excess GTP. In contrast, the addition of excess GDP resulted in the majority of CgtAE being found at the top of the gradient. Addition of excess GMP-PNP, however, resulted in a robust association with both the 30S and the 50S subunits. These data show that although CgtAE is normally a 50S-associated protein (Fig. 1A), purified CgtAE is capable of binding to both stripped 30S and 50S subunits (Fig. 1C), perhaps via an unmasked binding site. Importantly, these data also demonstrate that the GTP-bound form of CgtAE has a higher affinity for ribosomes than does the GDP-bound form. These data are also consistent with a recent report that showed that purified CgtAE bound to both 16S and 23S rRNA (60). It should be noted that, in contrast to rRNA studies that showed that a multimer form of purified CgtAE binds to rRNA (60), we found that only the CgtAE monomer bound to these ribosomal particles; multimers were found at the top of the gradient (data not shown).
A cgtAE(G80E D85N) mutant is impaired in ribosome assembly.
To determine whether CgtAE is required for 50S ribosome biogenesis, we examined the polysome profiles for a cgtAE(G80E D85N) mutant. An analogous temperature-sensitive obg allele was initially isolated in B. subtilis (36). The two mutated residues are in the N-terminal glycine-rich domain of the protein and are thought to be important for the structural stability of this domain (6) but do not seem to affect the GTPase activity or, by inference, the guanine nucleotide binding of the protein (79). The corresponding mutant allele in E. coli was originally constructed on a plasmid under the control of the PBAD promoter to complement a chromosomal cgtAE deletion (35), but the protein levels were not titratable by arabinose (data not shown). In these studies, we complemented a chromosomal cgtAE deletion mutant with plasmids harboring either the wild-type cgtAE or the cgtAE(G80E D85N) allele under the control of the cgtAE promoter on a low-copy-number plasmid [referred to below as cgtAE+ and cgtAE(G80E D85N), respectively]. The steady-state level of CgtAE protein in cells expressing cgtAE(G80E D85N) was approximately 10-fold that seen in cgtAE+ cells as determined by immunoblotting with an anti-CgtAE antibody (Fig. 2C). Importantly, in addition to a temperature-sensitive lethality at 42°C, cgtAE(G80E D85N) displays a growth defect at the permissive temperature of 30°C (1.8-fold-increased doubling time compared to that of the wild type). In these studies, the phenotype of cgtAE(G80E D85N) was examined at the permissive temperature.
FIG. 2.
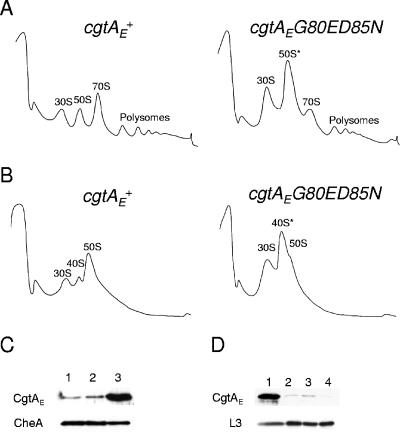
The polysome profile of a cgtAE(G80E D85N) mutant is aberrant. (A and B) Lysates from cgtAE+ (JM3903) and cgtAE(G80E D85N) (JM3907) cells were separated on 7 to 47% sucrose gradients under either high-Mg2+, low-salt conditions (10 mM MgCl2, 100 mM NH4Cl) (A) or low-Mg2+, high-salt conditions (1 mM MgCl2, 200 mM NH4Cl) (B). The positions of the ribosomal particles were detected by monitoring absorbance at 254 nm. (C) Protein extracts from wild-type (MG1655) (lane 1), cgtAE+ (JM3903) (lane 2), and cgtAE(G80E D85N) (JM3907) (lane 3) cells grown at 30°C were separated by SDS-PAGE and subjected to immunoblot analysis using antibodies against CgtAE or CheA (as a loading control). (D) Levels of CgtAE and L3 in 50S cgtAE+ (lane 1) and 50S* cgtAE(G80E D85N) (lane 2) cells from high-Mg2+, low-salt conditions and 50S cgtAE+ (lane 3) and 40S* cgtAE(G80E D85N) (lane 4) cells from low-Mg2+, high-salt conditions. Equivalent amounts of protein from relevant fractions, as determined by UV absorbance (A280), were separated by SDS-PAGE, and proteins were detected by immunoblotting using anti-CgtAE or anti-L3 antibodies.
We first examined the polysome profiles under standard high-Mg2+, low-salt (associating) conditions (10 mM MgCl2 and 100 mM NH4Cl in the gradients) that stabilize both loosely and tightly associated 70S ribosomes (Fig. 2A). Compared to levels with cgtAE+, there is a dramatic reduction in the levels of 70S subunits and polysomes, and a concomitant increase in the levels of 30S and 50S subunits, in cgtAE(G80E D85N). This phenotype is very similar to that seen for a mutant with a mutation of rrmJ, an rRNA methyltransferase modifying U2552 in the A-loop of 23S rRNA (5, 9), and agrees well with the hypothesis that CgtAE functions at a late stage in 50S maturation. We named the 50S particle from the cgtAE(G80E D85N) mutant 50S* to distinguish it from wild-type 50S.
To characterize the accumulated 50S subunits in the cgtAE(G80E D85N) mutant, we also analyzed polysome profiles obtained under low-Mg2+, high-salt (dissociating) conditions (1 mM MgCl2 and 200 mM NH4Cl in the gradients), which separate translating ribosomes into 30S and 50S subunits (Fig. 2B). Under these conditions, mature ribosomal subunits migrate at the 50S position. Precursor 50S subunits, however, can migrate differently, either due to the loss of loosely bound r-proteins and ribosome-associated proteins or due to a change in their conformation (5, 27, 57). As expected, the polysome profiles from cgtAE+ cell extracts showed predominantly 30S and 50S subunits. A small amount of a particle migrating at approximately the 40S position was also observed, perhaps derived from a small amount of pre-50S particles in this complemented strain. In the cgtAE(G80E D85N) mutant extracts, in addition to the 30S peak, there was a large peak, which we labeled 40S*, that almost masked the smaller 50S peak. These data suggest that the 50S* particles that accumulate in the cgtAE(G80E D85N) mutant under high-Mg2+, low-salt conditions are assembly intermediates and thus are subject to alteration under more stringent salt conditions. Furthermore, the association of CgtAE differed in these studies, as determined by immunoblot analysis of equal amounts of protein collected from wild-type (50S) and cgtAE(G80E D85N) (50S* and 40S*) particles (Fig. 2D). In cgtAE+ extracts, CgtAE associated with the 50S particles under high-Mg2+, low-salt but not low-Mg2+, high-salt conditions, indicating a loss of CgtAE from the particles under these conditions and confirming that CgtAE is a ribosome-associated factor and not a core r-protein. Interestingly, the mutant CgtAE(G80E D85N) protein did not bind well to either the 50S* (high-Mg2+, low-salt conditions) or the 40S* (low-Mg2+, high-salt conditions) particle.
Accumulation of rRNA precursors in the cgtAE(G80E D85N) mutant.
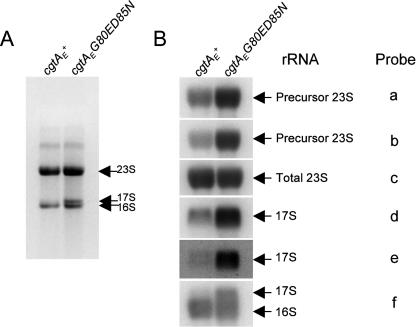
Previously, it was shown that a cgtAE(Ts) mutant is defective in 16S rRNA processing (60). The processing of the 23S rRNA, however, was not examined. Here we report that both 16S rRNA processing and 23S rRNA processing are defective in the cgtAE(G80E D85N) mutant strains. As expected, total cellular RNA separated on TBE-agarose gels revealed an accumulation of 17S rRNA (the precursor form of the 16S rRNA) in the cgtAE(G80E D85N) but not the cgtAE+ strain (Fig. 3). It is not possible, however, to resolve precursor 23S from mature 23S rRNA on these gels, since the pre-23S species generated by RNase III cleavage has stretches of only 3 to 7 and 7 to 9 nucleotides at its 5′ and 3′ ends, respectively (69). Therefore, to determine whether there was a defect in 23S rRNA processing, Northern blot analysis using either probes recognizing both mature and precursor species or probes specific for precursor sequences was performed (Fig. 3) (11). Consistent with the TBE-agarose gels, there was a significant increase in the level of 17S precursor in cgtAE(G80E D85N) compared to cgtAE+ extracts (Fig. 3). Significantly, the processing of the 23S rRNA was also impaired. We observed a three- to fourfold increase (varying by probe) in the level of precursor 23S in the cgtAE(G80E D85N) relative to the cgtAE+ extracts after normalization of loading according to probe c (Fig. 3). Thus, defects in the processing of both the small-subunit and large-subunit rRNAs were observed in cgtAE(G80E D85N), strongly implying that CgtAE is involved in ribosome assembly. Completion of 17S rRNA processing likely occurs on the translating ribosome and may be coregulated with protein synthesis (70, 71). Therefore, mutations in 50S subunit assembly proteins, such as SrmB and RluD, can result in processing defects in both small- and large-subunit rRNAs (11, 26). We predict that CgtAE is a bona fide 50S assembly factor and that the observed increase in 17S rRNA levels is a secondary consequence of the deficiency in generating 70S particles.
FIG. 3.
The cgtAE(G80E D85N) mutant accumulates rRNA precursors. (A) Total RNA (5 μg) isolated from cgtAE+ (JM3903) and cgtAE(G80E D85N) (JM3907) cells was separated on a 1.2% agarose-TBE gel and stained with ethidium bromide. The positions of the 23S, 16S, and 17S precursors are indicated. (B) Total-RNA samples were subjected to a denaturing 1.2% agarose-MOPS gel and probed with oligonucleotide probes against total 23S or total 16S and 17S rRNA (probes c and f, respectively), 5′ precursor sequences of 23S or 17S rRNA (probes a and d, respectively), or 3′ precursor sequences of 23S or 17S rRNA (probes b and c, respectively) (11). The radioactive signals were obtained by autoradiography and quantified using a PhosphorImager (Bio-Rad). The specific rRNA types are labeled.
Semiquantitative proteomic comparison of cgtAE+ and cgtAE(G80E D85N) mutant large ribosomal particles.
We predict that the 50S* subunit that accumulates in the cgtAE(G80E D85N) mutant is a pre-50S particle. To examine the differences in proteomic makeup between the 50S and 50S* particles found in cgtAE+ and cgtAE(G80E D85N) extracts, we turned to a new multiplex protein semiquantitative method, iTRAQ (59). In this method, isobaric tags modify the N terminus and lysyl side chains of every peptide in a complex mixture, allowing for relative quantitation of proteins. The power of this approach is that it allows for identification of a large number of proteins and the simultaneous comparison of protein levels in different protein samples.
Initially, we compared 50S subunits from cgtAE+ extracts to 50S* subunits isolated from cgtAE(G80E D85N) extracts, both obtained under high-Mg2+, low-salt conditions. Independently isolated duplicate mutant 50S* and cgtAE+ 50S samples were digested with trypsin and individually labeled with specific iTRAQ reagents. Samples were mixed and subjected to 2DLC-MS-MS. We assigned protein identifications to 4,166 peptides (≥95% confidence) and found 91 proteins with 2 or more identifying peptides (see Table S1 in the supplemental material). As expected, many of the proteins identified were ribosomal proteins (41) and known ribosome-associated proteins, such as CgtAE, CsdA, SrmB, RrmJ, RluB, RluC, and trigger factor. Approximately 25% of the peptides (a total of 983), however, were derived from AceE, AceF, and LpdA, the three proteins of the large pyruvate dehydrogenase complex. This oligomeric complex is approximately 4 MDa (2) and migrates on sucrose gradients with the 50S ribosomal particle. Thus, identification of these proteins was also expected.
The use of isobaric tags allows for the comparison of the relative levels of any identified protein, expressed as a ratio, in different samples. For example, we identified CgtAE protein in this study, and both immunoblotting and iTRAQ showed a reduction in the level of CgtAE in the cgtAE(G80E D85N) 50S* particle compared to the cgtAE+ 50S particle (compare Fig. 2D with Fig. 4A). It is worth noting that the difference in CgtAE levels observed by iTRAQ analysis (Fig. 4A) was not as dramatic as that seen by immunoblotting (Fig. 2D). One of the peculiarities of the iTRAQ methodology is that the dynamic range of the relative ratios is somewhat dampened due to the nature of isotope enrichment and the broad ion selector window of the model 4700 MALDI-TOF/TOF mass spectrometer. Thus, although iTRAQ is a robust method for identifying trends in relative protein levels with low variance, the magnitudes of differences observed by iTRAQ are somewhat less than the levels quantified by immunoblotting. Regardless, to date we have found that the ratios observed and their relative magnitudes as indicated by iTRAQ are supported by more conventional methods such as immunoblotting (Fig. 2D and 4A and B) (data not shown). Finally, with iTRAQ we have used a threshold of 25% deviation from a 1:1 ratio to distinguish significant changes, as reported previously (59).
As expected, the majority of large r-proteins were found at equivalent levels in the 50S* and 50S particles isolated from cgtAE(G80E D85N) and cgtAE+ strains, respectively, and therefore showed a 1:1 ratio (Fig. 4A). The large r-proteins L1 to L4, L7/L12, L22, L35, and L36 were not detected, either due to their small sizes (L35 and L36) or for other, unknown reasons. Significantly, levels of two r-proteins, L33 and L34, are moderately reduced and that of one r-protein, L16, is slightly reduced on the 50S* particle. The level of L23 is perhaps slightly reduced as well, although the degree of reduction is very close to the error threshold (25%) of the iTRAQ method. We predict that the efficient assembly of these r-proteins onto the maturing ribosome requires CgtAE function. Supporting this hypothesis, L16, L33, and L34 are assembled relatively late onto the maturing 50S particle, and none are required for the association of additional r-proteins in vitro (Fig. 4C) (31). Consistent with their association on a late 50S particle, these three r-proteins are absent or reduced in the pre-50S particles that accumulate in the srmB (11), csdA (10), and rrmJ (27) mutants. Thus, the reduction in the levels of these specific r-proteins provides strong evidence that the 50S* particle is an immature ribosomal subunit or represents a combination of mature 50S and immature pre-50S particles.
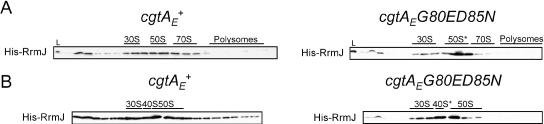
The accumulation of an intermediate particle in the cgtAE(G80E D85N) 50S* is further supported by the modest accumulation of two specific ribosome assembly factors, RrmJ and RluC. rrmJ mutants accumulate 50S particles that convert to ∼40S particles under low-Mg2+, high-salt conditions (5), strongly suggesting a role in 50S assembly. Overexpression of CgtAE suppresses both the growth and polysome defects of an rrmJ mutant, suggesting that CgtAE functions at the same time as or after RrmJ (72). To address this hypothesis, we examined by immunoblotting the migration of His-tagged RrmJ on sucrose gradients. In cgtAE+ extracts, His-RrmJ showed a broad distribution over several fractions, perhaps due to overexpression of this plasmid-borne gene. Regardless, the ribosome-bound RrmJ peaks with the 50S (Fig. 5A). In cgtAE(G80E D85N) extracts, however, His-RrmJ was predominantly associated with the 50S* peak (Fig. 5A), which could result either from an increase in binding sites for RrmJ due to the accumulation of 50S* or from an increased affinity of RrmJ for the 50S* particle.
FIG. 5.
His-RrmJ accumulates on the cgtAE(G80E D85N) 50S* particle. cgtAE+ and cgtAE(G80E D85N) cells expressing His-RrmJ (JM4711 and JM4714, respectively) were grown at 30°C, and the expression of His-RrmJ was induced by 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h. The cell lysates were separated on 7 to 47% sucrose gradients under either high-Mg2+, low-salt conditions (10 mM MgCl2, 100 mM NH4Cl) (A) or low-Mg2+, high-salt conditions (1 mM MgCl2, 200 mM NH4Cl) (B). The resulting fractions were subjected to immunoblot analysis with an anti-His antibody. The relative positions of the 30S, 50S, and 70S ribosomes and polysomes for each gradient are indicated.
In addition to identifying other known ribosome-associated proteins, such as the helicases CsdA and SrmB, the pseudouridine synthase RluB, and the chaperone Hsp15, we identified several uncharacterized proteins that comigrated with the 50S, including YhbY, YbeB, YibL, and YjgA. Interestingly, none of these proteins accumulated on the 50S* particle. We suggest that these proteins either are novel ribosome-associated proteins or associate with a different oligomeric complex that comigrates with ribosomal particles. For ribosome-associated proteins, we propose three possible reasons for a reduction on the 50S* particle. First, the protein may associate with a pre-50S particle early in the assembly pathway, dissociating before the formation of 50S*. Since there is a vast accumulation of 50S* particles in the cgtAE(G80E D85N) extracts, the contribution of pre-50S particles may be reduced in these extracts relative to that in cgtAE+ extracts, which have lower total levels of 50S particles. We envision that one or more of the bona fide early assembly proteins, such as the helicases SrmB and CsdA, and the pseudouridine synthase RluB, represent proteins that bind to and are released from a pre-50S particle prior to CgtAE function. This model is consistent with the association of these proteins with ∼40S particles and the observation that mutations in both srmB and csdA result in an accumulation of 40S particles under high-Mg2+, low-salt conditions (10, 11) (data not shown). Second, the proteins may bind only to 50S particles that are generated after the CgtAE maturation step. An example of the second category of proteins is Hsp15, which is virtually absent from the 50S* particle (32-fold reduction [Fig. 4A]). Hsp15 is a chaperone that is proposed to play a role in repairing and recycling erroneously dissociated ribosomes and thus binds only to the subset of mature 50S particles that have a nascent RNA chain bound to them (37). The absence of Hsp15 in the 50S* particle agrees well with a low level of mature 50S particles in that fraction. Finally, the proteins may simply have a reduced affinity for the 50S* particle.
We next compared 40S* particles from the cgtAE(G80E D85N) mutant with 50S particles from the cgtAE+ strain obtained under low-Mg2+, high-salt conditions using iTRAQ. In this study, duplicate samples from each strain were labeled with different isobaric tags, and the four samples were mixed and analyzed (a double duplex iTRAQ analysis). We identified 1,368 peptides (≥95% confidence) corresponding to 76 proteins (2 or more peptides) (see Table S1 in the supplemental material). The total number of peptides identified in this study was threefold lower than that in the study of particles obtained under high-Mg2+, low-salt conditions (see above), and the majority of these proteins were r-proteins (43) and proteins identified in the first study. Of note is the absence or severe reduction in the number of peptides for several proteins, likely due to the dissociation of these proteins from the ribosomes under more stringent salt concentrations. These include SrmB (25 peptides under high-Mg2+, low-salt [associating] conditions and none under low-Mg2+, high-salt [dissociating] conditions), RluC (31 and none, respectively), YhbY (42 and 3, respectively), YjgA (42 and none, respectively), and L31 (195 and none, respectively) (see Table S1 in the supplemental material). L31, in particular, is known to be loosely associated with the ribosome and dissociates from ribosomes under the conditions of most ribosome purification procedures (17). Other proteins, such as YbeB, YibL, and RrmJ, were also absent from the second iTRAQ study and may represent additional proteins that dissociate from ribosomes under these conditions. Since these proteins were identified with few (2 to 15) parent ions under high-Mg2+, low-salt conditions, it is also possible that their absence is due to the lower sample coverage (one-third fewer total parent ions identified in the low-Mg2+, high-salt study). This is certainly the case for RrmJ, since immunoblot studies revealed that His-RrmJ was strongly associated with the 40S* particle (Fig. 5B).
Consistent with what was observed under the high-Mg2+, low-salt conditions, the majority of the r-proteins were found at similar levels in the 40S* and 50S particles. Levels of both L17 and L7/12 showed modest increases in the 40S* particles. We do not currently have an explanation for this apparent enrichment of these two r-proteins. Importantly, levels of both L33 and L16 were modestly reduced in the 40S* particles. Curiously, L34 was found at similar levels in both particles (Fig. 4B). To verify the levels of L16, L33, and L34 on these particles, we examined their levels by immunoblotting. After normalizing the loading (according to L3 levels), we found that levels of L16, L33, and L34 were all reduced in the 40S* particle (Fig. 4D). The discrepancy for L34 is a mystery, although we note that in this study only four parent ions, each derived from the same peptide, were obtained for L34. One possibility, therefore, is that this particular peptide was differentially labeled by the iTRAQ reagent relative to other L34 peptides.
CsdA and RluB and, to a lesser degree, YhbY were enriched in the 40S* fractions. Since none of these proteins were enriched in the 50S* fractions, we conclude that they are not associated with the cgtAE(G80E D85N) intermediate. One possibility is that their enrichment here may be a consequence of a total enrichment of 40S particles in this study. In the first study (under high-Mg2+, low-salt conditions), similar fractions (50S and 50S*) were compared. Here (under low-Mg2+, high-salt conditions), earlier fractions representing the 40S* region were compared to 50S fractions. Therefore, any protein that is normally enriched in the 40S fractions, independently of CgtAE function, will be enriched in the 40S* particle. For CsdA and RluB this is certainly the case; these proteins fractionate with 40S particles in wild-type extracts (10) (data not shown).
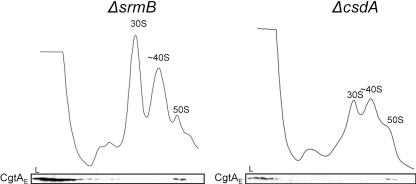
CgtAE does not associate with ∼40S precursors that accumulate in ΔsrmB and ΔcsdA mutants.
CgtAE binds to 50S particles and functions as a late 50S assembly factor. Unknown, however, is whether the CgtAE protein binds to an early pre-50S particle. It has been shown previously that mutations in either of the two DEAD-box RNA helicases, SrmB and CsdA, lead to the accumulation of distinct 50S intermediates, both of which migrate at ∼40S positions (10, 11). Each of these ∼40S particles is also distinct from the 40S* seen in the cgtAE(G80E D85N) mutant, as they accumulate under high-Mg2+, low-salt conditions and have different r-protein compositions from the 40S* particle (10, 11). We asked whether CgtAE could bind to these ∼40S particles. Whole-cell lysates from ΔsrmB or ΔcsdA mutants were fractionated over sucrose gradients designed to separate the 40S from the 50S particles. As expected, both ΔsrmB and ΔcsdA extracts showed the accumulation of a ∼40S precursor peak and a significant reduction in 50S particles (Fig. 6).In both mutants, the vast majority of CgtAE, as detected by immunoblotting, was found at the top of the gradient and was not bound to ribosomes (Fig. 6). Interestingly, the CgtAE protein that was bound to ribosomes was found exclusively associated with the 50S particles. The 40S particles that accumulate in the ΔsrmB and ΔcsdA mutants (10, 11) may represent pre-50S particles much earlier in the assembly pathway than those that accumulate in the cgtAE(G80E D85N) mutant. The finding that CgtAE did not associate with these early particles is consistent with a CgtAE association with the maturing ribosomal particle after SrmB and CsdA function.
FIG. 6.
CgtAE associates with the 50S but not the ∼40S particle from ΔsrmB or ΔcsdA mutant extracts. ΔsrmB or ΔcsdA cell extracts were sedimented through 7 to 47% 35-ml sucrose gradients, and the samples were monitored by UV absorbance at 254 nm. The subsequent fractions were analyzed by immunoblotting using an anti-CgtAE antibody. The relative positions of the 30S, ∼40S, and 50S peaks are indicated.
DISCUSSION
Ribosome biogenesis is a remarkably coordinated and hierarchical process. In eukaryotes, more than 170 nonribosomal accessory proteins are involved. In prokaryotes, in contrast, only a few accessory proteins have been identified. These include RNA-modifying enzymes (5, 26), RNA helicases (10, 11), chaperones (1, 18, 48, 49), and GTPases (32, 33, 50, 60, 72, 76). In this report, we further characterize the role of the late 50S ribosome assembly factor, CgtAE, and its temporal relationship with other known 50S assembly factors.
Several lines of evidence support a role for the bacterial Obg/CgtA proteins in late 50S biogenesis. First, in cell extracts, CgtA proteins are predominantly associated with the 50S particle and not significantly with 30S subunits, the mature 70S, or translating polysomes (Fig. 1A) (42, 60, 68, 80). The association of the C. crescentus and E. coli CgtA proteins with the 50S is sensitive to salt concentration (Fig. 2D) (42), consistent with a role in the ribosome assembly process but not as core r-proteins. Second, cgtAE mutants display a dramatically altered ribosomal profile, consisting of an accumulation of free 30S and 50S and a corresponding decrease in the 70S and polysomes (Fig. 2A) (60), in agreement with a role in late 50S assembly or subunit coupling. This profile is distinctly different from that seen in early 50S assembly mutants, such as ΔsrmB or ΔcsdA mutants, that accumulate ∼40S particles deficient in a number of r-proteins (10, 11) (Fig. 6). The free 50S particles accumulated in the cgtAE mutants are presumably immature ribosomal precursors, as shown by their conversion to ∼40S particles under more stringent salt conditions (Fig. 2B), an alteration also seen with the 50S particles that accumulate in the rrmJ mutant. Third, overexpression of CgtAE suppresses the slow growth and ribosome defects of the rrmJ mutant (72). Although RrmJ functions late in the assembly process at a point at which U2552 of the 23S rRNA is methylated (8), the suppression by CgtAE does not restore the missing modification (72), indicating that CgtAE does not simply replace the RrmJ methyltransferase function. It is likely, therefore, that CgtAE suppresses the assembly defect in the rrmJ mutant. One possibility is that a higher concentrations of CgtAE may overcome a weaker affinity of the protein for the unmethylated 50S particles.
Fourth, the 50S* particle that accumulates in the cgtAE(G80E D85N) mutant is not a wild-type 50S. We demonstrate here that levels of three r-proteins that assemble late onto the maturing 50S particle (31)—L33, L34, and L16—are reduced in the 50S* particle (Fig. 4), indicating that the accumulated 50S* particle may represent an immature 50S particle or a combination of mature 50S and immature pre-50S particles. Curiously, we did not observe a reduction in the level of L10, as has been reported with a different cgtAE mutant strain (60). Whereas L33 does not seem to play a major role in ribosome assembly (45-47) and not much is known about L34 function, L16 has been shown to accelerate the late steps of in vitro assembly (21) and to induce a conformational change in the 50S which may, in turn, affect the peptidyltransferase activity and subunit association of the ribosome (73). Moreover, L16 is also missing in the 50S intermediate particles accumulated in RbgA/YlqF-depleted cells in B. subtilis (50, 76). The reduction in protein levels of L33, L34, and L16 in the cgtAE(G80E D85N) mutant suggests that CgtAE either plays a direct role in the assembly pathway or is required for the efficient recruitment of these r-proteins. On the ribosome, L16 is found close to the peptidyltransferase center and may make contacts with aminoacylated tRNA in the A site (3, 53). L33 is also located close to this region and has previously been shown to cross-link with E site tRNA (64, 81). L34 as well as L23 (the levels of which are also potentially reduced in the cgtAE mutant) are found near the peptide exit channel and not in close proximity to either L16 or L33. Therefore, it is not likely that CgtAE directly recruits each of these proteins; rather, it may stabilize a 50S intermediate that facilitates late r-protein binding. It is also possible that the reduction of these r-proteins resulted from dissociation of the proteins from the ribosomes during the centrifugation steps due to decreased affinity for the 50S*.
Fifth, two ribosome assembly proteins, RrmJ and RluC, are enriched on the particle that accumulates in the cgtAE(G80E D85N) mutant, consistent with the accumulation of a pre-50S particle and not a mature 50S. Sixth, both 23S rRNA processing and 16S rRNA processing were impaired in the cgtAE(G80E D85N) mutant (Fig. 3) (60). In E. coli, rRNA processing and ribosome maturation are tightly coupled (71). In several cases, it has been demonstrated that a defect in 50S assembly also results in a defect in 16S processing, presumably due to a deficiency in mature 70S and polysomes (11, 26). Thus, the accumulation of 17S rRNA and the small-subunit assembly defect previously reported (60) (Fig. 3) may be the indirect consequences of an accumulation of uncoupled, immature 30S subunits. Finally, although CgtAE binds to a 50S particle, it does not bind to the early ∼40S intermediates that accumulate in the ΔsrmB and ΔcsdA mutants (Fig. 6). These data suggest that CgtAE binds to a late 50S assembly intermediate.
As in eukaryotes, the assembly of prokaryotic ribosomes is likely to be highly ordered and dynamic, with a series of intermediates accompanied by the association and dissociation of different assembly factors at various stages. Since only three major intermediates (sedimenting at 34S, 43S, and near 50S, respectively) of the 50S have been identified by the conventional sucrose gradients (30, 43), it is reasonable to suppose that most intermediates in 50S biogenesis are transient and present at low concentrations. Accumulating genetic evidence, however, has allowed for the identification and description of the relative temporal relationships of some of these assembly factors. For example, in 30S assembly, overexpression of RbfA suppresses the defects of a ΔrimM mutant (7), and overexpression of Era, in turn, suppresses the ribosome assembly defects in an rbfA deletion mutant (33). Thus, on the assembling 30S subunit, RimM is likely to function first, followed by RbfA and then Era. Similar functional relationships have been described for large-ribosome assembly. For example, overexpression of both CgtAE and EngA suppresses a methyltransferase rrmJ mutant (72), placing these two GTPases after RrmJ in the assembly pathway. In agreement with this, RrmJ was found to accumulate in the unassociated 50S* in the cgtAE(G80E D85N) mutant (Fig. 4A and Fig. 5A). Furthermore, CgtAE copurifies with CsdA, an ATP-dependent RNA helicase (60, 80) the deletion of which causes an accumulation of an intermediate ∼40S particle at low temperatures (10). Consistently, we found that CsdA was not enriched on the cgtAE(G80E D85N) mutant 50S* particle (Fig. 4A), although it was enriched in the mutant 40S* fractions (Fig. 4B), perhaps due to an enrichment of bona fide 40S assembly intermediates in these fractions. The ΔcsdA mutant accumulates a ∼40S particle which is deficient in several r-proteins, such as L6 and L25, that were present in the cgtAE(G80E D85N) mutant 50S* particle, placing the timing of ribosome assembly for these r-proteins between the CsdA and CgtAE assembly steps (10). Finally overexpression of CsdA suppresses a ΔsrmB mutant, consistent with CsdA functioning after SrmB (10). Clearly, additional studies to define and clarify the interrelationships among the ribosome assembly factors are warranted.
In cell extracts, the association of CgtAE with ribosomes was not affected by the guanine nucleotide concentration (Fig. 1B). This is also true for the C. crescentus CgtAC protein (42). In contrast, the guanine nucleotide occupancy of several other prokaryotic GTPases is critical for their association (13, 32, 50, 63, 82). Because the Obg/CgtA proteins exchange guanine nucleotides rapidly in vitro (40, 67, 79, 80), we predicted previously that the ribosome (or ribosome-associated proteins) might inhibit nucleotide exchange by CgtA in vivo (41). Here we showed that enhanced association of purified CgtAE with dissociated ribosomal subunits was observed in the presence of a nonhydrolyzable GTP analog (Fig. 1C). These data are consistent with a recent study showing that CgtAE binds to rRNA in a GTP-dependent manner (60). Therefore, it is likely that CgtAE bound to the ribosome, in contrast to purified CgtAE, does not freely exchange guanine nucleotides. Moreover, a ribosome-dependent inhibition of guanine nucleotide exchange would allow for GTP hydrolysis to play a critical role in CgtAE function. Such a role is consistent with the phenotypes of C. crescentus cgtAC mutants, which showed that GTP hydrolysis was critical for CgtAC function (41). This model, however, may not apply to the B. subtilis Obg protein, since it has been demonstrated that only a small amount of 5-guanylyl-imidodiphosphate (GIDP, a nonhydrolyzable GTP analog) can preserve the Obg-ribosome association in the crude cell lysates (82).
It is curious that, although Obg/CgtA proteins bind predominantly to the 50S subunits, as assayed by immunoblotting of cell extracts separated on sucrose gradients (42, 68, 80), purified CgtAE is capable of binding to both dissociated 30S and 50S subunits (Fig. 1C), and to both 16S and 23S rRNAs (60). CgtAE has also been shown to bind to DNA (35), and therefore, the in vitro binding to 16S rRNA and 30S particles may also be nonspecific binding. Since all CgtA proteins examined thus far associate predominantly with a 50S particle in vivo (Fig. 1A) (42, 60, 68, 80), we suggest that the 50S particle is the normal CgtAE substrate. It should be noted that an in vivo association with the 30S particle has also been reported (60). The CgtAE in these gradients, however, peaks in fractions earlier than those of the 30S subunit. Depending on the strains examined and the sucrose density centrifugation parameters used, we also occasionally observe some CgtAE migrating in fractions somewhat smaller than the 30S particle (data not shown). The nature of this particle is unknown. Alternatively, CgtAE may have an intermediate off-rate from the 50S, thereby distributing across the 30S peak merely by coincidence.
In addition to their ribosomal function, Obg/CgtA proteins have also been implicated in a variety of cellular processes including DNA replication, sporulation, morphological differentiation, and stress response (6, 20, 36, 55, 65, 77, 80). The relationship between the role of Obg/CgtA proteins in ribosome assembly and these other functions is unknown. In contrast to most ribosome assembly factors, however, all the bacterial Obg/CgtA proteins studied thus far are essential for cell growth (35, 44, 55, 68, 74). In C. crescentus, a temperature-sensitive allele of cgtAC causes a polysome defect at the permissive temperature, but this defect is not further perturbed at the nonpermissive temperature (15), implying that the essential function is independent of the ribosome assembly function. CgtAE interacts with SpoT, a ppGpp synthetase/hydrolyase critical for the stringent response in E. coli (80), and the B. subtilis Obg cocrystalized with ppGpp (6), the substrate/product of SpoT/RelA proteins. Furthermore, Obg interacts with several regulators (RsbT, RsbW, and RsbX) necessary for the stress activation of σB, the global controller of the stress regulon in B. subtilis (65). It is possible, therefore, that the involvement in the stress response is responsible for the essential nature of the Obg/CgtA proteins. Interestingly, both SpoT in E. coli (M. Jiang and J. R. Maddock, unpublished data) and the stress regulators in B. subtilis (66) are ribosome associated, raising the possibility that the role of Obg/CgtA proteins in the stress response is coupled to their ribosome association.
In this study we also demonstrated the utility of a new semiquantitative proteomic technique, iTRAQ, in identifying and quantifying the protein compositions of ribosomal particles. iTRAQ is an isobaric tagging system that allows quantitative analysis of as many as four different protein samples in a single experiment (59). This new approach has allowed for the simultaneous identification of many known and heretofore unknown ribosome-associated proteins. Among the unknown ribosome-associated proteins we have identified by iTRAQ, YbeB is a highly conserved 69-amino-acid protein, listed as one of the most common unknown proteins (23); it is similar to a plant protein, Iojap, that is involved in the stability of chloroplast ribosomes (78), and therefore, its association with the 50S subunit is not unexpected. YhbY is a 97-amino-acid protein with a crystal structure that reveals an RNA binding fold similar to that of the 30S binding protein IF-3 (56). Interestingly, yhbY is transcribed divergently from rrmJ. YibL and YjgA are 120- and 183-amino-acid proteins, respectively, whose association with the ribosome provides a first hint at their previously unknown functions.
Perhaps more importantly, iTRAQ analysis results in determination of the relative levels of identified proteins, as verified by immunoblotting. As with any high-throughput technology, however, iTRAQ is biased toward the detection and comparison of abundant proteins, and therefore, many interesting low-abundance proteins are not detected in the analysis. For example, by iTRAQ, we detected RrmJ on 50S particles under high-Mg2+, low-salt but not low-Mg2+, high-salt conditions (Fig. 4). By immunoblotting, however, His-RrmJ clearly associates with 50S particles under both conditions (Fig. 5). The discrepancy is due to our inability to detect RrmJ in the latter proteomics study. It should be noted, too, that the dynamic range of the relative ratios observed with the iTRAQ methodology may result in the underestimation of the changes in protein levels between different protein samples. For example, the potential reduction of L23 we observed suggests that other r-proteins were also reduced but were missed in the iTRAQ analyses. Despite the detection limitations, however, the use of isobaric tags coupled with 2DLC-MS-MS is clearly a robust method for performing comparative analysis of complex mixtures, such as the bacterial ribosome.
Supplementary Material
Acknowledgments
We are extremely grateful to Isabelle Iost for antibodies against L16, L33, and L34 and for the ΔsrmB and ΔcsdA strains, to James Bardwell for antibodies against CgtAE, to Ursula Jakob for the ΔrrmJ and isogenic wild-type control strains, and to V. James Hernandez for antibodies against L3. We also thank Susan Sullivan for both technical assistance and critical reading of the manuscript.
The proteome analyses were performed in collaboration with the National Resource for Proteomics and Pathways, funded by NCRR (1P41 RR 018627, to P.C.A.). This work was funded, in part, by a National Science Foundation grant (MCB-0316357) to J.R.M.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alix, J. H., and M. F. Guerin. 1993. Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:9725-9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelides, K. J., S. K. Akiyama, and G. G. Hammes. 1979. Subunit stoichiometry and molecular weight of the pyruvate dehydrogenase multienzyme complex from Escherichia coli. Proc. Natl. Acad. Sci. USA 76:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashan, A., I. Agmon, R. Zarivach, F. Schluenzen, J. Harms, R. Berisio, H. Bartels, F. Franceschi, T. Auerbach, H. A. Hansen, E. Kossoy, M. Kessler, and A. Yonath. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11:91-102. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bugl, H., E. B. Fauman, B. L. Staker, F. Zheng, S. R. Kushner, M. A. Saper, J. C. Bardwell, and U. Jakob. 2000. RNA methylation under heat shock control. Mol. Cell 6:349-360. [DOI] [PubMed] [Google Scholar]
- 6.Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure (Cambridge) 10:1581-1592. [DOI] [PubMed] [Google Scholar]
- 7.Bylund, G. O., L. C. Wipemo, L. A. Lundberg, and P. M. Wikstrom. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 180:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 9.Caldas, T., E. Binet, P. Bouloc, and G. Richarme. 2000. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271:714-718. [DOI] [PubMed] [Google Scholar]
- 10.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32:2751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48:1253-1265. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X., A. K. Walker, J. R. Strahler, E. S. Simon, S. L. Tomanicek-Volk, B. B. Nelson, M. C. Hurley, S. A. Ernst, J. A. Williams, and P. C. Andrews. 2006. Organellar proteomics. Analysis of pancreatic xymogen granule membranes. Mol. Cell Proteomics 5:306-312. [DOI] [PubMed] [Google Scholar]
- 13.Daigle, D. M., and E. D. Brown. 2004. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J. Bacteriol. 186:1381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta, K., J. L. Fuentes, and J. R. Maddock. 2005. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell 16:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta, K., J. M. Skidmore, K. Pu, and J. R. Maddock. 2004. The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol. Microbiol. 54:1379-1392. [DOI] [PubMed] [Google Scholar]
- 16.Deich, R. A., B. J. Metcalf, C. W. Finn, J. E. Farley, and B. A. Green. 1988. Cloning of genes encoding a 15,000-dalton peptidoglycan-associated outer membrane lipoprotein and an antigenically related 15,000-dalton protein from Haemophilus influenzae. J. Bacteriol. 170:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eistetter, A. J., P. D. Butler, R. R. Traut, and T. G. Fanning. 1999. Characterization of Escherichia coli 50S ribosomal protein L31. FEMS Microbiol. Lett. 180:345-349. [DOI] [PubMed] [Google Scholar]
- 18.El Hage, A., M. Sbai, and J. H. Alix. 2001. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet. 264:796-808. [DOI] [PubMed] [Google Scholar]
- 19.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 20.Foti, J. J., J. Schienda, V. A. Sutera, Jr., and S. T. Lovett. 2005. A bacterial G protein-mediated response to replication arrest. Mol. Cell 17:549-560. [DOI] [PubMed] [Google Scholar]
- 21.Franceschi, F. J., and K. H. Nierhaus. 1990. Ribosomal proteins L15 and L16 are mere late assembly proteins of the large ribosomal subunit. Analysis of an Escherichia coli mutant lacking L15. J. Biol. Chem. 265:16676-16682. [PubMed] [Google Scholar]
- 22.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 23.Galperin, M. Y., and E. V. Koonin. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 25.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 26.Gutgsell, N. S., M. P. Deutscher, and J. Ofengand. 2005. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 11:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hager, J., B. L. Staker, H. Bugl, and U. Jakob. 2002. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 277:41978-41986. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 29.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 30.Hayes, F., and D. H. Hayes. 1971. Biosynthesis of ribosomes in E. coli. I. Properties of ribosomal precursor particles and their RNA components. Biochimie 53:369-382. [DOI] [PubMed] [Google Scholar]
- 31.Herold, M., and K. H. Nierhaus. 1987. Incorporation of six additional proteins to complete the assembly map of the 50S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262:8826-8833. [PubMed] [Google Scholar]
- 32.Himeno, H., K. Hanawa-Suetsugu, T. Kimura, K. Takagi, W. Sugiyama, S. Shirata, T. Mikami, F. Odagiri, Y. Osanai, D. Watanabe, S. Goto, L. Kalachnyuk, C. Ushida, and A. Muto. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 32:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue, K., J. Alsina, J. Chen, and M. Inouye. 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48:1005-1016. [DOI] [PubMed] [Google Scholar]
- 34.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi, G., S. Moriya, and C. Wada. 2001. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 41:1037-1051. [DOI] [PubMed] [Google Scholar]
- 36.Kok, J., K. A. Trach, and J. A. Hoch. 1994. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J. Bacteriol. 176:7155-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korber, P., J. M. Stahl, K. H. Nierhaus, and J. C. Bardwell. 2000. Hsp15: a ribosome-associated heat shock protein. EMBO J. 19:741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 40.Lin, B., K. L. Covalle, and J. R. Maddock. 1999. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J. Bacteriol. 181:5825-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, B., J. M. Skidmore, A. Bhatt, S. M. Pfeffer, L. Pawloski, and J. R. Maddock. 2001. Alanine scan mutagenesis of the switch I domain of the Caulobacter crescentus CgtA protein reveals critical amino acids required for in vivo function. Mol. Microbiol. 39:924-934. [DOI] [PubMed] [Google Scholar]
- 42.Lin, B., D. A. Thayer, and J. R. Maddock. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindahl, L. 1975. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 92:15-37. [DOI] [PubMed] [Google Scholar]
- 44.Maddock, J., A. Bhatt, M. Koch, and J. Skidmore. 1997. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J. Bacteriol. 179:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maguire, B. A., and D. G. Wild. 1997. The effects of mutations in the rpmB,G operon of Escherichia coli on ribosome assembly and ribosomal protein synthesis. Biochim. Biophys. Acta 1353:137-147. [DOI] [PubMed] [Google Scholar]
- 46.Maguire, B. A., and D. G. Wild. 1997. Mutations in the rpmBG operon of Escherichia coli that affect ribosome assembly. J. Bacteriol. 179:2486-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maguire, B. A., and D. G. Wild. 1997. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol. Microbiol. 23:237-245. [DOI] [PubMed] [Google Scholar]
- 48.Maki, J. A., D. J. Schnobrich, and G. M. Culver. 2002. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell 10:129-138. [DOI] [PubMed] [Google Scholar]
- 49.Maki, J. A., D. R. Southworth, and G. M. Culver. 2003. Demonstration of the role of the DnaK chaperone system in assembly of 30S ribosomal subunits using a purified in vitro system. RNA 9:1418-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuo, Y., T. Morimoto, M. Kuwano, P. C. Loh, T. Oshima, and N. Ogasawara. 2006. The GTP-binding protein YlqF participates in the late step of 50S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 281:8110-8117. [DOI] [PubMed] [Google Scholar]
- 51.Nierhaus, K. H. 1991. The assembly of prokaryotic ribosomes. Biochimie 73:739-755. [DOI] [PubMed] [Google Scholar]
- 52.Nierhaus, K. H., and F. Dohme. 1974. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. USA 71:4713-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura, M., T. Yoshida, M. Shirouzu, T. Terada, S. Kuramitsu, S. Yokoyama, T. Ohkubo, and Y. Kobayashi. 2004. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J. Mol. Biol. 344:1369-1383. [DOI] [PubMed] [Google Scholar]
- 54.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okamoto, S., and K. Ochi. 1998. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol. Microbiol. 30:107-119. [DOI] [PubMed] [Google Scholar]
- 56.Ostheimer, G. J., A. Barkan, and B. W. Matthews. 2002. Crystal structure of E. coli YhbY: a representative of a novel class of RNA binding proteins. Structure 10:1593-1601. [DOI] [PubMed] [Google Scholar]
- 57.Pardo, D., C. Vola, and R. Rosset. 1979. Assembly of ribosomal subunits affected in a ribosomal mutant of E. coli having an altered L22 protein. Mol. Gen. Genet. 174:53-58. [DOI] [PubMed] [Google Scholar]
- 58.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 59.Ross, P. L., Y. N. Huang, J. N. Marchese, B. Williamson, K. Parker, S. Hattan, N. Khainovski, S. Pillai, S. Dey, S. Daniels, S. Purkayastha, P. Juhasz, S. Martin, M. Bartlet-Jones, F. He, A. Jacobson, and D. J. Pappin. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3:1154-1169. [DOI] [PubMed] [Google Scholar]
- 60.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10:393-408. [DOI] [PubMed] [Google Scholar]
- 61.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sayed, A., S. Matsuyama, and M. Inouye. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 264:51-54. [DOI] [PubMed] [Google Scholar]
- 64.Schuwirth, B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310:827-834. [DOI] [PubMed] [Google Scholar]
- 65.Scott, J. M., and W. G. Haldenwang. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. J. Bacteriol. 181:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott, J. M., J. Ju, T. Mitchell, and W. G. Haldenwang. 2000. The Bacillus subtilis GTP binding protein Obg and regulators of the σB stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 182:2771-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikora, A. E., K. Datta, and J. R. Maddock. 2006. Biochemical properties of the Vibrio harveyi CgtAV GTPase. Biochem. Biophys. Res. Commun. 339:1165-1170. [DOI] [PubMed] [Google Scholar]
- 68.Sikora, A. E., R. Zielke, K. Datta, and J. R. Maddock. 2006. The Vibrio harveyi GTPase CgtAV is essential and is associated with the 50S ribosomal subunit. J. Bacteriol. 188:1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sirdeshmukh, R., and D. Schlessinger. 1985. Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res. 13:5041-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srivastava, A. K., and D. Schlessinger. 1988. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. USA 85:7144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 72.Tan, J., U. Jakob, and J. C. Bardwell. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teraoka, H., and K. H. Nierhaus. 1978. Protein L16 induces a conformational change when incorporated into a L16-deficient core derived from Escherichia coli ribosomes. FEBS Lett. 88:223-226. [DOI] [PubMed] [Google Scholar]
- 74.Trach, K., and J. A. Hoch. 1989. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171:1362-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Traub, P., and M. Nomura. 1968. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. USA 59:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uicker, W. C., L. Schaefer, and R. A. Britton. 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59:528-540. [DOI] [PubMed] [Google Scholar]
- 77.Vidwans, S. J., K. Ireton, and A. D. Grossman. 1995. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177:3308-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walbot, V., and E. H. Coe, Jr. 1979. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl. Acad. Sci. USA 76:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Welsh, K. M., K. A. Trach, C. Folger, and J. A. Hoch. 1994. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J. Bacteriol. 176:7161-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 186:5249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wower, J., P. Scheffer, L. A. Sylvers, W. Wintermeyer, and R. A. Zimmermann. 1993. Topography of the E site on the Escherichia coli ribosome. EMBO J. 12:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, S., and W. G. Haldenwang. 2004. Guanine nucleotides stabilize the binding of Bacillus subtilis Obg to ribosomes. Biochem. Biophys. Res. Commun. 322:565-569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.