Abstract
The plant Polycomb-group (Pc-G) protein CURLY LEAF (CLF) is required to repress targets such as AGAMOUS (AG) and SHOOTMERISTEMLESS (STM). Using chromatin immunoprecipitation, we identify AG and STM as direct targets for CLF and show that they carry a characteristic epigenetic signature of dispersed histone H3 lysine 27 trimethylation (H3K27me3) and localised H3K27me2 methylation. H3K27 methylation is present throughout leaf development and consistent with this, CLF is required persistently to silence AG. However, CLF is not itself an epigenetic mark as it is lost during mitosis. We suggest a model in which Pc-G proteins are recruited to localised regions of targets and then mediate dispersed H3K27me3. Analysis of transgenes carrying AG regulatory sequences confirms that H3K27me3 can spread to novel sequences in a CLF-dependent manner and further shows that H3K27me3 methylation is not sufficient for silencing of targets. We suggest that the spread of H3K27me3 contributes to the mitotic heritability of Pc-G silencing, and that the loss of silencing caused by transposon insertions at plant Pc-G targets reflects impaired spreading.
Keywords: flowering, histone methylation, Polycomb
Introduction
In plants and animals, Polycomb-group (Pc-G) genes mediate mitotically stable repression of targets such as homeotic genes that are critical for developmental patterning and growth control. An antagonistic group of proteins, the trithorax-group (trx-G), act as activators. Both groups act to maintain on/off patterns of transcription defined early in development, rather than to set up these patterns, and are important for maintaining cell fates. Although the Pc-G and trx-G have long been thought to cause epigenetic changes in chromatin structure, due to the stable but ultimately reversible alterations in gene activity they promote, the mechanistic basis for their activity has been mysterious. Recent studies have implicated histone modifications as an important component of epigenetic changes. A variety of modifications on the amino-tails of histones have been characterised, of which methylation of lysine residues has been thought to be particularly stable (Jenuwein and Allis, 2001; Fischle et al, 2003). The consequence of lysine methylation can differ both according to which lysine residue is modified and also as to how many methyl groups are added—lysine residues can be mono-, di- or trimethylated (Bannister et al, 2002). For example, methylation of histone H3 at lysine 9 (H3K9) or lysine 27 (H3K27) is generally correlated with transcriptional repression, whereas methylation at lysine 4 (H3K4) is predominantly associated with transcriptional activity (Jenuwein and Allis, 2001; Peters et al, 2003; Ringrose and Paro, 2004). In addition, the level of methylation is important, for example H3K9me3 shows a different distribution from H3K9me1 and H3K9me2 in mammals (Peters et al, 2003). Several enzymes with histone lysine demethylase activity have now been identified, indicating that methylation can be rapidly reversed (Shi et al, 2004; Tsukada et al, 2006).
In animals, Pc-G proteins have been biochemically purified in at least two distinct complexes, Polycomb repressive complex 1 and 2 (PRC1 and PRC2). In accordance with their epigenetic role, both complexes have been recently shown to modify histones. Drosophila PRC2 consists of the four core members SUPPRESSOR OF ZESTE 12 (SU(Z)12), P55, EXTRA SEX COMBS (ESC) and ENHANCER OF ZESTE (E(Z)) (Ringrose et al, 2004). E(Z) carries a histone methyltransferase domain, the conserved SET domain, and can trimethylate lysine 9 and 27 of histone H3 but requires other PRC2 members to accomplish its catalytic activity (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Muller et al, 2002). A detailed examination of histone methylation at Pc-G targets revealed that silencing of Drosophila Pc-G target genes is in most cases correlated with both H3K27me3 and H3K9me3, whereas presence of only one modification was not an indicator of silencing (Ringrose et al, 2004). Subsequently, the marks set by PRC2 may create a binding site for the chromodomain protein POLYCOMB (PC), which then recruits PRC1 (Cao et al, 2002; Czermin et al, 2002). Inhibition of chromatin remodelling by SWI/SNF complexes and direct compaction of nucleosomes by PRC1 might then lead to stable, long-term silencing (Francis et al, 2004). So far, no DNA-binding activity of core Pc-G proteins has been determined; thus, it is largely unresolved how they are recruited to their target genes. However, in Drosophila, many DNA elements, the so-called Polycomb repressive elements (PREs), that are both required and sufficient for recruitment of Pc-G complexes have been identified (Ringrose et al, 2003, 2004).
In plants, only the PRC2 is structurally conserved and most of its members are encoded by small gene families. Genetic and molecular analysis suggests that their products act in several PRC2-like complexes with discrete but overlapping functions (Hsieh et al, 2003; Reyes and Grossniklaus, 2003; Chanvivattana et al, 2004; Schubert et al, 2005). In Arabidopsis, there are three E(z) homologues: MEDEA (MEA), CURLY LEAF (CLF) and SWINGER (SWN, also known as EZA1). MEA predominantly acts during seed development, whereas CLF and SWN are expressed more generally in plants. CLF is required to repress floral homeotic genes such as AGAMOUS (AG) and also the homeobox gene SHOOTMERISTEMLESS (STM) (Goodrich et al, 1997; Katz et al, 2004). It is likely that CLF regulates many other targets, as there is substantial redundancy between CLF and the related SWN gene (Chanvivattana et al, 2004). There are also three Su(z)12 homologues, FERTILISATION INDEPENDENT SEED2 (FIS2), VERNALISATION2 (VRN2) and EMBRYONIC FLOWER2 (EMF2). The action of FIS2, like MEA, is largely confined to the seed, whereas VRN2 and EMF2, like CLF and SWN, act more generally in development. VRN2 is required for repression of FLC, an inhibitor of flowering, in response to cold treatments (vernalisation). EMF2 has a similar role to CLF in repression of floral homeotic and other target genes. Double mutant analysis suggests that EMF2 and VRN2 also show redundancy, as double mutants have severe phenotypes and resemble clf swn doubles (Schubert et al, 2005). In contrast to the other Arabidopsis PRC2 homologues, ESC is represented by a single copy gene, FERTILISATION INDEPENDENT ENDOSPERM (FIE). Null fie mutants are embryonic lethal, like fis2 and mea mutants; however, depletion of FIE activity later in development, for example by co-suppression, reveals that FIE likely acts with CLF and EMF2 to repress common targets (Kinoshita et al, 2001; Katz et al, 2004). Thus, clf swn and emf2 vrn2 double mutants, as well as plants lacking FIE activity, have similar phenotypes and likely lack vegetative Pc-G activity.
Several studies suggest that the plant PRC2 may also act as an H3K27 methyltransferase. Immunostaining experiments show that in wild-type plants H3K27me3 localises to euchromatin, whereas H3K27me2 strongly labels heterochromatin and has weaker staining in euchromatin. In clf swn mutants and in transgenic plants with severely reduced FIE activity, H3K27me2 staining was reduced at euchromatin but not at heterochromatin; H3K27me3 staining in euchromatin was also reduced but frequently became re-distributed to heterochromatin (Lindroth et al, 2004). These results suggested that the plant Pc-G control H3K27me2 and H3K27me3 in euchromatin, but that other genes can also supply this function particularly in heterochromatin. In addition, chromatin immunoprecipitation (ChIP) experiments have shown that silencing of FLC by VRN2 is associated with H3K27me2 methylation (Bastow et al, 2004; Sung and Amasino, 2004). Despite this progress, the role of different histone methylation states in silencing of specific plant Pc-G targets remains poorly defined and for most targets it is not known whether the Pc-G act directly or indirectly. In addition, it is unclear how silencing of Pc-G target genes is inherited through mitosis and whether Pc-G proteins are persistently required to maintain repression.
Here, we show that CLF is a nuclear-localised protein that is persistently required for silencing of AG in leaves but is unlikely itself to constitute a heritable epigenetic mark. Using ChIP, we show that plant Pc-G targets are characterised by dispersed H3K27me3 methylation that colocalises with CLF protein on chromatin. We discuss the possible functions of H3K27me3 spreading for the inheritance and stability of epigenetic silencing in plants.
Results
The SET domain is necessary for CLF+ activity
The strongest similarity between the CLF and E(Z) proteins lies in their SET domains, suggesting that like E(Z), CLF also acts as an HMTase. To confirm that the SET domain was required for CLF+ activity, we characterised classical clf alleles to see if any had lesions within the SET domain. We found that the clf-81 allele (Kim et al, 1998), which phenotypically resembles the null clf-50 allele (Figure 1A), had a missense mutation that encoded the substitution R794H within the SET domain. Alignments indicated that the R794 residue is highly conserved between diverse SET domain proteins, including the human K4 H3 HMTase SET7/9 and the fission yeast K9 H3 HMTase CLR4 (Figure 1B). It lies in a helical region that is predicted from structural studies of the SET domain to be part of a groove that accommodates histone tails: for example, the corresponding residue (R258) of human SET7/9 is thought to bind the side chain of R2 on the histone H3 substrate (Xiao et al, 2003). The phenotype of the clf-81 allele (Figure 1B) may therefore reflect impaired histone binding by the CLF HMTase. Consistent with this, we found that histone methylation was reduced in clf-81 plants (see later results).
Figure 1.
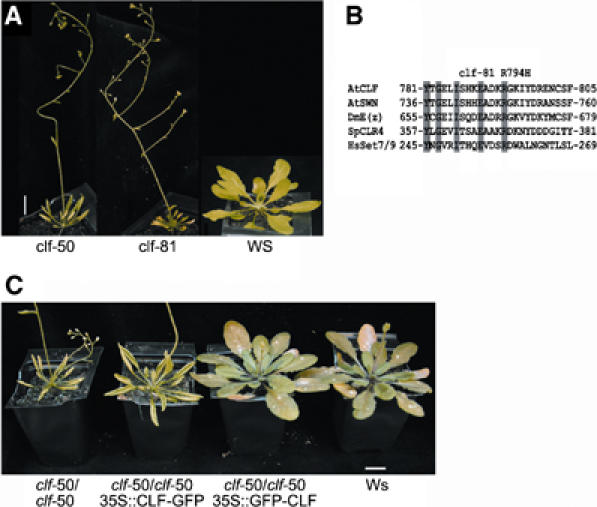
A severe clf allele carries a point mutation in the SET domain. (A) Alignment of a part of the SET domains of different SET domain proteins; residues conserved in all proteins are shaded, residue R794 that is mutated to H in clf-81 is marked. (B) clf-81 shows leaf curling and early flowering (compare to wild-type Ws (Wassilewskija)), similar to the clf-50 deletion allele. (C) clf-50 is complemented by a 35S∷GFP-CLF transgene but only partially by a 35S∷CLF-GFP transgene. All plants were grown under short day conditions; scale bar is 1 cm. AtCLF, Arabidopsis thaliana CURLY LEAF; AtSWN, A. thaliana SWINGER; DmE(z), Drosophila melanogaster ENHANCER OF ZESTE; SpCLR4, Schizosaccharomyces pombe CLR4; HsSET7/9, Homo sapiens SET7/9.
CLF protein is nuclear localised but is not present throughout mitosis
To localise the CLF protein, we made transgenes (35S∷GFP-CLF and 35S∷CLF-GFP) that expressed CLF as a fusion with GFP, under control of the cauliflower mosaic virus 35S promoter. The 35S∷GFP-CLF construct fully complemented the null clf-50 mutation in transgenic plants, whereas 35S∷CLF-GFP gave little or no complementation (Figure 1C). We did not observe any phenotypic abnormalities resulting from expressing CLF under the constitutive 35S promoter, perhaps because expression of the endogenous CLF gene is also fairly constitutive (Goodrich et al, 1997). Consistent with a role for CLF in chromatin modifications, microscopy indicated that the GFP-CLF fusion was predominantly localised to nuclei in transgenic plants (Figure 2). In contrast with the Arabidopsis Pc-G protein VRN1, which localises to metaphase chromosomes in root tips and is present throughout mitosis (Mylne et al, 2006), we did not observe any metaphase figures showing GFP-CLF fluorescence (compare Figure 2B and C). It is therefore unlikely that CLF itself constitutes a heritable epigenetic mark, as it is lost during mitosis.
Figure 2.
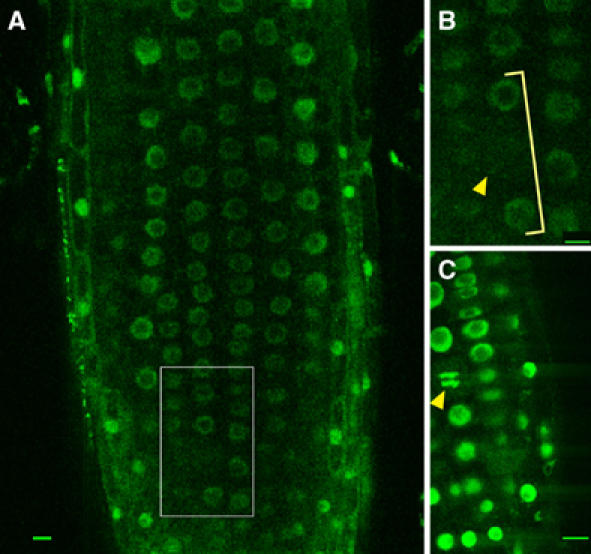
CLF protein is nuclear localised but is not present in nuclei throughout mitosis. (A) Root of a clf-50/clf-50 35S∷GFP-CLF plant showing GFP expression in most cells in the nuclei. (B) Close-up of the inset in (A) showing a cell without nuclear GFP expression (arrowhead). Adjacent cells in the same cell file show nuclear GFP (bracket). (C) Details of a VRN1∷VRN1-GFP root exhibiting a mitotic figure (arrowhead). At least 10 roots per line were analysed by confocal laser microscopy; in no case, GFP-stained mitotic figures were identified in clf/clf 35S∷GFP-CLF roots, whereas VRN1∷VRN1-GFP roots showed green-fluorescing mitotic figures in all roots. Scale bars, 10 μm.
The CLF protein is required persistently to silence AG
The CLF mRNA is expressed persistently during leaf and flower development (Goodrich et al, 1997). To test whether CLF protein is required persistently to maintain silencing, or can act in a ‘hit and run' fashion, we made a transgenic line (clf-2 pCLF∷CLF-GR; see Materials and methods for details) in which CLF+ activity is steroid dependent. Thus, plants that were supplied with dexamethasone (dex) steroid from germination onwards had a wild-type phenotype, whereas those grown in the absence of steroid had a clf mutant phenotype (Figure 3A–C). To monitor AG activity, we further introduced the pAG-I∷GUS reporter transgene, which carries the cis-acting sequences necessary for response to CLF (Sieburth and Meyerowitz, 1997), into this conditional CLF background. To test the requirement for CLF to maintain silencing of AG, we grew plants on agar plates containing steroid for 10 days, then transferred the seedlings to soil and either withdrew (−dex) or maintained (+dex) steroid supply. At the time of the shift, plants had 4–5 visible leaves (>3 mm in size) (Figure 3D) and microscopy indicated that all rosette and cauline leaves had been initiated and flowers were initiating at the shoot apex (Figure 3E). The morphology of the shifted plants was scored 17 days after the shift when they had bolted, and rosette leaves were fully expanded. Unlike the plants grown continuously on steroid, those that were removed from steroid developed curled rosette leaves, particularly in leaves 5 and 6 (Figure 3C). Staining the plants for pAG-I∷GUS activity confirmed that the rosette leaves 5 and 6 had strong staining compared to plants grown on steroid throughout (Figure 3F and G). This suggested that expression of CLF activity transiently during early leaf development is not sufficient to maintain silencing of AG in leaves.
Figure 3.
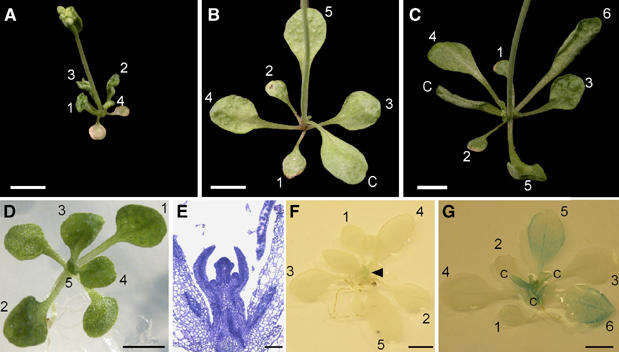
CLF is persistently required for repression of AG in leaves. (A–G) All plants are clf/clf and carry the pCLF∷CLF-GR and the pAG-I∷GUS transgenes. Plants were initially grown on MS medium under long-day condition with or without dex steroid and transferred at day 10 to soil and either sprayed or not sprayed with dex. (A) A 27-day-old plant grown without dex throughout. (B) A 27-day-old plant grown with dex throughout. (C) A 27-day-old plant grown for 10 days with dex, then transferred from soil and dex withdrawn. Note leaf curling of leaves 4 and 5 and cauline leaf (C). (D) Plant grown on dex for 10 days shows no leaf curling. (E) Longitudinal section through a 10-day-old seedling grown with dex. Note that all leaf primordia have formed and that the plant has started flowering, so that the influorescence bolt is visible. (F, G) GUS staining reflects AG expression. (F) A 27-day-old plant grown on dex throughout only shows GUS staining in the flowers (arrowhead). (G) A 27-day old plant shifted on day 10 from dex to no dex shows strong GUS staining in leaves 5 and 6 and cauline leaves. Numbers indicate leaf number emerging after germination. Scale bars: 5 mm (A–C, F, G), 2 mm (D), 50 μm (E).
AG and STM are direct targets of CLF
AG appears to be a principal target of CLF, as the clf mutant phenotype is largely dependent on AG+ activity (Goodrich et al, 1997). However, it was not known whether CLF acted directly on AG or indirectly, for example by repressing an activator of AG. To address this, we used ChIP to analyse the sequences bound by CLF in vivo. Because we lacked an antibody specific for CLF, we used an epitope-tag strategy: chromatin from 35S∷GFP-CLF clf-50 seedlings was immunoprecipitated (IP) using an α-GFP antibody that is reliable for ChIP (Vanoosthuyse et al, 2004). We found strong enrichment for sequences from multiple sites within the AG locus (Figure 4C and data not shown). However, we did not detect enrichment for the genes upstream or downstream of AG (Figure 4D) or for heterochromatic sequences such as TA3 (Figure 4B). In addition, we did not see enrichment for AG sequences in similar ChIP experiments using a control 35S∷GFP transgenic line (Supplementary Figure S3). Together, these results suggested that CLF binds AG in vivo, and is dispersed over multiple sites on AG chromatin.
Figure 4.
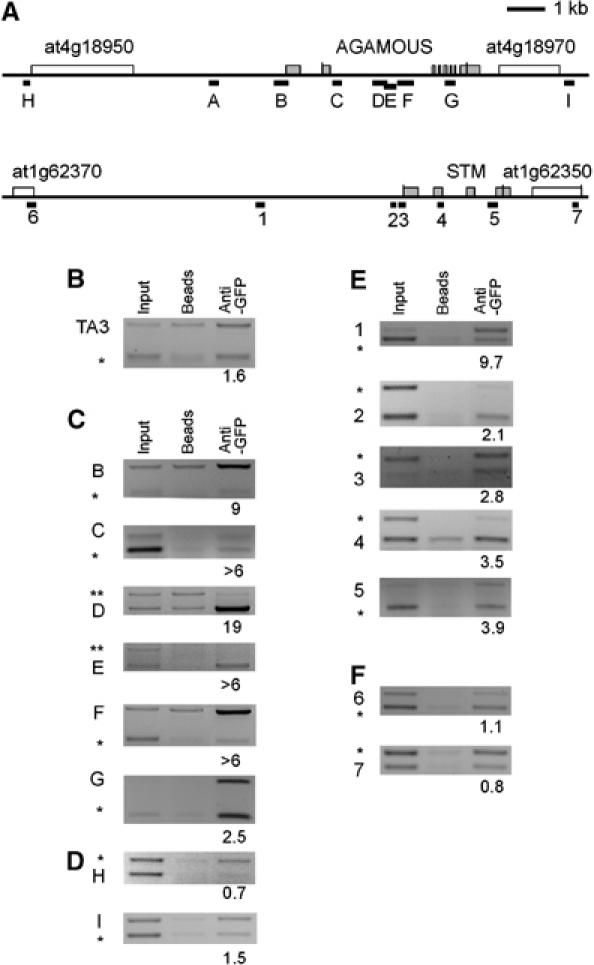
CLF is bound to AG and STM. (A) Schematic structure of the AG and STM loci including the 5′- and 3′-flanking genes. For AG and STM, the exon/intron structure (exons are shaded), and for the flanking genes, the transcribed regions are depicted. Black bars with letters (for AG) or numbers (for STM) indicate regions amplified in ChIP PCRs. (B, C) IP on chromatin preparations isolated from 10-day-old seedlings of a 35S∷GFP-CLF transgenic line was performed without antibody (‘beads') or anti-GFP antibodies and precipitated DNA was amplified by PCR. Semiquantitative ChIP duplex PCRs were used to amplify ACTIN 2/7 (marked with *) or PHOSPHOFRUCTOKINASE (PFK) sequences (marked with **) as internal controls and regions from TA3 retrotransposon (B) as negative control, from AG (C) and AG flanking genes (D), or from STM (E) and STM flanking genes (F). Numbers below the lanes indicate the ratio of the intensity of TA3, AG or STM products, respectively, compared to ACTIN or PFK intensity after IP normalised to the ratio before IP (‘input'). Each experiment was performed at least twice; representative experiment is shown.
We next asked whether the homeobox gene SHOOT MERISTEMLESS (STM), which is mis-expressed in clf mutants (Katz et al, 2004), is also bound by CLF. Indeed, further ChIP analyses revealed binding of CLF to STM. The binding of CLF at STM was weak relative to AG, but again appeared to be dispersed over the locus (Figure 4E) but not the neighbouring genes (Figure 4F).
The AG and STM loci are covered with the repressive mark H3K27me3 and lack the active mark H3K4me2
We next tested whether AG and STM carry the repressive histone methylation marks associated with Pc-G repression (H3K27me2, H3K27me3) using chromatin from whole seedlings, that is, tissues in which both genes are repressed in a Pc-G-dependent fashion. To confirm the specificity of the antibodies for different histone marks, we first tested heterochromatic sequences from the TA3 retrotransposon in ChIP assays. Consistent with previous reports (Lindroth et al, 2004; Mathieu et al, 2005; Naumann et al, 2005), we detected strong enrichment for H3K27me2 but not for H3K4me2 and H3K27me3 (Figure 5C). Next, we examined the CLF target genes: H3K27me3 covers the AG and STM loci but does not extend to the flanking genes, similar to the distribution observed for CLF (Figure 5A–D). Although microarray studies (see Genevestigator in Zimmermann et al, 2004) suggest that like AG and STM, the gene upstream of STM (At1g62370) is not detectably expressed in leaves, it notably lacked H3K27me3; thus, this is not a generic mark for transcriptionally inactive genes. Additionally, we detected weak, but reproducible enrichment of H3K27me2 in regions B, F and G of AG and regions 1 and 3 of STM (Figure 5A and C). B lies in the AG promoter region, whereas F is in a region of the AG second intron, which is highly conserved in different plant species (Hong et al, 2003). Thus, in contrast to H3K27me3, H3K27me2 appears to localise more specifically. Silencing of AG and STM was also reflected in reduced enrichment compared to ACT or PFK for H3K4me2, a mark that is predominantly found on active genes (Gendrel et al, 2002) (Figure 5A and C).
Figure 5.

Histone methylation patterns of AG and STM in wild-type and Pc-G mutant plants. (A–D) ChIPs were performed with antibodies against H3K4me2, H3K27me2 and H3K27me3 from wild-type (A–D), clf (A, C), emf2 (A, C), clf swn (C) and vrn2 emf2-3 (C) chromatin extracted from 10-day-old seedlings. Regions amplified are shown in Figure 4A and calculation of relative enrichment is described in Figure 4. Results of ChIP PCRs on the AG locus (A), on the AG flanking genes (B), the STM locus and TA3 retrotransposon (latter is H3K27me2control) (C) and on the STM flanking genes (D). (E) RT–PCRs with primers amplifying AG, STM or eIF2 coding regions on cDNAs generated from mRNA isolated from wild-type, clf and clf swn seedlings. EIF2 was used as a loading control.
The above ChIP experiments were performed on 12-day-old seedlings, in which most of the tissue is from young leaves. To test whether the methylation profiles changed during leaf development, we performed ChIP using fully expanded leaves from plants that were flowering. The results for H3K27me2 and H3K27me3 at AG and STM were similar to those for seedlings (Supplementary Figure S2). This suggested that the repressive H3K27 methylation marks persist throughout leaf development.
H3K27me3 is dependent on Pc-G activity
As CLF protein and H3K27me3 colocalise on AG, we asked whether this methylation is dependent on CLF+ activity. Indeed, coverage of AG with H3K27me3 was almost completely lost in clf mutants, whereas H3K27me2 was only slightly affected (Figure 5A). In emf2-10 mutants, which have a weak emf2 phenotype resembling that of clf mutants, AG is also de-repressed (Chanvivattana et al, 2004). Consistent with the proposed role of EMF2 and CLF in a common complex, emf2-10 mutants had very similar effects on H3K27 methylation to clf mutants (Figure 5A). In addition, regions E and F of AG gained H3K4me2 in the mutants, consistent with activation of AG (Figure 5). Importantly, H3K27me3 at the AG locus was also lost in clf-81, confirming the importance of the SET domain for CLF function and histone methylation (Supplementary Figure S1).
Unlike AG, there was little change in histone methylation at STM in clf or emf2-10 single mutants (Figure 5C). However, previous genetic studies suggested that CLF has partial redundancy with the related gene SWN, and similarly EMF2 does so with VRN2 (Chanvivattana et al, 2004; Lindroth et al, 2004; Schubert et al, 2005). Consistent with this, H3K27me3 was completely lost in all regions tested in both clf swn and emf2 vrn2 double mutants and H3K4me2 was correspondingly gained in regions 2–5 (Figure 5C). In addition, expression of STM is stronger in clf swn double mutants than in clf single mutants (Figure 5E). Thus, strong STM mis-expression in the double mutants is strictly correlated with loss of H3K27me3 and gain of H3K4me2, whereas weak mis-expression in the single mutants is accompanied by only slight changes in histone methylation at the STM locus.
Vernalisation induces VRN2-dependent gain of H3K27me3 on FLC but not its neighbours
Two previous studies have revealed that the FLC locus acquires VRN2-dependent H3K27me2 upon vernalisation (Bastow et al, 2004; Sung and Amasino, 2004). In addition, the FLC neighbouring genes at5g10130 and at5g10150 are downregulated during prolonged cold treatment, suggesting that silencing initiated at FLC might spread upstream and downstream (Finnegan et al, 2004). As we detected coverage of AG and STM with H3K27me3, we wondered whether coordinated downregulation of FLC and its neighbouring genes might reflect spread of H3K27me3 beyond FLC. Thus, we used ChIP to study changes in histone methylation before and after vernalisation (40 days at 4°C, then 15 days at 22°C under long-day conditions) in fca mutants (a vernalisation requiring background) and compared this to backgrounds in which vernalisation is impaired (vrn1 fca, vrn2 fca). Indeed, the FLC locus acquired significantly higher levels of H3K27me3 after vernalisation in regions 2, 3 and 4 (4- to 12-fold); thus, is likely covered with H3K27me3, similar to the pattern at AG and STM (Figure 6 and Supplementary Figure S4). Importantly, the gain of H3K27me3 was dependent on VRN2 but not VRN1: unlike vrn2, vrn1 mutations reduced but did not eliminate the rise in H3K27me3 after vernalisation (Figure 6 and Supplementary Figure S4). Although H3K27me3 covered FLC, it did not spread to the neighbouring genes: At5g10150 was devoid of H3K27me3 and At5g10130 showed only slight enrichment of H3K27me3 after vernalisation. We also observed enrichment of H3K27me2 after vernalisation at FLC in fca, but to a lower extent than H3K27me3 (two-fold; Figure 3 and Supplementary Figure S4).
Figure 6.
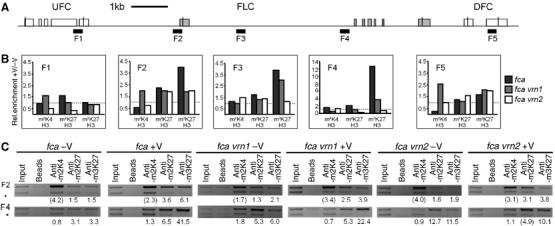
The entire FLC locus shows strong enrichment with H3K27me3 after vernalisation. (A) Schematic structure of the FLC locus including the flanking genes UFC and DFC. The FLC exons are shaded. Black bars indicate the regions amplified. (B) Summary of ChIP PCRs performed with antibodies against H3K4me2, H3K27me2 and H3K27me3 on chromatin extracted from the vernalisation-requiring background fca, vrn1 fca and vrn2 fca mutants before and after vernalisation. Plants were grown for either 2 days at 4°C (−V) or 40 days at 4°C (+V), then transferred to 22°C and harvested after 15 days. Relative enrichment for ChIPs was determined as described in Figure 4 and the ratio of +V/−V for each genotype and antibody was calculated. Dashed lines indicate relative enrichment of 1. (C) Example of ChIPs performed for the different genotypes, antibodies and growth conditions. The whole data set is available in Supplementary Figure S4.
H3K27me3 spreads on an AG transgene in a CLF-dependent manner
Because H3K27me3 and CLF cover large parts of AG, it is unlikely that they are recruited to all regions of AG in a DNA-sequence-specific manner. Thus, it is possible that initial recruitment of CLF and therefore H3K27me3 is guided by DNA-sequence specific factors and then spreads over the gene to achieve stable silencing. To reveal whether H3K27me3 can spread from AG sequences to neighbouring sequences, we performed ChIP on transgenic plants that carry a transgene (pAG-I∷GUS) that carries the cis-acting AG sequences necessary for regulation by CLF (Sieburth and Meyerowitz, 1997). The construct carries the 3′ portion of the gene (At4g18950) upstream of AG, the intergenic region between At4g18950 and AG, and the 5′-AG sequences extending to the third exon (Figure 7A). We amplified sequences that are only present on the transgene (β-GLUCURONIDASE (GUS) and NEOMYCIN PHOSPOTRANSFERASE (NPT)) and therefore are in close proximity to the AG regulatory regions (promoter and large intron) (Figure 7A). We detected strong enrichment for H3K27me3 on AG and GUS sequences in wild type but not in clf mutants carrying the transgene (Figure 7B). Because the GUS gene is not an endogenous Pc-G target, this suggests that H3K27me3 is initiated at AG sequences and spreads onto neighbouring sequences in a CLF-dependent manner. By contrast, H3K27me3 was only slightly enriched at the NPT gene, similar to At4g18950 in the native AG context, suggesting that it did not spread upstream of the AG gene. In addition, strong enrichment for H3K4me2 was observed in wild type at the NPT gene, but only little at GUS, consistent with the NPT gene being expressed and the GUS gene being silenced. In clf mutants, where pAG-I∷GUS is de-repressed, the GUS gene showed much stronger H3K4me2 enrichment.
Figure 7.
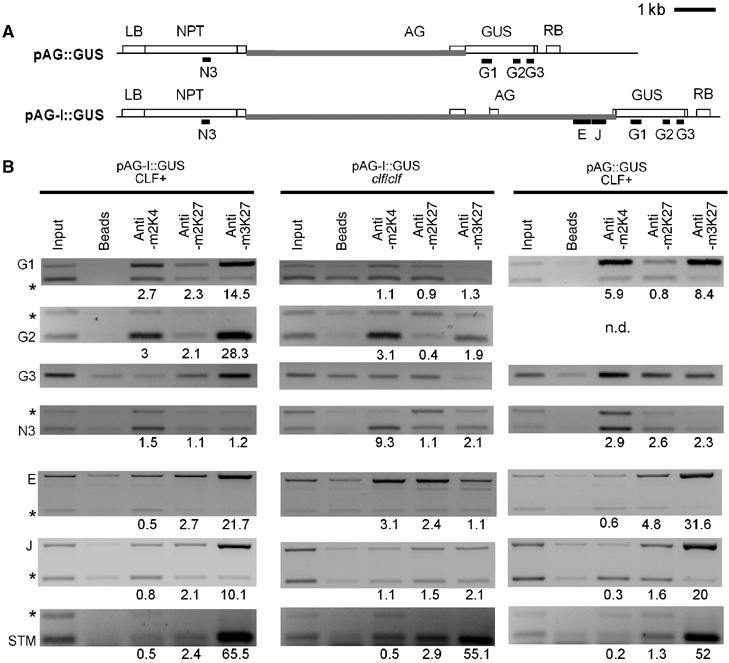
H3K27me3 spreads over AG transgenic sequences but is not sufficient for silencing. (A) Schematic structure of the pAG∷GUS and pAG-I∷GUS transgenes, constructed by Sieburth and Meyerowitz (1997). Black bars indicate regions amplified in ChIP PCRs. Grey bars indicate sequences homologous to AG. The pAG∷GUS T-DNA contains a NEOMYCIN PHOSPHOTRANSFERASE (NPT) resistance gene close to the left border (LB) of the T-DNA, around 6 kb of AG upstream region, the first AG exon, the β-GLUCURONIDASE (GUS) coding region and the right border (RB) of the T-DNA. The pAG-I∷GUS T-DNA consists of the same T-DNA backbone and AG upstream region but contains the first two exons and the first two introns of AG. The pAG-I∷GUS reporter reflects the endogenous AG expression pattern in wild-type and clf leaves, whereas pAG∷GUS is strongly mis-expressed in wild-type cotyledons and leaves (Sieburth and Meyerowitz, 1997). (B) Results of ChIP PCRs performed on IP with antibodies against H3K4me2, H3K27me2 and H3K27me3 on chromatin samples extracted from 10-day-old wild-type or clf seedlings carrying the pAG-I∷GUS transgene or wild-type seedlings carrying the pAG∷GUS transgene. Regions F and J of AG are present both at the endogenous AG locus and on the pAG-I∷GUS transgene. A region of STM (region 2 in Figure 4) served as an H3K27me3 control. Relative enrichment for ChIPs was determined as described in Figure 4.
H3K27me3 is not sufficient for silencing
The pAG∷GUS transgene lacks AG intragenic regulatory sequences and is expressed in leaves (Sieburth and Meyerowitz, 1997). Consistent with this, H3K4me2 enrichment at the GUS gene was greater than in the silenced pAG-I∷GUS (Figure 7B). Unexpectedly, we also found strong enrichment for H3K27me3 at GUS sequences. Thus, H3K27me3 is necessary for silencing of Pc-G targets but is not sufficient.
Discussion
Previous studies have suggested that CLF acts in a complex similar to PRC2 and that it likely regulates histone methylation. In particular, immunostaining experiments have shown global effects of the plant Pc-G genes on H3K27 methylation in euchromatin and heterochromatin. Here, we use ChIP to dissect how CLF and SWN control methylation patterns at specific targets. We show that CLF likely acts in several different complexes with different HMTase specificities. As with the animal PRC2, silencing by Pc-G requires H3K27me3; however, in plants, the H3K27me3 is spread throughout the chromatin of their targets. This may be significant for regulation of clusters of genes and for the disruption of silencing that occurs when transposons insert at Pc-G targets.
CLF and SWN mediate dispersed H3K27me3 methylation at their targets
Although CLF shows considerable redundancy with the related SWN gene, there is an absolute requirement for CLF+ activity for silencing of AG during vegetative development; thus, the effects of Pc-G on chromatin were most easily revealed for AG. We found a strong enrichment for H3K27me3 that extended over all regions of the AG locus that we tested, including the promoter, intron and exon regions. Importantly, this methylation was strictly dependent on CLF+ activity, as it was lost in clf mutants. It was also lost in emf2-10 mutants, consistent with the EMF2 and CLF proteins acting together in a common complex with HMTase activity (Chanvivattana et al, 2004). We also observed strong enrichment for H3K27me3 at two other Pc-G target genes, STM and FLC. Again, the methylation was not localised to specific regions of the targets but appeared to be dispersed throughout the promoter and transcribed regions. This contrasts with the punctate pattern of H3K27me3 that has been described in Drosophila, where it has been localised to discrete regulatory elements, the PREs (Cao et al, 2002; Ringrose et al, 2004; Wang et al, 2004). However, several recent animal studies have also revealed dispersed H3K27me3 methylation (Bernstein et al, 2006; Lee et al, 2006; Tolhuis et al, 2006).
The clf mutant phenotype is enhanced by swn mutations, suggesting that the two genes act redundantly (Chanvivattana et al, 2004). Presumably, there are some targets at which either CLF or SWN activity is sufficient for silencing so that they are only significantly mis-expressed in clf swn double mutants. Consistent with this, STM expression was much higher in clf swn double mutants that in the single mutants. This correlated well with the effects of the mutants on histone methylation at STM. The H3K27me3 enrichment at STM was depleted in clf swn double mutants, whereas the single mutants had similar levels to wild type. We observed similar redundancy between EMF2 and VRN2 for H3K27me3 enrichment at STM. We conclude that CLF/SWN and EMF2/VRN2 can act for the most part interchangeably in PRC2-like complexes with H3K27me3 HMTase activity. The catalytic activity of these complexes is likely provided by the SET domains of CLF and SWN. This is supported by our characterisation of the clf-81 allele, which demonstrates that a conserved residue in the SET domain that has been implicated in binding histones is necessary for H3K27me3 methylation at AG.
We were also able to directly localise CLF to AG and STM chromatin by ChIP analysis of transgenic lines that expressed epitope-tagged CLF (35S∷GFP-CLF) and observed a dispersed distribution. An obvious concern is that the distribution of GFP-CLF does not reflect that of the endogenous CLF protein, for example the 35S promoter might confer abnormally high levels of CLF protein that lead to spurious associations. We think this unlikely for two reasons. Firstly, the 35S∷GFP-CLF transgene fully complements the null clf-50 mutation but does not cause any gain-of-function phenotypes that might reflect ectopic CLF+ activity. Secondly, the distribution of the GFP-CLF fusion protein in our transgenic lines precisely matches that of H3K27me3 in wild-type (non-transgenic) backgrounds. Importantly, it does not spread any further from the AG locus than does H3K27me3 in wild type. We conclude that the distribution of GFP-CLF faithfully mimics that of the endogenous CLF protein. Therefore, AG and STM are direct targets of CLF and so CLF must act as a transcriptional repressor, at least for these two targets. The enrichment for GFP-CLF at STM was less than at AG, consistent with the redundancy between SWN and CLF for silencing of STM.
Pc-G targets show localised H3K27me2 enrichment
Immunostaining experiments have shown that H3K27me2 is chiefly found in heterochromatic regions, with a low level enrichment for euchromatin, whereas H3K27me3 is absent from heterochromatin (Lindroth et al, 2004; Mathieu et al, 2005; Naumann et al, 2005). Consistent with this, our ChIP experiments showed that TA3, a retrotransposon confined to heterochromatin, was enriched for H3k27me2 but not H3K27me3. In addition, we found enrichment for H3K27me2 at discrete regions of Pc-G targets, again suggesting that this mark is not exclusively heterochromatic. The levels of enrichment were modest relative to H3K27me2 at TA3 or H3K27me3 at AG. However, the results were very reproducible, for example we found H3K27me2 in region F of AG in more than 10 independent ChIP experiments. Unlike our results for H3K27me3, H3K27me2 methylation was not consistently eliminated in either clf swn or emf2 vrn2 double mutant backgrounds although it was reduced. This agrees with the immunostaining results, which showed that heterochromatic regions retain H3K27me2 in severe Pc-G mutants, whereas the staining in euchromatin is reduced (Lindroth et al, 2004). It is possible that the residual H3K27me2 methylation reflects further redundancy, for example between MEA and CLF/SWN. This is supported by the observation that MEA is mis-expressed in leaves in clf mutants (Katz et al, 2004; Jullien et al, 2006). However, in plants lacking FIE activity, H3K27me2 is also retained at heterochromatin (Lindroth et al, 2004). Because FIE is single copy, and therefore unlikely to be redundant, this suggests that proteins other than Pc-G can provide H3K27me2 HMTase activity in plants. In addition to CLF/SWN/MEA, the Arabidopsis genome encodes at least a further 26 SET domain proteins that could potentially contribute to this activity (Baumbusch et al, 2001).
The coverage with H3K27me3 at the CLF target loci argues against a DNA-sequence-specific recruitment of CLF to all modified histones. The DNA region in the AG intron that shows H3K27me2 is highly conserved between different plant species and carries sequences required for both activation and repression of AG (Busch et al, 1999; Deyholos and Sieburth, 2000; Hong et al, 2003). One possibility is that this corresponds to a plant PRE and is responsible for initial recruitment of a CLF-containing complex, after which H3K27me3 methylation spreads over the target (Figure 8). The specificity of CLF as a di- or trimethylase may vary according to its partners—for example, human EZH2 occurs in several complexes with subtly different composition and these have different HMTase specificities (Kuzmichev et al, 2004). It will be interesting to test whether this region is sufficient to confer Pc-G-mediated silencing in novel contexts, as this is a defining feature of animal PREs.
Figure 8.
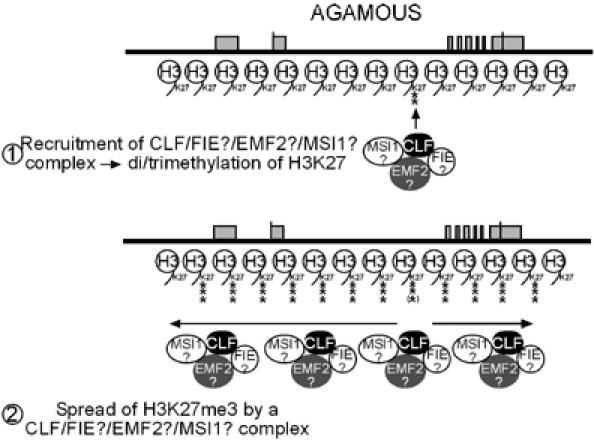
Model of the regulation of AG (and other Pc-G targets) by plant Pc-G proteins and H3K27 methylation. EMF2 and MSI1 are likely part of at least one of the putative complexes, as a reduction in their protein level causes phenotypes similar to clf and lines with reduced FIE levels.
H3K27me3 can spread beyond AG sequences but is not sufficient for silencing
If the dispersed methylation observed at CLF targets represents spreading from a localised initiation point, then methylation should spread beyond target sequences when they are juxtaposed in a novel context. To test this, we analysed histone methylation at the pAG-I∷GUS transgene, previously shown to contain all the cis-acting sequences required for regulation by CLF (Sieburth and Meyerowitz, 1997). Consistent with this, we found that H3K27me3 was recruited to the transgene in wild-type but not clf mutant leaves. Importantly, sequences exclusively present on the transgene (GUS) were enriched for H3K27me3, suggesting that H3K27me3 spreads from the AG intron to the 3′ transgenic sequences. The transgene also contained an NPT gene about 6 kb 5′ to the AG transcription start and this lacked H3K27me3. The gene At4g18950 is located in a similar position 5′ to the endogenous AG locus, and also lacks H3K27me3. Thus, there may be sequences 5′ of AG that prevent spreading into neighbouring genes, similar to insulators described in other systems (Scott et al, 2006). Alternatively, because transgenic plants were selected on the basis of NPT gene expression, this could have selected for lines in which methylation had not spread into the NPT gene. In general, none of the STM and AG 5′ or 3′ flanking genes carried H3K27me3, suggesting either that spreading is short range or that these target genes are flanked by insulators.
The pAG∷GUS transgene lacks important regulatory elements present in the large second AG intron and is de-repressed in leaves (Sieburth and Meyerowitz, 1997). Unexpectedly, we found that this transgene also recruits H3K27me3, which spreads into the GUS sequences. This suggests that H3K27me3 is necessary but not sufficient for Pc-G-mediated silencing. This is also supported by our analysis of the FLC gene. In addition to the changes in H3K27me2 described previously, vernalisation treatments resulted in increased H3K27me3 methylation at FLC. In vrn1 mutants, in which silencing of FLC after vernalisation is impaired, H3K27me3 was only slightly reduced.
Functional significance of H3K27me3 spreading
Although spreading of silencing from heterochromatic sequences into neighbouring sequences is well established in flies and yeast, there is less evidence for this occurring in plants, so it is not clear if the spread of H3K27me3 at Pc-G targets could mediate spreading of silencing to neighbouring sequences. It has been reported that vernalisation treatments cause the genes flanking FLC to be downregulated as well (Finnegan et al, 2004). However, the flanking genes did not acquire H3K27me3, which may account for the fact that, unlike FLC, their silencing is not stably maintained after vernalisation (Finnegan et al, 2005). Thus, vernalisation may induce a widespread remodelling of chromatin at the FLC locus including histone deacetylation (Finnegan et al, 2005), but maintenance of silencing and H3K27me3 is then restricted to FLC. Although for FLC, AG and STM, H3K27me3 was locus specific, other Pc-G targets might occur in clusters with dispersed H3K27me3. The Arabidopsis genome contains many islands of coexpressed, neighbouring genes, which might correspond to regions with a common chromatin structure and epigenetic mark such as H3K27me3 (Ma et al, 2005; Ren et al, 2005; Schmid et al, 2005; Zhan et al, 2006).
Within target loci, dispersed H3K27me3 is necessary for stable silencing. Thus, clf mutants retain localised H3K27me2 at AG, but lack dispersed H3K27me3 and AG silencing. Dispersed H3K27me3 chromatin may help propagation of chromatin modifications through mitosis: if parental nucleosomes are randomly distributed between daughter chromatids during mitosis, there would be a low fidelity of inheritance if only a few nucleosomes carry the modification (Henikoff et al, 2004). However, if many nucleosomes have the modification, fidelity and epigenetic stability would be ensured. It is also important to note that the PRC1, which is thought to confer stable, long-term silencing, is not structurally conserved in plants (Goodrich and Tweedie, 2002). Thus, coverage with H3K27me3, CLF and possibly other Pc-G proteins could manifest stable silencing through development independent of PRC1.
It is also notable that insertion of transposable elements at plant Pc-G target genes is often associated with a loss of silencing. This has been described for numerous gain-of-function alleles of the Antirrhinum AG and STM homologues PLENA and HIRZINA, and for the maize STM homologue KNOTTED (Bradley et al, 1993; Greene et al, 1994; Golz et al, 2002). A puzzling feature is that insertions at many different sites within the gene can all prevent silencing, so that it is unlikely that the failure simply results from disruption of a target site for a repressor. One possible explanation is that insertions prevent spreading of the H3K27me3 methylation at the target, and so disrupt silencing. Plant transposons are associated with distinct epigenetic marks, such as DNA methylation and H3K9me2 methylation (when silenced) or H3Kme2 (when active), that could inhibit spread of H3K27me3 methylation. It is striking that a recent study in mice revealed that large domains of H3K27me3 enrichment are significantly depleted for transposon sequences (Bernstein et al, 2006).
CLF is required persistently for silencing
Although epigenetic changes are by definition self-perpetuating, their propagation through cell division usually requires maintenance machinery—for example, the DNA methyltransferase MET1 is needed for maintenance of mCpG methylation in plants (reviewed by Chan et al, 2005). H3K27 histone methylation persists throughout leaf development, as it was present at AG in mature leaves as well as in young seedlings. In addition, H3K27me3 is necessary for silencing at Pc-G targets, as in all cases where it was lost, silencing was also lost. Using a line with steroid-dependent CLF activity, we found that loss of CLF activity resulted in leaf curling and ectopic AG expression in leaves that were recently emerged at the time that plants were withdrawn from steroid media. Older leaves were less affected, perhaps because cell division had finished by the time that steroid had declined below a critical level in the shifted plants. The results suggest that CLF is required persistently to keep AG silenced in leaves, presumably to restore histone methylation after DNA replication, when it is thought that parental histones and newly synthesised histones are randomly assorted between the two daughter DNA chains (Henikoff et al, 2004; Annunziato, 2005). It is unlikely that CLF itself provides an epigenetic tag, as it is lost from chromosomes during mitosis. Thus, histone methylation itself or other Pc-G proteins may confer mitotically heritable silencing.
In summary, we show that dispersed H3K27me3 is a signature of plant Pc-G target genes. Use of whole genome approaches, such as ChIP on chip, will help clarify the significance of spreading, for example by showing whether Pc-G targets can occur in clusters with common regulation. In addition, transgenic studies will help test whether regions of H3K27me2 correspond with PRE-like elements and whether distinct insulators occur to prevent spreading.
Materials and methods
Plant materials and growth conditions
The clf-81 mutation (Columbia background) was kindly provided by H Tsukaya. Other Pc-G mutants, reporter lines and growth conditions were as described previously (Goodrich et al, 1997; Chanvivattana et al, 2004).
Conditional and epitope-tagged CLF lines
To make the conditional CLF transgene pCLF∷CLF-GR, a 7.5k genomic clone that spanned the CLF locus was modified by site-directed mutagenesis (Quikchange, Stratagene) to introduce BamHI and EcoRI restriction sites immediately 5′ to the stop codon. A BamHI/EcoRI fragment of plasmid pBI-ΔGR (gift of A Lloyd) encoding the ligand binding domain of the rat glucocorticoid steroid receptor (residues 508–795) was then introduced as an in-frame fusion at the C-terminus of CLF. The resulting pCLF∷CLF-GR fusion construct was subcloned into a binary vector conferring glufosinate herbicide resistance in planta. Plant transformation of clf-2/+ heterozygotes by floral dip method gave rise to independent clf-2 pCLF∷CLF GR homozygotes, all of which showed steroid-dependent rescue of the clf mutant phenotype. To make the 35S∷GFP-CLF and 35S∷CLF-GFP constructs, we mutagenised the CLF cDNA clone pCDJ4 as described previously (Goodrich et al, 1997) to introduce restriction sites 5′ to the start and stop codons, respectively. We then introduced the GFP coding sequences from mGFP6 (gift of J Hasselhof) as in-frame fusions and subcloned the GFP-CLF fusion constructs into pART7/pART27 vectors so as to express them under control of CamV 35S promoter. We transformed clf-50/+ heterozygotes by floral dip method and thus derived clf-50 homozygotes that expressed GFP-tagged CLF. At least 10 independent transformants were evaluated for each construct.
Vernalisation treatment
For vernalisation treatments, seeds were surface sterilised, plated on solid MS medium and then kept at 4°C for 40 days. For non-vernalised samples, seeds were similarly treated but kept at 4°C for only 2 days. After moving to long-day conditions (16 h light, 8 h dark, 22°C), plants were grown for 15 days and then harvested.
ChIP and PCR analysis
ChIP was performed exactly as described by Gendrel et al (2002) (ChIPs in Figure 5) or with modifications included from Vanoosthuyse et al (2004) (Figures 4, 6 and 7). Plants were grown on soil or solid MS medium for 10–14 days, and nuclear enrichment and sonication were performed as described by Gendrel et al (2002). Antibodies (2 μg in 100 μl) were incubated for 1–4 h at 4°C with 10 μl of magnetic protein A beads (Dynal) on a roller shaker, which were washed three times with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.0, 167 mM NaCl) before and after incubation with the antibody. After removal of nuclear debris by centrifugation, the sonicated chromatin was diluted 1:10 with ChIP dilution buffer, 1/3–1/4 was added to the antibody-coated beads or beads without antibody (no ab control) and incubated on a roller shaker for 10–15 h at 4°C. Washes were performed as described by Gendrel et al (2002), except that only wash with TE was performed. Elution, reverse crosslinking and purification of the DNA were as described by Gendrel et al (2002). IP DNA was resuspended in 50–100 μl H2O and stored at −80°C.
IP DNA was analysed by duplex PCR using the primers described in Supplementary Table S1. Reactions were performed in 25 μl with 0.5 μl of IP DNA. PCR conditions were as follows: 94°C 2 min, 35 × (94°C 30 s, 60°C 30 s, 72°C 30 s), 72°C 5 min with varying amounts of primers. The amplified DNA was visualised on 2.5% agarose gels stained with ethidium bromide.
In all ChIP experiments, we amplified DNA using two primer pairs in duplex PCR reactions: one pair was specific for the gene of interest (e.g. AG), whereas the other pair was specific for a gene that is not expected to be enriched. Because different primer pairs seldom amplify with equal efficiency in duplex PCR, we compared the relative amounts of the two products amplified from IP DNA against the relative amounts amplified for input (pre-IP) DNA. The ratio reflects the enrichment for a sequence following IP and corrects for any differences in the efficiency of the two primers or of the amounts of chromatin isolated.
Fragment intensity was measured with the program Genetools (Syngene, Cambridge, UK). The relative enrichment was determined as follows: (intensity of band of fragment of interest (in IP)/intensity of control band (in IP; actin or PFK))/(intensity of band of fragment of interest (in input)/intensity of control band (in input; actin or PFK)). Several dilutions of input were used in amplification to reveal whether amplification was in a linear range. If band intensity was saturated owing to a high enrichment, the indicated numbers can give only approximate values. If PCR products were present in the ‘beads' control, intensities of amplifications were subtracted from the sample (IP with specific antibodies) values.
RT–PCR analysis
Total RNA was extracted with Trizol (Life Technologies) from leaves grown under long-day conditions (16 h light). The integrity of the RNA was verified on a gel before RT–PCR. Reactions were performed in 20 μl with 1 μg of RNA using the ImProm-II RT-System (Promega). Primers were designed to span introns or intron/exon borders to avoid amplification of genomic DNA. Primer sequences can be found in Supplementary Table S1 in Supplementary data.
Confocal microscopy
Plants were grown for 7 days on vertical agar plates containing ½ × MS and 1% sucrose. Roots were excised 7–10 days after germination, mounted in water and examined using a Bio-Rad Radiance 2100 confocal microscope (Hemel Hempstead, UK). GFP fluorescence was detected with 500-nm long-pass and 530-nm short-pass filters. Images were exported and treated using ImageJ software.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary data
Acknowledgments
DS was supported by a grant from BBSRC and a fellowship from the Deutsche Forschungsgemeinschaft. Work in JG's laboratory is supported by a BBSRC grant. We thank Ruth Bastow, Josh Mylne and Caroline Dean for help establishing ChIP and providing VRN1:GFP, vrn2-1 and vrn1-2 mutants.
Note added in proof Papp and Muller (Genes Dev 20: 2041–2054) and Kahn et al (J Biol Chem August 2, 2006) recently described dispersed H3K27me3 methylation at Pc-G targets in Drosophila.
References
- Annunziato AT (2005) Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 280: 12065–12068 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T (2002) Histone methylation: dynamic or static? Cell 109: 801–806 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95 [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E, Kovac KA, Jaligot E, Sheldon CC, James PW, Dennis ES (2005) The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J 44: 420–432 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Sheldon CC, Jardinaud F, Peacock WJ, Dennis ES (2004) A cluster of Arabidopsis genes with a coordinate response to an environmental stimulus. Curr Biol 14: 911–916 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang YM, Allis CD (2003) Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475–479 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL (2004) Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Golz JF, Keck EJ, Hudson A (2002) Spontaneous mutations in KNOX genes give rise to a novel floral structure in Antirrhinum. Curr Biol 12: 515–522 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Tweedie S (2002) REMEMBRANCE OF THINGS PAST: chromatin remodeling in plant development. Annu Rev Cell Dev Biol 18: 707–746 [DOI] [PubMed] [Google Scholar]
- Greene B, Walko R, Hake S (1994) Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T, Ahmad K (2004) Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet 20: 320–326 [DOI] [PubMed] [Google Scholar]
- Hong RL, Hamaguchi L, Busch MA, Weigel D (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Hakim O, Ohad N, Fischer RL (2003) From flour to flower: how Polycomb group proteins influence multiple aspects of plant development. Trends Plant Sci 8: 439–445 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F (2006) Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206: 175–183 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL (2001) Polycomb repression of flowering during early plant development. Proc Natl Acad Sci USA 98: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D (2004) Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 14: 183–193 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng XW (2005) Organ-specific expression of Arabidopsis genome during development. Plant Physiol 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J (2005) Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, Jenuwein T, Reuter G (2005) Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J 24: 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Ren XY, Fiers MW, Stiekema WJ, Nap JP (2005) Local coexpression domains of two to four genes in the genome of Arabidopsis. Plant Physiol 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Grossniklaus U (2003) Diverse functions of Polycomb group proteins during plant development. Semin Cell Dev Biol 14: 77–84 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R (2004) Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell 16: 641–653 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R (2004b) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura JM, Paro R (2003) Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell 5: 759–771 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schubert D, Clarenz O, Goodrich J (2005) Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 8: 553–561 [DOI] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF (2006) A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16: 119–129 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Muijrers I, de Wit E, Teunissen H, Talhout W, van Steensel B, van Lohuizen M (2006) Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Valsdottir R, Javerzat JP, Hardwick KG (2004) Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol Cell Biol 24: 9786–9801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS (2004) Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14: 637–646 [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ (2003) Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421: 652–656 [DOI] [PubMed] [Google Scholar]
- Zhan S, Horrocks J, Lukens LN (2006) Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J 45: 347–357 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary data


