Abstract
Fluctuations in base composition appear to be prevalent in Drosophila and mammal genome evolution, but their timescale, genomic breadth, and causes remain obscure. Here, we study base composition evolution within the X chromosomes of Drosophila melanogaster and five of its close relatives. Substitutions were inferred on six extant and two ancestral lineages for 14 near-telomeric and 9 nontelomeric genes. GC content evolution is highly variable both within the genome and within the phylogenetic tree. In the lineages leading to D. yakuba and D. orena, GC content at silent sites has increased rapidly near telomeres, but has decreased in more proximal (nontelomeric) regions. D. orena shows a 17-fold excess of GC-increasing vs. AT-increasing synonymous changes within a small (∼130-kb) region close to the telomeric end. Base composition changes within introns are consistent with changes in mutation patterns, but stronger GC elevation at synonymous sites suggests contributions of natural selection or biased gene conversion. The Drosophila yakuba lineage shows a less extreme elevation of GC content distributed over a wider genetic region (∼1.2 Mb). A lack of change in GC content for most introns within this region suggests a role of natural selection in localized base composition fluctuations.
BASE composition differences are well documented among both prokaryotic and eukaryotic genomes (Sueoka 1964) and prompted early proposals of neutral molecular evolution (i.e., biased base composition driven by asymmetric mutation pressure) (Freese 1962; Sueoka 1962). More recently, the accumulation of comparative sequence data from closely related species has allowed the identification of base composition changes on relatively short evolutionary timescales in mammalian (Bernardi 2000; Duret et al. 2002; Smith and Eyre-Walker 2002; Arndt et al. 2003; Webster et al. 2003; Belle et al. 2004; Hwang and Green 2004) and Drosophila (Akashi 1996; Takano-Shimizu 1999, 2001; Begun and Whitley 2002; Bachtrog 2003; Pérez et al. 2003; Kern and Begun 2005; Akashi et al. 2006) lineages.
Although differences in origination processes (mutation) are often invoked to explain genomewide differences in base composition and synonymous codon usage (e.g., Muto and Osawa 1987; Moran 1996; Rodríguez-Trelles et al. 2000a; Knight et al. 2001; Takano-Shimizu 2001; Chen et al. 2004), a number of studies have argued for lineage-specific fixation biases. Changes in biased gene conversion (Duret et al. 2002; Webster et al. 2003) and the intensity of natural selection (Bernardi 1995; Akashi 1996; Powell et al. 2003) can have widespread effects on base composition within genomes. Translational selection (Grantham et al. 1981), mutability (Singer and Ames 1970), costs of nucleotide synthesis (Rocha and Danchin 2002), and availability of nutrients (Giovannoni et al. 2005) have been proposed as selection pressures promoting GC content variation among genomes.
Recent changes in base composition may be best suited for determining the causes of GC content heterogeneity. Contrasts between functional classes of sites within coding DNA (silent and replacement) as well as between coding and noncoding regions can help to distinguish among the contributions of mutation biases, natural selection, and gene conversion. In addition, comparisons between polymorphic and fixed differences are sensitive to subtle differences in the fitness effects of mutations (Akashi 1999; Bustamante et al. 2001). Finally, associations between base composition changes and patterns of gene expression, replication timing, and recombination rates may prove critical for identifying mechanisms underlying genomic base composition change (e.g., Watanabe et al. 2002). Because expression patterns and recombination rates may be evolutionarily labile, meaningful tests of such associations require data for recent genomic evolution.
Nonstationary base composition may be prevalent in Drosophila genome evolution (Akashi 1996; Takano-Shimizu 1999, 2001; Rodríguez-Trelles et al. 1999, 2000a,b; Powell et al. 2003; Akashi et al. 2006). Studies so far suggest that declines in GC content are common, but Takano-Shimizu (2001) reported striking GC increases within the D. melanogaster species subgroup in the lineages leading to D. yakuba and D. orena. His data included 10 loci near the X chromosome telomere. The lineage leading to D. yakuba showed consistent GC increases in these genes. For the D. orena lineage, three genes showed strong elevations of GC content but the consistency of base composition changes was unclear because the numbers of inferred changes were small for the other genes. Interestingly, our recent analysis revealed strong and consistent declines of GC content in the D. yakuba and D. orena lineages among genes from regions of “normal” recombination from the autosomes and the X chromosome (Akashi et al. 2006). Takano-Shimizu's (2001) study did not include D. teissieri and D. simulans for most loci and employed a tree topology that differs from a strongly supported phylogeny in a more recent study (Ko et al. 2003).
We expanded available sequence data to test for region- and lineage-specific fluctuations in GC content and to identify its cause(s). We extended coding region data for some of the loci investigated by Takano-Shimizu (2001) and sequenced additional loci near the telomere to identify the limits of regional heterogeneity in base composition evolution. The addition of sequences of orthologs from additional D. melanogaster subgroup species doubles the number of lineages on which base composition changes can be assigned (from four to eight). Patterns of base composition evolution in 14 near-telomeric and 9 nontelomeric loci were studied from six D. melanogaster subgroup species: D. melanogaster, D. simulans, D. teissieri, D. yakuba, D. erecta, and D. orena. We reconstructed ancestral states at interior nodes in the species tree and inferred nucleotide changes on lineages leading to six extant species as well as on two ancestral lineages (D. teissieri–D. yakuba and D. erecta–D. orena). Base composition changes were localized both physically (with regard to genomic location) and in time (within the phylogenetic tree). Strong GC content increases appear to be confined to a narrow (∼130-kb) region near the telomeric end of the X chromosome in the D. orena lineage. In the D. yakuba lineage and its ancestral lineage, less extreme increases in GC content occurred in an overlapping, but considerably wider region (∼1.2 Mb). Changes in mutation processes may have contributed to base composition changes, but greater GC elevation in coding regions than within introns supports region- and lineage-specific changes in the fixation probabilities of GC-increasing mutations.
MATERIALS AND METHODS
DNA sequences and Drosophila strains:
We refer to an ∼1.5-Mb region near the tip of the D. melanogaster X chromosome (1A1 to 2B5 on the cytogenetic map) as “near telomeric.” Fourteen near-telomeric genes (6319 aligned codons total) and 9 “nontelomeric” X chromosome genes (4906 codons) are included in this study (see Table 1 for gene names and locations). Orthologs of these genes were compared among six species in the D. melanogaster species subgroup: D. melanogaster, D. simulans, D. yakuba, D. teissieri, D. erecta, and D. orena. Results for the 9 nontelomeric X chromosome loci are from Akashi et al. (2006). For near-telomeric genes, we examined a combination of available data from previous studies, genome projects, and sequences from our laboratory. Parts of the near-telomeric genes cin, y, sc, l(1)sc, ase, l(1)1Bb, EG:171D11.2, su(wa), and sta were examined previously by Takano-Shimizu (1999, 2001). We extended the sequenced regions for some of these genes and added data for D. melanogaster subgroup species that were not included in Takano-Shimizu's studies (see supplemental Table S1 available at http://www.genetics.org/supplemental/). Additional near-telomeric genes [CG2995, l(1)1Bi, CG11403, futsch, and dor] were sequenced to determine the genomic breadth of fluctuations in base composition. These genes were chosen on the basis of their cytogenetic locations, lengths (>400 codons), presence of introns, and low proportions of “simple” or repetitive regions (Wootton and Federhen 1996). The overall sequence length of the coding regions from near-telomeric regions is ∼2.5-fold larger than Takano-Shimizu's (2001) data set and the species sampling was increased to allow comparisons among eight lineages.
TABLE 1.
X chromosome near-telomeric and nontelomeric loci
| Locus | Full name | Codons | Map | Seq. map | Rec. | MCU | No. int | Int bp |
|---|---|---|---|---|---|---|---|---|
| Near telomeric | ||||||||
| CG2995 | 486 | 1A1 | 0.11 | 0.23 | 0.45 | 5 | 278 | |
| cin | cinnamon | 449 | 1A1 | 0.11 | 0.23 | 0.45 | 3 | 224 |
| y | yellow | 441 | 1A5 | 0.22 | 0.33 | 0.38 | ||
| sc | scute | 332 | 1A8 | 0.25 | 0.37 | 0.49 | ||
| l(1)sc | lethal of scute | 255 | 1B1 | 0.27 | 0.38 | 0.61 | ||
| ase | asense | 323 | 1B3 | 0.32 | 0.43 | 0.47 | ||
| l(1)1Bb | Exportin 6 | 613 | 1B4 | 0.33 | 0.44 | 0.64 | 5 | 438 |
| EG:171D11.2 | 467 | 1B5 | 0.35 | 0.46 | 0.51 | 4 | 295 | |
| l(1)1Bi | lethal l(1)1Bi | 578 | 1B12 | 0.49 | 0.60 | 0.57 | 3 | 165 |
| su(wa) | Suppressor of white-apricot | 515 | 1E1 | 0.90 | 0.96 | 0.55 | 5 | 257 |
| CG11403 | 577 | 1E4–5 | 1.09 | 1.13 | 0.56 | 2 | 108 | |
| futsch | futsch | 456 | 2A3 | 1.29 | 1.29 | 0.65 | 5 | 367 |
| sta | stubarista | 270 | 2B1 | 1.34 | 1.33 | 0.82 | 1 | 187 |
| dor | deep orange | 557 | 2B5 | 1.52 | 1.47 | 0.67 | 2 | 103 |
| Sum | 6319 | 35 | 2422 | |||||
| Nontelomeric | ||||||||
| per | period | 618 | 3B2–3 | 2.55 | 2.22 | 0.74 | 3 | 157 |
| Mcm3 | Minichromosome maintenance 3 | 496 | 4F5 | 5.17 | 3.59 | 0.65 | 2 | 72 |
| ND75 | NADH:ubiquinone reductase 75-kDa subunit precursor | 547 | 7E1 | 8.10 | 4.26 | 0.71 | 3 | 149 |
| RpII215 | RNA polymerase II 215kD subunit | 743 | 10C6–7 | 11.41 | 4.19 | 0.58 | 1 | 104 |
| Cyp28c1 | 429 | 10F1 | 11.67 | 4.16 | 0.65 | 4 | 109 | |
| g | garnet | 528 | 12B4–6 | 13.56 | 3.80 | 0.63 | 4 | 152 |
| Fur2 | Furin 2 | 540 | 14C1 | 16.21 | 3.07 | 0.61 | 6 | 278 |
| Zw | Zwischenferment | 517 | 18D13 | 19.50 | 1.92 | 0.82 | 2 | 140 |
| run | runt | 488 | 19E2 | 20.51 | 1.54 | 0.72 | 1 | 187 |
| Sum | 4906 | 26 | 1348 |
Gene symbols and cytogenetic map (map) and sequence map (Seq. map) positions in Drosophila melanogaster are from FlyBase (Drysdale et al. 2005) and Map Viewer of the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=7227&chr=X). Seq. map positions (megabases) are measured from the distal end of the chromosome (Wheeler et al. 2005). Codons are the number of aligned codons. Rec. represents recombination rates (centimorgans per megabase) estimated by fitting a polynomial regression of genetic map on physical map locations; the estimates are labeled “Rp” in Hey and Kliman (2002). Major codon usage (MCU) is the proportion of major codons in a gene. Codon preferences are from Akashi (1995). MCU values are the averages for the six extant D. melanogaster subgroup species. Note that map and Seq. map positions do not point to identical locations on the X chromosome ideograms in Figures 2 and 3. No. int and Int bp represent numbers of introns and numbers of aligned intronic nucleotides, respectively, for each locus.
Drosophila strains sequenced in our laboratory are from the National Drosophila Species Resource Center (Department of Biological Sciences, Bowling Green State University) and the Tucson Stock Center at the University of Arizona. The strains employed were: D. simulans (14021-0251.006), D. teissieri (14021-0257.00), D. yakuba (14021-0261.00), and D. orena (14021-0245.00). D. erecta (S-18) was provided by Michael Ashburner (Department of Genetics, University of Cambridge, Cambridge, United Kingdom).
DNA sequences were obtained using a combination of genomic and “vectorette” PCR followed by automated sequencing using Beckman (Fullerton, CA) CEQ 8800 and ABI Hitachi (San Jose, CA) 3730XL sequencers. Experimental protocols are described in Ko et al. (2003) and are available at http://www.bio.psu.edu/People/Faculty/Akashi/. The coding sequences of each gene were aligned using the CLUSTAL algorithm (Higgins and Sharp 1988) in the MegAlign application [DNASTAR (Madison, WI) software package] and adjusted manually. Codons aligning with gaps in any of the species were excluded from the analysis. Alignments of 35 introns for telomeric genes employed a combination of CLUSTAL and MCALIGN (Keightley and Johnson 2004) with extensive manual adjustment. All alignments are available upon request. Sequences obtained in this study were deposited in GenBank with accession nos. DQ184978–DQ185021.
DNA sequences from the D. yakuba and D. erecta draft genomes:
Sequence sources are provided in supplemental Table S1 available at http://www.genetics.org/supplemental/. The draft genome assembly of D. yakuba (released on April 17, 2004) was obtained from the Washington University School of Medicine Genome Sequencing Center (WUGSC) in St. Louis (http://genome.wustl.edu/projects/yakuba/) and the draft genome assembly of D. erecta produced by Agencourt Bioscience (released on October 28, 2004) was downloaded from Michael Eisen's website at the University of California at Berkeley (http://rana.lbl.gov/drosophila/multipleflies.html). Stand-alone BLAST (version 2.2.9, Altschul et al. 1990) was employed to search for orthologous regions of query sequences from D. melanogaster in each draft genome assembly. Identification of orthologs in these genomes appeared to be unambiguous; the BLAST bit scores of the best-matched sequences were far greater than those of second-best matches (bit scores <200). Regions or contigs containing best matches were extracted from the genome assemblies of D. yakuba and D. erecta. These sequences were annotated manually according to D. melanogaster annotations from FlyBase (Drysdale et al. 2005), using the Sequencher 4.0 application (Gene Codes, Ann Arbor, MI). For sequences from the D. erecta draft genome, bases with low-quality scores were confirmed in sequence traces from the Trace Archive at NCBI (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi).
Ancestral state reconstruction and classification of DNA changes:
We inferred ancestral nucleotides at internal nodes in a phylogeny using the BASEML program (version 2.0k) packaged in PAML (Yang 1997) as described in Akashi et al. (2006). We assume the phylogenetic relationships within the D. melanogaster subgroup given in Ko et al. (2003) (see Figure 1). BASEML takes gene sequences and a tree topology as inputs and employs the HKY85 substitution model (Hasegawa et al. 1985) to determine the joint probabilities of nucleotides at ancestral nodes in an unrooted tree. Base composition parameters are estimated from the extant sequences and likelihoods for parameter values for branch lengths and a transition/transversion rate ratio are maximized over the sequence data given the phylogenetic tree. Parameters are estimated separately for first, second, and third codon positions. This method assumes constancy of parameter values within the gene tree. At each site, probabilities of sets of nucleotides at ancestral nodes are determined given the maximum-likelihood estimates for parameters of the substitution model. Probabilities of ancestral and derived codons were calculated assuming independent evolution at the three nucleotide positions and these probabilities were treated as the counts for codon changes in eight lineages in the D. melanogaster subgroup. Each lineage is referred to by the upper node (e.g., the lineage connecting the common ancestor of D. teissieri and D. yakuba to D. teissieri is referred to as the D. teissieri lineage) and is abbreviated as follows: D. melanogaster (mel), D. simulans (sim), D. teissieri (tei), D. erecta (ere), and D. orena (ore) for the terminal lineages and D. teissieri–D. yakuba (teiyak) and D. erecta–D. orena (ereore) for the two ancestral lineages (note that the lineages ancestral to mel and sim and to teiyak and ereore are not examined because changes are inferred on an unrooted tree). Nucleotide reconstructions using this method can be considerably more reliable than maximum-parsimony inference when codon usage is biased or not at equilibrium (H. Akashi, P. Goel and A. John, unpublished data).
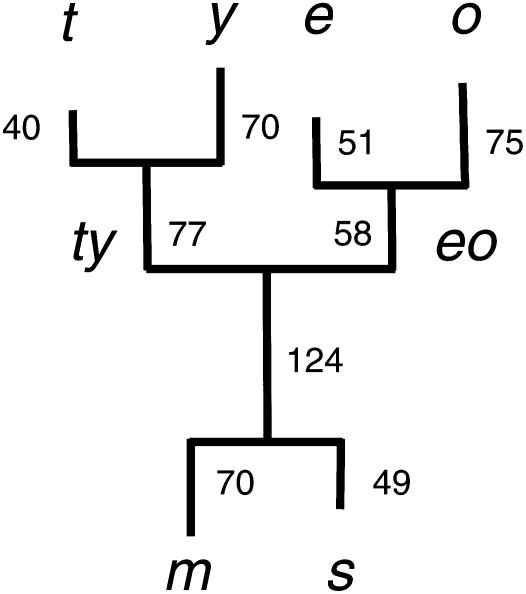
Figure 1.—
Unrooted synonymous distance tree of six species in the D. melanogaster species subgroup. Maximum-likelihood estimates of silent distances, ds, were calculated individually for 23 X chromosome loci using CODEML (F3 × 4 model; Yang 1997) under a codon-based substitution model. Branch lengths are averages across loci (×1000). The assumed tree topology (with a root on the lineage marked “124”) is based on Ko et al. (2003). Abbreviations for extant lineages are: m, D. melanogaster; s, D. simulans; t, D. teissieri; y, D. yakuba; e, D. erecta; and o, D. orena. Ancestral lineages are abbreviated D. teissieri–D. yakuba (ty) and D. erecta–D. orena (eo). Lineages are named according to their most recent node (i.e., ms refers to the internal branch from the internal node of the common ancestor of the subgroup to the internal node of the common ancestor of D. melanogaster and D. simulans).
Changes on each lineage were classified according to ancestral and derived codons. Silent changes were categorized into putative fitness categories under the major codon preference (MCP) model (reviewed in Ikemura 1985; Andersson and Kurland 1990; Sharp et al. 1995; Akashi 2001; Duret 2002). Under this model, synonymous codons vary in the rate and accuracy with which they are translated. A subset of synonymous codons that, in general, are recognized by abundant tRNA isoacceptors confer fitness benefits that increase with translation rates. Thus, major or “preferred” codons are abundant in highly expressed genes. Larger proportions of minor or “unpreferred” codons are maintained in less expressed genes by mutation pressure and genetic drift. Under this model, silent DNA changes can be classified into putative fitness classes (Akashi 1995). Unpreferred to preferred (up) changes are expected to confer fitness benefits especially in highly expressed genes and preferred to unpreferred (pu) changes are expected to be slightly deleterious. Codon preferences followed those in Akashi (1995). Major codon usage (MCU) = no. major/(no. major + no. minor) is used as a measure of codon bias. Scaled differences in the counts of up and pu changes, dup,pu = (up − pu)/(up + pu), are employed to compare the direction and magnitude of MCU differences among lineages. Positive and negative dup,pu indicates increases and decreases in MCU, respectively. Silent, replacement, and intron changes were classified according to their effect on GC content. Nucleotide ambiguity codes were used for abbreviations: W (weak) represents A or T and S (strong) represents G or C. “WS” is employed to represent A or T → G or C changes and G or C → A or T is abbreviated “SW.” dWS,SW = (WS − SW)/(WS + SW) measures the direction and magnitude of departures from equilibrium GC content.
Locations of X chromosome near-telomeric loci in D. yakuba, D. simulans, and D. erecta:
Most of the near-telomeric genes examined are located within the cin-su(wa)-sta region studied by Takano-Shimizu (1999, 2001). In addition to these genes, we examined one more distal (further from the centromere) gene, CG2995 (1A1), and one more proximal gene, dor (2B5), which is 180 kb from sta (2B1) in the D. melanogaster genome [only seven predicted genes are located between them (Wheeler et al. 2005)] (see Table 1 for gene locations). To determine the chromosomal positions of these genes in other D. melanogaster subgroup species, we performed BLAST (Altschul et al. 1990) searches against the D. simulans, D. yakuba, and D. erecta genome assemblies. We employed the consensus genome assembly (from six strains: c1674, md106ts, newc48, sim4, sim6, and w501) of D. simulans released on September 22, 2004. This assembly was obtained from the WUGSC website (http://genome.wustl.edu/projects/simulans/). The recent D. erecta draft genome assembly (released on August 1, 2005) contains longer scaffolds than the previously released version and was employed for determining gene locations.
RESULTS
Locations of X chromosome near-telomeric loci in the D. yakuba, D. simulans, and D. erecta genomes:
BLAST (Altschul et al. 1990) searches against the D. simulans and D. yakuba draft genomes showed that all 14 near-telomeric genes are found close to the distal end of the X chromosome with the same gene order and similar intergenic (base pair) distances as D. melanogaster (Drysdale et al. 2005). These genes were also found in the same scaffold from the recently released D. erecta genome assembly. Detailed results are given in supplemental Table S2 at http://www.genetics.org/supplemental/.
Lineage-specific codon bias evolution in nontelomeric regions of the X chromosome:
Equilibrium codon usage was tested for 9 nontelomeric X chromosome loci in eight lineages in the D. melanogaster subgroup. The patterns are similar to those observed in a set of 19 loci including these genes and autosomal loci from regions of relatively high recombination (Akashi et al. 2006). The mel, yak, and ore terminal lineages and the ereore ancestral lineage show strong declines in MCU (excesses of pu over up changes) and the tei lineage shows an increase in codon bias. These patterns appear to be consistent across genes in each of the five lineages (Table 2; Figure 2). The magnitudes of these departures from equilibrium are considerable: in tei, up outnumber pu by over twofold (dup,pu = 0.34), and the mel, yak, ore, and ereore lineages show greater than twofold excesses of pu (dup,pu < −0.4). The decline of MCU in mel is notable; pu changes outnumber up changes by greater than fivefold (dup,pu = −0.69). Among the other three lineages examined, teiyak shows an overall excess of up changes, but the pattern is not consistent across genes (Wilcoxon signed rank tests, WSR, P = 0.2). The sim lineage shows similar overall numbers of up and pu changes and similar numbers of genes with positive and negative dup,pu. Two genes (ND75 and Cyp28c1) in the ere lineage have experienced large increases in MCU, but there is no consistent trend across loci (Figure 2).
TABLE 2.
Lineage-specific silent changes for X chromosome loci in the D. melanogaster subgroup
| Lineage | Sil tot | up | pu | dup,pu | G | Gene dir | WSR P |
|---|---|---|---|---|---|---|---|
| mel | |||||||
| Near telomeric | 234.4 | 44.6 | 128.9 | −0.49 | 42.6*** | 1/13 | 0.0008** |
| Nontelomeric | 229.1 | 28.2 | 151.1 | −0.69 | 92.3*** | 0/9 | 0.0039** |
| sim | |||||||
| Near telomeric | 187.9 | 43.2 | 90.2 | −0.35 | 16.9*** | 2/12 | 0.0006** |
| Nontelomeric | 143.7 | 54.9 | 54.4 | 0.00 | 0.0 | 4/5 | 0.73 |
| tei | |||||||
| Near telomeric | 161.6 | 67.8 | 50.9 | 0.14 | 2.4 | 9/4 | 0.39 |
| Nontelomeric | 121.0 | 62.9 | 31.3 | 0.34 | 10.8** | 7/2 | 0.020* |
| yak | |||||||
| Near telomeric | 228.3 | 104.6 | 72.5 | 0.18 | 5.8* | 11/3 | 0.17 |
| yak-TRIMCU | 174.9 | 97.0 | 39.6 | 0.42 | 24.8*** | 11/0 | |
| Nontelomeric | 222.5 | 48.3 | 126.4 | −0.45 | 36.1*** | 1/8 | 0.0078* |
| teiyak | |||||||
| Near telomeric | 317.8 | 170.4 | 68.8 | 0.42 | 44.5*** | 9/5 | 0.042* |
| teiyak-TRIMCU | 231.2 | 145.2 | 26.2 | 0.69 | 90.7*** | 8/1 | |
| Nontelomeric | 173.4 | 74.5 | 48.0 | 0.22 | 5.8* | 6/3 | 0.20 |
| ere | |||||||
| Near telomeric | 189.1 | 53.6 | 82.4 | −0.21 | 6.1* | 3/11 | 0.050* |
| Nontelomeric | 176.1 | 77.5 | 48.7 | 0.23 | 6.6* | 4/5 | 0.50 |
| ore | |||||||
| Near telomeric | 302.4 | 179.4 | 63.6 | 0.48 | 57.4*** | 9/5 | 0.10 |
| ore-TRIMCU | 173.9 | 138.9 | 8.0 | 0.89 | 140.9*** | 6/0 | |
| Nontelomeric | 181.5 | 42.5 | 107.8 | −0.43 | 29.2*** | 0/9 | 0.0039** |
| ereore | |||||||
| Near telomeric | 205.5 | 66.0 | 95.1 | −0.18 | 5.3* | 4/10 | 0.10 |
| ereore-TRIMCU | 33.6 | 20.0 | 4.7 | 0.62 | 10.0** | 4/0 | |
| Nontelomeric | 188.6 | 42.9 | 105.3 | −0.42 | 27.0*** | 0/9 | 0.0039** |
Total counts of the preferred (up), unpreferred (pu), and total silent (Sil tot) changes across genes on the near-telomeric (6391 codons) and nontelomeric loci (4906 codons) for each lineage and on the near-telomeric regions of increasing MCU (TRIMCU) in yak, teiyak, ore, and ereore are shown. TRIMCU are defined as yak (cin-futsch, 5006 codons), teiyak (y-CG11403, 4101 codons), ore (y-EG:171D11.2, 2431 codons), and ereore (y-ase, 1351 codons). dup,pu, (up − pu)/(up + pu), indicates degrees of deviation from equal numbers of up and pu changes. The G-values of goodness-of-fit tests for the expected equal numbers of up and pu changes at equilibrium are shown. Gene dir represents the number of genes that show positive/negative dup,pu. Probabilities of Wilcoxon signed-rank test (WSR) are given except for TRIMCU. *P < 0.05, **P < 0.005, ***P < 0.0005. Abbreviations for lineages: mel, D. melanogaster; sim, D. simulans; tei, D. teissieri; yak, D. yakuba; teiyak, D. teissieri–D. yakuba; ere, D. erecta; ore, D. orena; and ereore, D. erecta–D. orena.
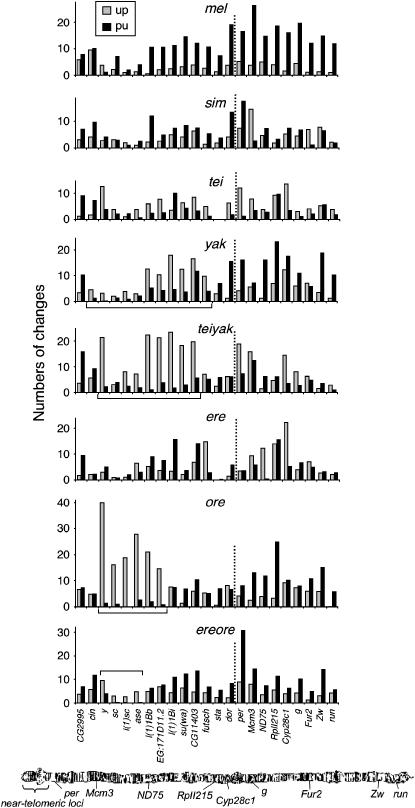
Figure 2.—
Codon bias evolution in near-telomeric and nontelomeric X chromosome regions in the D. melanogaster subgroup. Inferred numbers of preferred (up) and unpreferred (pu) silent changes are shown for 14 near-telomeric and 9 nontelomeric loci (separated by dashed lines). Distal genes from CG2995 to dor are all located within the “near-telomeric” region of the chromosome. The near-telomeric regions of increasing MCU (TRIMCU) in yak, teiyak, ore, and ereore are marked with brackets. Abbreviations for lineages are given in Table 2. The inset on the histogram for mel applies to each graph. The salivary gland X chromosome of D. melanogaster is shown (taken from the NCBI Map Viewer at http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=7227&chr=X) (Wheeler et al. 2005).
Heterogeneity in codon bias evolution between near-telomeric and nontelomeric X chromosome loci:
Codon bias evolution in X chromosome near-telomeric regions deviates strongly from patterns in nontelomeric regions in several lineages. Results are summarized in Table 2 and data from individual genes are provided in supplemental Table S3 at http://www.genetics.org/supplemental/. The strongest difference occurs in a clustered subset of near-telomeric genes in ore. Six loci, from y to EG:171D11.2, show strong excesses of up changes (each of these genes shows at least 10-fold higher up over pu). Across the six genes, up changes outnumber pu changes by >17-fold (138.9 up and 8.0 pu among 2431 codons) in contrast to the >2-fold excess in the opposite direction (excess pu changes) for nontelomeric X chromosome loci (Figure 2) and for autosomal loci (Akashi et al. 2006). Increases in MCU have also occurred in clustered sets of genes in near-telomeric regions in the yak and ereore lineages whereas nontelomeric genes showed decreases in MCU. Compared to patterns in ore, deviations are smaller, but occur across larger regions, in yak (97.0 up and 39.6 pu among 5006 codons for cin to futsch). Despite generally small numbers of changes in ereore, four clustered genes show excesses of up changes (20.0 up and 4.7 pu among 1351 codons for y to ase). Regions that show localized increases of MCU for each lineage are referred to as “near-telomeric regions of increasing MCU” (TRIMCU) (i.e., ore-TRIMCU from y to EG:171D11.2, ereore-TRIMCU from y to ase, and yak-TRIMCU from cin to futsch; also see Table 2 and Figure 2). The criteria used to define these regions were as follows. For the ore, ereore, and yak lineages, the TRIMCU region began with the gene with the highest dup,pu and adjacent genes were included until a gene was found for which pu + up > 8 and dup,pu < 0.1. For the teiyak lineage, where MCU appears to have increased throughout the genome, the cutoff criteria were changed to dup,pu < 0.5. These criteria were determined post hoc to compare intron and coding region changes within TRIMCUs. The ore, ereore, and yak lineages show 17.3-, 4.3-, and 2.5-fold excesses of up changes, respectively, within their TRIMCUs. The teiyak-TRIMCU (from y to CG11403) shows a 5.5-fold excess of up changes compared to a 1.6-fold excess of up changes in nontelomeric loci.
To test more generally for region-specific changes in parameters underlying codon bias evolution, we examined gene regions of similar MCU. Codon usage is generally less biased in the near-telomeric genes than in the nontelomeric loci examined (Table 1). The sensitivity of codon bias to fluctuations in parameter values is dependent on the values prior to the change (Akashi 1996). Uniform changes in selection intensity cause larger magnitudes of change in MCU at codons that were under stronger selection prior to the change (H. Akashi, P. Goel and A. John, unpublished data). For the data examined here, lower MCU (near-telomeric) genes are expected to show smaller changes in codon bias than higher MCU (nontelomeric) genes following genomewide changes in selection intensity (Nes). To control for regional variation in selection intensity, we restricted the analysis to relatively low-bias regions in both near-telomeric and nontelomeric genes. MCU was measured in sliding windows of 100 codons in each gene and codons located in any windows with MCU > 0.75 were excluded from the comparison (see Akashi et al. 2006 for details of the method). The cutoff was set to include sufficient numbers of codons from nontelomeric genes. Low MCU (MCUL) regions consisted of 5943 codons from 13 near-telomeric genes (average MCU = 0.54) and 2897 codons from 8 nontelomeric genes (average MCU = 0.61).
Among the lineages that did not show clusters of genes with increasing MCU (mel, sim, tei, and ere), mel and tei do not show differences in the magnitude of codon bias changes between MCUL regions in near-telomeric and nontelomeric loci (Table 3). sim shows a twofold excess of pu changes in near-telomeric regions (dup,pu = −0.33) and roughly equal numbers of pu and up changes in nontelomeric regions (dup,pu = 0.01), but the difference is only marginally statistically significant (G = 4.9, P = 0.026). ere shows an overall decline in MCU in near-telomeric regions (dup,pu = −0.21) and an overall increase in nontelomeric regions (dup,pu = 0.24). However, these patterns appear to reflect strong gene-specific effects in nontelomeric regions (large MCU increases in ND75 and Cyp28c1; also see Figure 2). yak, ore, and ereore show different directions of departures from equilibrium in TRIMCUs and nontelomeric regions (G = 33.1, 105.6, and 21.0, respectively; P < 10−5). Although both telomeric and nontelomeric regions have increased in GC content in the teiyak lineage, the teiyak-TRIMCU shows a considerably larger excess of up changes than nontelomeric regions (G = 13.6, P = 0.00023).
TABLE 3.
Lineage-specific silent changes for low-MCU regions of X chromosome near-telomeric and nontelomeric loci in the D. melanogaster subgroup
| Near telomeric (t)
|
TRIMCU (tr)
|
Nontelomeric (nt)
|
t vs. nt: | tr vs. nt: | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| up | pu | dup,pu | up | pu | dup,pu | up | pu | dup,pu | G | G | |
| mel | 43.0 | 119.7 | −0.47 | 19.5 | 91.5 | −0.65 | 3.0 | ||||
| sim | 41.3 | 81.6 | −0.33 | 31.5 | 30.8 | 0.01 | 4.9* | ||||
| tei | 65.2 | 50.9 | 0.12 | 40.1 | 22.8 | 0.28 | 1.0 | ||||
| yak | 98.5 | 64.0 | 0.20 | 93.9 | 38.1 | 0.42 | 36.1 | 69.9 | −0.30 | 18.2*** | 33.1*** |
| teiyak | 164.1 | 62.8 | 0.40 | 141.3 | 26.2 | 0.69 | 51.8 | 30.4 | 0.30 | 2.4 | 13.6*** |
| ere | 52.7 | 81.2 | −0.21 | 58.4 | 35.6 | 0.24 | 11.5** | ||||
| ore | 173.7 | 56.5 | 0.51 | 133.8 | 7.9 | 0.89 | 28.2 | 59.5 | −0.36 | 50.1*** | 105.6*** |
| ereore | 61.9 | 87.6 | −0.17 | 20.0 | 4.7 | 0.62 | 22.5 | 55.3 | −0.42 | 3.5 | 21.0*** |
Total counts of preferred (up) and unpreferred (pu) silent changes from near-telomeric (5943 codons), near-telomeric region of increasing MCU (TRIMCU), and nontelomeric (2897 codons) regions for low codon bias regions. Codons located within windows of 100 codons with MCU > 0.75 were eliminated from the analysis (see text for details). Total numbers of codons included in the TRIMCU regions are 4900, 3995, 2325, and 1351 for yak, teiyak, ore, and ereore, respectively. dup,pu, (up − pu)/(up + pu), indicates degrees of deviation from equal numbers of up and pu changes. G-values from 2 × 2 tests of independence for up and pu changes for near-telomeric vs. nontelomeric loci and for TRIMCU vs. nontelomeric loci are given. *P < 0.05, **P < 0.005, ***P < 0.0005. Definitions of TRIMCU regions and abbreviations for lineages are given in Table 2.
Base composition evolution in coding regions and introns:
Base composition evolution was compared between coding and adjacent intronic regions to identify mechanisms underlying region-specific silent DNA evolution. In Drosophila, all major codons are G- or C-ending; patterns of up and pu silent changes are similar to patterns of WS and SW changes (WS and SW changes in each lineage are shown in supplemental Table S4 at http://www.genetics.org/supplemental/). Inferred numbers of WS and SW changes at intronic and coding regions from near-telomeric loci are given in Table 4 for the four lineages (yak, teiyak, ore, and ereore) that show distinct patterns of silent changes in telomeric regions (results for other lineages are provided in supplemental Table S5 at http://www.genetics.org/supplemental/).
TABLE 4.
Base composition evolution among near-telomeric X chromosome loci in the D. melanogaster subgroup
|
yak
|
teiyak
|
ore
|
ereore
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| bp | WS | SW | WS | SW | WS | SW | WS | SW | |
| CG2995 | |||||||||
| Int | 278 | 0.1 | 6.4 | 2.0 | 6.3 | 1.8 | 0.2 | 5.0 | 3.4 |
| Sil | 1458 | 5.6 | 11.2 | 4.2 | 17.5 | 6.1 | 7.4 | 4.2 | 6.9 |
| Rep | 0.0 | 4.0 | 2.0 | 1.0 | 0.0 | 2.0 | 1.0 | 0.0 | |
| cin | |||||||||
| Int | 224 | 4.5 | 6.1 | 4.0 | 2.3 | 4.8 | 0.1 | 2.2 | 7.9 |
| Sil | 1347 | 5.6 | 2.3 | 6.5 | 11.6 | 5.9 | 5.9 | 8.2 | 12.8 |
| Rep | 1.0 | 3.6 | 0.0 | 3.7 | 3.0 | 3.6 | 5.2 | 4.0 | |
| y | |||||||||
| Int | |||||||||
| Sil | 1323 | 3.2 | 0.4 | 23.6 | 4.1 | 43.0 | 1.5 | 10.4 | 4.8 |
| Rep | 0.0 | 1.0 | 0.0 | 0.9 | 2.0 | 0.4 | 0.0 | 1.6 | |
| sc | |||||||||
| Int | |||||||||
| Sil | 996 | 2.1 | 4.8 | 3.2 | 2.7 | 16.3 | 1.1 | 4.9 | 1.8 |
| Rep | 1.0 | 1.8 | 0.0 | 0.0 | 2.0 | 0.0 | 1.6 | 0.0 | |
| l(1)sc | |||||||||
| Int | |||||||||
| Sil | 765 | 3.9 | 0.9 | 9.0 | 2.4 | 20.9 | 0.0 | 2.7 | 0.0 |
| Rep | 0.0 | 0.0 | 1.0 | 2.0 | 3.0 | 0.0 | 1.0 | 0.0 | |
| ase | |||||||||
| Int | |||||||||
| Sil | 969 | 4.0 | 3.2 | 7.7 | 2.2 | 31.2 | 3.0 | 6.6 | 0.8 |
| Rep | 1.8 | 0.0 | 2.0 | 1.0 | 11.0 | 1.0 | 2.2 | 0.2 | |
| l(1)1Bb | |||||||||
| Int | 438 | 18.6 | 10.7 | 14.5 | 6.4 | 15.1 | 7.8 | 17.1 | 18.4 |
| Sil | 1839 | 15.7 | 6.7 | 26.7 | 2.2 | 22.3 | 2.1 | 7.5 | 6.4 |
| Rep | 1.0 | 0.0 | 0.1 | 1.0 | 2.0 | 0.3 | 1.4 | 9.8 | |
| EG:171D11.2 | |||||||||
| Int | 295 | 6.1 | 2.0 | 15.2 | 3.8 | 5.6 | 4.1 | 4.6 | 6.5 |
| Sil | 1401 | 12.7 | 3.7 | 26.9 | 4.1 | 17.7 | 0.7 | 7.1 | 7.3 |
| Rep | 2.0 | 0.0 | 1.4 | 3.0 | 2.3 | 2.0 | 1.0 | 2.9 | |
| l(1)1Bi | |||||||||
| Int | 165 | 4.9 | 1.6 | 4.9 | 4.7 | 2.9 | 2.8 | 8.2 | 6.7 |
| Sil | 1734 | 21.5 | 4.7 | 29.2 | 1.9 | 8.8 | 7.0 | 4.4 | 13.3 |
| Rep | 2.5 | 1.0 | 10.4 | 3.0 | 9.4 | 11.2 | 2.9 | 1.3 | |
| Su(wa) | |||||||||
| Int | 257 | 4.0 | 6.3 | 10.0 | 0.4 | 4.1 | 6.9 | 4.0 | 6.8 |
| Sil | 1545 | 14.7 | 5.1 | 23.7 | 2.1 | 2.8 | 7.7 | 6.5 | 11.9 |
| Rep | 4.5 | 1.0 | 5.1 | 3.0 | 2.0 | 3.0 | 3.0 | 6.8 | |
| CG11403 | |||||||||
| Int | 108 | 2.1 | 5.0 | 4.1 | 3.1 | 1.0 | 1.0 | 0.0 | 1.0 |
| Sil | 1731 | 18.0 | 9.9 | 24.6 | 6.2 | 10.6 | 11.3 | 4.8 | 14.2 |
| Rep | 5.5 | 2.4 | 5.5 | 1.0 | 0.0 | 5.9 | 3.2 | 2.5 | |
| futsch | |||||||||
| Int | 367 | 5.0 | 6.0 | 7.8 | 7.2 | 8.7 | 6.2 | 2.4 | 3.4 |
| Sil | 1368 | 9.9 | 5.3 | 8.9 | 4.6 | 5.8 | 5.4 | 3.8 | 7.2 |
| Rep | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | |
| sta | |||||||||
| Int | 187 | 1.0 | 1.0 | 1.1 | 1.0 | 1.0 | 3.0 | 0.1 | 4.0 |
| Sil | 810 | 4.0 | 8.0 | 1.9 | 7.4 | 1.6 | 7.0 | 1.8 | 6.9 |
| Rep | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| dor | |||||||||
| Int | 103 | 1.0 | 2.0 | 2.6 | 2.4 | 3.9 | 3.3 | 1.2 | 2.5 |
| Sil | 1671 | 1.2 | 17.6 | 7.2 | 6.3 | 10.5 | 7.6 | 4.1 | 6.6 |
| Rep | 0.7 | 1.0 | 0.2 | 1.7 | 1.0 | 5.0 | 1.5 | 1.3 | |
| Sum | |||||||||
| Int | 2422 | 47.1 | 47.0 | 66.1 | 37.5 | 49.1 | 35.4 | 44.8 | 60.5 |
| Sil | 18957 | 122.0 | 83.7 | 203.4 | 75.4 | 203.3 | 67.6 | 77.0 | 100.7 |
| Rep | 21.0 | 21.8 | 27.7 | 22.2 | 37.7 | 34.4 | 24.8 | 30.3 | |
| TRIMCU | |||||||||
| Int | 45.1 | 37.6 | 48.6 | 18.3 | 20.7 | 12.0 | 0.0 | 0.0 | |
| Sil | 111.2 | 46.9 | 174.6 | 27.9 | 151.3 | 8.5 | 24.6 | 7.4 | |
| Rep | 20.3 | 10.8 | 25.4 | 14.9 | 22.3 | 3.7 | 4.7 | 1.7 | |
| Non-TRIMCU | |||||||||
| Int | 2.1 | 9.4 | 17.5 | 19.1 | 28.4 | 23.4 | 44.8 | 60.5 | |
| Sil | 10.8 | 36.8 | 28.8 | 47.4 | 52.0 | 59.1 | 52.4 | 93.3 | |
| Rep | 0.7 | 11.0 | 2.2 | 7.4 | 15.4 | 30.7 | 20.1 | 28.5 | |
Counts of AT → GC (WS) and GC → AT (WS) changes at intronic (Int), silent (Sil), and replacement (Rep) sites for each near-telomeric gene in four D. melanogaster subgroup species are shown. WS and SW changes from the loci located at the near-telomeric regions of increasing major codon usage (TRIMCU) are pooled. Total numbers of WS and SW changes for the near-telomeric loci outside TRIMCU regions (non-TRIMCU) are also shown. Definitions of TRIMCU regions and abbreviations for lineages are given in Table 2. Data from the D. melanogaster, D. simulans, D. teissieri, and D. erecta lineages are given in supplemental Table S5 at http://www.genetics.org/supplemental/.
Pooled numbers of WS and SW changes were compared between coding regions and introns within TRIMCU regions. Among the 14 near-telomeric loci, three of the genes in these regions [sc, l(1)sc, and ase] are intronless and we did not obtain data for the y intron. Within the TRIMCU region in ore, 9 introns (733 bp total) from l(1)1Bb and EG171:D11.2 were examined. These introns show an almost twofold excess of GC-increasing changes (WS/SW = 20.7/12.0), but the GC elevation is much larger (17.9-fold) in adjacent coding regions (G = 20.1, P < 10−5). The pattern is similar for the larger region of TRIMCU in teiyak. Nineteen introns (1263 bp) from five loci examined show a 2.7-fold excess of GC-increasing changes (WS/SW = 48.6/18.3), but the excess is slightly greater (6.3-fold excess of WS changes) in coding regions (G = 6.0, P = 0.015). The yak lineage shows similar numbers of GC-increasing and GC-decreasing changes for pooled intron data (WS/SW = 45.1/37.6) in its TRIMCU region (27 introns from seven loci, 1854 bp total). Evidence for greater GC increases in coding regions is weak (G = 5.8, P = 0.016). However, intron base composition evolution appears to be heterogeneous within the yak-TRIMCU region. Among the seven intron-containing genes in the region, three adjacent genes, l(1)1Bb, EG:171D11.2, and l(1)1Bi, show excesses of GC-increasing intron changes. Introns in the other four loci, cin, su(wa), CG11403, and futsch, experienced larger numbers of SW than WS changes (Table 4). In ereore, intron data are not available from the four genes that showed increases of codon bias.
Replacement changes within TRIMCU regions showed strong biases with respect to GC content. The ore-TRIMCU showed the strongest pattern, a sixfold excess of GC-increasing replacement changes (WS/SW = 22.3/3.7). Near-telomeric genes outside of this region show excess SW changes (WS/SW = 15.4/30.7). yak-, teiyak-, and ereore-TRIMCUs also show excesses of GC-increasing replacement changes: WS/SW = 20.3/10.8, 25.4/14.9, and 4.7/1.7, respectively. Although base composition skew at nonsynonymous sites is often attributed to mutational biases (Lobry 1997; Gu et al. 1998; Singer and Hickey 2000), changes in the efficacy of translational selection can also affect WS/SW ratios for amino acid-altering changes (Akashi 2003).
Nonstationary base composition and evolutionary rates:
Lineage- or region-specific departures from equilibrium MCU are often associated with differences in evolutionary rates (Akashi 1996; Takano 1998; Rodríguez-Trelles et al. 1999, 2000a,b). Ratios of the numbers of synonymous changes in near-telomeric and nontelomeric loci are heterogeneous among the eight lineages examined (2 × 8 contingency table, G = 39.6, P < 10−5; Table 2). Near-telomeric:nontelomeric rates are especially high in GC-elevated TRIMCU regions in teiyak and ore.
We also compared divergence between sister taxa to control for evolutionary distance (Table 5). The most notable difference is 2.7-fold higher synonymous divergence in the ore-TRIMCU region compared with ere (G = 52.8, P < 10−5). yak shows both a smaller GC elevation in its TRIMCU region than ore and a smaller rate difference (30% higher) relative to tei (G = 4.3, P = 0.038). Intron base composition is heterogeneous within the near-telomeric region in yak (discussed above) and intron divergence is ∼2.0-fold higher in yak than in tei (G = 20.2, P < 10−5). For nontelomeric X chromosome loci, mel shows a strong decline of codon bias and higher synonymous divergence than sim, which appears to be close to equilibrium (G = 19.7, P < 10−5). yak shows a decline of codon bias and higher rates of synonymous divergence than tei, which shows consistent increases in MCU (G = 30.4, P < 10−5; Table 5).
TABLE 5.
Molecular distances between sister species pairs in the Drosophila melanogaster subgroup
| Sp pairs | r | G | Gen dir | WSR | |||
|---|---|---|---|---|---|---|---|
| mel | sim | ||||||
| Nontelomeric | Sil | 229.1 | 143.7 | 1.6 | 19.7*** | 8/1 | 0.0078* |
| Near telomeric | 234.4 | 187.9 | 1.2 | 5.1* | 11/3 | 0.030* | |
| Nontelomeric | Rep | 22.7 | 27.0 | −1.2 | 0.4 | 4/4 | 1.00 |
| Near telomeric | 94.5 | 65.6 | 1.4 | 5.2* | 9/4 | 0.27 | |
| Nontelomeric | Int | 41.8 | 46.8 | −1.1 | 0.3 | 3/6 | 0.42 |
| Near telomeric | 93.9 | 81.7 | 1.1 | 0.9 | 6/4 | 0.37 | |
| tei | yak | ||||||
| Nontelomeric | Sil | 121.0 | 222.5 | −1.8 | 30.4*** | 0/9 | 0.0039** |
| Near telomeric | 161.6 | 228.3 | −1.4 | 11.5** | 3/11 | 0.030* | |
| yak-TRIMCU | 138.0 | 174.9 | −1.3 | 4.3* | 3/8 | 0.12 | |
| Nontelomeric | Rep | 26.8 | 19.4 | 1.4 | 1.2 | 3/2 | 0.25 |
| Near telomeric | 35.4 | 52.0 | −1.5 | 3.2 | 2/11 | 0.032* | |
| yak-TRIMCU | 32.1 | 39.3 | −1.2 | 0.7 | 2/8 | 0.083 | |
| Nontelomeric | Int | 44.5 | 70.0 | −1.6 | 5.7* | 4/5 | 0.50 |
| Near telomeric | 59.2 | 118.9 | −2.0 | 20.3*** | 0/10 | 0.0020** | |
| yak-TRIMCU | 50.4 | 102.5 | −2.0 | 18.1*** | 0/7 | 0.016* | |
| ere | ore | ||||||
| Nontelomeric | Sil | 176.1 | 181.5 | −1.0 | 0.1 | 3/6 | 0.65 |
| Near telomeric | 189.1 | 302.4 | −1.6 | 26.3*** | 4/10 | 0.035* | |
| ore-TRIMCU | 63.8 | 173.9 | −2.7 | 52.8*** | 0/6 | 0.031* | |
| Nontelomeric | Rep | 40.3 | 56.4 | −1.4 | 2.7 | 3/5 | 0.36 |
| Near telomeric | 62.6 | 102.5 | −1.6 | 9.7** | 4/10 | 0.15 | |
| ore-TRIMCU | 18.2 | 36.0 | −2.0 | 5.9* | 1/5 | 0.093 | |
| Nontelomeric | Int | 67.8 | 47.1 | 1.4 | 3.7 | 7/2 | 0.039* |
| Near telomeric | 80.4 | 98.1 | −1.2 | 1.7 | 4/6 | 0.23 | |
| ore-TRIMCU | 41.1 | 38.3 | 1.1 | 0.1 | 1/1 | 1.00 | |
Total counts of silent (Sil), replacement (Rep), and intronic (Int) changes from the near-telomeric loci (6391 codons), nontelomeric loci (4906 codons), and near-telomeric regions of increasing MCU (TRIMCU) are shown. Total lengths of aligned introns are 1348, 2422, 1854, and 733 bp for nontelomeric, near-telomeric, yak-TRIMCU, and ore-TRIMCU loci, respectively. Sp pairs, sister species pairs; r, ratio of changes between two sister species; the larger number of changes is used as the numerator in each comparison. Negative signs on the ratios indicate higher values in the right-column species (i.e., sim, yak, and ore) than in the left-column species (e.g., r = mel/sim if mel > sim and r = −sim/mel if sim > mel). The G-values of goodness-of-fit tests for the expected equal numbers of changes at equilibrium are shown. Gene dir represents the number of genes that show positive/negative d. Probabilities of Wilcoxon signed-rank test (WSR) are given. *P < 0.05, **P < 0.005, ***P < 0.0005. Definitions of TRIMCU regions and abbreviations for lineages are given in the Table 2 legend.
DISCUSSION
Local fluctuations in base composition evolution:
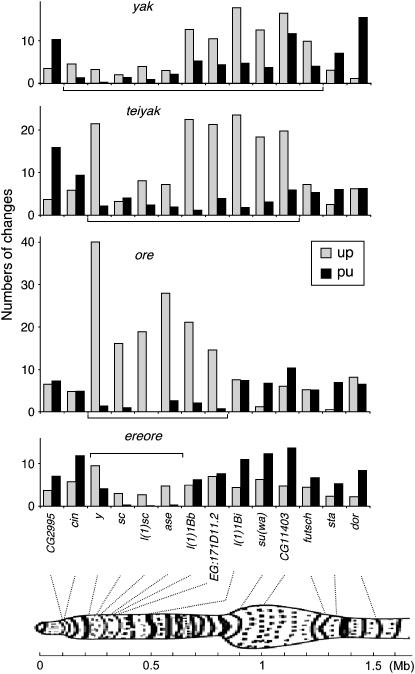
Dramatic region- and lineage-specific base composition fluctuations have occurred near the tip of the X chromosome within the D. melanogaster species subgroup. The most striking pattern is a >15-fold excess of GC-increasing over GC-decreasing changes in the D. orena lineage. This pattern appears to be limited to a small number of genes within an ∼130-kb region [from y (1A1) to EG:171D11.2 (1B5)]. Intron GC increases are consistent with either biased gene conversion, mutation, or selection for intron base composition. However, stronger elevations of GC at silent sites within coding regions than in introns suggest increased relative fixation probabilities of GC-increasing mutations (either biased gene conversion or natural selection). Because fixation probabilities are highly sensitive to selection coefficients (scaled to effective population size), seemingly reasonable parameter fluctuations can give rise to extreme patterns of molecular evolution. For example, an eightfold increase in selection intensity is sufficient to generate a WS:SW ratio of 17 (given u/v = 1.5 and an initial MCU of 0.5). Increases in MCU were observed in similar genomic regions in ore and its ancestral ereore lineage (small numbers of substitutions made the endpoints of the TRIMCU region difficult to determine in ereore). However, the excess of GC-increasing changes and the difference in rates of silent evolution relative to nontelomeric genes are less dramatic in ereore.
Although the D. yakuba lineage has experienced regional increases of GC content, the genomic breadth and magnitude of deviations differ considerably from patterns in ore. Smaller GC increases occurred in 11/14 near-telomeric loci spanning a wider genomic region [∼1.2 Mb, from cin (1A1) to futsch (2A3); see Figure 3]. Within this GC-increasing region, only three of seven intron-containing genes show GC increases within introns. Mutational changes or biased gene conversion may have contributed to GC elevation at these loci, but coding-region-specific GC increases at the other loci are consistent with a contribution of natural selection. The teiyak-TRIMCU appears to be similar in extent to the yak-TRIMCU and also shows stronger GC elevation at synonymous than at intronic sites.
Figure 3.—
Genomic breadths and magnitudes of regional MCU fluctuations near the tip of the X chromosome in the D. yakuba, D. teissieri–D. yakuba, D. orena, and D. erecta–D. orena lineages. The near-telomeric region of the salivary gland chromosome X of D. melanogaster is shown (Wheeler et al. 2005). The near-telomeric regions of increasing MCU (TRIMCU) in yak, teiyak, ore, and ereore are labeled by brackets. Abbreviations for lineages are given in Table 2. The inset on the histogram for ore applies to each graph. Chromosomal positions of loci are indicated by dashed lines.
Increases in the GC content of near-telomeric genes in the yak + teiyak and ore lineages confirm Takano-Shimizu's (2001) findings. However, Takano-Shimizu attributed base composition changes in yak to mutational biases because GC increases were similar in coding and noncoding regions [except for the y gene, where selection was invoked to explain coding-region-specific GC increases (Takano-Shimizu 1999)]. His data did not include sequences from D. teissieri for most of genes investigated and changes were assigned to a pooled yak + teiyak branch. Addition of data from D. teissieri reveals larger excesses of WS changes in the teiyak than in the yak TRIMCU and shows opposing directions of base composition evolution in nontelomeric regions, i.e., GC elevation in teiyak and decline in yak (Figure 2). Similar factors affecting base composition in the near-telomeric regions of the X chromosome may have operated within different contexts (genomewide increases and decreases in MCU, respectively) in these lineages.
Region-specific heterogeneity can complicate inference of the distribution of neutral, adaptive, and slightly deleterious mutations from comparisons of evolutionary rates between regions of high and low recombination (Charlesworth 1994). Munté et al. (2001) found faster rates of silent evolution of the y gene in the D. melanogaster subgroup than in other Drosophila lineages where the gene is located in regions of higher crossing over (within the ananassae subgroup and the obscura group). They attributed elevated divergence to reduced effectiveness of selection against slightly deleterious mutations under low recombination. Our findings suggest that rapid increases in codon bias explain at least some of the rate elevation within the D. melanogaster subgroup.
Our inference of changes in either natural selection or gene conversion in the coding regions of TRIMCU regions assumes uniform changes in parameters across loci and nucleotide classes as well as neutral intron base composition evolution. A number of studies (Mount et al. 1992; Bergman and Kreitman 2001; Halligan et al. 2004; Andolfatto 2005; Haddrill et al. 2005; Marais et al. 2005) have demonstrated functional constraints within Drosophila introns. Inference of mutational biases from intron data requires neutral evolution at variable sites. Strong constraint at a limited number of sites has little impact on our interpretations, but weak selection (perhaps favoring intermediate GC content) or adaptive evolution could lead to errors in attributing the causes of coding region evolution.
Inference of nucleotide changes within introns also depends strongly on sequence alignments. A considerable portion of intron alignments contains gaps and appears questionable (∼14.4% of total 2422 bp of aligned telomeric introns). Alignments appear to be improved when mel and sim are removed from the analysis since both species are more distantly related to the remaining four taxa. Our results remained similar for data excluding ambiguous alignments and for comparisons limited to tei, yak, ere, and ore (data not shown).
Ancestral state reconstructions can be reliable when lineages are short and when molecular evolution proceeds under a process resembling the substitution model employed in ancestral state inference (H. Akashi, P. Goel and A. John, unpublished data). Our ancestral reconstructions employed a stationary HKY85 substitution model. However, base composition evolution within the TRIMCU regions in the ore and teiyak lineages shows both strong departures from equilibrium and elevated rates of evolution. Because the departures from equilibrium are in the same direction, inference errors are likely to underestimate GC elevation in ore and teiyak and overestimate GC declines in ere in shared regions of increasing GC. This occurs because the relative rates of parallel up (in teiyak and ore) and pu (in ere and melsim) changes will be underestimated under a stationary model. Methods of ancestral state inference become critical when base composition has fluctuated strongly in multiple, long lineages. The use of more realistic substitution methods may be desirable in such cases, but parameter-rich models may not be appropriate for detecting gene-specific fluctuations on short lineages (Galtier and Gouy 1998).
Causes of regional fluctuations in base composition evolution:
Although strong patterns of heterogeneity in base composition evolution appear to be clear, the mechanism(s) underlying these fluctuations remain speculative. Takano-Shimizu (1999, 2001) showed ∼10-fold higher rates of recombination at the tip of the X chromosome in D. yakuba and D. erecta strains than in D. melanogaster and D. simulans. Recombination rates in near-telomeric regions may have increased in the ancestral lineage leading to the tei-yak-ere-ore clade (or decreased in the melsim lineage.). Changes in the recombination environment can have both selective and neutral effects on substitution patterns. Increased recombination improves the efficacy of natural selection (Hill and Robertson 1966; Felsenstein 1974; Li 1987; Kliman and Hey 1993; Comeron et al. 1999; McVean and Charlesworth 2000) and enhancing translational selection will elevate GC content in coding regions. In addition, GC-biased mutation and/or gene conversion appear to be correlated with recombination rates in Drosophila (Marais et al. 2001, 2003; Marais and Piganeau 2002). Such processes can elevate GC content across functional classes of nucleotide changes. It is notable that rates of gene conversion appear to be high despite reduced recombination in X chromosome telomeric regions in D. melanogaster (Langley et al. 2000; Andolfatto and Wall 2003; Braverman et al. 2005).
D. melanogaster telomeres consist of arrays of telomeric-specific non-long-terminal-repeat (non-LTR) retrotransposons (HeT-A, TART, and recently identified TAHRE) and are neighbored by long blocks of telomeric-associated complex DNA repeats (TAS) at subtelomeric regions (reviewed in Pardue and DeBaryshe 2003; Biessmann et al. 2005). The existence of homologous HeT-A, TART elements in D. yakuba and D. virilis supports their functional importance in Drosophila (Danilevskaya et al. 1998; Casacuberta and Pardue 2002, 2003a,b). These regions may be essential for protecting chromosomal ends from fusion and degradation owing to incomplete DNA replication at the tips of a linear chromosome (reviewed in Biessmann and Mason 1997; Pardue and DeBaryshe 2003; Biessmann et al. 2005; Melnikova and Georgiev 2005). The length and sequence composition of telomeric and subtelomeric regions vary among chromosomes and among strains of D. melanogaster, possibly reflecting the dynamics between transposition frequency and terminal nucleotide loss (Abad et al. 2004; Melnikova and Georgiev 2005). Interestingly, in D. orena, patterns of quinacrine staining near the telomere of the shared X chromosome arm differ from that in other melanogaster subgroup species and may correlate with densities of repetitive DNA (the D. orena X chromosome contains a large additional arm absent in other species) (Lemeunier et al. 1978).
Distributions and densities of repetitive sequences and/or transposable elements may play a role in heterogeneous base composition evolution. Switches in replication timing have been detected in a transition area between regions differing in GC percentage in the human genome (Tenzen et al. 1997; Watanabe et al. 2002). High densities of Alu clusters, polypurine/polypyrimidine tracts, and repetitive sequences may delay movement of the replication fork in the time-switched area. Mutation patterns may be related to replication timing in mammals (Wolfe et al. 1989). Heterochromatic telomeres generally replicate late in eukaryotic genomes (reviewed in Gilbert 2002). However, a recent analysis in D. melanogaster showed no delays in replication timing for euchromatin located proximal to heterochromatin (Schübeler et al. 2002).
In D. melanogaster telomeric regions, gene activity can be affected by nearby heterochromatic environments (Mikhailovsky et al. 1999; Boivin et al. 2003; Savitsky et al. 2003). Abundances and distributions of repeat elements are also associated with regional changes in gene expression in telomeric regions of the human X chromosome (Lyon 1998; Bailey et al. 2000; Carrel and Willard 2005). Such changes in expression patterns are expected to have a strong impact on the fitness advantage of major codons (reviewed in Akashi 2001).
Fine-scale investigations of nucleotide evolution among closely related species, in combination with greater knowledge of heterochromatin structure/function and its effect on adjacent euchromatic regions, may be critical to identifying patterns and underlying mechanisms of base composition evolution. Genome sequence comparisons among D. melanogaster, D. simulans, D. yakuba, and D. erecta will allow assignments of nucleotide changes to four lineages. However, our analyses suggest that fluctuations of evolutionary processes occur on very short timescales; the results presented here are strongly dependent on inclusion of sequence data from D. teissieri and D. orena. Pooling data from extant/parental lineages or from different chromosomal regions can obscure heterogeneity in genome evolution. In addition, recent changes in base composition evolution allow tests of associations with recombination rates, gene expression patterns, and replication timing in extant taxa and will be critical to advancing our understanding of a fundamental feature of genome evolution.
Acknowledgments
We thank two anonymous reviewers for their valuable suggestions that helped to improve this study. This work was supported by National Science Foundation grant DEB-0521964 to H.A.
References
- Abad, J. P., B. De Pablos, K. Osoegawa, P. J. De Jong, A. Martin-Gallardo et al., 2004. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 21: 1613–1619. [DOI] [PubMed] [Google Scholar]
- Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics 144: 1297–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1999. Inferring the fitness effects of DNA mutations from polymorphism and divergence data: statistical power to detect directional selection under stationarity and free recombination. Genetics 151: 221–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11: 660–666. [DOI] [PubMed] [Google Scholar]
- Akashi, H., 2003. Translational selection and yeast proteome evolution. Genetics 164: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., W.-Y. Ko, S. Piao, A. John, P. Goel et al., 2006. Molecular evolution in the Drosophila melanogaster species subgroup: frequent parameter fluctuations on the timescale of molecular divergence. Genetics 172: 1711–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andersson, S. G., and C. G. Kurland, 1990. Codon preferences in free-living microorganisms. Microbiol. Rev. 54: 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and J. D. Wall, 2003. Linkage disequilibrium patterns across a recombination gradient in African Drosophila melanogaster. Genetics 165: 1289–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt, P. F., D. A. Petrov and T. Hwa, 2003. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 20: 1887–1896. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., 2003. Protein evolution and codon usage bias on the neo-sex chromosomes of Drosophila miranda. Genetics 165: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. A., L. Carrel, A. Chakravarti and E. E. Eichler, 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 97: 6634–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and P. Whitley, 2002. Molecular population genetics of Xdh and the evolution of base composition in Drosophila. Genetics 162: 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle, E. M., L. Duret, N. Galtier and A. Eyre-Walker, 2004. The decline of isochores in mammals: an assessment of the GC content variation along the mammalian phylogeny. J. Mol. Evol. 58: 653–660. [DOI] [PubMed] [Google Scholar]
- Bergman, C. M., and M. Kreitman, 2001. Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11: 1335–1345. [DOI] [PubMed] [Google Scholar]
- Bernardi, G., 1995. The human genome: organization and evolutionary history. Annu. Rev. Genet. 29: 445–476. [DOI] [PubMed] [Google Scholar]
- Bernardi, G., 2000. The compositional evolution of vertebrate genomes. Gene 259: 31–43. [DOI] [PubMed] [Google Scholar]
- Biessmann, H., and J. M. Mason, 1997. Telomere maintenance without telomerase. Chromosoma 106: 63–69. [DOI] [PubMed] [Google Scholar]
- Biessmann, H., S. Prasad, M. F. Walter and J. M. Mason, 2005. Euchromatic and heterochromatic domains at Drosophila telomeres. Biochem. Cell Biol. 83: 477–485. [DOI] [PubMed] [Google Scholar]
- Boivin, A., C. Gally, S. Netter, D. Anxolabehere and S. Ronsseray, 2003. Telomeric-associated sequences of Drosophila recruit polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164: 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman, J. M., B. P. Lazzaro, M. Aguade and C. H. Langley, 2005. DNA sequence polymorphism and divergence at the erect wing and suppressor of sable loci of Drosophila melanogaster and D. simulans. Genetics 170: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C. D., J. Wakeley, S. Sawyer and D. L. Hartl, 2001. Directional selection and the site-frequency spectrum. Genetics 159: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel, L., and H. F. Willard, 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404. [DOI] [PubMed] [Google Scholar]
- Casacuberta, E., and M. L. Pardue, 2002. Coevolution of the telomeric retrotransposons across Drosophila species. Genetics 161: 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta, E., and M. L. Pardue, 2003. a Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc. Natl. Acad. Sci. USA 100: 3363–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta, E., and M. L. Pardue, 2003. b HeT-A elements in Drosophila virilis: retrotransposon telomeres are conserved across the Drosophila genus. Proc. Natl. Acad. Sci. USA 100: 14091–14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63: 213–227. [DOI] [PubMed] [Google Scholar]
- Chen, S. L., W. Lee, A. K. Hottes, L. Shapiro and H. H. Mcadams, 2004. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. USA 101: 3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., M. Kreitman and M. Aguade, 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya, O. N., C. Tan, J. Wong, M. Alibhai and M. L. Pardue, 1998. Unusual features of the Drosophila melanogaster telomere transposable element HeT-A are conserved in Drosophila yakuba telomere elements. Proc. Natl. Acad. Sci. USA 95: 3770–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby and the FlyBase Consortium, 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12: 640–649. [DOI] [PubMed] [Google Scholar]
- Duret, L., M. Semon, G. Piganeau, D. Mouchiroud and N. Galtier, 2002. Vanishing GC-rich isochores in mammalian genomes. Genetics 162: 1837–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1974. Uncorrelated genetic drift of gene frequencies and linkage disequilibrium in some models of linked overdominant polymorphisms. Genet Res. 24: 281–294. [DOI] [PubMed] [Google Scholar]
- Freese, E., 1962. On the evolution of base composition at DNA. J. Theor. Biol. 3: 82–101. [Google Scholar]
- Galtier, N., and M. Gouy, 1998. Inferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol. Biol. Evol. 15: 871–879. [DOI] [PubMed] [Google Scholar]
- Gilbert, D. M., 2002. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14: 377–383. [DOI] [PubMed] [Google Scholar]
- Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin et al., 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Grantham, R., C. Gautier, M. Gouy, M. Jacobzone and R. Mercier, 1981. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 9: r43–r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., D. Hewett-Emmett and W. H. Li, 1998. Directional mutational pressure affects the amino acid composition and hydrophobicity of proteins in bacteria. Genetica 102–103: 383–391. [PubMed] [Google Scholar]
- Haddrill, P. R., B. Charlesworth, D. L. Halligan and P. Andolfatto, 2005. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 6: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan, D. L., A. Eyre-Walker, P. Andolfatto and P. D. Keightley, 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, M., H. Kishino and T. Yano, 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hey, J., and R. M. Kliman, 2002. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics 160: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D. G., and P. M. Sharp, 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244. [DOI] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Hwang, D. G., and P. Green, 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. USA 101: 13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura, T., 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2: 13–34. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and T. Johnson, 2004. MCALIGN: stochastic alignment of noncoding DNA sequences based on an evolutionary model of sequence evolution. Genome Res. 14: 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A. D., and D. J. Begun, 2005. Patterns of polymorphism and divergence from non-coding sequences of D. melanogaster and D. simulans: evidence for non-equilibrium processes. Mol. Biol. Evol. 22: 51–62. [DOI] [PubMed] [Google Scholar]
- Kliman, R. M., and J. Hey, 1993. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol. Biol. Evol. 10: 1239–1258. [DOI] [PubMed] [Google Scholar]
- Knight, R. D., S. J. Freeland and L. F. Landweber, 2001. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2: RESEARCH0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W. Y., R. M. David and H. Akashi, 2003. Molecular phylogeny of the Drosophila melanogaster species subgroup. J. Mol. Evol. 57: 562–573. [DOI] [PubMed] [Google Scholar]
- Langley, C. H., B. P. Lazzaro, W. Phillips, E. Heikkinen and J. M. Braverman, 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier, F., B. Dutrillaux and M. Ashburner, 1978. Relationships within the melanogaster subgroup species subgroup of the genus Drosophila (Sophophora). III. The mitotic chromosomes and quinacrine fluorescent patterns of the polytene chromosomes. Chromosoma 69: 349–361. [Google Scholar]
- Li, W. H., 1987. Models of nearly neutral mutations with particular implications for nonrandom usage of synonymous codons. J. Mol. Evol. 24: 337–345. [DOI] [PubMed] [Google Scholar]
- Lobry, J. R., 1997. Influence of genomic G+C content on average amino-acid composition of proteins from 59 bacterial species. Gene 205: 309–316. [DOI] [PubMed] [Google Scholar]
- Lyon, M. F., 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80: 133–137. [DOI] [PubMed] [Google Scholar]
- Marais, G., and G. Piganeau, 2002. Hill-Robertson interference is a minor determinant of variations in codon bias across Drosophila melanogaster and Caenorhabditis elegans genomes. Mol. Biol. Evol. 19: 1399–1406. [DOI] [PubMed] [Google Scholar]
- Marais, G., D. Mouchiroud and L. Duret, 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. USA 98: 5688–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G., D. Mouchiroud and L. Duret, 2003. Neutral effect of recombination on base composition in Drosophila. Genet. Res. 81: 79–87. [DOI] [PubMed] [Google Scholar]
- Marais, G., P. Nouvellet, P. D. Keightley and B. Charlesworth, 2005. Intron size and exon evolution in Drosophila. Genetics 170: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcvean, G. A., and B. Charlesworth, 2000. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova, L., and P. Georgiev, 2005. Drosophila telomeres: the non-telomerase alternative. Chromosome Res. 13: 431–441. [DOI] [PubMed] [Google Scholar]
- Mikhailovsky, S., T. Belenkaya and P. Georgiev, 1999. Broken chromosomal ends can be elongated by conversion in Drosophila melanogaster. Chromosoma 108: 114–120. [DOI] [PubMed] [Google Scholar]
- Moran, N. A., 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount, S. M., C. Burks, G. Hertz, G. D. Stormo, O. White et al., 1992. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 20: 4255–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munté, A., M. Aguadé and C. Segarra, 2001. Changes in the recombinational environment affect divergence in the yellow gene of Drosophila. Mol. Biol. Evol. 18: 1045–1056. [DOI] [PubMed] [Google Scholar]
- Muto, A., and S. Osawa, 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA 84: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue, M. L., and P. G. Debaryshe, 2003. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 37: 485–511. [DOI] [PubMed] [Google Scholar]
- Pérez, J. A., A. Munté, J. Rozas, C. Segarra and M. Aguadé, 2003. Nucleotide polymorphism in the RpII215 gene region of the insular species Drosophila guanche: reduced efficacy of weak selection on synonymous variation. Mol. Biol. Evol. 20: 1867–1875. [DOI] [PubMed] [Google Scholar]
- Powell, J., E. Sezzi, E. Moriyama, J. Gleason and A. Caccone, 2003. Analysis of a shift in codon usage in Drosophila. J. Mol. Evol. 57: S214–S225. [DOI] [PubMed] [Google Scholar]
- Rocha, E. P., and A. Danchin, 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18: 291–294. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles, F., R. Tarrio and F. J. Ayala, 1999. Switch in codon bias and increased rates of amino acid substitution in the Drosophila saltans species group. Genetics 153: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Trelles, F., R. Tarrio and F. J. Ayala, 2000. a Fluctuating mutation bias and the evolution of base composition in Drosophila. J. Mol. Evol. 50: 1–10. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles, F., R. Tarrio and F. J. Ayala, 2000. b Evidence for a high ancestral GC content in Drosophila. Mol. Biol. Evol. 17: 1710–1717. [DOI] [PubMed] [Google Scholar]
- Savitsky, M., T. Kahn, E. Pomerantseva and P. Georgiev, 2003. Transvection at the end of the truncated chromosome in Drosophila melanogaster. Genetics 163: 1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler, D., D. Scalzo, C. Kooperberg, B. Van Steensel, J. Delrow et al., 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32: 438–442. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., M. Averof, A. T. Lloyd, G. Matassi and J. F. Peden, 1995. DNA sequence evolution: the sounds of silence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 349: 241–247. [DOI] [PubMed] [Google Scholar]
- Singer, C. E., and B. N. Ames, 1970. Sunlight ultraviolet and bacterial DNA base ratios. Science 170: 822–825. [DOI] [PubMed] [Google Scholar]
- Singer, G. A., and D. A. Hickey, 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17: 1581–1588. [DOI] [PubMed] [Google Scholar]
- Smith, N. G., and A. Eyre-Walker, 2002. The compositional evolution of the murid genome. J. Mol. Evol. 55: 197–201. [DOI] [PubMed] [Google Scholar]
- Sueoka, N., 1962. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. USA 48: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka, N., 1964. On the evolution of informational macromolecules, pp. 479–496 in Evolving Genes and Proteins, edited by V. Bryson and H. J. Vogel. Academic Press, New York.
- Takano, T. S., 1998. Rate variation of DNA sequence evolution in the Drosophila lineages. Genetics 149: 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Shimizu, T., 1999. Local recombination and mutation effects on molecular evolution in Drosophila. Genetics 153: 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Shimizu, T., 2001. Local changes in GC/AT substitution biases and in crossover frequencies on Drosophila chromosomes. Mol. Biol. Evol. 18: 606–619. [DOI] [PubMed] [Google Scholar]
- Tenzen, T., T. Yamagata, T. Fukagawa, K. Sugaya, A. Ando et al., 1997. Precise switching of DNA replication timing in the GC content transition area in the human major histocompatibility complex. Mol. Cell. Biol. 17: 4043–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., A. Fujiyama, Y. Ichiba, M. Hattori, T. Yada et al., 2002. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: disease-related genes in timing-switch regions. Hum. Mol. Genet. 11: 13–21. [DOI] [PubMed] [Google Scholar]
- Webster, M. T., N. G. Smith and H. Ellegren, 2003. Compositional evolution of noncoding DNA in the human and chimpanzee genomes. Mol. Biol. Evol. 20: 278–286. [DOI] [PubMed] [Google Scholar]
- Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese et al., 2005. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 33: D39–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., P. M. Sharp and W. H. Li, 1989. Mutation rates differ among regions of the mammalian genome. Nature 337: 283–285. [DOI] [PubMed] [Google Scholar]
- Wootton, J. C., and S. Federhen, 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266: 554–571. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]