Abstract
Pectins are a highly complex family of cell wall polysaccharides. As a result of a lack of specific mutants, it has been difficult to study the biosynthesis of pectins and their role in vivo. We have isolated two allelic mutants, named quasimodo1 (qua1-1 and qua1-2), that are dwarfed and show reduced cell adhesion. Mutant cell walls showed a 25% reduction in galacturonic acid levels compared with the wild type, indicating reduced pectin content, whereas neutral sugars remained unchanged. Immersion immunofluorescence with the JIM5 and JIM7 monoclonal antibodies that recognize homogalacturonan epitopes revealed less labeling of mutant roots compared with the wild type. Both mutants carry a T-DNA insertion in a gene (QUA1) that encodes a putative membrane-bound glycosyltransferase of family 8. We present evidence for the possible involvement of a glycosyltransferase of this family in the synthesis of pectic polysaccharides, suggesting that other members of this large multigene family in Arabidopsis also may be important for pectin biosynthesis. The mutant phenotype is consistent with a central role for pectins in cell adhesion.
INTRODUCTION
Pectins constitute a highly complex family of polysaccharides rich in d-galacturonic acid (GalA) in plant cell walls (Mohnen, 1999; Willats et al., 2001). Three classes of pectins can be distinguished based on two different backbone configurations: homogalacturonan (HGA), rhamnogalacturonan I (RG-I), and rhamnogalacturonan II (RG-II). HGA is a linear homopolymer of 100 to 200 (1→4)-α–linked GalA residues. RG-I consists of a backbone of up to 100 repeats of the disaccharide (1→2)-α-l-rhamnose-(1→4)-α-GalA, which carries side chains. Common side chains contain polymeric (1→4)-β–linked d-galactosyl or (1→5)-α–linked l-arabinosyl residues. RG-II is a highly complex but conserved molecule consisting of four characteristic side chains on a HGA backbone. The side chains contain 11 different sugars; of these, apiose, acetic acid, and 2-keto-3-deoxy-d-manno-octulosonic acid are found only in this molecule. RG-II can dimerize by means of borate diester links through the apiosyl residues (Kobayashi et al., 1996). The resistance of RG-II against hydrolases suggests that the molecule adopts a defined globular three-dimensional structure that may play an important structural role (Vidal et al., 1999).
Pectins form a gel matrix in the wall, the properties of which depend on the degree of cross-linking. HGAs are synthesized in the Golgi apparatus as methylated precursors, which then are secreted into the apoplast, where pectin methyl esterases (PME) can remove the methyl groups (Mohnen, 1999). The deesterified GalA residues form Ca2+ bonds that promote gel formation (Jarvis, 1984). In addition, the energy of the ester bond can be used to create PME-mediated cross-links between HGA chains, contributing to the gelatinization of the matrix (Gelineo-Albersheim et al., 2001).
Given their physical properties, pectins are thought to play an important role in the control of the porosity of the wall. Wall porosity in turn modulates the mobility of expansins and pectin and xyloglucan-modifying enzymes. The expansins promote the disruption of H bonds between xyloglucans and cellulose and promote the sliding apart of cellulose microfibrils in expanding cell walls. These activities control the extensibility of the cell wall and the incorporation of new polymers into the expanding cell wall. In addition to the effect on enzyme mobility, pectins also create an environment for the correct assembly of the cellulose-hemicellulose network, as was shown for synthetic composites (Chanliaud and Gidley, 1999). The ability to massively demethylate HGA creates the possibility of forming a highly Ca2+ cross-linked gel at defined locations in the cell wall. This is observed in the middle lamella, which is thought to control the adhesion between cells.
Transgenic plants that overexpress or underexpress pectin-modifying enzymes have been found to be useful in the study of the in vivo role of various pectins. Using this approach, polygalacturonase and PME were shown to play a role in cell wall breakdown during fruit ripening (Ridley et al., 2001). Inhibition of the expression of a PME in pea roots resulted in reduced root elongation, altered cell morphology, and reduced separation of root cap cells (Wen et al., 1999). A reduction in PME activity was correlated with enhanced elongation in shoot apical tissues in potato (Pilling et al., 2000). Overexpression of a fungal endo-β-1,4-d-galactanase in the cell wall in potato tubers led to a >70% loss of galactan side chains. The absence of a phenotype in these tubers showed that the removal of the galactan side chains in the apoplast did not affect the growth or development of the tuber.
Mutants affected in pectins have been isolated, although in some cases, the mutants were pleiotropic and not specifically pectin mutants. For example, the Arabidopsis emb30 mutants are affected in a gene that presumably functions in the secretory pathway (Shevell et al., 2000). They have a defective cell wall with abnormal localization of pectin but not of xyloglucan and arabinogalactan epitopes. These mutants display a loss of cell cohesion and defects in cell polarity and division. In tomato, the cnr fruit-ripening mutant is affected in the maturation process of HGA in the middle lamella of the fruit pericarp, leading to reduced cell cohesion (Orfila et al., 2001). Conversely, in Arabidopsis quartet mutants, microspores fail to separate during pollen development as a result of the persistence of pectic polysaccharides of the pollen mother cell wall (Rhee and Somerville, 1998). Thus, changes in pectins are correlated with changes in cell cohesion. Analysis of Arabidopsis mur1 mutants, which are deficient in GDP-l-Fuc, showed that their small stature is caused by the absence of fucosyl residues on RG-II, which affects its ability to dimerize through the formation of boron diester cross-links (O'Neill et al., 2001). This finding demonstrated that cell wall pectic organization is important in controlling plant growth. A more detailed knowledge of the function of pectins requires the identification of more mutants with defects in specific pectins.
Little is known about the biosynthesis of pectins. If one assumes that each linkage requires a specific enzyme, at least 53 glycosyltransferases would be required for the synthesis of all pectic polysaccharide structures (Mohnen, 1999). As with other cell wall polysaccharide polymerases, the isolation of the enzymes involved in pectin biosynthesis has been difficult. To date, only HGA synthase and pectin methyl transferase have been partially purified (Mohnen, 1999). The complete Arabidopsis sequence shows the presence of numerous potential glycosyltransferases; however, until now, none of the genes has been assigned to pectin synthesis.
Here, we identify two allelic mutants for a putative glycosyltransferase of family 8 (Campbell et al., 1997). The mutants were dwarfed; their cell walls showed a 25% reduction in GalA content. The reduced pectin content was corroborated further by immunofluorescence experiments using antibodies raised against specific pectic epitopes. These observations suggest that the encoded enzyme may be involved in the synthesis of pectic polysaccharides and raise the possibility that other members of this large multigene family in Arabidopsis may play related roles. Interestingly, cell adhesion in the mutants was reduced strongly, providing genetic evidence that links pectins to cell adhesion.
RESULTS
Dwarfism and Reduced Cell Adhesion Are Caused by Mutations in QUA1
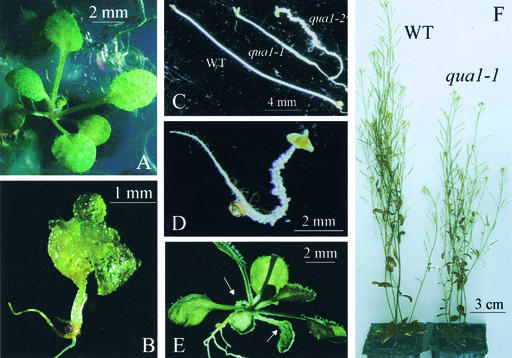
Two T-DNA insertion lines segregated in their progeny mutants with very similar misshapen in vitro phenotypes. These mutants were called quasimodo1 (qua1-1 and qua1-2). When grown in the light, severely affected mutants displayed a dwarf phenotype with an overall vitreous and rough aspect resulting from numerous cells protruding from their cotyledons, leaves, and hypocotyls (Figure 1B), in contrast to wild-type plants (Figure 1A). When grown in the dark, the mutants had shorter hypocotyls than wild-type plants and still exhibited lumps along the hypocotyl, resulting in a distorted shape of the etiolated plants (Figures 1C and 1D). qua1-1 mutants displayed a variable phenotype when grown in the light, ranging from an almost normal morphology (with protuberances barely visible and restricted to cotyledons or hypocotyls; Figure 1E) to a strongly perturbed development in which cells detached from all aerial organs (Figure 1B). The phenotype of the qua1-2 mutants was more pronounced than that of the qua1-1 mutants, with more severely dwarfed and misshapen morphology in both light- and dark-grown conditions than in qua1-1. The qua1-1 mutant phenotype segregated as a monogenic recessive locus (see the segregation analysis of a heterozygous qua1-1/+ plant in Table 1).
Figure 1.
Phenotypes of the qua1-1 and qua1-2 Mutants Grown in Vitro or in the Greenhouse.
(A) Wild-type plant grown in vitro for 17 days in a 16-h-light/8-h-dark regime.
(B) qua1-1 mutant with a severe phenotype grown in vitro for 17 days in a 16-h-light/8-h-dark regime. The qua1-2 mutant displayed the same morphology as the qua1-1 mutant shown here.
(C) Phenotype of the wild type (WT) and qua1-1 and qua1-2 mutants grown for 7 days in vitro in the dark in the absence of sugar.
(D) Close-up view of the qua1-2 mutant grown for 7 days in vitro in the dark.
(E) Phenotype of a qua1-1 mutant that displays an almost wild-type morphology except for the protuberances (arrows) on the cotyledons. This phenotype is never observed for the qua1-2 mutant.
(F) Wild-type and qua1-1 mutant plants grown in the greenhouse.
Table 1.
Genetic Analyses of the qua1-1 and qua1-2 Mutants
| Analysis | Percent Mutants |
n | χ2 |
|---|---|---|---|
| Complementation crosses | |||
| qua1-2/+ × qua1-1/qua1-1 | 52 | 140 | 0.26 (P = 0.61) |
| +/+ × qua1-1/+ | 0 | 50 | |
| +/+ × qua1-2/+ | 0 | 54 | |
| Segregation analyses | |||
| qua1-1/+ selfed | 19 | 398 | 0.8 (P = 0.37) |
| qua1-2/+ selfed | 4 | 370 | 86 (P < 10−20) |
| Ws selfed | 0 | 420 | |
| Analysis of green siliques | |||
| qua1-1/+ selfed | 1.6a | 309 | |
| qua1-2/+ selfed | 14 | 424 | |
| Ws selfed | 2.3 | 418 |
n refers to the size of the population (plantlets for complementation or segregation analyses, and seeds for green siliques analyses). For complementation crosses, χ2 is given for a ratio of 1:1 (wild type: mutant), and for segregation analyses, χ2 is given for a ratio of 3:1 (wild type:mutant). Ws, Wassilewskija.
Values in this column for green siliques represent percent aborted seeds.
By contrast, for qua1-2, a strong distortion in the segregation of the mutant phenotype was observed in the progeny of all heterozygotes tested (4% of mutants among 370 plantlets scored; χ2 = 86, P < 10−20) (Table 1). Analysis of green siliques from heterozygous plants (Table 1) revealed a high frequency of seed abortion for qua1-2/+ plants but not for wild-type or qua1-1/+ plants, which could account for the low number of qua1-2 mutants recovered. Complementation tests showed that both mutations were allelic (Table 1). After transfer to the greenhouse, most of the qua1-1 mutants continued their development, producing a slightly dwarfed plant with an inflorescence stem ∼25% shorter than that of wild-type plants (Figure 1F). The leaf color of mutants grown in soil was duller than that of the wild type; overall, the mutant plants were less rigid. In the case of qua1-2, the sole mutant (of 40 transferred) that survived for a few weeks in the greenhouse displayed an even more severely dwarfed stature than the qua1-1 mutants; it was extremely fragile.
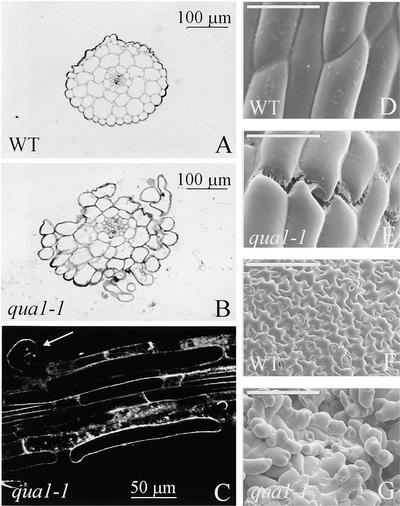
Transverse sections of hypocotyls of the qua1-1 mutants grown in the dark (Figure 2B) or in the light (data not shown) revealed an unaltered number of cell layers and cells compared with the wild type (Figure 2A). However, in contrast to control plants, which showed no lack of adhesion between cells, mutants displayed big gaps, with cells detaching from the hypocotyl (Figure 2B) and from roots (Figure 2C). Scanning electron microscopy confirmed the loss of cell cohesion in qua1-1. In the hypocotyl of light-grown mutants, rupture lines were apparent between epidermal cells in a direction transverse to the axis of elongation (Figure 2E), a situation that was not seen in control plants (Figure 2D). Furthermore, leaf epidermis exhibited numerous gaps surrounded by rounder cells (Figure 2G) than in the control (Figure 2F). Thus, the lumps visible in mutants grown in vitro are attributable to cells detaching from the organ surface.
Figure 2.
Structural Analyses of the Wild Type and qua1-1 Mutants.
(A) and (B) Cross-sections of the hypocotyls of wild-type (WT) and qua1-1 mutant plants stained with methylene blue after 7 days of growth in the dark in vitro in the absence of sugar.
(C) Loss of cell adhesion in roots of the qua1-1 mutant. Roots of 2-week-old plants grown in vitro with a 16-h-light/8-h-dark regime were stained with FM4-64 and visualized with a confocal microscope. The arrow indicates an epidermal cell that is partially detached from the roots.
(D) to (G) Scanning electron micrographs of wild-type and qua1-1 epidermal cells from hypocotyls ([D] and [E]) or leaves ([F] and [G]). Plants were grown for 2 weeks in vitro with a 16-h-light/8-h-dark regime before analysis. Bars = 50 μm ([D] and [E]) or 100 μm ([F] and [G]).
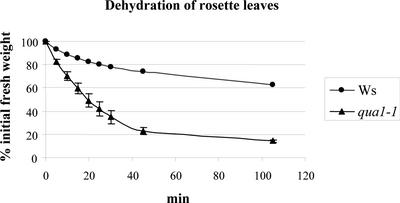
Mutant seedlings that were affected severely were very fragile when grown in vitro and tended to fall apart when manipulated, as a result of the altered cell cohesion. Mutants that displayed an almost normal phenotype (Figure 1E) were more sensitive to dehydration than wild-type plants grown in vitro (data not shown), probably reflecting the altered cell wall structure. Leaves of mutant plants grown in the greenhouse displayed a similar increased propensity to dehydrate compared with wild-type controls (Figure 3). Thus, after 30 min of dehydration under the laminar air unit, qua1-1 leaves were wilted, whereas wild-type leaves appeared to be much less affected.
Figure 3.
Dehydration of Rosette Leaves of Wild-Type (Wassilewskija [Ws]) and qua1-1 Plants Grown in the Greenhouse.
Rosette leaves of 1-month-old plants were collected, and their fresh weight was determined at different time intervals (time 0 is immediately after the transfer of the leaves in the hood). Four independent samples of three plants were used for each genotype. Bars indicate the standard error for each data point. For wild-type plants, the standard error was at most equal to 1.6%; therefore, the bars do not appear in the graph.
Mutant Cell Walls Show Reduced HGA Content
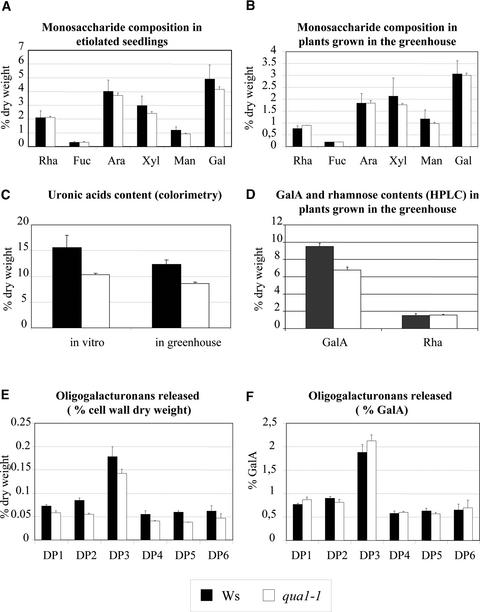
Chemical analyses of cell wall sugars were performed on qua1-1 and wild-type control plants grown in vitro or in the greenhouse. As shown in Figures 4A and 4B, neutral sugars (expressed as a percentage of cell wall dry weight) were present at similar levels in mutant and control plants under both growth conditions. Uronic acid content quantified by colorimetry in qua1-1 mutants, whether grown in vitro or in the greenhouse, was significantly lower than that in the wild type (25% less than in the wild type; Figure 4C). This decrease originated from the lower GalA content in the qua1-1 mutants, as shown by HPLC after methanolysis of cell walls prepared from mutant and control plants grown in the greenhouse (Figure 4D). Again, 25% less GalA was detected in mutant compared with wild-type plant leaves, whereas rhamnose levels remained unchanged. Thus, the qua1-1 mutants were deficient in homogalacturonan.
Figure 4.
Chemical Analyses of the Cell Walls of Wild-Type and qua1-1 Mutant Plants.
(A) and (B) Monosaccharide composition of wild-type (Wassilewskija [Ws]) and qua1-1 mutant cell walls grown in vitro in the dark (A) or in the greenhouse (B). Results are expressed as a percentage of cell wall dry weight. Plants were grown in vitro in the absence of sugar for 7 days in the dark or in the greenhouse for 1 month. Cell walls were prepared from total plant material (in vitro) or from rosette leaves (in the greenhouse). Bars indicate standard errors (n = 3). Similar results were obtained in another independent experiment (data not shown). Rha, rhamnose; Fuc, fucose; Ara, arabinose; Xyl, xylose; Man, mannose; Gal, galactose.
(C) Uronic acid content determined by colorimetry in wild-type and qua1-1 mutant cell walls grown in vitro in the dark or in the greenhouse. Results are expressed as a percentage of cell wall dry weight. Plants were grown in vitro in the absence of sugar for 7 days in the dark or in the greenhouse for 1 month. Cell walls were prepared from total plant material (in vitro) or from rosette leaves (in the greenhouse). Bars indicate standard errors (n = 3). Similar results were obtained in another independent experiment (data not shown).
(D) GalA and rhamnose contents determined by HPLC after methanolysis in the wild type and the qua1-1 mutant. Plants were grown for 1 month in the greenhouse. Results are expressed as a percentage of cell wall dry weight. Bars indicate standard errors (n = 4).
(E) Oligogalacturonans (DP1 to DP6) released by an endopolygalacturonase from wild-type and qua1-1 mutant cell walls. Results are expressed as a percentage of cell wall dry weight. Cell walls were prepared from rosette leaves of plants grown in the greenhouse for 1 month. Bars indicate standard errors (n = 3).
(F) Oligogalacturonans (DP1 to DP6) released by an endopolygalacturonase from wild-type and qua1-1 mutant cell walls. Results are expressed as a percentage of GalA. Cell walls were prepared from rosette leaves of plants grown in the greenhouse for 1 month. Bars indicate standard errors (n = 3).
Endopolygalacturonase treatment of cell walls prepared from greenhouse-grown plants resulted in the release of uronic acids at a lower level from HGA in the qua1-1 mutant than in the wild type (1.2% ± 0.21% of uronic acids released in the mutant versus 2.9% ± 0.1% in the wild type when expressed on a cell wall dry weight basis). Endopolygalacturonase preferentially degrades unmethylesterified HGA (Benen et al., 1999). Hence, these data can reflect a lower HGA content and/or a higher methylesterification of HGA in the mutant compared with the wild type. Analysis of the oligogalacturonans released by endopolygalacturonase revealed a lower amount of oligogalacturonans released in the qua1-1 mutant compared with the wild type for all degrees of polymerization (DP) studied (1 to 6; Figure 4E) when expressed on a cell wall dry weight basis. This decrease in oligogalacturonan release may reflect the lower HGA content of the qua1-1 mutant, because the amount of oligogalacturonans (DP1 to DP6) deriving from endopolygalacturonase treatment was similar in wild-type and mutant plants when expressed as a percentage of GalA (Figure 4F). Thus, data from the endopolygalacturonase treatment of cell walls corroborate the chemical analyses data, which show that the qua1-1 mutant is deficient in HGA.
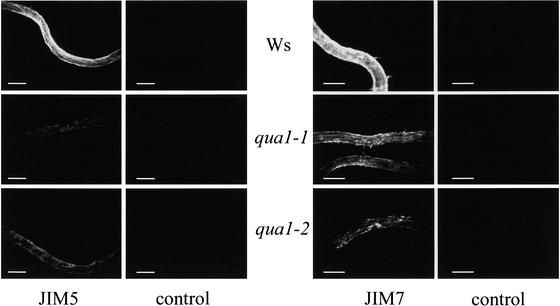
Given the reduced pectin content of mutant cell walls, we investigated the presence of pectic epitopes using specific antibodies. Nonfixed roots of 6-day-old mutant or wild-type plants grown in vitro were analyzed by immersion immunofluorescence using the JIM5 and JIM7 monoclonal antibodies (Willats et al., 2000) that recognize the HGA backbone. JIM5 recognizes poorly methylesterified epitopes, whereas JIM7 preferentially recognizes epitopes with higher degrees of esterification. A lower level of staining of the root was observed using both antibodies in qua1-1 and qua1-2 mutants compared with control plant roots (Figure 5). This finding suggests that either the surface HGA epitopes are specifically lost in the mutants during the sample preparation or qua1-1 and qua1-2 contain less HGA than the wild type, a result that would corroborate the chemical analysis data.
Figure 5.
Immersion Immunofluorescence of Wild-Type, qua1-1, and qua1-2 Roots Incubated with the Monoclonal Antibodies JIM5 and JIM7.
Roots of 6-day-old seedlings grown in vitro in a 16-h-light/8-h-dark regime were incubated with either the JIM5 or the JIM7 monoclonal antibody. After washes and incubation with a secondary fluorescein isothiocyanate–coupled antibody, roots were visualized with a microscope coupled to a charge-coupled device camera. Negative controls refer to roots incubated with the secondary antibody without previous exposure to the JIM5 or the JIM7 antibody. All images were recorded with the same settings (light intensity, filters, and camera settings) for wild-type, mutant, and background control samples. Ws, Wassilewskija. Bars = 200 μm.
Fourier transform infrared (FTIR) microspectroscopy analyses were performed on hypocotyls of etiolated plantlets grown in vitro. This powerful, noninvasive technique based on vibrational spectroscopy allows the quantitative detection of a range of functional groups, leading to a complex “fingerprint” of carbohydrate constituents and their organization from a small area of plant tissue (Chen et al., 1998). When used to analyze numerous cell wall mutants after data compression and hierarchical clustering, FTIR can lead to a highly relevant classification of numerous cell wall mutants, in which cellulose-deficient mutants form a distinct cluster from Fuc-deficient plants, which are clearly different from wild-type controls (G. Mouille, unpublished data). Interestingly, the qua1-1 and qua1-2 mutants form a cluster distinct from wild-type controls and from other cell wall mutants (data available at http://www-biocel.versailles.inra.fr/Anglais/06Paroi.html). These data demonstrate that the qua1-1 and qua1-2 mutants have common structural defects that are different from those of other dwarf mutants.
Determination of the QUA1 Locus
The progeny of plants heterozygous for qua1-1 showed a 3:1 segregation ratio for kanamycin resistance (542 resistant plants and 207 sensitive plants; χ2 = 2.77, P = 0.1), which is consistent with a single T-DNA insertion. DNA gel blot analyses also confirmed the occurrence of a single T-DNA insertion (data not shown). In addition, 100 kanamycin-resistant plants with wild-type phenotype produced in their progeny mutant-looking plants (1:4) and wild-type–looking plants (3:4). This finding suggests that the qua1-1 mutant phenotype is linked tightly to the T-DNA insertion (<1 centimorgan).
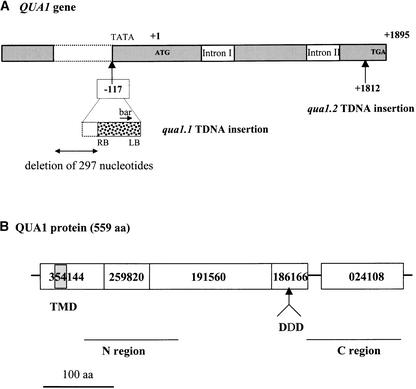
The molecular structure of the T-DNA insertion in qua1-1 was determined using PCR techniques. In this mutant, the T-DNA was found to be inserted in chromosome 3 with a concomitant deletion (297 nucleotides) of both the Arabidopsis genomic sequence and part of the left border of the T-DNA. The insertion site occurred 117 nucleotides upstream from the initiator ATG of a gene that codes for a putative protein of 559 amino acids (Q9LSG3 in EMBL, corresponding to entry At3g25140 from the MJL12 contig; Figure 6A).
Figure 6.
Structures of the QUA1 Gene and the QUA1 Protein.
(A) QUA1 gene structure and insertion sites of the T-DNAs in the qua1-1 and qua1-2 mutants. Numbers indicate the insertion sites of the T-DNAs (+1 being the A of the initiator ATG, +1895 being the A of the stop codon TGA). bar, Basta resistance gene; LB, left border of the T-DNA; RB, right border of the T-DNA; TATA, potential TATA box.
(B) QUA1 protein domains. ProDom protein domains of QUA1 are designated by their numbers. The ProDom 186166 domain is common to the members of the glycosyltransferase family 8 defined by Campbell et al. (1997) and contains the conserved DXD motif (DDD in QUA1). The gray box indicates the predicted transmembrane domain of QUA1. N and C regions indicate the regions of QUA1 that were used to build the phylogenetic trees in Figure 7. aa, amino acids.
cDNAs and ESTs corresponding to this gene were found by screening an Arabidopsis cDNA library (see Methods) and by database searching, respectively, indicating that this genomic sequence is expressed and confirming the introns predicted by annotation. The transcription start site was localized at least −26 nucleotides upstream from the initiator ATG, and the polyadenylation site was localized at 208 nucleotides behind the stop codon. Thus, the T-DNA insertion had occurred upstream of the initiator ATG, presumably close to the transcript 5′ end in qua1-1. A second allele, qua1-2, carrying a T-DNA insertion in QUA1, was isolated using reverse genetics (see Methods). The very similar but more severe phenotype is consistent with the insertion being located within the coding sequence (at position +1812 with respect to the A of the initiator ATG, in the 533rd codon). The identification of two independent alleles with very similar phenotypes that carry T-DNA insertions in the same gene indicates that the mutations in QUA1 are responsible for the mutant phenotype.
QUA1 Encodes a Glycosyltransferase That Is Conserved in Plants and Expressed Ubiquitously in Arabidopsis
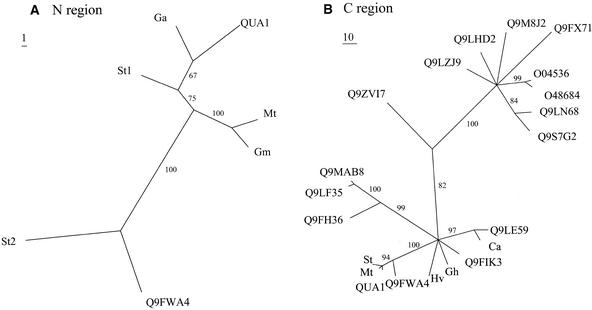
The QUA1 gene encodes a protein that belongs to glycosyltransferase family 8, as defined by Campbell et al. (1997). This is a family of anomeric carbon conformation–retaining glycosyltransferases that share a conserved domain (ProDom domain PD186166; Figure 6B) that includes the DXD motif that is represented in many glycosyltransferase families and is thought to be involved in metal ion binding (Unligil and Rini, 2000). Family 8 comprises proteins from organisms as diverse as plants, yeast, bacteria, and human; the function of some of these proteins is unknown. Several of them have been identified as being involved in the synthesis of lipopolysaccharides (in Neisseria gonorrhoeae and Salmonella typhimurium) or of glycogen (in human, mouse, and yeast). In Arabidopsis, the QUA1 gene belongs to a large multigene family of 37 members (Henrissat et al., 2001), all of which show significant sequence similarity to the protein encoded by the N. gonorrhoeae LGTC gene (Tavares et al., 2000). As noted by Tavares et al. (2000), in the proteins of this family, the sequence of the C terminus is highly conserved in a region that encompasses the ProDom domains PD186166 and PD024108 (Figure 6B) (boxes I to IV defined by Tavares et al. [2000]), whereas the N terminus is much less conserved. Based on a 150–amino acid region in either the N or C terminus of the QUA1 protein (Figure 6B), phylogenetic trees were built by parsimony, including proteins from the Arabidopsis family and potential proteins encoded by ESTs from other plants, such as cotton, tomato, and Medicago. Inspection of the tree built using the C-terminal region (Figure 7B) confirms that the Arabidopsis proteins that belong to glycosyltransferase family 8 are diverse and that the Q9FWA4 protein is the member most closely related to QUA1. However, some proteins encoded by ESTs from plant species other than Arabidopsis (Figures 7A and 7B) clearly are more similar to QUA1 than even Q9FWA4, suggesting that these proteins are orthologs of QUA1. Therefore, QUA1 must have specialized functions in plants.
Figure 7.
Phylogenetic Analysis of QUA1 and Homologous Proteins from Arabidopsis or Potentially Encoded by ESTs from Other Plant Species.
(A) Phylogenetic consensus tree using a 150–amino acid region that lies at the N-terminal end of QUA1 (amino acids 58 to 206). Bootstrap values >50% are shown. Homologous proteins used to build the tree are the Arabidopsis Q9FWA4 protein and potential proteins encoded by ESTs from Gossypium arboreum (Ga), Medicago truncatula (Mt), Glycine max (Gm), and Solanum tuberosum (St).
(B) Phylogenetic consensus tree using a 150–amino acid region that lies at the C-terminal end of QUA1 (amino acids 403 to 544). Bootstrap values >50% are shown. Homologous proteins used to build the tree are the Arabidopsis Q9FH36, Q9LF35, Q9MAB8, Q9FWA4, Q9FIK3, Q9LE59, O48684, O04536, Q9FX71, Q9M8J2, Q9LHD2, and Q9LZJ9 proteins and potential proteins encoded by ESTs or cDNAs from S. tuberosum (St), M. truncatula (Mt), Hordeum vulgare (Hv), Gossypium hirsutum (Gh), and Cicer arietinum (Ca). See Methods.
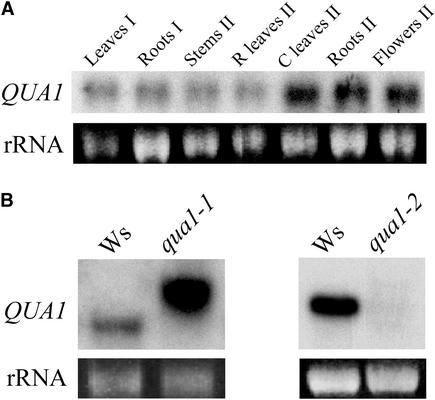
The expression of QUA1 was analyzed in different organs of plants grown in the greenhouse (Figure 8A). QUA1 mRNA was detected in all organs investigated at both the vegetative and floral stages. Levels of QUA1 mRNAs were studied in the wild type and qua1 mutants. Figure 8B shows that in qua1-1, the QUA1 probe cross-hybridized with an mRNA of higher molecular mass than predicted. Reverse transcriptase–mediated PCR analyses indicated that this transcript is a hybrid T-DNA–QUA1 transcript that probably results from read-through of the terminator near the left border of the T-DNA (data not shown). Hybrid transcripts arising from the Basta resistance gene just upstream of the left border of the T-DNA were observed frequently in T-DNA insertion lines and were the result of an inefficient termination of the highly expressed Basta mRNA under the control of the strong 35S promoter of Cauliflower mosaic virus (H. Höfte and D. Bouchez, personal communication). In the qua1-2 mutant, however, no mRNA cross-hybridizing with the QUA1 probe was detected either in plants grown in vitro (Figure 8B) or in the unique plant that survived for a few weeks in the greenhouse (data not shown). This finding indicates that qua1-2 is a null allele.
Figure 8.
RNA Gel Blot Analysis of QUA1 Expression.
(A) QUA1 expression in organs from wild-type plants grown in the greenhouse at the vegetative stage (I) or after floral induction (II). A total of 5 μg of RNAs was loaded in each lane. 25S rRNA fluorescence is shown as a control for lane loading.
(B) QUA1 mRNA levels were analyzed in wild-type (Wassilewskija [Ws]) and mutant (qua1-1 and qua1-2) plants grown in vitro for 2 weeks in a 16-h-light/8-h-dark regime. A total of 5 μg of RNAs was loaded in each lane. 25S rRNA fluorescence is shown as a control for lane loading.
DISCUSSION
The following lines of evidence show that mutations in the QUA1 gene are responsible for the mutant phenotype. First, two independent alleles carrying a T-DNA insertion in the QUA1 gene showed comparable phenotypes involving dwarfism and reduced cell adhesion. Second, for both mutants, the T-DNA insertion was linked closely to the mutant phenotype (<1 centimorgan for qua1-1). Third, in both mutants, the QUA1 mRNA was either absent (qua1-2) or produced primarily as a chimeric transcript from a promoter inside the T-DNA (qua1-1). In the case of qua1-1, the hybrid transcript, which is highly expressed presumably because of the strong 35S promoter present in the T-DNA, may not be translated efficiently, leading to a decreased amount of protein. Indeed, out-of-frame ATGs were found upstream from the initiator ATG in the hybrid transcript, which would hinder the translation of QUA1. In the case of qua1-2, the absence of the transcript could be attributable to the degradation of an incorrectly terminated transcript. In either case, no protein or a greatly reduced amount of QUA1 protein would be expected to be produced. In accordance with this, the phenotype of qua1-2 was stronger than that of qua1-1. The highly variable phenotype associated with the qua1-1 allele and not the qua1-2 allele could arise from the incomplete penetrance of the former allele as a result of the variable translation efficiency of the hybrid transcript. Fourth, among a collection of >200 dwarf mutants, we very rarely observed a reduced cell adhesion phenotype (H. Höfte, unpublished data). The occurrence of a very similar rare phenotype in two independent mutants carrying a single T-DNA insertion in the same gene, along with the close genetic linkage between the kanamycin resistance marker and the mutant phenotype, show that the phenotype is caused by mutations in QUA1. Additional evidence will be provided by the complementation of the mutant phenotype with a construct carrying the QUA1 gene. To date, our attempts to obtain a construct carrying the full-length QUA1 cDNA have been unsuccessful, possibly as a result of the instability of the recombinant clones in Escherichia coli (S. Bouton, unpublished data).
A defect in QUA1 led to reduced GalA content. In addition, the unaltered Rha, Ara, and Gal contents suggest that the qua1-1 mutation did not affect the synthesis of RG-I or its side chains. The simplest explanation for these observations is that qua1-1 affects HGA levels specifically. These results are corroborated by the root-immersion immunofluorescence experiments: labeling by either the JIM5 or the JIM7 antibody, which recognize HGA epitopes, was reduced strongly in the qua1-1 and qua1-2 mutants compared with control plants. Data from endopolygalacturonase treatment of cell walls also support the hypothesis that the qua1-1 mutant is deficient in HGA, although we cannot exclude a qualitative change in HGA (e.g., a higher degree of methylesterification) in this mutant. Furthermore, FTIR studies supported the idea that the qua1 mutants are novel cell wall mutants. Indeed, they form a cluster distinct from cellulose-deficient mutant and from other cell wall mutants with altered neutral sugar composition. Thus, the qua1 mutants represent a novel type of cell wall mutants affected specifically in pectin, and more precisely, HGA, content.
Several important conclusions can be drawn from the fact that the qua1 mutants are defective in pectin. First, the qua1-2 allele, which is the more severe of the two mutant alleles, led to defects in seed formation and thus to the observed lethality of the seeds. This finding suggests that pectins may be required for normal plant development, even at a very early stage. This is similar to the situation with Arabidopsis emb30 mutants, for which it was proposed that the altered pectin localization or synthesis possibly caused the pleiotropic phenotype of these mutants, resulting in the morphological defects of the mutant embryo (Shevell et al., 2000). Second, a unique aspect of the qua1-1 mutant phenotype is the reduced cell adhesion. This feature was observed in different organs, and the severity of the adhesion defect was correlated with the degree of dwarfism. The reduced cell adhesion is what can be expected for a pectin-deficient, in particular, a HGA-deficient, mutant. Indeed, unesterified HGA was shown to accumulate in the middle lamella and is thought to constitute the cement that holds cells together (Ridley et al., 2001). Moreover, in the qua1-1 mutant, leaf epidermal cells were rounded and protruded from the organ surface instead of forming the jigsaw cell shape observed in the wild type. This finding suggests that the adhesion to surrounding cells, in addition to cell wall strength (Burton et al., 2000), is required for the establishment of normal cell shape.
Several pieces of data from sequence analysis are consistent with the hypothesis that QUA1 may be involved in pectin synthesis. The protein encoded by QUA1 is a member of a large family of glycosyltransferases referred to as family 8 (Campbell et al., 1997). Analysis of QUA1 using the TMHMM transmembrane domain detection program (Sonnhammer et al., 1998) shows a predicted transmembrane domain near the N terminus. QUA1 is expected to behave as a type II membrane-anchored protein with a short cytosolic N terminus. This configuration is characteristic of glycosyltransferases localized in the Golgi apparatus, the site of synthesis of noncellulosic polysaccharides. This family is represented by 37 members in Arabidopsis that are predicted to retain the anomeric carbon conformation. Therefore, they may be involved in the synthesis of α-linked glycosidic linkages. This activity is consistent with a possible role of QUA1 in the synthesis of pectic polysaccharides. Furthermore, the presumed orthologs of QUA1 from Gossypium arboreum and Gossypium hirsutum are expressed in elongating fibers at a time when pectins are synthesized actively (7 to 10 days after anthesis), consistent with a possible role for this protein in pectin synthesis.
The complex structure of pectin requires the action of least 53 different enzymes (Mohnen, 1999). The closest related protein in Arabidopsis (Q9FWA4) shows 66% identity to QUA1 (79% homology). ESTs were found for both proteins that may have partially redundant roles in HGA synthesis. This could account in part for the recovery of qua1-2 mutants, albeit at a low frequency, in the progeny of heterozygous qua1-2/+ plants. Conversely, QUA1 and Q9FWA4 may have specialized roles, because, despite their similarity, each gene has a more closely related ortholog in other plant species. The demonstration of a possible role for QUA1 in the synthesis of pectins opens up the interesting possibility that other members of family 8 also may be involved in the synthesis of pectic polysaccharides. The functions of these other family members now can be investigated using reverse genetics combined with their expression in a heterologous host. In addition, screening for plantlets that show cell adhesion defects or that belong to the same FTIR cluster as qua1 mutants may allow the isolation of novel mutants defective in pectin. Indeed, we recently identified mutants that display a phenotype very similar to that of the qua1 mutants (S. Bouton, unpublished data). They cluster with the qua1 mutants after FTIR analyses (see http://www-biocel.versailles.inra.fr/Anglais/06Paroi.html) and yet belong to a different complementation group (S. Bouton, unpublished data).
However, we cannot exclude a more indirect role of QUA1 in pectin synthesis. Indeed, mutants have been isolated that display defects in pectins without being affected directly in pectin synthesis genes. For example, the Arabidopsis korrigan mutants affected in a membrane-bound endo-1,4-β-glucanase and plants deficient in cellulose contain higher amounts of pectins (Burton et al., 2000; His et al., 2001). Similarly, Arabidopsis cyt1 mutants contain fivefold less cellulose than the wild type while displaying an anomalous distribution of unesterified pectins (Lukowitz et al., 2001). However, these mutants are affected in a Man-1-P guanylyltransferase gene that generates the GDP-Man necessary for N-glycosylation and for Man/Fuc incorporation into cell walls. Thus, whether QUA1 is involved directly in pectin synthesis or not awaits the biochemical assessment of QUA1 glycosyltransferase activity. Doong and Mohnen (1998) recently described the solubilization and partial purification of a glycosyltransferase (α-1,4-galacturonosyltransferase) involved in the synthesis of HGA in tobacco. Interestingly, the enzyme preparation incubated with UDP-GalA was able to add a single GalA residue onto O-4 of the nonreducing end of oligomeric HGA acceptors but did not show processivity, in contrast to what was observed in intact pea Golgi or tobacco membrane preparations (Doong et al., 1995; Sterling et al., 2001). The authors speculated that full processivity requires either specific cofactors or substrate modifications or that the enzyme preparation has only a priming activity and other enzymes are required for the polymerization reaction. It will be interesting to determine whether QUA1 expressed in a heterologous system has one or the other activity.
Finally, the qua1 mutants were less affected than the Arabidopsis emb30 mutants (Shevell et al., 2000) while displaying pectin deficiency. The analysis of qua1 mutants, whether or not they are affected in a gene involved directly in pectin synthesis, will contribute to the further understanding of the role of pectin in cell cohesion and plant development.
METHODS
Plant Lines and Growth Conditions
The two T-DNA lines, qua1-1 and qua1-2, of Arabidopsis thaliana (both ecotype Wassilewskija) were isolated either during the systematic screening of the Versailles T-DNA insertion lines (qua1-1) or by reverse genetics (qua1-2) using the QUA1F3 oligonucleotide (5′-ACTATGGCACTCTTCCTCTTCACTCTCT-3′) that is specific to the QUA1 gene and the T-DNA–hybridizing oligonucleotide tag5 (5′-CTACAAATTGCCTTTTCTTATCGA-3′) on 37 hyperpools of DNA extracted from a total of 42,624 T-DNA lines, as described by Filleur et al. (2001). Sequencing of the amplified fragment led to the accurate determination of the T-DNA insertion site. Wild-type plants were control Wassilewskija plants.
Seeds were sown in vitro on a medium derived from that described by Estelle and Somerville (1987) containing 9 mM nitrate and 0.5% glucose or in the absence of sugar when mentioned. When plants were grown in vitro for cell wall analysis, sugar was omitted to avoid interference with subsequent analyses. After 2 days at 4°C, plates were transferred to a growth chamber for 6 to 17 days with a day/night regime of 16 h of light (150 μmol·m−2·s−1) at 20°C and 8 h of dark at 15°C. To measure d-galacturonic acid and neutral sugar contents, plates were placed for 4 h in the light and then transferred to the dark for 7 days before harvest.
When grown in the greenhouse, plants were watered with a nutrient solution (Coïc and Lesaint, 1975) and harvested ∼4 weeks after germination, at the vegetative stage, for dehydration experiments.
Dehydration Experiments
For dehydration experiments, rosettes of plants grown in the greenhouse were harvested and placed in a horizontal laminar air-flow unit (ESI Flufrance, Wissous, France) with pressure set at 200 Pa. Dehydration was followed by weighing four independent samples of three plants each per genotype at different times after the onset of the dehydration experiment.
Microscopy
Sections of hypocotyls of plantlets grown in vitro were obtained after fixing the samples in 2.5% glutaraldehyde in Hepes buffer and embedding in Historesin (Leica Microsystems, Rueil-Malmaison, France) according to the manufacturer's instructions. Sections 5 μm thick were cut on a Jung RM 2055 microtome (Leica), stained with methylene blue, and examined using a Nikon Microphot FXA microscope (Tokyo, Japan). Roots of 2-week-old plants grown in vitro under 16 h of white light per day were stained using the dye FM4-64 (Molecular Probes, Eugene, OR) and visualized with a Leica TCS-NT confocal microscope (Leica Microsystems, Heidelberg, Germany). Scanning electron micrographs were obtained as described by Desnos et al. (1996) on plantlets grown for 2 weeks in vitro under a 16-h-light/8-h-dark regime.
Chemical Analyses of Cell Walls
Cell walls were prepared from three or four independent samples of plants grown in vitro in the dark in the absence of sugar or from pooled rosette leaves of plants grown in the soil. Approximately 100 mg of plants was extracted in 1 mL of 96% ethanol and left to boil for 20 min. Samples then were rinsed five times for 30 min in 70% ethanol, followed by two washes in 96% ethanol and two washes in acetone. Subsequently, they were air-dried overnight at 40°C over Drierite (Grosseron, Nantes, France) under vacuum to remove all residual traces of humidity.
Identification and quantification of neutral sugars via gas-liquid chromatography was performed after sulfuric acid degradation (Hoebler et al., 1989). Insoluble samples were dispersed in 72% sulfuric acid for 30 min at 25°C before dilution to 1 M and hydrolysis (2 h at 100°C). For calibration, monosaccharide solutions as well as inositol (as an internal standard) were used.
Uronic acids were quantified by colorimetry using the automated m-phenyl-phenol-sulfuric acid method without tetraborate (Thibault, 1979). Galacturonic acid and rhamnose also were quantified by HPLC after methanolysis (Quemener and Thibault, 1990). In all cases, relative sugar composition or d-galacturonic acid content values were calculated on a cell wall dry weight percentage basis.
Oligogalacturonans were solubilized from cell walls using purified endopolygalacturonase from Aspergillus niger (lot 00801; Megazyme, Bray, Ireland). Cell walls were suspended (10 mg dry weight/1.5 mL) in 100 mM sodium acetate, pH 4, containing 0.02% thimerosal. Endopolygalacturonase (0.2 units) was added or not (blank), and the suspension was incubated for 40 h at 30°C under shaking. Endopolygalacturonase (0.2 units) was added again at 16 and 24 h of digestion. After incubation, the suspension was centrifuged at 9000 rpm for 10 min, and the supernatant was filtered through 0.45-μm filters (Millex; Millipore, St. Quentin-en-Yvelines, France) and boiled for 5 min. The uronic acid content of supernatant corresponding to solubilized oligogalacturonans was measured by colorimetry. Oligogalacturonans (degree of polymerization [DP] 1 to 6) were determined and quantified by high-performance anion-exchange chromatography using commercial standards (mono-, di-, and trigalacturonic acids) and purified oligogalacturonates from DP4 to DP6. A total of 30 μL of supernatant was injected on a Dionex analytical Carbopac PA-1 column (4 × 250 mm) equipped with a Carbopac PA-1 guard column (4 × 50 mm) under alkaline conditions (Sunnyvale, CA). Twenty percent of 500 mM NaOH was run during the 100-min analysis. NaOAc (1 M) was ramped from 0 to 20% for the first 40 min, from 20 to 50%, 50 to 70%, and 70 to 80% every 10 min until 70 min, and then ramped and stabilized to 80% for 5 min. The column then was reequilibrated for 25 min under the initial conditions (20% of 500 mM NaOH). High-performance anion-exchange chromatography was performed using a Waters (Milford, MA) 626 pump equipped with a Waters 600S controller, a Waters 717 autosampler, and a pulse amperometric detector (EC 2000; Thermo Separation Products, San Jose, CA). Borwin chromatography software (JMBS Developments, Le Fontanil, France) was used to both control chromatography and process the data. The mobile phases were degassed with helium to prevent the absorption of carbon dioxide and transformation to carbonate.
Fourier Transform Infrared Microspectroscopy of Plants Grown in Vitro
Fourier transform infrared microspectroscopy was performed as described by G. Mouille, M. Leconte, S. Robin, and H. Höfte (unpublished data) on hypocotyls of 4-day-old etiolated plants grown in vitro in the dark without sugar. Hypocotyls were pressed between two barium fluoride slides, rinsed in distilled water for 2 min, and dried at 37°C for 20 min. An area of 50 × 50 μm in the middle of the hypocotyls was selected for spectral collection on a ThermoNicolet Nexus (Nicolet Instrument, Madison, WI) spectrometer with a Continuum microscope accessory. Fifty interferograms were collected in transmission mode with an 8-cm−1 resolution and coadded to improve the signal-to-noise ratio of the spectrum. Spectra baseline correction, area normalization, data compression, and hierarchical clustering were performed as described by Mouille et al. (unpublished data).
Immersion Immunofluorescence
Immersion immunofluorescence was performed on plantlets that were grown in vitro for 6 days. Plantlets were picked from the culture medium, rinsed briefly, and then immersed for 1 h in 20 mM Hepes, pH 7.2, containing 5% (w/v) skim milk powder and monoclonal antibody (JIM5 or JIM7) (Knox et al., 1990; Willats et al., 2000) diluted 10-fold. After three rinses in 20 mM Hepes, plantlets were immersed for 1 h in 20 mM Hepes containing 5% (w/v) skim milk powder and the secondary antibody (anti-rat antibody conjugated to fluorescein isothiocyanate [Sigma reference F6257]) diluted 100-fold. After three rinses in 20 mM Hepes, plantlets were mounted in Citifluor antifade agent (Agar Scientific, Saclay, France) and observed with a Leica DMR microscope coupled to a charge-coupled device camera (XC-77CE; Sony, Tokyo, Japan) using a fluorescein isothiocyanate filter with an excitation filter set at 480 ± 20 nm and an emission filter set at 527 ± 15 nm. As a control for background fluorescence, plantlets were incubated in the presence of the secondary antibody without previous incubation with the JIM5 or JIM7 monoclonal antibody. All images were recorded with the same settings (light intensity, filters, and camera settings) for wild-type, mutant, and background control samples.
Molecular Analyses
The QUA1 cDNA clones were isolated by screening an Arabidopsis cDNA library (Minet et al., 1992) with a probe obtained by PCR walking (see below). Subcloning of the NotI fragment containing the cDNA into the NotI site of the pBluescript KS− vector (Stratagene) yielded the KSqua1 plasmid that was used to generate the QUA1 probe after digestion by NotI. For DNA gel blot analyses, the T-DNA left and right border probes were obtained after digesting the pGKB5 plasmid (Bouchez et al., 1993) with KpnI (left border) or SstI and EcoRV (right border).
RNA gel blot analyses were performed on 5 to 10 μg of total RNAs extracted with guanidinium hydrochloride (Logemann et al., 1987). Blots were made using Amersham Nylon N membranes, and hybridization was performed as recommended by the manufacturer using 32P-labeled probes (Pharmacia).
PCR walking was performed on DNA extracted from the qua1-1 mutant after digestion with EcoRV, essentially as described by Devic et al. (1997). The amplified fragment obtained after the successive nested PCR procedures was subcloned and sequenced. The right border of the T-DNA insertion was amplified using the QUA1-specific oligonucleotide 5′-CGTTCTGATATCCGAAATAAACATGTCTATTCA-3′ and the T-DNA right border–specific nucleotide 5′-CTGATACCAGACGTTGCCCGCATAA-3′ and then sequenced.
Reverse transcription was performed on 2.5 μg of total RNA treated with DNase using reverse transcriptase from Moloney murine leukemia virus (Invitrogen, Groningen, The Netherlands) as recommended by the manufacturer in a total volume of 20 μL. An aliquot (0.5 μL) of this reaction was used for subsequent PCR procedures using the Perkin-Elmer Taq enzyme in the supplied buffer (final concentration of 1.5 mM MgCl2, 200 μM deoxynucleotide triphosphate, and 400 nM of each oligonucleotide in a total volume of 25 μL with 2.5 units of Taq).
Phylogenetic Analyses
Database searching for ESTs encoding proteins homologous with QUA1 was performed with the BLAST algorithm (Altschul et al., 1990). Parsimony trees were generated from an alignment of the conserved regions with the Clustal W 1.8 program (Thompson et al., 1994) and the heuristic search algorithm of the PAUP 3.1 program (Swofford, 1991). Bootstrap values indicate the number of times that a particular node was found in trees generated from 1000 replicates.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences described in this article are as follows: Figure 7A, ESTs from Gossypium arboreum (BG445089), Medicago truncatula (BG648310), Glycine max (BE661625), and Solanum tuberosum (BG889490 and BG351005); Figure 7B, ESTs or cDNAs from S. tuberosum (BG888409), M. truncatula (BI272689), Hordeum vulgare (BG343589), G. hirsutum (AI725536), and Cicer arietinum (AJ276420).
Acknowledgments
We thank F. Vedele for encouraging this research project and C. Bellini for the initial isolation of the qua1-1 mutant. Our thanks also to E. Aletti, J. Kronenberger, O. Grandjean, and A.-M. Jaunet for help with the root immunofluorescence experiments, sections, fluorescence microscope, and scanning electron microscopy imaging, respectively. We are grateful to C. Meyer, H.-T. Truong, and K. Whitley for correcting the manuscript. This work was supported in part by the European Union fifth framework contracts BIO4 CT97-2231 and Europectin.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004259.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Benen, J.A.E., Kester, H.C.M., and Visser, J. (1999). Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur. J. Biochem. 259, 577–585. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Burton, R.A., Gibeaut, D.M., Bacic, A., Findlay, K., Roberts, K., Hamilton, A., Baulcombe, D.C., and Fincher, G.B. (2000). Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, J.A., Davies, G.J., Bulone, V., and Henrissat, B. (1997). A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326, 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanliaud, E., and Gidley, M.J. (1999). In vitro synthesis and properties of pectin/Acetobacter xylinus cellulose composites. Plant J. 20, 25–35. [DOI] [PubMed] [Google Scholar]
- Chen, L., Carpita, N.C., Reiter, W.-D., Wilson, R.H., Jeffries, C., and McCann, M.C. (1998). A rapid method to screen for cell-wall mutants using discrimination analysis of Fourier transform infrared spectra. Plant J. 16, 385–392. [DOI] [PubMed] [Google Scholar]
- Coïc, Y., and Lesaint, C. (1975). La nutrition minérale et en eau des plantes en horticulture avancée. Doc. Tech. Commerc. Potasses Alsace 23, 1–22. [Google Scholar]
- Desnos, T., Orbovic, V., Bellini, C., Kronenberger, K., Caboche, M., Traas, J., and Höfte, H. (1996). Procuste 1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively, in dark- and light-grown Arabidopsis seedlings. Development 122, 683–693. [DOI] [PubMed] [Google Scholar]
- Devic, M., Albert, S., Delseny, M., and Roscoe, T.J. (1997). Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem. 35, 331–339. [Google Scholar]
- Doong, R.L., Liljebjelke, K., Fralish, G., Kumar, A., and Mohnen, D. (1995). Cell free synthesis of pectin: Identification and partial characterization of polygalacturonate 4-α-galacturonosyltransferase and its products from membrane preparations of tobacco (Nicotiana tabacum L. cv Samsum) cell suspension cultures. Plant Physiol. 109, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong, R.L., and Mohnen, D. (1998). Solubilization and characterization of a galacturonosyltransferase that synthesizes the pectic polysaccharide homogalacturonan. Plant J. 13, 363–374. [Google Scholar]
- Estelle, M.A., and Somerville, C.R. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Filleur, S., Dorbe, M.-F., Cerezo, M., Orsel, M., Granier, F., Gojon, A., and Daniel-Vedele, F. (2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489, 220–224. [DOI] [PubMed] [Google Scholar]
- Gelineo-Albersheim, I., Darvill, A., and Davies, G.J. (2001). Structural studies of pectin. In 9th International Cell Wall Meeting. (Toulouse, France), p. 183.
- Henrissat, B., Coutinho, P.M., and Davies, G.J. (2001). A census of carbohydrate-active enzyme in the genome of Arabidopsis thaliana. Plant Mol. Biol. 47, 55–72. [PubMed] [Google Scholar]
- His, I., Driouich, A., Nicol, F., Jauneau, A., and Höfte, H. (2001). Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-beta-glucanase. Planta 212, 348–358. [DOI] [PubMed] [Google Scholar]
- Hoebler, C., Barry, J.L., David, A., and Delort-Laval, J. (1989). Rapid acid hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Jarvis, M.C. (1984). Structure and properties of pectin gels in plant cell walls. Plant Cell Environ. 7, 153–164. [Google Scholar]
- Knox, J.P., Linstead, P.J., King, J., Cooper, C., and Roberts, K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181, 512–521. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M., Matoh, T., and Azuma, J. (1996). Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 110, 1017–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to the requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98, 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.-E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Mohnen, D. (1999). Biosynthesis of pectins and galactomannans. In Comprehensive Natural Products Chemistry, Vol. 3, D. Barton, K. Nakanishi, and O. Meth-Cohn, eds (Amsterdam: Elsevier), pp. 497–527.
- O'Neill, M.A., Eberhard, S., Albersheim, P., and Darvill, A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294, 846–849. [DOI] [PubMed] [Google Scholar]
- Orfila, C., Seymour, G.B., Willats, W.G., Huxham, I.M., Jarvis, M.C., Dover, C.J., Thompson, A.J., and Knox, J.P. (2001). Altered middle lamella homogalacturonan and disrupted deposition of (1-5)-alpha-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol. 126, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling, J., Willmitzer, L., and Fisahn, J. (2000). Expression of a Petunia inflata pectin methylesterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta 210, 391–399. [DOI] [PubMed] [Google Scholar]
- Quemener, B., and Thibault, J.F. (1990). Assessment of methanolysis for the determination of sugars in pectins. Carbohydr. Res. 206, 277–287. [DOI] [PubMed] [Google Scholar]
- Rhee, S.Y., and Somerville, C.R. (1998). Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 15, 79–88. [DOI] [PubMed] [Google Scholar]
- Ridley, B.L., O'Neill, M.A., and Mohnen, D. (2001). Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967. [DOI] [PubMed] [Google Scholar]
- Shevell, D.E., Kunkel, T., and Chua, N.H. (2000). Cell wall alterations in the Arabidopsis emb30 mutant. Plant Cell 12, 2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. In Sixth International Conference on Intelligent Systems for Molecular Biology, J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen, eds (Menlo Park, CA: AAAI Press), pp. 175–182. [PubMed]
- Sterling, J.D., Quigley, H., Orellana, A., and Mohnen, D. (2001). The catalytic site of the pectin biosynthetic enzyme α-1-4 galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol. 127, 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (1991). Phylogenetic Analysis Using Parsimony (PAUP) Version 3.1. (Champaign, IL: Illinois Natural History Survey).
- Tavares, R., Aubourg, S., Lecharny, A., and Kreis, M. (2000). Organization and structural evolution of four multigene families in Arabidopsis thaliana: AtLCAD, AtLGT, AtMYST and AtHD-GL2. Plant Mol. Biol. 42, 703–717. [DOI] [PubMed] [Google Scholar]
- Thibault, J.F. (1979). Automatisation du dosage des substances pectiques par la méthode au méta-hydroxydiphenyl. Lebensm. Wiss. Technol. 12, 247–251. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unligil, U.M., and Rini, J.M. (2000). Glycosyltransferase structure and mechanism. Curr. Opin. Struct. Biol. 10, 510–517. [DOI] [PubMed] [Google Scholar]
- Vidal, S., Salmon, J.M., Williams, P., and Pellerin, P. (1999). Penicillium daleae, a soil fungus able to degrade rhamnogalacturonan II, a complex pectic polysaccharide. Enzyme Microbiol. Technol. 24, 283–290. [Google Scholar]
- Wen, F., Zhu, Y., and Hawes, M.C. (1999). Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11, 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats, W.G.T., Limberg, G., Bucholt, H.C., van Alebeek, G.-J., Benen, J., Christensen, T.M.I.E., Visser, J., Voragen, A.G.J., Mikkelsen, J.D., and Knox, J.P. (2000). Analysis of pectic epitopes recognized by conventional and phage display monoclonal antibodies using defined oligosaccharides and enzymatic degradation. Carbohydr. Res. 327, 309–320. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., McCartney, L., Mackie, W., and Knox, J.P. (2001). Pectin: Cell biology and prospects for functional analysis. Plant Mol. Biol. 47, 9–27. [PubMed] [Google Scholar]