Abstract
Recruiting matrix proteins with a peroxisomal targeting signal type 2 (PTS2) to the peroxisomal membrane requires species-specific factors. In Saccharomyces cerevisiae, the PTS2 receptor Pex7p acts in concert with the redundant Pex18p/Pex21p, whereas in Yarrowia lipolytica, Pex20p might unite the function of both S. cerevisiae peroxins. Herein, the genome of the filamentous fungus Neurospora crassa was analyzed for peroxin-encoding genes. We identified a set of 18 peroxins that resembles that of Y. lipolytica rather than that of S. cerevisiae. Interestingly, proteins homologous to both S. cerevisiae Pex7p and Y. lipolytica Pex20p exist in N. crassa. We report on the isolation of these PTS2-specific peroxins and demonstrate that NcPex20p can substitute for S. cerevisiae Pex18p/Pex21p, but not for ScPex7p. Like Pex18p, NcPex20p did not bind PTS2 protein or the docking proteins in the absence of ScPex7p. Rather, NcPex20p was required before docking to form an import-competent complex of cargo-loaded PTS2 receptors. NcPex7p did not functionally replace yeast Pex7p, probably because the N. crassa PTS2 receptor failed to associate with Pex18p/Pex21p. However, once NcPex7p and NcPex20p had been coexpressed, it proved possible to replace yeast Pex7p. Pex20p and Pex18p/Pex21p are therefore true orthologues, both of which are in need of Pex7p for PTS2 protein import.
INTRODUCTION
Peroxisome biogenesis has been studied in various model organisms ranging from yeast to human (for recent reviews, see Holroyd and Erdmann, 2001; Purdue and Lazarow, 2001; Sacksteder and Gould, 2000; Subramani et al., 2000). This has led to the identification of a number of PEX genes that are specifically implicated in that process. Many of the corresponding gene products, the so-called peroxins, are involved in peroxisomal matrix protein import, which can take place via two pathways, the first of which is dependent on the type 1 peroxisomal targeting signal (PTS1), and the second on the type 2 (PTS2). The cognate receptors for the PTS1 and the PTS2 signal are Pex5p and Pex7p, respectively, both of which bind their cargo proteins in the cytosol. On docking of the cargo-loaded receptors to the peroxisomal membrane, the two pathways seem to merge, because the peroxins of the docking complex, Pex14p and Pex13p, are mandatory for both routes.
Although several peroxins have been identified in all organisms analyzed, some of them seem to be species specific. One example is the class of proteins required to direct PTS2 cargo proteins to the peroxisomal membrane. In mammals, the PTS2 signal-recognition factor Pex7p acts in concert with Pex5pL, the long isoform of the PTS1 receptor (Braverman et al., 1998; Otera et al., 2000), whereas Saccharomyces cerevisiae uses Pex7p and the redundant Pex18p and Pex21p for that purpose (Purdue et al., 1998). In the case of Yarrowia lipolytica, which generally seems to carry a number of unique peroxins, Pex20p is essential for PTS2-dependent protein import (Titorenko et al., 1998), whereas a Pex7p orthologue has not been found in that organism.
On the other hand, evidence for a more conserved function of these divergent factors has recently emerged. The insertion within Pex5pL that is encoded by an additional exon and necessary for the Pex5pL interaction with Pex7p shows similarity to a region within Pex18p and Pex21p that is also involved in contacting Pex7p (Dodt et al., 2001; Otera et al., 2002). Even Pex20p contains such a Pex7p-binding domain and somewhat unexpectedly, has been shown to partially complement an S. cerevisiae pex18 Δpex21Δ mutant (Einwächter et al., 2001). However, Pex20p might have acquired functions in addition to those of Pex18p/Pex21p, because only YlPex20p 1) directly binds endogenous thiolase (Titorenko et al., 1998), 2) interacts with the S. cerevisiae docking proteins Pex13p and Pex14p in the absence of Pex7p (Einwächter et al., 2001; Stein et al., 2002), and 3) directly binds intraperoxisomal YlPex8p (Smith and Rachubinski, 2001). Pex20p might therefore compensate a possible absence of Pex7p in Y. lipolytica, although the lack of a completed Y. lipolytica genome sequence still leaves room for a Pex7p in that species.
The filamentous fungus Neursospora crassa has a long history as a model organism for addressing both genetic and biochemical questions. Although S. cerevisiae has proved to be a more tractable organism for many purposes, the release of the complete genome sequence of N. crassa might lay the foundation for a revival in Neurospora research, including research on peroxisome biogenesis. N. crassa harbors at least two compartments of the microbody family: 1) glyoxysomes, which house the fatty acid β-oxidation enzymes and two key enzymes of the glyoxylate cycle (Kionka and Kunau, 1985); and 2) the Woronin body, whose main function is to reseal membrane lesions and whose predominant matrix enzyme, Hex1p, contains a PTS1 (Jedd and Chua, 2000; Tenney et al., 2000). The peroxisomal marker enzyme catalase does not colocalize with either organelle and might thus be present in yet another microbody subclass (Thieringer and Kunau, 1991).
Herein, we report the identification of N. crassa peroxins and the isolation of both Pex7p and Pex20p from this fungus. We expressed the two proteins in S. cerevisiae and analyzed their ability to complement the yeast PTS2-specific counterparts. We provide evidence that Pex20p, just like Pex18p/Pex21p, only functions in a complex with Pex7p and discuss these results in terms of a mechanism for the early steps in PTS2 protein import that is conserved among species.
MATERIALS AND METHODS
Strains, Media, and General Methods
The Escherichia coli strain DH5α was used for all plasmid amplifications and isolations. The S. cerevisiae strains used in this study are listed in Table 1. Standard and oleic acid-containing media were prepared as described previously (Stein et al., 2002). Induction of CUP1 promoter-dependent myc-Pex7p was achieved by adding 25 mg/l CuSO4 to the medium. Standard recombinant DNA techniques were performed essentially as described previously (Sambrook et al., 1989).
Table 1.
S. cerevisiae strains used
| S. cerevisiae strain | Description | Source or reference |
|---|---|---|
| UTL-7A | MATα leu2-3, 112 ura3-52 trp1 | (Erdmann et al., 1989) |
| HF7c | MATa ura3-52 his3-200lys2-801 ade2-101 trp1-901 ssleu2-3/112 gal4-542 gal80-538 LYS2∷GAL1-HIS3 URA3∷(GAL4 17-mers)3-CYC1-LacZ | (Feilotter et al., 1994) |
| UTL-7A pex7Δ | pex7Δ∷LEU2 | (Marzioch et al., 1994) |
| UTL-7A pex14Δ | pex14Δ∷LEU2 | (Albertini et al., 1997) |
| HF7c pex7Δ | pex7Δ∷LEU2 | (Stein et al., 2002) |
| yKat36 | UTL-7Apex18Δ∷kanMX4 pex21Δ∷loxP | (Stein et al., 2002) |
| yKat110 | UTL-7Apex14Δ∷LEU2 pex18Δ∷kanMX4 pex21Δ∷loxP | (Stein et al., 2002) |
| yKat111 | UTL-7A pex18Δ∷kanMX4 pex21Δ∷loxP [pHPR131] | (Stein et al., 2002) |
| yHPR251 | UTL-7A [pHPR131] | (Stein et al., 2002) |
| yHPR252 | UTL-7A pex7Δ∷LEU2 [pHPR131] | (Stein et al., 2002) |
| yAS3 | UTL-7A pex7Δ PEX18-TAP TRP1 | (Stein et al., 2002) |
| yAS4 | UTL-7A pex21Δ PEX18-TAP TRP1 | (Stein et al., 2002) |
Plasmids and Oligonucleotides
Table 2 provides a list of plasmids and oligonucleotides used in this study. NcPEX7 (GenBank accession no. AY141206) and NcPEX20 (GenBank accession no. AY141207) were amplified from a N. crassa cDNA library by polymerase chain reaction with primer pair RE579/580 and RE575/576, respectively. The two products were subcloned into EcoRV-cut pBluescript SK+ (Stratagene, La Jolla, CA), resulting in pMAR28 (PEX7) and pMAR14 (PEX20). To express NcPex7p in S. cerevisiae, the NcPEX7 cDNA fragment was excised from pMAR28 with EcoRI/ClaI and cloned into the appropriately cut yeast expression vector pRS-FOXP-TERM (pMAR39). NcPex20p was similarly cloned by ligating an EcoRI/XhoI PEX20 cDNA fragment from pMAR14 into pRS-FOXP-TERM (pMAR22). The NcPEX20 expression construct was also subcloned into the TRP1-based expression vector YCplac22 as a BamHI/KpnI fragment (pAS5). The PEX18 open reading frame, including 292 base pairs of its promoter and 329 base pairs of its 3′ region, was amplified from S. cerevisiae genomic DNA by using primer pair RE221/222 and cloned into EcoRV-cut pBluescript SK+ (pAS1). This fragment was excised as an XbaI/HindIII fragment and cloned into YCplac33 (pAS3) and YCplac22 (pAS4).
Table 2.
Plasmids and oligonucleotides used
| Plasmid or oligonucleotide | Description | Source or reference |
|---|---|---|
| Plasmid | ||
| YCplac22 | TRP1-marked centromeric plasmid | (Gietz and Sugino, 1988) |
| YCplac33 | URA3-marked centromeric plasmid | (Gietz and Sugino, 1988) |
| pRS-FOXP-TERM | URA3-based yeast expression vector | C. Clayton, Heidelberg |
| pJR233 | MLS1pr-GFP-SKL | (Lametschwandtner et al., 1998) |
| pIH217 | LEU2-based MLS1pr-GFP-SKL | I. Heiland, Berlin |
| pHPR131 | ADH2pr-PTS2-DsRed | (Stein et al., 2002) |
| YEpmycPex7 | CUP1pr-myc-PEX7 | (Rehling et al., 1996) |
| pPC97-PEX7 | (Albertini et al., 1997) | |
| pPC97-PEX13 | (Albertini et al., 1997) | |
| pPC97-PEX14 | (Albertini et al., 1997) | |
| pPC97-FOX3 | (Rehling et al., 1996) | |
| pKat31 | pPC97-PEX13 (1–151) | (Stein et al., 2002) |
| pMar14 | pSK-NcPEX20 | This study |
| pMar22 | pRS-FOXP-TERM-NcPEX20 | This study |
| pMar26 | pPC86-NcPEX20 | This study |
| pMar28 | pSK-NcPEX7 | This study |
| pMar30 | pPC97-NcPEX7 | This study |
| pMar39 | pRS-FOXP-TERM-NcPEX7 | This study |
| pAS1 | pSK-PEX18pr-PEX18 | This study |
| pAS3 | YCplac33-PEX18pr-PEX18 | This study |
| pAS4 | YCplac22-PEX18pr-PEX18 | This study |
| pAS5 | YCplac22-NcPEX20 | This study |
| Oligonucleotide (5′–3′) | ||
| RE 221 | GTATAATCAGGTATGTAAGGG | |
| RE 222 | CGACAACTAAGTTCCAGAAAG | |
| RE 575 | ATAGATCTACGAATTCATGTCTGACAGCATGTGC | |
| RE 576 | ATGCGGCCGCCTCGAGTTAAGCGGAAGAAGCA | |
| RE 579 | ATGTCGACCGAATTCATGGCGTCCATGCTGGAATTT | |
| RE 580 | ATAGATCTATCGATCCTCTTTCAAACAACCTGCTTC | |
Yeast Two-Hybrid Assays
To analyze the interactions of NcPex20p and NcPex7p with S. cerevisiae peroxins in the yeast two-hybrid system, the NcPEX20 cDNA fragment was cloned as a BglII/NotI fragment from pMAR14 into a similarly digested pPC86 (pMAR26). The NcPEX7 cDNA fragment was excised from pMAR28 with SalI/BglII and cloned into pPC97 (pMAR30). All constructs containing S. cerevisiae peroxins or truncations thereof have been described previously (Stein et al., 2002). Two-hybrid plasmids were cotransformed into the HF7c wild-type or the otherwise isogenic pex7Δ strain and selected on SD plates without tryptophane and leucine. His-auxotrophy of transformed strains was determined by growth on selective plates lacking leucine, tryptophane, and histidine but containing 1 or 5 mM 3-aminotriazole.
Antibodies and Western Blotting
The antibodies used have either been described previously, namely, anti-Fox3p (Erdmann and Kunau, 1994), anti-Cta1p (Gurvitz et al., 1997), and anti-Pex7p (Stein et al., 2002), or purchased in the case of anti-protein A (Sigma-Aldrich, St. Louis, MO) and monoclonal anti-9E10 c-myc (Babco, Richmond, CA) antibodies. Immunoblotting was performed according to standard protocols. Horseradish peroxidase-coupled anti-rabbit or anti-mouse IgGs, in combination with the enhanced chemiluminscence system (Amersham Biosciences, Freiburg, Germany), were used to detect immunoreactive complexes.
Fluorescence Microscopy
Live cells were analyzed for DsRed and green fluorescent protein (GFP) fluorescence by using an Axioplan 2 microscope equipped with a Plan-Neofluar 1003/1.30 Ph3 oil objective and a 100-W mercury lamp and filter sets (Carl Zeiss, Jena, Germany). Images were recorded with a SPOT-cooled color digital camera (Diagnostic Instruments, Sterling Heights, MI) and processed with Lite MetaMorph imaging software (Universal Imaging, West Chester, PA). Immunofluorescence microscopy was carried out as described previously (Erdmann, 1994) by using anti-Fox3p or anti-Cta1p antibodies followed by an incubation with Cy3-conjugated donkey anti-rabbit IgGs (Jackson Immunoresearch Laboratories, West Grove, PA) for detection.
Coimmunoprecipitation and Subcellular Fractionation
Myc-Pex7p and Pex18p-TAP were immunoprecipitated from soluble protein extracts essentially as described previously (Stein et al., 2002). Strains were grown and induced in appropriate selective media to maintain extrachromosomal plasmids. Cells were broken open in solution A (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.2% Triton X-100) that contained the Complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) with a French press (Pex18p-TAP) or glass beads (myc-Pex7p). The cleared homogenates were incubated for 1 h at 4°C with IgG-Sepharose (Amersham Biosciences) in the case of Pex18p-TAP or with magnetic beads (Dynal, Hamburg, Germany) covered with monoclonal anti-myc antibodies (myc-Pex7p). After a repeated washing step with solution A, bound myc-Pex7p was released by cooking the beads in SDS sample buffer, whereas bound Pex18p was released from the beads by adding 5 U of tobacco etch virus (TEV) protease (Invitrogen, Karlsruhe, Germany) in solution A, adjusted to 10 mM dithiothreitol and 1 mM EDTA. All eluates were analyzed for the presence of Pex7p and Fox3p. Yeast lysates were prepared and fractionated by differential centrifugation as described previously (Erdmann et al., 1989).
Database Analysis
Homologues to known peroxins were identified by a TBLASTN search within the database of the Center for Genome Research at the Whitehead Institute (WICGR) by using the default search algorithm. Genes that were also annotated at Munich Information Center for Protein Sequences were compared for identity with the corresponding WICGR open reading frame. The query was carried out with the S. cerevisiae peroxins, except PEX8, PEX9, PEX20, PEX23, and PEX24 (Y. lipolytica) and PEX2 and PEX16 (Homo sapiens).
RESULTS
Predicted Peroxins in N. crassa
The release of the genomic sequence of N. crassa allowed us to search for the presence of potential orthologous peroxins in this organism by using the database of the Center for Genome Research at the Whitehead Institute, Cambridge, MA (Neurospora Sequencing Project. Whitehead Institute/MIT Center for Genome Research, assembly version 3; www-genome.wi.mit.edu/annotation/fungi/neurospora). The results from this search are summarized in Table 3. Proteins with significant similarity to 18 of the 25 currently known peroxins were identified. Interestingly, the search revealed that the N. crassa genome contains homologues to all known human peroxins. On the other hand, peroxins specific to S. cerevisiae and/or Pichia pastoris, i.e., Pex15p, Pex17p, Pex18p, Pex21p, Pex22p (Purdue and Lazarow, 2001), and Pex25p (Smith et al., 2002), are likely to be absent from N. crassa (Table 3). The set of N. crassa peroxins seems to resemble fairly closely that of Y. lipolytica, because it also includes Pex20p, Pex23p, and Pex24p, peroxins that had previously been found only in Y. lipolytica. A BLAST search with Pex9p, the remaining Y. lipolytica-specific peroxin did not yield hits with significant homologies and was therefore classified as being absent from N. crassa. From the peroxins that are specifically involved in the PTS2 branch of peroxisomal matrix protein import, N. crassa lacks Pex18p and Pex21p homologues, but, as outlined previously (Einwächter et al., 2001), it does contain stretches with strong similarity to both ScPex7p and YlPex20p (see below). So far, these two peroxins have not been identified together in any other organism.
Table 3.
In silico identification of peroxins in N. crassa
| Neurospora protein namea | Peroxin (organism)b | Description | S.c.c | Y.l.c | H.s.c | Neurospora contiga | Scored | Lengthd | Identityd (%) |
|---|---|---|---|---|---|---|---|---|---|
| NCU08118.1 | PEX1 (S. cerevisiae) | Member of the AAA-protein family | + | + | + | 3.487 (scaffold 46) | e-109 | 1139 | 30 |
| NCU02070.1 | PEX2 (H. sapiens) | Zinc-binding integral peroxisomal membrane protein | + | + | + | 3.91 (scaffold 5) | 2e-23 | 270 | 29 |
| NCU06175.1 | PEX3 (S. cerevisiae) | Integral peroxisomal membrane protein | + | − | + | 3.359 (scaffold 26) | 1e-14 | 251 | 27 |
| NCU02636.1 | PEX4 (S. cerevisiae) | Member of ubiquitin-conjugating protein family | + | − | − | 3.137 (scaffold 7) | 3e-18 | 110 | 42 |
| NCU02960.1 | PEX5 (S. cerevisiae) | 69-kDa protein containing tetratricopeptide repeat (TPR), PTS1-receptor | + | + | + | 3.153 (scaffold 8) | 3e-81 | 494 | 38 |
| NCU08373.1 | PEX6 (S. cerevisiae) | Member of the AAA-protein family | + | + | + | 3.501 (scaffold 50) | e-134 | 829 | 34 |
| NCU07662.1 (AY141206) | PEX7 (S. cerevisiae) | 40 kDa-protein containing WD40-repeats, PTS2-receptor | + | − | + | 3.458 (scaffold 41) | 1e-59 | 131 | 48 |
| NCU00032.1 | PEX8 (Y. lipolytica) | Integral peroxisomal membrane protein | + | + | − | 3.1 (scaffold 1) | 7e-30 | 590 | 24 |
| — | PEX9 (Y. lipolytica) | Integral peroxisomal membrane protein | − | + | − | — | — | — | — |
| NCU03277.1 | PEX10 (S. cerevisiae) | Zinc-binding integral peroxisomal membrane protein | + | + | + | 3.171 (scaffold 9) | 3e-35 | 420 | 26 |
| NCU04802.1 | PEX11 (S. cerevisiae) | Peroxisomal membrane protein, proliferation | + | − | + | 3.263 (scaffold 16) | 1e-15 | 147 | 37 |
| NCU05245.1 | PEX12 (S. cerevisiae) | Zinc-binding integral peroxisomal membrane protein | + | − | + | 3.295 (scaffold 19) | 2e-17 | 440 | 23 |
| NCU02618.1 | PEX13 (S. cerevisiae) | Peroxisomal membrane protein, contains SH3 domain | + | − | + | 3.135 (scaffold 7) | 3e-60 | 332 | 44 |
| NCU03901.1 | PEX14 (S. cerevisiae) | Component of peroxisomal import machinery | + | + | + | 3.212 (scaffold 12) | 3e-11 | 127 | 28 |
| — | PEX15 (S. cerevisiae) | Peroxisomal membrane protein | + | − | − | — | — | — | — |
| NCU01850.1 | PEX16 (H. sapiens) | Peroxisomal membrane protein | − | + | + | 3.82 (scaffold 5) | 5e-11 | 302 | 24 |
| — | PEX17 (S. cerevisiae) | Peroxisomal membrane protein | + | − | − | — | — | — | — |
| — | PEX18 (S. cerevisiae) | Involved in PTS2-protein import | + | − | − | — | — | — | — |
| NCU04301.1 | PEX19 (S. cerevisiae) | 40 kDa farnesylated protein associated with peroxisomes | + | + | + | 3.222 (scaffold 13) | 2e-11 | 177 | 25 |
| NCU04062.1 (AY141207) | PEX20 (Y. lipolytica) | Involved in PTS2-protein import | − | + | − | 3.214 (scaffold 12) | 8e-12 | 337 | 24 |
| — | PEX21 (S. cerevisiae) | Involved in PTS2-protein import | + | − | − | — | — | — | — |
| — | PEX22 (S. cerevisiae) | Peroxisomal membrane protein | + | − | − | — | — | — | — |
| NCU05564.1 | PEX23 (Y. lipolytica) | Peroxisomal membrane protein | − | + | − | 3.312 (scaffold 21) | 2e-58 | 283 | 33 |
| NCU06637.1 | PEX24 (Y. lipolytica) | Peroxisomal membrane protein | − | + | − | 3.384 (scaffold 30) | 2e-25 | 394 | 22 |
| — | PEX25 (S. cerevisiae) | Peroxisomal membrane protein, proliferation | + | − | − | — | — | — | — |
Annotated N. crassa protein names and contig numbers are according to Whitehead Institute/MIT Centre for Genome Research (http://www-genome.wi.mit.edu/annotation/fungi/neurospora).
Protein sequence of peroxin used as bait in BLAST search at the Whitehead Institute/MIT Center for Genome Research.
Peroxin present (+) or not identified (−) in indicated organism.
Score from BLAST search.
Isolation of PEX7 and PEX20 from N. crassa cDNA
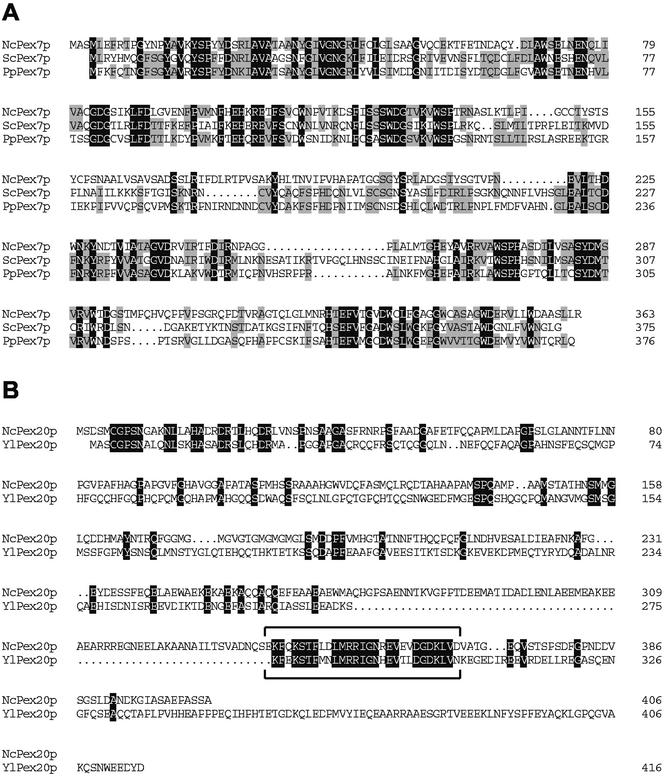
The homology search with Pex7p yielded two adjacent, nonoverlapping regions within the N. crassa genome, covering most of ScPex7p. The proper contig (3.458) was therefore analyzed for the presence of a potential start and a stop codon, both of which were found in proximity to the ends of the homologous stretches. The 5′ and 3′ ends found by this process were incorporated into the primer sequence designed to amplify the PEX7 open reading frame from N. crassa cDNA. The PEX20 gene was amplified according to the predictions quoted at the Munich Information Center for Protein Sequences (www.mips.gsf.de). Sequencing of the polymerase chain reaction products thus obtained for PEX7 and PEX20 revealed that they were identical to the respective genomic loci in the database with the exception of the presence of a single intron in each reading frame, ranging from base pairs 400–496 in PEX7 and from base pairs 121–197 in PEX20. An alignment of the deduced amino acid sequence of NcPex7p with Pex7p of both S. cerevisiae and P. pastoris is shown in Figure 1A. NcPex7p showed significant similarity to the two yeast proteins throughout its sequence, with 36.3% identical and 28.7% similar residues to the S. cerevisiae protein and 36.4% identical and 31.7% similar residues to Pex7p of P. pastoris. Likewise, the sequence of Pex20p was compared with that of YlPex20p. NcPex20p is less conserved, with a portion of 14.7% identical and 35.1% similar residues to YlPex20p; however, two short stretches of ∼30 amino acids in length are highly similar. Of these two, the one closer to the C terminus represents the region that is also conserved in the PTS2 auxiliary factors Pex5pL and Pex18p/Pex21p and that is required to interact with Pex7p (Figure 1B, box). The significance of the conserved N-terminal homologous region in Pex20p remains to be determined, but it might represent a second protein–protein interaction domain.
Figure 1.
Sequence alignment of Pex7p and Pex20p. (A) Amino acid sequences of Pex7p from N. crassa, S. cerevisiae, and P. pastoris were aligned with DNAMAN (Lynnon BioSoft, Quebec, Canada). Identical residues in all three sequences are shaded in black. Residues that are either similar in all three sequences or identical in two out of three sequences are shaded in gray. Similarity rule: D = E, R = K, Q = N, S = T, I = L = V = A. (B) Amino acid sequences of Pex20p from N. crassa and Y. lipolytica were similarly aligned, but only identical residues (shaded in black) are shown. The box marks the region within Pex20p that is conserved in Pex18p and Pex5pL.
Expression of NcPEX20 in S. cerevisiae Complements a pex18Δpex21Δ Mutation
The recent demonstration of a yeast pex18Δpex21Δ mutant being partially complemented by YlPex20p allowed bridging of the function of these two proteins (Einwächter et al., 2001). We therefore analyzed whether NcPex20p would be similarly proficient in substituting Pex18p/Pex21p. For that purpose, NcPEX20 was cloned behind the oleic acid-inducible FOX3 promoter and expressed in a pex18Δpex21Δ mutant. In addition, fluorescent marker proteins for both the PTS1 and the PTS2 pathway, whose fluorescence patterns reflect the ability of a strain to import PTS1 and PTS2 substrates respectively, were coexpressed in this strain. After 2 d of incubation on oleic acid plates, cells were inspected under the fluorescence microscope.
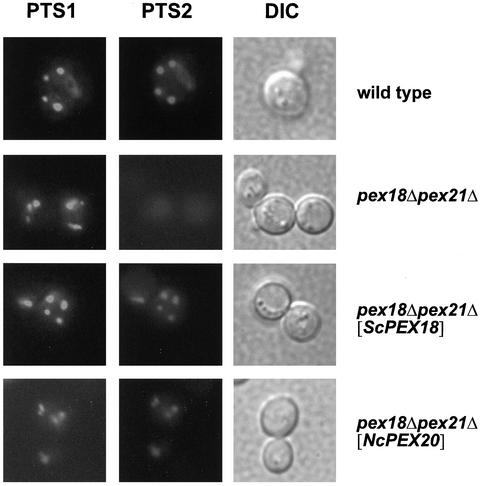
In the pex18Δpex21Δ mutant, the PTS2 marker protein PTS2-DsRed showed a diffuse fluorescence pattern, whereas the PTS1 protein GFP-SKL exhibited a punctate staining pattern, which was in line with the specific PTS2 protein import defect of the analyzed mutant (Figure 2). Expression of NcPex20p in the pex18Δpex21Δ mutant caused PTS2-DsRed to be localized in punctate structures that colocalized with GFP-SKL. Similar results were obtained when ScPex18p was expressed in this mutant strain. This result indicated that NcPex20p was able to restore the import of a synthetic PTS2-containing substrate in the pex18Δpex21Δ mutant.
Figure 2.
NcPex20p restores the PTS2 import deficiency of a pex18Δpex21Δ mutant in S. cerevisiae. The pex18Δpex21Δ strains expressing ScPex18p (pex18Δpex21Δ [PEX18]) or NcPex20p (pex18Δpex21Δ [NcPEX20]) together with PTS2-DsRed and GFP-PTS1 were analyzed for the fluorescence pattern of the PTS marker proteins. The wild-type and the untransformed mutant strain (pex18Δpex21Δ) served as controls. Before inspection, cells were kept on oleic acid-containing plates for 2 d. A punctate pattern indicates that protein import is enabled. Structural integrity of the cells is documented by differential interference contrast (DIC) microscopy.
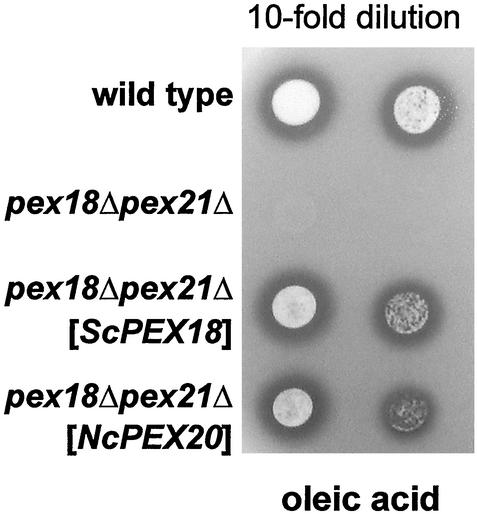
To now test whether NcPex20p is also able to physiologically replace Pex18p/Pex21p, these strains were spotted onto plates containing oleic acid as sole carbon source. As expected, it proved possible to rescue the growth phenotype of the pex18Δpex21Δ mutant on fatty acids by the expression of Pex18p in that strain (Figure 3). Importantly, also NcPex20p restored the mutant's ability to grow on oleic acid, thereby demonstrating the conserved function of these peroxins.
Figure 3.
Expression of NcPex20p restores growth of pex18Δpex21Δ cells on oleic acid. Tenfold dilutions of the wild-type, the untransformed pex18Δpex21Δ and the pex18Δpex21Δ strain expressing either ScPex18p or NcPex20p were spotted onto an oleic acid plate and incubated for 5 d at 30°C. Halos surrounding the spots indicate utilization of the fatty acid from the medium.
Interactions of NcPEX20 Resemble Those of PEX18
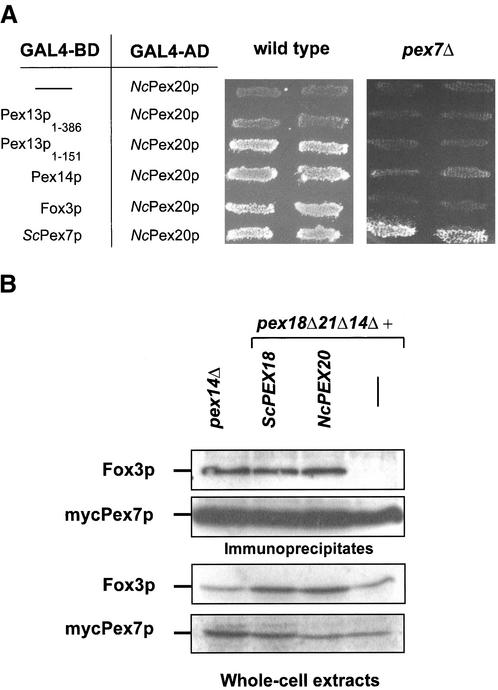
To nail down the function of NcPex20p in PTS2-dependent import, the protein's ability to interact with the known partners of Pex18p was addressed in a yeast two-hybrid assay (Stein et al., 2002). NcPex20p did indeed interact with Fox3p, Pex7p, and Pex13p1–151 (Figure 4A, left). Moreover, the previously observed dependency of the Pex18p–Fox3p and Pex18p–Pex13p1–151 interactions on Pex7p was also found to apply to the NcPex20p interactions (Figure 4A, right). In addition, NcPex20p interacted strongly with Pex14p in the wild-type strain, but only very weakly in the absence of Pex7p. The inability of NcPex20p to interact with Fox3p or the docking proteins Pex13p and Pex14p in the absence of Pex7p already implies that Pex7p is essential for NcPex20p to function in S. cerevisiae. In fact, growth of a pex7Δ mutant on oleic acid medium did not resume upon introduction of NcPex20p, and the PTS2-DsRed marker protein remained cytosolic in that strain (see below). These data favor a close link between Pex18p and NcPex20p.
Figure 4.
Function of NcPex20p resembles that of Pex18p. (A) Dependence of the NcPex20p interactions on ScPex7p. The indicated peroxins were tested in a two-hybrid assay for interaction with NcPex20p in the HF7c wild-type strain (left) and the otherwise isogenic pex7Δ strain (right). Interactions are indicated by the histidine prototrophy of two independent transformants at each case. (B) Accumulation of a Pex7p–Fox3p complex through substitution of Pex18p/Pex21p with NcPex20p in the docking mutant pex14Δ. The indicated strains were analyzed for the presence of Fox3p in the immunoprecipitate (20% of total) of myc-Pex7p by immunoblotting (top). The bottom panel shows 0.5% of the used total cell lysates.
We reported previously that Pex18p is instrumental in the formation of an import-competent complex containing the PTS2 receptor Pex7p and its cargo protein Fox3p (Stein et al., 2002). The accumulation of this complex in a pex14Δ mutant vanishes when Pex18p and Pex21p are concomitantly absent. Introduction of a plasmid-borne copy of PEX18 into a pex14Δpex18Δpex21Δ triple mutant caused Fox3p to be present in significant amounts in the immunoprecipitate of myc-Pex7p, indicating the reappearance of the accumulated complex as in a pex14Δ single mutant (Figure 4B). The same observation was made when a strain that had expressed NcPex20p in this triple mutant was used for analysis. Thus, like Pex18p, NcPex20p also possesses PTS2 complex-forming potential, which is required before docking because this complex is generated in a pex14Δ mutant.
NcPex7p Fails to Functionally Replace ScPex7p
The putative N. crassa PEX7 gene was cloned behind the FOX3 promoter and expressed in a pex7Δ mutant strain. The ability of NcPEX7 to complement the respective S. cerevisiae gene was again assessed by determining the cellular distribution of the PTS2-DsRed marker protein and the growth rate on oleic acid plates. The transformed strain did not lead to growth on oleic acid (see below). The PTS2 marker protein was dispersed in cells grown on oleic acid or ethanol, albeit on ethanol, punctate structures were discernible in rare cases (our unpublished data). We did not further investigate the identity of these structures because it became obvious that NcPex7p barely substituted the yeast protein. Rather, we inquired why NcPex7p was inefficient in functioning as a PTS2 receptor in yeast. Due to a fortuitous cross-reactivity of our anti-ScPex7p antibody, expression of NcPex7p could be analyzed immunologically. The protein was stably expressed at high levels under oleic acid-induction conditions, in agreement with NcPEX7 being under the control of the FOX3 promoter (Figure 5, bottom, lane 3). The failure of NcPex7p to functionally replace ScPex7p could therefore be due to the lack of interaction with a S. cerevisiae protein that is involved in PTS2 import.
Figure 5.
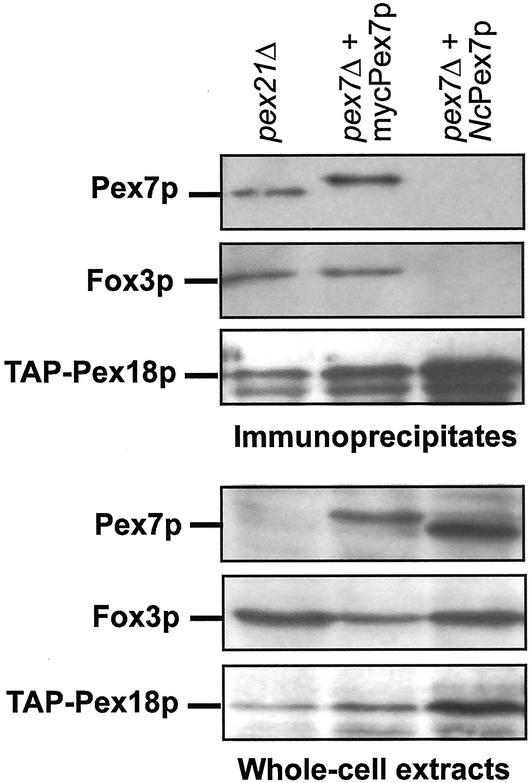
NcPex7p does not interact with ScPex18p. Pex18p-TAP was immunoprecipitated from whole-cell lysates of the strains pex21Δ PEX18-TAP as well as pex7Δ PEX18-TAP expressing myc-Pex7p or NcPex7p. Precipitates were analyzed for the presence of Pex7p (top, row 1), Fox3p (top, row 2), and Pex18p-TAP (top, row 3) by immunoblotting. The bottom panel shows 0.2% of the total cell lysates.
NcPex7p Does Not Interact with Pex18p
A fusion of NcPex7p with the Gal4p BD turned out to be autoactive in the yeast two-hybrid assay (our unpublished data). As a consequence, this method was not considered further for analyzing NcPex7p interactions, given that ScPex7p was previously found to interact with Fox3p as a Gal4p BD- but not as a Gal4p AD-fusion protein (Rehling et al., 1996). Instead, NcPex7p was tested biochemically for interaction with Pex18p by means of coimmunoprecipitation. For that matter, a functional TAP-tagged version of Pex18p was precipitated from the control strain pex21Δ as well as from pex7Δ strains overexpressing NcPex7p and myc-ScPex7p, respectively (Figure 5). The precipitate obtained from the pex21Δ strain did also contain ScPex7p (Figure 5, top, lane 1), demonstrating the interaction between endogenous ScPex7p and Pex18p. Likewise, it proved possible to coimmunoprecipitate myc-ScPex7p with Pex18p, as shown by the presence of the slightly slower migrating band representing myc-ScPex7p in the precipitate of this strain. In contrast, NcPex7p was not found in the respective Pex18p precipitate. In addition, only background levels of the PTS2 cargo protein Fox3p were detected in this precipitate, whereas a significant amount of Fox3p was found in the precipitates containing ScPex7p (Figure 5, top), in agreement with a Pex7p-mediated interaction between Pex18p and Fox3p (Stein et al., 2002). Taken together, these data indicate that NcPex7p does not interact with ScPex18p, which could explain the failure of the N. crassa PTS2 receptor to substitute for the S. cerevisiae counterpart.
Coexpression of NcPex7p and NcPex20p Rescues the PTS2 Import Defect of an S. cerevisiae pex7Δ Mutant
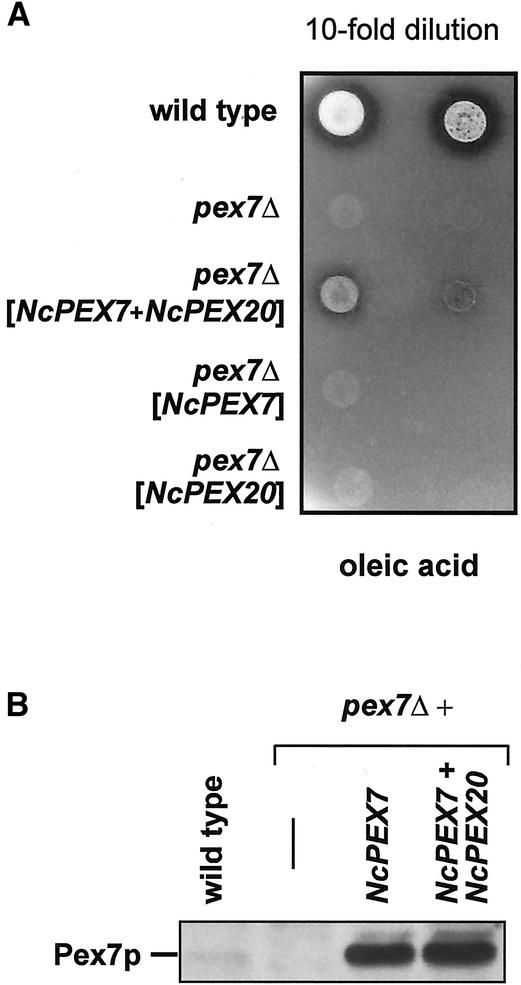
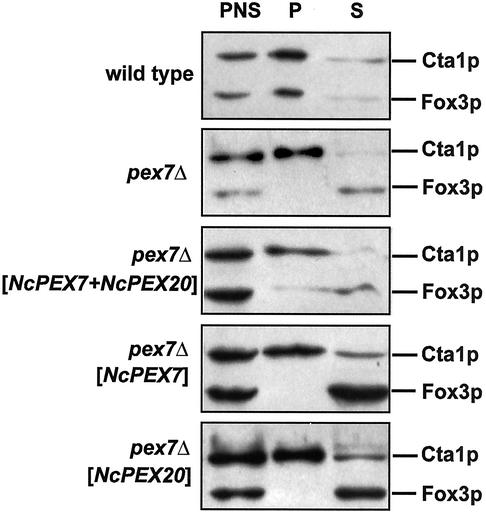
Our observation of NcPex20p being the orthologue of ScPex18p/Pex21p led us to suspect that an interaction between NcPex20p and NcPex7p should be relevant to PTS2-dependent import in N. crassa. Pursuing this train of thought further, by concomitantly expressing both peroxins in a S. cerevisiae pex7Δ strain, the requirement of NcPex7p to interact with Pex18p might be circumvented. Thus, a pex7Δ strain was transformed with expression plasmids carrying NcPEX7, NcPEX20, or both genes and the resulting strains were subjected to growth analysis on oleic acid plates. Compared with the untransformed pex7Δ strain, neither the expression of NcPex20p nor that of NcPex7p had any discernible effect. Interestingly, coexpression of NcPex7p and NcPex20p in pex7Δ did lead to a significant increase in cell mass and halos around the colonies were visible (Figure 6A). The concomitant presence of NcPex7p and NcPex20p did not result in higher levels of NcPex7p (Figure 6B), suggesting that NcPex20p actively converted NcPex7p into a functional PTS2 receptor that was able to translocate significant amounts of Fox3p into the peroxisomal lumen. We therefore separated postnuclear lysates of the same strains into a supernatant and an organellar pellet fraction that was enriched in peroxisomes and mitochondria. In all strains, the PTS1 protein Cta1p was predominately found in the pellet fraction, which was in line with the PTS2-specific defect of the pex7Δ mutation (Figure 7). The PTS2 protein thiolase was also preferentially present in the pellet fraction in the wild-type strain, but in the absence of ScPex7p, Fox3p was found in the supernatant fraction. This cytosolic distribution was not altered upon expression of NcPex7p or NcPex20p in the pex7Δ mutant. In contrast, coexpression of NcPex7p and NcPex20p resulted in the appearance of some thiolase in the pellet fraction (Figure 7).
Figure 6.
NcPex20p converts NcPex7p into a functional PTS2 receptor in S. cerevisiae. (A) Growth of a pex7Δ strain on oleic acid resumes in the concomitant presence of NcPex7p and NcPex20p. Tenfold dilutions of the wild-type, the untransformed pex7Δ as well as the pex7Δ strain transformed with either NcPEX7, NcPEX20, or NcPEX7 plus NcPEX20 were spotted onto oleic acid plates and incubated for 5 d at 30°C. Halo formation was observed in the strain coexpressing both N. crassa peroxins. (B) Expression of NcPex7p does not change in the presence of NcPex20p. Whole-cell extracts of the indicated strains that had been induced by oleic acid were analyzed for the expression level of NcPex7p using Pex7p-specific antibodies. The amount of NcPex7p exceeded that of native ScPex7p (lane 1) but was comparable in the absence or presence of NcPex20p (lanes 3 and 4).
Figure 7.
Redistribution of the PTS2 protein Fox3p into the organellar fraction in a pex7Δ strain in the concurrent presence of NcPex7p and NcPex20p. Postnuclear supernatants (PNS) of the indicated oleic acid-induced strains were subjected to differential centrifugation at 20,000 × g for 20 min. Equal portions of the resulting organellar pellet (P) and supernatant (S) fractions were analyzed for the distribution of Fox3p. The PTS1 protein Cta1p was monitored as control for the intactness of the isolated peroxisomes.
These strains were also analyzed for the location of native Fox3p by means of indirect immunofluorescence. Spheroplasts from oleic acid-induced cells were labeled with antibodies directed against the PTS1 protein Cta1p (Figure 8, left). The punctate staining pattern typical for peroxisomes was observed in all samples, including the untransformed pex7Δ strain, thereby demonstrating the occurrence of intact peroxisomal structures. Decoration with anti-Fox3p antibodies caused a diffuse staining in the pex7Δ strain, which reflects a nonperoxisomal location of Fox3p (Figure 8, right). The diffuse fluorescence pattern was maintained upon expression of NcPex20p or NcPex7p in that strain; however, the presence of both N. crassa proteins caused Fox3p to reappear in punctate structures. This result indicated that Fox3p relocated into peroxisomes only when NcPex20p and NcPex7p were present at the same time. Thus, the two N. crassa proteins are functionally linked and act together in PTS2-dependent matrix protein import.
Figure 8.
Fox3p relocates into peroxisomes in a pex7Δ mutant upon coexpression of NcPex20p and NcPex7p. The indicated strains were induced on oleic acid-containing medium for 12 h and subjected to immunofluorescence analysis. Right (PTS2), cells that had been decorated with antibodies against Fox3p and subsequently with CY3-coupled anti-rabbit antibodies. Left (PTS1), same strains but treated with anti-Cta1p antibodies. Only coexpression of NcPex7p and NcPex20p (pex7Δ [NcPEX7+NcPEX20]) restored the punctate fluorescence pattern of thiolase in a pex7Δ strain.
DISCUSSION
In essence, peroxisome biogenesis is evolutionary conserved. However, the degree of deviance seemed more pronounced for the PTS2 branch of the peroxisomal matrix protein import. In this report, we have shown that the filamentous fungus N. crassa possesses two PTS2-specific peroxins; the PTS2-signal recognition factor Pex7p that is common to many organisms, as well as Pex20p, which had hitherto been identified only in Y. lipolytica. These findings unequivocally dispose Pex20p into the group of PTS2-specific peroxins that are auxiliary to Pex7p, which includes Pex18p/Pex21p and human Pex5pL. Our observation that NcPex20p, when expressed in S. cerevisiae, substituted for Pex18p/Pex21p but not for Pex7p, supports this classification. The dependency of NcPex20p on Pex7p was corroborated in a two-hybrid assay because NcPex20p interacted with ScFox3p and the docking proteins Pex13p and Pex14p, but only in the presence of Pex7p.
In analogy to the function of NcPex20p, YlPex20p has been shown to interact with ScFox3p via ScPex7p. This led Einwächter et al. (2001) to propose the existence of a hitherto undetected Pex7p in Y. lipolytica. Our data on the interplay of N. crassa Pex7p and Pex20p clearly support this notion. On the other hand, the seemingly cross-functional YlPex20p and NcPex20p may still possess properties that are unique to each protein in its native environment. Particularly if Y. lipolytica were indeed found to lack Pex7p, the additional interactions of YlPex20p such as that with YlFox3p might be required to recruit and target thiolase to the docking complex, as proposed previously (Titorenko et al., 1998). It will be possible to give a definite answer by the time the genomic sequence of Y. lipolytica is released. In this respect, it should be noted that the genome of the nematode Caenorahabditis elegans does indeed lack an apparent orthologue of the PTS2 receptor Pex7p (Gurvitz et al., 2000; Motley et al., 2000). But in this case, C. elegans orthologues of PTS2-containing enzymes have gained a PTS1, suggesting that the PTS2 branch is entirely absent from the nematode (Motley et al., 2000), which is clearly not the case in Y. lipolytica.
The heterologous test system used in our study allowed us to demonstrate that the PTS2 receptor of N. crassa is not capable of importing PTS2 cargo proteins in the absence of Pex20p. This conclusion is based on the fact that on the one hand, NcPex7p failed to interact with Pex18p, and on the other it was not self-sufficient in doing the job. However, upon coexpression of NcPex20p and NcPex7p in pex7Δ cells, the import of Fox3p resumed. In these cells, thiolase was found in punctate structures and significant amounts were present in an organellar pellet. Strikingly, this strain was able to grow slowly on oleic acid plates, whereas strains harboring only NcPex7p or NcPex20p failed to do so at all. Since NcPex20p did not interact with Pex13p and Pex14p in the absence of ScPex7p in a two-hybrid assay, NcPex7p is the likely interaction partner of these S. cerevisiae peroxins upon docking and the same holds true for PTS2 signal recognition. Thus, by taking into account the multitude of interactions NcPex7p has to make with S. cerevisiae proteins, it is not surprising that the complementation observed was only partial.
The precise role of Pex20p in PTS2 import is not yet completely clear, but it is similar to that of Pex18p. NcPex20p was required for the formation of an import-competent complex containing thiolase and Pex7p. This complex accumulates in a pex14Δ mutant but not in a wild-type strain, suggesting that it represents an intermediate of the import process of PTS2 proteins (Stein et al., 2002). Because neither Pex18p nor Pex20p was able to interact directly with ScFox3p, these peroxins could promote the oligomerization or aggregation of cargo-loaded receptor complexes, which would be a prerequisite for the import of Fox3p. Similar ideas as to the role of Pex18p/Pex21p were recently put forward in a hypothesis that postulates the existence of such an oligomeric complex, a so-called preimplex (Gould and Collins, 2002). In mammals, Pex5pL would represent this oligomerization factor that might additionally undertake docking (Otera et al., 2000; Dodt et al., 2001). In the higher plant Arabidopsis thaliana, the PTS2 pathway depends on Pex5p just as in mammals; however, a single Pex5p isoform seems to be required for the import of both PTS1 and PTS2 proteins (Nito et al., 2002).
One final point that warrants discussion is our revelation of remarkably similar peroxin contents of N. crassa and Y. lipolytica. This indicates that the interplay of peroxins in Y. lipolytica is probably conserved to a larger degree than appreciated so far. In fact, our genome-wide search for peroxins also revealed a link between these two fungi and H. sapiens (Table 3). For instance, all three organisms but not S. cerevisiae contain Pex16p, a peroxin that is crucial for the early steps in peroxisome biogenesis in humans (South and Gould, 1999). On the other hand, N. crassa and H. sapiens probably lack several yeast-specific peroxins such as Pex15p and Pex17p (Table 3).
N. crassa only grows as a mycelium but Y. lipolytica can also undergo a transition to a mycelial form, which has been shown to require peroxisomes (Titorenko et al., 1997). N. crassa might therefore be the organism of choice to test whether intact peroxisomes are indeed mandatory for mycelial growth. Another challenging discovery stemming solely from work carried out with Y. lipolytica is the fusion of peroxisomal subpopulations in the course of peroxisome maturation (Titorenko and Rachubinski, 2001). Using N. crassa as a novel model organism to study peroxisome biogenesis might now allow an independent assessment of the more general validity of this and other hypotheses. Demonstrating herein the concerted action of NcPex7p and NcPex20p in PTS2-dependent protein import has shown that this could be indeed a promising approach.
ACKNOWLEDGMENTS
We thank F. Nargang for the N. crassa cDNA library, K. Stein for strains and plasmids, I. Heiland for plasmid pIH217, and H.F. Tabak and W.H. Kunau for antibodies. We also thank Phil Nelson for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft, grants ER178/2-3 and SFB449, and by the Fonds der Deutschen Chemischen Industrie. H.R. was supported by an EMBO long-term fellowship (ALTF255-2000).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0539. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0539.
REFERENCES
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JA, Veenhuis M, Kunau WH. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Braverman N, Dodt G, Gould SJ, Valle D. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum Mol Genet. 1998;7:1195–1205. doi: 10.1093/hmg/7.8.1195. [DOI] [PubMed] [Google Scholar]
- Dodt G, Warren D, Becker E, Rehling P, Gould SJ. Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J Biol Chem. 2001;276:41769–41781. doi: 10.1074/jbc.M106932200. [DOI] [PubMed] [Google Scholar]
- Einwächter H, Sowinski S, Kunau WH, Schliebs W. Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfill a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2001;2:1035–1039. doi: 10.1093/embo-reports/kve228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R. The peroxisomal targeting signal of 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. Yeast. 1994;10:935–944. doi: 10.1002/yea.320100708. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Kunau WH. Purification and immunolocalization of the peroxisomal 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. Yeast. 1994;10:1173–1182. doi: 10.1002/yea.320100905. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Mertens D, Kunau WH. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:5419–5423. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Collins CS. Opinion: peroxisomal protein import: is it really that complex? Nat Rev Mol Cell Biol. 2002;3:382–389. doi: 10.1038/nrm807. [DOI] [PubMed] [Google Scholar]
- Gurvitz A, Langer S, Piskacek M, Hamilton B, Ruis H, Hartig A. Predicting the function and subcellular location of Caenorhabditis elegans proteins similar to Saccharomyces cerevisiae β-oxidation enzymes. Yeast. 2000;17:188–200. doi: 10.1002/1097-0061(20000930)17:3<188::AID-YEA27>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz A, Rottensteiner H, Kilpelainen SH, Hartig A, Hiltunen JK, Binder M, Dawes IW, Hamilton B. The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-CoA reductase is encoded by the oleate-inducible gene SPS19. J Biol Chem. 1997;272:22140–22147. doi: 10.1074/jbc.272.35.22140. [DOI] [PubMed] [Google Scholar]
- Holroyd C, Erdmann R. Protein translocation machineries of peroxisomes. FEBS Lett. 2001;501:6–10. doi: 10.1016/s0014-5793(01)02617-5. [DOI] [PubMed] [Google Scholar]
- Jedd G, Chua NH. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat Cell Biol. 2000;2:226–231. doi: 10.1038/35008652. [DOI] [PubMed] [Google Scholar]
- Kionka C, Kunau WH. Inducible β-oxidation pathway in Neurospora crassa. J Bacteriol. 1985;161:153–157. doi: 10.1128/jb.161.1.153-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lametschwandtner G, Brocard C, Fransen M, Van Veldhoven P, Berger J, Hartig A. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J Biol Chem. 1998;273:33635–33643. doi: 10.1074/jbc.273.50.33635. [DOI] [PubMed] [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau WH. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13:4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF. Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 2000;1:40–46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Hayashi M, Nishimura M. Direct interaction, and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:355–366. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]
- Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, Fujiki Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p-PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J Biol Chem. 2000;275:21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- Otera H, Setoguchi K, Hamasaki M, Kumashiro T, Shimizu N, Fujiki Y. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol Cell Biol. 2002;22:1639–1655. doi: 10.1128/MCB.22.6.1639-1655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Marzioch M, Niesen F, Wittke E, Veenhuis M, Kunau WH. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- Sacksteder KA, Gould SJ. The genetics of peroxisome biogenesis. Annu Rev Genet. 2000;34:623–652. doi: 10.1146/annurev.genet.34.1.623. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA, Aitchison JD. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Rachubinski RA. A role for the peroxin Pex8p in Pex20p-dependent thiolase import into peroxisomes of the yeast Yarrowia lipolytica. J Biol Chem. 2001;276:1618–1625. doi: 10.1074/jbc.M005072200. [DOI] [PubMed] [Google Scholar]
- South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K, Schell-Steven A, Erdmann R, Rottensteiner H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol Cell Biol. 2002;22:6056–6069. doi: 10.1128/MCB.22.17.6056-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Koller A, Snyder WB. Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem. 2000;69:399–418. doi: 10.1146/annurev.biochem.69.1.399. [DOI] [PubMed] [Google Scholar]
- Tenney K, Hunt I, Sweigard J, Pounder JI, McClain C, Bowman EJ, Bowman BJ. Hex1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet Biol. 2000;31:205–217. doi: 10.1006/fgbi.2000.1230. [DOI] [PubMed] [Google Scholar]
- Thieringer R, Kunau WH. The β-oxidation system in catalase-free microbodies of the filamentous fungus Neurospora crassa. Purification of a multifunctional protein possessing 2-enoyl-CoA hydratase, L-3-hydroxyacyl-CoA dehydrogenase, and 3-hydroxyacyl-CoA epimerase activities. J Biol Chem. 1991;266:13110–13117. [PubMed] [Google Scholar]
- Titorenko VI, Ogrydziak DM, Rachubinski RA. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol. 1997;17:5210–5226. doi: 10.1128/mcb.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. The life cycle of the peroxisome. Nat Rev Mol Cell Biol. 2001;2:357–368. doi: 10.1038/35073063. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]