Abstract
Kv1 potassium channels are widely distributed in mammalian tissues and are involved in a variety of functions from controlling the firing rate of neurons to maturation of T-lymphocytes. Here we show that the newly described KCNE4 β-subunit has a drastic inhibitory effect on currents generated by Kv1.1 and Kv1.3 potassium channels. The inhibition is found on channels expressed heterologously in both Xenopus oocytes and mammalian HEK293 cells. mKCNE4 does not inhibit Kv1.2, Kv1.4, Kv1.5, or Kv4.3 homomeric complexes, but it does significantly reduce current through Kv1.1/Kv1.2 and Kv1.2/Kv1.3 heteromeric complexes. Confocal microscopy and Western blotting reveal that Kv1.1 is present at the cell surface together with KCNE4. Real-time RT-PCR shows a relatively high presence of mKCNE4 mRNA in several organs, including uterus, kidney, lung, intestine, and in embryo, whereas a much lower mRNA level is detected in the heart and in five different parts of the brain. Having the broad distribution of Kv1 channels in mind, the demonstrated inhibitory property of KCNE4-subunits could locally and/or transiently have a dramatic influence on cellular excitability and on setting resting membrane potentials.
INTRODUCTION
Voltage-gated Kv potassium channels are involved in a considerable number of physiological functions including neuronal excitability, synaptic transmission, and setting resting membrane potentials (Hille, 2001). Kv channels comprise a large family of proteins of quite diverse structural and functional properties and are widely expressed in both excitable and nonexcitable tissues. Nine subfamilies of Kv channels (Kv1–9) have been described in mammals (Hille, 2001; Chandy and Gutman, 1995; Roeper and Pongs, 1996).
Within the Kv1 (Shaker) subfamily, at least nine α-subunits (Kv1.1–Kv1.9) have been described. These are mostly non- or slowly inactivating with the exception of Kv1.4 and Kv1.7, which are fast inactivating A-type channels (Baumann et al., 1988; Chandy et al., 1990; Grupe et al., 1990; Kalman et al., 1998; Stuhmer et al., 1989; Tempel et al., 1988). The diversity of Kv1 channels is increased by the fact that all Kv1 subfamily members in vitro have been found to coassemble into functional channels, resulting in mixed biophysical properties (Isacoff et al., 1990; Ruppersberg et al., 1990). Many of these subunit combinations have also been identified in native tissue. For example, in the mammalian brain Kv1 channel α-subunits are mainly found as heteromultimer complexes, with Kv1.1 and Kv1.2 as the predominant partners in most channel complexes (Koch et al., 1997; Shamotienko et al., 1997). Kv1 channels are expressed with high abundancy in the brain (Grosse et al., 2000; Monaghan et al., 2001; Rhodes et al., 1997; Sheng et al., 1994; Veh et al., 1995; Wang et al., 1994), but also outside the CNS, Kv1 channels are found in both excitable tissue such as heart and smooth muscle, and in nonexcitable tissue and cell types such as colon, kidney, endothelial cells, epithelial cells, adipocytes, T-cells, and granulosa cells (Grunnet et al., 2002b; Grissmer et al., 1990; Adda et al., 1996; Mason et al., 2002; Dixon and McKinnon, 1994; Ouadid-Ahidouch et al., 1999; Pappone and Ortiz-Miranda, 1993).
Three different Kvβ-subunits, designated Kvβ1, Kvβ2, and Kvβ3 (notation according to Pongs et al., 1999), have been found to interact with Kv channels (Heinemann et al., 1995; Rettig et al., 1994; Leicher et al., 1998). All three Kvβ genes are subject to alternative splicing, resulting in three subunit subtypes for Kvβ1 (Kvβ1.1, Kvβ1.2, Kvβ1.3) and two subtypes for both Kvβ2 (Kvβ2.1, Kvβ2.2) and Kvβ3 (Kvβ3.1, Kvβ3.2) (Pongs et al., 1999). The presence of the Kvβ-subunits results in significant changes in the regulation of the Kv1 α-subunits. The most pronounced effect is found for Kvβ1.1, since this subunit confers rapid A-type inactivation to the otherwise noninactivating Kv1.1, Kv1.2, and Kv1.5 channels (Heinemann et al., 1996). Other functions, such as increased surface expression of Kv channels, are also reported for Kvβ-subunits (Fink et al., 1996; Shi et al., 1996).
The KCNE family forms another class of one-transmembrane proteins capable of modulating potassium channels. This family of K+-channel β-subunits consists of five members, designated KCNE1–5 (formerly known as minK, MiRP1–3, and KCNE1-like, respectively), all capable of modulating the KCNQ1 current in vitro (Abbott et al., 1999; Piccini et al., 1999; Grunnet et al., 2002a; Angelo et al., 2002; Tinel et al., 2000a; Schroeder et al., 2000). KCNE1–3 have furthermore been shown to modulate other members of the KCNQ family as well as ERG, HCN, and Kv K+ channels (McDonald et al., 1997; Yu et al., 2001; Zhang et al., 2001; Abbott et al., 1999, 2001; Schroeder et al., 2000; Tinel et al., 2000b). In a number of cases, currents recorded from native cells have been shown to be due to specific interactions between K+-channel α-subunits and β-subunits of the KCNE family. Several mutations within KCNE1–3 are associated with serious disease (Abbott et al., 1999, 2001; Splawski et al., 1997). For Kv channels, KCNE3 has been found to combine with the Kv3.4 α-subunit forming a subthreshold, A-type channel in skeletal muscle (Abbott et al., 2001) and KCNE2 assembles with Kv4.2 and is thereby potentially involved in shaping the Ito current in the heart or the native A-type currents (Zhang et al., 2001; Kim et al., 2001).
Until recently, no potential assembly partners of KCNE4 and KCNE5 have been discovered. We have reported interaction of KCNE4 and KCNE5 with KCNQ1, providing modulations of the KCNQ1 current (Grunnet et al., 2002a; Angelo et al., 2002). In the present study we demonstrate a drastic inhibition of Kv1.1 and Kv1.3 currents by the KCNE4-subunit in both Xenopus oocytes and mammalian HEK293 cells. KCNE4 mRNA is found at relative high levels in several peripheral tissues in which Kv1.1/Kv1.3 channels have also been identified. The interaction between KCNE4 and Kv1.1/Kv1.3 may thereby be important in certain physiological functions. These observations add to the understanding of the promiscuity of K+-channel β-subunits and support the idea that K+ currents in native cells reflect a complex interaction between pore-forming α-subunits and regulatory β-subunits.
METHODS
Molecular biology
cDNA coding for rKv1.1, rKv1.2, rKv1.4, rKv1.5, and hKv4.3 were kindly provided by O. Pongs. To obtain robust expression in Xenopus laevis oocytes rKv1.1, rKv1.3, rKv1.5, hKv4.3, and mKCNE4 (accession number 391797, The National Center for Biotechnology Information (NCBI)), cDNA were subcloned into the oocyte and mammalian expression vector pXOOM (Jespersen et al., 2002). Kv1.1SYG was generated by overlap extension PCR altering the GGA codon (Gly) into a TCG codon (Ser) of the rKv1.1 gene located in the pXOOM-rKv1.1 plasmid. Specific current could be obtained from Kv1.2 and Kv1.4 channels expressed from their original vectors, pAKS2-RCK5 and pAKS2-RCK4, respectively, and these channels were therefore not subcloned into the pXOOM vector.
For expression in HEK293 cells, the internal ribosome entry site (IRES) from EMCV (from pIRES-puro, ClonTech, Palo Alto, CA) and either Kv1.1 or Kv1.3 were PCR amplified, linked together by overlap extension, and subcloned into the mammalian expression vector pNS2n, a custom-designed derivative of pcDNA3neo (InVitrogen, Carlsbad, CA) expressing a neo-EGFP fusion gene, to give pIRES-Kv1.1 and pIRES-Kv1.3. The nucleotide linker between IRES and KCNQ1 was designed to have an optimal sequence for initiation of translation (ACACGATAATACCATG) (Morgan et al., 1992). pKCNE4-Kv1.1 and pKCNE4-Kv1.3 were constructed by inserting the KCNE4 gene upstream of the IRES element. Usage of an IRES element for bicistronic expression ensures expression of both KCNE4 and Kv proteins. The KCNE4-c-myc-tagged protein used in the confocal analyses was constructed by oligonucleotides and PCR, introducing nucleotides encoding a c-myc epitop sequence (EQKLISEEDL) in the C-terminal end of mKCNE4. The integrity of all PCR-generated constructs was confirmed by sequencing.
In vitro transcription and capping were performed using the mCAP mRNA capping kit (Stratagene, La Jolla, CA). mRNA was phenol/chloroform extracted, ethanol precipitated, and dissolved in TE buffer (10 mM Tris·Cl, 1 mM EDTA, pH 7.5) to a concentration of ∼0.1 μg/μl. The integrity of the transcripts was confirmed by agarose gel electrophoresis, and mRNA was stored at −80°C until injection.
Transient expression in Xenopus oocytes and HEK293 cells
Xenopus laevis surgery and oocyte treatment were done as previously described (Grunnet et al., 2001). Oocytes were kept in Kulori medium (90 mM NaCl, 1mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM HEPES, pH 7.4) for 24 h at 19°C before injection of 50 nl of mRNA (∼5 ng). For coexpression of Kv1- and KCNE4-subunits, cRNA was mixed in a 1:1 molar ratio. Since the absolute specific current level of oocytes expressing Kv1.1 and Kv1.3 channels were ∼10 times the current level of oocytes expressing Kv1.2 channels, all coexpression studies with Kv1.1 and Kv1.2 or Kv1.3 and Kv1.2 were performed after mRNA injection in a 1:10 ratio. Oocytes were kept at 19°C in Kulori medium for 2–7 days before measurements were performed.
HEK293 cells were grown in DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% FCS (Life Technologies) at 37°C, in 5% CO2. One day before transfection, 2 × 106 cells were plated in a cell culture T75 flask (Nunc). Cells were transfected with 2 μg of plasmid, using Lipofectamine (Life Technologies) according to the manufacturer's instructions. Electrophysiological studies were performed 48–72 h post transfection.
Electrophysiology
Oocytes. Current was recorded using a two-electrode voltage-clamp amplifier (Dagan CA-1B, Minneapolis, MN, USA). Electrodes were pulled from borosilicate glass capillaries on a horizontal patch electrode puller and had tip resistance between 0.3 and 2.0 MΩ when filled with 1 M KCl. During the experiments oocytes were placed in a small chamber (volume: 200 μl) connected to a continuous flow system (flow: 6 ml/min). Kv1 channels were activated by membrane depolarization, and channel activity was measured in Kulori solution consisting of (in mM): 90 NaCl, 1 KCl, 1 MgCl2, 1 CaCl2, 5 HEPES; pH was 7.4. All experiments were performed at room temperature.
HEK293 cells. Experiments were performed in whole-cell patch-clamp configuration at room temperature with an EPC-9 amplifier (HEKA Electronics, Lambrecht, Germany). Pipettes were pulled from thin-walled borosilicate glass and had a resistance between 1.5 and 2.5 MΩ. A custom-made perfusion chamber (volume 15 μl) with a fixed AgCl-Ag pellet electrode was mounted on the stage of an inverted microscope. Coverslips with transiently transfected HEK293 cells were transferred to the perfusion chamber and superfused with physiological solutions consisting of (in mM) 150 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2 and 10 HEPES (pH 7.4 with NaOH). Pipettes were filled with solutions consisting of (in mM) 144 KCl, 1 EGTA and 10 HEPES (pH 7.2 with KOH). CaCl2 and MgCl2 were added in concentrations calculated (EqCal, BioSoft, Cambridge, UK) to give a free Mg2+ concentration of 1 mM and free Ca2+ concentrations of 100 nM. Cell capacitance and series resistance was updated before each pulse application. Series resistance values were between 2.5 and 10.0 MΩ, and only experiments where the resistance remained constant during the experiments were analyzed. Current signals were low-pass filtered at 3 kHz and acquired using Pulse software (HEKA Electronics).
Analysis of electrophysiological data
Data analyses and drawings were performed using IGOR software (WaveMetrics, Lake Oswego, OR). All values are shown as mean ± SE. The degree of significance between the differences of two means was calculated using the t-test.
Fluorescence staining and confocal analysis
Cells were fixed in 4% (w/v) paraformaldehyde in PBS (136 mM NaCl, 2.5 mM KCl, 1.5 mM KH2PO4, 6.5 mM NaHPO4, pH 7.4) for 15 min, washed in PBS, and quenched for 20 min with 50 mM NH4Cl in PBS. After washing in PBS, cells were permeabilized with PBS containing 0.1% (w/v) Triton X-100 and 0.12% (w/v) fish skin gelatin for 8 min. To reduce nonspecific labeling, cells were blocked with 0.25% (w/v) gelatin in PBS for 30 min. The cells were then exposed to a primary antibody directed against Kv1.1 (1/100 dilution, Alomone Labs, Jerusalem, Israel) or c-myc (1/300 dilution, Boehringer Mannheim, Basel, Switzerland) for 1 h followed by incubation with Alexa 488-donkey anti-mouse IgG and Alexa 568-goat anti-rabbit IgG (1/800 dilutions, Molecular Probes, Leiden, The Netherlands) for 45 min. After washing, the coverslips were mounted in Prolong antifade (Molecular Probes, Leiden, the Netherlands).
The fluorescent-labeled samples were examined by sequential laser scanning confocal microscopy using a LEICA TCS SP2 equipped with argon and helium-neon lasers. The objective was 63×, NA1.2. Images were treated using Metamorph imaging software and Adobe Photoshop 5.5.
Cell surface biotinylation
HEK293 cells. The plasma membrane was labeled using the membrane-impermeable biotinylation reagent, sulfo-NHS-SS-biotin (Pierce Chemical, Rockford, IL). Kv1.1/KCNE4-expressing HEK293 cells were rinsed three times at 4°C with PBS (136 mM NaCl, 2.5 mM KCl, 1.5 mM KH2PO4, 6.5 mM NaHPO4, pH 7.4) containing 1 mM CaCl2 and 1 mM MgCl2 (PBS2+) and washed for 5 min. The buffer was removed, and biotinylation was performed by incubation in 1 mg/ml sulfo-NHS-SS-biotin in PBS2+ for 2 × 30 min at 4°C. The biotinylation was terminated by removal of the medium and by five washes (2 min each) in 2 mg/ml BSA in PBS2+ at 4°C.
Xenopus oocytes
Oocytes injected with KCNQ1 or KCNE4 and KCNQ1 cRNA were rinsed three times at 4°C in Kulori solution and washed for 5 min. The buffer was removed and biotinylation was performed in 35-mm dishes by incubation in 1.5 mg/ml sulfo-NHS-SS-biotin (Pierce) in Kulori solution for 2 × 20 min at 4°C. The biotinylation was terminated by a 20-min incubation in Kulori solution containing 100 mM glycine.
Preparation of membranes from Xenopus oocytes
Membranes from oocytes were prepared as described earlier (Grunnet et al., 2001). Batches of 50 oocytes injected with KCNQ1, KCNQ1 + KCNE4 or noninjected oocytes were homogenized in 10% sucrose dissolved in homogenization buffer (600 mM KCl, 5 mM MOPS, 100 μM PMSF, 1 μM pepstatin A, 1 μM p-aminobenzamide, aprotinine (1 μg/ml), and leupeptin (1 μg/ml), pH 6.8) in a volume of 10 μl/oocyte with 10 strokes at 1000 rpm in a glass/Teflon homogenizer (Braun-Melsungen) at 0°C. The homogenate was placed on top of a step gradient consisting of 7 ml 50% sucrose and 3.5 ml 20% sucrose in homogenization buffer and centrifuged at 67,000 × g for 30 min at 4°C in a Beckman SW 40 rotor. The interface (between 20% and 50% sucrose) was collected and subjected to centrifugation at 84,000 × g for 30 min at 4°C in a Beckman Ti 70.1 rotor. The supernatant was discarded and the pellet resuspended in 200 μl 300 mM sucrose, 100 mM KCl, 5 mM MOPS, pH 6.8 and stored at −80°C until use.
Streptavidin-coupled agarose adsorption
All steps were performed on ice unless otherwise indicated. Extracts of purified HEK293/oocyte membranes were diluted in 0.5 vol of a 1:1 slurry of streptavidin-coupled agarose beads (Sigma, St. Louis, MO) in binding buffer (1% TX-100, 150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCL, pH 7.5). Samples were incubated overnight at 4°C with end-over-end rolling mixing. The beads were then pelleted by centrifugation for 2 min at 367 × g. They were washed thrice in binding buffer, twice in binding buffer containing 0.5 M NaCl, and once in 50 mM Tris, pH 7.5. The pelleted beads were resuspended in 50 mM Tris, pH 7.5 and Laemmli sample buffer (0.0625 M Tris, 9.9% glycerol, 2% SDS, 5% beta-mercaptoethanol, 0.00125% bromphenol blue) added. Biotinylated proteins were eluted by heating for 4 min on a heating block at 100°C and were immediately collected using a syringe. The eluates were analyzed by SDS-PAGE followed by immunoblotting.
Western blotting
SDS-PAGE (8% acrylamide) was performed using the Bio-Rad Laboratories mini-gel system (Hercules, CA). Proteins were transferred onto a hybond-P PVDF transfer membrane (Amersham Biosciences, Freiburg, Germany, 0.45 μm) in 25 mM Tris base, 200 mM glycine, 20% methanol using a mini-transblot. After transfer, the membranes were incubated for 1 h at room temperature in blocking buffer (PBS containing 5% low-fat milk powder and 0.1% Tween-20). The membrane was incubated overnight in blocking buffer containing an antibody directed against Kv1.1 (1/100 dilution, Alomone Labs, Jerusalem, Israel). After washing, bound antibody was revealed with HRP-conjugated goat anti-rabbit IgG antibody (1/10000, Jackson Immunosearch Laboratories, Westgrove, PA) in blocking buffer for 30 min, followed by visualization with the ECLPlus detection system (Amersham Biosciences) according to manufacturer's instructions. Immunoblots were exposed on hyperfilm ECL (Amersham Biosciences).
Real-time RT-PCR
Tissues were isolated from 3-week-old female NMRI mice, 14-day-old embryo Swiss Webster mice (Ambion, Austin, TX), CHO-K1 cells, unstimulated and stimulated human CD4 positive T-lymphocytes prepared as previously described (Nielsen et al., 1996) (gift from N. Ødum), and total RNA extracted by the RNAqueous™-Midi kit (Ambion) according to the manufacturer's instructions. To avoid DNA contamination, the obtained RNA was treated with DNaseI followed by phenol/chloroform extraction. cDNA synthesis was performed with M-MLV reverse transcriptase (Promega) with a random hexanucleotide, according to the manufacturer's instructions. The chosen PCR primers give rise to fragments around 300 bp for mKCNE4, hKCNE4, mGADPH, and hGADPH. The real-time PCR was performed on a DNA Engine Opticon TM machine from MJ Research™ with the Opticon Monitor™ software. SYBR® Green PCR Master Mix (Applied Biosystems) was used, and the reactions were performed under the following conditions: 95°C for 30 s, 66°C for 15 s, and 72°C for 1 min, preceded by 5 min at 95°C and followed by 5 min at 72°C. Melting curves were performed after each reaction to verify the specificity of the product. As variations in cDNA quality within the different samples may occur, GADPH was included as an internal reference. By subtracting the GADPH CP (crossing point) value from the KCNE4 CP value of each sample, the cDNA quality of each sample is taken into account (Livak and Schmittgen, 2001). The relative quantification of the mRNA levels in each individual sample was done according to the model suggested by M. Pfaffl (Pfaffl, 2001). For each primer set a standard curve was made and the slope factor calculated. The corresponding real-time PCR efficiency (E) of one cycle in the exponential phase was calculated according to the equation: E = 10(−1/slope). The relative KCNE4 ratios were calculated according to the following equation: ratio = EΔCP(control − sample).
RESULTS
KCNE4 inhibits Kv1.1 and Kv1.3 channels
To investigate if KCNE4-subunits could affect the gating of Kv channels, Kv1.1 and Kv1.3 channels alone or together with the KCNE4-subunit were expressed in oocytes from Xenopus laevis and two-electrode voltage-clamp recordings were performed (Fig. 1). As expected, Kv1.1- and Kv1.3-expressing oocytes showed fast activating and slowly inactivating voltage dependent currents, which were activated at potentials more positive than −50 mV (Fig. 1 A). Coexpression of KCNE4 with either Kv1.1 or Kv1.3 resulted in a dramatic reduction of the specific Kv current to levels between a 10-fold decrease and a complete inhibition. In experiments where a very reduced Kv1.1 or Kv1.3 current could be detected, no change in the kinetics could be observed compared to Kv1.1 or Kv1.3 channels expressed in the absence of KCNE4. The average maximal current measured at +60 mV was determined to 28.1 ± 2.1 μA (n = 20) for Kv1.1 alone and to 2.3 ± 0.5 μA (n = 16) for Kv1.1 + KCNE4 (p < 0.0001). For Kv1.3 and Kv1.3 + KCNE4 the current was measured to be 12.9 ± 1.2 μA (n = 50) and 0.4 ± 0.1 μA (n = 33), respectively (p < 0.0001, illustrated in Fig. 1 B).
FIGURE 1.
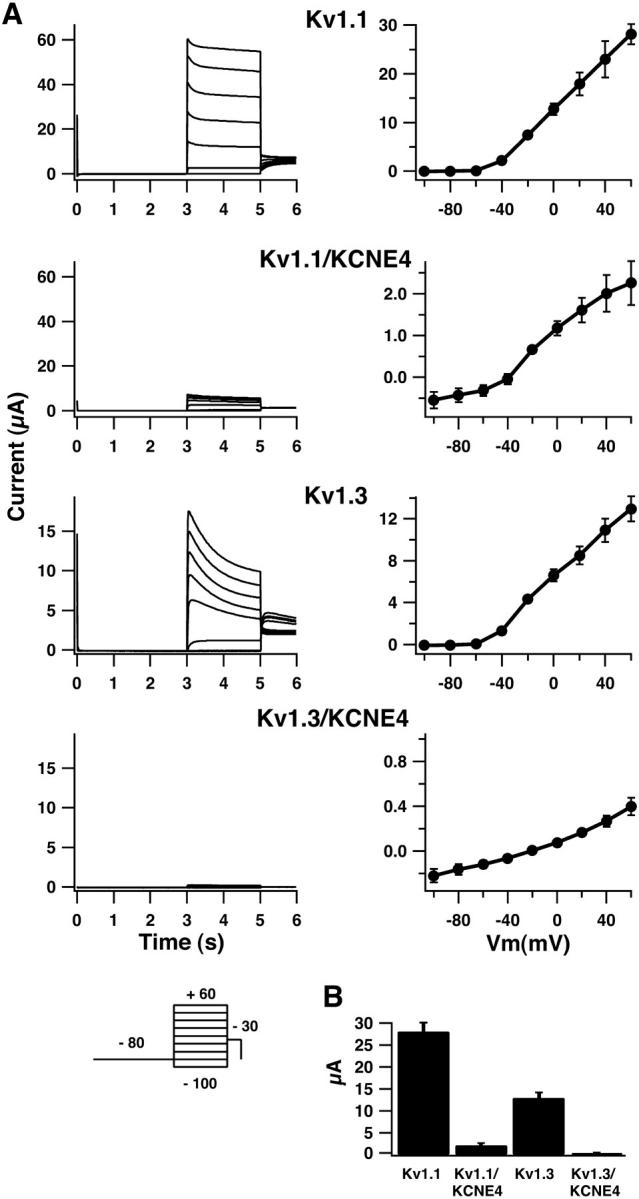
KCNE4 inhibits Kv1.1 and Kv1.3 currents in Xenopus laevis oocytes. (A) Current traces (left side) and averaged IV-curves (right side) obtained in oocytes expressing Kv1.1 or Kv1.3 alone or together with KCNE4. Oocytes were clamped at −80 mV for 3 s, current traces were elicited by 2-s voltage steps to potentials ranging from −100 mV to +60 mV in 20 mV increments, and tail currents were recorded at −30 mV (see inserted protocol). Oocytes expressing only Kv1.1 or Kv1.3 channels responded to this voltage protocol with a fast-activating and a slow-inactivating current. For oocytes coinjected with KCNE4, a drastic reduction in the overall current level was observed. (B) Mean current values of oocytes expressing either Kv1.1 or Kv1.3 alone or together with KCNE4. Peak current values were measured at +60 mV. For Kv1.1 and Kv1.1/KCNE4, an average current level of 28.1 ± 2.1 μA (n = 20) and 2.3 ± 0.5 μA (n = 16), respectively, was found. A similar drastic reduction in the overall current level was observed for Kv1.3 and Kv1.3/KCNE4 with values of 12.9 ± 1.2 μA (n = 50) and 0.4 ± 0.1 μA (n = 33), respectively.
Potential interactions between other KCNE-subunits and Kv1.1 or Kv1.3 were also investigated in the oocyte expression system. We did not observe any effect of KCNE1, KCNE2, or KCNE3 on Kv1.1 channels expressed in oocytes (n = 5, data not shown). Nor did we detect any effect of KCNE1 or KCNE2 on Kv1.3 (n = 5, data not shown), which, with respect to KCNE2, has been reported previously (Abbott et al., 1999). For unknown reasons coinjection of Kv1.3 and KCNE3 cRNA killed the oocytes shortly after injection, and electrophysiological experiments have therefore not been performed with this combination. A similar observation has been reported for KCNQ1/KCNE5-injected oocytes (Angelo et al., 2002).
To exclude the possibility that the inhibition by the KCNE4-subunit could be an artifact of the oocyte expression system, we performed whole-cell electrophysiology on HEK293 cells expressing Kv1.1 or Kv1.3 alone or expressing Kv1.1 or Kv1.3 together with KCNE4 (Fig. 2). Expression of both α- and β-subunits in the same cell is ensured by usage of a bicistronic vector where Kv1 translation is initiated after an internal ribosome entry site from EMCV (see Methods). As observed in oocytes, Kv1.1- and Kv1.3-expressing HEK293 cells showed fast activating and slowly inactivating voltage-dependent currents, which activated at potentials more positive than −50 mV (Fig. 2 A). Similarly, KCNE4 was capable of significantly reducing the current amplitude of both Kv1.1 and Kv1.3, although in a less drastic manner for Kv1.1 (Fig. 2 B). The average maximal current density measured at +60 mV was 195 ± 39 pA/pF (n = 18) for Kv1.1 alone and 75 ± 17 pA/pF (n = 19) for Kv1.1 + KCNE4 (p < 0.01). For Kv1.3 and Kv1.3 + KCNE4 the current density was determined to be 91 ± 21 pA/pF (n = 9) and 13 ± 4 pA/pF (n = 12), respectively (p < 0.001).
FIGURE 2.
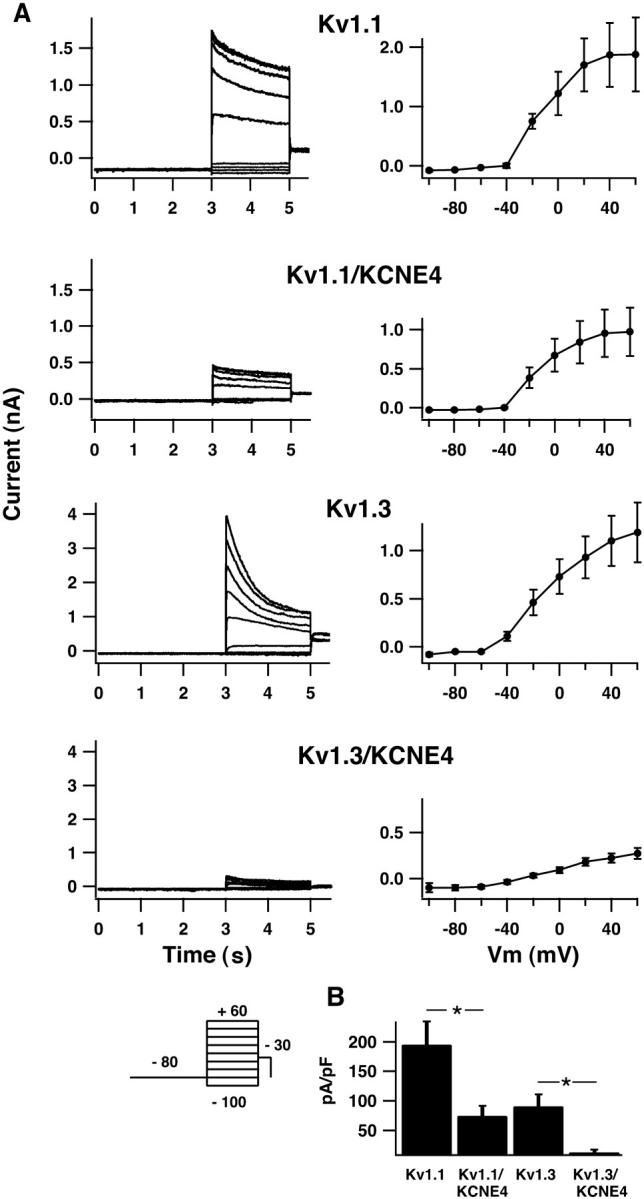
KCNE4 inhibits Kv1.1 and Kv1.3 currents in HEK293 cells. (A) Current traces from whole-cell recordings (left side) and averaged IV-curves (right side) for HEK293 cells expressing Kv1.1 or Kv1.3 alone or together with KCNE4. Transfected cells were clamped at −80 mV for 3 s, current traces were elicited by 2-s voltage steps to potentials ranging from −100 mV to +60 mV in 20-mV increments, and tail currents were recorded at −30 mV (see inserted protocol). Cells expressing Kv1.1 and Kv1.3 channels responded to this voltage protocol with a fast-activating and a slow-inactivating current. For cells expressing KCNE4 together with Kv1.1 or Kv1.3 α-subunits, a significant reduction in the overall current level was observed. (B) Mean current values of HEK293 cells expressing either Kv1.1 or Kv1.3 alone or together with KCNE4. Peak current values were measured at +60 mV. For Kv1.1 and Kv1.1/KCNE4 an average current density of 195 ± 39 pA/pF (n = 18) and 75 ± 17 pA/pF (n = 19), respectively, was found. An even more pronounced reduction in the overall current level was observed for Kv1.3 and Kv1.3/KCNE4 with values of 91 ± 21 pA/pF (n = 9) and 13 ± 4 pA/pF (n = 12), respectively.
Specificity of the KCNE4 inhibition among Kv channels
An important question is whether the inhibition by KCNE4 is a common phenomenon for Kv channels. To address this problem, Kv1.2, Kv1.4, Kv1.5, and Kv4.3 channels were expressed either alone or together with KCNE4 in the Xenopus oocytes. As demonstrated in Fig. 3, no significant alterations in kinetics or current amplitude were observed for Kv1.2, Kv1.4, Kv1.5, and Kv4.3 coexpressed with KCNE4 as compared to controls. The maximal current density measured at +60 mV was, for Kv1.2 alone, 1.71 ± 0.14 μA (n = 16); for Kv1.2/KCNE4, 1.53 ± 0.19 μA (n = 11); for Kv1.4, 1.26 ± 0.30 μA (n = 16); for Kv1.4/KCNE4, 1.61 ± 0.33 μA (n = 8); for Kv1.5 alone, 26.1 ± 8.4 μA (n = 3); for Kv1.5/KCNE4, 19.8 ± 2.3 μA (n = 5); for Kv4.3 alone, 26.4 ± 2.4 μA (n = 3); and for Kv4.3/KCNE4, 24.8 ± 2.0 μA (n = 3).
FIGURE 3.
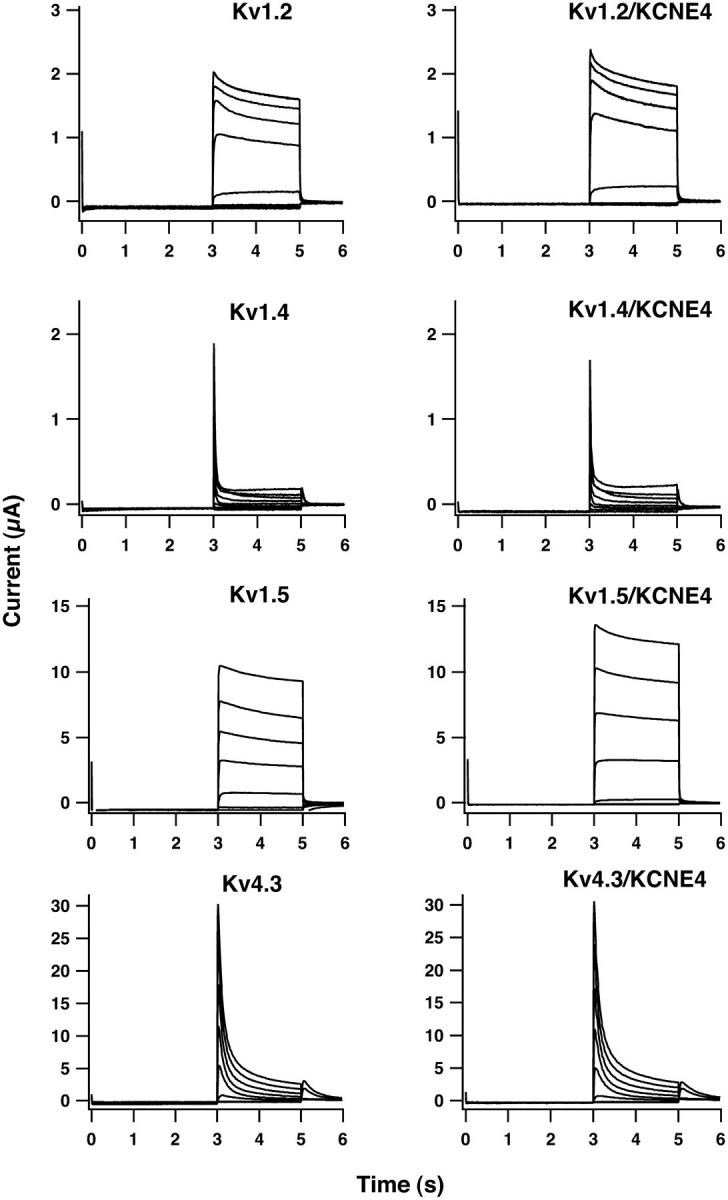
KCNE4 does not inhibit Kv1.2, Kv1.4, Kv1.5, or Kv4.3. Current traces from oocytes expressing Kv1.2, Kv1.4, Kv1.5, and Kv4.3 alone or together with KCNE4. Oocytes were clamped at −80 mV for 3 s, current traces were elicited by 2-s voltage steps to potentials ranging from −100 mV to +60 mV in 20-mV increments, and tail currents were recorded at −30 mV. Oocytes expressing Kv channels responded to this voltage protocol with a fast activation and a slow (Kv1.2, Kv1.5) or fast (Kv1.4, Kv4.3) inactivation. No difference in kinetics or current amplitude was found for oocytes coinjected with KCNE4 cRNA. Mean current values (measured at +60 mV) of oocytes expressing either Kv1 channels alone or together with KCNE4 were as follows: 1.71 ± 0.14 μA (n = 16) for Kv1.2; 1.53 ± 0.19 μA (n = 11) for Kv1.2/KCNE4; 1.26 ± 0.30 μA (n = 16) for Kv1.4; 1.61 ± 0.33 μA (n = 8) for Kv1.4/KCNE4; 26.1 ± 8.4 μA (n = 3) for Kv1.5; 19.8 ± 2.3 μA (n = 5) for Kv1.5/KCNE4; 26.4 ± 2.4 μA (n = 3) for Kv4.3; and 24.8 ± 2.0 μA (n = 3) for Kv4.3/KCNE4.
KCNE4 inhibition of heteromeric Kv1 channels
As Kv1 α-subunits both in vitro and in vivo are known to assemble into heteromeric complexes, we were interested in analyzing the effect of KCNE4 on channel complexes containing Kv1.1- or Kv1.3-subunits (Fig. 4). At least in mammalian brain, the most abundant Kv1 α-subunits are Kv1.1 and Kv1.2, and all heteromeric Kv1 channels contain either one or both of these subunits (Koch et al., 1997; Koschak et al., 1998). Different combinations of Kv1.1, Kv1.2, and Kv1.3 (Kv1.1/Kv1.2, Kv1.1/Kv1.3 and Kv1.2/Kv1.3) were injected in ratios calculated to obtain 50% contribution of each subunit to the overall current amplitude. To confirm the existence of heteromeric channel complexes, the following experiments were conducted. First, charybdotoxin (ChTX) was applied as a pharmacological tool to separate Kv1.1 and Kv1.2 currents. As previously reported, ChTX did not inhibit homomeric Kv1.1 channels in concentrations up to 30 nM (data not shown), whereas we found Kv1.2 channels to be blocked by ChTX with an IC50 of 1.7 ± 1.9 nM (n = 4), which is in the same range as previously reported (Grissmer et al., 1994). When ChTX was added to oocytes coexpressing functionally equal amounts of Kv1.1 and Kv1.2, it was found that around 85% of the overall current could be blocked with 30 nM ChTX (Fig. 4 C). A complete random assembly of Kv1.1- and Kv1.2-subunits into functional channels would imply that 6.25% of all channels should be homomeric Kv1.1, another 6.25% homomeric Kv1.2 channels, whereas the remaining complexes should consist of heteromeric Kv1.1/1.2 channels in different stoichiometry. These results obtained by ChTX inhibition thereby talks in favor of a small subpopulation of homomeric Kv1.1 channels that are insensitive to ChTX, whereas the remaining channels are made up of predominantly heteromeric Kv1.1/1.2 complexes and a small fraction of Kv1.2 homomeric channels that are all sensitive to ChTX. It could be argued that all ChTX-sensitive current originates from homomeric Kv1.2 channels, but if this was true it should be expected that the Kv1.1/1.2 dose-response curve was rightward-shifted compared to the Kv1.2 curve, due to the presence of some homomeric Kv1.1 channels. This is not observed, again indicating the presence of heteromeric Kv1.1/1.2 complexes.
FIGURE 4.
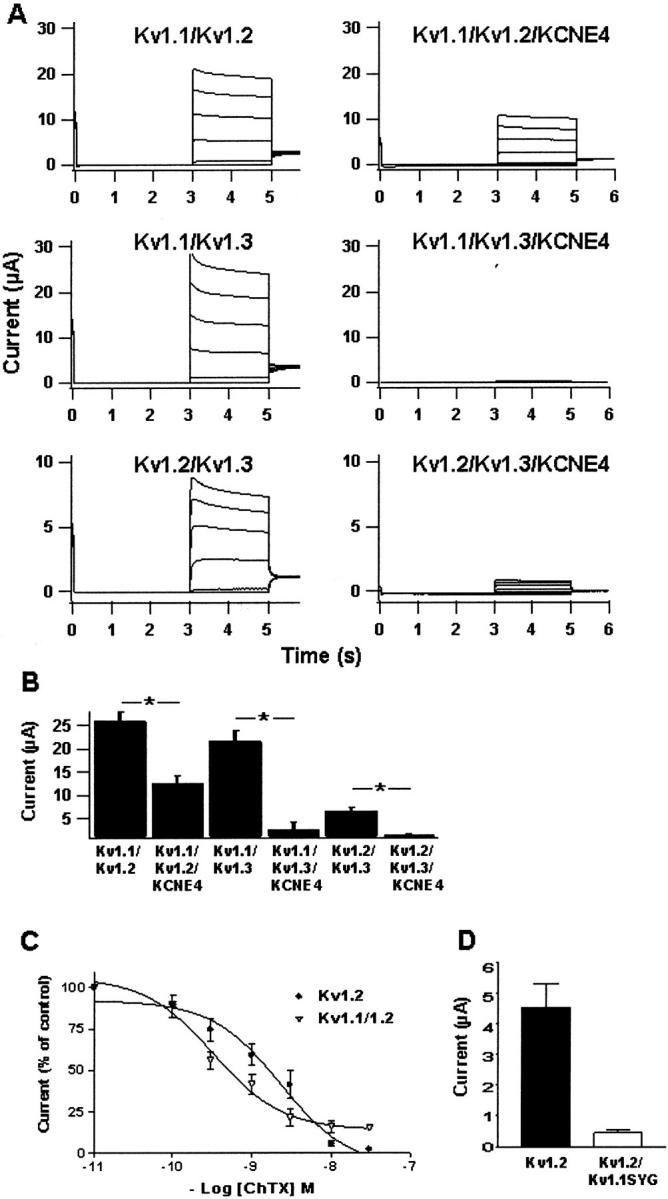
KCNE4 inhibition of heteromeric Kv1 channels. (A) Current traces from oocytes expressing Kv1.1/Kv1.2, Kv1.1/Kv1.3, and Kv1.2/Kv1.3 channels alone or together with KCNE4. Oocytes were clamped at −80 mV for 3 s, current traces were elicited by 2-s voltage steps for potentials ranging from −80 mV to +40 mV in 20-mV increments, and tail currents were recorded at −30 mV. (B) Mean current values of oocytes expressing Kv1 channels alone or of oocytes expressing Kv1 channels together with KCNE4. Peak current values, measured at +40 mV, were as follows: 26.1 ± 1.9 μA (n = 24) for Kv1.1/Kv1.2; 12.7 ± 1.7 μA (n = 24) for Kv1.1/Kv1.2/KCNE4; 21.9 ± 2.1 μA (n = 8) for Kv1.1/1.3; 2.5 ± 1.6 μA (n = 4) for Kv1.1/Kv1.3/KCNE4; 6.70 ± 0.70 μA (n = 11) for Kv1.2/Kv1.3; and 1.45 ± 0.30 μA (n = 12) for Kv1.2/Kv1.3/KCNE4. (C) Dose-response curve of charybdotoxin (ChTX) on Kv1.2- and Kv1.1/Kv1.2-expressing oocytes. Currents were measured at +60 mV. All data points are an average of four recordings on independent oocytes. (D) Current level of Kv1.2 and Kv1.2/Kv1.1SYG expressing oocytes measured at +60 mV.
Second, to analyze the potential complex formation between Kv1.1 and Kv1.2, a nonconducting dominant negative Kv1.1 mutant (Kv1.1SYG) was applied (Wollnik et al., 1997). Coexpression of Kv1.1SYG together with Kv1.2 resulted in a 10-fold current reduction as compared to the current obtained with Kv1.2 alone (4.51 ± 0.89 μA (n = 8) for Kv1.2 versus 0.43 ± 0.13 μA (n = 8) for Kv1.2/Kv1.1SYG) (Fig. 4 D). As for the ChTX experiments, this points toward formation of heteromeric Kv1.1/1.2 channels. Thereby, these experiments indicate expression of Kv1.1- and Kv1.2-subunits at a comparable level as well as heteromeric channel complex formation as reported by others (Isacoff et al., 1990; Ruppersberg et al., 1990; Christie et al., 1990). When KCNE4 was expressed together with either combination of the two α-subunits, a significant current reduction was observed. A current reduction of more than 50% was seen when KCNE4 was coexpressed with Kv1.1/Kv1.2 (12.7 ± 1.7 μA, n = 24) as compared to control levels (26.1 ± 1.9 μA, n = 24)(p < 0.0001, Fig. 4 B). As would be expected, oocytes expressing Kv1.1/Kv1.3/KCNE4 had a drastic reduction in current level (2.5 ± 1.6 μA, n = 4) compared to oocytes expressing only Kv1.1/Kv1.3 (21.9 ± 2.1 μA, n = 8) (p < 0.0001)(Fig. 4 B). When KCNE4 was expressed together with Kv1.2/Kv1.3, an almost fivefold reduction in the current level was found (1.45 ± 0.30 μA, n = 12) as compared to control oocytes expressing only Kv1.2/Kv1.3. (6.70 ± 0.70 μA, n = 11) (p < 0.0001).
Kv1.1 and Kv1.3 currents are modified by delayed injection of KCNE4 cRNA
Having established the specific inhibitory property of KCNE4 on Kv1.1 and Kv1.3 channels, a more detailed investigation of the KCNE4 inhibitory mechanism was performed. For other Kv-interacting β-subunits, it has been demonstrated that simultaneous translation of α- and β-subunits is necessary for the ability of the β-subunit to exert its effect on the α-subunit, indicating an interaction early in biosynthesis (Fink et al., 1996; Shi et al., 1996). To examine if this was true also for the inhibitory effect of KCNE4, we performed experiments with delayed injection of KCNE4 cRNA into oocytes already expressing Kv1.1 or Kv1.3 (Fig. 5). Fig. 5 A illustrates the current levels as a function of time in oocytes initially injected with Kv1.3 cRNA at time point 0. At 28 h later, when a current level of 6.6 ± 0.7 μA was reached, these oocytes were reinjected with either KCNE4 cRNA or H2O. At 32 h after the delayed injection, a complete block of the Kv1.3 current by KCNE4 was observed. At the same time point (60 h), Kv1.3-expressing oocytes, which had been reinjected with H2O, showed an almost twofold increase in the current level (11.6 ± 4.1 μA), which subsequently slowly declined until the end of the experiment after 164 h. The current in oocytes, in which the Kv1.3 channel had been blocked by delayed injection of KCNE4 or in which the channels were blocked by initial coexpression of KCNE4, remained blocked throughout the rest of the experiments (up to 100 h). As it may be speculated that injection of nonspecific cRNA can have an effect on the Kv1 current level, we injected KCNE2 cRNA into oocytes already expressing Kv1 channels. These oocytes showed a similar response as water-injected oocytes (data not shown).
FIGURE 5.
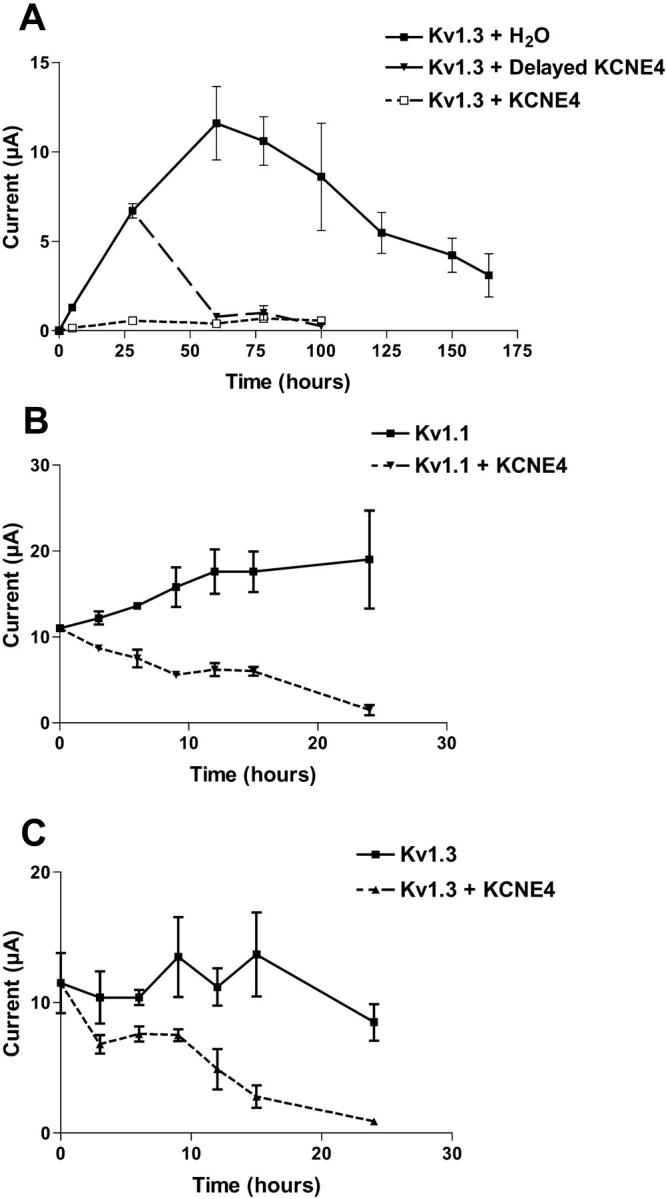
Kv current can be modified by delayed injection of KCNE4. (A) Oocytes injected with Kv1.3 cRNA were incubated 28 h, which resulted in a current level of 6.7 ± 0.7 μA recorded at +60°mV. The oocytes were then separated in two pools and subsequently injected with either H2O (filled squares) or KCNE4 cRNA (filled triangles). Injection of KCNE4 resulted in a complete inhibition of the Kv1.3 current, whereas H2O-injected oocytes continued to express Kv1.3 current. Control oocytes injected with both Kv1.3 and KCNE4 cRNA at time 0 (open squares) did not at any time provide Kv1.3 specific current. (B and C) To obtain a higher time resolution for the inhibitory property of KCNE4, oocytes expressing either Kv1.1 or Kv1.3 channels were injected with KCNE4 cRNA 72 h after Kv injection (time 0 at graph). Expression levels of Kv current were subsequently followed every third hour. For both Kv1.1- and Kv1.3-expressing oocytes, a complete inhibition of the current could be observed within 24 h. All data points are an average of four recordings on independent oocytes.
To obtain a higher time resolution of the ability of KCNE4 to inhibit Kv1.1 or Kv1.3 channels, additional delayed injecting experiments were performed (Fig. 5 B and Fig. 5 C). After 72 h when a current level of 11.0 ± 0.3 μA (Kv1.1) and 11.5 ± 4.0 μA (Kv1.3) was obtained (time 0 at graph), a fraction of the expressing oocytes was injected with KCNE4 cRNA. These experiments showed a significant reduction in the Kv1.1 and Kv1.3 current level within a few hours after delayed injection and a complete inhibition after 24 h. The results shown in Fig. 5 indicate a KCNE4 inhibitory mechanism on Kv1.1 and Kv1.3 channels similar to the mechanism we previously have reported on KCNQ1 where KCNE4 blocked KCNQ1 current within 24 h of delayed injection (Grunnet et al., 2002a).
Membrane expression of Kv1.1 is unaltered in the presence of KCNE4
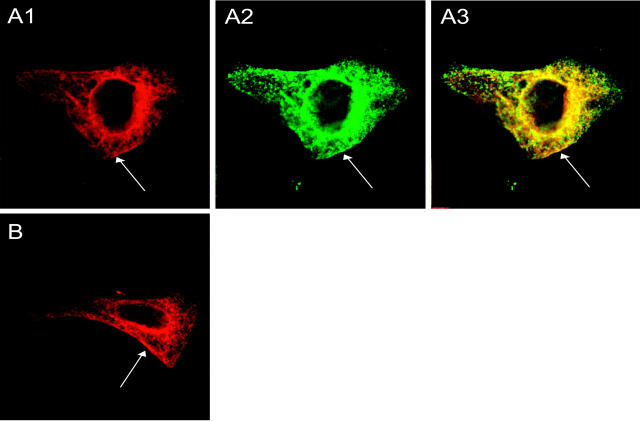
The results displayed in Fig. 5 leave some unanswered questions considering the mechanism of KCNE4 inhibition. It could be speculated that KCNE4 has the ability to withdraw functional Kv1 channels from the plasma membrane, or that a direct interaction in the plasma membrane or earlier in the translocation pathway is responsible for the inhibition. For KCNQ1 we previously observed that KCNE4-subunits do not alter the total amount of KCNQ1 proteins at the cell surface, demonstrating that the inhibition of KCNQ1 by KCNE4 takes place in the plasma membrane (Grunnet et al., 2002a). To analyze if KCNE4 also interacts with Kv1.1 in the plasma membrane, immunocytochemistry was performed on transiently transfected HEK293 cells. The cells were cotransfected with Kv1.1 or Kv1.1/KCNE4-c-myc, stained with antibodies raised against Kv1.1 channels and the c-myc epitop, and detected by confocal microscopy (Fig. 6). The functionality of the KCNE4 protein with a c-myc tag in the C-terminal tail was confirmed in the oocyte system, where no difference between wild-type and c-myc-tagged subunits was found (data not shown). Kv1.1 and KCNE4-c-myc protein could be detected both intracellularly and in what appears to be the plasma membrane, and the amount of Kv1.1 channels in the plasma membrane appeared to be the same both in the presence and absence of KCNE4 (Fig. 6, A1 and B). A relatively low amount of Kv1.1 proteins in the membrane is to be expected as Manganas and Trimmer (2000) previously showed that when Kv1.1 is expressed alone, a low percentage of the channels are found at the cell surface. When the staining pattern of Kv1.1 (Fig. 6 A1) and KCNE4-c-myc (Fig. 6 A2) were superimposed, a partly overlapping localization both in the membrane area and intracellular compartments could be detected (Fig. 6 A3).
FIGURE 6.
Kv1.1 and KCNE4 colocalize in the cell membrane. Confocal scans of a HEK293 cell transfected with Kv1.1/ KCNE4-c-cmyc (A) and Kv1.1 (B). After fixation the cells were labeled with anti-Kv1.1 antibody (A1 and B) and anti-c-myc antibody (A2). (A3) represents an overlay of the two labelings. For both proteins, large amounts of intracellular labeling were observed. However, some labeling could be detected in what appeared to be the plasma membrane (arrow). In many cases the pattern of staining of the two proteins overlapped (arrow). Scale bar 8 μm.
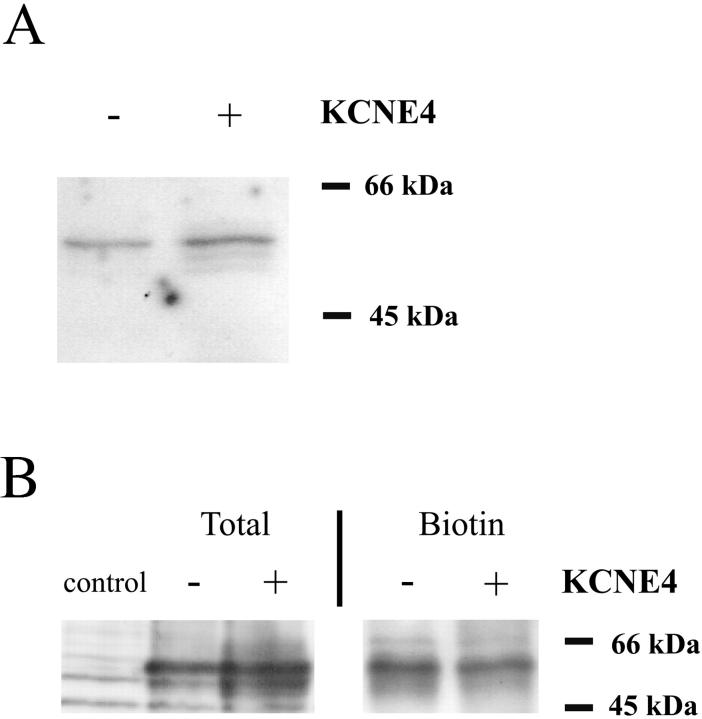
To confirm that Kv1.1 protein is located in the membrane when KCNE4-subunits are present, we performed Western blotting of membranes from oocytes and of HEK293 cells expressing Kv1.1, Kv1.1/KCNE4, and control plasmids (Fig. 7). Fig. 7 A is a Western blot of surface proteins from oocytes expressing Kv1.1 or Kv1.1/KCNE4. To obtain highly purified membranes, oocytes were initially biotinylated and membranes subsequently isolated by differential centrifugation. Finally, this membrane fraction was further purified by streptavidin-coupled agarose absorption. The Kv1.1 antibody detects a band of ∼60 kDa which corresponds with the previous reported size of the Kv1.1 protein obtained from cultured cells (Manganas and Trimmer, 2000). Whether KCNE4 is present or absent in the oocytes does not affect the surface expression level of Kv1.1, as comparable band intensities were detected in both cases. A similar result was obtained with Western blotting of biotinylated surface proteins from HEK293 cells expressing Kv1.1 or Kv1.1/KCNE4 (Fig. 7 B). Clear bands of ∼60 kDa were detected in both lanes of biotinylated surface proteins. As a control of the transfection efficiency, total protein from cells transfected with either Kv1.1 alone or KCNE4/Kv1.1 were analyzed. Comparable staining intensities were found, indicating that equal amounts of Kv1.1 protein were present in the cell-surface- and in the total-cell-preparation of cells expressing Kv1.1 and Kv1.1/KCNE4. In summary, the Western blotting and confocal microscopy demonstrate that KCNE4 does not alter the subcellular distribution of Kv1.1.
FIGURE 7.
KCNE4 does not hinder expression of Kv1.1 at the cell surface. (A) Western blot, stained with Kv1.1 antibody, of biotinylated surface proteins from oocytes expressing Kv1.1 or Kv1.1/KCNE4. (B) Western blot, stained with Kv1.1 antibody, of total protein and biotinylated surface proteins from HEK293 cells expressing Kv1.1, Kv1.1/KCNE4, and pXOOM (control). Expression of both Kv1.1 and KCNE4 in all transfected cells was obtained by usage of a bicistronic construct (see Methods). Kv1.1 was expressed at comparable levels in the plasma membrane in the absence and presence of KCNE4. + and − indicate the presence or absence of KCNE4 protein, respectively. Biotin indicates the loading of biotinylated proteins, whereas total indicates that total cell lysates were loaded.
Tissue quantification of KCNE4 mRNA levels
Kv1.1 and Kv1.3 are expressed in several organs and tissues. Therefore, to illuminate where a potential interaction between Kv1.1/Kv1.3 and KCNE4-subunits may take place in vivo, we performed real-time RT-PCR of KCNE4 on RNA obtained from a number of tissues (Table 1). Double-stranded DNA products were detected by the SYBR green fluorophore, and GADPH was used as an internal reference. The relative mKCNE4 mRNA expression level was normalized according to uterus tissue, which is the tissue with the highest expression level. Compared to uterus tissue, a relatively high expression level in embryo, lung, small intestine, colon, and kidney tissue was found by real-time RT-PCR. These results correspond well with the results we have previously obtained by RT-PCR (Grunnet et al., 2002a). In heart and brain tissue we detected ∼1% of the KCNE4 mRNA found in uterus tissue. When the brain was separated into different parts, a 1% expression level is also observed in the cerebellum, whereas a lower expression level was observed in the hippocampus, cortex, hypothalamus, and medulla spinalis. As the Kv1.3 channel is an important component in the maturation of T-lymphocytes (Grissmer et al., 1990), unstimulated and stimulated human T-lymphocytes were also tested for KCNE4 expression. No significant difference in KCNE4 expression was observed between unstimulated and stimulated T-cells, indicating that KCNE4 does not play a role in maturation of T-lymphocytes.
TABLE 1.
KCNE4 mRNA expression in mouse tissues and human T-cells.
| Tissue | CP(KCNE4) (Mean ± SD) | CP(GADPH) (Mean ± SD) | CP(KCNE4) – CP(GADPH) | Normalized (KCNE4) (relative to uterus) |
|---|---|---|---|---|
| Brain | 28.6 ± 0.4 | 20.2 ± 0.7 | 8.4 ± 0.9 | 0.01 |
| Cerebellum | 31.8 ± 0.5 | 20.9 ± 1.1 | 10.9 ± 1.2 | 0.01 |
| Hippocampus | 32.9 ± 0.5 | 21.3 ± 0.9 | 11.6 ± 1.0 | 0.0004 |
| Cortex | 32.4 ± 0.8 | 21.1 ± 0.8 | 11.2 ± 1.1 | 0.0006 |
| Hypothalamus | 31.2 ± 0.5 | 20.8 ± 0.8 | 10.4 ± 0.9 | 0.001 |
| Medulla spinalis | 31.9 ± 0.4 | 20.8 ± 0.9 | 11.1 ± 1.0 | 0.0007 |
| Uterus | 26.3 ± 0.3 | 23.5 ± 1.0 | 2.8 ± 1.1 | 1 |
| Kidney | 26.8 ± 0.1 | 20.9 ± 0.8 | 5.9 ± 0.8 | 0.11 |
| Lung | 29.6 ± 0.3 | 25.0 ± 1.1 | 4.7 ± 1.1 | 0.30 |
| Colon | 28.4 ± 0.09 | 22.8 ± 0.8 | 5.6 ± 0.8 | 0.15 |
| Small intestine | 28.0 ± 0.3 | 22.4 ± 1.1 | 5.6 ± 1.1 | 0.15 |
| Heart | 28.3 ± 0.08 | 19.8 ± 0.9 | 8.5 ± 0.9 | 0.009 |
| Embryo | 26.9 ± 0.4 | 22.7 ± 0.9 | 4.3 ± 1.0 | 0.60 |
| CHO-K1 | >45 | 32.0 ± 1.5 | >13 | <0.0001 |
| T-lymphocytes | ||||
| Unstimulated | 30.6 ± 0.3 | 21.2 ± 0.2 | 9.4 ± 0.4 | nd |
| Stimulated | 30.5 ± 0.3 | 21.9 ± 0.6 | 8.6 ± 0.6 | nd |
Real-time RT-PCR analyses of total RNA extracted from NMRI mice tissue, 14-day-old-embryo Swiss Webster mice (Ambion), CHO-K1 cells, and human T-lymphocytes. CP (crossing point) is the PCR-cycle number where a 0.01 fluorescent signal is detected (Opticon Monitor™ software). nd, not done.
DISCUSSION
In the present study we have shown that KCNE4 drastically inhibits Kv1.1 and Kv1.3 currents independently of the applied expression system. In Xenopus oocytes a more than 10-fold reduction and in HEK293 cells a 2.5-fold (Kv1.1) and a 7-fold (Kv1.3) reduction in the current amplitude was found. For most potassium channel β-subunits described until now, either a change in kinetics or an enhanced surface expression has been observed (Rettig et al., 1994; Shi et al., 1996; Barhanin et al., 1996; Sanguinetti et al., 1996; Abbott et al., 2001). However, we have recently shown that KCNE4 inhibits KCNQ1 current in both oocytes and mammalian cells, whereas others have reported that ERG1 and KCNQ4 currents are inhibited by KCNE3 in oocytes and that KCNE2 reduces KCNQ1 current in mammalian cells (Grunnet et al., 2002a; Tinel et al., 2000a; Schroeder et al., 2000).
It should be kept in mind that the choice of expression system for the coexpression of α- and β-subunits may seriously interfere with the results of experiments. For example, it has been described that KCNE2 does not affect KCNQ1 gating when coexpressed in oocytes, although KCNE2 apparently is associated with the KCNQ1 channel complex (Tapper and George, 2000; Abbott et al., 1999). To the contrary, coexpression of KCNE2 and KCNQ1 in mammalian cells resulted in a dramatic shift in the voltage-sensitivity of the KCNQ1 channels (Tinel et al., 2000a; Abbott et al., 1999). Until recently expression in both oocytes and mammalian cells suggested KCNE2 and hERG1 as the molecular component behind the cardiac Ikr current (Abbott et al., 1999; Isbrandt et al., 2002; Cui et al., 2001). However, this notion has now been challenged by the fact that KCNE2 and hERG1 expressed in mammalian cells might not recapitulate the native Ikr current (Weerapura et al., 2002). The fact that our experiments show that the inhibitory effect of KCNE4 on Kv1.1 and Kv1.3 channels is found after coexpression in oocytes as well as in mammalian cells suggests that this effect of KCNE4 may also be found in native cells, where KCNE4 β-subunits may be colocalized with voltage-regulated potassium channels of the Kv type.
The KCNE4 inhibition is not a general phenomenon among Kv channels, as KCNE4 proteins do not affect the kinetics or current levels of Kv1.2, Kv1.4, Kv1.5, or Kv4.3. In vivo Kv1 channels are often found to be heteromeric complexes in which up to four different Kv1-subunits form the functional channel (Shamotienko et al., 1997). As Kv1.2 is one of the major components in Kv1 heteromeric complexes, we coinjected Kv1.2 with either Kv1.1 or Kv1.3 into oocytes to analyze if KCNE4-subunits inhibit these heteromeric channels. When KCNE4 was expressed together with these complexes, a twofold and a fivefold reduction in the current level was observed for Kv1.1/Kv1.2 and for Kv1.2/Kv1.3 complexes, respectively. These results indicate that KCNE4 proteins somehow interact with Kv1.1- and Kv1.3-subunits present in the heteromeric complexes and thereby modulate the entire Kv1 channel complex.
KCNE4 has an overall 25–30% peptide sequence identity to KCNE1-3,5 and in the proposed transmembrane regions there is a 60–70% similarity between KCNE4 and the other KCNEs. For KCNE1, the transmembrane region has been shown to interact directly with the α-subunit (KCNQ1), and both the transmembrane segment and the C-terminal are responsible for modulation of kinetics (Romey et al., 1997; Tai and Goldstein, 1998; Tapper and George, 2000). Whether it is the transmembrane segment or other parts of the KCNE4 protein that is responsible for the interaction and modulation of Kv1.1/Kv1.3 and KCNQ1 channels is unknown. Abbott and Goldstein showed through mutant studies that similar mutations within the KCNE1,2,3-subunits result in modulation of three different α-subunits in a comparable manner (Abbott and Goldstein, 2002). This indicates that KCNEs interact by a similar mechanism with different α-subunits, which may explain why KCNE4 can inhibit such relatively different proteins as Kv1 and KCNQ1 channels (Grunnet et al., 2002a).
The mechanism behind KCNE4 and Kv1.1/Kv1.3 interactions was examined in a number of experiments. Western blotting and confocal microscopy showed that the cellular distribution of Kv1.1 and KCNE4 proteins was unchanged, even though either no current or a very reduced current could be detected, as compared to cells expressing only Kv1.1. This indicates that KCNE4 inhibition of Kv1.1/Kv1.3 is not due to an effect on the translation machinery or the transport pathway but rather caused by a direct effect on the gating of channels located in the cell surface membrane. However, this is only a proposition, as we cannot conclude from the conducted experiments that KCNE4 interacts directly with the channel complexes in the membrane. KCNE4 modulation of channel complexes already present in the cells opens up for the interesting aspect that KCNE4-subunits under certain conditions could be expressed and interact with Kv1.1 and Kv1.3 channels. Indeed, delayed injection experiments demonstrated that currents generated by Kv1.1 and Kv1.3 channels can be almost completely inhibited by KCNE4-subunits within 24 h of injection.
Since Kv1 potassium channels are expressed both in the brain and in many peripheral tissues, a subunit with inhibitory properties such as KCNE4 will potentially have a significant impact on membrane potential in many cell types. In excitable tissue such an effect would mean a general increase in cellular excitability as Kv1 channels normally reduce the firing rate. In other cell types, such as epithelia, the ability of these channels to perform transepithelial transport could be affected by KCNE4 expression. This potential of KCNE4-subunits will depend on colocalization of KCNE4 and Kv1.1 or Kv1.3. By real-time RT-PCR, a relatively high expression level of mKCNE4 mRNA was detected in uterus, kidney, lung, colon, and 14-day-old embryo. Lower expression level was found in heart, total-brain, and five different brain parts. These results fit very well with previous results generated by semiquantative RT-PCR (Grunnet et al., 2002a). The tissue distribution described above indicates that the functional role of KCNE4 may be found in epithelial tissues. Kv1 proteins have been described in several tissues in which we detect KCNE4 mRNA, such as lung, kidney, colon and uterus, but the exact location and function of the putative interaction cannot be concluded from this study.
As Kv1.1/Kv1.3 and KCNE4 may be coexpressed in several different cell types, it can be speculated that KCNE4 participates in a subacute (i.e., within hours) regulation of cellular functions by modulating the Kv1 current. What triggers the expression of KCNE4 is an open question, but hormones could be potential candidates. The time frame of the KCNE4 modulatory effect on Kv1.1/Kv1.3 channels is comparable to the time frame observed for proteins regulated by hormones, and, indeed, another member of the KCNE family, namely KCNE1, has been found to be regulated by hormones (Felipe et al., 1994). The function of KCNE4 could thereby be to down-regulate the conductance of Kv1.1/Kv1.3 channels already present in the cell surface.
We have previously reported that KCNQ1 is inhibited by KCNE4 (Grunnet et al., 2002a). The KCNE4 inhibition of KCNQ1 is comparable to the presented inhibition of Kv1.1/Kv1.3. For several of the other KCNE-subunits it has, as mentioned earlier, been shown that they may interact with more than one α-subunit family. β-subunits having more than one α-subunit partner give the possibility of having a very complex regulation of ion channels on the cellular level with a relatively low number of genes.
In summary, the results in this paper demonstrate the ability of KCNE4 to severely inhibit certain types of Kv1 channels. The growing number of reports showing interactions between KCNE proteins and different types of potassium channels points to this family of regulatory β-subunits as very important in a number of different physiological functions. However, the exact physiological function of the KCNE4 and Kv1.1/Kv1.3 interaction has to be elaborated in future studies.
Acknowledgments
We thank Birthe Lynderup, Tove Soland, Pia Hagman, and Inge Kjeldsen for competent technical assistance and Niels Ødum for providing the T-lymphocytes.
The Danish Heart Association, The John and Birthe Meyer Foundation, The Medical Research Council, The Carlsberg Foundation, and the Novo Nordisk Foundation supported this work.
References
- Abbott, G. W., M. H. Butler, S. Bendahhou, M. C. Dalakas, L. J. Ptacek, and S. A. Goldstein. 2001. MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell. 104:217–231. [DOI] [PubMed] [Google Scholar]
- Abbott, G. W., and S. A. Goldstein. 2002. Disease-associated mutations in KCNE potassium channel subunits (MiRPs) reveal promiscuous disruption of multiple currents and conservation of mechanism. FASEB J. 16:390–400. [DOI] [PubMed] [Google Scholar]
- Abbott, G. W., F. Sesti, I. Splawski, M. E. Buck, M. H. Lehmann, K. W. Timothy, M. T. Keating, and S. A. Goldstein. 1999. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 97:175–187. [DOI] [PubMed] [Google Scholar]
- Adda, S., B. K. Fleischmann, B. D. Freedman, M. Yu, D. W. Hay, and M. I. Kotlikoff. 1996. Expression and function of voltage-dependent potassium channel genes in human airway smooth muscle. J. Biol. Chem. 271:13239–13243. [DOI] [PubMed] [Google Scholar]
- Angelo, K., T. Jespersen, M. Grunnet, M. S. Nielsen, D. A. Klaerke, and S.-P. Olesen. 2002. KCNE5 induces time- and voltage-dependent modulation of the KCNQ1 current. Biophys. J. 83:1997–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhanin, J., F. Lesage, E. Guillemare, M. Fink, M. Lazdunski, and G. Romey. 1996. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 384:78–80. [DOI] [PubMed] [Google Scholar]
- Baumann, A., A. Grupe, A. Ackermann, and O. Pongs. 1988. Structure of the voltage-dependent potassium channel is highly conserved from Drosophila to vertebrate central nervous systems. EMBO J. 7:2457–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandy, K. G., and G. A. Gutman. 1995. CRC Handbook of Receptors and Channels. CRC Press, Boca Raton, FL. 1–75.
- Chandy, K. G., C. B. Williams, R. H. Spencer, B. A. Aguilar, S. Ghanshani, B. L. Tempel, and G. A. Gutman. 1990. A family of three mouse potassium channel genes with intronless coding regions. Science. 247:973–975. [DOI] [PubMed] [Google Scholar]
- Christie, M. J., R. A. North, P. B. Osborne, J. Douglass, and J. P. Adelman. 1990. Heteropolymeric potassium channels expressed in Xenopus oocytes from cloned subunits. Neuron. 4:405–411. [DOI] [PubMed] [Google Scholar]
- Cui, J., A. Kagan, D. Qin, J. Mathew, Y. F. Melman, and T. V. McDonald. 2001. Analysis of the cyclic nucleotide binding domain of the HERG potassium channel and interactions with KCNE2. J. Biol. Chem. 276:17244–17251. [DOI] [PubMed] [Google Scholar]
- Dixon, J. E., and D. McKinnon. 1994. Quantitative analysis of potassium channel mRNA expression in atrial and ventricular muscle of rats. Circ. Res. 75:252–260. [DOI] [PubMed] [Google Scholar]
- Felipe, A., T. J. Knittle, K. L. Doyle, D. J. Snyders, and M. M. Tamkun. 1994. Differential expression of Isk mRNAs in mouse tissue during development and pregnancy. Am. J. Physiol. 267:C700–C705. [DOI] [PubMed] [Google Scholar]
- Fink, M., F. Duprat, F. Lesage, C. Heurteaux, G. Romey, J. Barhanin, and M. Lazdunski. 1996. A new K+ channel beta subunit to specifically enhance Kv2.2 (CDRK) expression. J. Biol. Chem. 271:26341–26348. [DOI] [PubMed] [Google Scholar]
- Grissmer, S., B. Dethlefs, J. J. Wasmuth, A. L. Goldin, G. A. Gutman, M. D. Cahalan, and K. G. Chandy. 1990. Expression and chromosomal localization of a lymphocyte K+ channel gene. Proc. Natl. Acad. Sci. USA. 87:9411–9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissmer, S., A. N. Nguyen, J. Aiyar, D. C. Hanson, R. J. Mather, G. A. Gutman, M. J. Karmilowicz, D. D. Auperin, and K. G. Chandy. 1994. Pharmacological characterization of five cloned voltage-gated K+ channels, types Kv1.1, 1.2, 1.3, 1.5, and 3.1, stably expressed in mammalian cell lines. Mol. Pharmacol. 45:1227–1234. [PubMed] [Google Scholar]
- Grosse, G., A. Draguhn, L. Hohne, R. Tapp, R. W. Veh, and G. Ahnert-Hilger. 2000. Expression of Kv1 potassium channels in mouse hippocampal primary cultures: development and activity-dependent regulation. J. Neurosci. 20:1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunnet, M., B. S. Jensen, S. P. Olesen, and D. A. Klaerke. 2001. Apamin interacts with all subtypes of cloned small-conductance Ca2+-activated K+ channels. Pflugers Arch. 441:544–550. [DOI] [PubMed] [Google Scholar]
- Grunnet, M., T. Jespersen, H. B. Rasmussen, T. Ljungstrom, N. K. Jorgensen, S. P. Olesen, and D. A. Klaerke. 2002a. KCNE4 is an inhibitory subunit to the KCNQ1 channel. J. Physiol. 542:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunnet, M., H. G. Knaus, H. B. Rasmussen, A. Hay-Schmidt, and D. A. Klaerke. 2002b. The voltage-gated potassium channel Kv1.3 is expressed in epithelia. Biochim. Biophys. Acta. In press. [DOI] [PubMed]
- Grupe, A., K. H. Schroter, J. P. Ruppersberg, M. Stocker, T. Drewes, S. Beckh, and O. Pongs. 1990. Cloning and expression of a human voltage-gated potassium channel. A novel member of the RCK potassium channel family. EMBO J. 9:1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, S. H., J. Rettig, H. R. Graack, and O. Pongs. 1996. Functional characterization of Kv channel beta-subunits from rat brain. J. Physiol. 493:625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, S. H., J. Rettig, F. Wunder, and O. Pongs. 1995. Molecular and functional characterization of a rat brain Kv beta 3 potassium channel subunit. FEBS Lett. 377:383–389. [DOI] [PubMed] [Google Scholar]
- Hille, B. 2001. Ion Channels of Excitable Membranes. Sinauer Associates, Inc., Sunderland, MA, USA.
- Isacoff, E. Y., Y. N. Jan, and L. Y. Jan. 1990. Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature. 345:530–534. [DOI] [PubMed] [Google Scholar]
- Isbrandt, D., P. Friederich, A. Solth, W. Haverkamp, A. Ebneth, M. Borggrefe, H. Funke, K. Sauter, G. Breithardt, O. Pongs, and E. Schulze-Bahr. 2002. Identification and functional characterization of a novel KCNE2 (MiRP1) mutation that alters HERG channel kinetics. J. Mol. Med. 80:524–532. [DOI] [PubMed] [Google Scholar]
- Jespersen, T., M. Grunnet, K. Angelo, D. A. Klaerke, and S. P. Olesen. 2002. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques. 32:536–540. [DOI] [PubMed] [Google Scholar]
- Kalman, K., A. Nguyen, J. Tseng-Crank, I. D. Dukes, G. Chandy, C. M. Hustad, N. G. Copeland, N. A. Jenkins, H. Mohrenweiser, B. Brandriff, M. Cahalan, G. A. Gutman, and K. G. Chandy. 1998. Genomic organization, chromosomal localization, tissue distribution, and biophysical characterization of a novel mammalian Shaker-related voltage-gated potassium channel, Kv1.7. J. Biol. Chem. 273:5851–5857. [DOI] [PubMed] [Google Scholar]
- Kim, L. M., G. W. Abbott, A. Butler, and S. A. Goldstein. 2001. A role for the MiRP1-Kv4.2 channels in CNS A-type currents. Biophys. J. 80:437a. [Google Scholar]
- Koch, R. O., S. G. Wanner, A. Koschak, M. Hanner, C. Schwarzer, G. J. Kaczorowski, R. S. Slaughter, M. L. Garcia, and H. G. Knaus. 1997. Complex subunit assembly of neuronal voltage-gated K+ channels. Basis for high-affinity toxin interactions and pharmacology. J. Biol. Chem. 272:27577–27581. [DOI] [PubMed] [Google Scholar]
- Koschak, A., R. M. Bugianesi, J. Mitterdorfer, G. J. Kaczorowski, M. L. Garcia, and H. G. Knaus. 1998. Subunit composition of brain voltage-gated potassium channels determined by hongotoxin-1, a novel peptide derived from Centruroides limbatus venom. J. Biol. Chem. 273:2639–2644. [DOI] [PubMed] [Google Scholar]
- Leicher, T., R. Bahring, D. Isbrandt, and O. Pongs. 1998. Coexpression of the KCNA3B gene product with Kv1.5 leads to a novel A-type potassium channel. J. Biol. Chem. 273:35095–35101. [DOI] [PubMed] [Google Scholar]
- Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. [DOI] [PubMed] [Google Scholar]
- Manganas, L. N., and J. S. Trimmer. 2000. Subunit composition determines Kv1 potassium channel surface expression. J. Biol. Chem. 275:29685–29693. [DOI] [PubMed] [Google Scholar]
- Mason, D. E., K. E. Mitchell, Y. Li, M. R. Finley, and L. C. Freeman. 2002. Molecular basis of voltage-dependent potassium currents in porcine granulosa cells. Mol. Pharmacol. 61:201–213. [DOI] [PubMed] [Google Scholar]
- McDonald, T. V., Z. Yu, Z. Ming, E. Palma, M. B. Meyers, K. W. Wang, S. A. Goldstein, and G. I. Fishman. 1997. A minK-HERG complex regulates the cardiac potassium current I(Kr). Nature. 388:289–292. [DOI] [PubMed] [Google Scholar]
- Monaghan, M. M., J. S. Trimmer, and K. J. Rhodes. 2001. Experimental localization of Kv1 family voltage-gated K+ channel alpha and beta subunits in rat hippocampal formation. J. Neurosci. 21:5973–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, R. A., L. Couture, O. Elroy-Stein, J. Ragheb, B. Moss, and W. F. Anderson. 1992. Retroviral vectors containing putative internal ribosome entry sites: development of a polycistronic gene transfer system and applications to human gene therapy. Nucleic Acids Res. 20:1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M., A. Svejgaard, S. Skov, P. Dobson, K. Bendtzen, C. Geisler, and N. Odum. 1996. IL-2 induces beta2-integrin adhesion via a wortmannin/LY294002-sensitive, rapamycin-resistant pathway. Phosphorylation of a 125-kilodalton protein correlates with induction of adhesion, but not mitogenesis. J. Immunol. 157:5350–5358. [PubMed] [Google Scholar]
- Ouadid-Ahidouch, H., F. Van Coppenolle, B. X. Le, A. Belhaj, and N. Prevarskaya. 1999. Potassium channels in rat prostate epithelial cells. FEBS Lett. 459:15–21. [DOI] [PubMed] [Google Scholar]
- Pappone, P. A., and S. I. Ortiz-Miranda. 1993. Blockers of voltage-gated K channels inhibit proliferation of cultured brown fat cells. Am. J. Physiol. 264:C1014–C1019. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini, M., F. Vitelli, M. Seri, L. J. Galietta, O. Moran, A. Bulfone, S. Banfi, B. Pober, and A. Renieri. 1999. KCNE1-like gene is deleted in AMME contiguous gene syndrome: identification and characterization of the human and mouse homologs. Genomics. 60:251–257. [DOI] [PubMed] [Google Scholar]
- Pongs, O., T. Leicher, M. Berger, J. Roeper, R. Bahring, D. Wray, K. P. Giese, A. J. Silva, and J. F. Storm. 1999. Functional and molecular aspects of voltage-gated K+ channel beta subunits. Ann. N. Y. Acad. Sci. 868:344–355. [DOI] [PubMed] [Google Scholar]
- Rettig, J., S. H. Heinemann, F. Wunder, C. Lorra, D. N. Parcej, J. O. Dolly, and O. Pongs. 1994. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 369:289–294. [DOI] [PubMed] [Google Scholar]
- Rhodes, K. J., B. W. Strassle, M. M. Monaghan, Z. Bekele-Arcuri, M. F. Matos, and J. S. Trimmer. 1997. Association and colocalization of the Kvbeta1 and Kvbeta2 beta-subunits with Kv1 alpha-subunits in mammalian brain K+ channel complexes. J. Neurosci. 17:8246–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeper, J., and O. Pongs. 1996. Presynaptic potassium channels. Curr. Opin. Neurobiol. 6:338–341. [DOI] [PubMed] [Google Scholar]
- Romey, G., B. Attali, C. Chouabe, I. Abitbol, E. Guillemare, J. Barhanin, and M. Lazdunski. 1997. Molecular mechanism and functional significance of the MinK control of the KvLQT1 channel activity. J. Biol. Chem. 272:16713–16716. [DOI] [PubMed] [Google Scholar]
- Ruppersberg, J. P., K. H. Schroter, B. Sakmann, M. Stocker, S. Sewing, and O. Pongs. 1990. Heteromultimeric channels formed by rat brain potassium-channel proteins. Nature. 345:535–537. [DOI] [PubMed] [Google Scholar]
- Sanguinetti, M. C., M. E. Curran, A. Zou, J. Shen, P. S. Spector, D. L. Atkinson, and M. T. Keating. 1996. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 384:80–83. [DOI] [PubMed] [Google Scholar]
- Schroeder, B. C., S. Waldegger, S. Fehr, M. Bleich, R. Warth, R. Greger, and T. J. Jentsch. 2000. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 403:196–199. [DOI] [PubMed] [Google Scholar]
- Shamotienko, O. G., D. N. Parcej, and J. O. Dolly. 1997. Subunit combinations defined for K+ channel Kv1 subtypes in synaptic membranes from bovine brain. Biochemistry. 36:8195–8201. [DOI] [PubMed] [Google Scholar]
- Sheng, M., M. L. Tsaur, Y. N. Jan, and L. Y. Jan. 1994. Contrasting subcellular localization of the Kv1.2 K+ channel subunit in different neurons of rat brain. J. Neurosci. 14:2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, G., K. Nakahira, S. Hammond, K. J. Rhodes, L. E. Schechter, and J. S. Trimmer. 1996. Beta subunits promote K+ channel surface expression through effects early in biosynthesis. Neuron. 16:843–852. [DOI] [PubMed] [Google Scholar]
- Splawski, I., M. Tristani-Firouzi, M. H. Lehmann, M. C. Sanguinetti, and M. T. Keating. 1997. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat. Genet. 17:338–340. [DOI] [PubMed] [Google Scholar]
- Stuhmer, W., J. P. Ruppersberg, K. H. Schroter, B. Sakmann, M. Stocker, K. P. Giese, A. Perschke, A. Baumann, and O. Pongs. 1989. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO J. 8:3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, K. K., and S. A. Goldstein. 1998. The conduction pore of a cardiac potassium channel. Nature. 391:605–608. [DOI] [PubMed] [Google Scholar]
- Tapper, A. R., and A. L. George, Jr. 2000. MinK subdomains that mediate modulation of and association with KvLQT1. J. Gen. Physiol. 116:379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel, B. L., Y. N. Jan, and L. Y. Jan. 1988. Cloning of a probable potassium channel gene from mouse brain. Nature. 332:837–839. [DOI] [PubMed] [Google Scholar]
- Tinel, N., S. Diochot, M. Borsotto, M. Lazdunski, and J. Barhanin. 2000a. KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO J. 19:6326–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel, N., S. Diochot, I. Lauritzen, J. Barhanin, M. Lazdunski, and M. Borsotto. 2000b. M-type KCNQ2-KCNQ3 potassium channels are modulated by the KCNE2 subunit. FEBS Lett. 480:137–141. [DOI] [PubMed] [Google Scholar]
- Veh, R. W., R. Lichtinghagen, S. Sewing, F. Wunder, I. M. Grumbach, and O. Pongs. 1995. Immunohistochemical localization of five members of the Kv1 channel subunits: contrasting subcellular locations and neuron-specific co-localizations in rat brain. Eur. J. Neurosci. 7:2189–2205. [DOI] [PubMed] [Google Scholar]
- Wang, H., D. D. Kunkel, P. A. Schwartzkroin, and B. L. Tempel. 1994. Localization of Kv1.1 and Kv1.2, two K channel proteins, to synaptic terminals, somata, and dendrites in the mouse brain. J. Neurosci. 14:4588–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapura, M., S. Nattel, D. Chartier, R. Caballero, and T. E. Hebert. 2002. A comparison of currents carried by HERG, with and without coexpression of MiRP1, and the native rapid delayed rectifier current. Is MiRP1 the missing link? J. Physiol. 540:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollnik, B., B. C. Schroeder, C. Kubisch, H. D. Esperer, P. Wieacker, and T. J. Jentsch. 1997. Pathophysiological mechanisms of dominant and recessive KVLQT1 K+ channel mutations found in inherited cardiac arrhythmias. Hum. Mol. Genet. 6:1943–1949. [DOI] [PubMed] [Google Scholar]
- Yu, H., J. Wu, I. Potapova, R. T. Wymore, B. Holmes, J. Zuckerman, Z. Pan, H. Wang, W. Shi, R. B. Robinson, M. R. El Maghrabi, W. Benjamin, J. Dixon, D. McKinnon, I. S. Cohen, and R. Wymore. 2001. MinK-related peptide 1: A beta subunit for the HCN ion channel subunit family enhances expression and speeds activation. Circ. Res. 88:E84–E87. [DOI] [PubMed] [Google Scholar]
- Zhang, M., M. Jiang, and G. N. Tseng. 2001. minK-related peptide 1 associates with Kv4.2 and modulates its gating function: potential role as beta subunit of cardiac transient outward channel? Circ. Res. 88:1012–1019. [DOI] [PubMed] [Google Scholar]