Abstract
Selenium (Se) is an essential element for many organisms but is toxic at higher levels. CpNifS is a chloroplastic NifS-like protein in Arabidopsis (Arabidopsis thaliana) that can catalyze the conversion of cysteine into alanine and elemental sulfur (S0) and of selenocysteine into alanine and elemental Se (Se0). We overexpressed CpNifS to investigate the effects on Se metabolism in plants. CpNifS overexpression significantly enhanced selenate tolerance (1.9-fold) and Se accumulation (2.2-fold). CpNifS overexpressors showed significantly reduced Se incorporation into protein, which may explain their higher Se tolerance. Also, sulfur accumulation was enhanced by approximately 30% in CpNifS overexpressors, both on media with and without selenate. Root transcriptome changes in response to selenate mimicked the effects observed under sulfur starvation. There were only a few transcriptome differences between CpNifS-overexpressing plants and wild type, besides the 25- to 40-fold increase in CpNifS levels. Judged from x-ray analysis of near edge spectrum, both CpNifS overexpressors and wild type accumulated mostly selenate (SeVI). In conclusion, overexpression of this plant NifS-like protein had a pronounced effect on plant Se metabolism. The observed enhanced Se accumulation and tolerance of CpNifS overexpressors show promise for use in phytoremediation.
Selenium (Se) is an essential micronutrient for animals and bacteria but becomes toxic at higher concentrations. Accumulation of Se in the environment has been reported to be a cause of toxicity in aquatic organisms (Ohlendorf et al., 1988; Hamilton, 2004), livestock (Fessler et al., 2003), and humans (Hira et al., 2004). Selenate (SeO42−) is the most common soluble form of Se found in terrestrial systems. Seleniferous soils (>2 ppm Se) commonly found in the western United States arise from the weathering of Se-rich shale rock.
Se is essential for mammals and bacteria, where selenocysteine (SeCys) is an essential component for the formation of selenoenzymes such as glutathione peroxidase, a free radical scavenger (Stadtman, 1990). A diet enriched with Se can reduce the risk of cancer in humans (Ip et al., 2002). Thus, plants enriched with anticarcinogenic selenocompounds have enhanced nutritional value (Ellis et al., 2004). Whether or not Se is essential for higher plants themselves remains unproven. However, the green alga Chlamydomonas reinhardtii, which shares a common ancestor with land plants, was shown to contain selenoproteins. Labeling of C. reinhardtii cells with 75Se revealed four Se proteins, one of which is a glutathione peroxidase, thought to ameliorate oxidative stress (Novoselov et al., 2002).
Although certain plants tolerate and (hyper)accumulate Se to high concentrations (up to 0.5% of dry weight), generally Se uptake into plant tissue is thought to be inadvertent and potentially toxic (Anderson, 1993). The toxicity of selenate is thought to be due to its chemical similarity to sulfate, and, as such, proteins involved in sulfate transport and sulfur (S) metabolism can act on analogous Se compounds (Terry et al., 2000). When selenate is assimilated via the sulfate assimilation pathway, Se can be incorporated into the S amino acids Cys and Met, leading to the formation of SeCys and selenomethionine. The nonspecific incorporation of SeCys into proteins causes toxicity (Stadtman, 1990).
Attenuating the toxic effects of Se is of concern for public health and agriculture. Plants with enhanced Se tolerance and accumulation may potentially be used for environmental cleanup (Bañuelos et al., 2005). Several strategies have been used successfully to breed plants with enhanced Se tolerance and accumulation. Nonspecific incorporation of SeCys into proteins can be prevented by methylation of SeCys into nonprotein selenoamino acids via the enzyme SeCys methyl transferase (SMT; Neuhierl and Böck, 1996). Overexpression of this enzyme from the Se hyperaccumulator Astragalus bisulcatus in Arabidopsis (Arabidopsis thaliana) or Indian mustard (Brassica juncea) enhanced Se tolerance and accumulation (Ellis et al., 2004; LeDuc et al., 2004). Methylation of selenomethionine and SeCys can lead to the formation of volatile selenocompounds, another potential detoxification mechanism (Terry et al., 2000). Overexpression in Indian mustard of cystathionine-γ-synthase, the first enzyme in the conversion of SeCys to volatile Se, led to increased levels of dimethylselenide production and increased Se tolerance (Van Huysen et al., 2003). Overexpression of the key enzyme of the sulfate assimilation pathway, ATP sulfurylase, led to enhanced selenate reduction, Se accumulation, and Se tolerance (Pilon-Smits et al., 1999). In another study, barley (Hordeum vulgare) plants overexpressing thioredoxin h were capable of tolerating and accumulating more Se when grown on selenite (Kim et al., 2003) perhaps due to enhanced reduction of selenite to elemental Se (Se0). In another approach to prevent Se incorporation into protein, a mouse gene expressing a SeCys lyase (SL) was introduced into Arabidopsis and Indian mustard (Garifullina et al., 2003; Pilon et al., 2003). The enzyme proved capable of reducing the amount of Se in protein, probably by cleaving SeCys into alanine and Se0. The plants accumulated more Se when the gene was overexpressed in either the cytosol or the chloroplast, but tolerance to Se was enhanced only when expressed in the cytosol.
Recently, a chloroplastic protein, CpNifS, was discovered in Arabidopsis that has both Cys desulfurase and SL activity (Pilon-Smits et al., 2002), and thus potentially affects both S and Se metabolism. CpNifS can provide the necessary S to form iron (Fe)-S clusters for ferredoxin by cleaving Cys into elemental S and alanine (Ye et al., 2004; Abdel-Ghany et al., 2005), and possibly has a role in providing the elemental S for other S metabolites such as thiamine, in analogy with bacterial systems (Mihara and Esaki, 2002). However, the activity of this enzyme toward SeCys is almost 300 times higher than toward Cys (Pilon-Smits et al., 2002), suggesting CpNifS may affect plant Se metabolism as well. Since CpNifS converts SeCys into alanine and Se0, this enzyme may prevent Se incorporation into protein, which should protect plants from Se toxicity.
In this study, we overexpressed CpNifS in Arabidopsis to study the effects on Se and S metabolism. The effects on Se metabolism were explored by determining the ability of the transgenic plants to tolerate and accumulate Se, and the metabolic fate of the accumulated Se. Microarray analysis was performed to more fully understand how overexpression of CpNifS may affect Se tolerance and accumulation, and to investigate the transcriptome changes that occur when plants are grown on selenate.
RESULTS
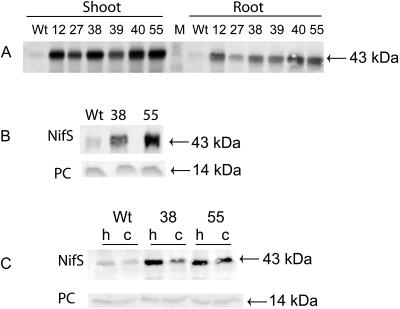
Generation and Characterization of Transgenic Lines
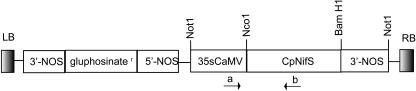
The CpNifS cDNA, including its plastid targeting sequence (Fig. 1), was constitutively expressed in Arabidopsis. The CpNifS protein expression level in the transgenic plant lines obtained was tested by immunoblotting using antibodies raised against purified CpNifS. A protein (43 kD) of the expected size for the mature protein after cleavage of the transit sequence was present at much higher levels in shoot and root tissues of the CpNifS lines (Fig. 2A) compared to wild type. The degree of overexpression varied among the CpNifS transgenic lines. In general, CpNifS expression was higher in shoot than root tissue in the transgenics, despite the use of the constitutive promoter (Fig. 2A). Transgenic lines CpNifS38 and CpNifS55 were chosen for further analysis. Judged from digital imaging, these two lines contained about 23-fold higher CpNifS protein concentration compared to wild type in the shoots. In root material, CpNifS levels were roughly 8-fold higher in the transgenics. To verify the localization of CpNifS in the chloroplasts, a western-blot analysis was performed on isolated chloroplasts from wild type, CpNifS38 and CpNifS55 (Fig. 2, B and C). The level of CpNifS protein in all plant types was slightly lower in the chloroplasts compared to homogenate, when compared on a chlorophyll basis. Since CpNifS was shown earlier to be chloroplastic (Pilon-Smits et al., 2002), some of the stromal proteins may be lost due to leakage in this procedure. The fraction of CpNifS in chloroplasts was similar in CpNifS55 compared to wild type, but somewhat lower in CpNifS38. Thus, it appears that the majority of the overproduced CpNifS was indeed targeted to the chloroplast, but it cannot be excluded that some of the protein was mistargeted, especially in CpNifS38.
Figure 1.
AtCpNifS construct used to transform Arabidopsis. Shown are the forward (a) and reverse (b) primers that were used to identify transgenic lines. 3′-NOS and 5′-NOS, Nopaline synthase terminator and promoter, respectively. LB and RB, Left border and right border of T-DNA, respectively.
Figure 2.
A, Immunoblot showing CpNifS overexpression in roots and shoots of six different homozygous CpNifS lines and wild type (Wt). Twenty micrograms of total protein was loaded per lane. Lines 38 and 55 were selected for further study. B and C, Immunoblot analysis of CpNifS and plastocyanin (PC) in homogenate (h) and isolated chloroplasts (c) from wild type and CpNifS-overexpressing lines 38 and 55. Three micrograms of chlorophyll was loaded per lane. Plastocyanin was visualized as a control for equal loading.
Effects of CpNifS Overexpression on Se and S Metabolism
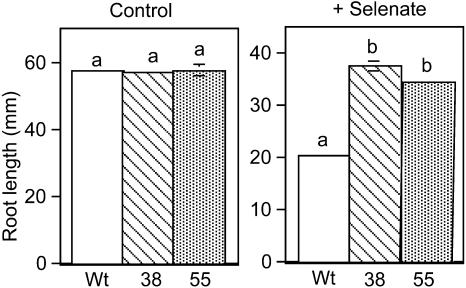
Since CpNifS can convert SeCys into Se0 and alanine, thereby potentially preventing the toxic process of nonspecific incorporation of Se into protein, we wanted to test whether CpNifS overexpression affects Se tolerance. The ability of the CpNifS overexpressors to tolerate selenate was investigated by measuring root growth on media with or without 40 μm selenate. The transgenics CpNifS38 and CpNifS55 had 1.7- to 2-fold longer roots after 10 d of treatment on selenate compared to wild type (P < 0.05; Fig. 3). Interestingly, the CpNifS transgenics did not show enhanced selenite tolerance (data not shown). There were also no differences in root length between wild-type and transgenic plants grown on control (Murashige and Skoog [MS]) media (Fig. 3) or on media supplied with cadmium or chromate (data not shown), indicating that CpNifS overexpression enhances selenate tolerance and the observed effect is selenate specific.
Figure 3.
Se tolerance as determined by measuring the root length of wild-type (Wt), CpNifS38, and CpNifS55 seedlings grown for 10 d on MS medium with and without 40 μm selenate. Shown are the mean (n = 25) and se of the mean. Lowercase letters above bars denote significant differences (P < 0.05).
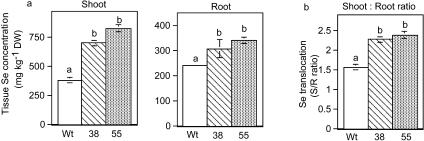
To determine if CpNifS overexpression alters Se accumulation, seedlings of wild type, CpNifS38, and CpNifS55 were grown on medium with 40 μm selenate. Transgenic plants accumulated more Se in shoot and root tissues than wild type (P < 0.05; Fig. 4a); the Se concentration in shoots was 2- to 3-fold higher in transgenics compared to wild type. The Se concentration in the shoots exceeded that in the roots for all three genotypes, agreeing with previous results that plants readily translocate Se to the shoot when supplied with selenate (de Souza et al., 1998; Pilon et al., 2003; LeDuc et al., 2004). Interestingly, the translocation of Se (the ratio of shoot Se to root Se) was more than 50% higher in CpNifS38 and CpNifS55 compared to wild type (P < 0.05; Fig. 4a).
Figure 4.
A, Shoot and root Se concentration in wild-type (Wt) and CpNifS-overexpressing (38 and 55) plants grown on MS medium containing 40 μm selenate for 20 d. Shown are the mean and se of the mean for the shoots and roots (n = 5 pooled samples from 20 seedlings each). B, Se translocation as determined by calculating the shoot to root ratio of all three lines. Lowercase letters above bars denote significant differences (P < 0.05).
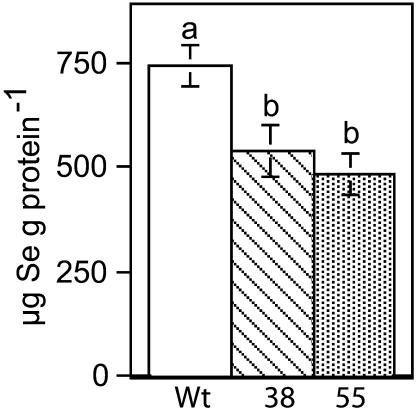
The observation that CpNifS overexpressors are more tolerant to Se, yet also accumulate more Se, suggests that their tolerance mechanism does not involve Se exclusion by the plant. Rather, overexpressors may have altered Se metabolism, resulting in accumulation of less toxic Se forms, or altered Se compartmentation. We hypothesized that these transgenics were better able to tolerate selenate because they are capable of shuttling Se away from protein incorporation by breaking down SeCys into Se0 and alanine. To test this hypothesis, the amount of Se in protein was determined. Indeed, the CpNifS transgenics incorporated approximately a third less Se in protein than the wild-type plants (P < 0.05; Fig. 5). Since nonspecific incorporation of Se into proteins is toxic, the capacity of the CpNifS overexpressors to prevent this toxic process likely caused the observed increased tolerance to Se.
Figure 5.
Se incorporation into protein in shoot tissue of wild-type (Wt) and CpNifS-overexpressing (38 and 55) seedlings grown for 14 d on 20 μm selenate. Shown are the mean and se of the mean (n = 5 pooled samples from 50 seedlings each). Lowercase letters above bars denote significant differences (P < 0.05).
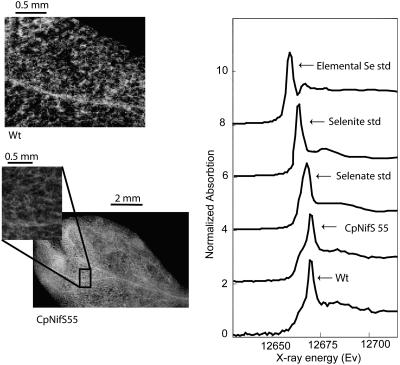
Micro x-ray fluorescence (μXRF) mapping and micro x-ray absorption spectroscopy (μXAS) were used to determine and compare the Se speciation in the leaves of wild type and the highest CpNifS overexpressor, CpNifS55. In addition to Se located inside cells, both plant types clearly show abundant levels of Se in their vascular tissue (Fig. 6, left). Judged from x-ray analysis of near edge spectra (XANES), there were no major differences between the transgenic and wild type with respect to the oxidation state of Se (Fig. 6, right). Both lines contained predominantly selenate with a small fraction of more reduced Se, as indicated by the shoulder at 12,660 eV. This possibly was an organic Se species (Pilon-Smits et al., 1999), Se0 or selenite. Judged from linear least-squares fitting to the three references, on average the fraction of Se present as selenate was 70% for wild type and 66% for CpNifS55. The chemical form of the remaining, more reduced Se cannot be determined with certainty from this limited number of references, but the fraction of more reduced Se may be a bit higher for CpNifS than for wild type, judged from the slightly more pronounced shoulder at 12,663 eV (Fig. 6) and the corresponding least-squares fitting analysis described above.
Figure 6.
Left, μXAS analysis showing spatial Se distribution in leaves of wild type and CpNifS55. Right, Representative XANES spectra obtained from the leaves shown, in comparison with Se standards selenate, selenite, and elemental (gray) Se.
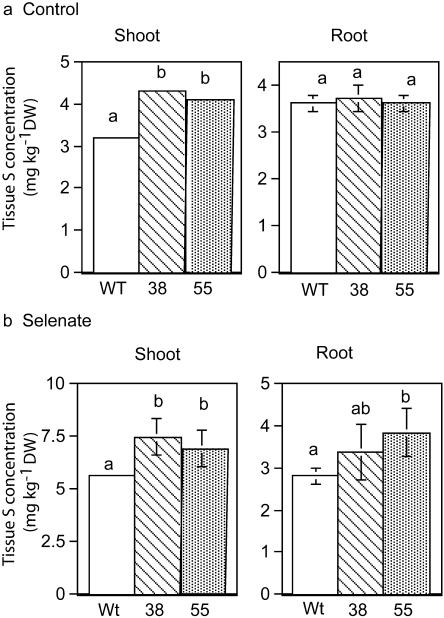
CpNifS can act on both Cys and SeCys, and as such its overexpression may affect both S and Se metabolism. Accumulation of S was therefore also analyzed. Under control conditions, CpNifS38 and CpNifS55 had higher levels of S in shoots, but not in roots, compared to wild type (P < 0.05; Fig. 7a). Additionally, CpNifS overexpression led to an increase in tissue S concentration in both shoots and roots compared to wild type when grown on selenate (Fig. 7b), suggesting a stimulating effect of CpNifS overexpression on S uptake. Selenate treatment also enhanced S uptake; compared to accumulation of S on control media, more S was accumulated in the shoots of all three genotypes grown on selenate.
Figure 7.
Shoot and root S concentrations in wild-type (Wt) and CpNifS-overexpressing (38 and 55) seedlings grown on medium with and without 40 μm selenate for 20 d. Shown are the means and se of the mean (n = 5 pooled samples from 20 seedlings each). Lowercase letters above bars denote significant differences (P < 0.05).
Microarray Analysis
GeneChip arrays (Affymetrix) were used to identify differences in Se-related transcriptome responses between CpNifS overexpressors and wild type, and to determine how growth on selenate changes the transcriptome of Arabidopsis. Transcriptome profiling was performed on roots of wild-type, CpNifS38, and CpNifS55 plants that were grown from seed on half-strength MS agar media with and without 40 μm selenate for 10 d.
We chose to focus on root tissue because we found clear differences between CpNifS plants and wild type with respect to root growth. Also, the transgenics differed in Se and S uptake and translocation, and we hypothesized that there would be differences in the expression of root sulfate transporters involved in sulfate/selenate uptake into the root symplast and the root xylem.
The microarray experiment confirmed the overexpression of CpNifS in the transgenic plants. Line CpNifS38 showed a 27-fold increase in CpNifS expression, and CpNifS55 had 40-fold higher CpNifS mRNA levels compared to wild type on control medium (Table I). These data confirm the overexpression observed at the protein level (Fig. 2). CpNifS transcription levels were not affected by the selenate treatment in wild type, i.e. the endogenous transcript level of CpNifS was not affected by selenate (Table I). However, preliminary results (H. Ye, M. Pilon, and E. Pilon-Smits, unpublished data) indicate that CpNifS is up-regulated by selenate at the protein level.
Table I.
Transcriptome differences between CpNifS overexpressors and wild-type plants
Wild-type (ecotype Ws) and CpNifS-overexpressing (NifS55 and NifS38) Arabidopsis plants were grown on control medium (MS) or with 40 μm selenate (Se) for 10 d, and transcriptomes of root RNA were analyzed using Affymetrix GeneChip arrays as described in “Materials and Methods.” Two sets of experiments (set 1 and set 2) were carried out separately to evaluate the reproducibility of the results. Genes showing more than 2-fold differences between the wild type and CpNifS overexpressors either on MS or Se conditions were selected by comparing the normalized signals and calculating their ratios, as shown in the NifS55/Ws and Nif38/Ws columns. The significance of the difference is represented by superscript letters, which describe the up and down effects of NifS overexpression on gene expression by fold change differences: a, >5.00; b, >2.00; c, >1.33; d, <0.75; e, <0.50. Regulation by Se (Se/MS ratio) is indicated using the same superscript letters. The absolute calls of each transcript (P, present; M, marginal; A, absent) are indicated on the right-hand side of the normalized values. The relative ratios are calculated for the pairs having “present” call at least in one sample.
| Affy Code
|
AGI Locus
|
Annotation
|
Set 1
|
Set 2
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ws
|
NifS55
|
NifS55/Ws
|
Ws
|
NifS38
|
NifS38/Ws
|
|||||||||||||
| Se | MS | Se/MS | Se | MS | Se/MS | Se | MS | Se | MS | Se/MS | Se | MS | Se/MS | Se | MS | |||
| 261744_at | AT1G08490 | NifS | 1.16 P | 1.03 P | 1.12 | 41.32 P | 47.44 P | 0.87 | 35.71a | 46.10a | 0.89 P | 0.75 P | 1.19 | 24.52 P | 20.01 P | 1.23 | 27.46a | 26.75a |
| 259161_at | AT3G01500 | Carbonic anhydrase 1 | 0.33 A | 9.36 P | 0.04e | 2.03 A | 0.42 A | 0.04e | 0.25 A | 8.47 P | 0.03e | 0.73 A | 3.30 A | 0.39e | ||||
| 258897_at | AT3G05730 | Expressed protein | 0.39 A | 1.08 P | 0.36e | 1.33 P | 0.41 P | 3.22b | 3.41b | 0.38e | 0.49 A | 1.07 P | 0.46e | 1.16 P | 1.75 P | 0.66d | 2.36b | 1.64c |
| 252537_at | AT3G45710 | Proton-dependent oligopeptide transporter family | 0.25 P | 1.16 P | 0.22e | 0.84 P | 1.49 P | 0.56d | 3.35b | 1.29 | 0.17 P | 0.82 P | 0.21e | 0.45 P | 0.66 P | 0.68d | 2.57b | 0.81 |
| 260535_at | AT2G43390 | Expressed protein | 0.61 P | 1.13 P | 0.54d | 1.56 P | 1.44 P | 1.09 | 2.55 | 1.28 | 0.71 A | 1.42 P | 0.50d | 1.50 P | 1.75 P | 0.86 | 2.10b | 1.23 |
| 251974_at | AT3G53200 | Myb family transcription factor (MYB27) | 0.64 M | 1.33 P | 0.48e | 1.46 P | 2.67 P | 0.55d | 2.27 | 2.00b | 0.84 P | 1.13 P | 0.74d | 1.78 P | 1.62 P | 1.10 | 2.11b | 1.43c |
| 264263_at | AT1G09155 | SKP1 interacting partner 3-related | 0.51 A | 0.69 P | 0.74d | 1.10 P | 1.66 P | 0.66d | 2.15 | 2.39b | 0.46 A | 1.58 P | 0.29e | 1.19 P | 0.90 P | 1.33 | 2.59b | 0.57d |
| 255054_s_at | AT4G23560 | Glycosyl hydrolase family 9 protein similar to cellulase | 1.44 P | 0.80 P | 1.80c | 0.73 P | 2.41 P | 0.30e | 0.51d | 3.02b | 0.45 A | 0.82 M | 0.99 P | 1.73 P | 0.57d | 2.21b | 2.11b | |
| 246340_s_at | AT3G44860 | S-Adenosylmethionine:carboxyl methyltransferase family | 15.18 P | 0.65 A | 23.50a | 5.93 P | 1.33 P | 4.45b | 0.39e | 2.06b | 10.71 P | 0.35 A | 30.34a | 7.94 P | 1.18 P | 6.75a | 0.74d | 3.33b |
| 247905_at | AT5G57400 | Hypothetical protein | 0.73 P | 1.00 A | 0.72d | 0.87 P | 2.02 P | 0.43e | 1.20 | 2.01b | 1.07 A | 0.76 M | 1.15 A | 1.70 P | 0.68d | 2.23b | ||
| 265400_at | AT2G10940 | Protease inhibitor/seed storage/LTP family | 0.30 A | 2.86 P | 0.10e | 0.33 A | 0.77 A | 0.27e | 0.64 A | 3.56 P | 0.18e | 0.91 A | 1.63 A | 0.46e | ||||
| 249390_at | AT5G39740 | 60S ribosomal protein L5 | 1.35 A | 1.09 P | 1.23 | 1.09 A | 0.30 A | 3.66b | 0.27e | 0.86 A | 1.36 P | 0.63d | 1.04 A | 0.29 A | 0.21e | |||
| 263369_at | AT2G20480 | Expressed protein | 1.38 P | 1.32 P | 1.05 | 1.05 P | 0.45 A | 2.32b | 0.76 | 0.34e | 0.89 P | 1.07 P | 0.83 | 0.44 A | 0.40 A | 0.49e | 0.37e | |
| 261197_at | AT1G12900 | Glyceraldehyde 3-phosphate dehydrogenase, putative | 0.99 P | 2.06 P | 0.48e | 0.69 A | 0.53 A | 0.70d | 0.26e | 0.88 A | 2.36 P | 0.37e | 0.89 A | 1.13 A | 0.48e | |||
| 256825_at | AT3G22120 | Protease inhibitor/seed storage/LTP family | 1.01 P | 1.11 P | 0.91 | 0.66 P | 0.51 P | 1.30 | 0.65d | 0.46e | 1.25 M | 1.12 P | 1.12 | 0.83 A | 0.10 A | 8.17a | 0.09e | |
| 262586_at | AT1G15480 | DNA-binding protein, putative | 1.32 P | 1.09 P | 1.21 | 0.61 P | 0.14 A | 4.42b | 0.46e | 0.13e | 1.59 P | 1.35 P | 1.18 | 0.33 P | 0.51 A | 0.64d | 0.21e | 0.38e |
| 253060_at | AT4G37710 | VQ motif-containing protein | 8.73 P | 0.90 A | 9.68a | 4.01 A | 0.63 A | 0.46e | 9.59 P | 0.49 A | 19.74a | 4.75 P | 0.38 A | 12.57a | 0.50e | |||
| 264718_at | AT1G70130 | Lectin protein kinase, putative | 4.86 P | 0.45 A | 10.93a | 1.08 A | 0.44 A | 0.22e | 6.94 P | 1.23 A | 5.64a | 2.99 A | 0.92 A | 0.43e | ||||
In both the wild type and CpNifS overexpressors, selenate treatment had a pronounced effect on the root transcript level of many genes involved in sulfate uptake and assimilation (Table II). Sulfate transporters were generally up-regulated when grown on Se, particularly SULTR1;1, SULTR2;1, and SULTR4;2. SULTR1;2 and SULTR4;1 were also up-regulated by Se. In the primary S-assimilation pathway, several ATP sulfurylase genes (APS1 and 3), APS reductase genes (APR1, 2, and 3), O-acetylserine (thiol) lyase (Bsas4;2), Ser acetyltransferase (Serat3;1), and a putative cystathionine γ-lyase (At1g64660) were up-regulated by selenate in all plant types. S-related genes that were down-regulated on selenate include APS2 and 4, an O-acetylserine (thiol) lyase (Bsas1;4), and γ-glutamylcysteine synthetase (GSH1). A homocysteine S-methyltransferase (HMT-3), homologous to the SMT of the Se hyperaccumulator A. bisulcatus (Neuhierl et al., 1999), was differentially regulated by selenate in wild type compared to the transgenics. While in wild type the HMT-3 transcript levels were not affected by Se, they decreased 2- to 3-fold in CpNifS plants. Apart from HMT3, a few other genes were differentially regulated by Se in the CpNifS roots compared to wild type (Table I). Other potentially Se-related genes that were regulated by selenate are two homologs of Se-binding proteins (At3g23800 and At4g14030/At4g14040; Table II). Both were regulated approximately 2.5-fold by Se treatment in all plant types. However, At3g23800 was down-regulated while At4g14030/At4g14040 was up-regulated.
Table II.
The effect of Se on S metabolism
Genes encoding sulfate transporters and genes for the synthesis of Cys, Met, and glutathione were selected from the transcriptome data of wild-type (Ws) and CpNifS overexpressor (NifS55 and NifS38) plants, described as set 1 and set 2 in Table I. The data of 24 h no-S treatment (−S) and the control S-replete culture at the same time point (+S) appear as references (set A and set B; Maruyama-Nakashita et al., 2005; http://arabidopsis.org/info/expression/ATGenExpress.jsp). Columbia-0 ecotype (Col) was used for the S limitation experiment. The up- and down-regulation by Se or by S limitation were calculated as relative ratios of normalized signals (Se/MS and −S/+S), and displayed using the same superscript letters as in Table I. The relative ratios are calculated for the pairs having “present” call at least in one sample.
| Affy Code
|
AGI Locus
|
Annotation
|
Gene Name
|
Set 1
|
Set 2
|
Set A
|
Set B
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ws
|
NifS55
|
Ws
|
Nifs38
|
Col
|
Col
|
||||||||||||||||
| Se | MS | Se/MS | Se | MS | Se/MS | Se | MS | Se/MS | Se | MS | Se/MS | −S | +S | −S/+S | −S | +S | −S/+S | ||||
| Sulfate transport | |||||||||||||||||||||
| 255105_at | AT4G08620 | Sulfate transporter | SULTR1;1 | 2.69 P | 0.29 P | 9.33a | 2.45 P | 0.59 P | 4.15b | 2.40 P | 0.21 P | 11.41a | 3.33 P | 0.25 P | 13.59a | 1.41 P | 0.16 P | 9.04a | 1.90 P | 0.13 P | 15.10a |
| 262133_at | AT1G78000 | Sulfate transporter | SULTR1;2 | 1.15 P | 0.65 P | 1.76c | 1.08 P | 0.98 P | 1.10 | 1.14 P | 0.69 P | 1.64c | 1.23 P | 0.69 P | 1.78c | 1.15 P | 0.73 P | 1.58c | 1.02 P | 0.72 P | 1.42c |
| 255958_at | AT1G22150 | Sulfate transporter | SULTR1;3 | 0.59 A | 1.74 A | 0.53 A | 0.49 A | 0.64 A | 0.27 A | 0.54 A | 0.90 A | 1.01 A | 0.77 A | 0.74 A | 0.20 A | ||||||
| 250475_at | AT5G10180 | Sulfate transporter | SULTR2;1 | 1.80 P | 0.34 P | 5.35a | 1.79 P | 0.23 P | 7.76a | 2.03 P | 0.27 P | 7.59a | 1.66 P | 0.27 P | 6.11a | 2.33 P | 0.24 P | 9.60a | 2.75 P | 0.31 P | 8.94a |
| 262134_at | AT1G77990 | Sulfate transporter | SULTR2;2 | 1.16 P | 1.14 P | 1.02 | 1.31 P | 1.04 P | 1.26 | 0.97 P | 0.89 P | 1.10 | 1.03 P | 0.72 A | 1.42c | 1.16 P | 0.87 P | 1.34c | 0.85 P | 0.76 A | 1.12 |
| 246310_at | AT3G51895 | Sulfate transporter | SULTR3;1 | 1.05 P | 1.03 P | 1.02 | 0.74 P | 0.66 P | 1.11 | 1.39 P | 1.05 P | 1.33c | 0.87 P | 0.89 P | 0.98 | 1.42 P | 0.97 P | 1.47c | 1.10 P | 0.65 P | 1.69c |
| 255443_at | AT4G02700 | Sulfate transporter | SULTR3;2 | 0.83 A | 0.75 A | 0.80 A | 1.01 A | 1.10 A | 1.00 A | 0.99 A | 1.06 A | 1.16 A | 1.00 A | 1.07 A | 0.75 M | ||||||
| 264901_at | AT1G23090 | Sulfate transporter | SULTR3;3 | 0.72 P | 1.41 P | 0.51d | 1.36 P | 1.82 P | 0.75d | 0.78 P | 1.32 P | 0.59d | 1.16 P | 1.43 P | 0.81 | 0.83 P | 0.84 P | 0.99 | 0.83 P | 0.84 P | 0.98 |
| 258287_at | AT3G15990 | Sulfate transporter | SULTR3;4 | 1.45 P | 0.91 P | 1.59c | 1.27 P | 1.07 P | 1.19 | 1.62 P | 1.14 P | 1.42c | 1.45 P | 0.93 P | 1.56c | 0.69 P | 0.46 P | 1.50c | 0.66 P | 0.48 P | 1.37c |
| 245912_at | AT5G19600 | Sulfate transporter | SULTR3;5 | 0.88 P | 0.96 P | 0.92 | 0.93 P | 1.02 P | 0.91 | 0.88 P | 0.98 P | 0.89 | 0.88 P | 1.19 P | 0.74d | 1.13 P | 1.36 P | 0.83 | 1.19 P | 1.28 P | 0.92 |
| 245855_at | AT5G13550 | Sulfate transporter | SULTR4;1 | 1.33 P | 0.81 P | 1.65c | 1.24 P | 0.88 P | 1.42c | 1.24 P | 0.73 P | 1.71c | 1.26 P | 0.63 P | 2.00c | 1.26 P | 0.81 P | 1.56c | 1.12 P | 0.60 P | 1.88c |
| 256244_at | AT3G12520 | Sulfate transporter | SULTR4;2 | 1.90 P | 0.58 P | 3.30b | 2.01 P | 0.67 P | 2.99b | 1.76 P | 0.67 P | 2.63b | 2.05 P | 0.56 P | 3.65b | 1.40 P | 0.34 P | 4.18b | 1.33 P | 0.41 P | 3.27b |
| Sulfate reduction | |||||||||||||||||||||
| 256835_at | AT3G22890 | ATP sulfurylase | APS1 | 1.05 P | 0.65 P | 1.60c | 0.96 P | 0.62 P | 1.54 | 0.99 P | 0.60 P | 1.67c | 1.01 P | 0.62 P | 1.61c | 1.70 P | 1.32 P | 1.29 | 1.97 P | 1.79 P | 1.10 |
| 255785_at | AT1G19920 | ATP sulfurylase | APS2 | 0.62 P | 1.12 P | 0.56d | 0.75 P | 1.00 P | 0.75 | 0.69 P | 1.27 P | 0.54d | 0.72 P | 1.17 P | 0.61d | 0.86 P | 1.36 P | 0.63d | 1.01 P | 1.28 P | 0.79 |
| 245254_at | AT4G14680 | ATP sulfurylase | APS3 | 1.99 P | 0.37 A | 5.40a | 1.27 P | 0.32 A | 4.00b | 2.12 P | 0.33 A | 6.34a | 1.69 P | 0.47 M | 3.59b | 1.19 P | 0.62 A | 1.94c | 1.39 P | 0.81 P | 1.72c |
| 249112_at | AT5G43780 | ATP sulfurylase | APS4 | 0.27 P | 1.14 P | 0.23e | 0.30 P | 1.14 P | 0.27e | 0.26 P | 1.03 P | 0.25e | 0.28 P | 1.04 P | 0.27e | 0.81 P | 1.14 P | 0.72d | 0.97 P | 1.46 P | 0.67d |
| 255284_at | AT4G04610 | APS reductase | APR1 | 1.02 P | 0.32 P | 3.24b | 1.08 P | 0.38 P | 2.87b | 0.94 P | 0.32 P | 2.96b | 1.33 P | 0.35 P | 3.80b | 1.02 P | 0.99 P | 1.03 | 1.22 P | 1.38 P | 0.88 |
| 264745_at | AT1G62180 | APS reductase | APR2 | 1.11 P | 0.50 P | 2.23b | 1.18 P | 0.40 P | 2.93b | 1.16 P | 0.39 P | 2.94b | 1.28 P | 0.43 P | 2.97b | 1.38 P | 0.77 P | 1.79c | 1.49 P | 0.89 P | 1.67c |
| 254343_at | AT4G21990 | APS reductase | APR3 | 2.10 P | 0.42 P | 5.06a | 1.91 P | 0.47 P | 4.11b | 2.41 P | 0.48 P | 5.07a | 2.30 P | 0.44 P | 5.18a | 1.60 P | 0.57 P | 2.82b | 1.38 P | 0.62 P | 2.21b |
| 267112_at | AT2G14750 | APS kinase | AKN1 | 1.11 P | 0.89 P | 1.26 | 1.10 P | 0.95 P | 1.16 | 0.76 P | 0.73 P | 1.03 | 1.05 P | 0.68 P | 1.55c | 1.17 P | 1.09 P | 1.07 | 0.81 P | 1.47 P | 0.55d |
| 252870_at | AT4G39940 | APS kinase | AKN2 | 1.17 P | 0.99 P | 1.19 | 1.10 P | 1.01 P | 1.09 | 0.99 P | 0.76 P | 1.30 | 1.02 P | 0.80 P | 1.27 | 0.83 P | 1.63 P | 0.51d | 0.67 P | 2.27 P | 0.30e |
| 259339_at | AT3G03900 | APS kinase | AKN3 | 0.74 P | 1.08 P | 0.69d | 0.94 P | 0.98 P | 0.96 | 1.14 P | 0.95 P | 1.20 | 1.11 P | 0.98 P | 1.13 | 1.18 P | 1.22 P | 0.97 | 0.95 P | 1.02 P | 0.93 |
| 247005_at | AT5G67520 | APS kinase | AKN4 | 0.98 P | 0.93 P | 1.05 | 1.02 P | 1.15 P | 0.89 | 0.98 P | 0.98 P | 1.00 | 0.89 P | 0.88 P | 1.01 | 1.20 P | 1.63 P | 0.74d | 1.31 P | 1.08 P | 1.21 |
| 250846_at | AT5G04590 | Sulfite reductase | SIR | 1.03 P | 0.90 P | 1.15 | 1.21 P | 0.82 P | 1.47c | 1.21 P | 0.90 P | 1.35c | 1.09 P | 0.85 P | 1.29 | 0.93 P | 1.05 P | 0.88 | 0.97 P | 1.21 P | 0.80 |
| Cys synthesis | |||||||||||||||||||||
| 245286_at | AT4G14880 | O-acetylserine (thiol) lyase | Bsas1;1 | 1.00 P | 1.11 P | 0.90 | 1.00 P | 1.35 P | 0.74d | 1.03 P | 1.10 P | 0.93 | 0.98 P | 1.11 P | 0.88 | 0.77 P | 0.90 P | 0.85 | 0.72 P | 0.79 P | 0.92 |
| 256930_at | AT3G22460 | O-acetylserine (thiol) lyase | Bsas1;4 | 0.95 P | 1.54 P | 0.62d | 1.23 P | 2.17 P | 0.57d | 0.69 P | 1.45 P | 0.48e | 1.05 P | 1.17 P | 0.90 | 0.81 P | 0.81 P | 1.00 | 0.94 P | 0.92 P | 1.03 |
| 260566_at | AT2G43750 | O-acetylserine (thiol) lyase | Bsas2;1 | 1.38 P | 1.00 P | 1.38c | 1.12 P | 0.90 P | 1.25 | 1.49 P | 0.95 P | 1.57c | 1.21 P | 1.00 P | 1.21 | 1.01 P | 0.87 P | 1.16 | 0.87 P | 0.91 P | 0.96 |
| 251487_at | AT3G59760 | O-acetylserine (thiol) lyase | Bsas2;2 | 1.01 P | 0.97 P | 1.04 | 0.96 P | 1.00 P | 0.96 | 0.94 P | 1.09 P | 0.86 | 1.06 P | 1.00 P | 1.06 | 0.85 P | 1.11 P | 0.77 | 0.84 P | 1.01 P | 0.83 |
| 251322_at | AT3G61440 | O-acetylserine (thiol) lyase | Bsas3;1 | 1.13 P | 1.18 P | 0.95 | 1.02 P | 1.24 P | 0.82 | 0.98 P | 1.11 P | 0.88 | 0.98 P | 1.04 P | 0.94 | 0.94 P | 0.94 P | 1.00 | 0.72 P | 0.88 P | 0.82 |
| 259094_at | AT3G04940 | O-acetylserine (thiol) lyase | Bsas4;1 | 1.03 P | 1.17 P | 0.88 | 0.84 P | 0.97 P | 0.87 | 0.86 P | 1.07 P | 0.81 | 0.76 P | 0.90 P | 0.84 | 1.15 P | 0.90 P | 1.27 | 1.03 P | 1.31 P | 0.79 |
| 246701_at | AT5G28020 | O-acetylserine (thiol) lyase | Bsas4;2 | 1.75 P | 0.98 P | 1.78c | 1.58 P | 1.15 P | 1.37c | 1.66 P | 1.02 P | 1.62c | 1.51 P | 0.87 P | 1.74c | 0.81 P | 0.83 P | 0.98 | 0.86 P | 0.65 P | 1.34c |
| 246700_at | AT5G28030 | O-acetylserine (thiol) lyase | Bsas4;3 | 0.24 A | 0.14 A | 1.77 A | 1.01 A | 1.60 A | 0.38 A | 1.06 A | 0.88 A | 1.24 A | 1.06 A | 0.99 A | 0.18 A | ||||||
| 259172_at | AT3G03630 | O-acetylserine (thiol) lyase | Bsas5;1 | 1.04 A | 0.71 P | 1.47c | 0.93 A | 1.00 A | 0.93 | 1.00 A | 1.14 P | 0.88 | 1.05 A | 0.85 A | 1.23 | 0.91 A | 0.93 A | 0.98 | 1.57 A | 1.09 P | 1.44c |
| 247982_at | AT5G56760 | Ser acetyltransferase | Serat1;1 | 0.92 P | 1.04 P | 0.89 | 0.91 P | 1.00 P | 0.91 | 0.87 P | 1.00 P | 0.87 | 0.94 P | 0.91 P | 1.04 | 1.99 P | 1.83 P | 1.09 | 2.00 P | 2.04 P | 0.98 |
| 260602_at | AT1G55920 | Ser acetyltransferase | Serat2;1 | 1.46 P | 1.22 P | 1.20 | 1.28 P | 1.52 P | 0.84 | 1.11 P | 0.94 P | 1.18 | 1.06 P | 0.86 P | 1.23 | 0.82 P | 0.63 P | 1.30 | 0.48 P | 0.48 P | 1.01 |
| 257194_at | AT3G13110 | Ser acetyltransferase | Serat2;2 | 1.23 P | 1.17 P | 1.05 | 0.93 P | 1.23 P | 0.76 | 0.78 P | 0.84 P | 0.93 | 0.75 P | 0.90 P | 0.83 | 0.98 P | 1.07 P | 0.91 | 1.02 P | 1.13 P | 0.90 |
| 264594_at | AT2G17640 | Ser acetyltransferase | Serat3;1 | 2.24 P | 0.98 P | 2.29b | 2.22 P | 0.95 P | 2.34b | 1.86 P | 0.97 P | 1.93c | 1.93 P | 0.72 P | 2.67b | 1.47 P | 0.79 P | 1.87c | 1.02 P | 0.51 P | 2.03b |
| GSH and Met synthesis | |||||||||||||||||||||
| 254270_at | AT4G23100 | γ-Glutamylcysteine synthetase | GSH1 | 1.02 P | 1.52 P | 0.67d | 1.07 P | 1.67 P | 0.64d | 0.92 P | 1.45 P | 0.64d | 0.92 P | 1.38 P | 0.66d | 0.75 P | 0.89 P | 0.84 | 0.69 P | 0.98 P | 0.71d |
| 246785_at | AT5G27380 | Glutathione synthetase | GSH2 | 1.22 P | 1.16 P | 1.05 | 1.12 P | 1.03 P | 1.09 | 1.11 P | 0.97 P | 1.14 | 1.07 P | 0.90 P | 1.19 | 0.70 P | 0.63 P | 1.10 | 0.66 P | 0.82 P | 0.80 |
| 259279_at | AT3G01120 | Cystathionine γ-synthase | CGS | 0.97 P | 1.12 P | 0.87 | 0.85 P | 1.07 P | 0.80 | 0.90 P | 1.03 P | 0.87 | 0.78 P | 1.12 P | 0.70d | 0.83 P | 1.04 P | 0.80 | 0.73 P | 1.05 P | 0.70d |
| 256531_at | AT1G33320 | Cystathionine γ-synthase, putative | 0.79 A | 1.52 A | 0.96 A | 1.40 A | 0.78 A | 0.72 A | 1.04 A | 0.90 A | 1.25 A | 1.43 A | 0.90 A | 1.29 A | |||||||
| 251666_at | AT3G57050 | Cystathionine β-lyase | CBL | 1.19 P | 1.19 P | 0.99 | 1.01 P | 1.03 P | 0.98 | 0.87 P | 0.99 P | 0.88 | 0.91 P | 0.94 P | 0.98 | 0.83 P | 1.06 P | 0.78 | 0.95 P | 1.32 P | 0.72d |
| 246185_at | AT5G20980 | Met synthase | 0.98 P | 0.86 P | 1.14 | 0.81 P | 0.78 P | 1.05 | 1.02 P | 0.88 P | 1.17 | 0.95 P | 1.17 P | 0.81 | 1.44 P | 1.54 P | 0.94 | 1.41 P | 1.22 P | 1.16 | |
| 259343_s_at | AT3G03780 | Met synthase | 0.95 P | 0.93 P | 1.02 | 0.88 P | 0.94 P | 0.94 | 0.99 P | 1.01 P | 0.97 | 0.99 P | 1.01 P | 0.98 | 1.80 P | 1.74 P | 1.04 | 1.45 P | 1.61 P | 0.90 | |
| Met metabolism | |||||||||||||||||||||
| 261957_at | AT1G64660 | Cystathionine γ-lyase, putative | 1.63 P | 1.06 P | 1.54c | 1.69 P | 1.09 P | 1.55c | 1.77 P | 0.94 P | 1.88c | 1.56 P | 0.94 P | 1.66c | 0.45 P | 0.26 P | 1.76c | 0.38 P | 0.21 P | 1.85c | |
| 260594_at | AT1G55880 | Cystathionine β-synthase, putative | 0.98 P | 1.32 P | 0.75d | 1.00 P | 1.11 P | 0.90 | 0.89 P | 1.09 P | 0.81 | 0.80 P | 0.94 A | 0.85 | 1.00 P | 0.80 A | 1.26 | 1.02 A | 1.04 P | 0.98 | |
| 260913_at | AT1G02500 | S-adenosylmethionine synthetase | SAM1 | 1.06 P | 1.16 P | 0.92 | 0.94 P | 1.31 P | 0.71d | 1.15 P | 1.09 P | 1.05 | 0.93 P | 1.19 P | 0.78 | 0.94P | 0.88 P | 1.06 | 0.75 P | 0.88 P | 0.85 |
| 255552_at | AT4G01850 | S-adenosylmethionine synthetase | SAM2 | 1.23 P | 1.38 P | 0.89 | 0.99 P | 1.25 P | 0.79 | 0.97 P | 1.10 P | 0.88 | 0.90 P | 1.15 P | 0.78 | 1.01P | 0.77 P | 1.32 | 0.76 P | 0.86 P | 0.89 |
| 263838_at | AT2G36880 | S-adenosylmethionine synthetase | SAM3 | 1.12 P | 1.24 P | 0.90 | 0.97 P | 1.19 P | 0.82 | 0.93 P | 1.06 P | 0.88 | 0.86 P | 0.99 P | 0.87 | 1.09P | 0.87 P | 1.26 | 0.85 P | 1.01 P | 0.84 |
| 258415_at | AT3G17390 | S-adenosylmethionine synthetase | 1.46 P | 1.05 P | 1.39c | 1.16 P | 0.89 P | 1.31 | 1.34 P | 0.94 P | 1.42c | 1.06 P | 0.89 P | 1.19 | 1.26P | 0.79 P | 1.59c | 0.95 P | 0.87 P | 1.10 | |
| 245356_at | AT4G13940 | S-adenosylhomocysteine hydrolase | 1.22 P | 1.22 P | 1.00 | 0.94 P | 1.16 P | 0.81 | 1.18 P | 1.05 P | 1.12 | 0.91 P | 1.12 P | 0.82 | 0.95P | 0.78 P | 1.21 | 0.71 P | 0.84 P | 0.84 | |
| 257173_at | AT3G23810 | S-adenosylhomocysteine hydrolase | 0.86 P | 1.00 P | 0.86 | 0.82 P | 0.92 P | 0.88 | 0.93 P | 1.00 P | 0.93 | 0.82 P | 1.13 P | 0.73d | 1.80P | 1.55 P | 1.16 | 1.60 P | 1.71 P | 0.94 | |
| 258075_at | AT3G25900 | Homocysteine S-methyltransferase | HMT-1 | 1.13 P | 0.89 P | 1.28 | 1.07 P | 1.02 P | 1.05 | 0.98 P | 0.83 P | 1.18 | 0.89 P | 0.83 P | 1.08 | 1.07P | 0.86 P | 1.25 | 1.20 P | 1.21 P | 0.99 |
| 251175_at | AT3G63250 | Homocysteine S-methyltransferase | HMT-2 | 1.17 P | 1.15 P | 1.02 | 1.05 P | 0.95 P | 1.11 | 1.11 P | 1.17 P | 0.95 | 0.91 P | 1.08 P | 0.84 | 0.71P | 0.76 P | 0.93 | 0.86 P | 0.84 P | 1.03 |
| 258322_at | AT3G22740 | Homocysteine S-methyltransferase | HMT-3 | 1.00 P | 1.10 P | 0.90 | 0.64 P | 1.58 P | 0.41e | 1.01 A | 0.97 P | 1.03 | 0.73 A | 1.33 P | 0.55d | 0.89A | 2.02 P | 0.44e | 0.93 A | 2.22 P | 0.42e |
| Se-binding protein | |||||||||||||||||||||
| 245285_s_at | AT4G14030 | Se-binding protein, putative | 1.102 P | 0.446 P | 2.47b | 1.165 P | 0.635 P | 1.83c | 0.986 P | 0.573 P | 1.72c | 1.28 P | 0.49 P | 2.61b | 1.314P | 1.081 P | 1.22 | 1.014 P | 0.882 P | 1.15 | |
| 245285_s_at | AT4G14040 | Se-binding protein, putative | |||||||||||||||||||
| 257197_at | AT3G23800 | Se-binding protein, putative | 0.59 P | 1.369 P | 0.43e | 0.743 P | 1.386 P | 0.54d | 0.505 P | 1.22 P | 0.41e | 0.64 P | 1.139 P | 0.56d | 0.861P | 1.215 P | 0.71d | 0.839 P | 1.629 P | 0.52d | |
DISCUSSION
Overexpression of CpNifS, a NifS-like chloroplast protein with SL activity, resulted in 2-fold enhanced tolerance to selenate and 2- to 3-fold higher Se accumulation. Because CpNifS mediates the breakdown of SeCys into alanine and Se0, we hypothesized that the transgenic plants increased selenate tolerance because of decreased SeCys incorporation into protein. Indeed, the transgenics showed significantly reduced incorporation of Se into protein. Therefore, this NifS-like endogenous plant protein clearly affects plant Se metabolism when overexpressed.
The CpNifS transgenics showed enhanced tolerance to selenate but not to selenite. This may be because the conversion from selenate to selenite is a very slow and rate-limiting step in Se assimilation, especially in Arabidopsis (de Souza et al., 1998; Pilon-Smits et al., 1999; Ellis et al., 2004). The slow production of SeCys in selenate-supplied plants may enable the CpNifS protein to keep up with its detoxification, preventing it from being incorporated into protein. Selenite-supplied plants were shown to quickly convert all Se into organic Se species (de Souza et al., 1998), and this flood of SeCys may have saturated the detoxification capacity of CpNifS in the transgenics.
In a previous study, a mouse SL was expressed in Arabidopsis (Pilon et al., 2003). When this mouse SL was expressed in the cytosol, the plants showed enhanced Se tolerance, but when expressed in the plastids the plants became less tolerant to Se. It was speculated that the produced Se0 may replace elemental S and interfere with Fe-S cluster assembly in the chloroplast. It is intriguing that overexpression of the mouse SL in the chloroplast decreased Se tolerance while overexpression of this endogenous Arabidopsis SL enhanced Se tolerance. This could be due to different SeCys/Cys substrate specificities of both enzymes. In vitro CpNifS showed a 300-fold higher activity toward SeCys compared to Cys, while the mouse SL had 5,000-fold higher activity toward the Se substrate (Esaki et al., 1982; Pilon-Smits et al., 2002). Also, there may be regulatory mechanisms in the chloroplast that modulate CpNifS (but not mouse SL) activity toward both substrates. These may allow CpNifS to protect against both Se incorporation into proteins and Fe-Se clusters. Finally, it cannot be excluded that not all CpNifS was targeted to the plastids in the transgenics, but that a small fraction was present in the cytosol, leading to a phenotype similar to that found when the mouse SL was expressed in the cytosol.
Another difference between the transgenics overexpressing the mouse SL in the chloroplast (cpSL) and the CpNifS transgenics described here was that the mouse cpSL plants did not show any difference in Se accumulation from selenate, while the CpNifS plants did. Again, this could be due to different SeCys/Cys substrate specificities of both enzymes. The mouse SL has negligible activity toward Cys, in contrast to CpNifS (Esaki et al., 1982; Pilon-Smits et al., 2002). Perhaps the increased uptake of selenate and sulfate are correlated with the breakdown of Cys.
CpNifS diverts Se away from protein incorporation, and it was hypothesized that this mechanism may allow transgenic plants to accumulate a different form of Se (e.g. Se0) compared to wild type. Indeed, there appeared to be a slightly higher fraction of more reduced Se in the transgenics judged from μXAS analysis. However, the majority of Se was present as SeVI in both wild type and CpNifS transgenics. It is possible that there was substantial accumulation of Se0 in the plastids, but this was masked by the abundance of selenate in other cellular compartments. In addition, the elevated S levels in the CpNifS transgenics may have contributed to their enhanced selenate tolerance, as they would allow S in cells to compete more successfully with Se for incorporation into S compounds.
In both wild type and CpNifS55, Se appeared to accumulate predominantly in the periphery of the cells and in the vascular tissue. The resolution does not enable us to discern between the cell wall, cytosol, or plastids, but it does not appear that Se was accumulated in the vacuoles. The high Se concentration in the vascular tissue may indicate that uptake into the shoot symplast was a limiting factor for Se accumulation in the leaf under these conditions.
Overexpression of CpNifS resulted in enhanced accumulation of both Se and S in selenate-treated plants; S was also accumulated more in shoots of CpNifS transgenics than in wild type under control conditions. This suggests that the breakdown of (Se)Cys enhances selenate and sulfate uptake. The enhanced Se accumulation in the CpNifS transgenics does not appear to be caused by more pronounced up-regulation of sulfate transporters at the transcriptional level, at least not in the root, since the levels of up-regulation were similar to wild type. Still, it is possible that S and Se fluxes through the plant differed in wild type and CpNifS transgenics via regulation at the protein level, or via differences in shoot transcription levels or transcriptional differences in certain cell types.
In all plant types, S levels were higher in selenate-treated plants compared to control plants, which can be explained by the observed up-regulation of various sulfate transporters. A similar up-regulation of sulfate transporters as well as of genes involved in sulfate assimilation (Leustek et al., 1994; Gutierrez-Marcos et al., 1996; Setya et al., 1996) and Cys synthesis (Hatzfeld et al., 2000; Kawashima et al., 2005) was observed in earlier studies where plants were subjected to S deficiency (Takahashi et al., 1997, 2000; Yoshimoto et al., 2002; Maruyama-Nakashita et al., 2003, 2005; Kataoka et al., 2004). Thus, selenate may be perceived by the plant as S deficiency, or even cause deficiency of certain S-containing metabolites. As mentioned above, the higher S levels in the CpNifS transgenics compared to wild type on Se may provide the plants with an additional mechanism to alleviate stress.
The discovery that an endogenous NifS-like protein with SL activity can affect plant Se metabolism is intriguing, in view of the current thought that Se is not an essential micronutrient for higher plants, as opposed to the green alga Chlamydomonas (Novoselov et al., 2002). The SL activity of CpNifS may be an evolutionary relic of essential Se metabolism or an unavoidable side reaction of a protein with Cys desulfurase function. It is feasible that in some species CpNifS has a function in Se metabolism by preventing nonspecific Se incorporation into protein, rather than providing Se0 for essential selenoprotein synthesis like it does in mammals and bacteria (Stadtman, 1990; Mihara and Esaki, 2002). However, this is probably not common as there are not many natural areas in the world where plants experience Se toxicity. Incidentally, the preference of CpNifS for SeCys over Cys may not be as pronounced in planta as it is in vitro (300-fold). Recently, a SufE-like chloroplast protein was shown to stimulate the Cys desulfurase activity of CpNifS 40-fold in vitro and to increase its affinity toward Cys 2-fold (H. Ye, S. Abdel-Ghany, E. Pilon-Smits, and M. Pilon, unpublished data). This protein likely regulates CpNifS activities toward its Se and S substrates in vivo.
Microarray analysis indicated that, apart from CpNifS, only the SMT homolog HMT-3 and a few other genes were differentially regulated by Se in the CpNifS roots compared to wild type. The higher CpNifS levels may explain the enhanced Se tolerance via prevention of Se incorporation into proteins, as discussed earlier. It is not known whether the HMT-3 protein has SMT activity. Higher SMT activity would be expected to also shuttle Se away from incorporation into proteins, via methylation of SeCys (Neuhierl et al., 1999; Ranocha et al., 2000). The HMT-3 levels were higher under control conditions in CpNifS roots than in wild type, but decreased in response to selenate in the CpNifS plants and not in wild type. Thus, HMT-3 did not appear to contribute to the enhanced Se tolerance in the CpNifS plants. None of the other differentially expressed genes offer an obvious explanation for the higher Se tolerance or accumulation properties of the CpNifS transgenics, but for many of them their functions are not quite known. Thus, from what is known currently it appears that CpNifS affects Se tolerance and accumulation directly rather than via pleiotropic effects on other transcript levels, illustrating the importance of this gene for selenate tolerance. However, as mentioned, it cannot be excluded that there were additional transcriptome differences in the shoot that caused the enhanced tolerance and accumulation in CpNifS plants.
In conclusion, overexpression of CpNifS mitigated Se toxicity and enhanced Se accumulation. The overexpression of CpNifS decreased the amount of nonspecific incorporation of Se in protein, allowing the transgenic plants to tolerate and accumulate more Se compared to wild-type Arabidopsis. Overexpression of CpNifS also affected S metabolism, enhancing S uptake. Microarray analysis helped explain some of the observed physiological effects of selenate. The paucity of transcripts that were differentially regulated in the transgenics compared to wild type on selenate suggests that overexpression of CpNifS is likely the direct reason why the transgenics are more tolerant to and accumulate more Se. CpNifS-overexpressing transgenic plants that have enhanced ability to tolerate and accumulate Se may ultimately be useful in phytoremediation—the use of plants to clean polluted soils. For that purpose, the same gene could be overexpressed in a high biomass species, such as Indian mustard. Plants with enhanced Se accumulation may also have value as fortified foods, since Se-enriched diets can help prevent cancer and other detrimental effects of oxidative stress.
MATERIALS AND METHODS
Overexpression of CpNifS
The CpNifS (At1g18490) coding sequence (Pilon-Smits et al., 2002) was cloned as a NcoI/BamHI fragment into pMOG18 (Sijmons et al., 1990) under control of the constitutive 35S cauliflower mosaic virus promoter, before transfer into the plant binary vector pBART containing the glufosinate-ammonium resistance marker. Sequence analysis confirmed the final construct in which the HindIII/EcoRI sites were replaced by NotI sites. The construct was then transferred into Agrobacterium tumefaciens (strain C58C1) and transformed into Arabidopsis (Arabidopsis thaliana; ecotype Wassilewskija [Ws]) using the floral dip method (Clough and Bent, 1998). Glufosinate-resistant lines were selected on half-strength MS (Sigma-Aldrich; Murashige and Skoog, 1962) agar media with 1% (w/v) Suc and 5 mg L−1 glufosinate-ammonium and transferred to soil. Plants were grown in Metromix potting soil under controlled conditions (light intensity of 40 μmol m−2 s−1, 16-h-light/8-h-dark cycle at 24°C). After 2 weeks, PCR analysis was used to confirm insertion of the gene construct by using 5′-ccttcgcaagacccttcctc and 5′-ggaagacagtttctcgtacaaat primers that hybridize to the 35S CaMV promoter and CpNifS sequence, respectively (Fig. 1). Six lines showed the expected PCR product, while no PCR product was detected in the wild-type control (data not shown). The six transgenic lines were propagated to homozygosity and did not show any phenotypic differences compared to wild type on control media and on soil.
Immunoblot analysis was used to determine the degree of overexpression of CpNifS in the transgenics. Shoot and root proteins from plants grown on half-strength MS media were separated by SDS-PAGE and transferred to nitrocellulose by electroblotting. The CpNifS protein was immunodetected using polyclonal antibodies raised in chicken against CpNifS protein (Pilon-Smits et al., 2002). Two transgenic lines, CpNifS38 and CpNifS55, were selected for further analysis. Protein concentration of wild type and the transgenics were determined using a desktop scanner and ImageJ imaging software (National Institute of Health; http://rsb.info.nih.gov/ij/).
Chloroplast isolation was performed as described by Rensink et al. (1998) from the two selected lines and wild type grown on soil for 3 weeks. A western blot was used to determine if CpNifS was localized to the stromal fraction of chloroplasts; 3 μg of chlorophyll were loaded per lane. Antibodies raised against plastocyanin were also used on blots from the same gel as controls for uniform loading.
Se and S Metabolism
Se tolerance was determined by measuring the root length of seedlings (n = 30) grown for 10 d on vertical plates containing half-strength MS agar media supplemented with 1% (w/v) Suc with or without selenate (40 μm Na2SeO4) as described by Pilon-Smits et al. (1999). Total Se accumulation was assayed by growing seedlings (n = 100) for 20 d on horizontal plates containing half-strength MS agar containing Suc media with or without 40 μm Na2SeO4. Roots and shoots were harvested, separated, washed to remove any external Se, and dried overnight at 70°C. Samples from pooled root samples (n = 3) and shoot samples (n = 5) were then acid digested and analyzed by inductively coupled plasma-atomic emission spectrometry as described (Pilon-Smits et al., 1999). Incorporation of Se in protein was determined in seedlings (n = 300) grown for 14 d on agar medium supplied with 20 μm SeO4 as described by Pilon et al. (2003).
GeneChip Hybridization and Microarray Analysis
Plants were grown in a growth chamber for 10 d on vertical plates with or without 40 μm Na2SeO4. For each line, total RNA was extracted from the roots using the RNeasy Plant Mini kit (Qiagen). The technical manual of the Arabidopsis Genome GeneChip array (Affymetrix) was used in preparing the labeled target cRNA. Double-stranded cDNA was prepared from 20 μg of total RNA using the Superscript Choice system (Invitrogen) and transcribed in vitro using the BioArray High Yield RNA transcript kit (Enzo). After purification and fragmentation, the labeled cRNA was hybridized to an Arabidopsis Genome Chip Array containing more than 22,500 probe sets representing approximately 24,000 genes and placed in a Hybridization Oven model 640 (Affymetrix). Washing and staining of the chips were carried out using GeneChip Fluidics Station model 400. Scanning was performed with the Gene Array Scanner (Agilent Technologies), and the Microarray Suite 5.0 (Affymetrix) and Gene Spring 7.2 (Silicon Genetics) were used for data analysis as described (Maruyama-Nakashita et al., 2003; 2005). Raw signals of each transcript were normalized with the median of all measurements on the chip, from which fold changes of signal intensities were calculated.
μXRF/μXAS
Whole-plant samples grown for 3 weeks on half-strength MS medium with 40 μm selenate were washed to remove any external Se, and shipped on dry ice to the Advanced Light Source at the Lawrence Berkeley Laboratory for analysis on Beamline 10.3.2 (Marcus et al., 2004). Intact mature leaves were mounted using silicone grease to a Peltier stage and kept at −30°C to reduce radiation damage. Crude mapping of the Se concentration was performed on one representative leaf for each plant type, with a 16 × 7 μm beam sampled in 12 × 12 μm pixels, followed by fine mapping (5 × 5 μm beam, 3 × 3 μm pixels) on at least five selected areas per leaf, all at 12,859 eV. Se K-edge XANES was measured to determine the Se oxidation state at specific points in the fine map (Pickering et al., 1999). A selenate solution was used as a calibration standard.
Acknowledgments
We thank Brady Hanson and Hale Tufan for their help in obtaining the CpNifS transgenics. The operations of the Advanced Light Source at Lawrence Berkeley National Laboratory are supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under contract number DE–AC02–05CH11231.
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant no. 2003–35318–13758 to E.A.H.P.-S. and M.P.) and by the National Science Foundation/Japan Society for the Promotion of Science (summer fellowship to D.V.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Elizabeth A.H. Pilon-Smits (epsmits@lamar.colostate.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068684.
References
- Abdel-Ghany SE, Ye H, Garifullina GF, Zhang L, Pilon-Smits EAH, Pilon M (2005) Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol 138: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JW (1993) Selenium interactions in sulfur metabolism. In LJ De Kok, ed, Sulfur Nutrition and Assimilation in Higher Plants—Regulatory, Agricultural and Environmental Aspects. SPB Academic Publishing, The Hague, The Netherlands, pp 49–60
- Bañuelos G, Terry N, LeDuc DL, Pilon-Smits EAH, Mackey B (2005) Field trial of transgenic Indian mustard plants shows enhanced phytoremediation of selenium-contaminated sediment. Environ Sci Technol 39: 1771–1777 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformations of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Souza MP, Pilon-Smits EAH, Lytle CM, Hwang S, Tai J, Hinma TSU, Yeh L, Terry N (1998) Rate limiting steps in selenium assimilation and volatilization by Indian mustard. Plant Physiol 117: 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DR, Sors TG, Brunk DG, Albrecht C, Orser C, Lahner B, Wood KV, Harris HH, Pickering IJ, Salt DE (2004) Production of Se-methylselenocysteine in transgenic plants expressing selenocysteine methyltransferase. BMC Plant Biol 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki N, Nakamura T, Tanaka H, Soda K (1982) Selenocysteine lyase, a novel enzyme that specifically acts on seleno-cysteine. Mammalian distribution and purification and properties of pig-liver enzyme. J Biol Chem 257: 4386–4391 [PubMed] [Google Scholar]
- Fessler AJ, Moller F, Talcott PA, Exon JH (2003) Selenium toxicity in sheep grazing reclaimed phosphate mining sites. Vet Hum Toxicol 45: 294–298 [PubMed] [Google Scholar]
- Garifullina GF, Owen JD, Lindblom SD, Tufan H, Pilon M, Pilon-Smits EAH (2003) Expression of a mouse selenocysteine lyase in Brassica juncea chloroplasts affects selenium tolerance and accumulation. Physiol Plant 118: 538–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL (1996) Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA 93: 13377–13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SJ (2004) Review of selenium toxicity in the aquatic food chain. Sci Total Environ 326: 1–36 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Ishizawa K, Saito K (2000) β-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol 123: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira CK, Partal K, Dhillon K (2004) Dietary selenium intake by men and women in high and low selenium areas of Punjab. Public Health Nutr 7: 39–43 [DOI] [PubMed] [Google Scholar]
- Ip C, Dong Y, Ganther HE (2002) New concepts in selenium chemoprevention. Cancer Metastasis Rev 21: 281–289 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Berkowitz O, Hell R, Noji M, Saito K (2005) Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol 137: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Garbisu C, Pickering IJ, Prince RC, George GN, Cho MJ, Wong JH, Buchanan BB (2003) Thioredoxin h overexpressed in barley seeds enhances selenite resistance and uptake during germination and early seedling development. Planta 218: 186–191 [DOI] [PubMed] [Google Scholar]
- LeDuc DL, Tarun AS, Montes-Bayon M, Meija J, Malit MF, Wu C, Abdel Samie M, Chiang CY, Tagmount A, deSouza M, et al (2004) Overexpression of selenocysteine methyltransferase in Arabidopsis and Indian Mustard increases selenium tolerance and accumulation. Plant Physiol 135: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Murillo M, Cervantes M (1994) Cloning of a cDNA encoding ATP sulfurylase from Arabidopsis thaliana by functional expression in Saccharomyces cerevisiae. Plant Physiol 105: 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus MA, MacDowell AA, Celestre R, Manceau A, Miller T, Padmore HA, Sublett RE (2004) Beamline 10.3.2 at ALS: a hard X-ray microprobe for environmental and materials sciences. J Synchrotron Radiat 11: 239–247 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling on sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42: 305–314 [DOI] [PubMed] [Google Scholar]
- Mihara H, Esaki N (2002) Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol 60: 12–23 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 437–497 [Google Scholar]
- Neuhierl B, Böck A (1996) On the mechanism of selenium tolerance in selenium-accumulating plants. Purification and characterization of a specific selenocysteine methyltransferase from cultured cells of Astragalus bisulcatus. Eur J Biochem 239: 235–238 [DOI] [PubMed] [Google Scholar]
- Neuhierl B, Thanbichler M, Lottspeich F, Böck A (1999) A family of S-methylmethionine-dependent thiol/selenol methyltransferases. Role in selenium tolerance and evolutionary relation. J Biol Chem 274: 5407–5414 [DOI] [PubMed] [Google Scholar]
- Novoselov SV, Rao M, Onoshko NV, Shi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN (2002) Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J 21: 3681–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf HM, Kilness AW, Simmons JL, Stroud RK, Hoffman DJ, Morrre JF (1988) Selenium toxicosis in wild aquatic birds. J Toxicol Environ Health 24: 67–92 [DOI] [PubMed] [Google Scholar]
- Pickering IJ, Prince RC, George GN, Rauser WE, Wickramasinghe WA, Watson AA, Dameron CT, Dance IG, Fairlie DP, Salt DE (1999) X-ray absorption spectroscopy of cadmium phytochelatin and model systems. Biochim Biophys Acta 1429: 351–364 [DOI] [PubMed] [Google Scholar]
- Pilon M, Owen JD, Garifullina GF, Kurihara T, Mihara H, Esaki N, Pilon-Smits EAH (2003) Enhanced selenium tolerance and accumulation in transgenic Arabidopsis expressing a mouse selenocysteine lyase. Plant Physiol 131: 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Garifullina GF, Abdel-Ghany S, Kato SI, Mihara H, Hale KL, Burkhead JL, Esaki N, Kurihara T, Pilon M (2002) Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol 130: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Hwang S, Lytle CM, Zhu Y, Tai JC, Bravo RC, Chen Y, Leustek T, Terry N (1999) Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction and tolerance. Plant Physiol 119: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, Bourgis F, Ziemak MJ, Rhodes D, Gage DA, Hanson AD (2000) Characterization and functional expression of cDNAs encoding methionine-sensitive and -insensitive homocysteine S-methyltransferases from Arabidopsis. J Biol Chem 275: 15962–15968 [DOI] [PubMed] [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P (1998) Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol 118: 681–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setya A, Murillo M, Leustek T (1996) Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA 93: 13383–13388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijmons PC, Dekker BM, Schrammeijer B, Verwoerd TC, van den Elzen PJ, Hoekema A (1990) Production of correctly processed human serum albumin in transgenic plants. Biotechnology (NY) 8: 217–221 [DOI] [PubMed] [Google Scholar]
- Stadtman TC (1990) Selenium biochemistry. Annu Rev Biochem 59: 111–127 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, de Almedia Engler J, Engler G, Van Montagu M, Saito K (1997) Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N, Zayed A, de Souza P, Tarun A (2000) Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol 51: 401–432 [DOI] [PubMed] [Google Scholar]
- Van Huysen T, Abdel-Ghany S, Hale KL, LeDuc D, Terry N, Pilon-Smits EAH (2003) Overexpression of cystathionine-γ-synthase in Indian mustard enhances selenium volatilization. Planta 218: 71–78 [DOI] [PubMed] [Google Scholar]
- Ye H, Garifullina GF, Abdel-Ghany SE, Zhang L, Pilon-Smits EAH, Pilon M (2004) The chloroplastic NifS-like protein of Arabidopsis thaliana is required for iron-sulfur cluster formation in ferredoxin. Planta 220: 602–608 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K (2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29: 465–473 [DOI] [PubMed] [Google Scholar]