Abstract
Anthocyanin accumulation is acknowledged as a phenotypic indicator of phosphate (Pi) starvation. However, negative regulators of this process and their molecular mechanisms remain largely unexplored. In this study, we demonstrate that WRKY33 acts as a negative regulator of phosphorus-status-dependent anthocyanin biosynthesis. WRKY33 regulates the expression of the gene encoding dihydroflavonol 4-reductase (DFR), a rate-limiting enzyme in anthocyanin production, both directly and indirectly. WRKY33 binds directly to the DFR promoter to repress its expression and also interferes with the MBW complex through interacting with PAP1 to indirectly influence DFR transcriptional activation. Under −Pi conditions, PHR1 interacts with WRKY33, and the protein level of WRKY33 decreases; the repression of DFR expression by WRKY33 is thus attenuated, leading to anthocyanin accumulation in Arabidopsis. Further genetic and biochemical assays suggest that PHR1 is also involved in regulating factors that affect WRKY33 protein turnover. Taken together, our findings reveal that Pi starvation represses WRKY33, a repressor of anthocyanin biosynthesis, to finely tune anthocyanin biosynthesis. This “double-negative logic” regulation of phosphorus-status-dependent anthocyanin biosynthesis is required for the maintenance of plant metabolic homeostasis during acclimation to Pi starvation.
Key words: WRKY33, phosphate signaling, PHR1, Arabidopsis thaliana, anthocyanins
This study reports that WRKY33 negatively regulates anthocyanin biosynthesis under +Pi conditions by binding directly to the DFR promoter to repress its expression and by interfering with the MBW complex to indirectly suppress DFR expression; while under −Pi conditions, WRKY33 physically interacts with and functions together with PHR1 to attenuate DFR repression and thus mediate acclimation to phosphate starvation in Arabidopsis thaliana.
Introduction
Phosphorus is an essential macronutrient that participates in vital biological processes in plants, such as biosynthesis of nucleic acids, ATP, and phospholipids and regulation of enzyme activity and photosynthesis (Kuo and Chiou, 2011; Wang et al., 2021). Plant roots absorb phosphorus from the soil primarily as free soluble phosphate (Pi). However, free Pi is easily fixed by calcium and aluminum ions in the soil, forming insoluble Pi, which cannot be absorbed by plant roots (Yuan and Liu, 2008). Pi deficiency is therefore one of the most common nutrient deficiencies that reduce plant productivity. Plants have evolved a number of Pi-starvation response mechanisms to adapt to low Pi stress over time, including remodeling of root structure, changes in metabolic flow, and regulation of gene expression (Chiou and Lin, 2011; Shi et al., 2021; Chen et al., 2022). Significant progress has been made in recent decades regarding components of the Pi signaling pathway. PHOSPHATE STARVATION RESPONSE1 (PHR1), a myeloblastosis (MYB) family transcription factor (TF), has been identified as the master TF that governs the Pi-starvation response by binding to the cis-element PHR1 binding site (P1BS; GNATATNC) in the promoters of Pi-starvation-induced (PSI) genes (Chiou and Lin, 2011; Shi et al., 2021; Li et al., 2022; Chen et al., 2022). SPX proteins function as negative regulators by interacting with PHR1 to inhibit its binding to PSI genes (Ried et al., 2021; Park et al., 2023).
Pi starvation can alter secondary metabolite biosynthesis in plants, especially anthocyanin biosynthesis (Khan et al., 2016). Mutation of PHR1 causes a decrease in anthocyanin biosynthesis during the Pi-starvation response (Pant et al., 2015). Anthocyanins are glycoside derivatives formed from anthocyanidin and saccharide via a glycosidic bond; they have a variety of functions in plant growth and resistance, including luring pollinators and seed dispersers, shielding plants from UV radiation, influencing auxin transport, and scavenging free radicals under stressful conditions (Xie et al., 2016; Naing and Kim, 2021). The biosynthetic pathway of anthocyanins has been well clarified. It begins with the precursor 4-coumaroyl coenzyme A, which is transformed into naringenin by chalcone synthase and chalcone isomerase (de Meaux et al., 2006; Tao et al., 2022a). Naringenin is acted upon by flavanone 3-hydroxylase, flavonoid 3′-hydroxylase, dihydroflavonol 4-reductase (DFR), anthocyanidin synthase, and flavonoid 3-o-glucosyl transferase to produce different anthocyanins (Shirley et al., 1992; Pelletier and Shirley, 1996; Nakatsuka et al., 2012). DFR serves as the rate-limiting enzyme and catalyzes the initial step of anthocyanin production (Liu et al., 2017; LaFountain and Yuan, 2021).
The regulation of anthocyanin biosynthesis has also been intensively studied. Anthocyanin biosynthesis is regulated at the transcriptional level by the MYB–basic helix-loop-helix (bHLH)–WD40 repeat domain regulatory complex (MBW complex). The main component of the MBW complex that stimulates anthocyanin biosynthesis by increasing the expression of anthocyanin biosynthetic genes is the R2R3 MYB protein PRODUCTION OF ANTHOCYANIN PIGMENTS1 (PAP1) (Borevitz et al., 2000; Tohge et al., 2005; Bhargava et al., 2010). TRANSPARENT TESTA8 (TT8), GLABROUS3 (GL3), and ENHANCER OF GLABRA3 (EGL3) are bHLH factors, and TRANSPARENT TESTA GLABRA1 (TTG1) is the most extensively researched WD40-repeat-containing protein (Gonzalez et al., 2008; Wang et al., 2020). Our previous publication showed that SPX4 sequesters PAP1 under +P conditions, thus preventing DFR expression (He et al., 2021).
The WRKY proteins are a large class of TFs that contain a WRKY domain and have been shown to play a vital role in resistance to biotic and abiotic stresses (Liu et al., 2015; Jiang et al., 2017). WRKY TFs regulate downstream gene expression by specifically binding to W-box (TTGACC/T) cis-elements in gene promoters (Eulgem et al., 2000; Ülker and Somssich, 2004). Several WRKY TFs have been reported to participate in the response to low-Pi stress. For example, overexpression of AtWRKY6 increases sensitivity to low-Pi stress and negatively regulates expression of the Pi exporter PHOSPHATE1 (PHO1) by binding to two W boxes in its promoter (Chen et al., 2009; Li et al., 2017). WRKY42 interacts with WRKY6 and binds directly to W-boxes in the PHO1 promoter to limit the production of PHO1 (Su et al., 2015). AtWRKY45 participates in Arabidopsis response to Pi deficiency by directly upregulating PHT1;1 expression (Hui et al., 2014). AtWRKY75 is induced by Pi deficiency, and Atwrky75 mutants are more susceptible to low-Pi stress (Devaiah and Raghothama, 2007). WRKY33 functions as a negative regulator by regulating the expression of ALMT1 to mediate the Pi-deficiency-induced modification of root architecture (Shen et al., 2021). The role of WRKY33 in plant secondary metabolite biosynthesis under biotic stress has also been studied in depth (Shen et al., 2021). However, the mechanism by which WRKY33 influences secondary metabolite biosynthesis in response to abiotic stress, especially Pi-deficient conditions, has received less attention. In this investigation, we found that WRKY33 negatively regulates anthocyanin biosynthesis during acclimation to Pi availability. Under Pi-sufficient conditions, WRKY33 binds directly to the promoter of DFR and interacts with PAP1 to suppress DFR expression, thus repressing anthocyanin biosynthesis. Under Pi-deficient conditions, PHR1 interacts with WRKY33 to inhibit the suppression of WRKY33 at the DFR promoter, leading to anthocyanin accumulation.
Results
WRKY33 represses Pi-starvation-induced anthocyanin accumulation
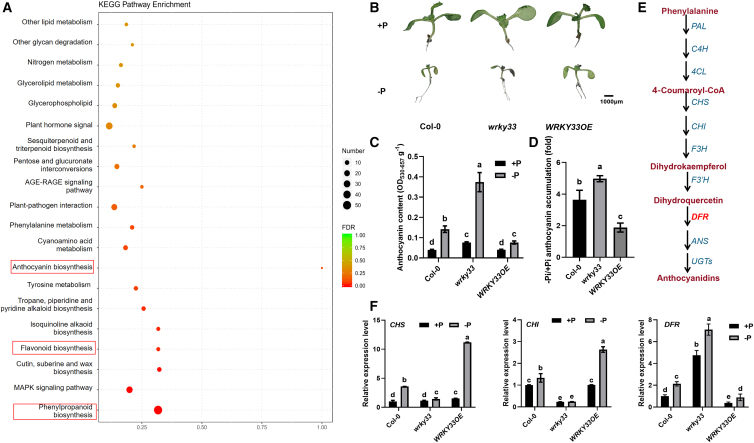
On the basis of our previous research showing that WRKY33 positively regulates indolic glucosinolate (IGS) biosynthesis in Arabidopsis and Brassica crops (Tao et al., 2022b), we wondered whether WRKY33 also participated in regulating the biosynthesis of other secondary metabolites. KEGG pathway enrichment analysis showed that WRKY33-regulated genes were significantly enriched in phenylpropanoid biosynthesis, flavonoid biosynthesis, and anthocyanin biosynthesis (Figure 1A). We therefore measured the contents of flavonoids and anthocyanins in wild-type (Columbia [Col-0]), wrky33 mutant, and WRKY33-OE Arabidopsis plants under +Pi and −Pi conditions. Consistent with a previous report, Pi starvation significantly induced the accumulation of flavonoids and anthocyanins (He et al., 2021). However, there were no significant differences in in situ flavonol and flavonoid contents between the WRKY33-related mutants and Col-0 (Supplemental Figure 1A and 1B). However, anthocyanin content was significantly higher in the wrky33 mutants and 50% lower in the WRKY33-OE plants compared with Col-0 under −Pi conditions (Figure 1B and 1C). The ratios of anthocyanin accumulation of Col-0, wrky33, and WRKY33-OE seedlings grown on −Pi and +Pi media were calculated (Supplemental Figure 2). It appeared that anthocyanin accumulation was significantly lower in WRKY33-OE than in Col-0. These results suggest that WRKY33 is involved in anthocyanin biosynthesis rather than flavonoid biosynthesis.
Figure 1.
WRKY33 represses Pi-deficiency-induced anthocyanin biosynthesis in Arabidopsis.
(A) KEGG enrichment analysis of 9-day-old wrky33 versus Col-0 grown on half-strength Murashige and Skoog medium (½ MS).
(B and C) Phenotypes (B) and anthocyanin contents (C) of 9-day-old Col-0, wrky33, and WRKY33-OE seedlings grown on ½ MS +P (1.25 mM Pi) and −P (0 mM Pi) media. The purple color indicates a high level of anthocyanin accumulation. Bar, 1000 μm. Different letters indicate significant differences (ANOVA, Fisher’s LSD test; P < 0.05).
(D) Ratios of the anthocyanin accumulation in −Pi/+Pi conditions.
(E) Simplified scheme of the anthocyanidin biosynthetic pathway in Arabidopsis.
(F) Expression levels of anthocyanidin biosynthesis genes (CHS, CHI, and DFR) in 9-day-old Col-0, wrky33, and WRKY33-OE plants grown on ½ MS +P (1.25 mM Pi) and −P (0 mM Pi) media. The transcripts were analyzed by RT–qPCR. Different letters indicate significant differences (ANOVA, Fisher’s LSD test; P < 0.05). Error bars indicate the SD of four biological replicates.
Numerous genes are involved in anthocyanin biosynthesis, including chalcone isomerase (CHI), chalcone synthase (CHS), and DFR (Kubasek et al., 1992; Burbulis and Winkel-Shirley, 1999; Nesi et al., 2000). DFR catalyzes the first step of anthocyanin biosynthesis and is sensitive to Pi-deficient and Pi-recovery conditions (He et al., 2021). Among these three genes, DFR showed an expression pattern similar to the pattern of anthocyanin content in WRKY33-related mutants (Figure 1D and 1F). This result suggested that WRKY33 repressed the DFR expression and anthocyanin accumulation induced by Pi deficiency in Arabidopsis.
WRKY33 directly and indirectly regulates DFR expression
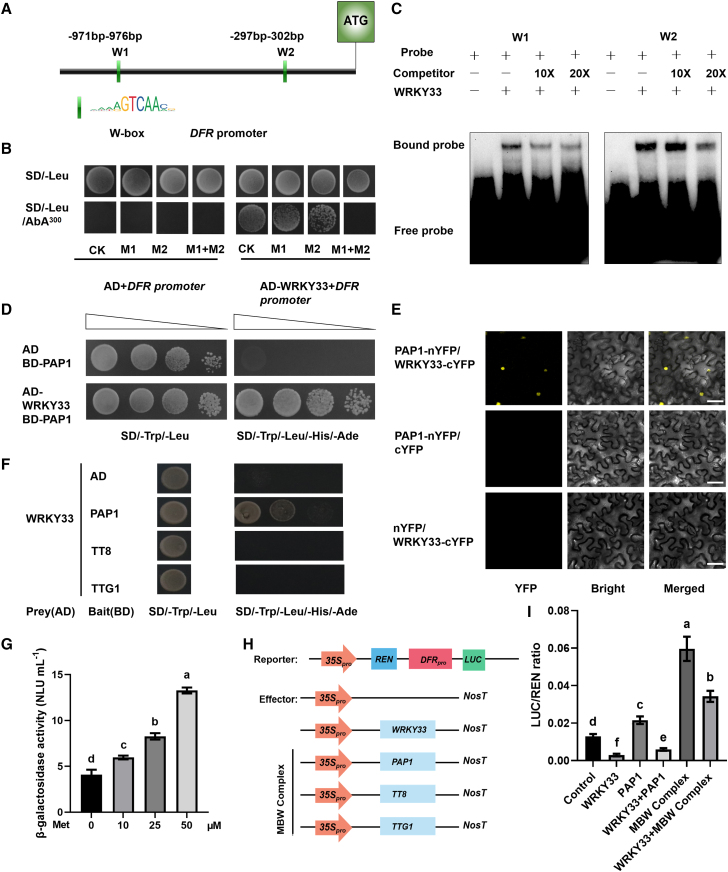
WRKY TFs specifically bind to the W box to regulate the expression of downstream genes (Eulgem et al., 2000). To better understand the role of WRKY33 in regulation of DFR expression and anthocyanin accumulation, we analyzed the DFR promoter and found that it contained two W boxes (Figure 2A). A yeast one-hybrid assay revealed that WRKY33 bound directly to the DFR promoter, but WRKY33 could not bind to DFR when W1 and W2 of the DFR promoter were mutated (Figure 2B). An electrophoretic mobility-shift assay (EMSA) showed that WRKY33 bound to both W1 and W2 of the DFR promoter (Figure 2C). These results indicated that WRKY33 binds directly to the DFR promoter.
Figure 2.
WRKY33 directly and indirectly regulates DFR expression.
(A) Schematic diagram of the DFR promoter and its W-box motifs.
(B) Yeast one-hybrid assay showing that WRKY33 directly binds to a specific region of the DFR promoter. CK, the original sequence of the DFR promoter; M1, sequence with a mutation of the W box in region 1 of the DFR promoter (−971 to −976 bp from ATG); M2, sequence with a mutation of the W box in region 2 of the DFR promoter (−297 to −302 bp from ATG). The W box was replaced with “AAAAA.”
(C) EMSA indicating the binding of WRKY33 to the DFR promoter in vitro.
(D) Yeast two-hybrid (Y2H) assay showing that PAP1 interacts with WRKY33. The empty vector pGADT7 was used as a negative control.
(E) Bimolecular fluorescence complementation (BiFC) assay indicating the interaction between PAP1 and WRKY33 in vivo. The N-terminal fragment of YFP (nYFP) was fused to PAP1 and the C-terminal fragment (cYFP) to WRKY33. Leaves from Nicotiana benthamiana were infiltrated with agrobacteria as indicated.
(F) Y2H assays to assess the interaction between WRKY33 and TTG1. Yeast cells cotransformed with WRKY33/AD and PAP1/BD, TT8/BD, or TTG1/BD were grown on SD/−Trp/−Leu and SD/−Trp/−Leu/−His/−Ade media. The empty vector pGADT7 was used as a negative control.
(G) Yeast three-hybrid (Y3H) assay to explore the influence of WRKY33 on the PAP1–TT8 interaction. The β-galactosidase activity represents PAP1–TT8 binding activities, and the promoter driving WRKY33 expression was suppressed by increasing concentrations of methionine (Met).
(H) Schematic diagram showing the reporter (DFRpro:LUC) and effector (35S:WRKY33, 35S:PAP1, 35S:TT8, and 35S:TTG1) constructs.
(I) Relative firefly LUC-to-REN ratios from transient expression assays. These represent the activity of the DFR promoter in the absence/presence of the MBW (PAP1–TT8–TTG1) complex and the combination of the MBW complex with WRKY33. Error bars indicate the SD of three biological replicates. Different letters above the bars indicate significant differences between groups (P < 0.05; ANOVA with Fisher’s LSD test).
To explore other TFs involved in the regulation of anthocyanin synthesis by WRKY33, we used WRKY33 as the bait in yeast two-hybrid (Y2H) assays and identified an interacting protein, PAP1 (Figure 2D). PAP1 is a key TF that regulates anthocyanin biosynthesis and binds directly to the DFR promoter to activate DFR expression (Sheng et al., 2005; Shin et al., 2015). A bimolecular fluorescence complementation (BiFC) assay was performed to confirm the interaction of WRKY33 and PAP1 in tobacco leaves, and coexpression of PAP1-nYFP and WRKY33-cYFP produced a fluorescent signal in the nucleus (Figure 2E). PAP1 functions as a central positive regulator among the components of the MBW complex in anthocyanin biosynthesis (Sheng et al., 2005). The best-studied bHLH factor and WD40 repeat domain protein are TRANSPARENT TESTA8 (TT8) and TRANSPARENT TESTA GLABRA1 (TTG1), respectively (Nesi et al., 2000; He et al., 2021; LaFountain and Yuan, 2021). To dissect the mechanism by which WRKY33 influences the MBW complex, we performed Y2H assays. The results showed that WRKY33 interacts with PAP1 but not with TT8 or TTG1 (Figure 2F). Next, we used PAP1 as the bait, and we subcloned WRKY33 into the pBridge vector under the control of a methionine (Met)-repressible promoter. With increasing Met concentration, WRKY33 levels were gradually reduced, causing increased growth of PAP1–TT8 yeast colonies. Yeast three-hybrid (Y3H) assays suggested that WRKY33 affects formation of the MBW complex (Figure 2G). PAP1, TT8, TTG1, and WRKY33 were used as effectors in a dual-luciferase (dual-LUC) experiment. Upon coexpression with the MBW complex (PAP1, TT8, and TTG1), the LUC activity of the DFR promoter increased by 4.6-fold compared with the control. WRKY33 inhibited the activating effect of the MBW complex on the DFR promoter by 43% (Figure 2H and 2I). These results indicated that WRKY33 might influence the MBW complex, indirectly suppressing its activation of the DFR promoter. Taken together, our findings suggest that WRKY33 regulates anthocyanin biosynthesis in both a direct and an indirect manner.
The binding affinity of WRKY33 for the DFR promoter is attenuated under Pi starvation
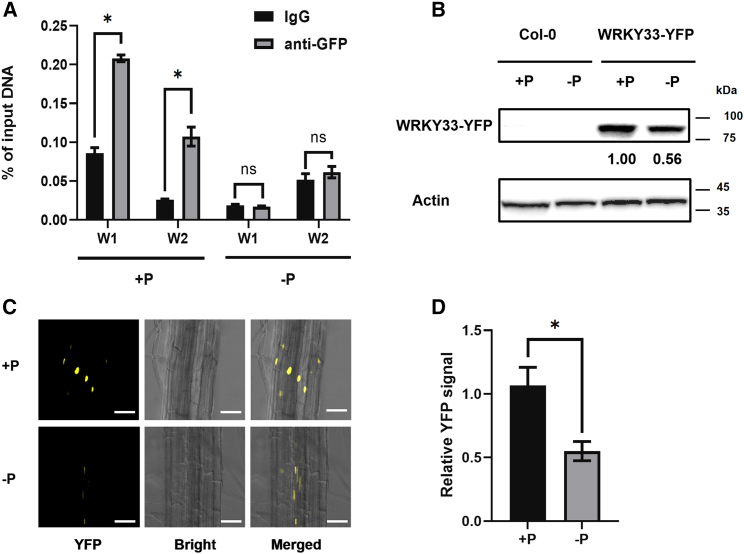
To further study the regulatory mechanism by which WRKY33 influences DFR expression and anthocyanin biosynthesis under different Pi conditions, we performed chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP–qPCR) on WRKY33-OE (with yellow fluorescent protein [YFP] tag) Arabidopsis and compared the DNA-binding ability of WRKY33 to DFR promoters between Pi-sufficient and Pi-starvation conditions. Under Pi-sufficient conditions, WRKY33 bound to the DFR promoter region containing the two W boxes. However, this binding was suppressed under Pi-starvation conditions (Figure 3A). To determine why enrichment of WRKY33 on the DFR promoter differed under Pi-sufficient and Pi-starvation conditions, we detected WRKY33 protein levels in Col-0 and WRKY33-YFP plants grown on +Pi and −Pi media. Western blot results showed that WRKY33-YFP protein levels were reduced under −Pi conditions (Figure 3B). We also used WRKY33-YFP lines for imaging analysis and observed the WRKY33 signal after plants had been transferred from adequate-Pi medium to Pi-deprived conditions for 72 h. Confocal microscopy showed that plants grown in the Pi-deficient medium exhibited weaker fluorescent signals (Figure 3C and 3D). These results suggested that WRKY33 protein levels and enrichment of WRKY33 on the DFR promoter were both reduced under −Pi conditions.
Figure 3.
Enrichment of WRKY33 on the DFR promoter is attenuated under Pi starvation.
(A) Binding of WRKY33 to DFR promoter regions. The chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP–qPCR) assay was performed on 9-day-old WRKY33-OE seedlings grown on ½ MS +P (1.25 mM Pi) and −P (0 mM Pi) media. Gene expression was quantified by quantitative reverse transcription PCR and calculated as a percentage of total input DNA. Fold enrichment represents the binding efficiency ratio of anti-GFP antibody/negative IgG antibody.
(B) Western blot of YFP-tagged WRKY33 in 9-day-old Col-0 and WRKY33-OE seedlings grown on +Pi and −Pi media.
(C and D) Images and quantitative analysis of WRKY33 signal in 9-day-old WRKY33OE plants grown on +Pi and −Pi media. Error bars indicate the SD of three biological replicates. Asterisks above the bars indicate significant differences between groups (P < 0.05; Student’s t-test).
PHR1 is involved in Pi-dependent regulation of DFR by WRKY33
Because regulation of DFR expression by WRKY33 was completely different under Pi-sufficient and Pi-deficient conditions, we hypothesized that WRKY33 might be related to the Pi pathway in Arabidopsis. WRKY33 has been reported to mediate the Pi-deficiency-induced modification of root architecture (Shen et al., 2021). Our results were consistent with the phenotypes of WRKY33-related mutants grown on +Pi and −Pi media (Supplemental Figure 3A and 3B). Moreover, expression levels of PSI genes, including IPS1, PHT1;4, and PHT2;1, were similar to that of DFR, indicating that WRKY33 is closely related to the Pi signaling pathway (Supplemental Figure 3C and 3D).
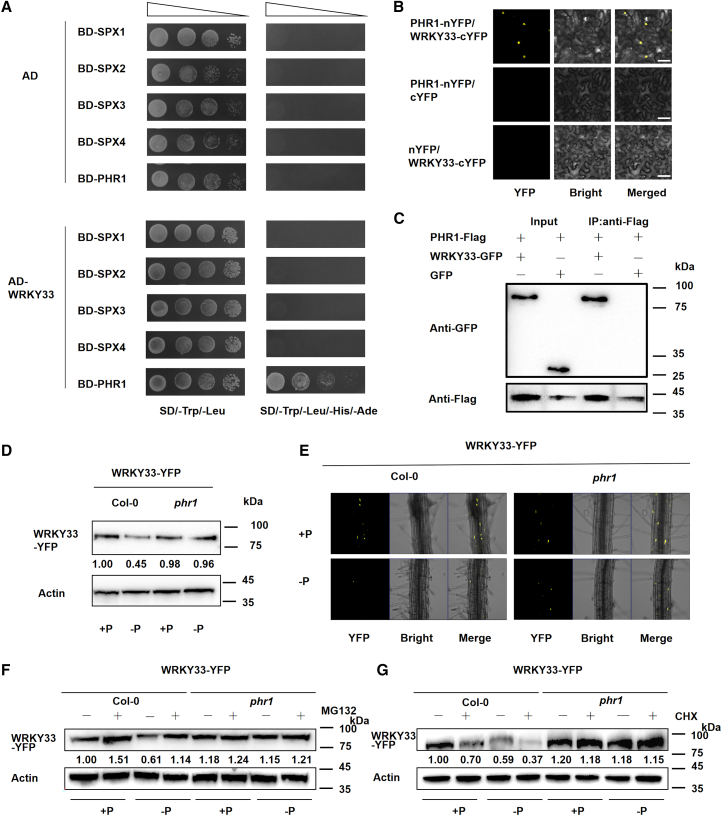
PHR1 and SPXs are vital regulators in the Pi signaling pathway (Ried et al., 2021), and Y2H assays revealed that WRKY33 interacted with PHR1 but not with SPX1–4 (Figure 4A). BiFC and coimmunoprecipitation assays confirmed the interaction between WRKY33 and PHR1 in vitro and in vivo (Figure 4B and 4C). Our previous research showed that DFR is prone to indirect regulation by PHR1, owing to the absence of a P1BS motif in its promoter region (He et al., 2021). To explore whether the interaction of WRKY33 and PHR1 would influence their binding to the DFR promoter, we coexpressed WRKY33 and PHR1 proteins with DFRPro-LUC in tobacco leaves. The increase in DFRPro-LUC expression revealed that PHR1 interacts with WRKY33 to regulate expression of DFR (Supplemental Figure 4). Thus, we propose a novel mechanism by which PHR1 regulates DFR by interacting with WRKY33.
Figure 4.
PHR1 is involved in Pi-dependent regulation of DFR by WRKY33.
(A) Y2H assays to assess interactions of WRKY33 with PHR1 and SPX1–4. Yeast cells cotransformed with WRKY33/AD and SPX1–4/BD or PHR1/BD were grown on SD/−Trp/−Leu and SD/−Trp/−Leu/−His/−Ade media. The empty vector pGADT7 was used as a negative control.
(B) BiFC assay indicating the interaction between PHR1 and WRKY33. The N-terminal fragment of YFP (nYFP) was fused to PHR1, and the C-terminal fragment (cYFP) was fused to WRKY33. Leaves from N. benthamiana were infiltrated with agrobacteria as indicated. The experiments had four biological replicates.
(C) Coimmunoprecipitation showed that PHR1 interacted with WRKY33 in N. benthamiana: WRKY33-GFP was coimmunoprecipitated with PHR1-FLAG.
(D) Western blot of YFP-tagged WRKY33 in WRKY33-OE and phr1 WRKY33OE plants grown under +Pi and −Pi conditions.
(E) YFP-tagged WRKY33 signal in WRKY33-OE and phr1 WRKY33OE plants after transfer from adequate Pi medium to Pi-deprived conditions for 72 h.
(F) Effects of MG132, a chemical inhibitor of the 26S proteasome, on the stability of WRKY33. WRKY33 protein levels were examined in WRKY33OE and phr1 WRKY33OE plants supplied with or without 50 μM MG132 for 24 h.
(G) Nine-day-old WRKY33OE and phr1 WRKY33OE plants were treated with 100 mM CHX for 4 h. Nuclear proteins were isolated and analyzed by immunoblotting.
To further examine the relationship between WRKY33 and PHR1 and explain the downregulation of WRKY33 protein levels under Pi-deficient conditions, we generated the homozygous WRKY33OE phr1 mutant in which WRKY33 was overexpressed in the phr1 mutant background (Supplemental Figure 5). WRKY33 protein levels scarcely decreased in phr1 mutants under −Pi conditions compared with +Pi conditions, but they decreased significantly in the Col-0 background (Figure 4B and 4E). To determine whether mutation of PHR1 promotes WRKY33 abundance, we examined the effect of MG132 on the stability of WRKY33. Western blot analysis showed that WRKY33 abundance increased upon MG132 treatment under +Pi conditions (Figure 4F). To further test the stability of WRKY33, we used cycloheximide (CHX) to block biosynthesis of new proteins. WRKY33 showed a decreased turnover rate in the phr1 mutant compared with Col-0 plants (Figure 4G). Anthocyanin accumulation was also observed in Col-0, phr1, WRKY33OE, and phr1 WRKY33OE plants under +Pi or −Pi conditions . The results showed that anthocyanin accumulation declined in both phr1 and WRKY33OE plants. The phenotype of phr1 WRKY33OE was similar to that of phr1 (Supplemental Figure 6). These results suggested that mutation of PHR1 increases WRKY33 protein stability and that PHR1 is involved in the regulation of factors that affect WRKY33 protein turnover.
PHR1 attenuates the inhibition of anthocyanin synthesis by WRKY33
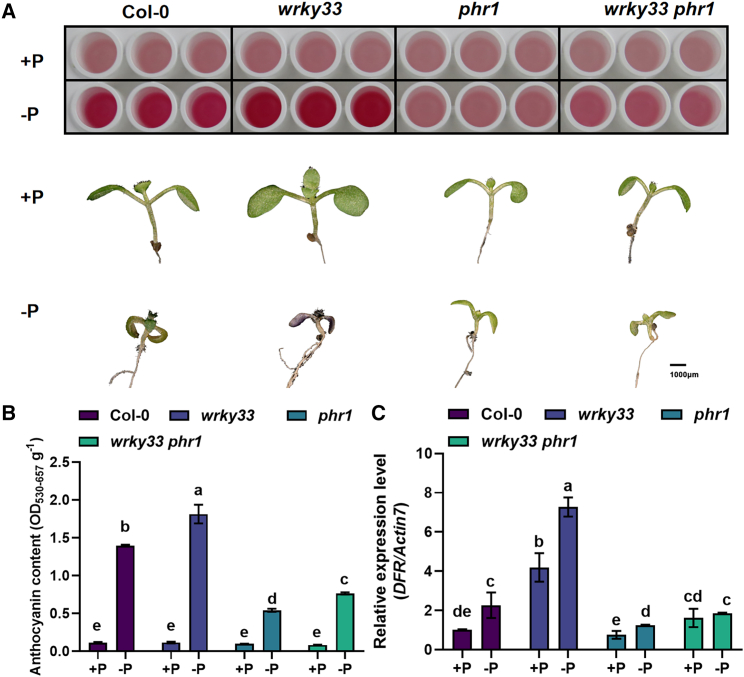
Our results showed that WRKY33 physically associates with PHR1 and that they function in the modulation of anthocyanidin biosynthesis. To understand whether WRKY33 and PHR1 genetically interact with each other in the mediation of anthocyanidin biosynthesis, we generated the wrky33 phr1 double mutant. The anthocyanidin content of wrky33 phr1 was higher than that of phr1 but lower than that of wrky33 (Figure 5A), and DFR expression levels were consistent with these phenotypes (Figure 5B and 5C).
Figure 5.
Genetic interaction between WRKY33 and PHR1.
(A) Anthocyanin accumulation and phenotypes of 9-day-old Col-0, wrky33, phr1, and wrky33 phr1 plants under +Pi and −Pi conditions.
(B and C) Anthocyanin contents (B) and relative expression levels of the anthocyanin biosynthesis gene DFR(C) in Col-0, wrky33, phr1, and wrky33 phr1 plants under +Pi and −Pi conditions. Different letters indicate statistically significant differences (ANOVA, Fisher’s LSD test; P < 0.05). All experiments had four biological replicates.
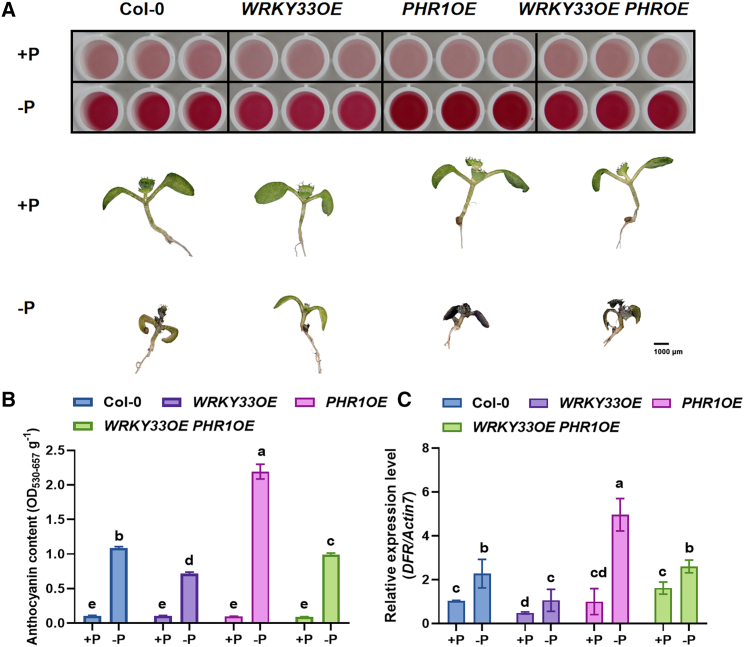
We also hybridized WRKY33OE with PHR1OE to generate WRKY33OE PHR1OE plants and analyzed the regulation of PHR1 by WRKY33. Anthocyanidin accumulation and DFR expression were increased in WRKY33OE PHR1OE plants compared with WRKY33OE plants under Pi-deficient conditions, indicating that PHR1 may attenuate the inhibition of anthocyanin synthesis by WRKY33 (Figure 6A–6C).
Figure 6.
PHR1 attenuates the inhibition of anthocyanin production by WRKY33.
(A) Anthocyanin accumulation and phenotypes of Col-0, WRKY33OE, PHR1OE, and WRKY33OE PHR1OE plants under +Pi and −Pi conditions.
(B and C) Anthocyanin content (B) and relative expression levels of the anthocyanin biosynthesis gene DFR(C) in Col-0, WRKY33OE, PHR1OE, and WRKY33OE PHR1OE plants under +Pi or −Pi conditions. Different letters indicate statistically significant differences (ANOVA, Fisher’s LSD test; P < 0.05). All experiments had four biological replicates.
Discussion
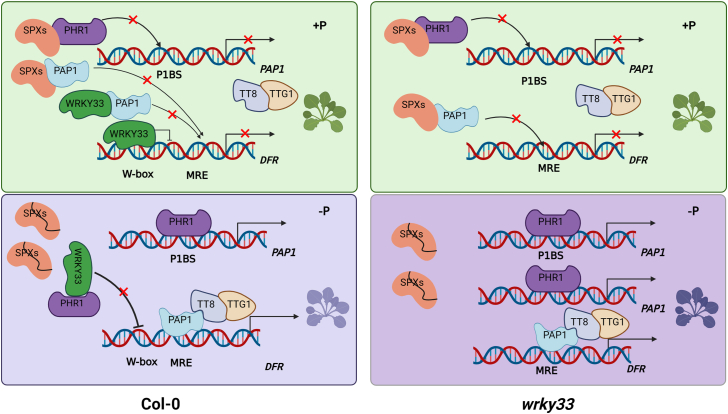
Anthocyanins are widely produced in plants, and their biosynthesis is sensitive to a variety of biotic and abiotic stressors (Xie et al., 2016; Li et al., 2020; Naing and Kim, 2021). It has long been held that plants produce excess anthocyanins under Pi-starvation conditions (Jiang et al., 2007; Lei et al., 2011). In this study, we provided evidence that WRKY33 inhibits anthocyanidin biosynthesis by directly binding to the promoter of DFR or by interacting with PAP1 under +Pi conditions. However, under −Pi conditions, the interaction of PHR1 and SPXs is weakened, releasing PHR1 to exert its function. The released PHR1 interacts with WRKY33 and represses binding of WRKY33 to the DFR promoter. Meanwhile, WRKY33 protein levels are downregulated, further weakening its repression of DFR. Ultimately, the inhibition of anthocyanin biosynthesis by WRKY33 is repressed, leading to anthocyanin accumulation under Pi-deficient conditions (Figure 7).
Figure 7.
A model for WRKY33-regulated phosphorus-status-dependent anthocyanin biosynthesis in Arabidopsis.
In Col-0, WRKY33 prevents the production of anthocyanidins by directly binding to the promoter of DFR or by interfering with the MBW complex through interacting with PAP1 under +Pi conditions. Under −Pi conditions, the interaction between PHR1 and SPXs is attenuated, enabling PHR1 to perform its role. PHR1 interacts with WRKY33 to prevent the binding of WRKY33 to the DFR promoter. WRKY33 protein abundance is also indirectly suppressed by PHR1, further impairing its ability to interact with PAP1. Sequestration by PHR1 may reduce access of WRKY33 to PAP1 and to the DFR promoter. The abundance of WRKY33 relative to PHR1 protein would shift that balance. In the end, WRKY33 inhibition of anthocyanin biosynthesis is further compromised, enabling anthocyanin accumulation, as well as more indirect effects on PSI gene expression and root system architecture under Pi-deficient conditions. The absence of WRKY33 results in a more pronounced phosphate-starvation response in the wrky33 mutant under −Pi conditions.
WRKY33 is a negative regulator of anthocyanin biosynthesis
Much progress has been made on the role of WRKY33 as the master regulator of gene expression dynamics and metabolic fluxes in Trp-derived secondary metabolism, including synthesis of IGS, camalexin, and 4OH-ICN, and in response to pathogens (Mao et al., 2012; Rajniak et al., 2015; Tao et al., 2022b). For example, WRKY33 mediates the production of 4-methoxyindole-3-ylmethyl glucosinolate, a type of IGS, conferring resistance to Alternaria brassicicola in Arabidopsis and Brassica crops (Tao et al., 2022b). In addition, WRKY33 positively regulates camalexin biosynthesis to improve plant resistance to Botrytis cinerea (Zheng et al., 2006; Birkenbihl et al., 2012). WRKY33 has also been reported to act as a condition-dependent master regulator in the 4OH–ICN pathway (Barco and Clay, 2020). These studies have focused on WRKY33-regulated plant secondary metabolism and its effects on plant resistance to pathogens, and most studies have shown that WRKY33 is a core positive regulator of responses to diverse stresses.
Massive metabolic changes occur when plants are subjected to phosphorus limitation, including changes in the synthesis of phenylpropanoids, flavonoids, and anthocyanins (Pant et al., 2015). The functions of flavonoids under biotic and abiotic stress have been widely reported (Lan et al., 2017; Bhatia et al., 2018; Sudheeran et al., 2020; Ferreyra et al., 2021). We wondered whether WRKY33 participated in the regulation of flavonoids. KEGG enrichment results suggested changes in the contents of flavonoids and anthocyanins in WRKY33-related mutants. Subsequent molecular biology experiments indicated that WRKY33 not only influenced flavonoids but also served as a negative regulator of anthocyanin biosynthesis under Pi-deficient conditions (Figure 1 and Supplemental Figure 1).
Anthocyanins have a wide range of functions in both vegetative and reproductive organs; plants have therefore evolved mechanisms to control when, where, and how much anthocyanin is produced in response to a variety of developmental, environmental, and hormonal stimuli (LaFountain and Yuan, 2021). There is a “double-negative logic” as to why plants need WRKY33 to repress anthocyanin biosynthesis. Pi deficiency, as an input signal, represses a repressor of the anthocyanin-activating complex, leading to activation of anthocyanin biosynthesis as the output. Such a negative feedback loop can avoid anthocyanin overshoot and is especially useful for maintaining anthocyanin concentration homeostasis in specific tissue types. Moreover, anthocyanins protect plants against UV radiation and scavenge free radicals (Ferreyra et al., 2021). Biosynthesis of plant secondary metabolites under normal growth conditions has ecological costs and can adversely affect plant growth (Wang et al., 2022). Therefore, WRKY33 protein level is higher under +Pi conditions compared with −Pi conditions, leading to less anthocyanin accumulation under +Pi conditions (Figures 3 and 4). The elaborate regulation of metabolic systems is required to maximize effectiveness while limiting the drawbacks of defense chemicals to achieve an optimal balance of growth and defense. The varied protein levels of WRKY33 under different Pi conditions suggest that plants can allocate resources economically and efficiently to respond to ever-changing environments.
Multiple roles of WRKY33 in adaption to biotic and abiotic stress
Plant defense responses are regulated by WRKY33, which is also implicated in responses to a variety of biotic and abiotic stresses (Jiang and Deyholos, 2009; Liu et al., 2018; Zheng et al., 2006). WRKY33 positively regulates plant resistance to biotic stress (Birkenbihl et al., 2012; Liao et al., 2016). To improve plant resistance to B. cinerea, MPK3/MPK6 and CPK5/CPK6 work together to regulate camalexin production via differential phosphoregulation of WRKY33 activity (Yang et al., 2020; Zhou et al., 2020). The ethylene and jasmonate pathways collaborate at several levels with the MPK3/MPK6–WRKY33 module to stimulate camalexin production in Arabidopsis upon pathogen infection (Zhou et al., 2022). Moreover, WRKY33 also acts as a positive regulator in plant resistance to abiotic stress. The interaction of WRKY33 and WRKY12 activates RAP2.2 expression during submergence to induce a hypoxia response in Arabidopsis (Tang et al., 2021), and the on-and-off module SR1–WRKY33–RAP2.2 regulates acclimation to submergence (Tang et al., 2021).
Pi is a macronutrient required by all living organisms (Kong et al., 2021). Here, we explored the role of WRKY33 in the plant response to Pi starvation. Disruption of WRKY33 has been reported to inhibit primary root growth and promote formation of root hairs under Pi-deficient conditions (Shen et al., 2021). We also demonstrated an association between WRKY33 and plant responses to Pi-deficient conditions, as WRKY33 affected PSI gene expression (IPS1, PHT1;4, and PHT2;1) and interacted with PHR1 to regulate Pi-deficiency-induced anthocyanin biosynthesis (Supplemental Figures 3 and 4). Unlike IPS1 and PHT2;1, PHT1;4 contains a W box in its promoter, which may explain its stronger response to +Pi conditions. Our results indicate that WRKY33 serves as a negative regulator of the response to Pi deficiency in A. thaliana. The multiple roles, especially the negative role, of WRKY33 in regulating secondary metabolite biosynthesis are a novel finding that expands our understanding of the mechanisms by which WRKY33 functions in abiotic stress. Excessive agricultural use of Pi fertilizer has resulted in accumulation of soil Pi, which has led to eutrophication (Pinckney et al., 2001; Jia et al., 2021). Because overexpression of WRKY33 could alleviate the Pi-deficiency response of plants with no significant abnormalities in physiological phenotype, WRKY33 shows significant potential for use in ameliorating negative environmental effects by reducing Pi fertilizer application.
Mechanisms by which WRKY33 regulates anthocyanin biosynthesis under +Pi or −Pi conditions
Anthocyanin accumulation is a phenotypic indicator of Pi-deficient conditions (Pinckney et al., 2001; Jia et al., 2021), and PHR1 is the master TF that governs the Pi-starvation response (Tang et al., 2022; Park et al., 2023). Our previous research showed that PHR1 indirectly regulates DFR, a key anthocyanin biosynthetic gene, because the DFR promoter lacks a P1BS motif (He et al., 2021). However, we found two W boxes in the DFR promoter, and molecular experiments verified that WRKY33 regulates DFR expression directly (Figure 2). In addition, WRKY33 also interacts with PAP1. The MBW complex has been reported to regulate anthocyanin biosynthesis at the transcriptional level (Gonzalez et al., 2008; Wang et al., 2020), and our previous research showed that SPX4 represses the function of PAP1 by interfering with the integrity of MBW transcription (He et al., 2021). Our Y2H assays showed that WRKY33 interacts with PAP1, not TT8 or TTG1, and Y3H assays showed that WRKY33 affects formation of the MBW complex. Combined with the dual-LUC assays, these results show that WRKY33 negatively regulates the expression of DFR by interfering with MBW formation (Figure 2). The transcript and protein levels of PHR1 are almost unchanged during induction of phosphorus starvation in A. thaliana (Osorio et al., 2019; Shi et al., 2021). However, SPXs interact with PHR1 to alter its binding to downstream PSI genes when inositol pyrophosphate levels change (Ried et al., 2021). Previous studies have shown that SPXs interact with PHR1 and PAP1 to influence anthocyanin synthesis during plant adaptation to Pi starvation (Duan et al., 2008; Bustos et al., 2010; Puga et al., 2014; He et al., 2021). However, absence of the P1BS motif in the promoters of anthocyanin biosynthesis genes suggests that PHR1 probably regulates these genes in an indirect manner. Our results showed that WRKY33 does not interact with SPX1–4 but does interact with PHR1 (Figure 4A). Moreover, PHR1 interacts with WRKY33 to regulate the expression of DFR (Figure 4 and Supplemental Figure 4). Genetic experiments also suggested that PHR1 attenuates the inhibition of anthocyanin synthesis by WRKY33 (Figure 6). Thus, we found a novel connection between PHR1 and anthocyanin biosynthesis.
Previous research reported that the mRNA level of WRKY33 decreased significantly under −Pi conditions (Shen et al., 2021). Our results also showed that the transcript and protein abundance of WRKY33 decreased under −Pi conditions, but this change was reversed in the phr1 mutant background. We therefore hypothesize that PHR1 regulates other transcriptional repressors or components of the ubiquitin–proteasome system, such as E3 ligases, and that mutation of PHR1 thus increases WRKY33 stability (Figure 4D–4G). In addition, sequestration by PHR1 may reduce the access of WRKY33 to PAP1 and the DFR promoter, which may be a dose-dependent response. SUBMERGENCE RESISTANT1 (SR1) has been reported to regulate the stability of WRKY33 to modulate the submergence response (Liu et al., 2021). Some ubiquitin E3 ligases involved in regulating WRKY33 degradation may be induced during Pi limitation, and this possibility needs further study.
In conclusion, WRKY33 binds directly to the DFR promoter and also interacts with PAP1 to indirectly regulate DFR expression, thereby suppressing anthocyanidin production under +Pi conditions. Under −Pi conditions, the interaction of PHR1 and SPXs is diminished, allowing free PHR1 to interact with WRKY33 and inhibit its binding to the DFR promoter, resulting in the accumulation of anthocyanin (Figure 7). Understanding of the mechanisms by which WRKY33 and PHR1 regulate anthocyanin biosynthesis under +Pi or −Pi conditions is critical for understanding plant adaptations to ever-changing environments.
Methods
Growth conditions and plant materials
Seeds were sterilized in 10% bleach for 12 min, washed 3–5 times with sterile water, stratified for 3 days at 4°C, and planted on ½ MS (Murashige and Skoog) growth medium. MS basal salts (MSP01-50LT; Caisson Labs, Smithfield, UT, USA) and MS without Pi (MSP11-50LT; Caisson Labs) were used to prepare the standard medium and Pi-deficient medium, respectively. Nine-day-old plants were harvested for root length measurement, anthocyanin determination, and gene expression analyses.
All A. thaliana plants used in this investigation were in the Col-0 background. Seeds of wrky33-2 (GABI_324B11), WRKY33-OE, phr1, and PAP1OE were used (He et al., 2021; Tao et al., 2022b). Genetic crosses yielded WRKY33OE PHR1OE, wrky33 phr1, and phr1 WRKY33OE, and homozygous lines were used for experimentation.
RNA-sequencing assay
Total RNA was extracted from 9-day-old plants (Col-0 and wrky33). Shanghai Personal Biotechnology sequenced the sequencing library on the Illumina HiSeq X platform. TAIR10 was used to map all reads to the Arabidopsis genome.
RNA extraction and RT–qPCR
Following the manufacturer’s instructions, total RNA was extracted from plant materials that had been homogenized in liquid nitrogen and mixed with TRIzol reagent (Invitrogen). The RNA samples were reverse transcribed into cDNA using Prime Script RT Master Mix (Takara). Arabidopsis ACTIN7 served as an internal control, and the expression levels of other genes were calculated using the 2−ΔΔCt method. The primers used in this study are listed in Supplemental Table 1.
In situ diphenylboric acid 2-aminoethyl ester staining
A method adapted from He was used for in situ flavonol visualization. Seedlings were bleached with ethanol overnight at room temperature, then stained for at least 10 min with a freshly produced aqueous solution of 0.25% (w/v) diphenylboric acid 2-aminoethyl ester and 0.00375% (v/v) Triton X-100. The roots were examined using a Leica TCS SP5 confocal laser-scanning microscope with an excitation wavelength of 475–504 nm for kaempferol and 577–619 nm for quercetin (He et al., 2021).
Anthocyanin determination
Anthocyanin quantification was performed as described by He et al. (2021) with a subtle change. Seedling samples (0.05 g) were incubated overnight in 500 μl of 0.1% HCl methanol solution, followed by the addition of 375 μl distilled water and 1000 μl chloroform for separation of anthocyanins from chlorophyll by vortexing and quick-spin procedures. Total anthocyanins in the aqueous phase were determined by measuring the absorbance at A530 and A657 using a spectrophotometer. Anthocyanin content was calculated by subtracting A657 from A530.
Yeast one-hybrid assays
Yeast one-hybrid assays followed the protocol of the Clontech yeast one-hybrid system. The promoter fragments of DFR were cloned into the pAbAi vector, and the coding sequence of WRKY33 was amplified and cloned into the pGADT7 vector to generate a prey construct. The empty vector (AD) served as the negative control. We used SD medium without Leu supplemented with 100 ng ml−1 AbA, and transformed yeast cells were dotted at 1, 10−1, and 100−1 dilutions on the SD/−Leu/AbA100 medium.
Dual-luciferase assay
Transient expression and dual-LUC assays were carried out as described previously (He et al., 2021). The promoter regions of DFR were subcloned into the pGreenII 0800-LUC vector as reporters. The cDNA sequences of WRKY33, PHR1, PAP1, TT8, and TTG1 driven by the 35S promoter served as effectors. The promoter region of DFR was cloned into the pGreenII 0800-LUC vector. The 35S::REN gene (Renilla luciferase [REN]) in the vector was used as an internal control. The ratio of firefly LUC to REN was measured using a dual-LUC reporter assay system.
Transient expression assays in Nicotiana benthamiana leaves
Transient expression assays were performed in N. benthamiana leaves as described previously with minor modifications (Tao et al., 2022b). The promoters of DFR were amplified and cloned into the pGreenII 0800-LUC vector to generate the reporter construct DFRpro:LUC. The coding sequence of WRKY33 was PCR amplified and cloned into pGreenII 0029-62-SK to generate the effector construct. We used a low-light cooled CCD imaging apparatus (NightOWL II LB983) to obtain the LUC image and measure the luminescence intensity. Each tobacco leaf was sprayed with 100 mM luciferin (Promega) and placed in darkness for 5 min before luminescence detection. The Dual-Luciferase Reporter Assay kit (Promega) was used to quantify the activities of LUC and REN.
Electrophoretic mobility-shift assay
Recombinant pET32a-WRKY33 was purified using His beads (70501; Beaver) according to the manufacturer’s instructions. After incubation in binding buffer at 4°C for 1 h, the recombinant His-WRKY33 proteins were washed three times and incubated with His beads (binding buffer with 50 mM imidazole; I5513; Sigma) elution buffer for another 2 h at 4°C. EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s protocol.
Chromatin immunoprecipitation–quantitative polymerase chain reaction
ChIP experiments were performed as described previously with some modifications. Two grams of 10-day-old 35S:AtWRKY33 seedlings were used for the ChIP assay. YFP antibody (Abcam) was used to immunoprecipitate protein–DNA complexes. Chromatin precipitated without antibody was used as the negative control, and the chromatin isolated before precipitation was used as the input control. Enrichment was calculated as a percentage of the input control. Primers used for ChIP–qPCR are listed in Supplemental Table 1.
Confocal microscopy
Fluorescence signals were detected using a Leica TCS SP5 confocal laser-scanning microscope. YFP was viewed at an excitation wavelength of 488 nm, and emission was collected at 518 nm.
Nuclear protein extraction and immunoblotting
Nine-day-old Arabidopsis plants grown on +Pi and −Pi media were ground and incubated in nuclear extraction buffer containing 50 mM Tris, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 100 mM MG132, and protease inhibitor cocktail. Lysates were incubated for 2 h with anti-GFP magnetic beads (Sigma-Aldrich) at 4°C. The supernatant was discarded, and the precipitate (which contained nuclear proteins) was retained for further experimentation.
WRKY33 protein levels were examined in 9-day-old WRKY33-OE and phr1 WRKY33OE plants supplied with 50 μM MG132 for 24 h or treated with 100 mM CHX for 4 h. Nuclear proteins were isolated and analyzed by immunoblotting according to the manufacturer’s protocol. Proteins were heated in sodium dodecyl sulfate (SDS) sample buffer for 5 min at 100°C. The samples were centrifuged at 12 000 g for 1 min and then analyzed on 10% SDS–PAGE gels. Anti-GFP antibody (Abcam) was used for western blotting, and bands were quantified using ImageJ.
Yeast two-hybrid assays
The Y2H assay was performed using the Matchmaker GAL4 Two-Hybrid System 3 following the manufacturer’s protocol (Clontech, Palo Alto, CA, USA). The pGBKT7 and pGADT7 constructs were cotransformed into yeast strain AH109 and selected on SD/−Leu/−Trp plates. Colonies were transferred to selective dropout SD medium (SD/−Ade/−Leu/−Trp/−His) supplemented with 20 mM 3-amino-1,2,4-triazole.
Yeast three-hybrid assays
WRKY33/AD and PAP1-TT8/pBridge were generated for use in Y3H assays. The pBridge vector in the Y3H system uses an inducible Met25 promoter. WRKY33 was subcloned into the pBridge vector and driven by the Met25 promoter. The constructs were cotransformed into AH109 yeast cells and selected on SD/−Leu/−Trp plates. Colonies were then moved to selective dropout liquid medium (SD/−Met/−Leu/−Trp/−His) with different concentrations of Met, and the β-galactosidase activity associated with PAP1–TT8 binding activities was quantified using the O-nitrophenyl-β-D-galactopyranoside liquid assay according to the manufacturer’s protocol (Clontech).
Bimolecular fluorescence complementation assay
The full-length cDNA sequences of WRKY33, PHR1, and PAP1 were cloned into the cYFP and nYFP vectors to generate the nYFP-PAP1, nYFP-PHR1, and cYFP-WRKY33 constructs. All the constructs were transformed into Agrobacterium tumefaciens strain GV3101 and then transiently coexpressed in N. benthamiana leaves. YFP fluorescence of tobacco leaves was imaged 48 h after infiltration using a Leica TCS SP5 confocal laser-scanning microscope. The excitation wavelength for YFP fluorescence was 488 nm, and fluorescence was detected at 500–542 nm.
Coimmunoprecipitation assay
N. benthamiana leaves were harvested and lysed with a buffer containing 50 mM Tris, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 100 mM MG132 (Sigma-Aldrich), and protease inhibitor cocktail (Roche). Lysates were incubated for 4 h with anti-FLAG M2 magnetic beads (Sigma-Aldrich) at 4°C. The beads were then washed three times and solubilized in 15 ml SDS sample buffer. Samples were analyzed by 10% SDS–PAGE. Proteins were transferred to a polyvinylidene fluoride membrane (Amersham), and blots were blocked for 1 h and incubated with an anti-GFP antibody (Abcam) for 1 h at room temperature. After incubation, blots were extensively washed and incubated with secondary antibody for 45 min at room temperature. Sensitive detection of the bound antibody was performed using an ECL Plus western blotting kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol.
Accession numbers
The following accession numbers apply to this work: WRKY33 (AT2G38470), CHS (AT5G13930), CHI (AT3G55120), DFR (AT5G42800), PAP1 (AT1G56650), TT8 (AT4G09820), TTG1 (AT5G24520), SPX1 (AT5G20150), SPX2 (AT2G26660), SPX3 (AT2G45130), SPX4 (AT5G15330), PHR1 (AT4G28610), IPS1 (AT3G09922), PHT1;4 (AT2G38940), and PHT2;1 (AT2G38940).
Data and code availability
The data that support the findings of this study are available from the corresponding author upon request.
Funding
This research was funded by the Zhejiang Provincial Natural Science Foundation of China under grant no. LR22C020003, the National Key Research and Development Program of China (2022YFD2200603), the National Natural Science Foundation of China under grant nos. 32000234 and 32172593, and the China Postdoctoral Science Foundation (2022M712831). We also are grateful for funding from the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products.
Author contributions
G.H. and H.T. conceived the project and designed the experiments; H.T. prepared the original draft and conducted the experiments; F.G. and H.T. revised the manuscript; L.L., Y.H., X.Z., M.W., J.W., Y.Z., and C.Z. interpreted and discussed the data; funding acquisition and writing, review, and editing were done by G.H., Q.W., and H.T. All authors commented on the manuscript before submission.
Acknowledgments
No conflict of interest is declared.
Published: January 16, 2024
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Qiaomei Wang, Email: qmwang@zju.edu.cn.
Gaojie Hong, Email: gjhong@126.com.
Supplemental information
References
- Barco B., Clay N.K. Hierarchical and dynamic regulation of defense-responsive specialized metabolism by WRKY and MYB transcription factors. Front. Plant Sci. 2020;10 doi: 10.3389/fpls.2019.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A., Mansfield S.D., Hall H.C., Douglas C.J., Ellis B.E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 2010;154:1428–1438. doi: 10.1104/pp.110.162735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia C., Pandey A., Gaddam S.R., Hoecker U., Trivedi P.K. Low temperature-enhanced flavonol synthesis requires Light-Associated regulatory components in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:2099–2112. doi: 10.1093/pcp/pcy132. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R.P., Diezel C., Somssich I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012;159:266–285. doi: 10.1104/pp.111.192641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis I.E., Winkel-Shirley B. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Tong S., Yang J., Qin J., Wang W., Chen K., Shi W., Li J., Liu J., Jiang Y. PtoWRKY40 interacts with PtoPHR1-LIKE3 while regulating the phosphate starvation response in poplar. Plant Physiol. 2022;190:2688–2705. doi: 10.1093/plphys/kiac404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.F., Li L.Q., Xu Q., Kong Y.H., Wang H., Wu W.H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low pi stress in Arabidopsis. Plant Cell. 2009;21:3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Lin S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011;62:185–206. doi: 10.1146/annurev-arplant-042110-103849. [DOI] [PubMed] [Google Scholar]
- de Meaux J., Pop A., Mitchell-Olds T. Cis-regulatory evolution of chalcone-synthase expression in the genus Arabidopsis. Genetics. 2006;174:2181–2202. doi: 10.1534/genetics.106.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B.N., Karthikeyan A.S., Raghothama K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Yi K., Dang L., Huang H., Wu W., Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008;54:965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Ferreyra M.L.F., Serra P., Casati P. Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol. Plant. 2021;173:736–749. doi: 10.1111/ppl.13543. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- He Y., Zhang X., Li L., Sun Z., Li J., Chen X., Hong G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2021;230:205–217. doi: 10.1111/nph.17139. [DOI] [PubMed] [Google Scholar]
- Jia X., Wang L., Zeng H., Yi K. Insights of intracellular/intercellular phosphate transport and signaling in unicellular green algae and multicellular land plants. New Phytol. 2021;232:1566–1571. doi: 10.1111/nph.17716. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Deyholos M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145:1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Ma S., Ye N., Jiang M., Cao J., Zhang J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017;59:86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- Khan G.A., Vogiatzaki E., Glauser G., Poirier Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016;171:632–644. doi: 10.1104/pp.16.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Wang G., Chen X., Li L., Zhang X., Chen S., He Y., Hong G. OsPHR2 modulates phosphate starvation-induced OsMYC2 signalling and resistance to Xanthomonas oryzae pv. Plant Cell Environ. 2021;44:3432–3444. doi: 10.1111/pce.14078. [DOI] [PubMed] [Google Scholar]
- Kubasek W.L., Shirley B.W., McKillop A., Goodman H.M., Briggs W., Ausubel F.M. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell. 1992;4:1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.F., Chiou T.J. The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 2011;156:1016–1024. doi: 10.1104/pp.111.175265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain A.M., Yuan Y.W. Repressors of anthocyanin biosynthesis. New Phytol. 2021;231:933–949. doi: 10.1111/nph.17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X., Yang J., Abhinandan K., Nie Y., Li X., Li Y., Samuel M.A. Flavonoids and ROS Play Opposing Roles in Mediating Pollination in Ornamental Kale (Brassica oleracea var. acephala) Mol. Plant. 2017;10:1361–1364. doi: 10.1016/j.molp.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Lei M., Zhu C., Liu Y., Karthikeyan A.S., Bressan R.A., Raghothama K.G., Liu D. Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol. 2011;189:1084–1095. doi: 10.1111/j.1469-8137.2010.03555.x. [DOI] [PubMed] [Google Scholar]
- Li D.D., Ni R., Wang P.P., Zhang X.S., Wang P.Y., Zhu T.T., Sun C.J., Liu C.J., Lou H.X., Cheng A.X. Molecular basis for chemical evolution of flavones to flavonols and anthocyanins in land plants. Plant Physiol. 2020;184:1731–1743. doi: 10.1104/pp.20.01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.Q., Huang L.P., Pan G., Liu L., Wang X.Y., Lu L.M. Identifying the genes regulated by AtWRKY6 using comparative transcript and proteomic analysis under phosphorus deficiency. Int. J. Mol. Sci. 2017;18:1046. doi: 10.3390/ijms18051046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li Y., Yao X., Wen Y., Zhou Z., Lei W., Zhang D., Lin H. Nitrogen-inducible GLK1 modulates phosphate starvation response via the PHR1-dependent pathway. New Phytol. 2022;236:1871–1887. doi: 10.1111/nph.18499. [DOI] [PubMed] [Google Scholar]
- Liao C.J., Lai Z., Lee S., Yun D.J., Mengiste T. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell. 2016;28:1662–1681. doi: 10.1105/tpc.16.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Jiang Y., Tang H., Tong S., Lou S., Shao C., Zhang J., Song Y., Chen N., Bi H., et al. The ubiquitin E3 ligase SR1 modulates the submergence response by degrading phosphorylated WRKY33 inArabidopsis. Plant Cell. 2021;33:1771–1789. doi: 10.1093/plcell/koab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li X., Wang M., Wen J., Yi B., Shen J., Ma C., Fu T., Tu J. Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 2018;16:911–925. doi: 10.1111/pbi.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kracher B., Ziegler J., Birkenbihl R.P., Somssich I.E. 2015. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Xiang M., Fan Y., Yang C., Zeng L., Zhang Q., Chen M., Liao Z. A Root-Preferential DFR-Like gene encoding dihydrokaempferol reductase involved in anthocyanin biosynthesis of purple-fleshed sweet potato. Front. Plant Sci. 2017;8:279. doi: 10.3389/fpls.2017.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23:1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naing A.H., Kim C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021;172:1711–1723. doi: 10.1111/ppl.13373. [DOI] [PubMed] [Google Scholar]
- Nakatsuka T., Saito M., Yamada E., Fujita K., Kakizaki Y., Nishihara M. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J. Exp. Bot. 2012;63:6505–6517. doi: 10.1093/jxb/ers306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio M.B., Ng S., Berkowitz O., De Clercq I., Mao C., Shou H., Whelan J., Jost R. SPX4 acts on PHR1-dependent and-independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 2019;181:332–352. doi: 10.1104/pp.18.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B.D., Pant P., Erban A., Huhman D., Kopka J., Scheible W.R. Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis , and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. 2015;38:172–187. doi: 10.1111/pce.12378. [DOI] [PubMed] [Google Scholar]
- Park S.H., Jeong J.S., Huang C.H., Park B.S., Chua N.H. Inositol polyphosphates-regulated polyubiquitination of PHR1 by NLA E3 ligase during phosphate starvation response in Arabidopsis. New Phytol. 2023;237:1215–1228. doi: 10.1111/nph.18621. [DOI] [PubMed] [Google Scholar]
- Pelletier M.K., Shirley B.W. Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol. 1996;111:339–345. doi: 10.1104/pp.111.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinckney J.L., Paerl H.W., Tester P., Richardson T.L. The role of nutrient loading and eutrophication in estuarine ecology. Environ. Health Perspect. 2001;109:699–706. doi: 10.1289/ehp.01109s5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga M.I., Mateos I., Charukesi R., Wang Z., Franco-Zorrilla J.M., de Lorenzo L., Irigoyen M.L., Masiero S., Bustos R., Rodríguez J., et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajniak J., Barco B., Clay N.K., Sattely E.S. A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature. 2015;525:376–379. doi: 10.1038/nature14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried M.K., Wild R., Zhu J., Pipercevic J., Sturm K., Broger L., Harmel R.K., Abriata L.A., Hothorn L.A., Fiedler D., et al. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 2021;12:384. doi: 10.1038/s41467-020-20681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N., Hou S., Tu G., Lan W., Jing Y. Transcription factor WRKY33 mediates the phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in Arabidopsis roots. Int. J. Mol. Sci. 2021;22:9275. doi: 10.3390/ijms22179275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao B., Zheng S., Zhang X., Wang X., Dong W., Xie Q., Wang G., Xiao Y., Chen F., et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell. 2021;184:5527–5540.e18. doi: 10.1016/j.cell.2021.09.030. [DOI] [PubMed] [Google Scholar]
- Shin D.H., Cho M., Choi M.G., Das P.K., Lee S.K., Choi S.B., Park Y.I. Identification of genes that may regulate the expression of the transcription factor production of anthocyanin pigment 1 (PAP1)/MYB75 involved in Arabidopsis anthocyanin biosynthesis. Plant Cell Rep. 2015;34:805–815. doi: 10.1007/s00299-015-1743-7. [DOI] [PubMed] [Google Scholar]
- Shirley B.W., Hanley S., Goodman H.M. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Xu Q., Zhang F.C., Chen Y., Li L.Q., Wu W.H., Chen Y.F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015;167:1579–1591. doi: 10.1104/pp.114.253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudheeran P.K., Ovadia R., Galsarker O., Maoz I., Sela N., Maurer D., Feygenberg O., Oren Shamir M., Alkan N. Glycosylated flavonoids: fruit's concealed antifungal arsenal. New Phytol. 2020;225:1788–1798. doi: 10.1111/nph.16251. [DOI] [PubMed] [Google Scholar]
- Tang H., Bi H., Liu B., Lou S., Song Y., Tong S., Chen N., Jiang Y., Liu J., Liu H. WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2021;229:106–125. doi: 10.1111/nph.17020. [DOI] [PubMed] [Google Scholar]
- Tang J., Wu D., Li X., Wang L., Xu L., Zhang Y., Xu F., Liu H., Xie Q., Dai S., et al. Plant immunity suppression via PHR1-RALF-FERONIA shapes the root microbiome to alleviate phosphate starvation. Embo J. 2022;41:e109102. doi: 10.15252/embj.2021109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Li L., He Y., Zhang X., Zhao Y., Wang Q., Hong G. Flavonoids in vegetables: improvement of dietary flavonoids by metabolic engineering to promote health. Crit. Rev. Food Sci. Nutr. 2022;1–15:1–15. doi: 10.1080/10408398.2022.2131726. [DOI] [PubMed] [Google Scholar]
- Tao H., Miao H., Chen L., Wang M., Xia C., Zeng W., Sun B., Zhang F., Zhang S., Li C., et al. WRKY33-mediated indolic glucosinolate metabolic pathway confers resistance against Alternaria brassicicola in Arabidopsis and Brassica crops. J. Integr. Plant Biol. 2022;64:1007–1019. doi: 10.1111/jipb.13245. [DOI] [PubMed] [Google Scholar]
- Tohge T., Nishiyama Y., Hirai M.Y., Yano M., Nakajima J.i., Awazuhara M., Inoue E., Takahashi H., Goodenowe D.B., Kitayama M., et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Ülker B., Somssich I.E. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Wang X.C., Wu J., Guan M.L., Zhao C.H., Geng P., Zhao Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020;101:637–652. doi: 10.1111/tpj.14570. [DOI] [PubMed] [Google Scholar]
- Wang H., Xu Q., Kong Y.H., Chen Y., Duan J.Y., Wu W.H., Chen Y.F., Chen, Jun-Ye., Duan Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014;164:2020–2029. doi: 10.1104/pp.113.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cai C., Li Y., Tao H., Meng F., Sun B., Miao H., Wang Q. Brassinosteroids fine-tune secondary and primary sulfur metabolism through BZR1-mediated transcriptional regulation. J. Integr. Plant Biol. 2023;65:1153–1169. doi: 10.1111/jipb.13442. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen Y.F., Wu W.H. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2021;63:34–52. doi: 10.1111/jipb.13053. [DOI] [PubMed] [Google Scholar]
- Xie Y., Tan H., Ma Z., Huang J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant. 2016;9:711–721. doi: 10.1016/j.molp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Yang L., Zhang Y., Guan R., Li S., Xu X., Zhang S., Xu J. Co-regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J. Integr. Plant Biol. 2020;62:1780–1796. doi: 10.1111/jipb.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Liu D. Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 2008;50:849–859. doi: 10.1111/j.1744-7909.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- Zhou J., Mu Q., Wang X., Zhang J., Yu H., Huang T., He Y., Dai S., Meng X. Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell. 2022;34:3066–3087. doi: 10.1093/plcell/koac139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X., He Y., et al. Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell. 2020;32:2621–2638. doi: 10.1105/tpc.19.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.