Abstract
Genetic variability significantly contributes to individual differences in skeletal muscle mass; however, the specific genes involved in that process remain elusive. In this study, we examined the role of positional candidates, Rps6ka6 and Pou3f4, of a chromosome X locus, implicated in muscle mass variability in CFW laboratory mice. Histology of hindlimb muscles was studied in CFW male mice carrying the muscle “increasing” allele C (n = 15) or “decreasing” allele T (n = 15) at the peak marker of the locus, rs31308852, and in the Pou3f4y/− and their wild-type male littermates. To study the role of the Rps6ka6 gene, we deleted exon 7 (Rps6ka6-ΔE7) using clustered regularly interspaced palindromic repeats-Cas9 based method in H2Kb myogenic cells creating a severely truncated RSK4 protein. We then tested whether that mutation affected myoblast proliferation, migration, and/or differentiation. The extensor digitorum longus muscle was 7% larger (P < 0.0001) due to 10% more muscle fibers (P = 0.0176) in the carriers of the “increasing” compared with the “decreasing” CFW allele. The number of fibers was reduced by 15% (P = 0.0268) in the slow-twitch soleus but not in the fast-twitch extensor digitorum longus (P = 0.2947) of Pou3f4y/− mice. The proliferation and migration did not differ between the Rps6ka6-ΔE7 and wild-type H2Kb myoblasts. However, indices of differentiation (myosin expression, P < 0.0001; size of myosin-expressing cells, P < 0.0001; and fusion index, P = 0.0013) were significantly reduced in Rps6ka6-ΔE7 cells. This study suggests that the effect of the X chromosome locus on muscle fiber numbers in the fast-twitch extensor digitorum longus is mediated by the Rps6ka6 gene, whereas the Pou3f4 gene affects fiber number in slow-twitch soleus.
Keywords: RSK4, myogenesis, muscle fibers
Introduction
Skeletal muscle tissue is responsible for vital functions such as locomotion and respiration. Additionally, it plays a role in maintenance of glucose homeostasis and thermoregulation, serves as a reservoir of amino acids during critical illness, and protects the bones and viscera. Reduced muscle mass due to aging and/or disease could affect these functions and lead to frailty, risk of falls and factures, and loss of independence (Cruz-Jentoft et al. 2010). There are substantial individual differences in muscle mass even before the onset of aging-related muscle mass decline. Heritability estimates indicate that a significant proportion of individual muscle mass differences, around 30–60%, are attributed to genetic variability (Silventoinen et al. 2008; Hernandez Cordero et al. 2019). While reverse genetics studies identified several key genes involved in the regulation of myogenesis (Rudnicki et al. 1992, 1993; Bladt et al. 1995; Seale et al. 2000; Millay et al. 2013; Bi et al. 2017) and muscle growth (McPherron et al. 1997), it remains unclear if naturally occurring variants of the same key genes could account for the variation in muscle mass and function observed in the population. A recent genome-wide association study (GWAS) indicated that most key genes implicated in regulation of myogenesis and muscle growth reside outside of ∼180 genomic loci linked to muscle mass in humans (Hernandez Cordero et al. 2019). Thus, a substantial pool of novel regulators of muscle mass that are responsible for the individual differences in a population remains to be determined.
The key index of muscle function, its contractile force, is positively associated with muscle mass. The muscle mass is determined by 3 main variables: the cross-sectional area of muscle fibers, fiber length, and the number of muscle fibers. The cross-sectional area of the fibers is adaptive and can change in response to the changing levels of physical activity and/or nutrition. Fiber length is a function of the size of the skeleton, increasing markedly during the axial growth of long bones and stabilizing after that (Gokhin et al. 2008). The number of muscle fibers appears to be the most rigid variable. In laboratory mice, the numbers of fibers are determined prenatally and remain stable for a substantial portion of the lifespan (Ontell et al. 1988). Similarly in humans, the number of fibers does not differ in elite body builders with years of training history compared to the age-matched controls, but body builders' fibers are over 50% larger in cross-sectional area (MacDougall et al. 1984). Therefore, it is important to understand whether the individual differences in muscle mass are due to the axial length differences, or an outcome of the adaptive changes in the fiber cross-sectional area, or if it results from differences in the muscle fiber number.
To investigate the role of the naturally occurring genetic variants in variability of muscle mass, we previously conducted a GWAS in outbred CFW mice (Nicod et al. 2016; Parker et al. 2016). That study identified genomic regions associated with the size of the individual muscles, albeit in limited resolution, as most of the identified regions contain more than one gene thus requiring further validation studies (Flint and Eskin 2012). The locus on chromosome X with a peak marker rs31308852 showed one of the strongest associations with the weight of fast-twitch tibialis anterior (TA), extensor digitorum longus (EDL), gastrocnemius, and plantaris muscles of CFW mice and attracted attention for the following reasons: (1) the confidence interval of the locus harbored only 3 positional candidates, Pou3f4, Cylc1, and Rps6ka6, and (2) all 3 genes were novel in the context of skeletal muscle development and growth. Pou3f4 is expressed in developing, but not adult, skeletal muscle in mice (Dominov and Miller 1996) and therefore emerged as a plausible candidate gene. The Rps6ka6 is ubiquitously expressed including in adult skeletal muscle tissue, albeit at a low level, whereas Cylc1 is expressed only in the testes. Because of these expression patterns of the 3 candidate genes, we focused on the Pou3f4 and Rps6ka6 genes. The aims of the present study were: (1) to examine if variation in the chromosome X locus affects muscle weight through the number or size of muscle fibers and (2) to test which of the 2 candidate genes, Pou3f4 and Rps6ka6, might be responsible for the effect of the locus on the muscle mass.
A preferred validation strategy for positional candidates is phenotyping a suitable knockout mouse model (Lionikas et al. 2023). However, in the instances where a knockout of the target gene does not exist, the cell cultures derived from relevant tissue offer an amenable and informative alternative (Hernandez Cordero et al. 2019; Chen et al. 2023). Because deletion of Rps6ka6 gene affects early embryonic development hampering derivation of a knockout (Cox et al. 2010), we employed both approaches in the present study.
Methods
Murine model
To examine the mechanisms of the effect of the locus on chromosome X, EDL muscle samples collected from CFW mice for previously published reports (Nicod et al. 2016; Parker et al. 2016) were used in the current study. We tested the hypothesis that variation in the locus could have affected (1) the number of muscle fibers constituting the muscle, (2) the cross-sectional area of the fibers, or (3) both. Male CFW mice of ∼90 days of age were selected for immunohistochemical analyses of the fibers in EDL muscle. The selected mice carried a C allele (n = 15) or T allele (n = 15) at a peak marker, rs31308852, of the locus which were associated with increased or decreased muscle weight, respectively. The EDL muscle was used because the genotype showed the strongest association with its weight compared with the TA, gastrocnemius, or plantaris muscles (Nicod et al. 2016). Samples were processed as previously described (Nicod et al. 2016; Parker et al. 2016).
To test the hypothesis that the association of the locus with muscle mass could be due to the effect of Pou3f4 variation, we studied a Pou3f4 knockout mouse model described previously (Coate et al. 2012; Brooks et al. 2020). The Pou3f4 knockout line (Jax, strain #036158) was maintained in accordance with the Georgetown University Institutional Animal Care and Use Committee protocol, #1147. Mice carrying the Pou3f4 null allele were maintained on C57Bl/6N background for over 5 generations before these experiments. Since Pou3f4 is located on the X chromosome, only males were used for these experiments to avoid possible variability in females due to X-inactivation. To generate Pou3f4y/− mice and littermate controls, Pou3f4 heterozygous females were crossed with C57Bl/6N wild-type (WT) males (purchased from Charles River Laboratories). The Direct PCR Lysis Reagent (Viagen Biotech) was used to prepare crude DNA samples from tail biopsies, and then, the following primer sets were used for PCR genotyping: TCCTTGCTTCCTCCAGTCAGAGATC and ACGTCCAGCGGCCAACCCCTCAATG (WT) and CAATGCTGTTTCACTGGTTATG and CATTGCCCCTGTTTCACTATC (Cre). Because the target phenotype, fiber number, remains stable over a substantial portion of the lifespan (Ontell et al. 1988), animals of various ages were used. Mice were sacrificed between the ages of 47 and 171 days, and carcasses of Pou3f4y/− (n = 14) and their WT littermates (Pou3f4y/+; n = 15) were frozen until dissection. On the day of muscle dissection, the samples were thawed and processed as previously described (Nicod et al. 2016; Parker et al. 2016). Briefly, the EDL and soleus muscles were dissected under a microscope, snap-frozen in isopentane chilled with liquid nitrogen, and stored at −70°C until histological processing.
Immunohistochemistry and imaging
Transverse cryosections from the middle part of the muscle belly were cut at a thickness of 10 µm with a cryotome (Leica CM1850UV) at −20°C and collected on a positively charged glass slide (SuperFrost Plus, Menzel Gläser). Sections were air dried and stored at −70°C. On the day of staining, the sections from CFW mice were blocked in 10% fetal calf serum in phosphate buffered saline (PBS) and incubated with primary antibodies against laminin 1:50 (L9393, Sigma-Aldrich) for 1–2 h followed by wash in PBS. After that, the samples were processed using the diaminobenzide (DAB) chromogenic staining kit SK-4100 (Vector Laboratories). Following incubation with biotinylated BA-100 secondary antibody (Vector Laboratories) at 1:200 concentration, samples were washed in PBS and incubated with avidin biotinylated horse radish peroxidase (HRP) reagent for 30 min. Following a wash in PBS, working solution of DAB was applied for no more than 5 min and the reaction was stopped by transferring the slides to distilled water. The secondary antibody, fluorescent Alexa goat antirabbit 594 (Invitrogen), was applied at 1:200 concentration for 1 h followed by wash in PBS. For the Pou3f4 model, EDL and soleus muscle sections were subjected to tetrazolium reductase staining [30 min incubation in 50 mM TRIS (pH7.6), 1 mM NADH, 1 mM nitroblue tetrazolium at 37°C] and ATPase staining [following a 5 min acid preincubation, pH 4.47 (Brooke and Kaiser 1970)], respectively. After staining, Mowiol 4–88 (Sigma-Aldrich) was added over the sections and covered with a coverslip sealed with nail varnish. Slides (1 per muscle) were scanned using the Axioscan Z1 slide scanner (Zeiss) with 10× magnification objective.
Cell culture
As no mouse Rps6ka6 gene knockout has been reported, to test the hypothesis that Rps6ka6 may be responsible for the effects of the X chromosome locus on muscle mass, we used the H2Kb myogenic cells. The H2Kb cell line was derived in Dr. Wells laboratory (Muses et al. 2011) from the EDL muscle of a H-2Kb-tsA58 immortomouse (CBA/Ca × C57BL/10 background) originally reported by Jat and colleagues (Jat et al. 1991). The immortomouse genome harbors temperature-sensitive (ts) large tumor antigen (Tag) gene from the tsA58 strain of the simian virus 40 (SV40). The expression of the immortalizing Tag gene driven by mouse major histocompatibility complex H2Kb promoter can be induced in vitro by interferons. Importantly, H2Kb cells were derived from a male mouse donor (Muses et al. 2011) and therefore contain 1 X chromosome and a single copy of the Rps6ka6 gene. A single copy of the X chromosome is advantageous for the generation of knockouts, and therefore, H2Kb cells were preferred to commonly used C2C12 myoblasts which are derived from a female mouse (Diokmetzidou et al. 2016) with 2 X chromosomes.
H2Kb myoblasts were maintained in growth medium containing 4 ng/mL of IFN-γ (R&D, 485-MI) in high-glucose Dulbecco's Modified Eagle's Medium (DMEM; Sigma, D6546), 20% fetal bovine serum, and 1% glutamine, at 33°C/10% CO2. To induce differentiation, the myoblasts were transferred to a high-glucose DMEM containing 5% horse serum and 1% glutamine and incubated at 37°C and 5% CO2.
Development of Rps6ka6-ΔE7 H2Kb clones
H2Kb cells lacking exon 7 of Rps6ak6 (Rps6ka6-ΔE7) were generated using clustered regularly interspaced palindromic repeats (CRISPR)-Cas9 genome editing. Guide RNAs (gRNA) flanking Rps6ak6 exon 7 (ENSMUSE00000152471) were designed using the Broad Institute web tool (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and, 2 sequences (5′-GATGGGGTTAGAACATACTG-3′ and 5′-AACTGTATTAACAATGCTAA-3′) were selected. Each gRNA was ligated in the pX459 plasmid (Addgene plasmid ID 48139) using Quick Ligation Kit (NEB, M2200L). The plasmid also contains the Cas9 endonuclease sequence (Ran et al. 2013). Following transformation, Stellar Competent Cells (Takara, 636763) were incubated in SOC medium (Takara, 2011234A) and plated on Luria-Bertani (LB) agar plates containing 80 μg/mL carbenicillin (Fisher Scientific, BP2648-1) for the overnight incubation at 37°C. The next day, colonies were isolated and expanded in LB broth overnight at 37°C on a shaker (250 rpm). The following morning, plasmids were purified using a miniprep kit (Qiagen, 27104). H2Kb cells were then transfected following a modified protocol described by Bauer and colleagues (Bauer et al. 2015). Briefly, H2Kb cell were seeded at 7,500 cells per well in a 24-well plate. The next day, dual transfection (with 25 µg of each plasmid) was carried out using Lipofectamine LTX (Invitrogen, A12621) following the manufacturer's protocol. After 16 h the medium was replaced, and the cells were subjected to puromycin (Merck, 540222) selection (2 μg/mL) for 72 h. The surviving cells were expanded and subjected to clonal selection as follows. The cells were trypsinized, and cell suspension containing 1 cell per 100 μL of growth medium was prepared through serial dilutions. Single cells were seeded in a 96-well plate, and after 1 week, 35% of the wells contained single cell origin clones. After another week, 30 clones were expanded and genotyped using PCR with a pair of primers flanking the gRNA target sites (forward primer 5′- GGAGGGTGTTGGCATTCTCA-3′, reverse primer 5′-TCCACCTCTCTAGAGCTAGGC-3′). Five clones generated amplicons migrating faster than the WT allele. Sanger sequencing indicated that 3 of the clones with fast migrating amplicons lacked exon 7 (Rps6ka6-ΔE7). The 3 clones that showed similar growth pattern to the Rps6ka6-ΔE7 clones at the start of the clonal selection but genotyped as WT allele (Rps6ka6-wt) were used as controls throughout the study.
Proliferation assay
Proliferation of H2Kb cells was measured using the Click-iT Plus EdU AlexaFluor 549 flow cytometry assay kit (Invitrogen, C10646). Cells at 100 cells per mm2 were seeded in a 96-well plate and incubated overnight at 33°C/10% CO2. The staining started with a 3 h incubation in 10 mM EdU-containing medium. Then, the cells were washed thrice in 1% bovine serum albumin (BSA) in PBS, Click-iT fixative was added to the cells for 15 min then washed in 1% BSA in PBS, and a 15 min incubation in Click-iT saponin-based permeabilization and wash reagent followed. At the end, the Click-iT Plus reaction cocktail was applied for 30 min followed by 3 5 min washes in Click-iT saponin-based permeabilization and wash reagent. Cell nuclei were stained with 300 nM DAPI solution applied for 5 min. After 3 washes in PBS, the samples were imaged. The experiment was repeated 3 times with consecutive passages of the cells.
Migration assay
The capacity of Rps6ka6-wt and Rps6ka6-ΔE7 myoblasts to migrate was assessed using an in vitro scratch assay as previously described (Liang et al. 2007). In brief, following an overnight incubation of 100,000 cells/well in a 6-well plate in growth medium, a scratch was performed on the bottom of the well longitudinally using a P200 pipette tip. The wound was left to regenerate overnight. To assess the migrative ability of myoblasts, 2 nonoverlapping images were taken immediately after the scratch was formed as well as 24 h postscratching using an inverted phase contrast light and fluorescence microscope (EVOS XL, Life Technologies). The experiment was repeated 3 times with consecutive passages of cells.
Differentiation assay
H2Kb cells were seeded in a 12-well plate at a density of 40,000 cells/well and incubated overnight in growth medium. The following day, the cells were washed in PBS, and differentiation medium was applied. Differentiation medium was replaced 3 days later and removed after 6 days, and the cells were fixed in 4% paraformaldehyde overnight at 4°C. The fixed cells were washed in PBS and permeabilized in 0.5% Triton X-100 in PBS for 15 min followed by a PBS wash. The cells were incubated in blocking buffer (10% fetal calf serum in PBS) for 1 h followed by an incubation in antimyosin, skeletal, fast antibody (M4276, Sigma-Aldrich) in PBS (1:400) for 2 h at room temperature. After 3 washes in PBS and Tween 20 0.025%, the secondary antibody (1:400 donkey antimouse Alexa 488, ab150109, abcam) and 300 nM DAPI in PBS were applied for 2 h at room temperature. Finally, the cells were washed 3 times in PBS before imaging. The experiment was repeated 4 times following consecutive passages. Six nonoverlapping images were acquired per well (2 images top, 2 middle, and 2 bottom of the well) with an inverted phase contrast light and fluorescence microscope (EVOS XL, Life Technologies).
Image analysis
The images were analyzed using the FIJI/ImageJ software (Schindelin et al. 2012). A semiautomatic process for the CFW samples involved setting the scale and splitting color channels. The laminin- and type IIA myosin-stained images were analyzed for the number and cross-sectional area of muscle fibers. The red channel image was converted to a binary image using the thresholding function and processed using the binary functions Dilate and Erode to enhance the boundaries of all muscle fibers. It was followed by the Analyze Particles function with a setting of 0.4–1.0 for circularity to exclude erroneous objects within the expected size range, 50–4,500 µm2, but not consistent with muscle fiber shape. The output was visually inspected. In the instances where adjacent fibers appeared incompletely separated, the connections were manually deleted before repeating the analysis. For Pou3f4 samples, muscle fibers were counted using the Cell Counter plugin.
In the analyses of the proliferation assay images, a threshold was set to select all the EdU-positive and DAPI-positive particles in the red and blue channel images, respectively, and convert them to a binary image. Subsequently, the number of nuclei was quantified using the Analyze Particles function. The percentage of proliferating cells was quantified by dividing the number of EdU-positive particles by the number of DAPI-positive particles and multiplying by 100%.
The quantification of the migrative capacity of the myoblasts was performed as previously described (Pijuan et al. 2019). In brief, following duplication of the wound region, the border of the cells was identified using the “find edges” function and the image was blurred using the “gaussian blur” function on FIJI/ImageJ. Using a threshold, the area of the well not covered in cells was selected and quantified through the Analyze Particles function. The percentage open wound was calculated by dividing the area devoid of cells at 24 h postwounding by the initial wound area at 0 h postwounding and multiplied by 100%. The percentage wound closure was calculated by subtracting the percentage open wound from 100%.
In the analyses of differentiation assay images, we quantified the total number of myosin-positive cells and myosin-positive cells containing more than 2 nuclei (fusion index) using the “multi-point” tool. The counts were corrected against the total area (mm2) of 6 captured images per well. To measure the area of myosin-positive cells, reflecting the size of differentiating cells, a threshold was set to create a binary mask highlighting the myosin-positive cells. The Analyze Particles function was then used to calculate the average area (μm2) per myosin-expressing cell.
Statistical analyses
Statistical analysis and data visualization was performed on the Prism5 software (GraphPad, version 5.04). A t-test was used in the analyses of histology results. A 2-way ANOVA followed by Bonferroni posttest was employed to assess the effects of genotype (2 levels) and time (3 or 4 levels) in the analyses of in vitro data. P-values are stated in-text, and the level of significance is denoted on the graphs as *P < 0.05, **P < 0.01, and ***P < 0.001. Data are presented as mean and standard error of the mean (SEM). The mean and SEM presented in text for the H2kb myoblasts-related data is a result of averaging the means and SEMs obtained from the technical replicates of each experiment.
Results
The X chromosome locus is associated with fiber number in fast-twitch muscle
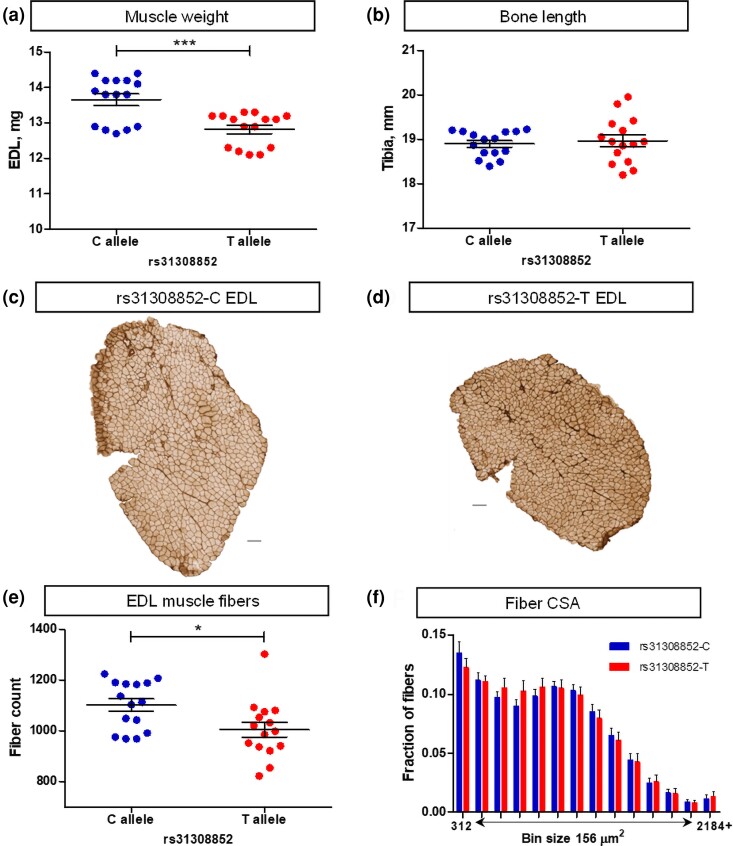
To examine the cellular mechanisms of the effect of the X chromosome locus on muscle weight, we studied the histological properties of the fast-twitch EDL muscle in the CFW carriers of the divergent alleles at a peak marker of the locus, rs31308852. The muscles of the rs31308852-C mice were 7% heavier (P = 0.0003) compared with the rs31308852-T allele mice (Fig. 1a). The length of the tibia was similar between the 2 genotypes, 18.9 ± 0.1 vs 19.0 ± 0.1 mm, respectively (P = 0.6431) (Fig. 1b). These data demonstrate that the difference in weight was due to the girth of muscle, not the length. The girth can be affected by the number of muscle fibers, their cross-sectional area, or both. The analysis of the histological samples (Fig. 1c and d) revealed that this difference in muscle weight was mediated by 10% more fibers in rs31308852-C mouse muscles compared with rs31308852-T (1,102 ± 25 vs 1,005 ± 30, P = 0.0176) (Fig. 1e). The size of the fibers, as assessed by the distribution of fiber cross-sectional area, was similar (Fig. 1f). Thus, the difference in the fast-twitch muscle weight between the genotypes at this X chromosome locus was due to the difference in fiber number.
Fig. 1.
The genotype of the X chromosome locus rs31308852 is associated with the number of EDL muscle fibers in CFW male mice. a) EDL weight. b) Tibia length. c and d) Laminin-stained transverse sections of EDL muscle from the rs31308852-C and rs31308852-T mouse, respectively. Horizontal scale bars 100 µm. e) Fiber number in the EDL muscle. f) Distribution of the cross-sectional area (CSA) of EDL fibers across 156 µm2 bins. Data are presented as mean and SEM (n = 15 per genotype).
Rps6ka6 affects differentiation of myogenic cells
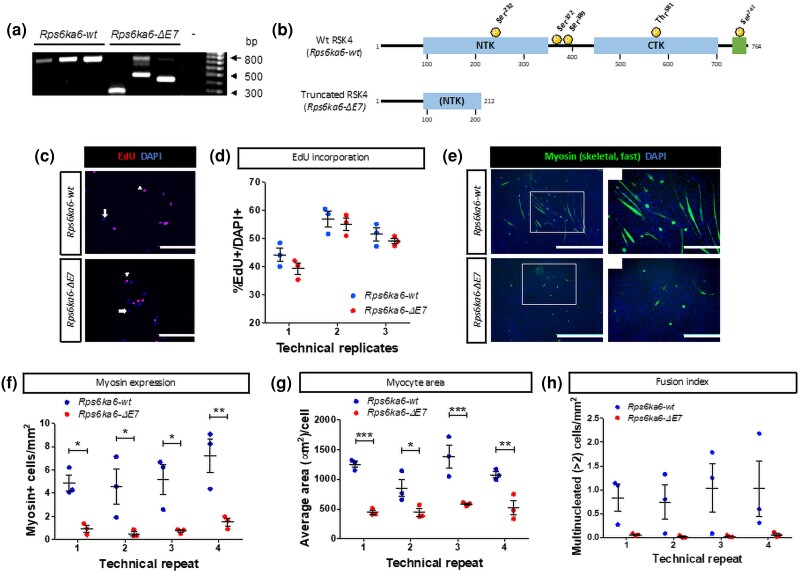
The rs31308852 is a missense SNP resulting in the Arg45His substitution in the Rps6ka6 encoded protein. To study the effects of Rps6ka6 on myogenic lineage, we targeted exon 7 of the gene for excision in H2Kb cells. Three clones of cells with deletion, Rps6ka6-ΔE7, and 3 WT clones, Rps6ka6-wt, were generated for further analyses (Fig. 2a). Excision of exon 7 creates a premature stop codon (Supplementary Fig. 1), resulting in a severely truncated protein (Fig. 2b). We first quantified the EdU-positive cells to compare the rate of proliferation between the 2 genotypes (Fig. 2c). We were not able to identify a statistically significant difference between Rps6ka6-wt and Rps6ka6-ΔE7 cells, with cumulative average across all 3 repeats of the EdU-positive cells over the total number of cells of 50.8 ± 2.2% vs 47.8 ± 2.5%, respectively (2-way ANOVA, genotype factor, P = 0.11) (Fig. 2d). The effect of time (between the 3 consecutive passages of the cells) was statistically significant (P < 0.0001) because of the lower proportion of the EdU-positive cells of both genotypes at the first time point (Fig. 2d), but the interaction term was not statistically significant (P = 0.7748). The results of this assay reveal no Rps6ka6 effect on proliferation of the myogenic cells.
Fig. 2.
Rps6ka6 is essential for myoblast differentiation but is not required for myoblast proliferation. a) PCR genotyping results of the 3 Rps6ka6-wt and 3 Rps6ka6-ΔE7 clones used in our study (- denotes negative control). b) Schematic representation of the truncated RSK4 protein resulting from the Rps6ka6-ΔE7 gene. NTK, N-terminal kinase domain; CTK, C-terminal kinase domain; green region, ERK-binding domain. c) Representative images of EdU-treated Rps6ka6-wt and Rps6ka6-ΔE7 myoblasts. Scale bars: 200 μm. d) Quantification of %EdU-positive, proliferating Rps6ka6-wt and Rps6ka6-ΔE7 myoblasts. e) Representative images of myosin-stained myotubes following differentiation of Rps6ka6-wt and Rps6ka6-ΔE7 myoblasts. Scale bars: 1,000 μm in large fields of view and 400 μm in insets. f) Quantification of myosin-positive Rps6ka6-wt and Rps6ka6-ΔE7 cells. g) Quantification of the average area of myosin-positive Rps6ka6-wt and Rps6ka6-ΔE7 cells. h) Quantification of the multinucleated Rps6ka6-wt and Rps6ka6-ΔE7 myosin-positive cells. Data are presented as mean ± SEM (n = 3 per genotype).
Additionally, we assessed the migrative capacity of Rps6ka6-wt and Rps6ka6-ΔE7 myoblasts using an in vitro scratch assay (Supplementary Fig. S2a). We were not able to identify any statistically significant difference between the motility of the Rps6ka6-wt (cumulative average of 41.6 ± 4.4% wound closure) and Rps6ka6-ΔE7 cells (cumulative average of 42.9 ± 2.7% wound closure) (2-way ANOVA, genotype factor, P = 0.74) (Supplementary Fig. S2b). Nevertheless, there was some technical variation between consecutive passages (2-way ANOVA, technical repeat factor, P = 0.0099) (Supplementary Fig. S2b). These data suggest that variation in the Rps6ka6 gene does not affect the migrative ability of myoblasts in vitro.
We then compared myogenic differentiation between the 2 genotypes (Fig. 2e). Counting the myosin-expressing cells across the 4 time points indicated that they were ∼5-fold more numerous in the Rps6ka6-wt cultures compared with the Rps6ka6-ΔE7 cell cultures (5.4 ± 1.2 vs 0.91 ± 0.22 cell/mm2; 2-way ANOVA genotype factor, P < 0.0001) (Fig. 2f). The effects of time or the time-by-genotype interaction were not statistically significant (P = 0.2384 and P = 0.7517, respectively). Myosin-expressing cells were ∼2-fold larger in Rps6ka6-wt cell cultures compared with Rps6ka6-ΔE7 (1,141.5 ± 110.2 vs 499.5 ± 59.4 µm2; 2-way ANOVA genotype factor, P < 0.0001) (Fig. 2g). Although the myosin-expressing area varied among the consecutive passages (effects of time P = 0.0364), the time-by-genotype interaction was not statistically significant (P = 0.1943). We then examined the fusion index characterized by the number of cells with more than 2 myonuclei per unit of area. The number of the multinucleated cells in the Rps6ka6-wt cell cultures, albeit variable among the clones, was larger compared with the Rps6ka6-ΔE7 cell cultures (0.91 ± 0.43 vs 0.040 ± 0.018 cell/mm2; 2-way ANOVA genotype factor, P = 0.0013) (Fig. 2h). The effects of time or the time-by-genotype interaction were not statistically significant (P = 0.9464 and P = 0.9563, respectively). These data demonstrate that disruption of Rps6ka6 significantly impairs the differentiation potential of the myogenic cells.
Pou3f4 only affects fiber number in slow-twitch muscles
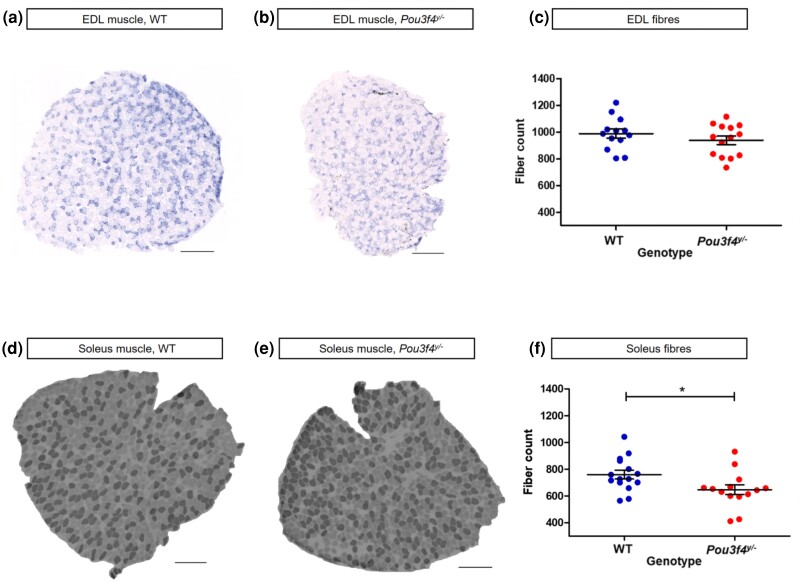
To study if Pou3f4 could be associated with the effect of the X chromosome locus, we analyzed muscle samples of the Pou3f4y/− mice and their WT littermates. We focused on the number of muscle fibers because this trait was primarily affected by the rs32308852 genotype in CFW mice and because it is not influenced by the wide age range of mice used in this analysis (Ontell et al. 1988). Fiber number in the EDL muscle was similar between the Pou3f4y/− and WT mice (938 ± 32 and 987 ± 34, respectively; P = 0.2947) (Fig. 3a). The analysis of the slow-twitch soleus muscle showed a 15% reduction in fiber number in the Pou3f4y/− muscles compared with the WT (646 ± 36 vs 760 ± 33, P = 0.0268) (Fig. 3b). Hence, Pou3f4 deletion reduces the number of fibers in the slow-twitch soleus muscle but does not affect muscle fiber number in the fast-twitch EDL.
Fig. 3.
Pou3f4 ablation leads to a reduction in fiber number in slow- but not fast-twitch muscles. a and b) Micrographs of tetrazolium reductase–stained fast-twitch EDL muscle of WT and Pou3f4y/− mice, respectively. c) Fiber number in EDL muscle samples. d and e) Micrographs of ATPase-stained slow-twitch soleus muscle of WT and Pou3f4y/− mice, respectively. f) Fiber number in soleus muscle samples. Horizontal scale bars 200 µm. Data are presented as mean ± SEM (WT, n = 15; Pou3f4y/−, n = 14).
Discussion
In this study, we examined the cellular mechanisms of the effect of the chromosome X locus associated with variability in fast-twitch muscle mass of CFW mice (Nicod et al. 2016) and studied the role of 2 positional candidates, Rps6ka6 and Pou3f4. We found that the locus was primarily associated with ∼10% difference in the number of muscle fibers constituting fast-twitch EDL muscle. The Rps6ka6 gene played an enabling role in the differentiation of myogenic cells and emerged as the primary candidate for explaining the effect of the locus on the fast-twitch muscle mass. The Pou3f4 gene did not affect the number of muscle fibers in the fast-twitch muscle but did so in the slow-twitch muscle. Both positional candidates impacted the properties of myogenic tissue.
Genetic variability affects the number of muscle fibers
In a seminal study, Lexell and colleagues (Lexell et al. 1988) documented a 2-fold individual difference in the number of muscle fibers in vastus lateralis of young males. This observation is of clinical importance because the number of fibers declines with aging (Lexell et al. 1988) and fewer fibers at a young age may signal an increased risk of sarcopenia, the aging-related decline in muscle mass and function, which erodes the quality of life of the elderly and increases the risk of mortality (De Buyser et al. 2016; Beaudart et al. 2017). However, that study did not investigate if the difference in the number of fibers is a congenital outcome of, for example, availability of nutrients during prenatal development as demonstrated in rodents (Dwyer and Stickland 1994) or due to inheritance of variants of genes that are relevant in the development, growth, and maintenance of muscle tissue. The findings of our study that the number of fibers is affected by the genotype at the rs31308852 marker suggest that individual differences observed in humans may be an outcome of inherited variants of relevant genes.
Rps6ka6 plays a role in myogenic differentiation
The Rps6ka6 gene encodes the p90 ribosomal S6 kinase 4 (RSK4) protein, 1 of the 4 members of this gene family (Lara et al. 2013). The RSK proteins are downstream targets of the extracellular signal regulated kinase (Ras/ERK) pathway. The RSK1–3 proteins have been well characterized, but the function of RSK4 remains poorly understood (Romeo et al. 2012). Unlike the 3 other isoforms, RSK4 exhibits a growth factor independent constitutive activity (Dummler et al. 2005). The gene has been studied in the context of cancer (Xu et al. 2021), but this is the first report on its role in myogenesis of skeletal muscle. A low level of Rps6ka6 transcript is found in skeletal muscle in humans (Supplementary Fig. S3). This is consistent with the low levels of RSK4 protein in mouse skeletal muscle (Dummler et al. 2005). It is primarily a cytosolic protein with a small fraction showing nuclear localization (Dummler et al. 2005) and low level of expression in most tissues (Romeo et al. 2012). The gene, however, is expressed at a substantial level during embryonic development, and in the organogenesis stage, its expression exceeds the combined level of the RSK1-3 transcripts (Romeo et al. 2012). Deletion of Rps6ka6 severely affects the morphology of the embryo, highlighting its importance during development (Cox et al. 2010). A SIFT score for missense SNP rs31308852 in CFW population is predicted to be tolerated for protein function (Ng and Henikoff 2003); however, additional studies are required to establish if this substitution could explain subtle phenotypic effects.
The C2C12 myogenic cell line has been derived from satellite cells and is used as an in vitro model of myogenesis (Yaffe and Saxel 1977; Blau et al. 1983). Expression of Rps6ka6 in C2C12 cells at a myoblast stage is low but increases during differentiation into myotubes, the precursors of muscle fibers (Chen et al. 2006). This temporal pattern of expression parallels the findings of our study where mutation of Rps6ka6 did not affect proliferation of H2Kb myoblasts but did affect their differentiation. Hence, despite the low level of Rps6ka6 expression in adult tissues, Rps6ka6 may play a role during embryogenesis and fetal development, at the time when muscle fibers are formed in mice (Ontell et al. 1988). The molecular mechanisms of the effect of RSK4 remain unclear.
Muscle-type specific effects of Pou3f4
The Pou3f4 gene encodes a transcription factor (Mathis et al. 1992) implicated in X-linked deafness (DNFX2) in humans (de Kok et al. 1995). It was targeted in the present study because in a GWAS on muscle weight in CFW mice, Pou3f4 was the only annotated gene in the 95% confidence interval of the locus for the TA and EDL muscles (Nicod et al. 2016). Furthermore, a previous report implicated this gene in in vivo myogenesis (Dominov and Miller 1996). In the present study, however, deletion of the gene did not significantly affect the number of fibers in the fast-twitch EDL muscle but did decrease the number of fibers, albeit only by 15%, in the slow-twitch soleus muscle. It is peculiar though that no association of soleus weight with the X chromosome linked locus was detected in the CFW mice GWAS (Nicod et al. 2016). This could be due to a combination of the modest effect of the gene emerging from the Pou3f4y/− model analysis and the size of the soleus muscle which is ∼40% smaller than EDL in CFW males (Nicod et al. 2016). The latter would limit the accuracy of measurements and the statistical power for detection of associations in a GWAS.
The Pou3f4 gene is expressed in mouse muscle tissue during embryonic days 15–18 (Dominov and Miller 1996) when muscle fibers are actively forming (Chal and Pourquié 2017). The rodent muscle fiber type identity between the slow-twitch and fast-twitch types is acquired at around the same time of fetal development (Schiaffino and Reggiani 2011). The temporal overlap of this developmental process with the expression of Pou3f4 might be linked to the slow-twitch muscle-specific effect in the Pou3f4y/− model; however, specific mechanisms remain to be explored. Expression of Pou3f4 declines postnatally and becomes undetectable in adult muscle, suggesting its primary role in the developmental stage (Dominov and Miller 1996). This temporal pattern of expression may explain why POU3F4 mutations in DFNX2 patients do not appear to cause noticeable muscle abnormalities (Dominov and Miller 1996).
Conclusion
We found that the X chromosome locus implicated in the variation of the fast-twitch muscle mass in CFW mice affects the number of muscle fibers. The 2 positional candidate genes, Rps6ka6 and Pou3f4, impact the properties of myogenic tissue. Rps6ka6 affects the differentiation of myogenic cells and might be responsible for the effect of the locus on the weight of fast-twitch muscle. Pou3f4 affects fiber number in slow-twitch muscle. Hence, both positional candidates of the locus influence skeletal muscle mass through impacting the number of muscle fibers.
Supplementary Material
Acknowledgments
Authors are grateful for Mr James Archibald for assistance with histological analysis of muscle samples and for the Microscopy and Histology Core Facility at the University of Aberdeen for the help with imaging.
Contributor Information
Konstantinos Lekkos, School of Medicine, Medical Sciences and Nutrition, University of Aberdeen, Aberdeen AB25 2ZD, UK.
Afra A Bhuiyan, School of Medicine, Medical Sciences and Nutrition, University of Aberdeen, Aberdeen AB25 2ZD, UK.
Abdullah M K Albloshi, School of Medicine, Medical Sciences and Nutrition, University of Aberdeen, Aberdeen AB25 2ZD, UK; Department of Anatomy and Histology, School of Medicine, University of Albaha, Alaqiq 65779, Saudi Arabia.
Paige M Brooks, Department of Biology, Georgetown University, Washington, DC 20057, USA.
Thomas M Coate, Department of Biology, Georgetown University, Washington, DC 20057, USA.
Arimantas Lionikas, School of Medicine, Medical Sciences and Nutrition, University of Aberdeen, Aberdeen AB25 2ZD, UK.
Data availability
The H2Kb cell clones are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.
Funding
A.L. was supported by awards AR052879 and AR056280 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, award 249156 from the FP7-PEOPLE-2009-RG programme, award CGA/18/05 from the Chief Scientist Office, award 21/019 from the NHS Grampian, and award 204815/Z/16/Z from the Wellcome Trust. T.M.C. was supported by NIH R01 DC016595. P.M.B. was supported by NIH R01 DC016595, NIH F31 DC019824, and TL1TR001431. Research reported in this article was supported by the National Center for Advancing Translational Sciences under award number TL1TR001431. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
A.L. conceived the study and analyzed the data. K.L. phenotyped H2Kb cells and produced figures. A.A.B. carried out CRISPR/Cas9-mediated genomic modification of H2Kb cells. A.M.K.A. conducted immunohistochemistry and image analyses of muscle samples. T.M.C. and P.M.B. maintained and supplied Pou3f4 model mice. The manuscript was drafted by A.L. with input from all coauthors.
Literature cited
- Bauer DE, Canver MC, Orkin SH. 2015. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J Vis Exp. (95):e52118. doi: 10.3791/52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudart C, Biver E, Reginster J, Rizzoli R, Rolland Y, Bautmans I, Petermans J, Gillain S, Buckinx F, Dardenne N, et al. . 2017. Validation of the SarQoL®, a specific health-related quality of life questionnaire for sarcopenia. J Cachexia Sarcopenia Muscle. 8(2):238–244. doi: 10.1002/jcsm.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sánchez-Ortiz E, Bassel-Duby R, Olson EN. 2017. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 356(6335):323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. 1995. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376(6543):768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Blau HM, Chiu CP, Webster C. 1983. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32(4):1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Brooke MH, Kaiser KK. 1970. Three “myosin adenosine triphosphatase” systems: the nature of their pH lability and sulfhydryl dependence. J Histochem Cytochem. 18(9):670–672. doi: 10.1177/18.9.670. [DOI] [PubMed] [Google Scholar]
- Brooks PM, Rose KP, MacRae ML, Rangoussis KM, Gurjar M, Hertzano R, Coate TM. 2020. Pou3f4-expressing otic mesenchyme cells promote spiral ganglion neuron survival in the postnatal mouse cochlea. J Comp Neurol. 528(12):1967–1985. doi: 10.1002/cne.24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chal J, Pourquié O. 2017. Making muscle: skeletal myogenesis in vivo and in vitro. Development 144(12):2104–2122. doi: 10.1242/dev.151035. [DOI] [PubMed] [Google Scholar]
- Chen IB, Huber M, Guan T, Bubeck A, Gerace L. 2006. Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis. BMC Cell Biol. 7(1):38–38. doi: 10.1186/1471-2121-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Pan L, Zeng Y, Zhu X, You L. 2023. CPNE1 regulates myogenesis through the PERK-eIF2α pathway mediated by endoplasmic reticulum stress. Cell Tissue Res. 391(3):545–560. doi: 10.1007/s00441-022-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate TM, Raft S, Zhao X, Ryan AK, Crenshaw EB 3rd, Kelley MW. 2012. Otic mesenchyme cells regulate spiral ganglion axon fasciculation through a Pou3f4/EphA4 signaling pathway. Neuron 73(1):49–63. doi: 10.1016/j.neuron.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BJ, Vollmer M, Tamplin O, Lu M, Biechele S, Gertsenstein M, van Campenhout C, Floss T, Kühn R, Wurst W, et al. . 2010. Phenotypic annotation of the mouse X chromosome. Genome Res. 20(8):1154–1164. doi: 10.1101/gr.105106.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Jentoft AJ, Baeyens JP, Bauer JM, Boirie Y, Cederholm T, Landi F, Martin FC, Michel J-P, Rolland Y, Schneider SM, et al. . 2010. Sarcopenia: European consensus on definition and diagnosis: report of the European Working Group on Sarcopenia in Older People. Age Ageing. 39(4):412–423. doi: 10.1093/ageing/afq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buyser SL, Petrovic M, Taes YE, Toye KRC, Kaufman J, Lapauw B, Goemaere S. 2016. Validation of the FNIH sarcopenia criteria and SOF frailty index as predictors of long-term mortality in ambulatory older men. Age Ageing. 45(5):602–608. doi: 10.1093/ageing/afw071. [DOI] [PubMed] [Google Scholar]
- de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers H-H, Cremers FPM. 1995. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 267(5198):685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- Diokmetzidou A, Tsikitis M, Nikouli S, Kloukina I, Tsoupri E, Papathanasiou S, Psarras S, Mavroidis M, Capetanaki Y. 2016. Strategies to study desmin in cardiac muscle and culture systems. Meth Enzymol. 568:427–459. doi: 10.1016/bs.mie.2015.09.026. [DOI] [PubMed] [Google Scholar]
- Dominov JA, Miller JB. 1996. POU homeodomain genes and myogenesis. Dev Genet. 19(2):108–118. doi:. [DOI] [PubMed] [Google Scholar]
- Dummler BA, Hauge C, Silber J, Yntema HG, Kruse LS, Kofoed B, Hemmings BA, Alessi DR, Frödin M. 2005. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J Biol Chem. 280(14):13304–13314. doi: 10.1074/jbc.M408194200. [DOI] [PubMed] [Google Scholar]
- Dwyer CM, Stickland NC. 1994. Supplementation of a restricted maternal diet with protein or carbohydrate alone prevents a reduction in fetal muscle fibre number in the guinea-pig. Br J Nutr. 72(2):173–180. doi: 10.1079/BJN19940022. [DOI] [PubMed] [Google Scholar]
- Flint J, Eskin E. 2012. Genome-wide association studies in mice. Nat Rev Genet. 13(11):807–817. doi: 10.1038/nrg3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Ward SR, Bremner SN, Lieber RL. 2008. Quantitative analysis of neonatal skeletal muscle functional improvement in the mouse. J Exp Biol. 211(6):837–843. doi: 10.1242/jeb.014340. [DOI] [PubMed] [Google Scholar]
- Hernandez Cordero AI, Gonzales NM, Parker CC, Sokolof G, Vandenbergh DJ, Cheng R, Abney M, Sko A, Douglas A, Palmer AA, et al. . 2019. Genome-wide associations reveal human-mouse genetic convergence and modifiers of myogenesis, CPNE1 and STC2. Am J Hum Genet. 105(6):1222–1236. doi: 10.1016/j.ajhg.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jat PS, Noble MD, Ataliotis P, Tanaka Y, Yannoutsos N, Larsen L, Kioussis D. 1991. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc Natl Acad Sci U S A. 88(12):5096–5100. doi: 10.1073/pnas.88.12.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara R, Seckl MJ, Pardo OE. 2013. The p90 RSK family members: common functions and isoform specificity. Cancer Res. 73(17):5301–5308. doi: 10.1158/0008-5472.CAN-12-4448. [DOI] [PubMed] [Google Scholar]
- Lexell J, Taylor CC, Sjostrom M. 1988. What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. J Neurol Sci. 84(2-3):275–294. doi: 10.1016/0022-510X(88)90132-3. [DOI] [PubMed] [Google Scholar]
- Liang C, Park AY, Guan J. 2007. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Hernandez Cordero AI, Kilikevicius A, Carroll AM, Bewick GS, Bunger L, Ratkevicius A, Heisler LK, Harboe M, Oxvig C. 2023. Stanniocalcin-2 inhibits skeletal muscle growth and is upregulated in functional overload-induced hypertrophy. Physiol Rep. 11(15):e15793. doi: 10.14814/phy2.15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall JD, Sale DG, Alway SE, Sutton JR. 1984. Muscle fiber number in biceps brachii in bodybuilders and control subjects. J Appl Physiol Respir Environ Exerc Physiol. 57(5):1399–1403. doi: 10.1152/jappl.1984.57.5.1399. [DOI] [PubMed] [Google Scholar]
- Mathis JM, Simmons DM, He X, Swanson LW, Rosenfeld MG. 1992. Brain 4: a novel mammalian POU domain transcription factor exhibiting restricted brain-specific expression. EMBO J. 11(7):2551–2561. doi: 10.1002/j.1460-2075.1992.tb05320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. 1997. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387(6628):83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Millay DP, O'Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. 2013. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499(7458):301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muses S, Morgan JE, Wells DJ. 2011. A new extensively characterised conditionally immortal muscle cell-line for investigating therapeutic strategies in muscular dystrophies. PLoS One 6(9):e24826. doi: 10.1371/journal.pone.0024826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod J, Davies RW, Cai N, Hassett C, Goodstadt L, Cosgrove C, Yee BK, Lionikaite V, McIntyre RE, Remme CA, et al. . 2016. Genome-wide association of multiple complex traits in outbred mice by ultra low-coverage sequencing. Nat Genet. 48(8):912–918. doi: 10.1038/ng.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontell M, Hughes D, Bourke D. 1988. Morphometric analysis of the developing mouse soleus muscle. Am J Anat. 181(3):279–288. doi: 10.1002/aja.1001810306. [DOI] [PubMed] [Google Scholar]
- Parker CC, Gopalakrishnan S, Carbonetto P, Gonzales NM, Leung E, Park YJ, Aryee E, Davis J, Blizard DA, Ackert-Bicknell CL, et al. . 2016. Genome-wide association study of behavioral, physiological and gene expression traits in commercially available outbred CFW mice. Nat Genet. 48(8):919–926. doi: 10.1038/ng.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan J, Barceló C, Moreno DF, Maiques O, Sisó P, Marti RM, Macià A, Panosa A. 2019. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front Cell Dev Biol. 7:107. doi: 10.3389/fcell.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo Y, Zhang X, Roux PP. 2012. Regulation and function of the RSK family of protein kinases. Biochem J. 441(2):553–569. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. 1993. Myod or Myf-5 is required for the formation of skeletal muscle. Cell 75(7):1351–1359. doi: 10.1016/0092-8674(93)90621-V. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71(3):383–390. doi: 10.1016/0092-8674(92)90508-A. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. 2011. Fiber types in mammalian skeletal muscles. Physiol Rev. 91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell 102(6):777–786. doi: 10.1016/S0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Silventoinen K, Magnusson PK, Tynelius P, Kaprio J, Rasmussen F. 2008. Heritability of body size and muscle strength in young adulthood: a study of one million Swedish men. Genet Epidemiol. 32(4):341–349. doi: 10.1002/gepi.20308. [DOI] [PubMed] [Google Scholar]
- Xu J, Jia Q, Zhang Y, Yuan Y, Xu T, Yu K, Chai J, Wang K, Chen L, Xiao T, et al. . 2021. Prominent roles of ribosomal S6 kinase 4 (RSK4) in cancer. Pathol Res Pract. 219:153374. doi: 10.1016/j.prp.2021.153374. [DOI] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270(5639):725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The H2Kb cell clones are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.