Abstract
Heparan sulfate (HS) is a linear polysaccharide that plays a key role in cellular signaling networks. HS functions are regulated by its 6-O-sulfation, which is catalyzed by three HS 6-O-sulfotransferases (HS6STs). Notably, HS6ST2 is mainly expressed in the brain and HS6ST2 mutations are linked to brain disorders, but the underlying mechanisms remain poorly understood. To determine the role of Hs6st2 in the brain, we carried out a series of molecular and behavioral assessments on Hs6st2 knockout mice. We first carried out strong anion exchange-high performance liquid chromatography and found that knockout of Hs6st2 moderately decreases HS 6-O-sulfation levels in the brain. We then assessed body weights and found that Hs6st2 knockout mice exhibit increased body weight, which is associated with abnormal metabolic pathways. We also performed behavioral tests and found that Hs6st2 knockout mice showed memory deficits, which recapitulate patient clinical symptoms. To determine the molecular mechanisms underlying the memory deficits, we used RNA sequencing to examine transcriptomes in two memory-related brain regions, the hippocampus and cerebral cortex. We found that knockout of Hs6st2 impairs transcriptome in the hippocampus, but only mildly in the cerebral cortex. Furthermore, the transcriptome changes in the hippocampus are enriched in dendrite and synapse pathways. We also found that knockout of Hs6st2 decreases HS levels and impairs dendritic spines in hippocampal CA1 pyramidal neurons. Taken together, our study provides novel molecular and behavioral insights into the role of Hs6st2 in the brain, which facilitates a better understanding of HS6ST2 and HS-linked brain disorders.
Keywords: Dendritic spine, heparan sulfate, heparan sulfate 6-O-sulfation, HS6ST2, memory
Introduction
Heparan sulfate (HS) is a linear polysaccharide that presents at the cell surface and extracellular matrix of animal cells. HS can interact with hundreds of proteins and acts as a key regulator of the cellular signaling network (Xu and Esko 2014). HS 6-O-sulfation, a chemical modification on HS, regulates the interactions between HS and its binding proteins to impact HS biological functions (Xu and Esko 2014). HS 6-O-sulfation is catalyzed by three HS 6-O-sulfotransferases (HS6ST1, HS6ST2, and HS6ST3) in humans and mice.
HS plays a critical role in the brain, a tissue that carries out its function largely depending on the extracellular and cell surface signaling. Notably, mutations in HS genes cause multiple brain disorders in humans. For instance, mutations in exostosin glycosyltransferase 1 (EXT1), which encodes an enzyme that is indispensable for HS biosynthesis, cause autism, intellectual disability, epilepsy, and brain abnormalities in patients (Li et al. 2002; Wuyts et al. 2002; Maas et al. 2015). Similarly, mutations in EXT2 cause developmental delay, intellectual disability, autism, seizures, macrocephaly, neurological regression, and behavioral issues in patients (Farhan et al. 2015; El-Bazzal et al. 2019; Gupta et al. 2019). Furthermore, a single nucleotide polymorphism of N-deacetylase/N-sulfotransferase 3 (NDST3), which encodes an enzyme that catalyzes N-deacetylation and N-sulfation of HS, is associated with schizophrenia and bipolar disorder (Lencz et al. 2013; Zhang et al. 2016). In addition, knockout of Ext1 in mice resulted in axon guidance deficits, brain development deficits, social interaction impairments, and repetitive behavior (Inatani et al. 2003; Irie et al. 2012). HS is also essential for neuronal patterning and synaptogenesis (Saied-Santiago and Bulow 2018; Zhang et al. 2018). However, despite these advances, the role and molecular mechanisms of HS in the mammalian brain are not fully understood.
HS6STs also play an important role in the brain. Mutations in HS6ST1 cause Kallmann syndrome (Tornberg et al. 2011), a disorder that is mainly caused by deficits in the gonadotropin-releasing hormone expressing neurons and olfactory neurons (Schwanzel-Fukuda et al. 1989; Truwit et al. 1993; Yousem et al. 1993). In addition, Hs6st genes have been shown to play an essential role in axon guidance and embryonic brain development in mice (Pratt et al. 2006; Conway et al. 2011; Clegg et al. 2014; Tillo et al. 2016). However, except for these studies in the embryonic brain, the role and molecular mechanisms of HS6STs and HS 6-O-sulfation in the adult mammalian brain remain unknown.
HS6ST2 is an X-linked gene that is mainly expressed in the brain (Moon and Zhao 2021). Mutations in HS6ST2 cause intellectual disability, adaptive behavior, and severe neurodevelopmental impairment in human patients (Paganini et al. 2019). HS6ST2 mutations are also associated with neuroticism (Luciano et al. 2021). However, despite these genetic associations, the role and molecular mechanisms of HS6ST2 in the brain remain largely unknown. HS6ST2 mRNA has two splicing isoforms, a long isoform and a short isoform. Although a study using the Northern blot approach reported that the long isoform is enriched in the brain (Habuchi et al. 2003), the tissue-specific expression patterns of the two isoforms and their quantification need further investigation.
In this study, we analyzed the splicing patterns of Hs6st2 in different mouse tissues and investigated the protein structures of the long and short Hs6st2 isoforms using machine learning-based predictions. To determine the role of Hs6st2 in the adult mouse brain, we carried out a series of molecular and behavioral assessments of the Hs6st2 knockout mice (KO), including strong anion exchange-high performance liquid chromatography (SAX-HPLC), behavioral tests, RNA sequencing (RNA-seq), immunohistochemistry, and microscopy. We found that male Hs6st2 KO mice exhibit decreased HS 6-O-sulfation levels, increased body weight, behavioral deficits, impaired hippocampal transcriptome, and impaired hippocampal dendritic spines. Together, our study demonstrates the role of Hs6st2 in the adult mouse brain, which facilitates a better understanding of HS6STs and HS-linked brain disorders.
Results
Brain specifically expresses the long isoform of Hs6st2
To quantitatively assess the abundance of the long and short mRNA isoforms of Hs6st2 among tissues (Fig. 1A), we analyzed the splicing patterns of Hs6st2 from high-throughput RNA-seq data that were generated from different tissues in adult mice (Consortium 2012; Moon and Zhao 2021). We selected the cerebral cortex, spleen, ovary, and adrenal gland, because our previous study showed that Hs6st2 is specifically expressed in these tissues in adult mice (Moon and Zhao 2021). We used RNA-seq data because RNA-seq technology can unbiasedly capture mRNA transcripts spanning exon-exon junctions, which are the direct evidence for exon splicing and mRNA isoform detection. Together, we analyzed 1.4 billion sequencing reads from the four adult mouse tissues. To assess splicing patterns, we also developed a computational pipeline to identify and quantify exon-exon junction reads (Fig. 1A).
Fig. 1.
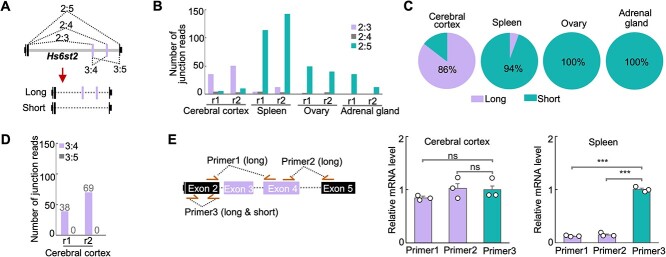
Brain specifically expresses the long isoform of Hs6st2. A) Diagrams showing the gene structure and the two isoforms of Hs6st2 mRNA. The dashed lines with numbers represent the junction reads spanning two exons. B) Bar plot showing the numbers of junction reads from the mouse RNA-seq data. r1, replicate 1. C) Pie charts showing the percentages of Hs6st2 long and short mRNA isoforms in four mouse tissues. D) Bar plot showing the numbers of junction reads joining exon 3 to exon 4 (3:4) and to exon 5 (3:5) in the cerebral cortex. E) qPCR results of the expression levels of the long isoform and short isoform of Hs6st2. Statistics: numbers on the line, P-values of two-tailed t-test; ns, P-value > 0.05; ***, P-value < 0.001.
To quantify the abundance of the long and short isoforms of Hs6st2, we calculated the number of RNA-seq junction reads that join exon 2 to exon 3 (2:3, long isoform) and to exon 5 (2:5, short isoform) (Fig. 1A). We found that 80% of the exon 2 junction reads are 2:3 in the cerebral cortex, while only 13% are 2:5 in the same tissue (Fig. 1B and Supplementary Fig. S1A). These results suggest that the long isoform of Hs6st2 is the major isoform (86%) in the brain (Fig. 1C). In contrast, 92%, 96%, and 100% of the exon 2 junction reads are 2:5 in the spleen, ovary, and adrenal gland (Fig. 1B and Supplementary Fig. S1A), suggesting that the short isoform of Hs6st2 is the major isoform (>94%) in these tissues (Fig. 1C). Taken together, these results demonstrate that the brain specifically expresses the long isoform of Hs6st2, while the non-brain tissues preferentially express the short isoform.
Given that both exon 3 and exon 4 of Hs6st2 are alternatively spliced, we next asked to what extent the two exons are included simultaneously in the Hs6st2 long isoform. To determine the extent to which exon 3 is included in the long isoform, we compared the numbers of junction reads that join exon 2 to exon 3 (2:3) and to exon 4 (2:4) (Fig. 1A). We found that the numbers of 2:3 junction reads are 13.8-fold higher than the numbers of 2:4 junction reads (Fig. 1B and Supplementary Fig. S1A) from the cerebral cortex RNA-seq data, suggesting that exon 3 is extensively included (>93%) in the long isoform and rarely has been skipped. To determine the extent to which exon 4 is included in the long isoform, we compared the numbers of junction reads that join exon 3 to exon 4 (3:4) and to exon 5 (3:5) (Fig. 1A). We identified 107 3:4 junction reads and zero 3:5 junction reads (Fig. 1D and Supplementary Fig. S1A) from the cerebral cortex RNA-seq data, suggesting that exon 4 is always included in the long isoform. Taken together, these results demonstrate that the long isoform of Hs6st2 contains both exon 3 and exon 4.
To validate the expression pattern of the long and short isoforms of Hs6st2, we next carried out isoform-specific real-time quantitative reverse transcription PCR (qPCR) to examine the expression levels of the two isoforms in the cerebral cortex and spleen. We designed three pairs of qPCR primers. Two of them, Primer1 and Primer2, are specific to the long isoform of Hs6st2, while Primer3 is for both the long and short isoforms (Fig. 1E). We found that the expression levels of the long isoform were high in the cerebral cortex but low in the spleen (Fig. 1E, Primer1, 2), whereas the expression levels of the short isoform were high in the spleen but low in the cerebral cortex (Fig. 1E, Primer3). These results suggest that the Hs6st2 long isoform is the major isoform in the cerebral cortex and the Hs6st2 short isoform is the major isoform in the spleen (Fig. 1E), which are consistent with the RNA-seq results (Fig. 1B–D). Together, these results indicate that the mouse brain specifically expresses the long isoform of Hs6st2.
Hs6st2 long isoform retains the sulfotransferase domain and may acquire a novel sub-structure
To determine the functional differences between the long and short isoforms of Hs6st2 proteins, we investigated their protein structures. We used AlphaFold2 (Jumper et al. 2021), a machine learning-based software, to predict the structures of the short and long isoforms of the Hs6st2 proteins. We found that the short isoform of Hs6st2 protein contains a transmembrane region and a sulfotransferase domain (Supplementary Fig. S2A). Notably, the structure, hydrophobicity, and electrostatic potential of the sulfotransferase domain of the Hs6st2 short isoform (Supplementary Fig. S2A) are similar to that of the sulfotransferase domains in zebrafish Hs6st3 (Xu et al. 2017) and in human HS6ST1 (Moon and Zhao 2022), suggesting that the enzymatic activities of HS 6-O-sulfotransferases are well conserved. We next used the AlphaFold2 to predict the protein structure of the Hs6st2 long isoform (Supplementary Fig. S2B). We found that the transmembrane region and the sulfotransferase domain are highly similar between the Hs6st2 short and long isoforms (Supplementary Fig. S2), suggesting that both protein isoforms may have similar cellular localization and enzymatic activities. The Hs6st2 long isoform contains 40 more amino acids compared to the short isoform. Intriguingly, these 40 amino acids, although localizing within the sulfotransferase domain, form a unique sub-structure (indicated by the black arrows in Supplementary Fig. S2B) without disrupting the sulfotransferase domain, which is consistent with a previous report that the Hs6st2 long isoform has the HS 6-O-sulfotransferase activity (Habuchi et al. 2003). Notably, this sub-structure of the extended loop, which is predicted by AlphaFold2, needs further validation using experimental approaches such as crystallography. Taken together, these results suggest that Hs6st2 long isoform retains the sulfotransferase domain and may acquire a novel sub-structure.
Knockout of Hs6st2 gene in mice
Despite the genetic link between HS6ST2 mutations and human brain disorders such as intellectual disability (Paganini et al. 2019), the role and molecular mechanisms of HS6ST2 in the adult mammalian brain remain unknown and could be advanced using a mouse model. Therefore, we used frozen sperm and the cryo-recovery approach to re-generate the Hs6st2 knockout (KO) mice, which were originally generated by the Koji Kimata laboratory (Sugaya et al. 2008). In this KO mouse line, the exon 2 of Hs6st2 is deleted (Fig. 2A), which removes DNA sequences that encode amino acids spanning the two critical domains in Hs6st2 protein (green rectangle in Fig. 2A). We only focused on male mice because all the known patients carrying HS6ST2 mutations are male.
Fig. 2.
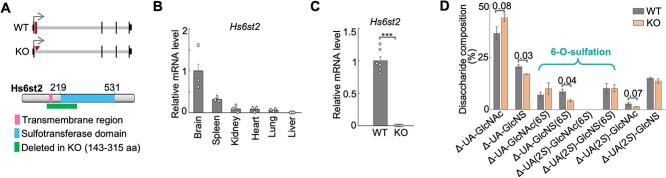
Knockout of Hs6st2 gene in mice. A) Diagrams showing the Hs6st2 gene structures of WT and KO mice (top) and the protein domains of WT Hs6st2 (bottom). Exon 2 is shown as a rectangle and is deleted in KO (top). The rectangle at bottom represents the amino acid (aa) sequences encoded by exon 2 (143-315 aa). B) qPCR results showing the mRNA levels of Hs6st2 (n = 3) in six tissues in 9-week-old WT mice. Brain expression was normalized to one. C) qPCR results showing the mRNA levels of Hs6st2 in the brain of 14-week-old WT (n = 6) and KO (n = 6) mice. ***, P-value < 0.001, one-tailed t-test. D) Bar plot showing the percentages of disaccharides in the brain of Hs6st2 KO mice and WT littermates. Numbers on the lines, P-values of one-tailed t-test.
To validate the KO mouse line, we cloned the corresponding DNA sequences from the KO mice and carried out Sanger sequencing. We found the joining of the upstream and downstream introns of the Hs6st2 exon 2 (Supplementary Fig. S1B), thus confirming the deletion of the exon 2 genomic sequences from the genome of the KO mice.
To determine which tissues were directly affected by the Hs6st2 KO, we first assessed the expression levels of Hs6st2 in different tissues in adult mice. We performed qPCR from six tissues of 9-week-old male wild type (WT) mice. In agreement with our previous findings using RNA-seq (Moon and Zhao 2021), Hs6st2 was highly expressed in the brain, moderately expressed in the spleen, and very lowly expressed in the other four tissues (Fig. 2B). These expression profiles suggest that knockout of Hs6st2 may primarily affect the brain. Therefore, we focused on the brain of the Hs6st2 KO mice in our subsequent investigation.
To determine the extent to which the Hs6st2 mRNAs were disrupted in the KO mice, we performed qPCR from the brains of 14-week-old male mice. We found that Hs6st2 mRNA levels were decreased by 99% in KO mice compared to their WT littermates (Fig. 2C), suggesting that Hs6st2 mRNAs are almost eliminated in the KO mice. However, we were unable to measure Hs6st2 protein levels, because none of the commercial antibodies we tested gave a specific band to Hs6st2 in Western blotting (data not shown). Taken together, these results demonstrate that Hs6st2 mRNAs are disrupted in the Hs6st2 KO mice.
Knockout of Hs6st2 in mice decreases HS 6-O-sulfation levels in the brain
To determine the role of Hs6st2 KO on HS 6-O-sulfation in the brain, we used the strong anion exchange-high performance liquid chromatography (SAX-HPLC) to measure the levels of HS 6-O-sulfation (Supplementary Fig. S3A). Briefly, glycosaminoglycans (GAGs) were isolated from the brain of 14-week-old WT and Hs6st2 KO mice and were digested into disaccharides by heparinase. The disaccharides were then separated by SAX-HPLC, and the eluents were analyzed by post-column derivatization to obtain the disaccharide components (Supplementary Fig. S3A and B). We found that Hs6st2 KO mice showed a significant decrease in the 6-O-sulfation levels on Δ-UA-GlcNS(6S), while the 6-O-sulfation levels on Δ-UA-GlcNAc(6S) and Δ-UA(2S)-GlcNS(6S) were well maintained (Fig. 2D). In addition, we found an increase in Δ-UA-GlcNAc, a decrease in Δ-UA-GlcNS, and a decrease in Δ-UA(2S)-GlcNAc in the KO mice (Fig. 2D). Notably, the 2-O-sulfation levels on Δ-UA(2S)-GlcNS were well maintained in the brains of Hs6st2 KO mice. Taken together, these results demonstrate that knockout of Hs6st2 decreases HS 6-O-sulfation levels on Δ-UA-GlcNS(6S) in the brain.
Hs6st2 KO mice exhibit higher body weight, normal food intake, and abnormal metabolic pathways
During our handling of the mice, we noticed that Hs6st2 KO mice were bigger compared to their WT littermates. To further investigate this phenotype, we carried out a systematic assessment of body weight for WT and KO mice from four weeks of age to 14 weeks of age (Fig. 3A). We found that the KO mice exhibited higher body weight compared to their WT littermates starting from five weeks of age (Fig. 3A). Notably, our findings of body weight are consistent with a previous study, which also found an increase of body weight in male Hs6st2 null mice (Nagai et al. 2013).
Fig. 3.
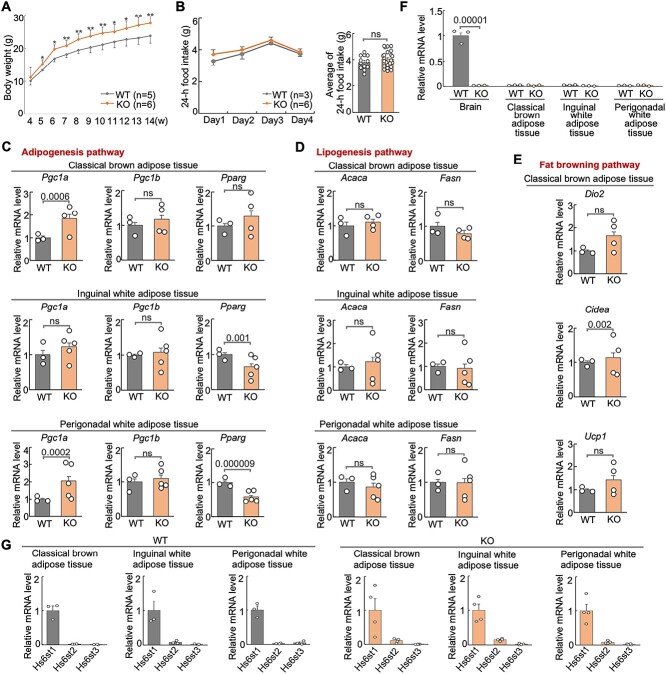
Hs6st2 KO mice show higher body weight, normal food intake, and abnormal metabolic pathways. A) Body weight of WT and KO mice. B) Food intake of WT and KO mice. C–E) qPCR results of the expression of genes in three metabolic pathways in the three adipose tissues. F) qPCR results of the expression of Hs6st2 in brain and three adipose tissues in WT and KO mice. G) qPCR results of the expression of Hs6st1, Hs6st2, and Hs6st3 in the three adipose tissues in WT and KO mice. Statistics: numbers on the line, P-values of two-tailed t-test. ns, P-value > 0.05, two-tailed t-test.
To determine the mechanisms underlying higher body weight, we first tested whether Hs6st2 KO mice consume more food than their WT littermates. We measured 24-h food intake of WT and KO mice for four consecutive days. As a result, we found that there was no difference in food intake between WT and KO mice (Fig. 3B), suggesting that Hs6st2 KO mice consume a similar amount of food compared to their WT littermates.
We next asked whether metabolic pathways were dysregulated in Hs6st2 KO mice. To test this, we examined the gene expression of multiple key genes in the adipogenesis pathway, the lipogenesis pathway, and the fat browning pathway in three types of fat tissues, including the classical brown adipose tissue (BAT), the inguinal white adipose tissue (iWAT), and the perigonadal white adipose tissue (pWAT) (Fig. 3C–E). For the three genes in the adipogenesis pathway, Pgc1a is upregulated in the BAT and pWAT in KO mice, while Pparg is downregulated in iWAT and pWAT (Fig. 3C). The two genes in the lipogenesis pathway showed no expression changes in KO mice in the three adipose tissues (Fig. 3D). For the three genes in the fat browning pathway, Cidea was upregulated in BAT in KO mice; Dio2 and Ucp1 showed no expression changes (Fig. 3E). Notably, consistent with our previous study (Moon and Zhao 2021), Hs6st2 is not expressed in adipose tissues (Fig. 3F). In addition, based on the mRNA expression levels (Fig. 3G), we found that Hs6st1, not Hs6st2, is the major HS 6-O-sulfotransferase in the three adipose tissues. Therefore, dysregulation of metabolic pathways in adipose tissues in KO mice is likely a secondary effect of the Hs6st2 disruption in the brain. Taken together, these results demonstrate that Hs6st2 KO mice exhibit higher body weight, normal food intake, and abnormal metabolic pathways.
Hs6st2 KO mice exhibit a trend of hyperactivity
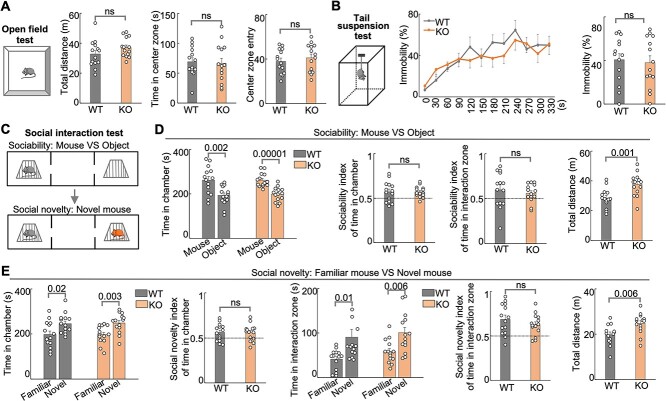
Individuals with HS6ST2 mutations present with severe psychomotor symptoms (Paganini et al. 2019). To determine the extent to which Hs6st2 KO mice recapitulate the patient-related symptoms, we carried out a series of mouse behavioral tests on the KO mice. First, we carried out the open field test (Fig. 4A, left panel), which assesses the general locomotor activity, exploratory behavior, and anxiety-like behavior in mice. We found that the KO mice showed a trend to travel a longer distance compared to their WT littermates (Fig. 4A, middle panel, P-value = 0.08), suggesting that Hs6st2 KO mice might be hyperactive. We also found that the KO and WT mice spent an equal amount of time in the center zone and entries to the center zone (Fig. 4A, right panel), indicating that Hs6st2 KO mice exhibit no anxiety-like behavior. Second, we carried out a tail suspension test (Fig. 4B, left panel), which assesses antidepressant-like behavior in mice. We found no difference between WT and KO mice (Fig. 4B), suggesting that Hs6st2 KO mice exhibit no impairment in antidepressant-like activity. In addition, to assess the social ability of Hs6st2 KO mice, we carried out the social interaction tests to determine the extent to which a mouse prefers a mouse over an object or prefers a novel mouse over a familiar mouse (Fig. 4C). We found no difference in the sociability test and social novelty test between Hs6st2 KO and WT mice (Fig. 4D and E and Supplementary Fig. S4A), suggesting that Hs6st2 KO mice have normal social ability. Notably, we found that Hs6st2 KO mice traveled a longer distance in both types of social interaction tests (Fig. 4D and E), suggesting that Hs6st2 KO mice are hyperactive. Together, these results indicate that Hs6st2 KO mice exhibit a trend of hyperactivity.
Fig. 4.
Male Hs6st2 KO mice (n = 14) exhibit hyperactivity compared to their WT littermates (n = 14). A) The open field test diagram, the total distance traveled, the time in the center zone, and the center zone entry. m, meter. s, second. B) The tail suspension test diagram (left), the immobility percentages per 30 s (middle), and the average immobility percentages (right). C–E) the social interaction test diagram (C), the sociability test results (D), and the social novelty test results (E). Statistics: numbers on the lines, P-values of two-tailed t-test. ns, P-value > 0.05, two-tailed t-test.
Hs6st2 KO mice exhibit memory deficits
Given that human HS6ST2 mutations are linked to intellectual disability (Paganini et al. 2019), we next carried out learning and memory-related behavioral tests on Hs6st2 KO mice, including Y-maze test, contextual fear conditioning test, and novel object recognition test.
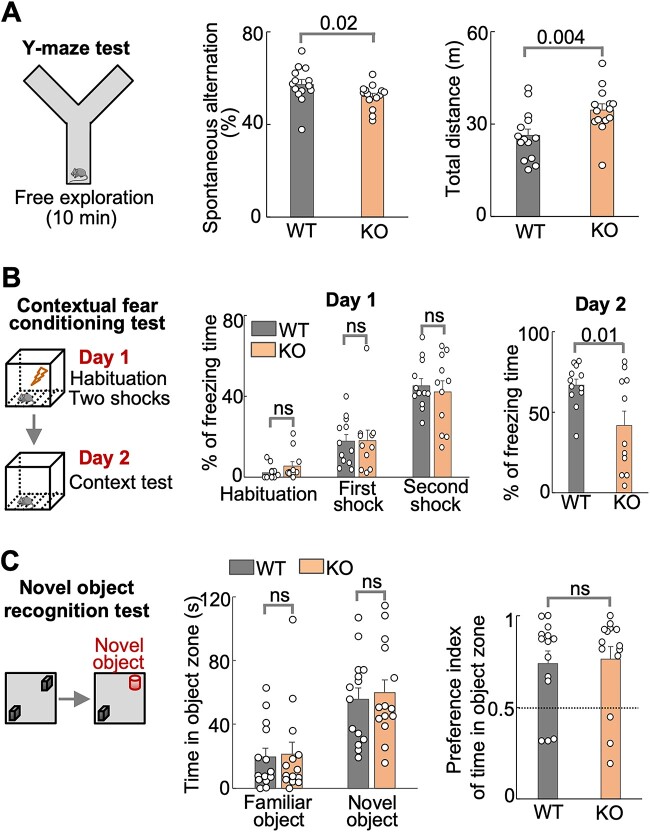
The Y-maze test assesses spatial working memory in mice (Fig. 5A, left panel). Mice normally remember the arm they recently visited and spontaneously visit the other arm. We found that Hs6st2 KO mice exhibited lower levels of spontaneous alternation in Y-maze test compared to their WT littermates (Fig. 5A and Supplementary Fig. S4B), suggesting that Hs6st2 KO mice have deficits in spatial working memory. We also found that KO mice traveled a longer distance compared to their WT littermates (Fig. 5A, right panel), further supporting that Hs6st2 KO mice might be hyperactive.
Fig. 5.
Male Hs6st2 KO mice (n = 14) exhibit memory-related deficits compared to their WT littermates (n = 14). A) The Y-maze test diagram (left), the spontaneous alternation (middle), and the total distance traveled (right). B) The contextual fear conditioning test diagram (left), the percentages of freezing times in day 1 (middle), and the percentages of freezing times in day 2 (right). C) The novel object recognition test diagram (left), the time in object zones (middle), and the discrimination indexes (right). Statistics: numbers on the lines, P-values of one-tailed t-test. ns, P-value > 0.05, one-tailed t-test.
The contextual fear conditioning test is a widely used behavioral paradigm to assess associative fear learning and memory in mice (Fig. 5B, left panel). Mice normally freeze when they are given an electronic foot shock, and they learn from this experience and will freeze more when given a second foot shock. We found that both WT and KO mice showed increased freezing time after the first and second shocks in the Day 1 test (Fig. 5B, middle panel), suggesting that WT and KO mice have normal ability in fear learning. For the second day, mice normally remember the chamber. When putting the mice into the same chamber, they will freeze. Intriguingly, in the Day 2 test, the KO mice exhibited less freezing time compared to their WT littermates (Fig. 5B, right panel), suggesting that Hs6st2 KO mice have deficits in associative fear memory.
The novel object recognition test is a commonly used behavioral test to assess short-term recognition memory in mice (Fig. 5C, left panel). Mice normally spend more time with the novel object compared to the familiar object. We found that both the WT and KO mice preferred the novel object over the familiar object (middle panel in Fig. 5C and Supplementary Fig. S4C). However, we found no difference between WT and KO mice in the time spent in the object zone (Fig. 5C, middle panel) or in discrimination indexes (Fig. 5C, right panel), suggesting that Hs6st2 KO mice have no deficits in short-term recognition memory. Notably, KO mice traveled a longer distance compared to their WT littermates in both tests (Supplementary Fig. S4C and D).
Taken together, these results suggest that Hs6st2 KO mice exhibit deficits in spatial working memory and associative fear memory, but not in short-term recognition memory.
Knockout of Hs6st2 alters transcriptome in the hippocampus
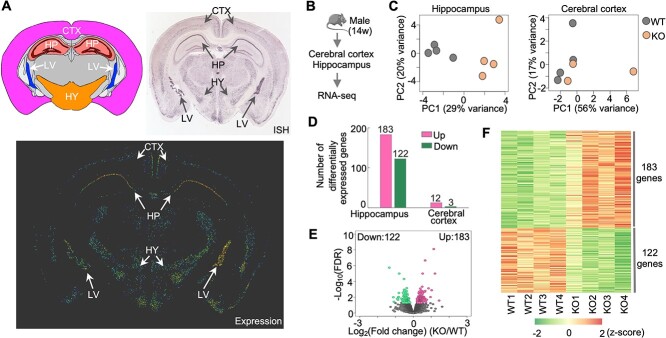
To determine the molecular mechanisms underlying the memory deficits in Hs6st2 KO mice, we first analyzed the expression profiles of Hs6st2 in adult mouse brains. Based on in situ hybridization (ISH) results from the Allen Mouse Brain Atlas (Lein et al. 2007), we found that Hs6st2 is highly expressed in the hippocampus, hypothalamus, lateral ventral, moderately expressed in the cerebral cortex, and not expressed in other brain regions (Fig. 6A). Given that the hippocampus and cerebral cortex are highly associated with memory, and to reveal the molecular mechanisms underlying memory deficits, we assessed transcriptome changes in the hippocampus and cerebral cortex of Hs6st2 KO mice. We extracted RNAs from the cerebral cortex and hippocampus of 14-week-old male KO mice and their WT littermates and assessed transcriptomes using RNA-seq (Fig. 6B). We obtained 912 million high-quality sequencing reads (Supplementary Fig. S5A), aligned them to the mouse genome, and quantified the numbers of reads mapped to the exonic regions of all mouse genes. We then performed a principal component analysis on the transcriptomes and found a clear separation of the WT and KO datasets, particularly in the hippocampus datasets (Fig. 6C). To identify genes that were affected by the Hs6st2 KO, we used the DESeq2 method (Love et al. 2014) to compare gene expression profiles between the WT and KO datasets. We found that 305 genes were differentially expressed (False Discovery Rate (FDR) < 0.05) in the hippocampus of Hs6st2 KO mice (Fig. 6D–F and Supplementary Table 1), suggesting that knockout of Hs6st2 alters transcriptome in the hippocampus. In contrast, only 15 genes were differentially expressed (FDR < 0.05) in the cerebral cortex (Fig. 6D and Supplementary Fig. S5B and C), indicating that the transcriptome of the cerebral cortex was well maintained in the Hs6st2 KO mice. Together, these results demonstrate that knockout of Hs6st2 alters transcriptome in the hippocampus.
Fig. 6.
Knockout of Hs6st2 alters transcriptome in the hippocampus. A) Hs6st2 expression profile in adult mouse brain measured by in situ hybridization (ISH). Allen mouse brain atlas, mouse.brain-map.org/gene/show/30274. CTX, cerebral cortex; HP, hippocampus; HY, hypothalamus; LV, lateral ventral. B) Experiment diagram. C) Principal component (PC) analysis results of hippocampus and cerebral cortex RNA-seq data. D) Numbers of upregulated genes (up) and downregulated genes (down) in the hippocampus and cerebral cortex of Hs6st2 KO mice. E) Volcano plot of hippocampus RNA-seq data. F) Heatmap of the 305 differentially expressed genes in the hippocampus.
No compensation of other HS biosynthesis enzymes and HSPG core proteins in Hs6st2 KO mice
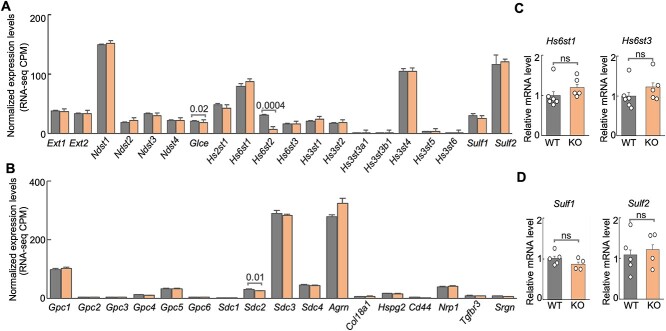
To determine the extent to which other HS-related genes may compensate for the knockout of Hs6st2 in mice, we analyzed the expression profiles of 20 genes encoding HS biosynthesis enzymes and 17 genes encoding HSPG core proteins in the hippocampus of WT and Hs6st2 KO mice. We first analyzed the expression levels of the 37 genes in our RNA-seq datasets. We found that none of these genes were upregulated in the Hs6st2 KO mice (Fig. 7A and B), suggesting that there is no compensation of other HS biosynthesis enzymes and HSPG core proteins for the knockout of Hs6st2 at the gene expression level. Notably, except for Hs6st2, we found that the expression levels of Glce and Sdc2 were also decreased in the KO mice (Fig. 7A and B). Furthermore, to independently validate our RNA-seq results, we used qPCR to measure the expression levels of Hs6st1 and Hs6st3 in the hippocampus of WT and Hs6st2 KO mice. We found no increase in gene expression for Hs6st1 and Hs6st3 (Fig. 7C), which is consistent with our RNA-seq results (Fig. 7A). We also used qPCR to measure the expression levels of Sulf1 and Sulf2, two genes encoding the HS 6-O-sulfation sulfatases, in the hippocampus of WT and Hs6st2 KO mice. We found no increase in gene expression for Sulf1 and Sulf2 (Fig. 7D), which is consistent with our RNA-seq results (Fig. 7A). Taken together, these results suggest that there is no gene expression-level compensation of other HS biosynthesis enzymes and HSPG core proteins for the knockout of Hs6st2.
Fig. 7.
Expression of genes encoding HS biosynthesis enzymes and HSPG core proteins in WT and Hs6st2 KO mice. A) Normalized expression levels of 20 genes encoding HS biosynthesis enzymes from our RNA-seq data. CPM, count per million unique mapped read pairs. B) Normalized expression levels of 17 genes encoding HSPG core proteins from our RNA-seq data. C) qPCR results of Hs6st1 and Hs6st3 in the hippocampus of WT and Hs6st2 KO mice. D) qPCR results of Sulf1 and Sulf2 in the hippocampus of WT and Hs6st2 KO mice. Statistics: numbers on the line, P-values of two-tailed t-test; ns, P-value > 0.05.
Knockout of Hs6st2 inhibits genes in the dendrite and synapse pathways and promotes genes in the ribosome and protein translation pathways
To determine the biological pathways affected by the Hs6st2 knockout, we performed DAVID pathway enrichment analysis (Huang et al. 2007) and gene set enrichment analysis (GSEA) (Subramanian et al. 2005) on the differentially expressed genes identified in the hippocampus.
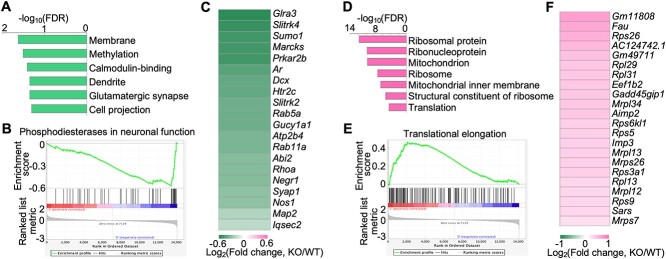
DAVID pathway enrichment analysis showed that the downregulated genes are enriched in neuronal pathways, such as dendrite, glutamatergic synapse, and cell projection (Fig. 8A). In addition, GSEA analysis also showed the downregulation of neuronal pathways such as phosphodiesterases in neuronal function (Fig. 8B). Notably, 20 of the differentially expressed genes in the hippocampus are involved in dendrite and glutamatergic synapse pathways. To further investigate these 20 genes, we analyzed their values of expression changes and found that all of them were downregulated in the hippocampus of Hs6st2 KO mice (Fig. 8C). Notably, the downregulation of neuronal pathways in the hippocampus may directly contribute to the memory deficits of Hs6st2 KO mice.
Fig. 8.
Knockout of Hs6st2 inhibits genes in the dendrite and synapse pathways and promotes genes in the ribosome and protein translation pathways in the hippocampus. A) The top enriched DAVID pathways of the downregulated genes in the hippocampus. B) GSEA results showing the downregulation of the phosphodiesterases in neuronal function pathway. C) Heatmap of the expression changes of the 20 differentially expressed genes that are involved in the dendrite and glutamatergic synapse pathways. D) The top enriched DAVID pathways of the upregulated genes in the hippocampus. E) GSEA results showing the upregulation of the translational elongation pathway. F) Heatmap of the expression changes of the 22 differentially expressed genes that are involved in the ribosome and protein translation pathways.
DAVID pathway enrichment analysis showed that the upregulated genes are enriched in ribosome pathways and protein translation pathways, such as ribosomal protein, ribonucleoprotein, ribosome, and translation (Fig. 8D). In addition, GSEA analysis also showed the upregulation of protein translation-related pathways such as translational elongation (Fig. 8E). Notably, 22 of the differentially expressed genes in the hippocampus are involved in ribosome and protein translation pathways. To further investigate these 22 genes, we analyzed their values of expression changes and found that all of them were upregulated in the hippocampus of Hs6st2 KO mice (Fig. 8F). Given that protein translation pathways in the hippocampus have long been shown to play a critical role in memory formation and consolidation (Miller et al. 2002; Sutton and Schuman 2006; Hernandez and Abel 2008; Santini et al. 2014; Cho et al. 2015; Conde et al. 2021), these changes in ribosome and protein translation pathways in Hs6st2 KO mice likely contribute to the memory-related deficits in these KO mice.
Together, these results indicate that knockout of Hs6st2 inhibits genes in the dendrite and synapse pathways and promotes genes in the ribosome and protein translation pathways in the hippocampus.
Knockout of Hs6st2 decreases HS levels and impairs dendritic spines of pyramidal neurons in the hippocampus CA1 region
To determine the mechanisms underlying Hs6st2 KO, we next examined in which cell type Hs6st2 is expressed in the mouse brain. We analyzed the expression levels of Hs6st2 in seven cell types in the mouse brain from the Brain RNA-Seq database (https://www.brainrnaseq.org/) (Zhang et al. 2014). We found that Hs6st2 is highly expressed in neurons and oligodendrocyte progenitor cells and is lowly expressed in astrocytes, newly formed oligodendrocytes, myelinating oligodendrocytes, microglia, and endothelial cells (Supplementary Fig. S6). The high expression levels of Hs6st2 in neurons indicate that knockout of Hs6st2 might directly affect neurons in the mouse brain. Given that Hs6st2 is highly expressed in neurons (Supplementary Fig. S6) and in the hippocampus CA1 region (Fig. 6A), we next focused on neurons in the hippocampus CA1 region, particularly the HS levels and dendritic spines.
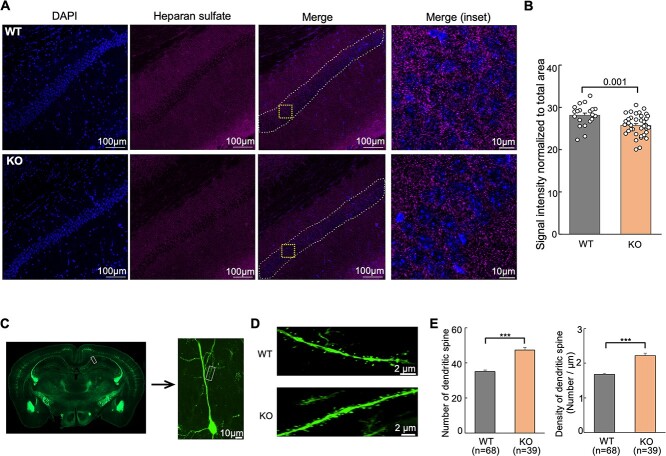
To further determine the extent to which knockout of Hs6st2 affects HS levels, we carried out immunohistochemistry experiments using an HS-specific antibody, 10E4. We focused on the hippocampal CA1 pyramidal neurons (Fig. 9A) because Hs6st2 is highly expressed in these neurons (Fig. 6A). Notably, we found a significant decrease in HS levels in hippocampal CA1 pyramidal neurons in the Hs6st2 KO mice (Fig. 9B), suggesting that knockout of Hs6st2 decreases HS levels in hippocampal CA1 pyramidal neurons.
Fig. 9.
Knockout of Hs6st2 decreases HS levels and impairs dendritic spines in hippocampal CA1 pyramidal neurons. A) Immunohistochemistry results using the anti-HS antibody, 10E4. The white dashed lines in the merge images indicate the quantified regions. The orange squares in the merge images indicate the merge (inset) regions. B) Quantification of immunohistochemistry results. C) Representative images of the hippocampus, a hippocampal CA1 pyramidal neuron, and an apical dendrite of the Thy1-YFP-H mice. D) Representative images of dendritic spines of hippocampal CA1 pyramidal neurons in the WT and Hs6st2 KO mice. E) Quantification of the numbers and densities of dendritic spines in hippocampal CA1 pyramidal neurons. Statistics: number on the line, P-value of two-tailed t-test; ***, P-value < 0.001, two-tailed t-test.
Given that our RNA-seq results showed that genes affected by the Hs6st2 KO are enriched in the dendrite and synapse pathways (Fig. 8A), we next investigated the changes of dendritic spines in hippocampal CA1 pyramidal neurons in the Hs6st2 KO mice. We crossed our Hs6st2 KO mice with Thy1-YFP-H transgenic mice (Feng et al. 2000), which express yellow fluorescent protein at high levels in selected pyramidal neurons in the hippocampus CA1 region (Fig. 9C) and have been widely used to characterize dendritic spine morphology (Lin et al. 2023). We focused our analyses on the apical dendrites of hippocampal CA1 pyramidal neurons (Fig. 9C) and measured the dendrite diameter, spine numbers, and spine densities in WT and Hs6st2 KO mice (Fig. 9D). We found no difference in the dendrite diameter between WT and Hs6st2 KO mice (Supplementary Fig. S7). Intriguingly, we found that the numbers and densities of dendritic spines in hippocampal CA1 pyramidal neurons are significantly higher in the Hs6st2 KO mice compared to the WT mice (Fig. 9E). Together, these results demonstrate that knockout of Hs6st2 decreases HS levels and impairs dendritic spines in hippocampal CA1 pyramidal neurons.
Discussion
Mutations in HS genes cause multiple brain disorders in humans, but the underlying molecular mechanisms remain largely unknown. In this study, we demonstrated that the brain specifically expresses the long isoform of Hs6st2, which agrees with a previous report (Habuchi et al. 2003). The long isoform of Hs6st2 encodes a protein that retains the sulfotransferase domain and may acquire a novel sub-structure. We also carried out a series of behavioral assessments on the Hs6st2 KO mice and found that male Hs6st2 KO mice exhibit higher body weight, hyperactivity, and memory deficits. To determine the underlying mechanisms, we first carried out SAX-HPLC and found moderately decreased levels of HS 6-O-sulfation in the brain. We also performed RNA-seq and identified transcriptome changes in the hippocampus, particularly the downregulation of genes in the dendrite and synapse pathways and the upregulation of genes in the ribosome and protein translation pathways, which likely contribute to the memory deficits. Furthermore, we carried out immunohistochemistry and microscopy experiments and found that knockout of Hs6st2 decreases HS levels and impairs dendritic spines in hippocampal CA1 pyramidal neurons. Together, our study for the first time demonstrates the role of Hs6st2 in the adult mammalian brain and provides new insights into the role of HS in brain health and disease.
The differences between the long and short Hs6st2 isoforms remain unknown. An early study reported that the human long and short HS6ST2 isoforms catalyzed the same 6-O-sulfation reaction but preferred different substrates (Habuchi et al. 2003). We also found that the Hs6st2 long isoform retains the sulfotransferase domain and may acquire a novel sub-structure predicted by AlphaFold2 (Supplementary Fig. S2). This novel sub-structure needs further experimental validation and has the potential to recognize or generate specific patterns or motifs on the HS chain. For instance, knockout of Hs6st2 in mice decreased HS 6-O-sulfation levels in Δ-UA(2S)-GlcNS(6S) in the ear and fat pad (Anower et al. 2013; Nagai et al. 2013), which express the short isoform, but not in the brain (Fig. 2D), which expresses the long isoform. These results indicate that disruption of Hs6st2 long and short isoforms may have different effects on HS 6-O-sulfation. In addition, the Hs6st2 long isoform may have other functions besides being a 6-O-sulfotransferase. Thus, further studies are needed to illustrate the precise biochemical properties of the Hs6st2 long isoform, such as the enzymatic activities, substrate specificity, and interacting proteins. However, due to the lack of specific antibodies, it is difficult to directly investigate the Hs6st2 short and long protein isoforms. To overcome this antibody limitation, one way is to ectopically express a tagged version of the two proteins followed by molecular characterization, which may help to reveal the differences between the two isoforms.
Knockout of Hs6st2 in mice only moderately decreased HS 6-O-sulfation levels in the brain, which is likely due to three potential reasons. First, Hs6st1 and Hs6st3 were stably expressed in Hs6st2 KO mice (Fig. 7A and C), which may help to maintain the HS 6-O-sulfation profiles in Hs6st2 KO mice. Similarly, the decreases of HS 6-O-sulfation levels in the brain of Hs6st1 KO were also moderate (Habuchi et al. 2007). Notably, double knockout of Hs6st1 and Hs6st2 in mouse embryonic fibroblasts and double knockout of HS6ST1 and HS6ST2 in human hepatoma cells significantly decreased HS 6-O-sulfation levels compared to the single knockout of the two genes (Sugaya et al. 2008; Anower et al. 2019). Second, brain is a highly heterogeneous organ, and Hs6st2 is specifically expressed in some brain regions (Fig. 6A). Thus, HS 6-O-sulfation levels in brain regions with high Hs6st2 expression may be significantly reduced in the KO mice, and such reduction might be diluted when using the whole brain for disaccharide analysis (Fig. 2D). Third, the long isoform of Hs6st2 may have other functions instead of HS 6-O-sulfotransferase. Therefore, further studies are needed to directly test these different possibilities.
Hs6st2 KO mice exhibit higher body weight (Fig. 3A). This increase in body weight is not due to food intake (Fig. 3B), but likely because of the abnormal metabolic pathways (Fig. 3C–E). A previous study also found that the increased body weight of Hs6st2 null mice was associated with impaired glucose metabolism, impaired insulin resistance, reduced energy metabolism in brown adipose tissue, higher levels of serum thyroid-stimulating hormone, and lower levels of serum thyroxine levels (Nagai et al. 2013). However, given that Hs6st2 is highly expressed in the brain but not expressed in the adipose tissues (Fig. 3F), defects in the brain are likely the primary effects of Hs6st2 knockout and the metabolic phenotypes are likely secondary effects. Notably, Hs6st2 is also expressed in the hypothalamus (Fig. 6A), a brain region that plays a critical role in glucose homeostasis, brown and white adipose tissue metabolism, long-term energy balance, and body weight (Coll et al. 2007; Zhang and Bi 2015; Timper and Bruning 2017). Therefore, it will be crucial to focus on the hypothalamus to further dissect these body weight-related phenotypes, such as hypothalamus-specific knockout of Hs6st2 in mice or re-expression of Hs6st2 in the hypothalamus of the Hs6st2 KO mice.
Knockout of HS genes in mice causes brain-related behavioral deficits. For instance, knockout of Ext1 in mouse forebrain excitatory neurons resulted in social interaction impairments and repetitive behavior (Irie et al. 2012); knockout of Ndst3 in mice resulted in reduced anxiety-related behavior (Pallerla et al. 2008); knockout of Sulf1 and Sulf2 in mice resulted in motor deficits (Aizawa et al. 2019). Here we show that Hs6st2 KO mice exhibit memory-related deficits. We found that these deficits are associated with transcriptome alternations in the hippocampus, particularly the downregulation of genes in the dendrite and synapse pathways (Fig. 8A–C). Furthermore, we found that knockout of Hs6st2 decreases HS levels and impairs dendritic spines in hippocampal CA1 pyramidal neurons (Fig. 9). We also found the upregulation of genes in ribosome pathways and protein translation pathways, which have long been shown to play a critical role in memory formation and consolidation (Miller et al. 2002; Cho et al. 2015; Conde et al. 2021). It remains unclear how Hs6st2 in the brain alters the transcriptome. One potential mechanism is via the FGF signaling, which is because FGF signaling has been shown as a major downstream signaling for HS 6-O-sulfation in the brain (Clegg et al. 2014; Chan et al. 2017; Yamada et al. 2017; Moon and Zhao 2022). In addition, FGFR mutations have been shown to promote ribosome pathways (Neben et al. 2014; Neben et al. 2017; Holmes et al. 2020). Therefore, it will be interesting to determine to what extent that knockout of Hs6st2 is via the FGF/FGFR signaling to inhibit the dendrite and synapse pathways and promote the ribosome and protein translation pathways.
This study also highlights multiple areas for further investigation. First, given that this study only examined male mice, the extent to which female KO mice show the same molecular and behavioral deficits remains unknown. Thus, further characterization of female Hs6st2 KO mice is required to comprehensively reveal the role of Hs6st2 in the brain. Second, brain-specific knockout of Hs6st2 in mice is needed to further dissect the behavioral deficits. In addition, it will be intriguing to determine the extent to which ectopic expression of Hs6st2 short and long isoforms in the hippocampus of Hs6st2 brain-specific knockout mice can restore the behavioral and molecular deficits. Third, compensation among Hs6st gene members is a confounding factor for single-knockout mice. Simultaneous knockout of all three Hs6st genes in the mouse brain is a better approach to further determine the role of HS 6-O-sulfation in the brain. Lastly, the detailed mechanisms underlying the memory deficits of Hs6st2 KO mice remain unclear. Given that our findings highlight the dendrite and synapse pathways (Figs 8 and 9), it will be intriguing to further characterize neurons and oligodendrocyte progenitor cells in Hs6st2 KO mice, particularly the hippocampus CA1 region. Collectively, these further experiments will help to reveal the molecular mechanisms of HS and HS 6-O-sulfation in the brain and may provide insights into potential treatments for HS and HS 6-O-sulfation associated brain disorders.
Materials and methods
Animals
The Hs6st2 KO mice in C57BL/6 genetic background were generated using the cryo-recovery approach. The frozen sperm from the Hs6st2−/y mice (Nagai et al. 2013) were obtained from Dr. Jeffrey Esko’s laboratory (University of California San Diego, USA) with the permission from Dr. Koji Kimata (Aichi Medical University, Japan). The Thy1-YFP-H transgenic mice were purchased from the Jackson Laboratory (Strain #:003782). All mice used in this study were maintained in the C57BL/6 genetic background. Mice were kept on a regular 12-hour (h) light/12-h dark cycle in a constant environment with ambient temperature of 21 ± 2 °C and humidity of 50 ± 10%. All animal studies, including handling and housing, were carried out in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals recommendations and according to the approved Institutional Animal Care and Use Committee (IACUC) protocol of the New York Institute of Technology (#2019-JZ-01A1).
RNA isolation and quality assessment
RNAs were isolated from the tissues of 9–17 weeks-old Hs6st2 KO mice and their WT littermates. The samples were snap-frozen using liquid nitrogen and stored at −80 °C until use. The Trizol reagent (Invitrogen, #15596026) was used to extract total RNAs following the manufacturer’s instruction. The Nanodrop spectrometer and the Bioanalyzer (Agilent 2100) using RNA 6,000 Nano Chip were used to assess the quality, concentration, and integrity of the isolated RNAs. RNA samples with a RIN value of eight or higher were included for the downstream experiments.
RNA-seq library preparation, quality control, and high-throughput sequencing
RNA-seq libraries were prepared according to the manufacturer instructions. The Stranded Total RNA Library Prep kit (Zymo, #R3003) was used to generate RNA-seq libraries for the cerebral cortex samples. The Invitrogen™ Collibri™ Stranded RNA Library Prep Kit for Illumina™ Systems (Thermo Fisher Scientific, # A38994096) was used to generate RNA-seq libraries for the hippocampus samples. 1,000 ng of total RNAs were used to prepare the RNA-seq libraries. The Bioanalyzer (Agilent 2100) using DNA 7,500 chip was used to assess the quality of the RNA-seq libraries to make sure that the fragment sizes of all libraries were within the range of 300–500 bp. The Illumina NovaSeq 6,000 sequencer was used to sequence the RNA-seq libraries.
RNA-seq alignment, quantification, and analyses
The detailed steps of the RNA-seq data analyses were described previously (Johnson et al. 2017; Zhao et al. 2018; Moon and Zhao 2022). Briefly, STAR 2.7.7a (Dobin et al. 2013) was used to align the RNA-seq FASTQ files to the mouse mm10 genome using the parameters of “--runThreadN 40 --outFilterMultimapNmax 1 --outFilterMismatchNmax 3 --outFilterScoreMinOverLread 0.25 --outFilterMatchNminOverLread 0.25”. A Perl script was used to quantify the number of reads aligned to the exons of each gene. The raw read counts for RNA-seq data were normalized, processed, and compared using DESeq2 (Love et al. 2014). Briefly, the table of read counts was imported into R into a data.frame using the read.table command. The data.frame was then converted into a DESeqDataSet data object by the DESeqDataSetFromMatrix command. Lowly expressed genes (less than 10 read counts in four or more samples) were excluded from the downstream analysis. The DESeq command was used to control features for estimating size factors using the estimateSizeFactors command, to estimate the dispersions using the estimateDispersions command, to fit a Negative Binomial GLM model, and to compare gene expression levels using the nbinomWaldTest command and the Wald significance tests. The cutoff for a significant difference in gene expression was False Discovery Rate (FDR, adjusted P-value) < 0.05. The CPM values for GSEA analysis were obtained using the median of ratios method by the command of “round(fpm(dds, robust = TRUE),3)”. The regularized log transformation and the rlog command were used to transform the DESeqDataSet data object into log2 scale, to minimize differences between samples for rows with small counts, and to normalize to library size. The transformed data were used to perform the principal component analysis (PCA) to check the similarity among samples using the plotPCA command and the top 500 highly variable genes. The script is available at our GitHub repository (https://github.com/Jerry-Zhao/Hs6st2KO2022).
Identification of exon-exon junction reads for Hs6st2
The RNA-seq SAM files were used to identify exon-exon junction reads for Hs6st2. The CIGAR strings in SAM files were used to identify alignment locations based on the left and right mapping loci of a gap in the X chromosome. Specifically, Left=51471407 and Right=51679890 are for the exon2:exon3 junction reads; Left=51428745 and Right=51679890 are for the exon2:exon4 junction reads; Left=51390362 and Right=51679890 are for the exon2:exon5 junction reads; Left=51428745 and Right= 51471374 are for the exon3:exon4 junction reads; Left= 51390362 and Right=51471374 are for the exon3:exon5 junction reads; and Left=51390362 and Right=51428658 are for the exon4:exon5 junction reads. The script is available from our GitHub repository (https://github.com/Jerry-Zhao/Hs6st2KO2022).
Protein structure prediction and visualization
The AlphaFold v2.0 (Jumper et al. 2021) was used to predict the protein structures of the mouse Hs6st2 short isoform and long isoform. The amino acid sequences of the short and long Hs6st2 protein isoforms were obtained from the National Center for Biotechnology Information (NCBI) database by the accession numbers of NP_001277396.1 and NP_001070670.1. The AlphaFold docker was installed, and the default parameters were used except for the “-max_template_date = 2020-05-14”. The “ranked_0.pdb” protein structure was selected and visualized using the UCSF Chimera (Pettersen et al. 2004).
Real-time quantitative reverse transcription PCR (qPCR) and quantification
The Prime script reverse transcription kit (Takara, #RR037A) was used to synthesize cDNAs from the isolated RNAs. The SYBR Green PCR Master Mix (Applied Biosystems, #4309155) was used to carry out the real-time PCR. The endogenous reference was 18S rRNA. The primers used were listed as follows: Hs6st2 forward, 5′-CCTGGTGCGCAACATCC-3′; Hs6st2 reverse, 5′-TGCATTTCTTCTGCCCCAC-3′; Hs6st1 forward, 5′-TGGACCGAACTCACCAACTGTG-3′; Hs6st1 reverse, 5′-CATTCACTCAGGTAGCGGGATAC-3′; Hs6st3 forward, 5′-CTGGACCGAGCTCACCAAC-3′; Hs6st3 reverse, 5′-CGTCGCACATATGGAGAGAGGT-3′; Pgc1a forward, 5′-CCCTGCCATTGTTAAGACC-3′; Pgc1a reverse, 5′-TGCTGCTGTTCCTGTTTTC-3′; Pgc1b forward, 5′-CAGGGTGGGGACTCTGGA-3′; Pgc1b reverse, 5′-AGTCAAAGTCACTGGCGTCC-3′; Pparg forward, 5′-TGTTATGGGTGAAACTCTGGG-3′; Pparg reverse, 5′-AGAGCTGATTCCGAAGTTGG-3′; Acaca forward, 5′-AAGGCTATGTGAAGGATGTGG-3′; Acaca reverse, 5′-CTGTCTGAAGAGGTTAGGGAAG-3′; Fasn forward, 5′-CCCCTCTGTTAATTGGCTCC-3′; Fasn reverse, 5′-TTGTGGAAGTGCAGGTTAGG-3′; Cidea forward, 5′-GAATAGCCAGAGTCACCTTCG-3′; Cidea reverse, 5′-AGCAGATTCCTTAACACGGC-3′; Dio2 forward, 5′-CCTCCTAGATGCCTACAAACAG-3′; Dio2 reverse, 5′-TGATTCAGGATTGGAGACGTG-3′; Ucp1 forward, 5′-TCTCTGCCAGGACAGTACCC-3′; Ucp1 reverse, 5′-AGAAGCCACAAACCCTTTGA-3′; 18S rRNA forward, 5′-GTAACCCGTTGAACCCCATT-3′; 18S rRNA reverse, 5′-CCATCCAATCGGTAGTAGCG-3′; Hs6st2 Primer1 forward, 5′-CGGCGGTGGTGGATGGCAAG-3′; Hs6st2 Primer1 reverse, 5′-GGCTTTGTGGAGGATGGAGAGTTGG-3′; Hs6st2 Primer2 forward, 5′-ACTCTCCATCCTCCACAAAGCC-3′; Hs6st2 Primer2 reverse, 5′-CCAAGTTGCTCCTCTCTGGACA-3′; Hs6st2 Primer3 forward, 5′-CCTGGTGCGCAACATCC-3′; Hs6st2 Primer3 reverse, 5′-TGCATTTCTTCTGCCCCAC-3′; Sulf1 forward, 5′-ATGCTCACTGGGAAGTACGT-3′; Sulf1 reverse, 5′-CTGTAGCCGGTGTTGTTGAG-3′; Sulf2 forward, 5′-GACAAGGCGTATCATGGAGC-3′; Sulf2 reverse, 5′-CGTACTTGCCGGTGAGAATG-3′. To carry out qPCR amplification reactions, the following cycling parameters were used: AmpliTaq activation at 95 °C for 10 min, denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min (40 cycles). The qPCR quantification was described in detail in our previous work (Moon and Zhao 2022). Briefly, the cycle threshold (Ct) values were obtained from the StepOnePlus real-time PCR system (Applied Biosystems) and each selected gene was normalized by Ct value the 18S rRNA and the relative expression values were calculated according to the 2−ΔΔCt method. The mean value of the 2−ΔΔCt and standard error were used to generate the bar plot. The ΔCt values and one-tailed t-test were used to calculate the P-value for the gene expression differences between WT and Hs6st2 KO samples.
Measure of food intake
The 16-week-old male Hs6st2 KO mice (n = 6) and their WT littermates (n = 3) were single-housed in mouse cages (1 mouse/cage) and were isolated in a different room for three days before the food intake measurement. The weights of food consumed and newly provided were measured for each mouse daily at 9:00 AM for 4 consecutive days.
Animals for behavioral tests
The 7-week-old mice were housed in mouse cages (5 mice/cage) and were isolated in a different room for a week before the behavioral tests. On the day of the behavioral test, the experimental mice were placed in the behavioral test room to habituate for 1 h before the test. The time interval between two behavioral tests was one week.
Open field test
A square-shaped high-density plastic apparatus was used for the Open field test (50 cm in length, 50 cm in width, 30 cm in height). First, the apparatus was put in a bright open area. Each mouse was then gently placed in the center of the apparatus and was allowed to explore freely for 8 min. A video camera was placed above the apparatus to monitor, record, and track each mouse. The camera was connected to a laptop with the ANY-maze video tracking system (version 6.1). The ANY-maze software was used to calculate the total distance traveled and the time spent in the center. The software also provided a result report. The apparatus was cleaned with 70% ethanol between two tests. The total distance traveled and the time in the center zone were used to calculate the mean and standard error values for the bar plots and to perform the one-tailed t-test to get the P-values.
Tail suspension test
Each mouse was suspended by the end of its tail using scotch tape to a suspension bar (47 cm above the floor) and recorded for 6 min in a closed and lighted chamber. A lightweight plastic cylinder was placed around the tail base to prevent the bending of the mouse’s tail during the test. A video camera was placed behind the chamber. The immobility percentages were calculated using the FreezeFrame software (version 5). Mice were considered immobile when there was no voluntary body or limb movement. The percentages of immobility per 30 s were measured for a total of 360 s, which means a total of 12 consecutive time periods. For the line plot, the average percentages of immobility of the WT and KO mice were calculated for each of the 12 time periods, and these average percentages were used to calculate the mean and standard error values for the line plot and to perform the one-tailed t-test to get the P-values. For the bar plot, the average percentage of immobility of the 12 time periods was calculated for each mouse, and these average percentages were used to calculate the mean and standard error values for the bar plot and to perform the one-tailed t-test to get the P-value.
Social interaction test
The Social interaction test was conducted in a gray rectangular three-chambered apparatus (60 cm in length, 40 cm in width, 22 cm in height). The apparatus was divided into three equal-sized chambers, which were separated by walls with openings between the chambers. These openings on the walls allow the mice to move freely among the three chambers. A video camera was placed above the apparatus to monitor, record, and track each mouse. The camera was connected to a laptop with the ANY-maze video tracking system. For the habituation trial on day 1, the left and right chambers were put into a cylindrical wire cage (10 cm in diameter, 20 cm in height, 2 cm spacing between the bars). Each experimental mouse was placed into the center chamber and habituated to the apparatus for 10 min. For the social interaction test on day 2, the left and right chambers were put into a cylindrical wire cage. For the sociability test, a trained male WT (C57BL/6) unfamiliar mouse was placed inside the wire cage in the left chamber, while the wire cage in the right chamber was empty. The experimental mouse was placed in the center chamber and was allowed to explore freely for 10 min. Afterward, both mice were removed from the apparatus and were placed into separate holding cages. The apparatus was cleaned with 70% ethanol. After the sociability test, the experimental mouse was familiar with the trained WT mouse. The social novelty test was carried out next. The familiar trained WT mouse was put into the wire cage in the left chamber, and another trained male WT (C57BL/6) mouse novel to the testing mouse was put into the wire cage in the right chamber. The experimental mouse was then placed into the center chamber and was allowed to explore freely for 10 min. The ANY-maze software was used to calculate the time spent in each chamber and zone and the total distance traveled. The times in each zones and total distance traveled were used to calculate the mean and standard error values for the bar plots and to perform the one-tailed t-test to get the P-values. For the Sociability test, the sociability index for each mouse was calculated as the Mouse_zone_time/(Mouse_zone_time + Object_zone_time). These sociability indexes were used to calculate the mean and standard error values for the bar plot and to perform the one-tailed t-test to get the P-value. For the Social novelty test, the social novelty index for each mouse was calculated as the Novel_Mouse_zone_time/(Novel_Mouse_zone_time + Familiar_mouse_zone_time). These social novelty indexes were used to calculate the mean and standard error values for the bar plot and to perform the one-tailed t-test to get the P-value.
Y-maze test
A Y-shaped three-arm grey plastic apparatus was used for the Y-maze test. The three arms were 28 cm long, 10 cm wide, and 15 cm high and were intersected at 120°. A video camera was placed above the apparatus to monitor, record, and track each mouse. The camera was connected to a laptop with the ANY-maze video tracking system. For the test, each mouse was placed at the end of an arm and was allowed to explore freely through the maze for 10 min. The ANY-maze software was used to measure the numbers of entries, total distance traveled, and the spontaneous alternations. The spontaneous alternation was calculated using the ANY-maze software by [(number of alternations)/(total number of arm entries −2)] × 100. The spontaneous alternation values and total distance traveled were used to calculate the mean and standard error values for the bar plots and to perform the one-tailed t-test to get the P-values.
Contextual fear conditioning test
A fear conditioning chamber (The HABITEST Modular Behavioral Test System from the Coulbourn Instruments) was used for this test. A light was located at the top left wall of the chamber. The grid stainless steel bars on the floor of the chamber were connected to an electric foot shock stimulus. A video camera was placed behind the chamber and was connected to a laptop with the FreezeFrame software. For the Day 1 test, each mouse was habituated in the chamber for 120 s followed by two foot-shocks (0.6 mA for 2 s) occurring at the 120-s time point and the 270-s time point. After 360 s of recording, the mouse was removed from the chamber. For the Day 2 test (after 24 h), the mouse was placed into the same fear conditioning chamber without foot-shocks for 240 s. The percentages of freezing time were measured using the FreezeFrame software.
For the Day 1 test, the freezing percentages per 30 s were measured for a total of 360 s, which means a total of 12 consecutive time periods. The time period five is the first foot shock, and the time period ten is the second foot shock. The average freezing percentages of the time periods of one to four for a mouse was calculated as the habituation freezing percentage; the average freezing percentages of the time periods six to nine for a mouse was calculated as the first-shock freezing percentage; and the average freezing percentages of the time periods of 11 and 12 for a mouse was calculated as the second-shock freezing percentage. These habituation freezing percentages, first-shock freezing percentages, and second-shock freezing percentages were used to calculate the mean and standard error values for the bar plots and to perform the one-tailed t-test to get the P-values. For the Day 2 test, the freezing percentages per 30 s were measured for a total of 240 s, which means a total of eight consecutive time periods. The average freezing percentages of the eight time periods for a mouse was calculated as the mean freezing percentage. These mean freezing percentages were used to calculate the mean and standard error values for the bar plot and to perform the one-tailed t-test to get the P-value.
Novel object recognition test
A square-shaped high-density plastic apparatus, the same one as used for the Open field test, was used for the Novel objection recognition test. A video camera was placed above the apparatus to monitor, record, and track each mouse. The camera was connected to a laptop with the ANY-maze video tracking system. First, two identical objects were put into the apparatus, one at the left bottom corner and the other one at the right top corner. Each experimental mouse was placed in the center of the apparatus and was allowed to explore freely for 8 min. After the exploration, the experimental mouse was removed from the apparatus and was placed in a holding cage for 5 min. The apparatus was cleaned with 70% ethanol. Next, one of the objects was replaced with a different-shaped novel object. The experimental mouse was then placed back in the apparatus and was allowed to explore freely for 4 min. The ANY-maze software was used to measure the time spent in each object zone. The times in object zones were used to calculate the mean and standard error values for the bar plots and to perform the one-tailed t-test to get the P-values. The preference index for each mouse was calculated as Novel_object_time/(Novel_object_time + Familiar_object_time). These discrimination indexes were used to calculate the mean and standard error values for the bar plot and to perform the one-tailed t-test to get the P-value.
SAX-HPLC
The SAX-HPLC experiments were performed by the Complex Carbohydrate Research Center at the University of Georgia using methods as described in previous publications (Chen et al. 2018; Zhang et al. 2018; Casasnovas et al. 2021). The percentages of HS modifications in GAGs isolated from the brain tissue were used to calculate the mean values and standard error values and to generate the bar plot. The one-tailed t-test was used to calculate the P-values.
DAVID pathway enrichment analyses and gene set enrichment analysis (GSEA)
The DAVID web tool (Huang et al. 2007) was used to perform the pathway enrichment analyses using the lists of upregulated genes and downregulated genes, and FDR < 0.05 was used as the cutoff for enriched pathways. The GSEA software (Subramanian et al. 2005) was used to perform the GSEA analysis. The input file for GSEA was the file containing the read count per million uniquely mapped reads (CPM) values from DESeq2. The gmt file was the msigdb.v7.4.symbols.gmt. The chip file was the Mouse_Gene_Symbol_Remapping_MSigDB.v7.0.chip. The parameters were “1000 permutation”, “collapse” and “permutation type: gene_set”.
Immunohistochemistry for heparan sulfate
The brains were dissected from 16-week-old male Hs6st2 KO mice (n = 5) and their WT littermates (n = 3). The brains were fixed in 10% paraformaldehyde for 48 h and immersed in 30% sucrose in Phosphate-buffered saline (PBS, Sigma) to cryoprotect the tissue structure. After 24 h, the tissues were transferred into new 30% sucrose in PBS and stored at 4 °C until use. The whole brains were embedded and frozen in a block of optimal cutting temperature compound (Fisher scientific) and sectioned with a thickness of 50 μm of coronal cryosections. Consecutive sections were collected and stored in the PBS until use.
Immunohistochemical staining was performed using mouse monoclonal anti-HS antibody (10E4, AMSBIO #370255-B-50-H). Briefly, the brain sections were washed with PBS for 3 min at room temperature, permeabilized with 1% Triton X-100 in PBS for 15 min, and washed for 5 min in PBS on a rocker at room temperature. The tissues were then blocked with 5% goat serum with 0.1% Triton X-100 in PBS for 1 h at room temperature and incubated with 10E4 (1:200 dilution in 5% goat serum with 0.1% Triton X-100 in PBS) overnight at 4 °C on a rocker. On the following day, the tissues were washed with PBS for 5 min for 3 times and incubated with streptavidin, Alexa Fluor 647 conjugate AlexaFluor (1:500 dilution in 5% goat serum with 0.1% Triton X-100 in PBS) for 2 h at room temperature. The tissues were washed with PBS for 5 min for 3 times and were then stained with DAPI. Finally, the tissues were mounted with cover glass, sealed with nail polish, and stored at 4 °C. The 10E4 staining was imaged by the Nikon confocal microscope using the 20× magnification. The images were saved in TIFF format.
Immunohistochemistry image analyses
The ImageJ was used to quantify the IHC staining. For each image, we calculated the average fluorescence intensity (intensity/area) for the hippocampal CA1 pyramidal neuron area and for five background areas (areas that show no fluorescence). Briefly, we drew lines along the hippocampus CA1 pyramidal neuron area, set the parameters by Analyze - > Set Measurements, and calculated the fluorescence measurements by Analyze - > Measure. Similarly, we selected a small area of the image that has no fluorescence as a background area, calculated the fluorescence measurements by Analyze - > Measure, and repeated this background measurement for five different background areas. The average fluorescence intensity for the image was calculated by the average fluorescence intensity of the hippocampal CA1 pyramidal neuron area subtracting the average fluorescence intensity of the five background areas.
Microscope and quantification for dendritic spines
The whole brains were dissected from 8-week-old male Thy1-YFP-H;Hs6st2 KO mice (n = 3) and their WT littermates (n = 3). The brains were fixed in 10% paraformaldehyde for 48 h and immersed in 30% sucrose in PBS to cryoprotect the tissue structure. After 24 h, the tissues were transferred into new 30% sucrose in PBS and stored in 4 °C. The whole brains were embedded and frozen in a block of optimal cutting temperature compound (Fisher scientific) and sectioned with a thickness of 50 μm of coronal cryosections. Consecutive sections were collected and stored in the PBS. Lastly, the brain sections were mounted with cover glass, sealed with nail polish, and stored at 4 °C until imaging.
The brain sections were first scanned using Zeiss slide scanner with 514 nm laser and 10× objective. We next used the Zeiss LSM 980 confocal microscope to image the hippocampus CA1 pyramidal neurons and their apical dendrites. For the hippocampus CA1 pyramidal neurons, they were identified and imaged using Zeiss LSM 980 confocal microscope with airyscan mode, fast airyscan sheppard sum (SR-4Y), 514 nm laser, 60×, 1.4 NA oil-immersion objective, and 2,992 × 2,992 pixels, and the series of optical sections in the z-axis were acquired at 0.2 μm intervals. For the apical dendrites of these neurons, they were identified and imaged using Zeiss LSM 980 confocal microscope with airyscan mode of super resolution, 514 nm laser, 60×, 1.4 NA oil-immersion objective with 3× zoom, and 978 × 978 pixels, and the series of optical sections in the z-axis were acquired at 0.04 μm intervals.
To analyze the spine numbers and densities, 3–10 neurons per mouse were randomly selected, and 2–3 apical dendrites per neuron (length of 20 μm) were analyzed. The numbers of dendritic spines were manually quantified using imageJ software and the dendritic spine counter plugin. The densities of dendritic spines were calculated by the numbers of dendritic spines normalized to the dendrite length.
Statistical analysis and figure plotting
All the statistical analyses and figure plotting were performed using R 4.0.4 (www.r-project.org) and Microsoft Excel.
Supplementary Material
Acknowledgments
We thank Dr. Jeffery Esko at the University of California San Diego for providing frozen sperm of the Hs6st2 KO mice. We acknowledge the New York Institute of Technology Center for Biomedical Innovation and the Imaging Center for support, training, and assistance in image acquisition and processing.
Contributor Information
Sohyun Moon, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Hiu Ham Lee, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Stephanie Archer-Hartmann, Complex Carbohydrate Research Center, 315 Riverbend Road, University of Georgia, Athens, GA 30602, United States.
Naoko Nagai, Institute for Molecular Science of Medicine, Aichi Medical University, 1-1 Yazakokarimata, Nagakute, Aichi 480-1195, Japan.
Zainab Mubasher, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Mahima Parappurath, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Laiba Ahmed, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Raddy L Ramos, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Koji Kimata, Multidisciplinary Pain Center, Aichi Medical University, 1-1 Yazakokarimata, Nagakute, Aichi 480-1195, Japan.
Parastoo Azadi, Complex Carbohydrate Research Center, 315 Riverbend Road, University of Georgia, Athens, GA 30602, United States.
Weikang Cai, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Jerry Yingtao Zhao, Department of Biomedical Sciences, New York Institute of Technology College of Osteopathic Medicine, Northern Boulevard, P.O. Box 8000, Old Westbury, New York 11568, United States.
Author contributions
Sohyun Moon (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Investigation [equal], Methodology [equal], Project administration [equal], Validation [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Hiu Ham Lee (Data curation [equal], Methodology [equal], Resources [equal], Software [equal], Writing—review & editing [equal]), Stephanie Archer-Hartmann (Data curation [equal], Formal analysis [equal], Methodology [equal], Writing—review & editing [equal]), Naoko Nagai (Resources [equal], Writing—review & editing [equal]), Zainab Mubasher (Data curation [equal], Writing—review & editing [equal]), Mahima Parappurath (Data curation [equal], Formal analysis [equal], Writing—review & editing [equal]), Laiba Ahmed (Data curation [supporting], Formal analysis [supporting], Writing—review & editing [supporting]), Raddy Ramos (Data curation [supporting], Formal analysis [supporting], Resources [supporting], Writing—review & editing [supporting]), Koji Kimata (Resources [equal], Writing—review & editing [equal]), Parastoo Azadi (Data curation [equal], Formal analysis [equal], Methodology [equal], Resources [equal], Software [equal], Writing—review & editing [equal]), Weikang Cai (Data curation [equal], Formal analysis [equal], Methodology [equal], Resources [equal], Software [equal], Writing—review & editing [equal]), and Yingtao Zhao (Conceptualization [equal], Data curation [equal], Formal analysis [equal], Funding acquisition [equal], Investigation [equal], Methodology [equal], Project administration [equal], Resources [equal], Software [equal], Supervision [equal], Validation [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal])
Funding
This work was supported by the National Institutes of Health [R15NS130456 to J.Y.Z., R24GM137782 to P.A., R01MH125903 and R03AG083363 to W.C.]; the Mizutani Foundation for Glycoscience [Reference No. 230014 to J.Y.Z.]; and the Japan Society for the Promotion of Science (JSPS) KAKENHI grant [21K08588 to N.N.]
Conflict of interest statement. The authors declare no competing financial interests.
Data availability
The RNA-seq data of raw and processed files generated in this study have been deposited with the NCBI Gene Expression Omnibus under the accession number of GSE213745. The accession numbers for publicly available RNA-seq data are GSE90205, GSE90182, GSE90190, and GSE90185. The protein structure PDB files are available from our GitHub repository (https://github.com/Jerry-Zhao/Hs6st2KO2022).
Code accessibility
All computational scripts used in this study are available in the GitHub repository (https://github.com/Jerry-Zhao/Hs6st2KO2022).
References
- Aizawa S, Okada T, Keino-Masu K, Doan TH, Koganezawa T, Akiyama M, Tamaoka A, Masu M. Abnormal pyramidal decussation and bilateral projection of the corticospinal tract axons in mice lacking the heparan sulfate endosulfatases, Sulf1 and Sulf2. Front Mol Neurosci. 2019:12:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anower EKMF, Habuchi H, Nagai N, Habuchi O, Yokochi T, Kimata K. Heparan sulfate 6-O-sulfotransferase isoform-dependent regulatory effects of heparin on the activities of various proteases in mast cells and the biosynthesis of 6-O-sulfated heparin. J Biol Chem. 2013:288(6):3705–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anower EKF, Singh G, Deng Y, Gordts P, Esko JD. Triglyceride-rich lipoprotein binding and uptake by heparan sulfate proteoglycan receptors in a CRISPR/Cas9 library of Hep3B mutants. Glycobiology. 2019:29(8):582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasnovas J, Damron CL, Jarrell J, Orr KS, Bone RN, Archer-Hartmann S, Azadi P, Kua KL. Offspring of obese dams exhibit sex-differences in pancreatic heparan sulfate glycosaminoglycans and islet insulin secretion. Front Endocrinol (Lausanne). 2021:12:658439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Price DJ, Pratt T. FGF8 morphogen gradients are differentially regulated by heparan sulphotransferases Hs2st and Hs6st1 in the developing brain. Biol Open. 2017:6(12):1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ambadapadi S, Wakefield D, Bartee M, Yaron JR, Zhang L, Archer-Hartmann SA, Azadi P, Burgin M, Borges C, et al. Selective deletion of heparan sulfotransferase enzyme, Ndst1, in donor endothelial and myeloid precursor cells significantly decreases acute allograft rejection. Sci Rep. 2018:8(1):13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Yu NK, Choi JH, Sim SE, Kang SJ, Kwak C, Lee SW, Kim JI, Choi DI, Kim VN, et al. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015:350(6256):82–87. [DOI] [PubMed] [Google Scholar]
- Clegg JM, Conway CD, Howe KM, Price DJ, Mason JO, Turnbull JE, Basson MA, Pratt T. Heparan sulfotransferases Hs6st1 and Hs2st keep Erk in check for mouse corpus callosum development. J Neurosci. 2014:34(6):2389–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll AP, Farooqi IS, O'Rahilly S. The hormonal control of food intake. Cell. 2007:129(2):251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Dusman MJ, Dey PN, Elía-Zudaire Ó, Rabaneda LG, García-Lira C, Grand T, Briz V, Velasco ER, Andero R, Niñerola S, et al. Control of protein synthesis and memory by GluN3A-NMDA receptors through inhibition of GIT1/mTORC1 assembly. elife. 2021:10:e71575. 10.7554/eLife.71575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012:489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway CD, Price DJ, Pratt T, Mason JO. Analysis of axon guidance defects at the optic chiasm in heparan sulphate sulphotransferase compound mutant mice. J Anat. 2011:219(6):734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013:29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bazzal L, Atkinson A, Gillart AC, Obeid M, Delague V, Megarbane A. A novel EXT2 mutation in a consanguineous family with severe developmental delay, microcephaly, seizures, feeding difficulties, and osteopenia extends the phenotypic spectrum of autosomal recessive EXT2-related syndrome (AREXT2). Eur J Med Genet. 2019:62(4):259–264. [DOI] [PubMed] [Google Scholar]
- Farhan SM, Wang J, Robinson JF, Prasad AN, Rupar CA, Siu VM, FORGE Canada Consortium, FORGE Canada Consortium, Hegele RA. Old gene, new phenotype: mutations in heparan sulfate synthesis enzyme, EXT2 leads to seizure and developmental disorder, no exostoses. J Med Genet. 2015:52(10):666–675. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000:28(1):41–51. [DOI] [PubMed] [Google Scholar]
- Gupta A, Ewing SA, Renaud DL, Hasadsri L, Raymond KM, Klee EW, Gavrilova RH. Developmental delay, coarse facial features, and epilepsy in a patient with EXT2 gene variants. Clin Case Rep. 2019:7(4):632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi H, Miyake G, Nogami K, Kuroiwa A, Matsuda Y, Kusche-Gullberg M, Habuchi O, Tanaka M, Kimata K. Biosynthesis of heparan sulphate with diverse structures and functions: two alternatively spliced forms of human heparan sulphate 6-O-sulphotransferase-2 having different expression patterns and properties. Biochem J. 2003:371(1):131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007:282(21):15578–15588. [DOI] [PubMed] [Google Scholar]
- Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008:89(3):293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G, Gonzalez-Reiche AS, Lu N, Zhou X, Rivera J, Kriti D, Sebra R, Williams AA, Donovan MJ, Potter SS, et al. Integrated transcriptome and network analysis reveals spatiotemporal dynamics of calvarial suturogenesis. Cell Rep. 2020:32(1):107871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007:8(9):R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003:302(5647):1044–1046. [DOI] [PubMed] [Google Scholar]
- Irie F, Badie-Mahdavi H, Yamaguchi Y. Autism-like socio-communicative deficits and stereotypies in mice lacking heparan sulfate. Proc Natl Acad Sci U S A. 2012:109(13):5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Zhao YT, Fasolino M, Lamonica JM, Kim YJ, Georgakilas G, Wood KH, Bu D, Cui Y, Goffin D, et al. Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nat Med. 2017:23(10):1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021:596(7873):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007:445(7124):168–176. [DOI] [PubMed] [Google Scholar]
- Lencz T, Guha S, Liu C, Rosenfeld J, Mukherjee S, DeRosse P, John M, Cheng L, Zhang C, Badner JA, et al. Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat Commun. 2013:4(1):2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Momoi MY. Association of autism in two patients with hereditary multiple exostoses caused by novel deletion mutations of EXT1. J Hum Genet. 2002:47(5):262–265. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Huang TN, Hsueh YP. Quantification of the density and morphology of dendritic spines and synaptic protein distribution using Thy1-YFP transgenic mice. STAR Protoc. 2023:4(2):102290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Davies G, Summers KM, Hill WD, Hayward C, Liewald DC, Porteous DJ, Gale CR, McIntosh AM, Deary IJ. The influence of X chromosome variants on trait neuroticism. Mol Psychiatry. 2021:26(2):483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SM, Shaw AC, Bikker H, Ludecke HJ, van der Tuin K, Badura-Stronka M, Belligni E, Biamino E, Bonati MT, Carvalho DR, et al. Phenotype and genotype in 103 patients with tricho-rhino-phalangeal syndrome. Eur J Med Genet. 2015:58(5):279–292. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002:36(3):507–519. [DOI] [PubMed] [Google Scholar]
- Moon S, Zhao YT. Spatial, temporal and cell-type-specific expression profiles of genes encoding heparan sulfate biosynthesis enzymes and proteoglycan core proteins. Glycobiology. 2021:31(10):1308–1318. [DOI] [PubMed] [Google Scholar]
- Moon S, Zhao YT. Convergent biological pathways underlying the Kallmann syndrome-linked genes Hs6st1 and Fgfr1. Hum Mol Genet. 2022:31(24):4207–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Habuchi H, Sugaya N, Nakamura M, Imamura T, Watanabe H, Kimata K. Involvement of heparan sulfate 6-O-sulfation in the regulation of energy metabolism and the alteration of thyroid hormone levels in male mice. Glycobiology. 2013:23(8):980–992. [DOI] [PubMed] [Google Scholar]
- Neben CL, Idoni B, Salva JE, Tuzon CT, Rice JC, Krakow D, Merrill AE. Bent bone dysplasia syndrome reveals nucleolar activity for FGFR2 in ribosomal DNA transcription. Hum Mol Genet. 2014:23(21):5659–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neben CL, Tuzon CT, Mao X, Lay FD, Merrill AE. FGFR2 mutations in bent bone dysplasia syndrome activate nucleolar stress and perturb cell fate determination. Hum Mol Genet. 2017:26(17):3253–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganini L, Hadi LA, Chetta M, Rovina D, Fontana L, Colapietro P, Bonaparte E, Pezzani L, Marchisio P, Tabano SM, et al. A HS6ST2 gene variant associated with X-linked intellectual disability and severe myopia in two male twins. Clin Genet. 2019:95(3):368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallerla SR, Lawrence R, Lewejohann L, Pan Y, Fischer T, Schlomann U, Zhang X, Esko JD, Grobe K. Altered heparan sulfate structure in mice with deleted NDST3 gene function. J Biol Chem. 2008:283(24):16885–16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004:25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pratt T, Conway CD, Tian NM, Price DJ, Mason JO. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. J Neurosci. 2006:26(26):6911–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saied-Santiago K, Bulow HE. Diverse roles for glycosaminoglycans in neural patterning. Dev Dyn. 2018:247(1):54–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, Klann E. Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog Mol Biol Transl Sci. 2014:122:131–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res. 1989:6(4):311–326. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005:102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008:283(16):10366–10376. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006:127(1):49–58. [DOI] [PubMed] [Google Scholar]
- Tillo M, Charoy C, Schwarz Q, Maden CH, Davidson K, Fantin A, Ruhrberg C. 2- and 6-O-sulfated proteoglycans have distinct and complementary roles in cranial axon guidance and motor neuron migration. Development. 2016:143(11):1907–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timper K, Bruning JC. Hypothalamic circuits regulating appetite and energy homeostasis: pathways to obesity. Dis Model Mech. 2017:10(6):679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornberg J, Sykiotis GP, Keefe K, Plummer L, Hoang X, Hall JE, Quinton R, Seminara SB, Hughes V, van Vliet G, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci U S A. 2011:108(28):11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truwit CL, Barkovich AJ, Grumbach MM, Martini JJ. MR imaging of Kallmann syndrome, a genetic disorder of neuronal migration affecting the olfactory and genital systems. AJNR Am J Neuroradiol. 1993:14(4):827–838. [PMC free article] [PubMed] [Google Scholar]
- Wuyts W, Roland D, Lüdecke HJ, Wauters J, Foulon M, van Hul W, Van Maldergem L. Multiple exostoses, mental retardation, hypertrichosis, and brain abnormalities in a boy with a de novo 8q24 submicroscopic interstitial deletion. Am J Med Genet. 2002:113(4):326–332. [DOI] [PubMed] [Google Scholar]
- Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014:83(1):129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moon AF, Xu S, Krahn JM, Liu J, Pedersen LC. Structure based substrate specificity analysis of Heparan Sulfate 6-O-sulfotransferases. ACS Chem Biol. 2017:12(1):73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Kerever A, Yoshimura Y, Suzuki Y, Nonaka R, Higashi K, Toida T, Mercier F, Arikawa-Hirasawa E. Heparan sulfate alterations in extracellular matrix structures and fibroblast growth factor-2 signaling impairment in the aged neurogenic niche. J Neurochem. 2017:142(4):534–544. [DOI] [PubMed] [Google Scholar]
- Yousem DM, Turner WJ, Li C, Snyder PJ, Doty RL. Kallmann syndrome: MR evaluation of olfactory system. AJNR Am J Neuroradiol. 1993:14(4):839–843. [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bi S. Hypothalamic regulation of Brown adipose tissue thermogenesis and energy homeostasis. Front Endocrinol (Lausanne). 2015:6:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014:34(36):11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lu W, Wang Z, Ni J, Zhang J, Tang W, Fang Y. A comprehensive analysis of NDST3 for schizophrenia and bipolar disorder in Han Chinese. Transl Psychiatry. 2016:6(1):e701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Lu H, Peixoto RT, Pines MK, Ge Y, Oku S, Siddiqui TJ, Xie Y, Wu W, Archer-Hartmann S, et al. Heparan Sulfate organizes neuronal synapses through neurexin partnerships. Cell. 2018:174(6):1450–1464.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YT, Kwon DY, Johnson BS, Fasolino M, Lamonica JM, Kim YJ, Zhao BS, He C, Vahedi G, Kim TH, et al. Long genes linked to autism spectrum disorders harbor broad enhancer-like chromatin domains. Genome Res. 2018:28(7):933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data of raw and processed files generated in this study have been deposited with the NCBI Gene Expression Omnibus under the accession number of GSE213745. The accession numbers for publicly available RNA-seq data are GSE90205, GSE90182, GSE90190, and GSE90185. The protein structure PDB files are available from our GitHub repository (https://github.com/Jerry-Zhao/Hs6st2KO2022).