Abstract
FOXA1 is a pioneer transcription factor that is frequently mutated in prostate, breast, bladder, and salivary gland malignancies. Indeed, metastatic castration-resistant prostate cancer (mCRPC) commonly harbour FOXA1 mutations with a prevalence of 35%. However, despite the frequent recurrence of FOXA1 mutations in prostate cancer, the mechanisms by which FOXA1 variants drive its oncogenic effects are still unclear. Semaphorin 3C (SEMA3C) is a secreted autocrine growth factor that drives growth and treatment resistance of prostate and other cancers and is known to be regulated by both AR and FOXA1. In the present study, we characterize FOXA1 alterations with respect to its regulation of SEMA3C. Our findings reveal that FOXA1 alterations lead to elevated levels of SEMA3C both in prostate cancer specimens and in vitro. We further show that FOXA1 negatively regulates SEMA3C via intronic cis elements, and that mutations in FOXA1 forkhead domain attenuate its inhibitory function in reporter assays, presumably by disrupting DNA binding of FOXA1. Our findings underscore the key role of FOXA1 in prostate cancer progression and treatment resistance by regulating SEMA3C expression and suggest that SEMA3C may be a driver of growth and tumor vulnerability of mCRPC harboring FOXA1 alterations.
Subject terms: Prostate cancer, Cancer genomics, Transcriptional regulatory elements
Introduction
FOXA1, a member of the forkhead box family of transcription factors, acts as a pioneer factor that binds to and de-compacts condensed chromatin to facilitate access of other transcription factors and transcriptional machinery to initiate the transcriptional cascade1–6. FOXA1 is well-documented to influence AR activity and can dramatically affect its cistrome7–10 leading to the up- or down-regulation of a large number of genes. However, the relationship between the AR and FOXA1 is complex. In prostate cancer, FOXA1 is known to have AR-dependent and AR-independent activities10,11; moreover, in those that are AR-dependent, FOXA1 can exert both positive and negative effects on expression of AR-regulated genes11–13. The nuances of FOXA1-AR interplay are likely dependent on biological context as well as proximity and arrangement of genomic FOXA1 motifs in relation to AR motifs.
Landmark genomic studies have revealed that FOXA1 is among the most frequently-mutated genes in prostate cancer patients9,14–17. These mutations have been documented in multiple independent clinical cohorts and associated with poor prognosis suggesting that FOXA1 lesions may be a biomarker with potential diagnostic or prognostic significance. Recent coinciding reports have begun to unravel the clinical importance of FOXA1 mutations in PCa18,19. Adams et al. identified two mutation hotspots in FOXA1 and demonstrated that FOXA1 variants trigger phenotypic changes by promoting growth and inducing epithelial-to-mesenchymal transition (EMT). Parolia et al. define three unique classes of FOXA1 variants that behave differently with respect to its effects on FOXA1 transcriptional activity and cell growth. However, molecular mechanisms by which FOXA1 variants mediate these tumor-promoting activities remain poorly understood.
To address this gap, we identified semaphorin 3C (SEMA3C) as a candidate FOXA1 target gene that might mediate the pro-oncogenic effects such as increased proliferation and EMT linked with FOXA1 variants in prostate cancer. SEMA3C is a secreted member of the semaphorin family of extracellular signaling proteins that has been implicated in various cancers including prostate, breast, gastric, lung, and pancreatic cancer, as well as glioblastoma20–27. We have previously shown that SEMA3C is regulated by both FOXA1 and AR in prostate cancer28. Furthermore, SEMA3C and its receptor, Plexin B1, drive cancer growth by transactivating multiple receptor tyrosine kinases including EGFR, HER2 and MET20,29–31. Additionally, SEMA3C has been linked to the induction of EMT and stemness of prostate epithelial cells as well as increased prostate cancer invasion and migration21,29,32. Importantly, since FOXA1-altered prostate cancer cell lines [AR-dependent LAPC4 (class-2 frameshift) and AR-independent DU145 (FOXA1-low)] display a dependence on SEMA3C for proliferation, this suggests that cancer cells bearing FOXA1 alterations may exhibit a tumor vulnerability and growth pathway addiction to SEMA3C. Collectively, these findings suggest that SEMA3C may be a putative driver of the cancer-promoting activities associated with FOXA1 variants, implying that the FOXA1 alterations commonly observed in clinical prostate cancer samples might mediate their oncogenic effects via SEMA3C.
To extend these findings we sought to better understand FOXA1’s control over SEMA3C expression. Given repeated observations of FOXA1 mutations in prostate cancer, we were also motivated to investigate the implications of clinically-observed mutations in FOXA1 on SEMA3C expression. Functional validation of genomic alterations to FOXA1 may be of clinical significance. Our results indicate that alterations to FOXA1 lead to increased SEMA3C expression levels in prostate cancer specimens and also in vitro. Additionally, we show that FOXA1 is negative regulator of SEMA3C expression through intronic elements, and that missense mutations in FOXA1 alter its transcriptional activities in reporter assays. We posit that these effects may be operationally due to modified protein-DNA interactions in FOXA1 mutants.
Results
FOXA1 alterations correlate with elevated SEMA3C expression in cBioPortal datasets
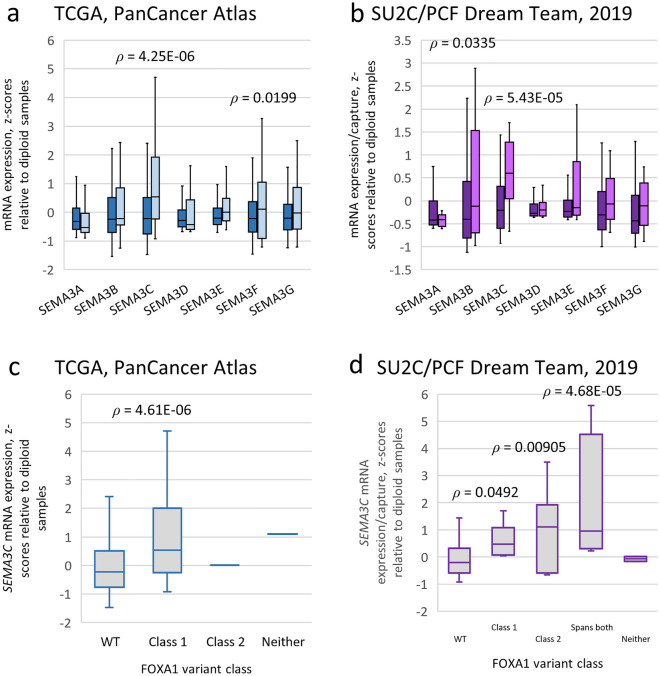
Considering FOXA1’s known control over SEMA3C expression combined with FOXA1’s predisposition to genomic alterations, we set out to determine if these two phenomena displayed any correlation. Using publicly-available TCGA and SU2C/PCF expression datasets, class 3 semaphorin levels in FOXA1-wild-type and FOXA1-altered (missense, insertions, deletions) specimens were compared (Fig. 1). This investigation revealed that, in aggregate, patients with FOXA1 alterations have elevated levels of SEMA3C compared to their non-altered counterparts. Of note, among all class 3 semaphorins, differential expression between wild-type and altered FOXA1 was most pronounced in SEMA3C. Parolia et al. classified class-1 FOXA1 mutations as those confined to the FKHD and class-2 as those c-terminal to the FKHD. When stratified in this way, SEMA3C expression was found to be elevated in both class-1 and class-2 variants compared to WT (Fig. 1c,d). The expression of other androgen receptor targets in wild-type and altered FOXA1 samples within the TCGA and SU2C/PCF cohorts were also examined (Supplementary Figure S1).
Figure 1.
Class 3 semaphorin expression levels in wild-type versus altered FOXA1 prostate cancer specimens using cBioPortal. Boxplots of class 3 semaphorin mRNA levels in wild-type (shaded dark) versus altered FOXA1 specimens (shaded light) from the Prostate Adenocarcinoma cohort (TCGA, PanCancer Atlas) (a) and the Metastatic Prostate Adenocarcinoma cohort (SU2C/PCF Dream Team, PNAS 2019) (b) obtained from cBioPortal. Boxplots of class 3 semaphorin mRNA levels in the TCGA (c) and the SU2C/PCF (d) cohorts as a function of FOXA1 class variants. Class-1 variants included those in the FKHD (residues 168 to 269); class-2 variants included all variants c-terminal to residue 269; ‘Spans both’ in the SU2C/PCF cohort includes one or more variants that span residues used to define both class-1 and class-2 variants; ‘Neither’ indicates a variant in FOXA1 that was outside of the definition of both class-1 and class-2 variants. The number of samples are as follows: for TCGA, WT n = 461, Class 1 n = 25, Class 2 n = 1, Neither n = 1; for SU2C/PCF, WT n = 187, Class 1 n = 9, Class 2 n = 6, Spans both n = 4, Neither n = 2. Boxes span the interquartile range. Horizontal line within the box represents the median SEMA3C mRNA levels. Bars represent the 95th percentile range of readings. Statistical analysis was performed using the unpaired two-tailed Student’s t-test relative to WT samples. TCGA, mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM). SU2C/PCF 2019, mRNA expression z-scores relative to diploid samples (FPKM capture).
FOXA1 mutants alter the chromatin at the SEMA3C locus
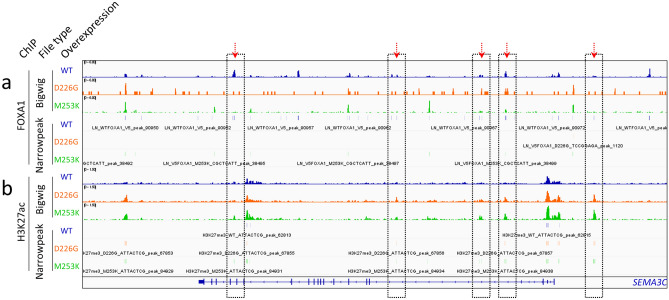
To better understand if the tumor-level correlation between class-1 FOXA1 alterations and SEMA3C expression is retained in in vitro systems, we first leveraged existing ChIP-Seq datasets from multiple recently-published reports. In a study by Gao et al., the authors overexpressed wild-type FOXA1 and the class-1 FOXA1 variants p.D226G and p.M253K in LNCaP and conducted FOXA1 ChIP-Seq (subseries GSE133386) or H3K27ac ChIP-Seq (subseries GSE133387). Examining this dataset using Integrative Genomics Viewer33 showed reduced recruitment of FOXA1 variants to the SEMA3C locus as compared to wild-type FOXA1 (Fig. 2a). This was concomitant with an increase in H3K27ac peaks in LNCaP overexpressing FOXA1 variants as compared to LNCaP overexpressing wild-type FOXA1 (Fig. 2b). Areas with pronounced redistribution of FOXA1 and H3K27ac at the SEMA3C locus between WT and variants are highlighted by red arrows and boxes. Taken together this would suggest that mutations to FOXA1 impair FOXA1’s interaction with the SEMA3C locus and increases acetylation marks on nearby histones. Adams et al. overexpressed wild-type Foxa1 and the class-1 Foxa1 variants p.F254_E255del and p.R219S in primary mouse organoids followed by FOXA1 or AR ChIP-Seq (subseries GSE128867). These too indicated differential recruitment of wild-type versus variants of FOXA1 to the Sema3c locus (Supplementary Figure S2). Parolia et al. overexpressed wild-type FOXA1 and the class-1 FOXA1 variants p.I176M and p.R261G in 22Rv1 cells followed by FOXA1 or AR ChIP-Seq (subseries GSE123618). These studies similarly showed reduced recruitment of FOXA1 to the SEMA3C locus by FOXA1 variants as compared to wild-type FOXA1 (Supplementary Figure S3). The impact that FOXA1 variants had on recruitment of AR to SEMA3C was inconsistent.
Figure 2.
FOXA1 and H3K27ac ChIP-Seq in FOXA1 wild-type versus altered cells at the SEMA3C locus. To identify differential recruitment of FOXA1 (a) and H3K27ac (b) to SEMA3C imposed by FOXA1 alterations, bigWig (upper) and narrowPeak (lower) ChIP-Seq files produced by Gao et al., were examined in Integrative Genomics Viewer. FOXA1 and H3K27ac ChIP-Seqs in wildtype (blue), FOXA1 p.D226G (orange), and FOXA1 p.M253K (green) are displayed as indicated. The SEMA3C gene is shown below. Y-axes are fixed at 6.0 for the FOXA1 bigWig files and 1.5 for the H3K27ac bigWig files. Regions with a particularly dramatic shift in chromatin constituents between WT and variants are highlighted by red arrows and boxed.
FOXA1 mutants have altered gene expression
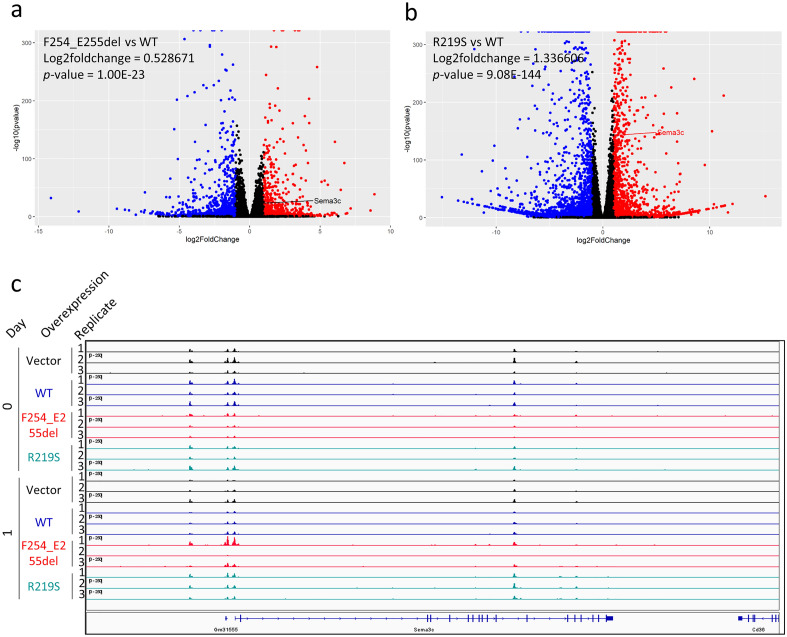
Leveraging the RNA-Seq dataset by Adams et al. (subseries GSE128666), we were able to evaluate the effects of Foxa1 alterations on endogenous expression of Sema3c. In this study, Adams et al. stably transduced murine organoids with dox-inducible Foxa1 constructs or empty vector as control. We compared the Sema3c expression levels of the two Foxa1 variants (p.F254_E255del and p.R219S) to those of the wild-type line at the day 1 (of dox induction) time point. Overexpression of p.F254_E255del and p.R219S led to a 0.53 and 1.34 log2-fold induction of Sema3c, respectively, compared to wild-type Foxa1 (Fig. 3a,b). The associated p-values for p.F254_E255del and p.R219S were 1 × 10−23 and 9 × 10−144, respectively. These variants were also associated with states of more accessible Sema3c chromatin in ATAC-Seq analyses (subseries GSE128421; Fig. 3c). Examining the RNA-Seq data by Gao et al. (subseries GSE133384) revealed that the RPKM for SEMA3C changed only modestly in the p.D226G and p.M253T variants compared to wild-type (data not shown). Collectively, this work supports the notion that in in vitro systems, alterations to FOXA1 impart functional changes with regard to its regulation of SEMA3C.
Figure 3.
Global expression in murine organoids overexpressing Foxa1 constructs. Volcano plots comparing p.F254_E255del to WT (a) and p.R219S to WT (b) in the RNA-Seq data by Adams et al. (subseries GSE128666). X-axis is log2-fold change and y-axis is − log10 p-value. Day 1 triplicate data from Adams et al. was used. Expression is relative to wild-type Foxa1. Red is ≥ twofold upregulation and p < 0.05; blue is ≥ twofold downregulation and p < 0.05. The expression of Sema3c is indicated. Within the plot, Sema3c is highlighted. (c) ATAC-Seq bigWig files (subseries GSE128421) were visualized in Integrative Genomics Viewer. Day 0 and 1 in empty vector (black), wildtype (blue), Foxa1 p.F254_E255del (red), and Foxa1 p.R219S (cyan) are displayed. The Sema3c gene is shown below. Y-axes are fixed at 250.
FOXA1 and AR DNA motifs overlap at the intron 2 of human SEMA3C
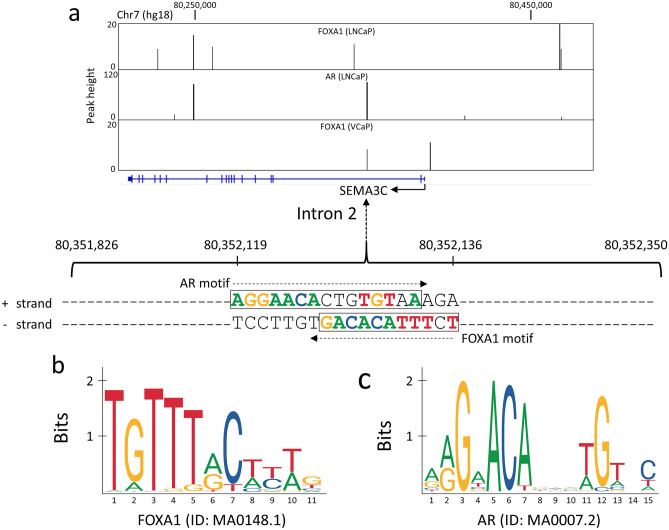
We previously demonstrated that the AR regulates SEMA3C expression through an androgen response element (ARE) located in intron 2 of SEMA3C28. The presence of this ARE was first identified in a ChIP-Seq study by Yu et al.34 that also reported a total of 35,107 and 11,774 genomic regions (or peaks) bound by the FOXA1 protein in LNCaP cells and VCaP cells, respectively. As illustrated in Fig. 4a, two such ChIP-Seq peaks are located at the intron 2 of SEMA3C on chromosome 7. The DNA sequences within these ChIP-Seq peaks were searched for the presence of FOXA1 (Fig. 4b) and AR DNA motifs (Fig. 4c), using two different motif scanning programs, RSAT35 and JASPAR36. A FOXA1 DNA motif was found within the ChIP-Seq peak at intron 2 (p = 6.4e−04), and an AR DNA motif was found within the same peak (p = 2.5e−05) through RSAT. Both the FOXA1 and AR motifs were also identified by the JASPAR program as the highest scoring motifs in that ChIP-Seq DNA region with relative scores of 0.88 and 0.94, respectively. The AR motif on the positive strand and the FOXA1 motif on the minus strand overlap on the same DNA region that spans from 80,352,119 and 80,352,136 (Fig. 4). We postulated that these DNA motifs were in some way responsible for regulation of SEMA3C expression in specimens containing FOXA1 variants.
Figure 4.
FOXA1 and AR DNA motifs at the human SEMA3C locus. (a) The ChIP-Seq peak data derived from the three experiments on FOXA1 in LNCaP, AR in LNCaP and FOXA1 in VCaP cells34 are shown as vertical black bars overlaid on the SEMA3C locus (blue horizontal line: exons are shown as vertical blue bars) using the UCSC Genome Browser (hg18, chromosome 7). The DNA sequences containing the FOXA1 and AR motifs, as predicted by the RSAT and JASPAR programs, are shown within the peak that spans from genomic positions 80,351,826 to 80,352,350 within intron 2 of SEMA3C. DNA sequences that match the consensus motifs are colored accordingly. The AR motif is located on the positive ( +) strand, and the FOXA1 motif is located on the minus (−) strand, both shown in a rectangular box respectively. (b) A consensus FOXA1 motif as reported in the JASPAR database is shown by a sequence logo. (c) A consensus AR motif as reported in the JASPAR database is shown by a sequence logo.
FOXA1 acts through a motif in the second intron of SEMA3C
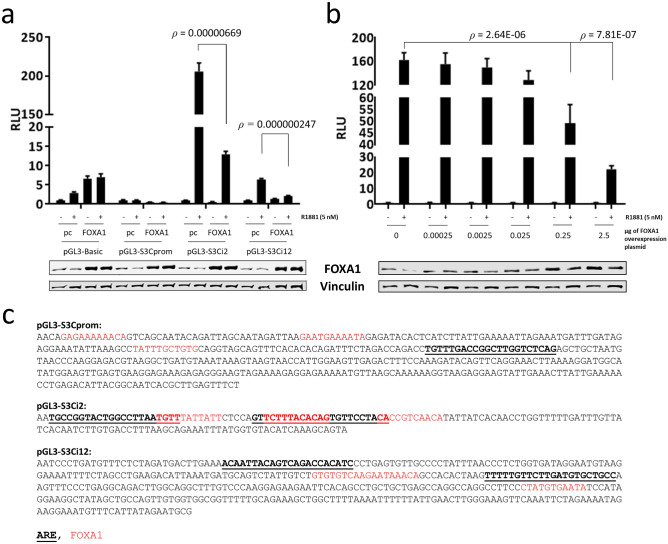
To ascertain which cis element(s) within the SEMA3C locus might be regulated by FOXA1, FOXA1-binding regions in the promoter, intron 2, and intron 12 were cloned into luciferase reporter vectors to generate pGL3-S3Cprom, pGL3-S3Ci2, and pGL3-S3Ci12, respectively. Cloned sequences are displayed along with top-ranked FOXA1 motifs and AREs as determined by RSAT and JASPAR (Fig. 5c). LNCaP were co-transfected with the reporter vector and FOXA1 overexpression plasmids and then treated with R1881. Overexpression of FOXA1 profoundly suppressed R1881-induced induction of pGL3-S3Ci2 and, to a lesser extent, pGL3-S3Ci12 (Fig. 5a). Neither R1881 nor FOXA1 overexpression affected transactivation of pGL3-S3Cprom or empty vector (pGL3-Basic) to the same degree as pGL3-S3Ci2. Overexpression of FOXA1 was verified by Western blot (Fig. 5a, lower). LNCaP were co-transfected with pGL3-S3Ci2 and increasing amounts of the FOXA1 overexpression plasmid and then treated with R1881. These experiments showed that FOXA1-mediated attenuation of R1881-induced transcription is dose-dependent (Fig. 5b). Sequence analysis of the second intron of murine Sema3c showed a stretch with high similarity to the region in the second intron of human SEMA3C containing the FOXA1 and AR motifs (Supplementary Figure S4). Collectively, these findings align with our previous observations indicating that FOXA1 is a potent negative regulator of SEMA3C expression28 and suggest that FOXA1 operates through cis elements located in the second intron of SEMA3C.
Figure 5.
Cis elements in the FOXA1-binding regions of the SEMA3C locus. FOXA1 motif-containing regions of the SEMA3C promoter, second intron, and twelfth intron were cloned into the reporter vector, pGL3-Basic. LNCaP were co-transfected with reporter plasmids and either empty overexpression vector (‘pc’) or a FOXA1 overexpression plasmid (‘FOXA1’), stimulated with R1881 (5 nM), and then harvested to determine luciferase activity (a). Overexpression of FOXA1 was verified by Western blot. pGL3-S3Ci2 reporter construct was titrated against increasing amounts of FOXA1 overexpression plasmid (b). Western blot was used to monitor FOXA1 levels. Cloned regions are displayed and FOXA1 motifs (red) and AR elements (bold, underlined) are indicated (c). Minus strands are shown.
FOXA1 variants have reduced transcriptional repressive activity in reporter gene assays
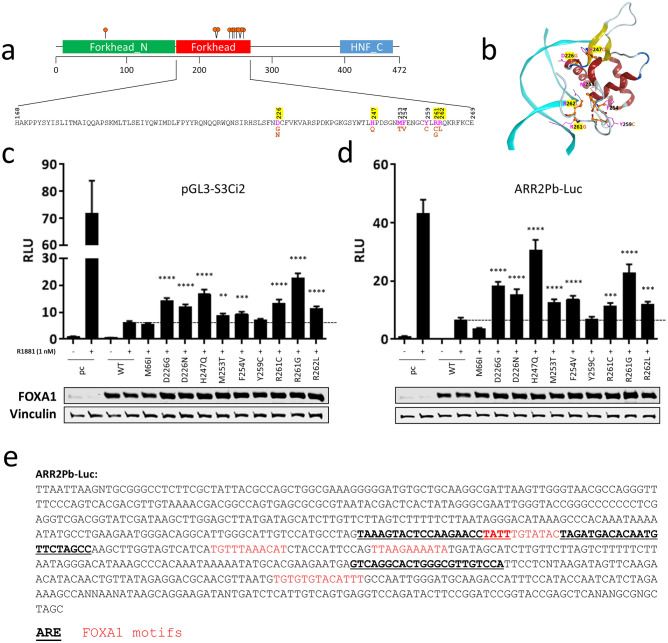
We next set out to understand the functional impacts of FOXA1 mutations in our reporter assays. While many FOXA1 mutations have been annotated to date, nine class-1 variants and one non-class-1 variant were selected for this study as illustrated schematically in Fig. 6a. Notably, numerous FOXA1 mutants investigated in this study contain amino acid substitutions located at the interface between the FOXA1 forkhead domain and DNA (Fig. 6b). These substitutions are predicted to weaken the protein-DNA interactions as residues such as Arg261 and Arg262, which are known to interact with the nucleotides and DNA backbone37, have been changed to non-interacting Gly261 and Leu262, respectively. To functionally characterize the FOXA1 mutants, we employed reporter gene assays using the pGL3-S3Ci2 construct due to its robust response in Fig. 5a. When FOXA1 constructs were co-transfected with pGL3-S3Ci2, wild-type FOXA1 suppressed R1881-induced luciferase activity whereas the missense mutations: D226G, D226N, H247Q, R261C, R261G, and R262L, showed impaired suppression compared to wild-type FOXA1 (Fig. 6c). This was also the case when another androgen-responsive construct, ARR2Pb-Luc, was used. Indeed, wild-type FOXA1 suppressed R1881 induction of luciferase activity but mutants D226G, D226N, H247Q, M253T, F254V, R261C, R261G, and R262L showed decreased suppression compared to wild-type FOXA1. M66I and Y259C showed enhanced and no altered suppression, respectively (Fig. 6d). Cloned sequence in ARR2Pb-Luc is displayed along with top-ranked FOXA1 motifs and AREs (Fig. 6e). Through modelling, we note that those variants whose mutated residues face the DNA have the most profound loss of activity (Fig. 6b). In particular, mutations from charged residues to hydrophobic ones were associated with the biggest effects. Furthermore, mutations at charged residues that reside in close proximity to the DNA impacted FOXA1’s transcriptional activity the greatest, possibly by hindering DNA binding. We acknowledge the fact that reporter assays present a limited view of the endogenous events that take place during gene expression and that events like chromatin re-modelling and pioneering events, which also influence gene expression, are not properly captured in reporter assays. Nonetheless, our data demonstrate that alterations to FOXA1 have functional consequences and may be responsible for altered SEMA3C expression in prostate cancer specimens.
Figure 6.
The effect of FOXA1 variants on androgen-responsive reporter constructs. (a) Schematic illustration of the FOXA1 gene with amino acid substitutions chosen for this study shown below. Nine of the constructs generated had a mutation at the c-terminus of the FKHD; one construct, M66I, contained a mutation n-terminal to the FKHD DNA-binding domain. Charged residues are highlighted in yellow. (b) Numerous mutations selected for our studies are located at the FOXA1-DNA interface. Amino acid changes, as a result of DNA mutations investigated in this study, are shown on the forkhead domain of the FOXA1 protein (alpha-helix in red and beta-strand in yellow ribbons). DNA is shown in cyan ribbons. Wild-type amino acids are shown in pink, with substitutions shown in orange. To determine the impact of missense mutations to FOXA1 in reporter gene assays, wild-type (WT) and FOXA1 mutants were co-transfected into cells along with AR-responsive reporter vectors for 2 days and then stimulated with R1881 in 0.2% CSS Opti-MEM for 24 h (1 nM) for an additional 24 h. Reporter constructs used included pGL3-S3Ci2 which harbours the intronic SEMA3C androgen response element (c) and ARR2Pb-Luc (d). Empty vector (pcDNA3.1 or ‘pc’) was transfected as a negative control. Relative Luminescence Units (RLU, y-axis) was scaled to sample pc without R1881, which was set to 1. Luminescence was read on a TECAN Infinite M200. Data represent mean, ± SD; **p < 0.01, ***p < 0.001, ****p < 0.0001. Significance is compared to sample: WT + R1881 (horizontal bar) by a Student’s t-test. Overexpression was confirmed by Western blot analysis; vinculin served as a loading control. (e) The cloned sequence within ARR2Pb-Luc is shown with AREs and FOXA1 motifs indicated in bold/underline and red, respectively. The sequence within pGL3-S3Ci2 was shown previously.
Discussion
Interrogation of clinical datasets revealed that patients with FOXA1 alterations have higher SEMA3C expression levels. Similar findings were discovered using in vitro ChIP-Seq, RNA-Seq, and ATAC-Seq datasets. Our reporter assays indicate that FOXA1 negatively regulates SEMA3C by way of cis elements in the second intron of SEMA3C. Furthermore, the variants D226G, D226N, H247Q, R261C, R261G, and R262L seemingly diminished FOXA1’s repressive effects leading to increased luciferase activity in constructs bearing FOXA1 motifs. FOXA1’s inhibitory effects on AR in reporter assays may relate to DNA occupancy by FOXA1 given the fact that ChIP-Seq analyses showed reduced binding of FOXA1 mutants to the SEMA3C locus. Moreover, point mutations mapping to the DNA-binding residues of FOXA1 forkhead domain strongly affected its repressive activity.
Our work adds to the body of knowledge amassing around FOXA1 in prostate cancer. We demonstrate that at least some of the clinically-observed FOXA1 alterations have functional consequences and, aided by computer modelling, infer that the structural basis for functional changes are related to altered protein-DNA interaction. These findings add support to the existing belief that FOXA1 mutations have tangible and measurable repercussions. Undoubtedly, biological context will influence the manifestation of each variant. Whether these mutations drive disease progression and the clinical interpretations of FOXA1 variants remains to be fully understood.
Adams et al. and Parolia et al. illuminate numerous clinical and biological ramifications of mutations to FOXA1 in PCa. In agreement with our findings, Adams et al. show that mutations to the residues D226, H247, and R261 are among the most impactful to FOXA1 in reporter assays compared to wild-type FOXA1. Our work is further substantiated by Parolia et al. who show that the R261G variant has more potent AR-driven transcriptional activity than wild-type FOXA1. Interestingly, Adams et al. discovered the enrichment of a non-canonical FOXA1 binding motif, resembling the GTAAA(C/T) motif but with a substitution of (G/A) for (C/T) at position 6, among FOXA1 p.R219S ChIP-Seq and ATAC-Seq peaks. The FOXA1 motif in intron 2 of SEMA3C conforms to the non-canonical motif opening up the possibility that variants of FOXA1 p.R219 play a unique role in SEMA3C regulation.
The data presented here builds on existing work and shows that elevated SEMA3C levels in FOXA1-alterated prostate cancer patients may be linked to intronic FOXA1 elements in SEMA3C. We note that the FOXA1 motif in the second intron of SEMA3C overlaps with the ARE (Fig. 4) which we previously determined to confer positive AR regulation of SEMA3C. It is therefore possible that FOXA1 normally co-occupies this ARE or displaces AR from this region with the net effect of dampened SEMA3C expression. If FOXA1 incurs a mutation, then FOXA1’s repression of AR-mediated SEMA3C expression may be lifted giving rise to elevated SEMA3C expression and acquisition of cell motility, invasiveness, and mesenchymal or stem-like phenotypes. Further analyses are warranted to prove that the proposed interaction between FOXA1 and AR at the second intron of SEMA3C is actually occurring and whether or not this particular type of interplay between FOXA1 and AR occurs elsewhere in the genome. It should also be noted that while we have drawn a link between FOXA1, AR, and SEMA3C in prostate cancer, a direct causal relationship cannot be concluded without further studies.
Work presented here outlines one avenue of SEMA3C regulation and is intended to inspire future work which will be needed to fully understand the mechanisms governing elevated SEMA3C levels in prostate cancer patients containing FOXA1 alterations. Our reporter assays are a measure of transcription initiation, but numerous other mechanisms likely underpin heightened SEMA3C expression which may or may not include cis-elements examined in our investigations. As an example, a notable FOXA1 ChIP-Seq peak upstream of SEMA3C should also be investigated for its roles in regulating SEMA3C expression. In addition, rescue experiments should be conducted to determine if a wild-type FOXA1 cistrome at SEMA3C can be restored at peaks lost due to FOXA1 mutations. Also, despite its effects on SEMA3C chromatin, overexpression of FOXA1 variants in LNCaP did not alter SEMA3C expression based on findings by Gao et al. and also in our own hands (data not shown). This indicates that the functional impacts of mutations to FOXA1 in luciferase assays alone, do not account for changes in SEMA3C expression or that endogenous expression of wild-type FOXA1 may mask changes induced by ectopic expression of FOXA1 variants. Additional studies will be needed to fully dissect the details and extent of FOXA1 regulation of SEMA3C. In particular, interrupting endogenous FOXA1 through genome editing, rather than through exogenous FOXA1, would be highly informative. Yet other future studies to carry out include examining other variants of the same residue (eg H247Q vs. H247R). In addition, numerous other missense mutations have been annotated in FOXA1 and should also be characterized. This work sets the precedent for the regulation of semaphorins by FOXA1 and the potential effects of FOXA1 mutations in this context. As FOXA1 is seen to be mutated in other forms of cancer, such as breast cancer, mutational analysis of FOXA1 should be examined in this area as well.
The multifaceted role of FOXA1 in regulating gene expression highlights the distinction between oncogenic driver mutations and tumor suppressor loss-of-function mutations. While FOXA1 is traditionally recognized as a pioneer factor that enhances gene transcription, our findings show it can also act as a repressor for certain growth-promoting genes. In particular, FOXA1 seems to restrain the expression of the SEMA3C growth factor by binding to its intronic cis elements. Intriguingly, class-1 variants of FOXA1 appear to dampen this repressive action. This dynamic may help reconcile seemingly divergent roles of FOXA1 as both an oncogene and a tumor suppressor. Our data, which highlights FOXA1's inhibitory influence on the SEMA3C growth pathway and the gain-of-function activities of FOXA1 mutations that diminish its repressive activities on SEMA3C, offers a framework that bridges these two contrasting views.
Materials and methods
Bioinformatics and ChIP-Seq data analysis
Previous ChIP-Seq data from Yu et al.34 was extracted from the NCBI Gene Expression Omnibus (GEO; RRID:SCR_005012) database38. In particular, the file (‘GSM353633_20A9RAAXX_C425.allregions.txt.gz’), which contained 35,107 DNA regions (i.e. peaks) bound by the FOXA1 protein in LNCaP cells (GEO accession: GSM353633), was parsed to a bedGraph format and visualized in the UCSC Genome Browser39 to identify FOXA1 binding sites nearby to the SEMA3C (RefSeq accession number NM_006379) locus on chromosome 7 of the human reference genome (hg18). Same analysis was done on the file (‘GSM353630_20A9RAAXX_C421.allregions.txt.gz’), which contained 11,774 DNA regions bound by the FOXA1 protein in VCaP cells (GEO accession: GSM353630), and the file (‘GSM353644_jy10s123.allregions.txt.gz’), which contained 44,536 DNA regions bound by the AR protein in LNCaP cells treated with R1881 (GEO accession: GSM353644). The actual DNA sequences that compose each binding peak region (~ 500 bps) at the SEMA3C locus were extracted and scanned for any presence of the FOXA1 (11 bps) and AR motif (15 bps), using DNA motif scanning programs, RSAT35 and JASPAR36. The DNA frequency matrices that define the FOXA1 motif (ID: MA0148.1) and AR motif (ID: MA0007.2) were obtained from the JASPAR database36. ChIP-Seq, RNA-Seq, and ATAC-Seq in FOXA1 wild-type versus variant-expressing cells was obtained at the GEO accession numbers: GSE133455, GSE128667, and GSE123625. bigWig and narrowPeak files were visualized in Integrative Genomics Viewer33 for ChIP-Seq and ATAC-Seq analyses. Differential gene expression analysis was carried out using DESeq2 (RRID:SCR_015687) using default parameters. Log2-fold expression is versus wild-type. Clustal Omega40 and ClustalW were used for sequence alignment.
FOXA1 protein structure modeling
The forkhead domain of the human FOXA1 protein structure was modeled based on the same domain of human FOXA3 (PDB ID: 1VTN37) that shares 96% sequence identity across the 100 amino acids (residue #168 to #267). The four amino acid differences were modified to match the FOXA1 sequence (G188S, E209Q, A245T, S249D) using the MOE protein modeling tool41.
DNA sequences
In reporter gene assays, genomic sequence encompassing highly-ranked FOXA1 motifs were cloned into the luciferase reporter backbone pGL3-Basic (Promega, E1751) to generate reporter constructs. Sequences are displayed in Fig. 5. Other AR-responsive reporter vectors were a kind gift from Dr. Paul S. Rennie (Department of Urologic Sciences, University of British Columbia). FOXA1 overexpression was achieved using a wild-type FOXA1 overexpression plasmid was obtained from Genscript (Clone ID: OHu23484, NM_004496.3). Mutant FOXA1 constructs were derived from this plasmid using QuikChange II Site-Directed Mutagenesis Kit (Stratagene, 200,523). pcDNA3.1 (RRID:Addgene_79663) served as an empty vector control. Plasmids were transfected using TransIT-2020 (Mirus, MIR 5404).
Cell culture
LNCaP (ATCC, CRL-1740, RRID:CVCL_0395) were cultured in RPMI 1640 supplemented with 10% FBS. Cells were treated at the indicated concentrations of androgen or 0.05% ethanol as a vehicle control in 0.2% charcoal-stripped serum (CSS) in Opti-MEM (Gibco, 11,058–021).
Luciferase assay
5 × 105 LNCaP cells were transiently transfected in full serum-containing media in triplicate in 12-well format with 0.6 µg of reporter vector or reporter vector control and 0.6 µg of wild-type or mutant FOXA1 or empty overexpression plasmid control as well as 30 ng of renilla plasmid (phRL-SV40) kindly provided by the Mui lab (Immunity and Infection Research Centre, Vancouver Coastal Health Research Institute, Vancouver, British Columbia). Two days following transfection, unless otherwise stated cells were treated with EtOH or R1881 in 0.2% CSS Opti-MEM for 24 h at which time cell lysates were harvested for luciferase assay using the Dual-Luciferase Reporter Assay System (Promega, E1960) and read on a TECAN Infinite M200 PRO. ARR2Pb-Luc is an androgen-responsive construct containing the probasin promoter. In all luciferase assays, firefly luciferase luminescence was normalized to renilla luciferase luminescence.
Western blot
Whole cell extracts were prepared in 50 mM Tris–HCl, 150 mM NaCl, 1% NP40, 10 mM NaF, 10% Glycerol, supplemented with protease inhibitor cocktail (Roche, 04,693,116,001) and quantitated using a BCA approach. 60 µg of protein was run on 10% acrylamide gels and transferred onto nitrocellulose membrane. Western blots were imaged on a LI-COR Odyssey system. Vinculin served as a loading control. Primary antibodies: FOXA1 (Santa Cruz Biotechnology, sc-6553, RRID:AB_2104865) and vinculin (Sigma-Aldrich, V4505, RRID:AB_477617). Secondary antibodies: anti-mouse alexa fluor 680 (Invitrogen, A21058, RRID:AB_2535724), and anti-goat alexa fluor 680 (Invitrogen, A21084, RRID:AB_2535741).
Clinical datasets
The Prostate Adenocarcinoma cohort (TCGA, PanCancer Atlas) and Metastatic Prostate Adenocarcinoma cohort (SU2C/PCF Dream Team, PNAS 2019) were obtained from cBioPortal42,43. Gene expression in patients with mutated FOXA1 were compared to patients with unaltered FOXA1 within the same dataset.
Statistical analyses
Statistical analysis was performed using the Student’s two-tailed t-test. Data are represented as mean ± SD unless otherwise stated. Data presented a representative of three biological replicates.
Supplementary Information
Acknowledgements
This research was proudly funded by grants from the Michael Smith Foundation for Health Research (Grant # 17318), Prostate Cancer Canada (Grant #s TAG2014-06 and GS2015-06), the Terry Fox Foundation (Grant # TFF-116129), the Canadian Cancer Society Research Institute (Grant # 700347), Cancer Research Society (Grant # F09-60564), the NIH Pacific Northwest Prostate Cancer SPORE (Grant # NCI P50 CA097186), Networks of Centres of Excellence of Canada (CECR PC-TRIADD), Prostate Cancer Foundation BC, and NSERC.
Author contributions
K.J.T., L.L., M.H., K.D., D.T., J.W.P., and C.J.O. designed the experiments; K.J.T., L.L., M.H., K.D., D.T., B.M., P.Y., S.B., and J.W.P. performed the experiments and analysis; K.J.T., L.L., M.H., K.D., D.T., and C.J.O. interpreted the results; K.J.T., L.L., M.H., K.D., D.T., Y.W., A.C., P.S.R., M.E.G., and C.J.O. wrote or provided input toward the manuscript.
Data availability
The data analyzed in this study were obtained from cBioPortal and Gene Expression Omnibus (RRID:SCR_005012) at GSM353633, GSM353630, GSM353644, GSE133455, GSE128667, and GSE123625.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-57854-w.
References
- 1.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/S1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 2.Shim EY, Woodcock C, Zaret KS. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/Gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He HSHS, et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010;42:343–U101. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakshatri H, Badve S. FOXA1 in breast cancer. Expert Rev. Mol. Med. 2009 doi: 10.1017/S1462399409001008. [DOI] [PubMed] [Google Scholar]
- 6.Bernardo GM, Keri RA. FOXA1: A transcription factor with parallel functions in development and cancer. Biosci. Rep. 2012;32:113–130. doi: 10.1042/Bsr20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao N, et al. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 8.Yu XP, et al. Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann. NY. Acad. Sci. 2005;1061:77–93. doi: 10.1196/annals.1336.009. [DOI] [PubMed] [Google Scholar]
- 9.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin HJ, Zhao JC, Ogden I, Bergan RC, Yu JD. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73:3725–3736. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahu B, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, Lolla L. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltran H, et al. Impact of therapy on genomics and transcriptomics in high-risk prostate cancer treated with neoadjuvant docetaxel and androgen deprivation therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017;23:6802–6811. doi: 10.1158/1078-0432.CCR-17-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580:93–99. doi: 10.1038/s41586-020-2135-x. [DOI] [PubMed] [Google Scholar]
- 18.Adams EJ, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019 doi: 10.1038/s41586-019-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parolia A, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019 doi: 10.1038/s41586-019-1347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peacock JW, et al. SEMA3C drives cancer growth by transactivating multiple receptor tyrosine kinases via Plexin B1. Embo Mol. Med. 2018 doi: 10.15252/emmm.201707689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman JG, Meadows GG. Increased class 3 semaphorin expression modulates the invasive and adhesive properties of prostate cancer cells. Int. J. Oncol. 2007;30:1231–1238. [PubMed] [Google Scholar]
- 22.Esselens C, et al. The cleavage of semaphorin 3C induced by ADAMTS1 promotes cell migration. J. Biol. Chem. 2010;285:2463–2473. doi: 10.1074/jbc.M109.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik MFA, Satherley LK, Davies EL, Ye L, Jiang WG. Expression of semaphorin 3C in breast cancer and its impact on adhesion and invasion of breast cancer cells. Anticancer Res. 2016;36:1281–1286. [PubMed] [Google Scholar]
- 24.Miyato H, Tsuno NH, Kitayama J. Semaphorin 3C is involved in the progression of gastric cancer. Cancer Sci. 2012;103:1961–1966. doi: 10.1111/Cas.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Satue M, Blanco J. Identification of semaphorin E gene expression in metastatic human lung adenocarcinoma cells by mRNA differential display. J. Surg. Oncol. 1999;72:18–23. doi: 10.1002/(SICI)1096-9098(199909)72:1<18::AID-JSO5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, et al. Increased semaphorin 3c expression promotes tumor growth and metastasis in pancreatic ductal adenocarcinoma by activating the ERK1/2 signaling pathway. Cancer Lett. 2017;397:12–22. doi: 10.1016/j.canlet.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Man JH, et al. Sema3C promotes the survival and tumorigenicity of glioma stem cells through Rac1 activation. Cell Rep. 2014;9:1812–1826. doi: 10.1016/j.celrep.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam KJ, et al. Androgen receptor transcriptionally regulates semaphorin 3C in a GATA2-dependent manner. Oncotarget. 2017;8:9617–9633. doi: 10.18632/oncotarget.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin L, et al. MAOA promotes prostate cancer cell perineural invasion through SEMA3C/PlexinA2/NRP1-cMET signaling. Oncogene. 2021;40:1362–1374. doi: 10.1038/s41388-020-01615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swiercz JM, Kuner R, Offermanns S. Plexin-B1/RhoGEF-mediated RhoA activation involves the receptor tyrosine kinase ErbB-2. J. Cell Biol. 2004;165:869–880. doi: 10.1083/jcb.200312094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giordano S, et al. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat. Cell Biol. 2002;4:720–724. doi: 10.1038/Ncb843. [DOI] [PubMed] [Google Scholar]
- 32.Tam KJ, et al. Semaphorin 3 C drives epithelial-to-mesenchymal transition, invasiveness, and stem-like characteristics in prostate cells. Sci. Rep. 2017;7:11501. doi: 10.1038/s41598-017-11914-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina-Rivera A, et al. RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 2015;43:W50–56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan A, et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 38.Barrett T, et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madeira F, et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022;50:W276–W279. doi: 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chemical Computing Group. Molecular Operating Environment., <http://www.chemcomp.com/> (
- 42.Gao JJ, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data (vol 2, pg 401, 2012) Cancer Discov. 2012;2:960–960. doi: 10.1158/2159-8290.CD-12-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study were obtained from cBioPortal and Gene Expression Omnibus (RRID:SCR_005012) at GSM353633, GSM353630, GSM353644, GSE133455, GSE128667, and GSE123625.