Abstract
In the absence of cell migration, plant architecture is largely determined by the direction and extent of cell expansion during development. In this report, we show that very-long-chain fatty acid (VLCFA) synthesis plays an essential role in cell expansion. The Arabidopsis thaliana eceriferum10 (cer10) mutants exhibit severe morphological abnormalities and reduced size of aerial organs. These mutants are disrupted in the At3g55360 gene, previously identified as a gene coding for enoyl-CoA reductase (ECR), an enzyme required for VLCFA synthesis. The absence of ECR activity results in a reduction of cuticular wax load and affects VLCFA composition of seed triacylglycerols and sphingolipids, demonstrating in planta that ECR is involved in all VLCFA elongation reactions in Arabidopsis. Epidermal and seed-specific silencing of ECR activity resulted in a reduction of cuticular wax load and the VLCFA content of seed triacylglycerols, respectively, with no effects on plant morphogenesis, suggesting that the developmental phenotypes arise from abnormal sphingolipid composition. Cellular analysis revealed aberrant endocytic membrane traffic and defective cell expansion underlying the morphological defects of cer10 mutants.
INTRODUCTION
Very-long-chain fatty acids (VLCFAs; fatty acids with chain lengths >18 carbons) are essential biological components found in cuticular waxes, seed storage triacylglycerols (TAGs), and sphingolipids in plants, but their precise biological functions remain poorly characterized. VLCFAs are synthesized by sequential addition of C2 moieties from malonyl-CoA to preformed C18 acyl groups derived from the de novo fatty acid synthesis pathway of the plastid. Four enzymatic reactions are involved in the elongation process: condensation of C18-CoA with malonyl-CoA to form a 3-ketoacyl-CoA, reduction of 3-ketoacyl-CoA to 3-hydroxy-CoA, dehydration to an enoyl-CoA, and reduction of the enoyl-CoA (Fehling and Mukherjee, 1991). The enzymes comprising the fatty acid elongation pathway are membrane bound and organized in a complex referred to as the elongase (von Wettstein-Knowles, 1982).
In Saccharomyces cerevisiae, genes encoding enzymes of the elongase have been identified, with the exception of the dehydratase (Oh et al., 1997; Kohlwein et al., 2001; Han et al., 2002). Subcellular localization studies indicated that the Elo2p/Elo3p condensing enzymes are evenly localized in the endoplasmic reticulum (ER) (David et al., 1998; Kohlwein et al., 2001), whereas the Tsc13p enoyl-reductase is enriched in a specific ER domain at the nuclear–vacuolar interface (Kohlwein et al., 2001). Disruption of the elongation enzymes inhibits VLCFA biosynthesis required for sphingolipid assembly in yeast and arrests yeast growth (Oh et al., 1997; Kohlwein et al., 2001; Han et al., 2002). Sphingolipids are a minor but important class of lipids most abundant in the plasma membrane, but also found in the trans-Golgi network and recycling endosomes (Simons and Toomre, 2000). They are composed of a long-chain sphingosine base typically 18 carbons long, an amide-linked VLCFA at the C2-amino group, and a polar head group, generally a sugar residue, present at the C1 carbon. Because of their extended hydrophobic chain, VLCFA moieties are essential components of sphingolipids that determine their physical properties and characteristics of the membranes that harbor these lipids. In yeast and in mammalian cells, sphingolipids interact with sterols to trigger the formation of highly ordered, detergent-resistant plasma membrane microdomains known as lipid rafts. Lipid rafts are involved in cellular trafficking of certain plasma membrane proteins, such as glycosylphosphatidylinositol (GPI)-anchored proteins, protein stability at the cell surface, and endocytosis (Simons and Toomre, 2000; Ikonen, 2001; Wang and Chang, 2002). These processes play important roles in signal transduction at the cell surface and the generation and maintenance of cell polarity, which are crucial in proper execution and coordination of cell activities, including cell expansion.
In plants, information on the molecular and cellular organization of VLCFA biosynthesis and regulation is also starting to emerge. The condensing enzyme has been recognized as the key component of the elongase that determines substrate and tissue specificities of VLCFA elongation (Millar and Kunst, 1997). A large plant-specific FATTY ACID ELONGATION1 (FAE1)-like condensing enzyme gene family and an unrelated ELO-like gene family of putative condensing enzymes (Dunn et al., 2004) have been identified in Arabidopsis thaliana. However, with the exception of FAE1, a seed-specific condensing enzyme involved in C20 and C22 fatty acid biosynthesis for seed storage lipids (Kunst et al., 1992), and CER6, a condensing enzyme that catalyzes the elongation of C22 fatty acyl-CoAs or longer (O. Rowland, T.R. Blacklock, and L. Kunst, unpublished data) in the epidermal cells of Arabidopsis (Millar et al., 1999; Fiebig et al., 2000), the functions of other condensing enzymes in VLCFA elongation are not known. For example, genetic studies of FAE1-like genes such as KCS1, FDH, and HIC demonstrated that they are important for Arabidopsis cell and organ morphogenesis and adaptation to the environment (Todd et al., 1999; Yephremov et al., 1999; Gray et al., 2000; Pruitt et al., 2000), but a clear relationship between defective VLCFA synthesis and the cellular, developmental, and physiological phenotypes of kcs1, fdh, and hic mutants has not been established.
In contrast with the condensing enzymes, it has been proposed that the other three enzyme activities of the elongase have broad substrate specificities and are shared by all the plant VLCFA biosynthetic pathways (Millar and Kunst, 1997). This hypothesis has not been tested because the genes encoding the two reductases and the dehydratase have not been isolated from Arabidopsis. Recently, a single Arabidopsis gene, At3g55360, was identified as an enoyl-CoA reductase (ECR) candidate based on its similarity to the yeast TSC13 gene encoding the ECR (Kohlwein et al., 2001). Heterologous expression of At3g55360 resulted in complementation of the temperature-sensitive lethality of yeast tsc13-1elo2Δ cells (Gable et al., 2004). The putative Arabidopsis ECR gene is ubiquitously expressed, and the protein physically interacts with the Elo2p and Elo3p condensing enzymes when expressed in yeast (Gable et al., 2004). In this study, the in planta functions of the ECR were examined using Arabidopsis mutants disrupted in the ECR gene. We demonstrate that the ECR gene product is involved in VLCFA elongation required for the synthesis of all the VLCFA containing lipids, including cuticular waxes, seed TAGs, and sphingolipids. Furthermore, disruptions in the ECR gene have profound effects on shoot growth and morphogenesis. Biochemical and cellular analyses suggested that changes in sphingolipids may be an underlying cause of the observed morphological defects and revealed impaired endocytic membrane traffic and defective cell expansion in the mutants.
RESULTS
Isolation and Genetic Characterization of Mutants Disrupted in the ECR Gene
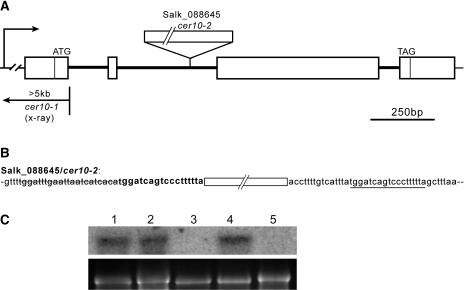
We initiated a reverse-genetics approach in Arabidopsis to address the in planta function of the ECR. Two Salk lines, Salk_088645 (Figure 1A) and Salk_138092, obtained from the Arabidopsis Biological Resource Center were found to carry T-DNA insertions in the second intron of the ECR gene. The Salk_088645 line also had a small 22-bp deletion and an 18-bp duplication of genomic DNA flanking the T-DNA insertion site (Figure 1B). No visible developmental phenotypes were observed in homozygous Salk_138092 plants. By contrast, homozygous Salk_088645 plants exhibited abnormal organ morphology (described in the following section) and stem glossiness. These phenotypes cosegregated with the T-DNA insertion as a single Mendelian recessive locus (n = 150), suggesting that a single T-DNA insertion in the ECR gene is genetically linked to the observed phenotypes.
Figure 1.
Identification of cer10-1 and cer10-2 Mutations in the ECR Gene.
(A) The ECR gene structure and mutation sites of cer10 alleles. White boxes represent exons. cer10-1 has at least a 5-kb deletion and/or rearrangement located in the promoter region, the 5′-untranslated region, and at least 90 bp downstream of the translation start of the gene, whereas cer10-2 (Salk_088645) carries a T-DNA insertion in the second intron of the gene.
(B) The genomic DNA sequence flanking the T-DNA insertion in cer10-2. White box represents T-DNA insertion site. A small deletion of 22 bp (strikethrough) and a small duplication (bold) of an 18-bp genomic DNA fragment (underline) flanking the T-DNA insertion in cer10-2 are detected.
(C) RNA gel blot analysis of steady state ECR mRNA in wild-type Col-0 (lane 1), Salk_138092 (lane 2), cer10-2 (lane 3), wild-type Ler (lane 4), and cer10-1 (lane 5). The bottom panel shows ethidium bromide–stained 18S rRNAs.
Changes in shoot morphology and the stem glossiness of the Salk_088645 homozygous mutant resembled that of the eceriferum10 (cer10) mutant (Koornneef et al., 1989). Furthermore, the cer10 mutant was mapped to chromosome 3, 5.7 centimorgans north of BGL1 (Lolle et al., 1998), close to the ECR gene. A complementation test performed between the cer10 and the Salk_088645 mutant (data not shown) demonstrated that cer10 is allelic to the Salk_088645. We therefore refer to the original cer10 mutant allele as cer10-1 and designate the T-DNA insertional allele Salk_088645 as cer10-2.
The cer10-1 mutant was obtained from an x-ray–mutagenized population (Koornneef et al., 1989). PCR amplifications in the ECR gene region indicated that a deletion and/or rearrangement greater than 5 kb is present in cer10-1, which spans the promoter region, the 5′-untranslated region, and at least 90 bp downstream of the translation start of the ECR gene (Figure 1A).
Transcription of the ECR Gene in the cer10 Mutants
RNA gel blot analysis was used to assess steady state levels of ECR gene transcript in the cer10-1, cer10-2, and Salk_138092 lines. Total RNA was isolated from flower buds of each mutant and hybridized with a radiolabeled full-length ECR cDNA. A single band of ∼1.2 kb was present in wild-type Columbia-0 (Col-0), Salk_138092, and wild-type Landsberg erecta (Ler) (Figure 1C, lanes 1, 2, and 4), but no band was detected in either cer10-1 or cer10-2 (Figure 1C, lanes 3 and 5), indicating that both mutants are transcriptional knockouts. The abundance of ECR transcript in Salk_138092 was comparable to that in wild-type Col-0, suggesting that the T-DNA insertion in this line does not affect the ECR transcription, consistent with the observation that the plant developed normally. We therefore used cer10-1 and/or cer10-2 mutants for subsequent analysis.
The cer10 Mutation Affects Organ Morphogenesis throughout Shoot Development
The cer10 mutant was originally isolated because of its glossy stem phenotype indicative of decreased cuticular wax accumulation (Koornneef et al., 1989). In addition to the wax phenotype, we found that both cer10-1 and cer10-2 exhibited abnormal morphogenesis of all the shoot organs. The earliest phenotypic change observed in the cer10 mutant was downward pointing cotyledons in 3- to 4-d-old seedlings (Figure 2B). Twelve-day-old cer10 seedlings developed true leaves, but the leaves were crinkled and smaller than those of the wild type (cf. Figures 2C and 2D). Despite their small stature and morphological abnormalities, the cer10 plants continued to grow and generated inflorescences, floral organs, and a normal number of leaves without an apparent time delay in comparison with the wild type as shown for 22-d-old plants in Figure 2E. Fully expanded leaves in cer10 were approximately one-third the size of wild-type leaves and they curled along the long axis (cf. Figures 2F and 2G). Inflorescences of the mutant were considerably shorter than those of wild-type plants and had a slender, zigzag appearance (Figure 2H). Flower buds of cer10 were also smaller and fused together (cf. Figures 2I and 2J). Mutants exhibiting floral organ fusions like the cer10 mutant have been reported as having reduced fertility, depending on the severity of the fusion event. The cer10 line was described as male semisterile but female fertile (Lolle et al., 1998). Unlike cer1 and cer6 wax-deficient mutants where problems with pollen hydration cause conditional male sterility (Aarts et al., 1995; Millar et al., 1999), high humidity did not restore fertility in the cer10 mutants. Examination of stamens revealed that cer10 stamen filaments were short and crooked and unable to position anthers at the height of the stigma (cf. Figures 2K and 2L). In addition, most cer10 pollen grains were shriveled, and Alexander's staining (Alexander, 1969) showed that they were inviable (cf. Figures 2M and 2N).
Figure 2.
Abnormal Organ Morphogenesis in the cer10 Mutant.
(A) and (B) Four-day old seedlings of wild-type Col-0 (A) and cer10-2 (B).
(C) and (D) Twelve-day-old seedlings of wild-type Col-0 (C) and cer10-2 (D).
(E) Twenty-two-day-old wild-type Col-0 (left) and cer10-2 (right) plants.
(F) and (G) Expanded adult leaves of wild-type Col-0 (F) and cer10-2 (G).
(H) Stems of adult wild-type Col-0 (left) and cer10-2 (right) plants.
(I) and (J) Flowers of wild-type Col-0 (I) and cer10-2 (J).
(K) and (L) Mechanically opened 11 to 13 stage flowers of wild-type Col-0 (K) and cer10-2 (L).
(M) and (N) Pollen grains of wild-type Col-0 (M) and cer10-2 (N) labeled with Alexander's stain. Bar = 10 μm for (M) and (N).
Green Fluorescent Protein–ECR Complements the Developmental Defects of the cer10 Mutant and the Growth Defect of Yeast tsc13-1Δ Cells and Is Localized to the ER
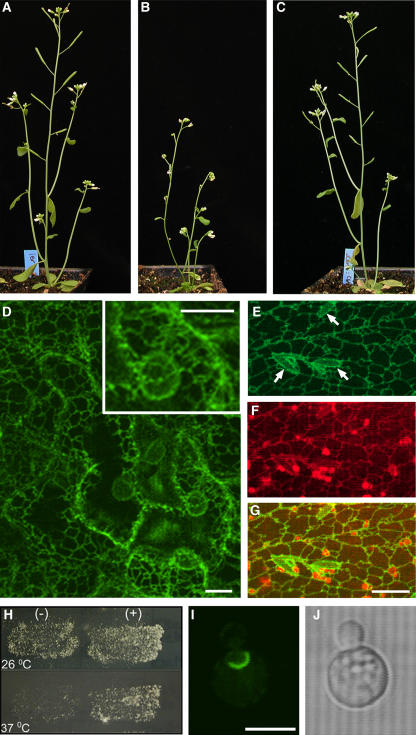
To exclude the possibility that mutations in other genes contribute to defective organ morphogenesis in the cer10 lines, the wild-type ECR cDNA was cloned, fused with green fluorescent protein (GFP) at its 5′ end, and expressed in cer10-1 mutant plants under the control of the 35S promoter. Ten out of 12 independent transgenic lines recovered had wild-type appearance, indicating that all the morphological abnormalities in leaves, stems, and flowers described above were completely restored (Figure 3C). These results demonstrated that the mutation in the ECR gene is indeed responsible for the observed morphological defects.
Figure 3.
Complementation of the cer10 Mutant and Subcellular Localization of the ECR.
(A) to (C) Four-week-old wild-type Ler (A), cer10-1 (B), and cer10-1:GFP-ECR plants (C).
(D) Projection of 12 × 1-μm optical sections of GFP fluorescence through GFP-ECR expressing cer10-1 leaf epidermal cells. Note the similar fluorescence intensity between cortical ER network and nuclear envelopes. Bars = 5 μm.
(E) to (G) Colocalization of GFP-ECR labeled network (E) and the ER network stained by hexyl rhodamine B (F). (G) was obtained by merging images (E) and (F). The arrows in (E) identify the fusiform bodies of the ER outlined in negative contrast by GFP-ECR. Bar = 10 μm for (E) to (G).
(H) The yeast tsc13-1elo2Δ cells transformed with the plasmid p426-ADH without GFP-ECR growing at 26°C (top left) and at 37°C (bottom left) and the tsc13-1elo2Δ cells transformed with the plasmid p426-ADH:GFP-ECR growing at 26°C (top right) and 37°C (bottom right).
(I) and (J) Confocal GFP fluorescence (I) and bright-field (J) images of a yeast tsc13-1elo2Δ cell expressing GFP-ECR. Bar = 5 μm.
Expression of the fully functional GFP-ECR fusion protein in transgenic cer10 plants allowed us to investigate the subcellular localization of the ECR in Arabidopsis. In all 10 transgenic lines with different levels of GFP-ECR expression, the GFP fluorescence labeled a reticulate network typical of the ER in all examined organs, as illustrated in leaves (Figure 3D). The reticulate network labeled by GFP-ECR was also marked by hexyl rhodamine B (Figures 3E to 3G), a dye that stains the ER in plants (Boevink et al., 1996). Furthermore, in some cells, the fusiform bodies, specific ER subdomains (Hawes et al., 2001), were also outlined by the fluorescence of GFP-ECR (Figure 3E, arrows), confirming that GFP-ECR is localized to the ER membrane. Unlike yeast ECR, which is enriched at the nuclear envelope–vacuole junction (Kohlwein et al., 2001), the Arabidopsis ECR is not, as indicated by the similar fluorescence intensity of the GFP-ECR between nuclear envelopes and the cortical ER network (Figure 3D and inset). This uniform distribution of ECR on the ER membrane was also verified by transiently expressing GFP-ECR under the native CER10 promoter in Arabidopsis leaves (data not shown).
Expression of the Arabidopsis GFP-ECR fusion protein in the yeast tsc13-1elo2Δ mutant under the control of the ALCOHOL DEHYDROGENASE (ADH) promoter rescued its temperature-sensitive lethality, indicating that the fusion protein is also functional in the yeast elongase (Figure 3H). As found with the native yeast ECR Tsc13p, the plant GFP-ECR was enriched in a specific ER domain reminiscent of the nuclear–vacuole junction (Figure 3I). These results suggest a different localization of the ECR in Arabidopsis and yeast.
Direction and Extent of Cell Expansion in the cer10 Mutants
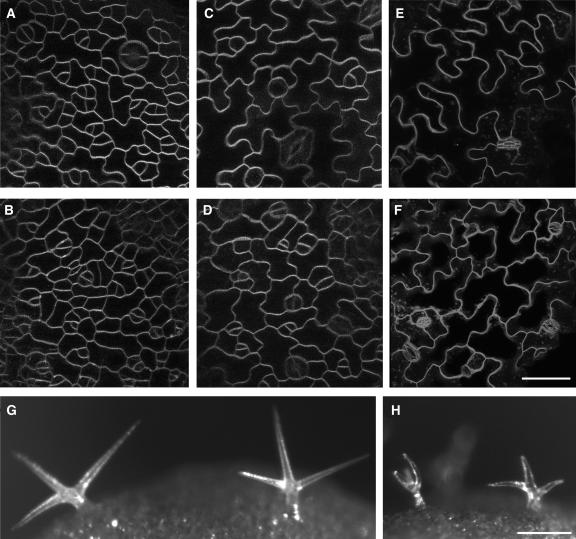
Most if not all organs of cer10 mutants were shrunken and smaller than normal. We reasoned that such organ abnormalities might be a result of a common cellular defect(s) in cell division and/or cell expansion. To explore this, we investigated the number, size, and morphology of leaf epidermal cells in the mutant. Expanded leaves from wild-type plants and cer10 mutants were stained with FM4-64, a lipophilic styryl fluorescent dye known to intercalate the plasma membrane so that the cell shapes can be easily outlined and examined in vivo by confocal microscopy (Zheng et al., 2004). It was immediately apparent that cer10 cells were considerably smaller than wild-type cells, and they had less pronounced lobe structures than those found in the wild-type leaf epidermis (cf. Figures 4E and 4F). In an area of 100 × 100 μm, fully expanded cer10 leaf epidermis had 13 to 15 cells (Figure 4F) compared with only four to six cells found in the epidermis of the wild-type leaves (Figure 4E). Thus, the cer10 leaf cells are approximately three times smaller than those in wild-type plants, consistent with the leaf size reduction of cer10 (Figure 2G). A similar reduction in the length of the stem and of the stem epidermal cells in the cer10 mutant was also found (data not shown). These well-correlated reductions in cell size and organ size illustrated that organ abnormalities seen in the cer10 mutants have likely arisen from aberrant cell expansion. A detailed examination of epidermal pavement cell development demonstrated that initiation of new cells was not compromised in the cer10 leaf epidermis (cf. Figures 4A and 4B), but the direction and extent of cell expansion in both the early stages (stages I and II) and the late stage (after stage II) of leaf development (Fu et al., 2002) were affected (cf. Figures 4C and 4D).
Figure 4.
Defective Cell Expansion in the cer10 Mutants.
(A) to (F) Cell shape was defined by FM4-64 staining, and all images were projections of 10 × 1-μm optical sections of FM4-64 fluorescence through leaf epidermal cells. Bar = 25 μm.
(A) and (B) Newly formed epidermal cells of a wild-type Ler leaf (A) and a cer10-1 mutant leaf (B).
(C) and (D) Expanding epidermal cells of a wild-type Ler leaf (C) and a cer10-1 mutant leaf (D).
(E) and (F) Epidermal cells in a fully expanded wild-type Ler leaf (E) and a cer10-1 mutant leaf (F).
(G) Stereomicrograph of wild-type Ler leaf trichomes.
(H) Stereo images of leaf trichomes in a cer10-1 mutant plant. Bar = 100 μm for (G) and (H).
This conclusion was further substantiated by examination of leaf trichomes. Wild-type trichomes normally consist of a long stalk (∼150 μm long) and several branches (∼150 μm) (Figure 4G). We noted that in the cer10 mutants, although the basic body plan of trichomes was established, they appeared to have short, crooked, and abnormally swollen stalks and branches (Figure 4H), indicating that the cell expansion required for generation of normal trichome stalk and branches is impaired in the cer10 mutant.
Reduced Cuticular Wax Load and Decreased VLCFA Content in Seed Storage Lipids and Sphingolipids Demonstrate, in Planta, the Role of the ECR in VLCFA Elongation
ECR is an enzyme that catalyzes the last step of VLCFA elongation, reduction of the enoyl-CoA, in eukaryotic cells. To investigate the biochemical basis for the observed morphological phenotype as well as to confirm in planta the suggested role of the Arabidopsis ECR in VLCFA elongation (Gable et al., 2004), we evaluated the VLCFA content of complex lipids known to contain these acyl groups or require them for their synthesis.
VLCFAs are precursors of all the aliphatic components of cuticular wax. In both cer10-1 and cer10-2, a reduction of ∼60% in total stem cuticular wax, with a similar reduction in each major wax component, was detected (Table 1). The VLCFA level measured in seed TAGs was also lower (∼30% reduction) in both cer10 lines in comparison with the wild type (Table 2), indicating that mutations in ECR affected VLCFA production for TAG assembly. In yeast, a mutation in the TSC13 gene resulted in shorter acyl chain lengths in sphingolipids, and it was proposed to be the major cause of growth defects of tsc13-1elo2Δ cells (Kohlwein et al., 2001).
Table 1.
Cuticular Wax Loads on Stems of Wild-Type, cer10, and ProCER6:ECRia Plants (μg/g Dry Weight)
| Primary Alcohols
|
Fatty Acid
|
Aldehydes
|
Alkane
|
2° Alcohol
|
Ketone
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Line | Total | C26 | C28 | C30 | C30 | C28 | C30 | C29 | C29 | C29 |
| Col-0b | 8,531 ± 115 | 360 ± 59 | 710 ± 24 | 254 ± 18 | 123 ± 15 | 214 ± 17 | 884 ± 61 | 3,493 ± 63 | 1,747 ± 68 | 747 ± 26 |
| cer10-2b | 3,197 ± 223 | 115 ± 7 | 243 ± 16 | 193 ± 28 | 62 ± 3 | 66 ± 12 | 246 ± 34 | 1,388 ± 141 | 607 ± 66 | 278 ± 17 |
| Lerb | 10,104 ± 726 | 366 ± 51 | 654 ± 79 | 286 ± 48 | 144 ± 19 | 248 ± 26 | 956 ± 60 | 4,329 ± 334 | 2,093 ± 202 | 1,031 ± 77 |
| cer10-1b | 4,016 ± 101 | 132 ± 6 | 269 ± 32 | 251 ± 20 | 84 ± 26 | 73 ± 6 | 455 ± 64 | 1,682 ± 84 | 761 ± 54 | 311 ± 17 |
| Col-0c | 11,435 ± 1,639 | 467 ± 105 | 868 ± 199 | 309 ± 102 | 126 ± 14 | 317 ± 47 | 985 ± 133 | 5,121 ± 729 | 2,272 ± 361 | 971 ± 193 |
| ProCER6:ECRic | 6,652 ± 179 | 223 ± 53 | 512 ± 57 | 330 ± 41 | 71 ± 7 | 110 ± 19 | 532 ± 59 | 3,017 ± 149 | 1,368 ± 72 | 489 ± 26 |
Each value is the mean of six independent measurements ±sd.
Transgenic Arabidopsis expressing an ECR RNAi gene construct driven by the epidermis-specific CER6 promoter.
Two independent wax load measurements on 5-week-old (b) and 7-week-old plants (c).
Table 2.
Altered Fatty Acid Composition of Seed Storage Lipids in cer10 and ProFAE1:ECRia Plants (mol %)
| Line | LCFA | VLCFA | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 20:2 | 22:1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Col-0 | 75.8 | 24.2 | 7.9 ± 0.26 | 2.9 ± 0.16 | 13.5 ± 1.12 | 29.5 ± 1.18 | 19.6 ± 1.44 | 1.8 ± 0.12 | 18.0 ± 0.57 | 2.1 ± 0.14 | 1.6 ± 0.13 |
| cer10-2 | 83.5 | 16.5 | 8.0 ± 0.12 | 3.1 ± 0.06 | 17.8 ± 0.34 | 31.0 ± 0.54 | 21.7 ± 0.43 | 1.1 ± 0.06 | 12.5 ± 0.09 | 1.8 ± 0.02 | 0.7 ± 0.13 |
| ProFAE1:ECRi | 83.9 | 16.1 | 8.6 ± 0.08 | 3.4 ± 0.06 | 20.0 ± 0.98 | 31.9 ± 0.28 | 19.7 ± 0.55 | 1.2 ± 0.09 | 12.9 ± 0.86 | 1.2 ± 0.13 | 0.6 ± 0.08 |
| Ler | 77.3 | 22.7 | 6.9 ± 0.12 | 2.8 ± 0.02 | 20.7 ± 0.3 | 27.6 ± 0.29 | 17.2 ± 0.17 | 2.0 ± 0.03 | 17.1 ± 0.15 | 1.6 ± 0.03 | 1.4 ± 0.03 |
| cer10-1 | 83.4 | 16.6 | 7.1 ± 0.51 | 3.1 ± 0.04 | 28.1 ± 0.34 | 26.5 ± 0.51 | 17.2 ± 0.42 | 1.5 ± 0.01 | 13.1 ± 0.18 | 1.1 ± 0.07 | 0.6 ± 0.04 |
Each value is the mean of six independent measurements ±sd.
Transgenic Arabidopsis expressing an ECR RNAi gene construct driven by the seed-specific FAE1 promoter.
To determine if the cer10 mutation affected VLCFA composition of sphingolipids, we first examined the VLCFA content of glucosylceramides (GlcCers), an abundant subclass of sphingolipids that are important constituents of the plasma membrane and tonoplast in plants (Dunn et al., 2004). Similar to sphingolipids in rye (Secale cereale) leaves (Cahoon and Lynch, 1991) and maize (Zea mays) roots (Bohn et al., 2001), we found that hydroxylated fatty acids are the predominant fatty acids in GlcCers isolated from Arabidopsis shoots. In wild-type (Ler) plants, hydroxylated C16:0 and C24:0 accounted for ∼53 and ∼28% of GlcCer fatty acids, respectively (Table 3). In the cer10-1 mutant, the hydroxylated C16:0 fatty acid content was increased to 64% at the expense of hydroxylated VLCFAs (Table 3, note the C24:0-OH reduction to ∼21% in the mutant).
Table 3.
Altered Fatty Acyl Chain Length of GlcCers in the cer10 Plants (mol %)
| Line | LCFA-OH | VLCFA-OH | 16:0-OH | 18:0-OH | 20:0-OH | 22:0-OH | 23:0-OH | 24:0-OH | 25:0-OH | 26:0-OH |
|---|---|---|---|---|---|---|---|---|---|---|
| Ler | 52.8 | 47.2 | 52.8 ± 1.18 | Trace | 0.8 ± 0.15 | 7.3 ± 0.57 | 2.0 ± 0.72 | 27.6 ± 0.16 | 3.3 ± 0.22 | 6.5 ± 0.82 |
| cer10-1 | 63.7 | 36.3 | 63.7 ± 0.08 | Trace | 0.9 ± 0.13 | 7.2 ± 0.76 | 1.1 ± 0.11 | 21.4 ± 1.04 | 1.7 ± 1.2 | 4.4 ± 0.12 |
In plants, in addition to GlcCers, inositol phosphorylceramides are thought to be another major subclass of complex sphingolipids (Dunn et al., 2004). Analyses of long-chain bases in GlcCers isolated from 8-week-old Arabidopsis (Imai et al., 2000) and in total tissues of 19-d-old Arabidopsis (Sperling et al., 1998) suggested that other complex sphingolipids, such as inositol phosphorylceramides, might be relatively more abundant than GlcCers (Imai et al., 2000). However, the actual molecular species composition of Arabidopsis sphingolipids has not been established for any tissue or isolated membrane domain because, at present, methods for purification of complex sphingolipids, other than GlcCers, are not available. In an attempt to assess if the cer10 mutation affected the VLCFA composition of other complex sphingolipids besides the GlcCers, we examined the profile of hydroxylated fatty acyl chains in total lipids extracted from shoot tissues because only sphingolipids have been reported to contain hydroxylated fatty acids (Cahoon and Lynch, 1991; Millar et al., 1998; Bohn et al., 2001; Bonaventure et al., 2003). As indicated in Table 4, the profile of major hydroxylated fatty acids was different from that found in GlcCers (Table 3). Thus, hydroxylated fatty acids released from complex sphingolipids other than GlcCers were included. Similar to GlcCers, hydroxylated long fatty acid (C16:0) content in total shoot lipids was increased at the expense of VLCFAs (Table 4). These data suggest that VLCFA composition of complex sphingolipids besides GlcCers was also affected in the cer10 mutant. Furthermore, we found no significant difference in absolute levels of major hydroxylated fatty acids between wild-type and cer10 plants (Table 4). Even though the determination of absolute levels of sphingolipids by measuring hydroxylated fatty acids is not as accurate as long-chain base analyses (Sperling et al., 1998) because all acyl chains in sphingolipids may not be hydroxylated, our results suggest that total sphingolipid content of the mutant was similar to the wild type.
Table 4.
Altered Profile (mol %) but Not Quantity (μg/g Fresh Shoot Tissues) of Major Hydroxylated Fatty Acyl Chains Released from Complex Sphingolipids in cer10 Plants
| Line | Quantity | LCFA-OH | VLCFA-OH | 16:0-OH | 24:1-OH | 24:0-OH |
|---|---|---|---|---|---|---|
| Ler | 101.2 ± 12.5 | 35.7 | 64.3 | 35.7 ± 0.8 | 21.1 ± 0.3 | 43.2 ± 0.5 |
| cer10-1 | 97.7 ± 21.1 | 41.0 | 59.0 | 41.0 ± 0.9 | 22.9 ± 0.7 | 36.1 ± 0.2 |
Collectively, these results confirmed that the ECR gene product is involved in the synthesis on all VLCFAs in Arabidopsis, required for cuticular wax, storage lipid, and sphingolipid metabolism. However, defects in cuticular wax biosynthesis or seed storage lipid VLCFA accumulation do not typically affect organ morphogenesis during plant development. For example, the most dramatic reduction in cuticular wax load was observed in CER6-silenced plants that contained <7% of wild-type wax levels, without any visible consequences on plant morphology (Millar et al., 1999). Similarly, an abolishment of VLCFA in seed TAGs has no effect on the morphology of vegetative or floral parts of the plant (Kunst et al., 1992). To directly verify that loss of the ECR activity in the epidermis or the embryo did not cause morphological changes observed in cer10 mutants, we silenced the ECR gene specifically in these tissues by introduction of RNA interference (RNAi) constructs targeted against the ECR gene in the epidermal cells (ProCER6:ECRi) or the embryo (ProFAE1:ECRi). Transgenic Arabidopsis ProCER6:ECRi lines exhibited a ∼40% reduction in total wax load (Table 1), whereas ProFAE1:ECRi plants showed an ∼30% reduction in the seed storage lipid VLCFA content (Table 2), respectively. No morphological defects in transgenic plants were observed in either case. Therefore, the reduction the VLCFA content of sphingolipids is the most likely cause of the morphological abnormalities seen in the cer10 mutants.
Defective Endocytic Membrane Traffic in the cer10 Mutants
The presence of the natural C26 VLCFA in yeast sphingolipids is an essential prerequisite for maintaining lipid raft domains (Eisenkolb et al., 2002) that play important roles in endomembrane organization and dynamics in yeast and animal cells (Ikonen, 2001). To determine if detected changes in sphingolipid acyl chain length in the cer10 mutant affect the endomembrane structure and function, we examined the endomembrane system in leaf epidermal pavement cells using various fluorescent markers.
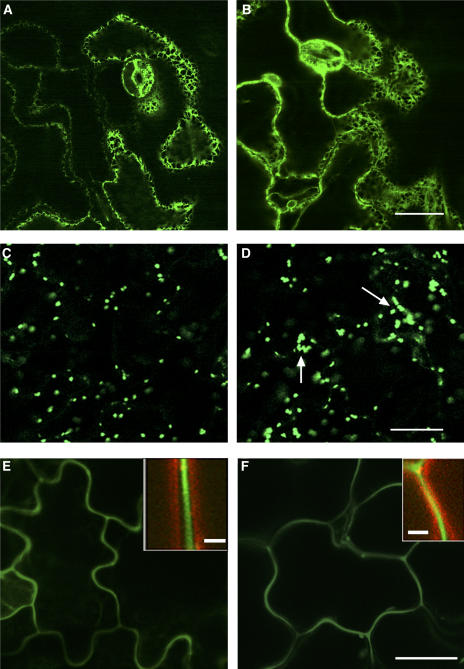
Crossing the cer10-1 mutant with a line containing the ER marker GFP-HDEL (Zheng et al., 2004) allowed ER visualization that indicated that there was no significant difference in the overall ER morphology between wild-type (Figure 5A) and cer10 cells (Figure 5B). Examination of Golgi stacks marked by sialyltransferase (ST)-GFP (Saint-Jore et al., 2002; Zheng et al., 2004) demonstrated that they were similar in number (Figures 5C and 5D) and mobility in the wild type and cer10. However, in the cer10 mutant, the Golgi appeared larger and had a tendency to form ring-like clusters comprised of 8 to 15 Golgi stacks (Figure 5D, arrows). Despite this change in Golgi stack organization, the steady state secretion evaluated by transport of the fluorescent secretory cargo molecule secretory GFP (secGFP) (Zheng et al., 2004) to the apoplast was not impaired in the cer10 cells (cf. Figures 5E and 5F).
Figure 5.
Morphology of the ER and Golgi and Secretion of secGFP to the Apoplast.
(A) and (B) GFP-HDEL–labeled ER in wild-type (A) and cer10-1 (B) leaf epidermal cells. Bar = 10 μm.
(C) and (D) ST-GFP–labeled Golgi stacks in wild-type (C) and cer10-1 (D) leaf epidermal cells. The arrows point to ST-GFP clusters consisting of ∼15 Golgi stacks. Bar = 10 μm.
(E) and (F) secGFP fluorescence distribution in wild-type (E) and cer10-1 (F) leaf epidermal cells. The insets in (E) and (F) show secGFP fluorescence (green) in the apoplast defined by the plasma membrane labeling with FM4-64 (red) dye. Bar = 10 μm.
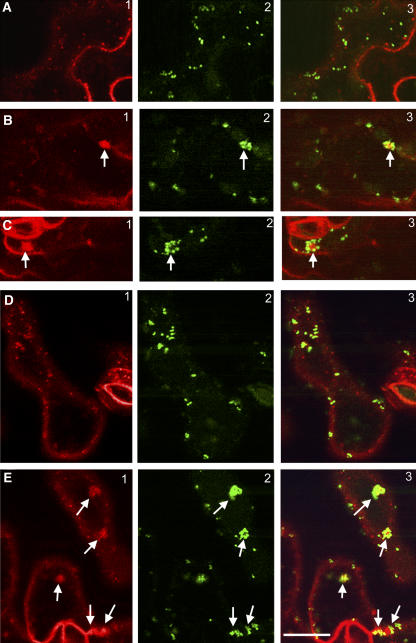
We then used FM4-64 to track endocytic transport in the mutant cells. As in yeast, the uptake of the dye in plant cells is ATP and time dependent (Bolte et al., 2004), and it serves as a good marker for studying membrane internalization, recycling, and endocytosis (Geldner et al., 2003). In wild-type epidermal pavement cells, the fluorescence of FM4-64 first labeled the plasma membrane and was subsequently internalized, giving rise to various intracellular fluorescent punctate structures, which were distinct from the ST-GFP–labeled Golgi (Figure 6A). Prolonged staining (>1 h) usually outlined a large vacuolar structure as described (Bolte et al., 2004). In the cer10-1 mutant, in addition to the labeling of punctate structures, we observed that the internalized FM4-64 aggregated into one or two large clusters per cell (Figures 6B and 6C, arrows). Longer labeling revealed several smaller vacuoles in most cells, rather than a single large vacuole. Strikingly, the FM4-64–labeled clusters were surrounded with an ST-GFP–labeled ring of Golgi stacks (Figures 6B and 6C, arrows), resembling the pattern described for the defective endocytic membrane traffic in brefeldin A (BFA) or cytochalasin D (cyt-D)–treated cells (Baluska et al., 2002; Geldner et al., 2003; Grebe et al., 2003). These results suggested abnormalities in the endocytic membrane organization and transport in the cer10 mutant. We found that FM4-64 internalization in the cer10 mutant was also hypersensitive to BFA treatment, as indicated in Figure 6E, showing more apparent clusters (two to three) of internalized FM4-64 surrounded by ST-GFP–labeled Golgi stacks in cer10 cells treated with BFA (10 μM) for 30 min (cf. Figures 6E to 6B and 6C). No clear FM4-64 aggregates surrounded by Golgi stacks were seen in wild-type cells after the identical BFA treatment (Figure 6D). These results further support the hypothesis that FM4-64 clusters observed in the cer10 mutant have likely arisen from defective endocytic membrane traffic.
Figure 6.
Defective Endocytic Membrane Traffic in the cer10 Mutants.
(A) FM4-64–labeled endosomal compartments (A1) and ST-GFP fluorescence (A2) in wild-type leaf epidermal cells. A3 is a merged image of A1 and A2.
(B) and (C) FM4-64–labeled endosomal compartments (B1 and C1) and ST-GFP–labeled Golgi stacks (B2 and C2) in cer10-1 leaf epidermal cells. B3 and C3 are merged images of B1 and B2, and C1 and C2, respectively. The arrows indicate clustered FM4-64–labeled endosomal compartments surrounded by ST-GFP–labeled Golgi.
(D) FM4-64–labeled endosomal compartments (D1) and ST-GFP–labeled Golgi (D2) in wild-type leaf epidermal cells treated with 10 μM BFA for 0.5 h. D3 is a merged image of D1 and D2.
(E) FM4-64–labeled endosomal compartments (E1) and ST-GFP–labeled Golgi (E2) in cer10-1 leaf epidermal cells treated with 10 μM BFA for 0.5 h. E3 is a merged image of E1 and E2. The arrows indicate FM4-64–labeled endosomal compartments surrounded by ST-GFP–labeled Golgi rings.
Bar = 10 μm for (A) to (E).
DISCUSSION
ECR catalyzes the final step of VLCFA synthesis, the reduction of a double bond that is in the α,β position to a carbonyl group to yield a fully saturated VLCFA. A bioinformatics survey of the Arabidopsis genome identified the gene At3g55360 as the sole ECR candidate (Kohlwein et al., 2001; Dunn et al., 2004). In this study, we have identified two Arabidopsis cer10 mutants, cer10-1 and cer10-2, carrying a deletion and a T-DNA insertion in the ECR gene, respectively, which abolish ECR transcript accumulation. The two mutants exhibit identical biochemical and developmental phenotypes that reveal the functions of ECR in VLCFA production and plant development.
ECR Catalyzes VLCFA Biosynthesis throughout the Plant
Analyses of Arabidopsis cer10 mutants disrupted in the ECR gene described in this report provide in planta evidence that the ECR is involved in VLCFA biosynthesis for the production of wax, seed TAGs, and sphingolipids. This result is consistent with our prediction (Millar and Kunst, 1997) that, unlike the condensing enzyme, the ECR is not a unique component of individual ER-associated fatty acid elongation systems, but instead participates in VLCFA elongation reactions in all cells to produce substrates of diverse chain lengths used to make different classes of lipids.
Because of the universal role of ECR in plant VLCFA production and evidence that VLCFAs are essential for normal embryo and postembryonic development of Arabidopsis (Baud et al., 2004) and yeast viability (Kohlwein et al., 2001), we anticipated that knockout mutations in the single Arabidopsis ECR gene would be lethal. However, the isolation of the cer10 mutants demonstrated that this was not the case. Curiously, in the absence of the ECR gene product, cer10 mutants still accumulated considerable amounts of cuticular wax, as well as VLCFAs in sphingolipids and seed TAGs. It is possible that the ECR identified on the basis of sequence similarity to the yeast Tsc13p is not the sole ECR in Arabidopsis. Alternatively, enzymes functionally similar to the ECR may complement the cer10 deficiency by supporting VLCFA synthesis. The C terminus of the yeast ECR Tsc13p has homology to steroid-5-reductases that, like the ECR, catalyze the reduction of a double bond adjacent to a carbonyl group (Kohlwein et al., 2001). Arabidopsis has four coding sequences containing the conserved steroid-5-reductase domain in addition to the ECR (Arabidopsis Genome Initiative, 2000), so it is conceivable that in the absence of a functional ECR, one or more of the gene products encoded by these genes might act as ECRs in VLCFA elongation.
The Arabidopsis ECR Is Localized throughout the ER
In plants, the cellular organization of VLCFA biosynthesis has not been characterized in detail. However, CER6, a condensing enzyme (Millar et al., 1999) that catalyzes the elongation of C22 fatty acyl-CoAs or longer in epidermal cells (O. Rowland, B. Blacklock and L. Kunst, unpublished data), is distributed evenly on the ER (H. Zheng and L. Kunst, unpublished data). Visualization of the functional GFP-ECR fusion protein in transgenic plants by confocal microscopy revealed that the Arabidopsis ECR is also distributed throughout the ER.
VLCFA biosynthesis in yeast is also catalyzed by the ER-bound enzymes (Kohlwein et al., 2001). However, the yeast ECR protein is highly enriched in the nuclear–vacuolar junction, a specific ER subdomain (Kohlwein et al., 2001). Interestingly, when expressed in S. cerevisiae, the Arabidopsis ECR is also enriched in a specific ER domain reminiscent of the nuclear–vacuole junction. This demonstrates a fundamental difference in ECR location between plants and yeast that might be functionally significant. It has been hypothesized that localized synthesis of VLCFAs in yeast might facilitate the formation and subsequent traffic of lipid rafts (Kohlwein et al., 2001). Indeed, in yeast, detergent-resistant lipid microdomains begin to be formed in the ER, and GPI-anchored cargo proteins may be sorted in the ER (Muniz et al., 2001). Thus, the localized VLCFA synthesis in the ER might be required for a physical separation of cargo molecules into lipid rafts at the level of the ER.
In mammalian cells, formation of lipid rafts occurs in the Golgi (Ikonen, 2001) and sorting of raft-associated proteins might take place in the trans-Golgi network and endosomes (Ikonen, 2001; Nabi and Le, 2003). This difference in the cellular location of lipid raft formation between yeast and mammals may be related to the difference in the length of the very long fatty acyl chains of their sphingolipids (Ikonen, 2001). Yeast sphingolipids accumulate C26 acyl groups as their major VLCFAs, whereas mammalian sphingolipids primarily contain C24 VLCFA. Arabidopsis sphingolipids also mostly contain C24 VLCFA and resemble sphingolipids in mammals. It is thus plausible that biosynthetic and trafficking events involving sphingolipids and formation of lipid rafts in Arabidopsis are different from S. cerevisiae but analogous to mammals.
ECR Is Required for Normal Shoot Development and Cell Expansion
Despite our prediction that the ECR-deficient plants may not be viable, cer10 mutants survived embryogenesis but exhibited severe morphological abnormalities and reduced size of all the aerial organs. A detailed examination of epidermal pavement cells in developing leaves illustrated that cer10 cells were approximately three times smaller than wild-type cells, a good correlation to the threefold reduction in leaf size. Initiation of new cells was not compromised in the cer10 leaf epidermis, but aberrant cell expansion detected in both the early and the late stages of leaf development was the likely cause of reduced organ size (Fu et al., 2002).
Molecular and cellular mechanisms that regulate plant cell expansion during development remain poorly understood. Characterization of Arabidopsis cer10 mutants implicated VLCFAs as important factors in this process. As discussed above, VLCFAs serve as precursors of cuticular wax and accumulate in TAGs and sphingolipids in higher plants. At present, conclusive experiments to determine whether a sole reduction of VLCFA levels in sphingolipids causes cer10 phenotypes, such as rescuing cer10 phenotypes by specifically adjusting VLCFA composition in sphingolipids to the wild type, are not feasible. Nevertheless, several lines of evidence point to altered VLCFA composition of sphingolipids as the underlying cause of developmental abnormalities observed in ECR-deficient cer10 lines. Previous research on cuticular wax and seed storage lipid production demonstrated that a decrease in wax load to below 7% of wild-type levels and a complete absence of VLCFAs in TAGs are tolerated by plants without any consequences on their growth and development (Kunst et al., 1992; Aarts et al., 1995; Millar et al., 1999). Similarly RNAi-induced reduction in cuticular wax accumulation and VLCFA content of TAGs that phenocopied those of the cer10 mutants, performed in this study, had no effect on shoot size or morphology. By contrast, in yeast cells, the chain length of the sphingolipid VLCFA constituent is known to be of critical importance. Yeast elo2Δ elo3Δ double mutants with lesions in condensing enzymes catalyzing C24/C26 fatty acid synthesis are inviable (Oh et al., 1997). More specifically, the natural C26 fatty acid is an essential structural determinant of sphingolipids required for their association into lipid raft domains (Eisenkolb et al., 2002). The presence of C26 VLCFA is also required for proper functioning of lipid rafts in protein trafficking and protein stability of plasma membrane proteins in vivo (David et al., 1998; Eisenkolb et al., 2002). In plants, the existence of lipid rafts has not been demonstrated in vivo, but detergent-resistant plasma membrane domains that are greatly enriched in sphingolipids identified as GlcCers and specifically contain H+-ATPases, heterotrimetric G-proteins, and GPI-anchored proteins have been identified in tobacco (Nicotiana tabacum) leaves (Peskan et al., 2000; Mongrand et al., 2004) and BY-2 cells (Mongrand et al., 2004). Recently, detergent-resistant, sphingolipid-rich, and sterol-rich membrane domains enriched in H+-ATPases and GPI-anchored proteins were also isolated from cultured Arabidopsis callus (Borner et al., 2005). Thus, if lipid rafts indeed exist in plants, it is conceivable that VLCFA composition of sphingolipids plays an important role in their formation, as is the case in yeast, and that substitution of C24 acyl groups of sphingolipids with shorter chain lengths in cer10 mutants resulted in aberrant execution and coordination of raft-related activities in membrane traffic leading to abnormal cell expansion, as suggested for mutants with altered sterol composition (Schrick et al., 2000; Willemsen et al., 2003).
Aberrant Endocytic Membrane Traffic in ECR-Deficient Mutants
Based on the visualization of GFP-HDEL in both wild-type and cer10 mutant plants, it appears that the overall morphology of the ER between wild-type and mutant plants is not markedly different. However, an ST-GFP fluorescence-based investigation of the cer10 mutant revealed that Golgi stacks in the mutant appeared to aggregate and form ring-like structures. Using FM4-64 as an internalization marker, we showed that endocytic membrane traffic in expanding leaf pavement cells of the cer10 was defective, resulting in clustering of internalized FM4-64–labeled membrane surrounded by a ring of Golgi stacks. This cellular phenotype resembles the changes observed in BFA- or cyt-D–treated elongating root cells (Geldner et al., 2003; Grebe et al., 2003).
Although endocytosis has not been well studied in plants, there is enough information available to suggest that the identified defect in this process could account for the aberrant cell expansion in the cer10 mutants. Endocytosis is known to be very active during cell expansion (Samuels and Bisalputra, 1990). In addition, mutations in Arabidopsis dynamin-like genes (Kang et al., 2003a, 2003b) result in defects in plasma membrane recycling and cell expansion. Dynamins are large GTP binding proteins involved in both clathrin-dependent and raft-mediated endocytic traffic (Nabi and Le, 2003). In plants, cell expansion is driven by turgor pressure and directly depends on local modification of the cell wall. Recently, Baluska et al. (2002) showed that cell wall pectin recycling necessary for cell wall remodeling is dependent on endocytic membrane traffic/recycling. Finally, auxin transport important for cell expansion requires proper plasma membrane localization of auxin transporters via BFA- and cyt-D–sensitive endocytosis (Friml et al., 2002; Geldner et al., 2003; Grebe et al., 2003).
Recently, using the fluorescent sterol binding antibiotic filipin, Grebe et al. (2003) demonstrated that sterols and certain plasma membrane proteins such as PIN2 actually undergo endocytosis/recycling that is sensitive to BFA and cyt-D. This sterol-related process is selectively involved in correct distribution of PM proteins and appears to function independently of the GNOM-dependent endocytosis (Grebe et al., 2003; Willemsen et al., 2003). We speculate that the sterol-related endocytic membrane traffic (Grebe et al., 2003) and the sphingolipid-sensitive endocytic membrane traffic described here might represent a distinct endocytic process analogous to raft-dependent endocytosis in yeast and mammals, defined by its clathrin independence and dynamin dependence (Nabi and Le, 2003).
Unlike the obvious endocytosis defects, changes in the steady state secretion of secGFP were not detected in cer10 mutants. At present, it is not clear if this is an indication that sphingolipids are not involved in the secretion of secGFP in plants or if more pronounced changes in sphingolipid composition are required to affect this process. Because sphingolipids are essential for transport of raft-associated proteins, it would be interesting to follow trafficking of GPI-anchored proteins (Borner et al., 2003) as well as plant homologs of yeast Pma1 H+-ATPase, whose conformation and distribution require proper maintenance of lipid rafts (Eisenkolb et al., 2002; Wang and Chang, 2002) in the cer10 mutants, to gain a clearer picture of the potential roles of sphingolipids in membrane transport processes in plants.
In summary, the functional characterization of the plant ECR impaired in cer10 mutants provides genetic evidence for the involvement of VLCFAs in cell expansion during plant organogenesis. The availability of cer10 mutants with altered sphingolipid composition offers new opportunities for the identification of lipid rafts and elucidation of their in vivo function, formation, and regulation in plants.
METHODS
Plant Materials and Growth Conditions
Salk T-DNA insertional mutant lines and cer10-1 mutant seed were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). cer10-1 plants expressing GFP-HDEL, ST-GFP, and secGFP were made by crossing transgenic Arabidopsis thaliana homozygous for the GFP fusions (pollen donors) to the mutant (egg donor). Plants on soil (Sunshine Mix 5; SunGro, Vancouver, British Columbia, Canada) or on AT-agar plates (Somerville and Ogren, 1982) were grown at 20 to 22°C under continuous light (100 to 120 μE m−2 s−1 photosynthetically active radiation).
RNA Extraction and Gel Blot Analysis
The RNA extractions were performed as described by Zheng et al. (2004). Approximately ten micrograms of RNA/lane was separated on a 1.1% denaturing agarose gel, transferred onto a nylon membrane, and hybridized with 32P-labeled ECR DNA, followed by detection with a Storm860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Plasmid Construction and Plant and Yeast Transformation
The plasmid pENTR/SD/D-TOPO containing the ECR cDNA (U17245) was obtained from the Arabidopsis Biological Resource Center. To generate GFP-fused ECR, the cDNA of the ECR gene was amplified from pENTR/SD/D-TOPO with two primers, ECRBS5 (5′-CGGGATCCGTCGACATGAAGGTCACCGTCGTCTCCCCGC-3′) and ECRSTOP3 (5′-CGGGATCCTAAAGGAATGGAGGAAGTATCAC-3′), and was subcloned into the binary vector pVKH18-GFPN as a SalI-BamHI fragment resulting in pVKH18-GFP-ECR. The plasmid pVKH18-GFPN had been assembled using the following steps. The coding sequence of GFP5 in pVKH18-GFP-HDEL (Zheng et al., 2004) was amplified with primers GFP5N5 (5′-GCTCTAGAACAATGAGTAAAGGAGAAGAACTTTTCACTGG-3′) and GFP5N3 (5′-CGGGATCCTGCGTCGACTTTGTATAGTTCATCCATGCCATGTG-3′) and subcloned into pVKH18-sGFP (Zheng et al., 2004), replacing secGFP to yield pVKH18-GFPN.
To engineer the RNAi ECR gene constructs ProCER6:ECRi and ProFAE1:ECRi, the ECR gene was amplified as a BamHI/XhoI/XbaI-EcoRI fragment with primers ECRXX5 (5′-CGGGATCCCTCGAGTCTAGAATGAAGGTCACCGTCGTCTCCGC-3′) and ECREcoRI3 (5′-CGGAATTCAAGGAATGGAGGAAGTATCAC-3′) and as a BamHI/SalI-EcoRI fragment with primers ECRBS5 and ECREcoRI3. These PCR products were then inserted into pHANNIBAL (Wesley et al., 2001) using XhoI-EcoRI and BamHI-HindIII sites to create the intron-spliced ECR gene construct pHANNIBAL-ECRi. The generated ECRi was cleaved as an XbaI-SalI fragment and used to replace GFPN in the pVKH18- ProCER6:GFPN and pVKH18-ProFAE1:GFPN, yielding pVKH18-ProCER6:ECRi and pVKH18-ProFAE1:ECRi, respectively. The plasmids pVKH18- ProCER6:GFPN and pVKH18-ProFAE1:GFPN were generated by replacing the 35S promoter in expression vector pVKH18-GFPN with the CER6 promoter in pLK379 (S. Clemens and L. Kunst, unpublished data) and the FAE1 promoter in pMS128 (Rossak et al., 2001). All PCR products were first cloned into pBluescript KS+ and sequenced.
Transgenic Arabidopsis cer10-1 lines expressing GFP-ECR, ProCER6:ECRi, and ProFAE1:ECRi were generated by transformation of the plasmid pVKH18-GFP-ECR into Arabidopsis cer10-1 mutant plants and pVKH18-ProCER6:ECRi and pVKH18-ProFAE1:ECRi into wild-type Col-0 plants using the floral dipping method (Clough and Bent, 1998).
For expression of GFP-ECR in yeast tsc13-1elo2Δ cells, the coding region of GFP-ECR in pVKH18-GFP-ECR was released as a XbaII-Ecl136II fragment and then subcloned between SpeI and SmaI of the yeast expression vector p426-ADH (Mumberg et al., 1995), yielding p426-ADH:GFP-ECR. Saccharomyces cerevisae strain TDY2057 (tsc13-1elo2Δ) (Kohlwein et al., 2001) was transformed and grown on selective media lacking uracil for complementation analysis.
Extraction and Analysis of Cuticular Wax, Seed Storage Lipids, and Sphingolipids
Cuticular wax was extracted and analyzed by gas chromatography (GC) according to Millar et al. (1999). Seed fatty acid methyl esters were prepared and analyzed as described by Rossak et al. (2001). The extraction of total lipids from shoots of 6-week-old Arabidopsis plants, after the addition of C17:1 as internal standard, and isolation of GlcCers from the total lipid extract were performed according to Cahoon and Lynch (1991). The total lipids and isolated GlcCers were transmethylated in 3M methanolic-HCl (Supelco, Bellefonte, PA) at 80°C for 18 h followed by extraction with hexane. The solvent was removed by a gentle stream of N2, then all samples were treated with bis-N,N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich, St. Louis, MO) in pyridine (Fluka, Buchs, Switzerland) for 30 min at 70°C. The qualitative analyses of hydroxylated fatty acids in total lipids and fatty acids from GlcCers were performed using a capillary gas chromatograph (6890N; Agilent, Palo Alto, CA) with mass spectrometric detector (70 eV, mass-to-charge ratio 50 to 750, 5973N; Agilent). The quantitative analysis of the mixtures was performed by capillary GC (Agilent; 30 m DBHP-1, 0.32-mm i.d., df = 1 μm, J&W Scientific, Folsom, CA) using a flame ionization detector under the same GC conditions as above.
Stereomicroscopy
A WildM5A stereomicroscope (Wild Heerbrugg, Gais, Switzerland) fitted with a digital SPOT camera (Diagnostic Instruments, Sterling Heights, MI) was used to take close-up pictures of seedlings, leaves, trichomes, flowers, stamens, and anthers. Pollen grains were mechanically released from anthers and were stained for 10 to 15 min with Alexander's stain (Alexander, 1969) for morphology and viability examination. Images were captured with a Zeiss Axioskop 2 light microscope (Jena, Germany) equipped with a digital SPOT camera.
FM4-64 and Hexyl Rhodamine B Staining, BFA Treatment, and Confocal Laser Scanning Microscopy
Leaves of Arabidopsis plants were immersed in FM4-64 (8.2 μM) solution for 10 min for plasma membrane staining and 30 to 60 min for membrane internalization analysis. For ER staining, seedlings of transgenic cer10 expressing GFP-ECR were immersed in hexyl rhodamine B solution (1.6 μM) for 10 to 30 min. BFA treatment was performed by immersion of expanding leaves of Arabidopsis plants in BFA (10 μM) solution. The GFP, FM4-64, and hexyl rhodamine B fluorescence were examined with a Radiance 2000 confocal laser scanning microscope (Bio-Rad, Hercules, CA). The excitation wavelength for GFP was 488 nm with the emission filter set at 500 to 530 nm. FM4-64 and hexyl rhodamine B were excited with a 543-nm argon ion laser line and a 600-nm long-pass emission filter. All confocal images obtained were processed with ImageJ (http://rsb.info.nih.gov/ij) and Adobe Photoshop 5.0 (Mountain View, CA) software.
Acknowledgments
We are grateful to Reinhard Jetter for his help with GC–mass spectrometry analysis of total lipids and gluocosylceramides, to Chris Hawes for the ST-GFP line, to Ian Moore for kindly allowing us to use the GFP-HDEL and secGFP lines, to Teresa Dunn for the yeast tsc13-1elo2Δ mutant, to Peter Waterhouse for the pHannibal vector, and to the Salk Institute Genomic Analysis Laboratory (La Jolla, CA) for T-DNA insertional lines and the plasmid pENTR/SD/D-TOPO containing the ECR cDNA. We also thank Gillian Dean, George Haughn, Tanya Hooker, Lacey Samuels, and Geoffrey Wasteneys for insightful comments on the manuscript. This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to L.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ljerka Kunst (kunst@interchange.ubc.ca).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.030155.
References
- Aarts, M.G.M., Keijzer, C.J., Stiekema, W.J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7, 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Hlavacka, A., Samaj, J., Palme, K., Robinson, D.G., Matoh, T., McCurdy, D.W., Menzel, D., and Volkmann, D. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol. 130, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud, S., Bellec, Y., Miquel, M., Bellini, C., Caboche, M., Lepiniec, L., Faure, J.D., and Rochat, C. (2004). gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep. 5, 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P., Cruz, S., Hawes, C., Harris, N., and Oparka, K.J. (1996). Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J. 10, 935–941. [Google Scholar]
- Bohn, M., Heinz, E., and Lüthje, S. (2001). Lipid composition and fluidity of plasma membranes isolated from corn (Zea mays L.) roots. Arch. Biochem. Biophys. 387, 35–40. [DOI] [PubMed] [Google Scholar]
- Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N.D., and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214, 159–173. [DOI] [PubMed] [Google Scholar]
- Bonaventure, G., Salas, J.J., Pollard, M.R., and Ohlrogge, J.B. (2003). Disruption of the FATB gene in Arabidopsis demonstrated an essential role of saturated fatty acids in plant growth. Plant Cell 15, 1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H., Lilley, K.S., Stevens, T.J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., MacAskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistent membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon, E.B., and Lynch, D.V. (1991). Analysis of glucocerebrosides of rye leaf and plasma membrane. Plant Physiol. 95, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- David, D., Sundarababu, S., and Gerst, J.E. (1998). Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J. Cell Biol. 143, 1167–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, T.M., Lynch, D.V., Michaelson, L.V., and Napier, J.A. (2004). A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. (Lond) 93, 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenkolb, M., Zenzmeier, C., Leitner, E., and Schneiter, R. (2002). A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol. Biol. Cell 13, 4414–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling, E., and Mukherjee, K.D. (1991). Acyl-CoA elongase from a higher plant (Lunaria annua): Metabolic intermediates of very-long-chain acyl-CoA products and substrate specificity. Biochim. Biophys. Acta 1082, 239–246. [DOI] [PubMed] [Google Scholar]
- Fiebig, A., Mayfield, J.A., Miley, N.L., Chau, S., Fischer, R.L., and Preuss, D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12, 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator AtPIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable, K., Garton, S., Napier, J.A., and Dunn, T.M. (2004). Functional characterisation of the Arabidopsis thaliana ortholog of Tsc13p, the enoyl reductase of the yeast microsomal fatty acid elongating system. J. Exp. Bot. 55, 543–545. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230. [DOI] [PubMed] [Google Scholar]
- Gray, J.E., van der Lee, F.M., Bahrami, A.R., Sijmons, P.C., Woodward, F.I., Schuch, W., and Hetherington, A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408, 713–716. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Mobius, W., Ueda, T., Nakano, A., Geuze, H.J., Rook, M.B., and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387. [DOI] [PubMed] [Google Scholar]
- Han, G., Gable, K., Kohlwein, S.D., Beaudoin, F., Napier, J.A., and Dunn, T.M. (2002). The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J. Biol. Chem. 277, 35440–35449. [DOI] [PubMed] [Google Scholar]
- Hawes, C., Saint-Jore, C., Martin, B., and Zheng, H. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 6, 245–246. [DOI] [PubMed] [Google Scholar]
- Ikonen, E. (2001). Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13, 470–477. [DOI] [PubMed] [Google Scholar]
- Imai, H., Morimoto, Y., and Tamura, K. (2000). Sphingoid base composition of monoglucosylceramides in Brassicaceae. J. Plant Physiol. 157, 453–456. [Google Scholar]
- Kang, B.H., Busse, J.S., and Bednarek, S.Y. (2003. a). Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.H., Rancour, D.M., and Bednarek, S.Y. (2003. b). The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J. 35, 1–15. [DOI] [PubMed] [Google Scholar]
- Kohlwein, S.D., Eder, S., Oh, C.S., Martin, C.E., Gable, K., Bacikova, D., and Dunn, T. (2001). Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and Thiel, F. (1989). A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis thaliana. J. Hered. 80, 118–122. [Google Scholar]
- Kunst, L., Taylor, D.C., and Underhill, E.W. (1992). Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol. Biochem. 30, 425–434. [Google Scholar]
- Lolle, S.J., Hsu, W., and Pruitt, R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., Clemens, S., Zachgo, S., Giblin, E.M., Taylor, D.C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11, 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Kunst, L. (1997). Very long chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., Wrischer, M., and Kunst, L. (1998). Accumulation of very-long-chain fatty acids in membrane glycerolipids is associated with dramatic alterations in plant morphology. Plant Cell 11, 1889–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J.P., Hartmann, M.A., Bonneu, M., Simon-Plas, S., Lessire, R., and Bessoule, J.J. (2004). Lipid rafts in higher plant cells. J. Biol. Chem. 279, 36277–36286. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Muniz, M., Morsomme, P., and Riezman, H. (2001). Protein sorting upon exit from the endoplasmic reticulum. Cell 104, 313–320. [DOI] [PubMed] [Google Scholar]
- Nabi, I.R., and Le, P.U. (2003). Caveolae/raft-dependent endocytosis. J. Cell Biol. 161, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C.S., Toke, D.A., Mandala, S., and Martin, C.E. (1997). ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272, 17376–17384. [DOI] [PubMed] [Google Scholar]
- Peskan, T., Westermann, M., and Oelmüller, R. (2000). Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 267, 6989–6995. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.E., Vielle-Calzada, J.P., Ploense, S.E., Grossniklaus, U., and Lolle, S.J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 97, 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossak, M., Smith, M., and Kunst, L. (2001). Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol. Biol. 46, 717–725. [DOI] [PubMed] [Google Scholar]
- Saint-Jore, C.M., Evins, J., Batoko, H., Brandizzi, F., Moore, I., and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29, 661–678. [DOI] [PubMed] [Google Scholar]
- Samuels, A.L., and Bisalputra, T. (1990). Endocytosis in elongating root cells of Lobella erinus. J. Cell Sci. 97, 157–166. [Google Scholar]
- Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., Schmidt, J., and Jurgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Simons, K., and Toomre, D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- Somerville, C.R., and Ogren, W.L. (1982). Isolation of photorespiratory mutants of Arabidopsis. In Methods in Chloroplast Molecular Biology, R.B. Hallick and N.H. Chua, eds (New York: Elsevier), pp. 129–139.
- Sperling, P., Zähringer, U., and Heinz, E. (1998). A sphingolipid desaturase from higher plants. J. Biol. Chem. 273, 28590–28596. [DOI] [PubMed] [Google Scholar]
- Todd, J., Post-Beittenmiller, D., and Jaworski, J.G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130. [DOI] [PubMed] [Google Scholar]
- von Wettstein-Knowles, P.M. (1982). Elongase and epicuticular wax biosynthesis. Physiol. Veg. 20, 797–809. [Google Scholar]
- Wang, Q., and Chang, A. (2002). Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, PMA1. Proc. Natl. Acad. Sci. USA 99, 12853–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., Van Den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11, 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Kunst, L., Hawes, C., and Moore, I. (2004). A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 37, 398–414. [DOI] [PubMed] [Google Scholar]