Abstract
The regulation of light harvesting in higher plant photosynthesis, defined as stress-dependent modulation of the ratio of energy transfer to the reaction centers versus heat dissipation, was studied by means of carotenoid biosynthesis mutants and recombinant light harvesting complexes (LHCs) with modified chromophore binding. The npq2 mutant of Arabidopsis thaliana, blocked in the biosynthesis of violaxanthin and thus accumulating zeaxanthin, was shown to have a lower fluorescence yield of chlorophyll in vivo and, correspondingly, a higher level of energy dissipation, with respect to the wild-type strain and npq1 mutant, the latter of which is incapable of zeaxanthin accumulation. Experiments on purified thylakoid membranes from all three mutants showed that the major source of the difference between the npq2 and wild-type preparations was a change in pigment to protein interactions, which can explain the lower chlorophyll fluorescence yield in the npq2 samples. Analysis of the xanthophyll binding LHC proteins showed that the Lhcb5 photosystem II subunit (also called CP26) undergoes a change in its pI upon binding of zeaxanthin. The same effect was observed in wild-type CP26 upon treatment that leads to the accumulation of zeaxanthin in the membrane and was interpreted as the consequence of a conformational change. This hypothesis was confirmed by the analysis of two recombinant proteins obtained by overexpression of the Lhcb5 apoprotein in Escherichia coli and reconstitution in vitro with either violaxanthin or zeaxanthin. The V and Z containing pigment-protein complexes obtained by this procedure showed different pIs and high and low fluorescence yields, respectively. These results confirm that LHC proteins exist in multiple conformations, an idea suggested by previous spectroscopic measurements (Moya et al., 2001), and imply that the switch between the different LHC protein conformations is activated by the binding of zeaxanthin to the allosteric site L2. The results suggest that the quenching process induced by the accumulation of zeaxanthin contributes to qI, a component of NPQ whose origin was previously poorly understood.
INTRODUCTION
Oxygenic photosynthesis inevitably leads to the production of highly reactive oxygen species. When the input light intensity exceeds the photosynthetic saturation limit, photoinhibition occurs as a result of the overproduction of these toxic oxygen species (Aro et al., 1993a, 1993b). Chloroplasts have developed many protective systems to diminish the effects of photodamage, including scavenging of reactive oxygen species (ROS) from both the lipid phase (Havaux and Niyogi, 1999) and the chloroplast stroma (Asada and Takahashi, 1987). Other mechanisms are aimed at preventing the formation of the harmful oxygen species and are collectively known as nonphotochemical energy quenching (NPQ) that functions by thermally deactivating chlorophyll molecules in photosystem II (PSII) in their first singlet excited state, the precursor species to highly reactive singlet oxygen (Demmig-Adams and Adams, 2000) through the intermediate of a triplet chlorophyll state. Although the mechanism(s) of NPQ is/are still unknown, many of its characteristics are commonly accepted. One is the involvement of the conversion of violaxanthin (Viola) to zeaxanthin (Zea), by means of an increase in the trans-thylakoid pH gradient generated by high light that triggers the violaxanthin deepoxidase enzyme. The interchange of these carotenoids via the intermediate antheraxanthin (Anthera), reversible in the dark by zeaxanthin epoxidase, is called xanthophyll cycle. Low lumenal pH also results in protonation of specific light-harvesting complexes (LHCs) (Dominici et al., 2002; Li et al., 2002a; Li et al., 2002b; Pesaresi et al., 1997; Walters et al., 1996), which, synergistically with zeaxanthin binding, leads to a conformational change necessary for NPQ (Moya et al., 2001). The PSII subunit S (PsbS) was found to be essential for NPQ (Li et al., 2000) and may trigger the quenching in a series of steps involving protonation and, possibly, zeaxanthin binding (Li et al., 2002b; Holt et al., 2005). It was also shown that Lhcb (light-harvesting complexes of PSII) proteins replace violaxanthin for zeaxanthin in the allosteric L2 site that induces quenching of chlorophyll fluorescence in vitro (Formaggio et al., 2001). The quenching occurs by increasing the relative amplitude of the short (1 ns) fluorescence lifetime component at the expense of the long (4 ns) component found in these Lhcb (Crimi et al., 2001), corresponding to two distinct conformations of the Lhcb proteins (Moya et al., 2001). The short lifetime component conformation is correlated with the dissipative state (Gilmore et al., 1998). Interconversion of the protein conformation between these two states would regulate the efficiency of energy transfer to the PSI I reaction centers (Holt et al., 2004).
Xanthophyll cycle and protonation of lumen exposed protein sites lead to the full expression of NPQ, which has been shown to contain several components (Ruban and Horton, 1999). Thus, the npq1 mutant is unable to accumulate zeaxanthin and yet retains part of its capacity for rapidly reversible, nigericin-sensitive quenching, defined as qE (Niyogi, 1999), whereas the npq4 mutant, although completely lacking PsbS, still undergoes a limited fluorescence quenching upon exposure to excess light (Li et al., 2000). This suggests that PsbS and zeaxanthin might act through distinct, although interconnected, mechanisms.
In this work, we have analyzed two Arabidopsis thaliana mutants: npq1, unable to produce zeaxanthin, and npq2, which constitutively accumulates this xanthophyll species (Niyogi et al., 1998). We show that under dark adapted conditions the npq2 mutant exhibits sustained quenching while npq1 and the wild type do not and that the source of the sustained quenching is the lower fluorescence yield of the xanthophyll binding LHC proteins. Purification and analysis of LHC proteins from the npq2 mutant showed that one antenna subunit, called CP26, is present in a new isoform; it suggests, in light of CP26 being encoded by a single gene (Arabidopsis Genome Initiative, 2000), that this effect results from a conformational change, undetectable in the dark-adapted wild type and npq1. CP26 undergoes the same pI shift in wild-type plants only after treatment inducing accumulation of zeaxanthin. Recombinant proteins, obtained by in vitro refolding of the CP26 apoprotein, with either violaxanthin or zeaxanthin, also differ in their isoelectric point, implying that the xanthophyll species bound determines the protein conformation and fluorescence yield. The capacity for photoprotection of plants lacking PsbS or zeaxanthin was assayed by growth in excess light conditions and showed that at least two different components contribute to fluorescence quenching: one of which (qE) is directly dependent on PsbS and light, while a second component, described here, is based on the binding of zeaxanthin to Lhcb proteins but does not require a trans-thylakoid pH gradient once zeaxanthin is bound to LHC proteins. Throughout this article, these two mechanisms will be referred to as PsbS-dependent and pH-independent quenching, respectively.
RESULTS
To elucidate the mechanism by which fluorescence quenching by zeaxanthin occurs, we have analyzed Arabidopsis wild-type (ecotype Columbia) and mutants: npq1, unable to synthesize zeaxanthin; npq2, which lacks violaxanthin and instead accumulates zeaxanthin; npq4, which is blocked in the PsbS-dependent fluorescence quenching mechanism (Li et al., 2000); lhcb5, which lacks PSII antenna CP26. In the dark, the wild-type strain contains violaxanthin, a fraction of which is transformed into zeaxanthin during exposure to high light.
In Vivo Measurements
Effect of High Light Treatment in Vivo
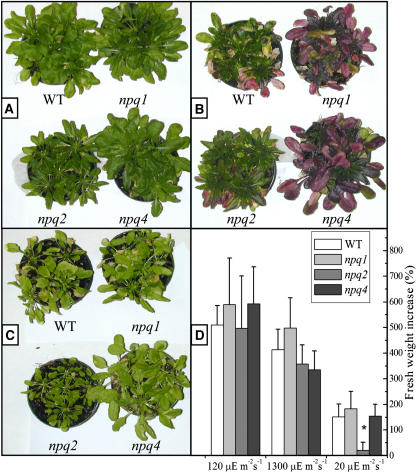
To assess the importance of zeaxanthin and PsbS in photoprotection, we have studied the effect of growing wild-type and npq1, npq2, and npq4 mutant plants under low light conditions (20 μE m−2 s−1, 20°C), control conditions (120 μE m−2 s−1, 20°C), and under excess light (1300 μM m−2 s−1, 20°C) for 3 weeks. In control conditions, all genotypes showed a similar growth rate (Figure 1D). In high light conditions (Figure 1B), older leaves at the periphery of the rosettes of wild-type plants showed bleaching of chlorophyll accompanied by reddening of leaves as a result of accumulation of the purple flavonoid anthocyanin, an indicator of stress in Arabidopsis (Dixon and Paiva, 1995; Xiang et al., 2001). The younger leaves at the center of rosettes, however, were normally green and did not accumulate anthocyanins nor show other stress effects. These symptoms were much stronger in npq1 and npq4 mutants, leading to chlorophyll loss and reddening extending to the younger leaves, while older leaves were almost completely depleted of chlorophyll. npq1 genotype was clearly more light sensitive than npq4, the damage extending to very young leaves. npq2 did not undergo significant photodamage, only showing a limited reddening of older leaves but not chlorophyll bleaching (Figure 1B). Growth in limiting light conditions (20 μE m−2 s−1, 20°C) showed a higher mean fresh weight for the wild type, npq1, and npq4 than for npq2, the growth of which was severely impaired (Figure 1C). The plant growth rate was evaluated from fresh weight both at the beginning and at the end of the treatment at different light conditions (Figure 1D); it clearly appears that in limiting light the growth of npq2 plants was only 15% of the wild type, while at high light conditions, growth of npq2 is not significantly different from the wild type. The absence of violaxanthin in the npq2 mutant results in deficiency in the hormone abscisic acid (ABA) (Koornneef et al., 1982). Consequently, the aba mutants cannot fully regulate stomatal opening, exhibit a wilty phenotype, and require water-saturated atmosphere. We have used an allele of aba1, npq2-1 (Niyogi et al., 1998), which is not a complete loss-of-function mutation: ABA is synthesized in sufficient amounts so that the mutant is not particularly sensitive to wilting. Nevertheless, because the absence of ABA could affect growth, we have confirmed our results in npq2 plants supplemented with ABA.
Figure 1.
Effect of Different Light Conditions during Growth of Wild-Type and Mutant Arabidopsis Plants.
(A) to (C) Each pot contained five seedlings. Control plants (A) (120 μE m−2 s−1); high light–grown plants (B) (1300 μE m−2 s−1); plants grown in limiting light conditions (C) (20 μE m−2 s−1). In low light, npq2 plants show a lower growth, consistent with sustained dissipation of excitation energy. In high light, npq2 plants appear to be highly resistant with respect to npq1 and npq4. npq1 plants, lacking zeaxanthin, appear to be more severely affected than npq4 plants, lacking PsbS.
(D) Fresh weight increase measured on populations of Arabidopsis wild-type and mutant genotypes growth for 3 weeks in different light conditions. During this treatment, the npq2 plant had been daily sprayed with 10 μM abscisic acid. Data are expressed as means ± sd, n = 15. Data sets with a significance level of P < 0.05 according to Student's t test are marked with an asterisk.
The above results are consistent with a sustained dissipation of excitation energy in npq2 plants, which have both PsbS and high zeaxanthin. In fact, they are limited in growth at low light and show the best level of photoprotection in excess light conditions. A contribution to this latter effect can also be due to a specific antioxidant property of zeaxanthin (Havaux and Niyogi, 1999). Conversely, the phenotype of npq1 and npq4 plants is consistent with impaired capacity for energy dissipation, allowing for high growth rates in limiting light and photodamage in excess light with respect to wild-type plants. Nonetheless, the phenotype of npq1 is not the same as npq4: the light sensitivity of npq1 plants is higher than that of npq4 plants, suggesting that at least in part different mechanisms might be affected by the two mutations.
Leaf Fluorescence Yield
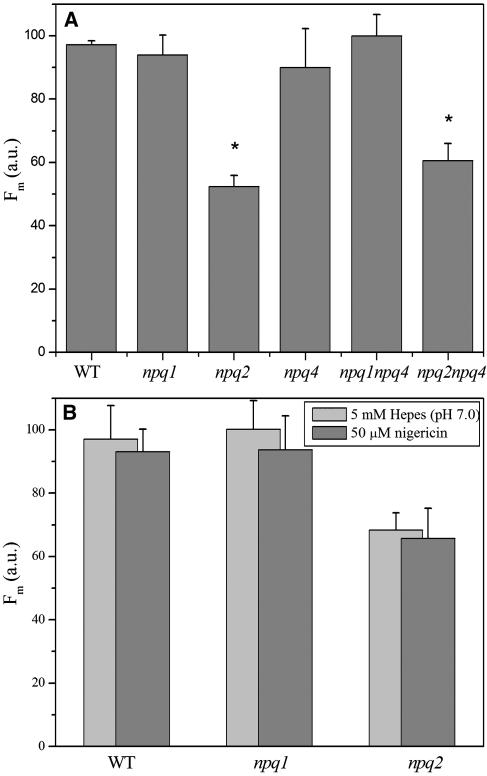
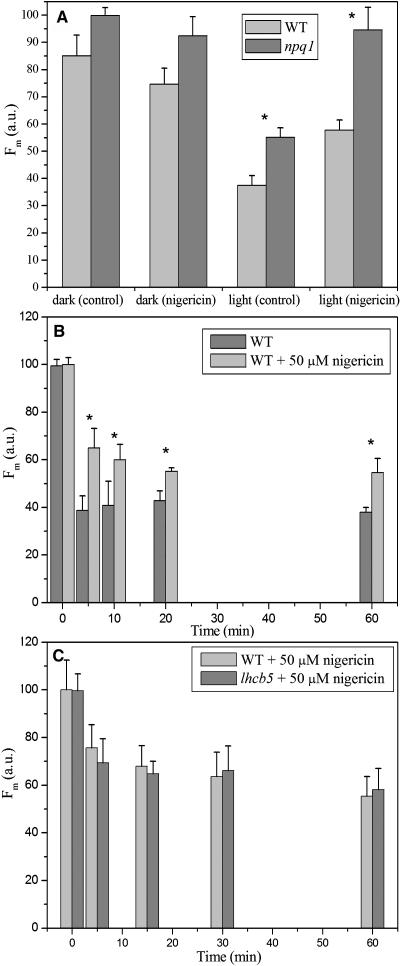
Dark adapted plants were analyzed for fluorescence yield upon illumination with saturating light. Figure 2A shows a histogram of the maximal fluorescence (Fm) after 1.8 s of LED light exposure for six genotypes. It clearly appears that after normalization for minor differences in chlorophyll concentration per leaf surface, the relative fluorescence yield of npq2 and npq2 npq4 leaves was ∼40% lower than fluorescence of leaves from the rest of the genotypes. Pigment composition obtained by whole-leaf acetone extraction (Table 1) shows that the two genotypes with the lowest fluorescence yield are characterized by high zeaxanthin content. The difference in fluorescence yield of npq2 with respect to the wild type, npq1, and npq4 was maintained upon infiltration of leaves with the uncoupler nigericin (Figure 2B), thus implying that this effect was not dependent on a residual trans-thylakoid pH gradient.
Figure 2.
Relative Fluorescence Yield of Leaves from Arabidopsis Wild Type and npq Mutants.
Fluorescence amplitude was measured at Fm on intact leaves (A) and on leaves infiltrated (B) with either 5 mM Hepes, pH 7.0, (light gray) or 50 μM nigericin (dark gray). We used a video imaging apparatus for Fm determination (see Methods) and normalized Fm to the chlorophyll content per leaf surface. Values are the average of measurements performed on four leaves. These results have been confirmed using a PAM fluorimeter and leaf discs. Data sets with a significance level of P < 0.05 according to Student's t test are marked with an asterisk. a.u., arbitrary units.
Table 1.
Pigment Composition of Thylakoid Membranes Isolated from Wild-Type and npq Mutants Dark Adapted (Lanes 1 to 6) and after Light Stress (1200 μE m−2 s−1, 15 min) on the Wild-Type Genotype (Lane 7)
| Sample | Neo | Viola | Lute | Zea | Anthera | β-Car | Chlorophyll b | Chlorophyll a |
|---|---|---|---|---|---|---|---|---|
| Wild type | 3.4 | 2.8 | 8.0 | nd | nd | 5.4 | 22.4 | 77.6 |
| npq1 | 4.1 | 2.5 | 9.4 | nd | nd | 5.8 | 20.9 | 79.1 |
| npq2 | nd | nd | 8.4 | 10.2 | nd | 5.3 | 22.1 | 77.9 |
| npq4 | 3.7 | 2.3 | 9.1 | nd | nd | 7.6 | 22.0 | 78.0 |
| npq1 npq4 | 4.3 | 3.5 | 10.0 | nd | nd | 5.8 | 22.1 | 77.9 |
| npq2 npq4 | nd | nd | 11.1 | 12.0 | nd | 6.1 | 21.0 | 79.0 |
| Wild type/light | 4.2 | 1.4 | 8.3 | 2.3 | 0.4 | 5.7 | 24.0 | 76.0 |
The data are normalized to 100 chlorophyll a + b molecules. β-Car, β-carotene; nd, not detectable. In all cases, error was lower than 4%.
Nonphotochemical Quenching
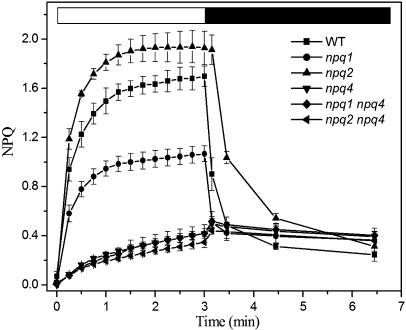
Leaf fluorescence was measured during illumination with saturating light and NPQ was calculated as (Fm − F′m)/F′m (see Methods). The wild-type strain showed a rapid quenching phase in the first minute to a value of 1.5, followed by a slower phase, leading to an NPQ value of 1.7 after 3 min of illumination (Figure 3). The npq2 mutant behaved similar to the wild type during the first onset of illumination then became faster and reached saturation more rapidly, reaching NPQ values of 1.8 and 1.9 after 1 and 3 min, respectively. NPQ induction kinetics of the npq1 mutant were slower than that of the wild type and npq2 in both the first and the second phases of the rise, reaching a value of only 1.1 at the end of the light period. The npq4 genotype showed a very low level of light induced quenching (0.35), which did not relax upon returning plants to dark, consistent with previous reports (Li et al., 2000).
Figure 3.
Kinetics of NPQ of Wild Type Arabidopsis and Mutants.
In vivo NPQ of chlorophyll fluorescence was measured with a PAM fluorimeter, and it was induced by saturating actinic light (1200 μE m−2 s−1, white bar) on wild-type and mutant leaves. Dark-relaxation kinetic was followed (black bar). The values of NPQ were calculated as described in Methods. Values are the average of measurements performed on four distinct leaves.
In Vitro Measurements
Step Solubilization of Thylakoid Membranes
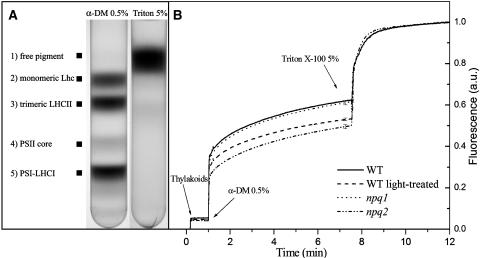
To elucidate the origin of the lower fluorescence yield in the npq2 mutant with respect to the wild type and npq1, thylakoid membranes were purified either from dark adapted leaves of the three genotypes or from light-treated wild-type plants, and their fluorescence yield measured in a cuvette with continuous stirring. According to the in vivo results, the relative fluorescence yield of wild-type and npq1 thylakoids was higher than that from npq2 thylakoids (Figure 4B). The membranes were solubilized in two steps. During the first treatment, 0.5% (w/v) final concentration of the mild detergent dodecyl-α-d-maltoside (α-DM) was added to the samples. This treatment lead to solubilization of the membranes and dissociation of the pigment binding proteins from each other without release of chlorophyll from their protein moiety (Caffarri et al., 2001). The second treatment with 5% (w/v) Triton X-100 denatured the pigment–proteins and yielded free pigments, generating the maximum quantum yield of fluorescence obtainable for the system (Giuffra et al., 1997) (Figure 4A). Figure 4B shows the changes in steady state fluorescence yield that the membranes from the different genotypes undergo upon the above described treatments: the increase in fluorescence induced by 0.5% α-DM is higher for the wild type and npq1 with respect to npq2, while further treatment with Triton X-100 brought fluorescence yield of the three samples to the same level. This implies that the fluorescence quantum yield of chlorophyll extracted is the same, as expected, while pigment–protein interactions (defined as Qpi, see Methods), retained in 0.5% α-DM, are responsible for the higher quenching observed in npq2 with respect to the wild type or npq1. Protein–protein and lipid–proteins interactions in the membrane (defined as Qm) also decrease the fluorescence yield of chlorophyll, but this effect appears to be similar in the membranes from the different genotypes (Figure 4B). For a more quantitative picture, we measured Qm+pi = 22.86 (npq2), 17.33 (wild type), and 17.12 (npq1), in agreement with the in vivo measurements that showed lower chlorophyll fluorescence in npq2. However, whereas Qm was very similar in all the samples, Qpi was much higher in npq2 (1.00) versus the wild type (0.61) and npq1 (0.62), as shown in Table 2. Step solubilization of thylakoids from light-treated wild-type leaves yields a quenching effect (Qpi = 0.88, Qm+pi = 19.94) similar to that observed in the npq2 mutant.
Figure 4.
Fluorescence Yield of Thylakoid Membranes upon Step Solubilization.
(A) Sucrose density gradient profiles of thylakoids solubilized with 0.5% α-DM (right) and 5% Triton X-100 (left). The same level of solubilization and separation into component green bands was obtained using thylakoids from npq1, npq2, and npq4 genotypes. Composition of each band is indicated.
(B) Thylakoid membranes were purified from dark-adapted leaves of wild-type, npq1, and npq2 genotypes and from wild-type leaves after light treatment (see Methods for details); their fluorescence yield was measured after solubilization with 0.5% α-DM and 5% Triton X-100. Chlorophyll concentration of isolated thylakoids was set to 0.1 μg/mL. The samples were measured at room temperature under continuous stirring. Data are expressed as means ± sd of four measurements. a.u., arbitrary units.
Table 2.
Release of Chlorophyll Fluorescence Quenching by Step Solubilization of Thylakoid Membranes
| Wild Type | Wild Type/Light-Treated | npq1 | npq2 | |
|---|---|---|---|---|
| Qm | 10.42 ± 0.06 | 10.12 ± 0.16 | 10.17 ± 0.26 | 10.92 ± 0.43 |
| Qpi | 0.61 ± 0.01 | 0.88 ± 0.02a | 0.62 ± 0.02 | 1.00 ± 0.03ab |
| Qm+pi | 17.33 ± 0.08 | 19.94 ± 0.14a | 17.12 ± 0.36 | 22.86 ± 0.76ab |
Parameters Qm, Qpi, and Qm+pi were calculated as described in Methods and are the average of four independent measurements.
Data sets with a significance level of P < 0.05 according to Student's t test with respect to the wild type.
Data sets significantly different (P < 0.05) with respect to the light-treated wild type.
When wild-type plants are exposed to high light, they accumulate zeaxanthin. It can be asked if newly formed zeaxanthin induces a quenching effect in wild-type thylakoids similar to that observed in npq2. To answer this question, we have treated wild-type plants with strong light, thus obtaining a deepoxidation state (Z + 0.5A)/(V + A + Z) of 0.68 ± 0.02 and isolated thylakoids using a pH 5.2 buffer to avoid reepoxidation. When analyzed for fluorescence yield during step solubilization, dark adapted and light adapted wild-type thylakoids closely matched the behavior of the wild type and npq2 (Figure 4B), respectively, thus implying that the fluorescence quenching effect as a result of zeaxanthin accumulation in LHC proteins is activated as a consequence of xanthophyll cycle operation in wild-type plants.
Analysis of Pigment–Protein Complexes
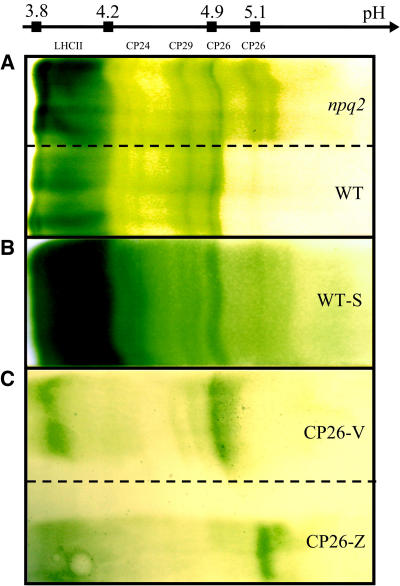
Thylakoid membranes were mildly solubilized and fractionated by sucrose gradient ultracentrifugation, which revealed several yellow/green bands. As assessed by SDS-PAGE analysis (see Supplemental Figure 1 online) the separation of Arabidopsis thylakoids is very similar to that previously described for maize (Zea mays) (Caffarri et al., 2001). Thus, xanthophyll containing LHC proteins migrated in the second and third bands in their monomeric and trimeric forms respectively, while the fourth band contained the PSII core complex and the fifth band contained the photosystem I–LHCI complex (Figure 4A; see Supplemental Figure 1 online). Because fluorescence in vivo is mainly derived from PSII, we analyzed the fluorescence yield of the PSII components in bands 2 to 4. We found that whereas the PSII core complex (band 4) had the same fluorescence yield for both the wild type and npq2, LHC proteins from npq2 had lower fluorescence yield than the wild type by 21% (band 2) and 14% (band 3) (Table 3). Band 2 fractions from the wild type and npq2 were concentrated and further fractionated by preparative isoelectric focusing (IEF), yielding the separation shown in Figure 5A. The green bands were identified by their SDS-PAGE profiles and absorption spectra (Dainese and Bassi, 1991). Whereas LHCII (the major LHC of PSII), CP24, and CP29 comigrated in the lanes from the wild type and npq2, CP26 from npq2 formed a new band at a pH value more basic by 0.2 units than CP26 from the wild type. This effect was not complete because approximately one-third of total CP26 still migrated at the original pI value of 4.9. HPLC analysis of acetone extracts from the CP26 bands eluted from the IEF gel showed differences in xanthophyll composition: both CP26 isoforms bound two xanthophyll molecules per polypeptide; however, CP26 from wild type–bound lutein (Lute), Viola, and neoxanthin (Neo) in a ratio of 2:1:1, whereas the same protein from npq2 binds only Lute and Zea in approximately equal amounts. The same procedure was repeated with wild-type plants after 1 h of light treatment that induced a deepoxidation state of 0.69 ± 0.03 (average of three experiments). Isolation of thylakoids from these plants required some time, during which the deepoxidation state decreased to 0.25. To maximize Zea accumulation, we thus isolated thylakoids using Mes buffer at pH 5.2, which maintained the deepoxidation state at 0.6 (see Methods for details). These thylakoids were washed and fractionated as described above for npq2 thylakoids. The IEF pattern is shown in Figure 5B: the band at pI = 5.1 is reproduced in the sample from wild-type thylakoids, although to a somewhat lower extent with respect to the case of npq2, probably because of the lower zeaxanthin content. SDS-PAGE analysis showed that, although CP26 was the major band in the pI shifted band, small amounts of CP29 were present (see Supplemental Figure 1 online) in the sample from npq2. In the Zea-containing wild-type sample, small amounts of CP29 are also present (data not shown). These results show that not only CP26 but also CP29 undergo pI shift upon treatment inducing accumulation of Zea and that this shift is constitutively present in npq2, while in the wild type appears only after treatment inducing accumulation of zeaxanthin.
Table 3.
Relative Fluorescence Yield of PSII Components from Bands 2 to 4 of Sucrose Gradient Fractionation
| Sample | Band 2 | Band 3 | Band 4 |
|---|---|---|---|
| Wild type | 276.50 ± 5.90 | 252.38 ± 8.59 | 117.71 ± 4.58 |
| npq1 | 297.40 ± 3.39* | 273.58 ± 2.93* | 109.69 ± 0.66 |
| npq2 | 219.18 ± 4.80* | 218.07 ± 2.97* | 121.62 ± 2.99 |
Band 2, monomeric Lhcb proteins; band 3, trimeric LHCII complex; band 4, PSII core complex. Data are the average of four independent measurements. The chlorophyll concentration was 0.1 μg/mL. Data sets with a significance level of P < 0.05 according to Student's t test are marked with an asterisk.
Figure 5.
Flatbed IEF Fractionation.
Profiles obtained after IEF of monomeric Lhcb proteins (A) isolated from npq2 (top) and wild-type (bottom) thylakoids by sucrose gradient fractionation, monomeric Lhcb complexes (B) isolated from light-treated wild-type plants (WT-S), whose thylakoids were isolated in acidic buffer to maintain deepoxidation level, and recombinant Lhcb5 proteins (C) reconstituted in vitro with either zeaxanthin (top, CP26-Z) or violaxanthin (bottom, CP26-V).
Recombinant CP26 Proteins with Distinct Xanthophyll Compositions Have Distinct pI Values
Changes in the pI of a membrane protein may reveal conformational (Ben Or and Chrambach, 1983) and/or posttranslational modifications, such as phosphorylation (Bergantino et al., 1995), which change the exposed charges of proteins. To verify if zeaxanthin binding was the only cause for the change in pI, recombinant holoproteins were produced by overexpressing At-Lhcb5 cDNA in Escherichia coli and reconstituting in vitro the apoprotein with chlorophyll a and chlorophyll b plus either a mixture of Lute + Viola + Neo or Lute + Zea. Figure 5C shows the results of IEF experiments on the two recombinant proteins. For both proteins, two green bands were obtained, the more acidic band containing free pigments and the less acidic band containing the CP26 pigment–protein complexes. The less acidic band reproduced the shift in pI observed for native CP26 from npq2 with respect to the wild type, confirming that the only reason for the pI shift of CP26 from npq2 membranes was the binding of Zea versus Viola.
CP26 Isoforms Show Different Fluorescence Yields
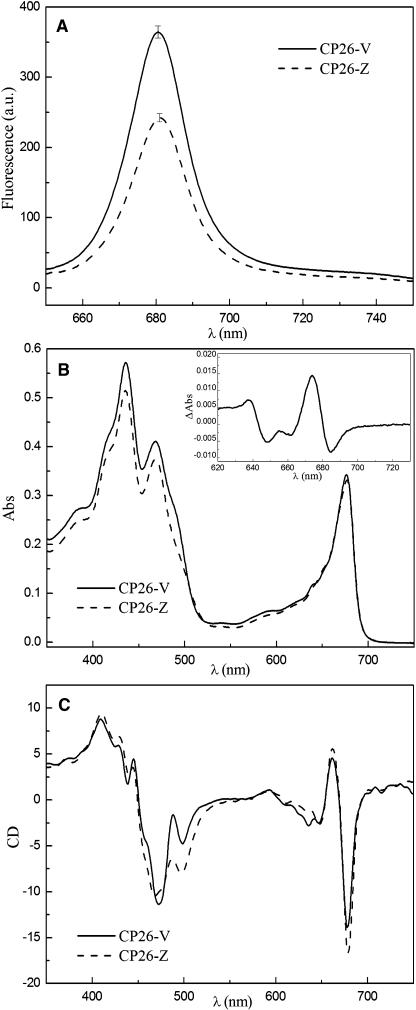
The green bands were eluted from the IEF gel and submitted to sucrose gradient ultracentrifugation to eliminate carrier ampholytes and free pigments. Pigment analysis (Table 4) showed that the two CP26 isoforms had the same chlorophyll a and chlorophyll b content, whereas their xanthophyll complement was either Lute + Zea or Lute + Viola + Neo for the low and high fluorescence forms, respectively. Figure 6A shows the relative fluorescence yield of the two CP26 isoforms: the Lute + Zea binding protein (to be referred to as CP26-Z) had a 35% lower fluorescence yield with respect to the Lute + Viola + Neo binding protein (to be referred to as CP26-V).
Table 4.
Pigment Composition of the Lhcb5 Isoforms
| Sample | Neo | Viola | Lute | Zea | Chlorophyll b | Chlorophyll a | Chlorophyll a/b |
|---|---|---|---|---|---|---|---|
| CP26-V | 0.7 | 0.3 | 1.2 | nd | 3.1 | 5.9 | 1.9 |
| CP26-Z | nd | nd | 0.7 | 1.2 | 3.1 | 5.9 | 1.9 |
Both isoforms were isolated by preparative IEF and further purified by sucrose gradient ultracentrifugation. The values were normalized to the number of chlorophylls per monomeric protein. nd, not detectable.
Figure 6.
Spectral Properties at Room Temperature of Lhcb5 Isoforms Purified by IEF (See Figure 5B).
(A) Comparison of fluorescence yield of CP26-V (solid line) and CP26-Z (dotted line) upon excitation at 625 nm. Data are the average of four independent measurements. a.u., arbitrary units.
(B) Absorption spectra of CP26-V (solid line) and CP26-Z (dotted line) normalized to the chlorophyll content. Inset, difference spectrum in the Qy transition region.
(C) Circular dichroism spectra of CP26-V (solid line) and CP26-Z (dotted line). All the spectra were normalized to the same chlorophyll content.
Spectral Properties of CP26 Isoforms Suggest That Pigment–Pigment Interactions Are Affected
Although the chlorophyll a and chlorophyll b content was the same in the two samples, the absorption spectra differed in the chlorophyll Qy spectral region where carotenoids do not absorb (Figure 6B), besides the obvious spectral differences in the blue spectral region where the absorption of xanthophylls and chlorophylls is superimposed. In particular, the different absorption spectrum suggests that a chlorophyll b form peaking at 638 nm is shifted to 648 nm and a chlorophyll a form is shifted from 674 to 685 nm. Circular dichroism (CD) spectra, which reveal the mutual organization of chromophores within the protein, show two peaks at 636 and 648 nm, the former lacking in the CP26-Z sample. In the chlorophyll a Qy region, the 661(+)/677(−) signal has larger amplitude and is red shifted by 2 nm (Figure 6C). These spectral changes suggest that, upon binding of Zea versus Viola/Neo to the protein, the local environment of the chlorophyll a and chlorophyll b chromophores is modified and thus support their undergoing a conformational change.
Relations with the PsbS-Mediated qE Quenching
To clarify the interactions between qE (PsbS-dependent quenching) and the pH-independent quenching induced by zeaxanthin binding to Lhcb proteins, we have produced npq1 npq4 and npq2 npq4 double mutants, both lacking PsbS and having either no zeaxanthin or no violaxanthin, respectively. The fluorescence yield of the two mutants was analyzed both in the dark and upon high light treatment. Only light-dependent quenching needs the presence of PsbS because the chlorophyll fluorescence yield of the npq1 and npq1 npq4 mutants in the dark was similar to that of the wild type, whereas the yield for the npq2 npq4 mutant was similar to that of npq2 (Figure 2A). Under high light illumination, the npq4, npq1 npq4, and npq2 npq4 genotypes have the same kinetics and amplitude of NPQ, which differ from the kinetics and amplitude obtained for the npq1, npq2, and wild-type genotypes (Figure 3).
pH-Independent Quenching in the Wild Type upon Illumination
To check the relevance of the pH-independent quenching in the wild type, we exposed leaves to high light conditions (1200 μE m−2 s−1), then rapidly infiltrated with nigericin and measured fluorescence yield and pigment composition as compared with leaves kept in the dark. A set of leaves was also infiltrated with buffer without the uncoupler to correct for changes in optical properties that leaves underwent during infiltration (see Methods). The results (Figure 7A) showed that the treatment with the uncoupler was unable to fully restore fluorescence yield of light treated leaves (which accumulated 2.3 mmol zeaxanthin/100 mmol of total chlorophyll, corresponding to a deepoxidation state of 0.6, after 15 min of light treatment; see Table 1) to the same level obtained in wild-type leaves left in the dark and not accumulating zeaxanthin. When the same experiment was performed with the npq1 mutant, unable to accumulate zeaxanthin, the nigericin treatment was very efficient in fully restoring fluorescence yield, as shown in the case of 15 min of illumination, thus implying that accumulation of zeaxanthin in the wild type upon light treatment was responsible for the pH-independent quenching. Because of the 1 min period needed for nigericin infiltration, the qE component is in part relaxed and appears of smaller amplitude than in Figure 3. These results were confirmed using thylakoid membranes and the PAM 101 fluorimeter rather than the fluorescence video imaging apparatus. The amplitude of the pH-independent quenching in wild-type thylakoids was essentially the same as obtained in leaves upon normalization for Zea content, whereas no pH-independent quenching was observed in npq1 thylakoids (data not shown).
Figure 7.
Fluorescence Analysis of Nigericin-Sensitive and Nigericin-Resistant NPQ Components in Wild-Type and npq1 Genotypes of Arabidopsis.
(A) Wild-type and npq1 leaves were either left in the dark or illuminated (1200 μE m−2 s−1) for 15 min at room temperature to induce NPQ and zeaxanthin accumulation (in the wild type). Leaves were then rapidly infiltrated with either 5 mM Hepes, pH 7.0, or 50 μM nigericin to abolish ΔpH. Fm level was measured using a video imaging fluorescence system as described in Methods. The time required for infiltration and measurement was ∼1 min. a.u., arbitrary units.
(B) Chlorophyll fluorescence intensity determinations in Arabidopsis wild-type leaves for Fm and Fm′. After each light window, the Fm′ value was measured on leaves rapidly infiltrated with 50 μM nigericin.
(C) Comparison of nigericin-resistant NPQ component in wild-type and lhcb5 leaves, measured as in Figure 7B. Data sets with a significance level of P < 0.05 according to Student's t test are marked with an asterisk.
The time course of the pH-independent quenching was studied by exposing leaves to high light conditions for different periods and then measuring fluorescence with or without nigericin infiltration. The results are shown in Figure 7B: it appears that the half-time is ∼2 min versus 0.5 of qE (see Figure 3) and is saturated after ∼20 min of illumination at 1200 μE m−2 s−1.
Because the effect of zeaxanthin accumulation was particularly evident in CP26 complexes, we have analyzed an Arabidopsis strain lacking the CP26 complex (lhcb5 mutant; see Supplemental Figure 3 online) obtained by T-DNA insertion into the Lhcb5 coding region. When fluorescence yield was measured upon illumination and nigericin infiltration, the CP26-less strain showed a pH-independent fluorescence quenching similarly to the wild type (Figure 7C).
DISCUSSION
Photoprotection from excess light is operated by a complex network of molecular mechanisms: from changes in the orientation of leaves and chloroplast movements to decreased light interception to detoxification from ROS produced when excess redox energy is diverted from NADP+ to O2. Of particular interest are the mechanisms, collectively known as NPQ, that prevent ROS formation by finely regulating the level of excited singlet state of chlorophyll in the antenna complex of PSII. Through NPQ, the energy in excess to the amount used in electron transport is dissipated into heat. Physiological analysis has resolved multiple components in NPQ: a rapidly reversible, nigericin-sensitive qE, the slowly relaxing components qI, attributed to photoinhibition, and qT, attributed to state I–state II transitions (Horton, 1996). Genetic analysis has identified PsbS as a gene product indispensable for qE, while zeaxanthin and lutein appear as modulators of qE amplitude (Niyogi et al., 1998; Pogson et al., 1998). Zeaxanthin accumulation also has been involved in long term (overwinter) downregulation of photosynthesis (Verhoeven et al., 1999; Gilmore and Ball, 2000). Current understanding of energy dissipation suggests that PsbS catalyzes qE upon protonation of lumenal-exposed residues (Li et al., 2002b) and binding of zeaxanthin. An alternative mechanism of quenching has been proposed based on the observation that Viola, Anthera, and Zea bind to LHC proteins, particularly CP26 and CP24 (Bassi et al., 1993) to the L2 binding site that has been shown to have allosteric properties (Formaggio et al., 2001). Upon Zea binding, LHC proteins show fluorescence quenching as measured by steady state and time resolved methods (Moya et al., 2001).
Decreased Growth Rate in Limiting Light in npq2 Is Consistent with Sustained Energy Dissipation
In this work, we have studied the behavior of Arabidopsis mutants exposed to either excess light or limiting light with respect to control conditions. It clearly appears that the highest capacity for photoprotection is performed by the npq2 mutant that did not show photodamage even at high light conditions (Figure 1B). This mutant has normal levels of PsbS and constitutively high levels of zeaxanthin, the two identified components for excitation energy dissipation. Consistent with sustained energy dissipation, npq2 has decreased growth with respect to the wild type at low light conditions (Figures 1C and 1D) and shows its best performance in excess light that causes photobleaching in the older leaves of the wild type, npq1, and npq4. npq1 and npq4 lack zeaxanthin and PsbS, respectively. The extent of damage in npq1 and npq4, however, is not the same as it would be implied if zeaxanthin and PsbS would contribute to the same reaction; rather, npq1 is more severely affected and yet conserves ∼50% of qE, whereas npq4 has none. On the other hand, the reduced growth of npq2 in limiting light implies that some kind of sustained energy dissipation is activated deriving from a more prompt and intense qE (Figure 3) and from a lower dark adapted fluorescence yield (Figure 2A).
This implies that zeaxanthin, in addition to its probable action through PsbS, also contributes to an additional energy dissipating mechanism. This mechanism should be different from qE because the level of residual light-induced energy dissipation in npq1 is higher than in npq4 (Figure 3) consistently with previous reports (Niyogi et al., 1998; Li et al., 2000) and should be active in npq2, which has constitutively high photoprotection and low growth. Exposure to excess light, in addition to short-term energy dissipation, induces ROS synthesis, leading to upregulation of many genes (Rossel et al., 2002) and growth retardation (Wagner et al., 2004). Although these effects might, in principle, be relevant to the phenotypes we observed in high light conditions, we do not think these long term effects are responsible for the reduced growth of npq2 because this phenotype is most evident in very low light, a condition that is unlikely to promote ROS formation; moreover, zeaxanthin accumulation has been shown to further decrease the photooxidative damage (Baroli et al., 2003). Putative carotenoid cleaving enzymes have been shown to be involved in the synthesis of signal molecules affecting branching in Arabidopsis, implying that accumulation of its substrate might affect plant development (Sorefan et al., 2003). Nevertheless, the light dependence of the growth phenotype and the homogeneous branching habit suggest this is not the case.
pH-Independent Quenching Is Not Sensitive to Uncouplers and Is Maintained in Isolated Lhcb Proteins
We have identified this mechanism in the pH-independent quenching of chlorophyll fluorescence, present in npq2 and absent in the violaxanthin containing npq1 genotype (Figure 2B). The reduced fluorescence yield typical of npq2 is associated with isolated, detergent-solubilized LHC proteins as clearly shown by the experiments of step solubilization of chlorophyll in Figure 4: first solubilization using 0.5% α-DM disrupts protein–protein interactions but not pigment–protein and pigment–pigment interactions within individual chlorophyll binding proteins. Fluorescence quenching in LHC proteins has been reported as a result of binding of Zea versus Viola to the inner allosteric site L2 (Formaggio et al., 2001), whereas binding to the external site V1 (Caffarri et al., 2001) did not change the fluorescence yield of the complexes. The photoprotection effect of zeaxanthin in npq2 would then result from two components: (1) the increased singlet chlorophyll excited state dissipation suggested from the decreased fluorescence yield and slow growth and (2) the previously reported antioxidant effect of zeaxanthin released into the thylakoid membranes (Havaux and Kloppstech, 2001; Havaux and Niyogi, 1999; Baroli et al., 2003). This latter mechanism is clearly distinct from the fluorescence quenching effect described here because chlorophyll–carotenoid energy transfer requires van der Vaals distance between chromophores (Dexter, 1953), whereas xanthophylls bound to the site V1 cannot even operate energy transfer to chlorophyll (Caffarri et al., 2001), a process active at much longer distances (Forster, 1965). Our results (Figure 4B) clearly show that the quenching effect observed in npq2 is also active in the wild type upon exposure to excess light and accumulation of zeaxanthin.
What Is the Molecular Basis of This Quenching Effect?
IEF allows for detection of small changes in the pI of a protein, which often result from conformational changes, such as ligand binding in both soluble (Ek et al., 1980) and membrane proteins (Ben Or and Chrambach, 1983), or posttranslational modifications, such as phosphorylation in the case of CP29 (Bergantino et al., 1995). The analysis of monomeric LHC proteins in npq2 clearly shows that CP26 migrates at two different pH values, whereas in dark-adapted wild type and npq1 only the more acidic band could be detected (Figure 5A). Because CP26 is the product of a single gene in Arabidopsis (Jansson, 1999), the dual location might be either due to an unknown covalent modification of CP26 or to a conformational change. The first hypothesis was discarded by the experiment in which the CP26 complex was reconstituted, from the apoprotein overexpressed in bacteria, with either Viola or Zea (Figure 5C). Because the apoprotein was the same in both cases, a change in the pI could only be ascribed to the binding of Viola versus Zea. Neither of the two xanthophylls contain ionizable groups, which could confer such a large pI shift to the pigment–protein complex. We therefore conclude that Viola versus Zea binding to CP26 induces a conformational change involving either a shift in the pKa value of the exposed residues and/or changes in their solvent accessibility. CP26 is a protein binding DCCD (N,N′-dicyclohexylcarbodiimide) to two lumen exposed Glu residues (Walters et al., 1996), suggesting that changes in the exposure of one or both E116 and E224 residues, located in the amphiphylic helix D and in the helix B to helix C loop, respectively, might be the modification responsible for the pI shift.
The finding of a conformational change in CP26 is consistent with the recent proposal, based on spectroscopic evidence, that LHC proteins can assume two conformations with different fluorescence yield (Moya et al., 2001), and the ratio between the two forms is controlled by the binding of Zea versus Viola to the allosteric L2 site (Formaggio et al., 2001). This finding of two biochemically distinct conformations supports the proposed model (Morosinotto et al., 2003) that a function of the xanthophyll cycle is to provide regulation of the light harvesting through the action of allosteric sites in LHC proteins.
The less acidic CP26 isoform (pI = 5.1) binds Zea and has lower fluorescence yield. Conversely, the more acidic isoform (pI = 4.9) binds Viola and has the higher fluorescence yield (Figure 6A). The Zea binding and Viola binding isoforms thus correspond to the short (1.2 ns) and long (4 ns) fluorescence lifetime conformations described previously (Moya et al., 2001). Additional evidence for two different conformations is provided by absorption and CD spectroscopy. Although the two CP26 isoforms have the same pigment complement, except for the Viola-to-Zea substitution, distinct changes were observed in both the CD (Figure 6C) and absorption spectra (Figure 6B) extending into the chlorophyll Qy transition region where there is no direct absorption from carotenoids. It should be noted that these spectral changes are fully consistent with those previously described in CP26 containing either Viola or Zea as the only xanthophyll species (Croce et al., 2002).
Is pH-Independent Quenching Restricted to CP26?
Because CP26 only accounts for 4% of total chlorophyll in the PSII-LHCII complex, it might be asked if the zeaxanthin quenching effect is restricted to this complex or is a common property of LHC proteins. It was shown that not only CP26 but also CP29 (Crimi et al., 2001) and LHCII (Formaggio et al., 2001; Polivka et al., 2002) display fluorescence quenching when Zea is bound to the L2 site, and yet these complexes do not show a pI shift in Figure 5A. We conclude that pI shift, evident in CP26, may merely be due to a singularity of protein sequences: a structural change may not appear as a pI shift if it does not imply a change in the exposure of ionizable residues. DCCD binding residues in CP26 (Walters et al., 1996), which are not conserved in all LHC proteins, could be responsible for pI shift. In addition, pI shifts possibly occurring in LHCII might be hidden by the presence of multiple gene products focusing in a large pH range from 3.8 to 4.2 (Dainese et al., 1990) (see Figure 5A). The hypothesis that the pH-independent quenching is not restricted to CP26 but extends to all LHC proteins is consistent with the analysis of CP26-less insertion mutants (Figure 7C), which exhibits pH-independent quenching similar to wild-type plants. The involvement of several LHCs in conformational change/pH-independent quenching could explain the reduced fitness of LHC-deficient plants (Ganeteg et al., 2004). Conservation of each antenna subunit could provide ecological flexibility for adaptation of green algae (Elrad et al., 2002) and plants (Kulheim et al., 2002) to variable environmental conditions.
Previous work with spinach (Spinacia oleracea) involved IEF analysis of wild-type thylakoids treated to accumulate zeaxanthin (Ruban et al., 1994; Färber et al., 1997), and yet no report of a pI-shifted CP26 was made. We think this is probably due to the use of different procedures: in earlier work, direct IEF of detergent solubilized membranes was applied, whereas this work includes enrichment of the sample before IEF by sucrose gradient ultracentrifugation. The monomeric LHC fraction, containing CP26 and CP29, represents ∼8% of the total chlorophyll amount in the different gradient fractions.
Relevance of Zeaxanthin-Induced, pH-Independent Quenching in Wild-Type Plants
It is relevant to ask if the process described here in npq2 plants is acting in wild-type plants as well during photoprotective quenching accompanied by zeaxanthin accumulation. The experiments described in Figure 7 show that this is indeed the case. In fact, treatment with nigericin, which abolishes the transmembrane pH gradient and qE, is effective in restoring high fluorescence yield only in plants that did not previously accumulate zeaxanthin, whereas only ∼60% of the quenching was released in high light–treated wild-type leaves. It should be underlined that high light treatment in the npq1 mutant, unable to accumulate zeaxanthin, makes the nigericin treatment fully effective in restoring high fluorescence yield. This implies that in npq1, qE was the only quenching effect active upon high light treatment, whereas in the wild type a pH-independent quenching mechanism was also at work. This mechanism is likely to be the basis of the previously observed decrease in fluorescence in dark adapted npq2 single cells of Chlamydomonas reinhardtii with respect to wild-type control (Holub et al., 2000). The step solubilization experiment in Figure 4B shows that the effect of zeaxanthin in the wild type is the same as in npq2.
An additional observation is that the level of pH-independent quenching induced in high light–treated wild-type plants is not very different from that observed in npq2, although the former had a lower zeaxanthin content than the latter (Table 1). This result implies that the zeaxanthin effect is saturated at rather low concentrations. Consistently, the half-time of this quenching component was found to be of ∼2 min, thus longer than that of qE but much shorter than the time required for full V-to-Z conversion (Demmig-Adams et al., 1989). This is likely to be due to energy equilibration within the PSII antenna, which is energetically shaped as a shallow funnel (Jennings et al., 1993). The ultrafast chlorophyll–chlorophyll energy transfer between LHC protein subunits (Gradinaru et al., 1998) would extend the effect of a limited number of quenching centers within the pigment bed to the overall PSII.
Relation between pH-Independent and qE Quenching Types
At least two types of chlorophyll fluorescence quenching can thus be activated during excess light stress: one is persistent in the dark and maintained in isolated pigment–protein complexes, implying that it is not dependent on low lumenal pH. These characteristics are similar to those found in long term overwinter quenching (Verhoeven et al., 1999; Gilmore and Ball, 2000). Previous results demonstrated the existence of a form of sustained (Z + A)-dependent energy dissipation, detected in cold-acclimated leaves and retained even upon warming; this mechanism is correlated with slow Z + A to violaxanthin conversion and is not associated to nocturnal maintenance of low lumenal pH (Verhoeven et al., 1998). Leaves naturally acclimated to freezing temperature display a cold-hard band feature of the chlorophyll a fluorescence spectra; this mechanism leads reduction of PSII quantum yield by competing with antenna excitation, thus allowing both PSII energy dissipation and protective storage of chlorophylls during winter (Matsubara et al., 2002; Gilmore et al., 2003), and is positively correlated with decrease of a nonphosphorylated form of D1 (Ebbert et al., 2001, 2005).
The second type of quenching, known as qE, is pH dependent and requires protonation of lumenal exposed acidic residues in the PsbS subunit (Li et al., 2002b), which has DCCD binding sites (Dominici et al., 2002). Both types of quenching have been proposed to imply conformational changes in LHC proteins (Gilmore and Ball, 2000; Horton et al., 2000). Our data show that such a conformational change actually occurs in the case of dark-resistant, pH-independent quenching. It is interesting to consider if these two quenching processes are due to the same underlying conformational change or not. It is worth noting that, upon light exposure, npq2 develops faster quenching (Figure 3) with respect to the wild type and npq1,and yet its fluorescence yield was already decreased by 40% before the onset of high light treatment. After 8 min of exposure to excess light, the residual chlorophyll fluorescence yield of npq2 is 18.7% with respect to the yield in dark-adapted wild type versus 27.7% in the wild type and 44.8% in npq1 (corresponding to NPQ values of 4.3, 2.6, and 1.2, respectively, as calculated using the Fm level of the wild type). These results indicate that the pH-independent quenching process induced by Zea binding in the dark adds to the qE induced by low pH upon illumination and is further evidence that the two types of quenching are distinct. This is confirmed by the behavior of the double mutant npq2 npq4, which lacks PsbS and qE and yet shows decreased fluorescence yield (Figure 2A). Moreover, the kinetics of NPQ in the light are faster in npq2 with respect to npq1 and the wild type,suggesting that the presence of Zea instead of Viola in the complex before illumination allows for faster transition to the fully quenched state. These findings are consistent with an earlier report (Demmig-Adams et al., 1990; Ruban et al., 1993). It is not clear if the constitutive presence of Zea, effective in increasing the rate and amplitude of light-induced quenching in npq2, belongs to Lhcb proteins or to other subunits, such as PsbS (Li et al., 2002a). The analysis of npq4, npq1 npq4, and npq2 npq4 mutants clearly shows that the modulation of NPQ rate by Zea fully depends on the presence of PsbS and therefore suggests that the site sensitive to the presence of zeaxanthin might be located in this gene product. Evidences for binding of pigments to PsbS is controversial (Funk et al., 1995; Aspinall-O'Dea et al., 2002; Dominici et al., 2002) but has been proposed to be a step in the mechanism of qE. The increased NPQ rate in npq2, but not in npq2 npq4, is consistent with the proposal that Zea binds to PsbS. It should, however, be noted that lack of Zea in npq1 does not prevent NPQ to the same extent as the lack of PsbS in npq4, thus implying that PsbS can catalyze energy dissipation even in the absence of Zea, which either implies a different quenching mechanism in this mutant or the binding of other xanthophylls, such as lutein (Pogson et al., 1998). Therefore, Zea appears to be an allosteric modulator of both qE and the pH-independent quenching. Previous work with plants lacking CP26 because of antisense inhibition showed little or no effect on qE, while the presence of a pH-independent quenching was not assessed (Andersson et al., 2001).
What Is the Physiological Role of pH-Independent Quenching in Vivo?
The binding of Zea to Lhcb proteins and to PsbS appears to provide a more efficient response in conditions of repetitive excess light exposure, such as under sun flecks deriving from the overcasting of the canopy, in two different ways: first, binding of Zea to LHC proteins provides an offset regulation of the efficiency of light harvesting; second, availability of Zea in the dark, as released from LHC proteins that had accumulated this pigment during excess light exposure, allows for faster PsbS-dependent dissipation upon exposure to light.
The kinetics of Zea incorporation into individual LHC proteins has been shown to be pH dependent and to vary widely between the different gene products (Morosinotto et al., 2002). This may serve to ensure a wide range of temporal responses to light and to modify the plant future response to light in accordance with information obtained from previous stresses (Demmig-Adams and Adams, 1996). Thus, qE and pH-independent quenching are connected through the availability of Zea, as released from Lhcb proteins or produced by violaxanthin deepoxidase enzyme activity, for activation of PsbS. In this way, a plant that has stored Zea upon previous exposure to high light is able to trigger qE more promptly (Demmig-Adams et al., 1990) by avoiding the temporal lag necessary to release Viola from the V1 site (Caffarri et al., 2001) and its deepoxidation when no Zea is present. Published results suggest a more general role for xanthophyll cycle: it was shown that downregulation of nitrate reductase activity, and thus lower NADPH use, increases zeaxanthin levels (Foyer et al., 1994). Consistently, inhibition of transketolase activity increases starch accumulation and zeaxanthin levels in dark adapted leaves, allowing for faster and stronger excess energy dissipation in the event of excess light (Henkes et al., 2001). This suggests that zeaxanthin accumulation is a tool for preventive modulation of the amplitude and rate of heat dissipation response in relation to the cell metabolism and redox poise with higher NADPH/NADP+ ratios being an indication of stronger sensitivity to excess light.
Conclusions
We report on a quenching mechanism induced by the synthesis of zeaxanthin in high light and independent from PsbS. This mechanism is based on the conformational change to a low fluorescence yield state induced on LHC proteins upon binding of zeaxanthin to the allosteric site L2. This can be detected in CP26 (Lhcb5) by a pI shift. This mechanism is thought to interact with the PsbS-dependent mechanism (qE) by their common intermediate: the zeaxanthin produced in excess light conditions. Differently from qE, this quenching component is insensitive to nigericin and corresponds, at least in part, to the slowly relaxing component of the quenching previously reported as qI (Horton, 1996).
METHODS
Plant Material
Wild-type plants of Arabidopsis thaliana ecotype Columbia and mutants npq1, npq2-1 allelic to aba1 (Niyogi et al., 1998), psbS-1.3 (Grasses et al., 2002) allelic to npq4 (Li et al., 2000), and lhcb5 (The Arabidopsis Information Resource stock number SALK_014869, accession number 4505017; see Supplemental Figure 3 online) were obtained from the Arabidopsis Stock Center, from Carlo Soave (Milan, Italy), and from Dario Leister (Max Planck Institute for Plant Breeding Research, Köln, Germany). For sake of simplicity, we will use the notation of npq2 for npq2-1 and npq4 for psbs-1.3. Genotypes npq1 npq4 and npq2 npq4 were obtained by crossing single mutant plants. Plants were grown for 3 weeks in controlled conditions (∼120 μE m−2 s−1, 20°C, 8 h light/16 h dark). For long-term treatment, 3-week-old seedlings were exposed to light conditions of 20, 120, or 1300 μE m−2 s−1 for 3 weeks (20°C, 8 h light/16 h dark). Light was provided by halogen lamps and filtered through a 2-cm recirculation water layer to remove infrared radiation. Short term high light treatment was performed for 1 h at 1200 μE m−2 s−1; each set of npq2 plants had been daily sprayed with 10 μM ABA to avoid possible differences in growth rate because of ABA lack. For growth rate determinations, 15 rosettes from each population before and after exposure for 3 weeks to different light conditions were harvested and immediately weighed. Growth rates were expressed as percentage of fresh weight increase, according to the equation (FW3weeks − FWto)*100/FWto, where FW3weeks is mean fresh weight of rosettes bulk harvested after 3 weeks of light treatment and FWto is mean fresh weight of rosettes bulk harvested before light treatment.
In Vivo Fluorescence and NPQ Measurements
Fm was measured on leaves dark-adapted for 1 h, with a homemade video imaging apparatus previously described (Swiatek et al., 2001) upon induction by a 1.8-s pulse of 600 μE m−2 s−1 LED light at 590 nm and fluorescence detection at λ >680 nm with a CCD camera. This fluorescence level was the same (within 5%) as obtained by treating leaves with 50 μM DCMU (Sigma-Aldrich, St. Louis, MO), thus corresponding to Fm. Nonphotochemical quenching of chlorophyll fluorescence was measured with the same apparatus by exposing leaves to white light (actinic lamp) of intensity of 1200 μE m−2 s−1. These measurements were repeated with a PAM fluorimeter (WALZ, Effeltrich, Germany): minimum fluorescence (F0) was measured with a 0.15 μE m−2 s−1 beam, Fm was determined with a 1.5-s light pulse (5000 μE m−2 s−1), white continuous light (1200 μE m−2 s−1) was supplied by a KL1500 halogen lamp (Schott, Mainz, Germany). NPQ was calculated according to the equation (Van Kooten and Snel, 1990) NPQ = (Fm − F′m)/F′m, where Fm is the maximum chlorophyll fluorescence from dark-adapted leaves and F′m the maximum chlorophyll fluorescence under actinic light exposition. After fluorescence determination, leaves were weighted, leaf area was measured, and chlorophylls were extracted with N,N-dimethylformamide (Sigma-Aldrich); chlorophyll concentration was determined according to Porra et al. (1989). In some experiments, nigericin (Sigma-Aldrich) was used at 50 μM for inhibition of qE by infiltrating leaves in a syringe; control plants were infiltrated with 5 mM Hepes, pH 7.0, to correct for changes in the optical properties of leaves upon infiltration.
Fluorescence measurements on stacked thylakoids were performed in a PAM stirred cuvette as previously described (Gilmore et al., 1995). Thylakoids for fluorescence measurements were isolated from dark-adapted Arabidopsis wild-type leaves as previously described (Gilmore et al., 1998) and were diluted before fluorescence measurements to a final concentration of 0.05 mg chlorophyll/mL. Buffer for Fm measurements contained 0.1 M sorbitol, 5 mM MgCl2, 10 mM NaCl, 10 mM KCl, 10 mM Tricine, pH 7.8, 0.2% BSA, 50 mM sodium ascorbate, and 50 μM methyl viologen.
Thylakoid Isolation and Sample Preparation
Unstacked thylakoids were isolated from leaves as previously described (Bassi et al., 1988), with some modifications for light-treated wild-type plants. To maintain maximum deepoxidation state in isolated membranes, unstacked thylakoids were rapidly isolated by grinding leaves in 5 mM EDTA, 50 mM Mes, pH 5.2, and 50 mM sodium ascorbate at 4°C; thylakoids were filtered through a sieve, centrifuged (10 min at 10,000g, 4°C), and then resuspended in a buffer with 5 mM EDTA and 10 mM Hepes, pH 7.8.
Membranes (obtained either from dark-adapted or light-treated leaves) corresponding to 500 μg of chlorophylls were washed with 5 mM EDTA and then solubilized in 1 mL with 0.6% α-DM and 10 mM Hepes, pH 7.5. Solubilized samples were then fractionated by ultracentrifugation in a 0.1 to 1 M sucrose gradient containing 0.06% α-DM and 10 mM Hepes, pH 7.5 (22 h at 280,000g, 4°C).
Monomeric Lhcb proteins were further fractionated by flatbed IEF at 4°C as previously described (Dainese et al., 1990). Green bands were harvested and eluted from a small column with 10 mM Hepes, pH 7.5, and 0.06% α-DM, and further fractionated on a 0.1 to 1 M sucrose gradient containing 0.06% α-DM and 10 mM Hepes, pH 7.5, for 23 h at 280,000g, at 4°C.
Pigment Analysis
The pigments were extracted from thylakoids and isolated complexes with 80% acetone, then separated and quantified by HPLC (Gilmore and Yamamoto, 1991) and by fitting of the spectrum of the acetone extract with the spectra of individual pigments (Croce et al., 2002).
Gel Electrophoresis
SDS-PAGE analysis was performed with the Tris-Tricine buffer system as previously described (Schagger and von Jagow, 1987).
Spectroscopy
Spectra were obtained using samples in 10 mM Hepes, pH 7.5, 0.06% α-DM, and 0.2 M sucrose. Absorption measurements were performed using an SLM-Aminco DW-2000 spectrophotometer at room temperature. CD spectra were recorded with a Jasco 600 spectropolarimeter at room temperature. Fluorescence emission spectra were measured at room temperature using a Jasco FP-777 spectrofluorimeter excited at 440 nm with a 5-nm bandwidth beam for excitation and 3 nm for emission. The chlorophyll concentration was 0.1 μg/mL.
Experiments of fluorescence dependence on step solubilization of thylakoids (Figure 4) were performed using a Jasco PF-777 spectrofluorimeter equipped with a homemade system of sample injection into a stirring cuvette. The excitation light at the sample level was of 16 μE m−2 s−1 (excitation wavelength, 440 nm; emission wavelength, 680 nm; bandwidth, 5 nm for excitation, 3 nm for emission). Unstacked thylakoids corresponding to a final chlorophyll concentration of 0.1 μg/mL were added to the stirring solution, and two different detergents were added: first, α-DM at final 0.5% (w/v) concentration from a 10% stock solution, then Triton X-100 at final 5% (w/v) concentration from a 50% stock solution. After each addition, time for fluorescence level stabilization was allowed. Quenching parameters Qm, Qpi, and Qm+pi were calculated according to the equations Qm = (Fd − Ft)/Ft, Qpi = (Fx − Fd)/Fd, and Qm+pi = (Fx − Ft)/Ft, where Ft is thylakoid fluorescence (unsolubilized membranes) at t = 1 min from the start of measurement, Fd is pigment–protein complexes fluorescence (in α-DM 0.5%) at t = 7.5 min, and Fx is free-pigment fluorescence (in 5% Triton X-100) measured at t = 12 min (see Figure 4, Table 2). Qm is the quenching of chlorophyll–protein complexes fluorescence provided by their incorporation in the thylakoid lipid bilayer; Qpi is the quenching of free chlorophyll provided by their binding into chlorophyll–protein complexes; Qm+pi is the quenching of free pigment fluorescence provided by their incorporation into chlorophyll–proteins and in the thylakoids.
DNA Cloning and Isolation of Overexpressed Lhcb5 Apoprotein
Plasmids were constructed using standard molecular cloning procedures (Sambrook et al., 1989). cDNA of Lhcb5 from Arabidopsis (GenBank accession number AF134129) was supplied by the ABRC at The Ohio State University. The coding region for the putative mature polypeptide was amplified by PCR (forward primer, 5′-CGCGGATCCAAGAAAAAGCCAGCTCCTGC-3′, reverse primer, 5′-GGTCTGCAGTGAGAGTGGGAGCTCTCTCG-3′). The amplified region was cloned into pQE-50 (Qiagen, Valencia, CA) expression vector. Lhcb5 apoprotein was expressed and isolated from Escherichia coli, using SG13009 strains, following a protocol previously described (Paulsen and Hobe, 1992).
Reconstitution and Purification of Lhcb5 Pigment–Protein Complexes
Refolding procedure was performed as previously described (Giuffra et al., 1996) with the following modifications: chlorophyll a/b ratio in the pigments mixture used for reconstitution was 3, and Lhcb5 was refolded with either a mixture of Lute, Viola, and Neo or Lute and Zea. All the pigments used were purified from thylakoids of Arabidopsis.
Supplementary Material
Acknowledgments
We thank T. Morosinotto (Verona, Italy) for discussion and N. Holt (Berkeley, CA) for critically reading the manuscript. S. Cazzaniga and G. Bonente are thanked for technical support. This work was funded by MIUR FIRB Grants RBAU01E3CX and RBNE01LACT.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Roberto Bassi (bassi@sci.univr.it).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.030601.
References
- Andersson, J., Walters, R.G., Horton, P., and Jansson, S. (2001). Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. Plant Cell 13, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aro, E.-M., McCaffery, S., and Anderson, J.M. (1993. a). Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol. 103, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro, E.-M., Virgin, I., and Andersson, B. (1993. b). Photoinhibition of photosystem-2. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Asada, K., and Takahashi, M. (1987). Production and scavenging of active oxygen in photosynthesis. In Photoinhibition, D.J. Kyle, C.B. Osmond, and C.J. Arntzen, eds (Amsterdam: Elsevier), pp. 227–287.
- Aspinall-O'Dea, M., Wentworth, M., Pascal, A., Robert, B., Ruban, A., and Horton, P. (2002). In vitro reconstitution of the activated zeaxanthin state associated with energy dissipation in plants. Proc. Natl. Acad. Sci. USA 99, 16331–16335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli, I., Do, A.D., Yamane, T., and Niyogi, K.K. (2003). Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlorophyllamydomonas reinhardtii from photooxidative stress. Plant Cell 15, 992–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi, R., Pineau, B., Dainese, P., and Marquardt, J. (1993). Carotenoid-binding proteins of photosystem-II. Eur. J. Biochem. 212, 297–303. [DOI] [PubMed] [Google Scholar]
- Bassi, R., Rigoni, F., Barbato, R., and Giacometti, G.M. (1988). Light-harvesting chlorophyll a/b proteins (LHCII) populations in phosphorylated membranes. Biochim. Biophys. Acta 936, 29–38. [Google Scholar]
- Ben Or, S., and Chrambach, A. (1983). Heterogeneity of the glucocorticoid receptors: Molecular transformations during activation, detected by electrofocusing. Arch. Biochem. Biophys. 221, 343–353. [DOI] [PubMed] [Google Scholar]
- Bergantino, E., Dainese, P., Cerovic, Z., Sechi, S., and Bassi, R. (1995). A post-translational modification of the photosystem II subunit CP29 protects maize from cold stress. J. Biol. Chem. 270, 8474–8481. [DOI] [PubMed] [Google Scholar]
- Caffarri, S., Croce, R., Breton, J., and Bassi, R. (2001). The major antenna complex of photosystem II has a xanthophyll binding site not involved in light harvesting. J. Biol. Chem. 276, 35924–35933. [DOI] [PubMed] [Google Scholar]
- Crimi, M., Dorra, D., Bosinger, C.S., Giuffra, E., Holzwarth, A.R., and Bassi, R. (2001). Time-resolved fluorescence analysis of the recombinant photosystem II antenna complex CP29. Effects of zeaxanthin, pH and phosphorylation. Eur. J. Biochem. 268, 260–267. [DOI] [PubMed] [Google Scholar]
- Croce, R., Canino, G., Ros, F., and Bassi, R. (2002). Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41, 7334–7343. [DOI] [PubMed] [Google Scholar]
- Dainese, P., and Bassi, R. (1991). Subunit stoichiometry of the chloroplast photosystem-II antenna system and aggregation state of the component chlorophyll-a/b binding proteins. J. Biol. Chem. 266, 8136–8142. [PubMed] [Google Scholar]
- Dainese, P., Hoyer-hansen, G., and Bassi, R. (1990). The resolution of chlorophyll a/b binding proteins by a preparative method based on flat bed isoelectric focusing. Photochem. Photobiol. 51, 693–703. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams, B., and Adams III, W.W. (1996). The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1, 21–26. [Google Scholar]
- Demmig-Adams, B., and Adams, W.W. (2000). Harvesting sunlight safely. Nature 403, 371–374. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams, B., Adams, W.W., Heber, U., Neimanis, S., Winter, K., Krüger, A., Czygan, F.-C., Bilger, W., and Björkman, O. (1990). Inhibition of zeaxanthin formation and of rapid changes in radiationless energy dissipation by dithiothreitol in spinach leaves and chloroplasts. Plant Physiol. 92, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams, B., Winter, K., Kruger, A., and Czygan, F.-C. (1989). Light stress and photoprotection related to the carotenoid zeaxanthin in higher plants. In Photosynthesis. Plant Biology, Vol. 8, W.R. Briggs, ed (New York: Alan R. Liss), pp. 375–391.
- Dexter, D.L. (1953). A theory of sensitized luminescence in solids. J. Chem. Phys. 21, 836–850. [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici, P., Caffarri, S., Armenante, F., Ceoldo, S., Crimi, M., and Bassi, R. (2002). Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J. Biol. Chem. 277, 22750–22758. [DOI] [PubMed] [Google Scholar]
- Ebbert, V., Adams III, W.W., Mattoo, A.K., Sokolenko, A., and Demmig-Adams, B. (2005). Up-regulation of a photosystem II core protein phosphatase inhibitor and sustained D1 phosphorylation in zeaxanthin-retaining, photoinhibited needles of overwintering Douglas fir. Plant Cell Environ. 28, 232–240. [Google Scholar]
- Ebbert, V., Demmig-Adams, B., Adams III, W.W., Mueh, K.E., and Staehelin, L.A. (2001). Correlation between persistent forms of zeaxanthin-dependent energy dissipation and thylakoid protein phosphorylation. Photosynth. Res. 67, 63–78. [DOI] [PubMed] [Google Scholar]
- Ek, K., Gianazza, E., and Righetti, P.G. (1980). Affinity titration curves: Determination of dissociation constants of lectin-sugar complexes and of their pH-dependence by isoelectric focusing electrophoresis. Biochim. Biophys. Acta 626, 356–365. [DOI] [PubMed] [Google Scholar]
- Elrad, D., Niyogi, K.K., and Grossman, A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Färber, A., Young, A.J., Ruban, A.V., Horton, P., and Jahns, P. (1997). Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants. Plant Physiol. 115, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaggio, E., Cinque, G., and Bassi, R. (2001). Functional architecture of the major light-harvesting complex from higher plants. J. Mol. Biol. 314, 1157–1166. [DOI] [PubMed] [Google Scholar]
- Forster, T. (1965). Delocalized excitation and excitation transfer. In Modern Quantum Chemistry, Part III: Action of Light and Organic Crystals, O. Sinanoglu, ed (New York: Academic Press), pp. 93–137.
- Foyer, C.H., Lescure, J.C., Lefebvre, C., Morot-Gaudry, J.F., Vincentz, M., and Vaucheret, H. (1994). Adaptations of photosynthetic electron transport, carbon assimilation, and carbon partitioning in transgenic Nicotiana plumbaginifolia plants to changes in nitrate reductase activity. Plant Physiol. 104, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk, C., Schröder, W.P., Napiwotzki, A., Tjus, S.E., Renger, G., and Andersson, B. (1995). The PSII-S protein of higher plants: A new type of pigment-binding protein. Biochemistry 34, 11133–11141. [DOI] [PubMed] [Google Scholar]
- Ganeteg, U., Kulheim, C., Andersson, J., and Jansson, S. (2004). Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 134, 502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, A.M., and Ball, M.C. (2000). Protection and storage of chlorophyll in overwintering evergreens. Proc. Natl. Acad. Sci. USA 97, 11098–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, A.M., Hazlett, T.L., and Govindjee, G. (1995). Xanthophyll cycle-dependent quenching of photosystem II chlorophyll a fluorescence: Formation of a quenching complex with a short fluorescence lifetime. Proc. Natl. Acad. Sci. USA 92, 2273–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, A.M., Matsubara, S., Ball, M.C., Barker, D.H., and Itoh, S. (2003). Excitation energy flow at 77 K in the photosynthetic apparatus of overwintering evergreens. Plant Cell Environ. 26, 1021–1034. [Google Scholar]
- Gilmore, A.M., Shinkarev, V.P., Hazlett, T.L., and Govindjee, G. (1998). Quantitative analysis of the effects of intrathylakoid pH and xanathophyll cycle pigments on chlorophyll a fluorescence lifetime distributions and intensity in thylakoids. Biochemistry 37, 13582–13593. [DOI] [PubMed] [Google Scholar]
- Gilmore, A.M., and Yamamoto, H.Y. (1991). Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra, E., Cugini, D., Croce, R., and Bassi, R. (1996). Reconstitution and pigment-binding properties of recombinant CP29. Eur. J. Biochem. 238, 112–120. [DOI] [PubMed] [Google Scholar]
- Giuffra, E., Zucchelli, G., Sandona, D., Croce, R., Cugini, D., Garlaschi, F.M., Bassi, R., and Jennings, R.C. (1997). Analysis of some optical properties of a native and reconstituted photosystem II antenna complex, CP29: Pigment binding sites can be occupied by chlorophyll a or chlorophyll b and determine spectral forms. Biochemistry 36, 12984–12993. [DOI] [PubMed] [Google Scholar]
- Gradinaru, C.C., Özdemir, S., Gülen, D., van Stokkum, I.H.M., van Grondelle, R., and Van Amerongen, H. (1998). The flow of excitation energy in LHCII monomers: Implications for the structural model of the major plant antenna. Biophys. J. 75, 3064–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasses, T., Pesaresi, P., Schiavon, F., Varotto, C., Salamini, F., Jahns, P., and Leister, D. (2002). The role of delta pH-dependent dissipation of excitation energy in protecting photosystem II against light-induced damage in Arabidopsis thaliana. Plant Physiol. Biochem. 40, 41–49. [Google Scholar]
- Havaux, M., and Kloppstech, K. (2001). The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 213, 953–966. [DOI] [PubMed] [Google Scholar]
- Havaux, M., and Niyogi, K.K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 96, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkes, K., Sonnewald, U., Badur, R., Flachmann, R., and Stitt, M. (2001). A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13, 535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, N.E., Fleming, G.R., and Niyogi, K.K. (2004). Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43, 8281–8289. [DOI] [PubMed] [Google Scholar]
- Holt, N.E., Zigmantas, D., Valkunas, L., Li, X.-P., Niyogi, K.K., and Fleming, G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307, 433–436. [DOI] [PubMed] [Google Scholar]
- Holub, O., Seufferheld, M.J., Gohlke, C., Govindjee, G., and Clegg, R.M. (2000). Fluorescence lifetime imaging (FLI) in real-time: A new technique in photosynthesis research. Photosynthetica 38, 581–599. [Google Scholar]
- Horton, P. (1996). Nonphotochemical quenching of chlorophyll fluorescence. In Light as an Energy Source and Information Carrier in Plant Physiology, R.C. Jennings, ed (New York: Plenum Press), pp. 99–111.
- Horton, P., Ruban, A.V., and Wentworth, M. (2000). Allosteric regulation of the light-harvesting system of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240. [DOI] [PubMed] [Google Scholar]
- Jennings, R.C., Bassi, R., Garlaschi, F.M., Dainese, P., and Zucchelli, G. (1993). Distribution of the chlorophyll spectral forms in the chlorophyll/protein complexes of photosystem-II antenna. Biochemistry 32, 3203–3210. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-Van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Kulheim, C., Agren, J., and Jansson, S. (2002). Rapid regulation of light harvesting and plant fitness in the field. Science 297, 91–93. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Bjorkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Gilmore, A.M., and Niyogi, K.K. (2002. a). Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J. Biol. Chem. 277, 33590–33597. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Phippard, A., Pasari, J., and Niyogi, K.K. (2002. b). Structure-function analysis of photosystem II subunit S (PsbS) in vivo. Funct. Plant Biol. 29, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Matsubara, S., Gilmore, A.M., Ball, M.C., Anderson, J.M., and Osmond, B. (2002). Sustained downregulation of photosystem II in mistletoes during winter depression of photosynthesis. Funct. Plant Biol. 29, 1157–1169. [DOI] [PubMed] [Google Scholar]
- Morosinotto, T., Baronio, R., and Bassi, R. (2002). Dynamics of chromophore binding to Lhc proteins in vivo and in vitro during operation of the xanthophyll cycle. J. Biol. Chem. 277, 36913–36920. [DOI] [PubMed] [Google Scholar]
- Morosinotto, T., Caffarri, S., Dall'Osto, L., and Bassi, R. (2003). Mechanistic aspects of the xanthophyll dynamics in higher plant thylakoids. Physiol. Plant. 119, 347–354. [Google Scholar]
- Moya, I., Silvestri, M., Vallon, O., Cinque, G., and Bassi, R. (2001). Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry 40, 12552–12561. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Grossman, A.R., and Björkman, O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, H., and Hobe, S. (1992). Pigment-binding properties of mutant light-harvesting chlorophyll-a/b-binding protein. Eur. J. Biochem. 205, 71–76. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Sandona, D., Giuffra, E., and Bassi, R. (1997). A single point mutation (E166Q) prevents dicyclohexylcarbodiimide binding to the photosystem II subunit CP29. FEBS Lett. 402, 151–156. [DOI] [PubMed] [Google Scholar]
- Pogson, B.J., Niyogi, K.K., Björkman, O., and DellaPenna, D. (1998). Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 95, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polivka, T., Zigmantas, D., Sundstrom, V., Formaggio, E., Cinque, G., and Bassi, R. (2002). Carotenoid S(1) state in a recombinant light-harvesting complex of Photosystem II. Biochemistry 41, 439–450. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Rossel, J.B., Wilson, I.W., and Pogson, B.J. (2002). Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 130, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban, A.V., and Horton, P. (1999). The xanthophyll cycle modulates the kinetics of nonphotochemical energy dissipation in isolated light-harvesting complexes, intact chloroplasts, and leaves of spinach. Plant Physiol. 119, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban, A.V., Young, A.J., and Horton, P. (1993). Induction of nonphotochemical energy dissipation and absorbance changes in leaves. Plant Physiol. 102, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban, A.V., Young, A.J., Pascal, A.A., and Horton, P. (1994). The effects of illumination on the xanthophyll composition of the photosystem II light-harvesting complexes of spinach thylakoid membranes. Plant Physiol. 104, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Sorefan, K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., Chatfield, S., Ward, S., Beveridge, C., Rameau, C., and Leyser, O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek, M., Kuras, R., Sokolenko, A., Higgs, D., Olive, J., Cinque, G., Muller, B., Eichacker, L.A., Stern, D.B., Bassi, R., Herrmann, R.G., and Wollman, F.A. (2001). The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13, 1347–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kooten, O., and Snel, J.F.H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25, 147–150. [DOI] [PubMed] [Google Scholar]
- Verhoeven, A.S., Adams, W.W., and Demmig-Adams, B. (1998). Two forms of sustained xanthophyll cycle-dependent energy dissipation in overwintering Euonymus kiautschovicus. Plant Cell Environ. 21, 893–903. [Google Scholar]
- Verhoeven, A.S., Adams, W.W., Demmig-Adams, B., Croce, R., and Bassi, R. (1999). Xanthophyll cycle pigment localization and dynamics during exposure to low temperature and light stress in Vinca major. Plant Physiol. 120, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Przybyla, D., Op den Camp, R., Kim, C., Landgraf, F., Lee, K.P., Wursch, M., Laloi, C., Nater, M., Hideg, E., and Apel, K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185. [DOI] [PubMed] [Google Scholar]
- Walters, R.G., Ruban, A.V., and Horton, P. (1996). Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl. Acad. Sci. USA 93, 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Werner, B., Christensen, E., and Oliver, D. (2001). The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 126, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.