Keywords: disease progression, early growth response 1, inflammatory response, respiratory disease, therapeutic target
Abstract
Early growth response 1 (EGR1), which is involved in cell proliferation, differentiation, apoptosis, adhesion, migration, and immune and inflammatory responses, is a zinc finger transcription factor. EGR1 is a member of the EGR family of early response genes and can be activated by external stimuli such as neurotransmitters, cytokines, hormones, endotoxins, hypoxia, and oxidative stress. EGR1 expression is upregulated during several common respiratory diseases, such as acute lung injury/acute respiratory distress syndrome, chronic obstructive pulmonary disease, asthma, pneumonia, and novel coronavirus disease 2019. Inflammatory response is the common pathophysiological basis of these common respiratory diseases. EGR1 is highly expressed early in the disease, amplifying pathological signals from the extracellular environment and driving disease progression. Thus, EGR1 may be a target for early and effective intervention in these inflammation-associated lung diseases.
INTRODUCTION
Early growth response 1 (EGR1), first identified as nerve growth factor-induced protein A (NGFI-A), is a member of the EGR family of early response transcription factors (1). This protein is not only involved in neuronal differentiation and transcriptional regulation but also appears active in various life activities. External stimuli such as neurotransmitters, cytokines, hormones, endotoxins, hypoxia, and oxidative stress activate EGR1. Moreover, a variety of biological processes in cells are regulated by EGR1, including cell proliferation, differentiation, apoptosis, adhesion, migration, immune response, and inflammation.
Being expressed in the early stages of various diseases, EGR1 amplifies pathological signals from the extracellular environment and drives disease progression. Furthermore, EGR1 belongs to the zinc finger transcription factor family and can regulate the downstream activation of hundreds or thousands of target genes. EGR1 can also activate the expression of PTEN and TP53 and is considered an oncogene (2, 3). EGR1 is involved in cell proliferation, the cell cycle, apoptosis, angiogenesis, invasion, and the tumor microenvironment (4) and has been found to promote tumor progression (5), as summarized in two reviews (4, 6). EGR1 is also an important signal communication molecule, existing as a third messenger that amplifies pathological signals from the extracellular environment, thereby driving the progression of, for example, cardiovascular diseases (7–9), osteoarthropathy (10, 11), diabetic nephropathy (12), inflammatory diseases (13), cholestatic liver injury (14), neuroinflammatory diseases (15), and tumors (6, 16, 17).
Inflammatory lung diseases, including acute lung injury/acute respiratory distress syndrome (ALI/ARDS), chronic obstructive pulmonary disease (COPD), asthma, pulmonary fibrosis, coronavirus disease 2019 (COVID-19), and pneumonia, are a constant threat to human life and are a major public health problem with a huge economic cost. The inflammatory response is the common pathophysiological basis for these respiratory diseases. Inflammation is primarily caused and maintained by physical and chemical stimuli, pathogenic microbial infection, and host cell injury or death. EGR1 is one of the key molecules upregulated in the expression of several common respiratory diseases. EGR1 is the gatekeeper of human macrophage inflammation promoters, and its inhibition by the NuRD corepressor complex suppresses inflammation genes. (13). Two recent reviews summarized the activation of EGR1 in the initial phase of host-pathogen interactions (18) and its role in viral infections (19). In summary, current research suggests that EGR1 is inextricably linked to inflammation-associated diseases.
EGR1 plays a crucial role in inflammatory lung diseases by acting as a “master switch” for transcription factors that activate genes involved in signaling and inflammatory immune cell activation. However, the specific mechanisms by which EGR1 regulates these biological processes have not been fully elucidated. This review provides an overview of research into the EGR1 gene and associated inflammation-related lung diseases. Our insights offer a theoretical basis for future clinical diagnosis prediction and drug therapy target discovery.
BIOLOGICAL CHARACTERISTICS OF EGR1
Discovery History of EGR1
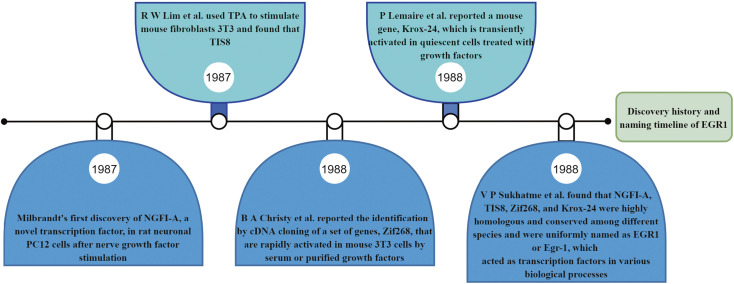
Milbrandt (1) first isolated and identified NGFI-A (the previous name for EGR1) in 1987. Differential hybridization was used to screen rat neuronal PC12 cell lines, and a differential cDNA library was obtained. Cloning of cDNA rapidly induced by nerve growth factor (NGF) and treatment with actinomycin (cycloheximide, CHX) revealed that EGR1 has a nucleotide sequence that encodes a protein that is highly homologous to other transcriptional regulatory proteins. This suggests that EGR1 may have similar functions and be involved in neuronal differentiation and transcriptional regulation. The gene sequence was subsequently cloned by four independent research groups. Lim et al. (20) cloned tetradecanoyl phorbol acetate (TPA) induction sequence 8 (TIS8) from TPA-stimulated mouse fibroblast 3T3 cells, which rapidly and strongly induced early growth response gene expression; as a result, Egr1 was also named TIS8. TIS8 was activated by TPA treatment in mouse 3T3 cell lines. Sukhatme et al. (21) found that EGR1 and Egr1, growth factors stimulating resting fibroblasts, can induce expression and that Egr1 may function as a transcription factor in various biological processes. Lemaire et al. (22) reported a mouse gene, Krox-24, that encodes a protein with three DNA-binding domains with coordinated zinc ions expressed during the G0/G1 transition of the cell cycle. Christy et al. (23) reported a gene identified through cDNA cloning, Zif268, that is rapidly activated by serum or purified growth factors in mouse 3T3 cells and encodes a protein with the three tandem zinc finger sequences of eukaryotic transcription factors. NGFI-A and Egr1 share 98% homology at the amino acid level, and encode rat and mouse proteins, respectively. Interestingly, Zif268 and Krox-24 cDNA clones are homologs of mouse Ngfi-A and were later gradually unified with Egr1 nomenclature (21). The historical timeline of the discovery of EGR1 is shown in Fig. 1.
Figure 1.
Historical timeline of the discovery of EGR1. EGR1, early growth response 1; NGFI-A, nerve growth factor-induced protein A; TPA, tetradecanoyl phorbol acetate.
As studies progressed, it was found that EGR1 is not only involved in neuronal differentiation and transcriptional regulation but also appears to be active in various biological processes. EGR1 is commonly expressed in eukaryotes and can alter a variety of cellular biological processes; this is strongly related to the fact that EGR1, as a transcription factor, can regulate the activation of thousands of downstream target genes. EGR1 is a Cys2-His2-type zinc finger transcription factor that can bind to the DNA promoter sequence of the specific sequence 5′-GCG(T/G)GGGCG-3′. The binding of EGR1 to this sequence is independent of whether the CpG island is methylated or not but depends on the form of cytosine modification (24, 25). Since any gene with this sequence information in the promoter sequence could be an EGR1 binding site, by searching the database of transcription factor EGR1 binding target genes, we found that there are thousands of such downstream target genes that can be regulated in different diseases.
EGR1 is also considered a tumor suppressor gene since it activates PTEN and TP53 expression (2, 3) and is involved in DNA damage, apoptosis, cell cycle, cell differentiation and death, and immune and inflammatory responses. EGR1 also directly promotes the transcription of IL-6, tissue factor, TNF-α, COX2, mPGES1, MCP-1, and intercellular adhesion molecule-1 (ICAM-1) inflammation-related mediators. The 5′- GCGGGGGCG-3′ site may belong to the transcriptional activation region but may also be a transcriptional repressor region. Studies on EGR1 repression of downstream target gene transcription are scarce, and reports on inflammatory lung diseases are particularly limited. However, some studies have reported that Egr1 binds to the sequence 5′- GCGGGGGCG-3′ within the promoter of the human stathmin gene. As a result of either endogenous or exogenous expression of Egr1, stathmin gene promoter activity and expression levels are downregulated, causing an oncogenic effect (26). ALAS1 is the first rate-limiting enzyme in heme biosynthesis, and its promoter sequence ( GCGGGGCG) is the binding site for EGR1. When EGR1 is expressed in cells, ALAS1 mRNA levels and intracellular heme levels are reduced (27). COL2A1 proximal promoter activity is inhibited by IL-1β-induced activation of Egr-1 (28). It acts as a transcriptional repressor of calcium chelator protein promoters (29). Overexpression of sodium calcium exchanger-1 is thought to be detrimental to calcium efflux from cardiomyocytes, whereas ChIP experiments demonstrated that Egr-1 is essential for NCX1 promoter binding and repression of NCX1 expression (30). Besides acting as a transcriptional activator, Egr-1 also acts as a repressor to control downstream target gene expression (31). Egr1/egr1/EGR1 is highly conserved among mice, rats, chickens, zebrafish, chimpanzees, dogs, cattle, and humans (32).
Structural Characteristics of EGR1
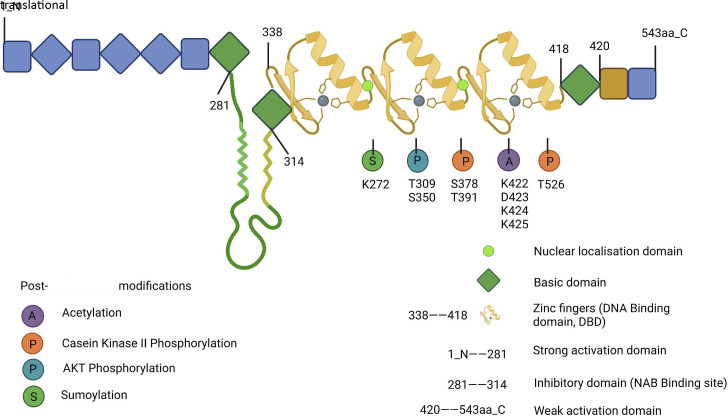
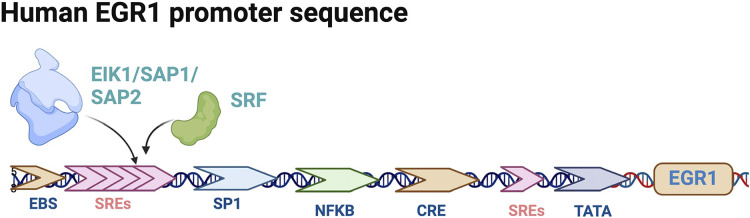
There are four members of the EGR family, EGR1, EGR2, EGR3, and EGR4, which share a highly homologous DNA-binding domain that recognizes the same DNA response elements. EGR1 is by far the most widely studied family member, and expression of egr1 is seen in most eukaryotes. EGR1 (21) has also been known as NGFI-A (1), TIS8 (20), KROX-24 (22), and ZIF-268 (23). The EGR1 gene is localized on human chromosome Chr5q23-31 and encodes 543 amino acids. It consists of three highly conserved DNA structural domains of Cys2-His2-type zinc finger proteins, encoding transcription factors with molecular mass predicted to be 58 kDa, whereas EGR1 detected by immunoblotting showed an apparent molecular mass of 80–100 kDa, probably owing to posttranslational modifications (21, 33–35). The structural domain region of the EGR1 protein is mainly divided into three different biological functional regions: DNA-binding, transcriptional activation, and transcriptional repression. The DNA-binding region has three repetitive zinc finger structures that bind to homologous DNA-binding sites: the transcriptional repression region located between the transcriptional activation region and the DNA-binding domain is identified as the binding site of two transcription factors, NGFI-A binding protein 1 (NAB1) and NGFI-A binding protein 2 (NAB2), both of which inhibit the biological activity of EGR1 (36–38). EGR1 and its family members EGR2 and EGR3 can also increase the expression of NAB2, which in turn inhibits EGR1 expression, forming a negative feedback loop regulated by EGR1 itself (38). EGR1 can also downregulate EGR1 transcription by binding to its own 5′-GCG(T/G)GGGCG-3′ promoter sequence (39). The molecular mechanism of this inhibitory activity is not known, but it is an additional negative feedback loop beyond the induction of NAB2 synthesis by EGR1, allowing a transient but not continuous synthesis of EGR1. The human EGR1 promoter region has five serum reaction elements (SREs), and SRE-mediated biological activity requires the involvement of two types of transcription factor: a ternary complex (EIK1, SAP1, or SAP2) and serum response factor (SRF), which together mediate EGR1 biological activity. The EGR1 promoter region also has two cyclic adenosine phosphate (cAMP) response element binding proteins (CREBs), an activator protein 1 (AP1), and two gene-specific activator protein 1 (Sp1) binding sites are identified (40, 41). Schematic diagrams of the structural domain and posttranslational modification sites of the EGR1 protein are shown in Fig. 2, and the human EGR1 gene promoter sequence is shown in Fig. 3.
Figure 2.
Schematic diagram of the structural domain and posttranslational modification sites of EGR1 protein. EGR1, early growth response 1. [Image created with BioRender.com and published with permission.]
Figure 3.
Pattern map of human EGR1 gene promoter sequence. EGR1, early growth response 1; SREs, serum reaction elements. [Image created with BioRender.com and published with permission.]
EGR1 Activation Is Mainly Dependent on PKA and MAPK Pathway Activation
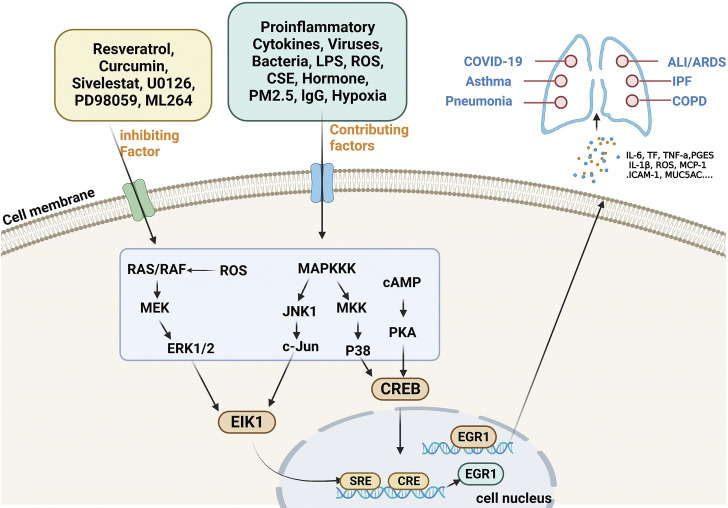
The earliest EGR1 was found to be induced by NGF (1), which is a nuclear protein, and elevated expression of EGR1 can be activated in many different ways. EGR1 can be activated transiently and rapidly by different agonists and external conditions; therefore, many signaling cascades converge on the Egr1 gene. The mouse Egr1 gene has ELK1, EGR1, CGbox, two CREs, and five SREs on its promoter sequence, and SREs can bind to serum response factors (27). In addition, EGR1 is considered a third messenger kinase, and its activation is influenced by multiple upstream signaling pathways, most notably involving protein kinase A (PKA) and mitogen-activated protein kinase (MAPK). When the cell is stimulated by the outside world, the signal is transmitted to the cell through the receptors on the surface of the cell membrane. cAMP participates in signaling as a second messenger and activates PKA, and the activated PKA then activates cyclic adenosine effector element binding protein (CREB) to undergo phosphorylation. CREB enters the nucleus and binds to CRE response elements to form a complex that initiates the transcription of EGR1 (42, 43), forming a cAMP/PKA/EGR1 signaling axis involved in various life activities. The MAPK pathway is a general term for a large class of signaling pathways, including the c-Jun N-terminal protein kinase (JNK) pathway, the extracellular signal-regulated kinase extracellular signal-regulated kinase (ERK)1/2 pathway, and the p38 MAPK pathway. The mammalian MAPK protein kinase signaling pathway is mainly activated by stress and inflammation (44, 45), and the JNK pathway is mainly regulated by stress. Activation of the JNK-c-Jun signaling axis can further activate EIK1, and the activated EIK1 can bind to the SRE response element of the EGR1 promoter sequence to initiate EGR1 transcription and participate in life activity processes through the JNK/c-Jun/EIK1/EGR1 pathway (46). The ERK1/2 pathway regulates EGR1 transcription, and EGR1 activation is also regulated by EIK1-dependent ERK1/2 phosphorylation. When lipopolysaccharide (LPS) stimulates cells, it signals through the RAS/RAF/MEK/ERK1/2/EIK1 signaling pathway to activate EGR1 expression; moreover, it promotes EGR1 transcription through the MAPKKK/MKK/p38/CREB signaling axis (46). Cytokines, viruses, bacteria, oxidative stress, cigarette smoke, IgG immune complexes (47), chemotherapeutic drugs, neurotransmitters, growth factors, cytotoxic metabolites, hormones, endotoxins, hypoxia, hyperoxia, ischemia-reperfusion, zinc oxide nanoparticles (48), PM2.5 (49), oxidative stress (50), mechanical damage and shear stress (51), and other extracellular signals stimulate activation (8, 52), the progression of inflammatory lung diseases, and increased expression of EGR1. Modern molecular biology techniques can also be applied to overexpress the EGR1 gene, thereby activating EGR1-related functions. EGR1 may be involved in life activities by interlinking multiple upstream signals and downstream target genes.
Inhibition of EGR1
The signaling pathway involved in EGR1 contributes significantly to the development of human diseases and is a promising target for drug development; drugs targeting the signaling mechanisms involved in EGR1 have been investigated to treat diseases. Dyson et al. (53) found that resveratrol inhibits Kaposi’s sarcoma-associated herpesvirus (KSHV) reactivation by reducing cellular EGR1 levels. The findings demonstrated that in KSHV-infected cells, resveratrol reduces ERK1/2 activity and EGR1 expression, thereby inhibiting virus reactivation during latency. Moreover, these findings contribute to the study of pathogenesis due to KSHV. Muthusamy et al. (54) showed that blocking ERK1/2 by inhibitor U0126 terminated PMA-induced Egr1 expression, demonstrating that PMA stimulates NHE2 expression by activating PKCδ; the MEK–ERK1/2 signaling pathway is then activated followed by stimulation of Egr-1 expression. Cubero et al. (55) found that ethanol acts synergistically with arachidonic acid to induce ERK1/2 phosphorylation and nuclear translocation of Egr1, and that PD98059 (ERK1/2 inhibitor) and curcumin (EGR1 inhibitor) blocked arachidonic acid-mediated TNFα upregulation in Kupffer cells. Sakashita et al. (56) reported that neutrophil elastase inhibitor, sivelestat, attenuated ventilator-induced lung injury (VILI) in mice by downregulating attenuated JNK/c-Jun/Egr-1 expression. A novel small molecule compound, ML264, has also been shown to inhibit the expression of EGR1 and EGR1 upstream and downstream proteins (11, 57). ML264, resveratrol, curcumin, inhibitor of neutrophil elastase sivelestat, inhibitor against MEK–ERK U0126, and inhibitor against ERK1/2 pathway PD98059 all act as inhibitors of the EGR1 activation pathway. These molecules target specific steps in the EGR1 upstream and downstream pathways or directly inhibit the activation of EGR1 itself. Using molecular biology techniques, EGR1 can be knocked down or knocked out, which is a common gene manipulation tool. A recent review summarizing the use of small interfering RNAs (siRNAs) for ALI/ARDS studies showed that proinflammatory cytokines and chemokines can be altered by siRNAs (58).
Potential Therapeutic Target of EGR1 in Human Lung Disease
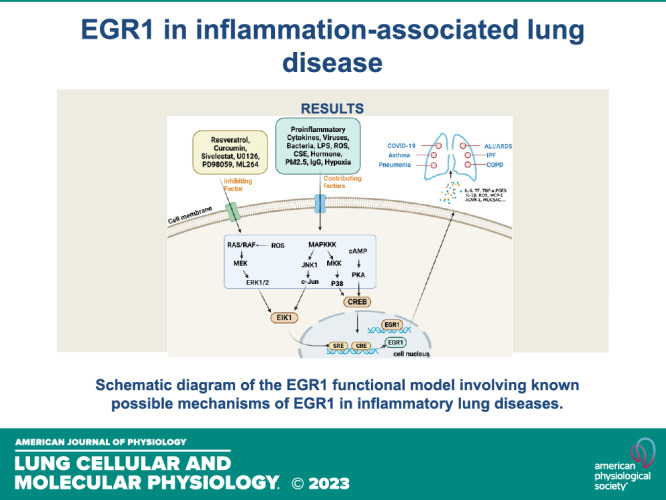
EGR1 is also a tumor suppressor gene, and studies have reported EGR1 targets in lung cancer, involving issues such as angiogenesis, signaling, proliferation, invasion, apoptosis, metastasis, EMT, and chemotherapy resistance. For example, a major component of turmeric, curcumin, provides antioxidant and anti-inflammatory properties and can modulate the EGR1 gene to inhibit tumor angiogenesis to exert antitumor effects (59). NT157 compound, a synthetic tyrosine, is a multi-target drug that increases EGR1 expression and facilitates the inhibition of signaling networks important in lung carcinogenesis and development (60). Cyantoin exerts its inhibitory effects on lung cancer through the negative regulation of α7 nicotinic acetylcholine receptors (α7-nAChR) (nicotinic and its metabolite receptors) and upregulation of EGR1 expression (61). MG624 is an α7-nAChR antagonist that inhibits angiogenesis in lung cancer through the Egr-1/FGF2 pathway (62). Cyclooxygenase inhibitor mediates TSP1 (an antiangiogenic and anti-invasive protein) through enhanced EGR1 expression to inhibit tumor cell invasion (63). The herbal formulation Yang Yin Jiedu induces apoptosis in lung cancer through EGR1 activation (64). By inhibiting phosphatidylinositol-dependent protein kinase 1 expression through Egr-1, selegiline represents a new approach to inhibit lung cancer cell growth (65). Egr1 has a facilitative effect on Oct4 overexpression in metastasis of lung cancer (66). In combination with EZH1 and LSD2, the pseudogene DUXAP8 may promote proliferation and invasion of non-small cell lung cancer (NSCLC) cells, which may provide a new therapeutic target for this disease (67). Mutant p53 and Egr-1 activate histone L to promote NSCLC EMT (68). EGR1 plays an important role in lung injury caused by various etiologies. Rukosaponin element has anti-inflammatory and antithrombotic effects, inhibits phosphorylation of MEK1/2 and ERK1/2, and reduces Egr-1 expression in the lung to play a role in pulmonary embolic disease. Trametinib attenuates LPS-induced ALI/ARDS by inhibiting the MEK–ERK–Egr-1 pathway (69). Carbon monoxide prevents VILI by peroxisome proliferator activated receptor-γ (PPAR-γ) and preventing early ventilation-dependent (Egr-1) upregulation (70). Carbon monoxide cannot be promoted for clinical use owing to its strong hemoglobin-binding capacity, competitive inhibition of oxygen-hemoglobin binding, and neurotoxicity. However, it has provided sufficient evidence that targeted inhibition of the RRAP-γ/EGR1 proinflammatory axis may be a new strategy for the treatment of VILI. The PPAR-γ/EGR-1–proinflammatory mediator axis plays a critical role in ALI (47). EGR1 can also exacerbate VILI by upregulating prostaglandin synthesis (71), and EGR1 contributes to the VILI process. Hemorrhagic shock induces ALI in rats, which can be attenuated by valproic acid through the regulation of p-ERK1/2–EGR-1 (72). As a key regulator of inflammatory mediator expression, EGR1 plays an important role in lung ischemia-reperfusion injury and allograft rejection during lung transplantation (73–76). Through the recognition of de novo endothelial remodeling, EGR1 may be an important therapeutic target in the treatment of pulmonary hypertension in humans (77). The mechanistic pattern of EGR1 in inflammation-associated lung diseases is shown in Fig. 4.
Figure 4.
Schematic diagram of the EGR1 functional model involving known possible mechanisms of EGR1 in inflammatory lung diseases. EGR1, early growth response 1; PKA, protein kinase A; CREB, cAMP-response element binding protein. [Image created with BioRender.com and published with permission.]
Genetics and Polymorphisms of EGR1 in Lung Disease
EGR1 is expressed in the human airways and regulates inflammation. Chan et al. (78) examined the effect of Chinese marker EGR1 single nucleotide polymorphisms on 298 Chinese asthmatic children, revealing that the association between Egr-1 polymorphisms and total plasma IgE in asthmatics is significant. Another study, which examined 214 Chinese adults with allergic rhinitis, found a 2.3-fold and 1.9-fold increased risk of plasma total IgE and atopy, respectively, in patients purely congenic for the A allele compared with G allele carriers of EGR1, revealing that EGR1 polymorphisms are associated with total IgE and atopy in adults with allergic rhinitis. An exploration of Egr-1 inhibitor pharmacogenetics may be beneficial (79). Another study reported genotypic analysis of 151 male patients who smoked with COPD and 100 male smokers without COPD. The study found that an increased risk of COPD was associated with the G allele of Egr-1 gene polymorphism [odds ratio (OR), 2.05; 95% confidence interval (CI), 1.15–3.72]; COPD risk increased with G allele polymorphism (GG or GA genotype; OR 2.56; 95% CI, 1.31–5.16). This suggests that polymorphisms in the Egr-1 gene are an important risk factor for COPD susceptibility (80).
EGR1 AND INFLAMMATION-ASSOCIATED LUNG DISEASES
EGR1 in ALI/ARDS
ARDS is a heterogeneous clinical syndrome. ARDS is a clinical diagnosis, whereas ALI is a histopathological diagnosis (81). The typical pathology of ALI is uncontrolled inflammation and alveolar inflammatory cell infiltration. In recent decades, humans have gained an understanding of the complex issues involved in the pathogenesis and repair of lung injury; however, as of now, only therapies that target VILI are effective. Morbidity and mortality in ARDS remain high, and preclinical studies have shown that many promising therapeutic candidates are ineffective in human trials. Human ARDS is likely to respond differently to treatment owing to clinical and biological heterogeneity (82). Therefore, the advancement of precision medicine in ARDS is urgent. As early as 2007, Ngiam et al. (83) summarized the role of EGR1 in ALI as a nuclear transcription factor that preferentially binds to the 5′-GCGGGGGCG-3′ sequence in the promoter and directly mediates the transcriptional elevation of downstream target genes, including, but not limited to, ICAM1, IL-1β, coagulation factor III (tissue factor), growth factors platelet-derived growth factor-α and β-polypeptides (PDGF), fibroblast growth factor-2 (FGF-2), vascular endothelial growth factor (VEGF), CD44, transforming growth factor-β (TGF-β1), fibronectin, matrix metalloproteinases, serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), and member 1 (SERPINE1). ICAM1, IL-1β, and TF are proinflammatory mediators. Increased TF also promotes fibrin deposition, and PDGF and FGF-2 mediate fibroblast proliferation, causing lung fibrotic injury. VEGF promotes vascular permeability in ALI/ARDS (84). CD44 mediates fibroblast migration and fibrin matrix invasion in ALI (85); however, the role of CD44 in lung injury is controversial. The activation of TGF-β1 exacerbates apoptosis, inflammatory response, and fibrosis and increases lung epithelial permeability (84, 86). Increased fibronectin is considered a marker of tissue injury, and matrix metalloproteinase overexpression accelerates tissue destruction and degrades extracellular matrix components (87). Elevated SERPINE1 inhibits fibrinolysis and is an important factor in thrombosis (88), and the above gene-mediated effect is a major cause of ALI. EGR1 may exacerbate lung injury through downstream target gene-mediated inflammation, thrombosis, matrix formation, pulmonary edema, fibrosis, and apoptosis. Hoetzel et al. (70) reported that VILI is governed by EGR1, which acts as a key proinflammatory regulator, and carbon monoxide produced by endogenous heme oxygenase has shown promise as an anti-inflammatory gas therapy. Carbon monoxide can protect against VILI through peroxisome proliferator-activated receptor-γ (PPAR-γ) and inhibition of Egr-1, and inhibition of PPAR-γ using GW9662 eliminates the protective effect of carbon monoxide. Faller et al. (89) further described the mechanisms and prospects of treating lung injury with carbon monoxide, which inhibits inflammation, apoptosis, and proliferation. The molecular mechanisms underlying lung protection mediated by carbon monoxide exposure are yet to be fully understood but may involve several signaling pathways, including the MAPK, NF-κB, AP-1, AKT, PPAR-γ, EGR-1, HIF-1α, Bcl-2 family members, heat shock proteins, or fibrinolytic molecules. At present, clinical trials have not been conducted owing to limitations associated with carbon monoxide safety.
Trametinib, a variant MAPK inhibitor, inhibits the production of Egr-1 and proinflammatory mediators, including IL-1β, MCP-1, and TNF-α, as well as Egr-1-dependent genes, comprising ICAM-1, TF, and prostaglandin E synthase (PTGES). Chen et al. (69) found that LPS-induced ALI was inhibited by trametinib by inhibiting the MEK–ERK–Egr1 pathway; these findings indicate that trametinib could be used clinically to treat ALI. The deposition of airway IgG immune complexes is associated with ALI. Yan et al. (47) described a negative correlation between PPARγ and Egr1 expression; during inflammation of lungs stimulated by IgG immune complexes, as a result of PPAR activation, EGR-1 expression was suppressed, attenuating the inflammatory response. These findings suggest that regulation of the PPARγ–Egr1 proinflammatory mediator axis may be a novel strategy for blocking ALI. Akhter et al. (90) identified that S1PR1 mediates Egr1-dependent ERK expression during ALI, and that by binding to and activating the promoter of sphingosine kinase 1 (S1P), EGR1 promotes S1P production, vascular repair, and endothelial barrier restoration, which are essential for reconstitution of the endothelial barrier. Dong et al. (91) compared the potential mechanisms and key gene changes of ALI induced by mechanical ventilation, LPS, and LPS + mechanical ventilation in three groups of conditions by bioinformatics methods and found that EGR1 and ATF3 expression was elevated in each group compared with the control group. Sakashita et al. (56) reported that neutrophil elastase inhibitor (sivelestat) attenuated VILI in mice and that mechanical ventilation increased JNK/c-Jun and Egr-1 mRNA expression. These changes could be reversed by sivelestat, which completely inhibited the ventilation-increased neutrophil elastase and myeloperoxidase activity and the histopathological extent of lung injury, neutrophil infiltration, and lung water content. Furthermore, MIP-2, IL-6, and TNF-α concentrations were attenuated. Together, the evidence suggests that inhibition of EGR1 may be an effective treatment for reducing lung injury, but the specific functions and mechanisms of EGR1 in ALI/ARDS require further investigation.
EGR1 in COPD
COPD is an irreversible lung disease that progresses over time and is characterized by inflammation of the airways, narrowing of the airways, and loss of elasticity of the airways, which result in chronic restriction of airflow. Ning et al. (92) revealed that the expression of EGR1 protein is induced by cigarette smoke extracts in alveolar epithelial, airway epithelial, and stromal cells of smokers. Similarly, through bioinformatics analysis, Huang et al. (93) identified 14 hub genes positively or negatively associated with COPD. These included the EGR1 gene, which is involved in mediating lung inflammation injury and plays a key role in the pathogenesis of COPD. Reynolds et al. (94) reported that EGR1 is induced by cigarette smoke and has a proinflammatory function, which may help to understand the development of COPD in the lungs of smokers. Smoking is a major risk factor for COPD, and imbalance between proteases and antiproteases is thought to contribute to the pathogenesis of emphysema. Elevated matrix metalloproteinase-2 (MMP-2) in the lung is one of the pathogenic mechanisms of COPD protease/antiprotease imbalance.
Zhang et al. (95) found that EGR1 is persistently upregulated in advanced emphysema and that EGR1 has the ability to regulate genes associated with the pathophysiology of emphysema, extracellular matrix formation and remodeling, and thrombosis and genes encoding cytokines, chemokines, and growth factors, providing new insights into the pathogenesis of this irreversible lung disease. Ning et al. (96) reported that chemical inhibitors of ERK1/2 MAPK, but not p38 and JNK, reduced fibroblast MMP-2 activity induced by cigarette smoke extract. Cigarette smoke extract-induced MMP-2 and membrane type 1 matrix in lung fibroblasts metalloproteinase (MT1–MMP) expression were EGR-1 dependent, revealing a molecular mechanism of matrix remodeling in cigarette smoke-associated emphysema. Wu et al. (97) reported that the ROS/MAPK (ERK-1/2)/EGR1 axis stimulates placental growth factor (PIGF) release from human bronchial epithelial cells after exposure to cigarette smoke extracts, and that PlGF is aberrantly expressed in COPD.
Apoptosis of airway epithelial cells and emphysema progression are believed to be caused by its downstream signaling molecules. EGR1 mediates the lung inflammatory response leading to the development of COPD in smokers’ lungs; however, it is unknown which genes directly downstream of EGR-1 regulate this process. Ning et al. (98) identified a novel target gene of EGR-1, geranylgeranyl diphosphate synthase (GGPPS), which can regulate proinflammatory function by activating MAPK signaling, revealing a new ERK/EGR-1/GGPPS/ERK-positive feedback loop that exacerbates cigarette smoke-induced lung inflammation, increasing our understanding of the mechanism underlying cigarette smoke-induced COPD. Chen et al. (99) reported that, in vitro and in vivo, Egr-1 promotes autophagy and apoptosis upon exposure to cigarette smoke. Li et al. (100) found that the MAPK/EGR1/HSP70 pathway regulates cigarette smoke-induced inflammatory processes. Wang et al. (101) reported that in human bronchial epithelial cells, EGR1 is essential for MUC5AC expression, which is a key molecule regulating mucus production in the airway epithelium. Hattori et al. (102) reported that aryl hydrocarbons (e.g., amino anthracene) induce EGR1 expression via the aryl hydrocarbon receptor (AHR), leading to apoptosis, which can be prevented by using antagonists of AHR. Reynolds et al. (103) showed that Egr-1 regulates the expression of the receptor for advanced glycation end-products (RAGE) during development in lung epithelial cells during cigarette smoke exposure and that a positive feedback mechanism between Egr-1 and RAGE may be involved. An important function of autophagy is to maintain cellular homeostasis and control inflammatory immune responses; a study (80) of men with COPD showed that Egr-1 gene polymorphisms and autophagy-related genes were associated with higher rates of developing the disease (OR, 2.05; 95% CI, 1.15–3.72). Use of β-linked protein inhibitor FH535 increases CSC-induced cytotoxicity and inhibits β-linked protein expression, also blocking the expression of EGR-1 target genes, fibronectin, and phosphatase; EGR-1 nuclear accumulation is not affected (104). Role of EGR1 in COPD is reported in the literature as demonstrated in Table 1.
Table 1.
Involvement of EGR1 in COPD (see text for more detail)
| Disease or Model of Disease | Interactions/Mechanism | References |
|---|---|---|
| Cigarette smoke induces human bronchial epithelium (16-HBE) | Cigarette smoke extract induces PIGF release from human bronchial epithelial cells via the ROS/MAPK (ERK-1/2)/EGR1 axis; EGR1 binds to the PIGF promoter, promoting high PlGF expression to further mediate apoptosis and exacerbate emphysema. | Wu et al. (97) |
| Cigarette smoke stimulates C57BL/6 mice and BEAS-2B cells | GGPPS is a target gene of EGR1 and MAPK ERK/EGR1/GGPPS/MAPK form a positive feedback loop that exacerbates cigarette smoke-induced lung inflammation | Shen et al. (98) |
| Cigarette smoke extract (CSE) treatment of lung fibroblasts | Emphysema caused by cigarette smoke induces the expression of MMP-2 and MT1-MMP via the EGR1 signaling pathway | Ning et al. (96) |
| Cigarette smoke induced HBE and BEAS-2B in patients with COPD | In vitro and in vivo exposure of lung epithelial cells to cigarette smoke EGR1 mediates E2F to promote autophagy and apoptosis | Chen et al. (99) |
| Cigarette smoke induced NL9 cells | An inflammatory factor and chemokine pathway is activated by tobacco smoke by MAPK–EGR1–HSP70 | Li et al. (100) |
| CSE-stimulated lung epithelial cells A549 and mouse lung | High expression of CSE proinflammatory EGR1 further mediates elevated expression of inflammatory factors IL-1β and TNF-α, exacerbating airway inflammation | Reynolds et al. (94) |
| CSE stimulates HBE | CSE promotes the interaction of EGR1 and AP-1 to further mediate the expression of the target gene MUC5AC, a key molecule regulating mucus production in the airway epithelium | Wang et al. (101) |
| Lung tissue from patients undergoing lung surgery for advanced emphysema | EGR1 is persistently upregulated in advanced emphysema and regulates genes associated with the pathophysiological progression of emphysema | Zhang et al. (95) |
| Bioinformatics analysis | Ning et al. (92); Huang et al. (93) | |
| CSE in epithelial (rat ATI cell lines R3/1 and A549) and macrophage (RAW264.7) cell lines | Positive feedback mechanism between Egr-1 and RAGE promotes sustained high expression of RAGE during cigarette smoke exposure | Reynolds et al. (103) |
| One hundred fifty-one male smoking patients with COPD and 100 male smoking controls | Having the G allele of the Egr-1 gene polymorphism increases the risk of COPD | Chen et al. (80) |
| Cigarette smoke condensate (CSC) induces BEAS-2B | FH535 increases CSC-induced cytotoxicity and inhibits EGR1 expression, but does not affect EGR-1 nuclear accumulation | Polk (104) |
| Cigarette smoke stimulation BEAS-2B | CS induces EGR1 expression via AHR, leading to apoptosis | Hattori et al. (102) |
AHR, aryl hydrocarbon receptor; BEAS-2B, bronchial epithelium transformed with Ad12-SV40 2B; COPD, chronic obstructive pulmonary disease; EGR1, early growth response 1; ERK, extracellular signal-regulated kinase; GGPPS, geranylgeranyl diphosphate synthase; HBE, human bronchial epithelium; IL-1β, interleukin-1 beta; MAPK, mitogen-activated protein kinase; MMP-2, matrix metalloproteinase-2, MT1-MMP, membrane type 1 matrix metalloproteinase; PIGF, placental growth factor; TNF-α, tumor necrosis factor-alpha.
Role of EGR1 in Asthma
As a chronic inflammatory disease of the airways, asthma causes coughing, wheezing, shortness of breath, and chest tightness. It can also trigger processes such as mucus production, airway wall remodeling, and bronchial hyperresponsiveness (105). Asthma can occur at all ages, especially in children, and atopy, low lung function, and respiratory infections are the main risk factors for persistent asthma (105). Hamed et al. (106) reported that formoterol treatment of all epithelial cell models reduced cAMP-dependent protein kinase-mediated phosphorylation of basal ERK1/2, and formoterol also inhibited basal expression of EGR1, which can control cell growth and repair in the airways.
Asthma is characterized by airway remodeling and allergic inflammation, but their interrelationships are unclear. Wang et al. (107) explored the effect of increased smooth muscle layer thickness and allergy on airway hyperresponsiveness using TGF-α and conditionally induced Egr-1-knockout mice. The results showed a positive correlation between smooth muscle layer thickness and airway responsiveness, with increased airway responsiveness during allergy. Similarly, using transforming growth factor (TGF)-alpha (TGF-α) lung-specific induction in Egr-1-knockout mice [TGF-α Tg/Egr-1 (ko/ko)], Kramer et al. (108) showed that rapamycin reduced airway remodeling and airway hyperresponsiveness, demonstrating mTOR signaling in TGF-α-induced/EGFR-mediated reactive airway disease in mice.
TGF-α Tg/Egr-1 (ko/ko) mice are one of the models used in asthma studies, with the first report appearing in 2009 (109). Using TGF-α-transgenic mice crossed to Egr-1-knockout mice to study the role of Egr-1 in TGF-α-induced lung disease, it was found that a lack of Egr-1 significantly increased the severity of TGF-α-induced lung disease, increased pulmonary fibrosis, and led to increased airway hyperresponsiveness to acetylcholine, smooth muscle hyperplasia, and no increase in lung inflammation. These changes make it an ideal model for the study of airway remodeling diseases, including pulmonary fibrosis.
Role of EGR1 in Pneumonia
Pneumonia is an acute respiratory infection that affects the alveoli and distal airways in all age groups. It is a major health problem associated with high mortality (110). As an opportunistic pathogen, Pseudomonas aeruginosa causes hospital-acquired pneumonia. Zheng et al. (111) reported that P. aeruginosa infection rapidly and transiently induces Egr-1 expression, and in acute P. aeruginosa lung infection, Egr-1 promotes systemic inflammation and negatively regulates nitric oxide production, which contributes to bacterial clearance. The same authors subsequently reported that macrophages overexpressing Egr-1 were inhibited in their phagocytic activity, and the autophagy activator rapamycin reversed the enhanced activation of transcription factor NRF2 displayed by macrophages knocked down for Egr-1. As a result, Egr-1 inhibits macrophage phagocytosis of P. aeruginosa via autophagy and NRF2 signaling (112). Wang et al. (113) reported that the anti-inflammatory effects of ATF3 were reduced by the overexpression of Egr-1. The effects of ATF3 on Mycoplasma pneumonia-induced expression, release, and airway inflammation were associated with its negative regulation of the Egr-1/Fyn signaling pathway. Bea et al. (114) found that through TLR4 signaling and activation of the MEK–ERK1/2 pathway, Egr-1 induced macrophage TF expression, even increasing the occurrence of acute coronary events.
Role of EGR1 in COVID-19
COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is currently a global health threat. Yuan et al. (115) reported that four different coronaviruses activated the EGR family, including gamma coronavirus infectious bronchitis virus, alpha coronavirus porcine epidemic diarrhea virus, human coronavirus-229E, and beta coronavirus. In addition, they found that knockdown of EGR1 decreased c-JUN and c-FOS expression, and knockdown of c-JUN and/or c-FOS decreased EGR1 expression. These results suggest that two early response gene families may be cross-activated and reciprocally regulated, and that upregulation of EGR1 regulates viral replication, apoptosis, and antiviral responses in different ways. Alexander et al. (116) reported on SARS-CoV-2 infection in human Calu-3 cells and identified posttranscriptional regulation and several transcription factors (ATF3, JUN, ZBTB20, HIVEP2, and EGR1) as key factors in infection. In a recent study using gene expression profiles of bronchoalveolar lavage fluid, a bioinformatic approach was used to investigate the underlying mechanisms in patients with COVID-19 and asthma. Five genes, EEF1A1, EGR1, UBA52, DDX5, and IRF8, were identified as key common host factors for COVID-19 and asthma, and several drugs with therapeutic potential were predicted (117). Another bioinformatics analysis (118) identified 52 pivotal genes, including EGR1, associated with infection in lung samples from patients infected with SARS-CoV-2. Hoque et al. (119) applied machine learning and bioinformatics analysis to reveal differential gene expression profiles of potential biomarkers and pharmacological compounds targeting SARS-CoV-2, and differentially expressed gene transcription factor interactions in patients with COVID-19. According to network analysis, E2F1, MAX, EGR1, YY1, and SRF are highly expressed transcription factors that may be therapeutic targets for COVID-19.
CONCLUSIONS
The signaling pathways involved in EGR1 play a role in promoting early disease development in several inflammatory respiratory diseases, including ALI/ARDS, COPD, asthma, pneumonia, and COVID-19. EGR1 may be an effective therapeutic target for drug development for the treatment of lung diseases caused by inflammatory responses, for which the summary in this paper provides an underlying theoretical basis.
GRANTS
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81760351 and 82060361). The design of the study and writing of the manuscript were in accordance with the rules of the funding.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.Z. prepared figures; Z.Z. drafted manuscript; Z.Z. edited and revised manuscript; Z.Z. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Editage (www.editage.cn) for English language editing. The graphical abstract was created with BioRender.com.
REFERENCES
- 1. Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science 238: 797–799, 1987. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 2. Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol 3: 1124–1128, 2001. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 3. Nair P, Muthukkumar S, Sells SF, Han SS, Sukhatme VP, Rangnekar VM. Early growth response-1-dependent apoptosis is mediated by p53. J Biol Chem 272: 20131–20138, 1997. doi: 10.1074/jbc.272.32.20131. [DOI] [PubMed] [Google Scholar]
- 4. Li TT, Liu MR, Pei DS. Friend or foe, the role of EGR-1 in cancer. Med Oncol 37: 7, 2019. doi: 10.1007/s12032-019-1333-6. [DOI] [PubMed] [Google Scholar]
- 5. Li L, Ameri AH, Wang S, Jansson KH, Casey OM, Yang Q, Beshiri ML, Fang L, Lake RG, Agarwal S, Alilin AN, Xu W, Yin J, Kelly K. EGR1 regulates angiogenic and osteoclastogenic factors in prostate cancer and promotes metastasis. Oncogene 38: 6241–6255, 2019. doi: 10.1038/s41388-019-0873-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang B, Guo H, Yu H, Chen Y, Xu H, Zhao G. The role of the transcription factor EGR1 in cancer. Front Oncol 11: 642547, 2021. doi: 10.3389/fonc.2021.642547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fasolo F, Jin H, Winski G, Chernogubova E, Pauli J, Winter H, Li DY, Glukha N, Bauer S, Metschl S, Wu Z, Koschinsky ML, Reilly M, Pelisek J, Kempf W, Eckstein HH, Soehnlein O, Matic L, Hedin U, Bäcklund A, Bergmark C, Paloschi V, Maegdefessel L. Long noncoding RNA MIAT controls advanced atherosclerotic lesion formation and plaque destabilization. Circulation 144: 1567–1583, 2021. doi: 10.1161/CIRCULATIONAHA.120.052023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khachigian LM. Early growth response-1, an integrative sensor in cardiovascular and inflammatory disease. J Am Heart Assoc 10: e023539, 2021. doi: 10.1161/JAHA.121.023539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fan K, Huang W, Qi H, Song C, He C, Liu Y, Zhang Q, Wang L, Sun H. The Egr-1/miR-15a-5p/GPX4 axis regulates ferroptosis in acute myocardial infarction. Eur J Pharmacol 909: 174403, 2021. doi: 10.1016/j.ejphar.2021.174403. [DOI] [PubMed] [Google Scholar]
- 10. Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI, Lotz MK. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage 26: 1531–1538, 2018. doi: 10.1016/j.joca.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun X, Huang H, Pan X, Li S, Xie Z, Ma Y, Hu B, Wang J, Chen Z, Shi P. EGR1 promotes the cartilage degeneration and hypertrophy by activating the Kruppel-like factor 5 and β-catenin signaling. Biochim Biophys Acta Mol Basis Dis 1865: 2490–2503, 2019. doi: 10.1016/j.bbadis.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 12. Dai X, Liao R, Liu C, Liu S, Huang H, Liu J, Jin T, Guo H, Zheng Z, Xia M, Ling W, Xiao Y. Epigenetic regulation of TXNIP-mediated oxidative stress and NLRP3 inflammasome activation contributes to SAHH inhibition-aggravated diabetic nephropathy. Redox Biol 45: 102033, 2021. doi: 10.1016/j.redox.2021.102033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trizzino M, Zucco A, Deliard S, Wang F, Barbieri E, Veglia F, Gabrilovich D, Gardini A. EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci Adv 7: eaaz8836, 2021. doi: 10.1126/sciadv.aaz8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J, Zhu X, Zhang M, Zhang Y, Ye S, Leng Y, Yang T, Kong L, Zhang H. Limb expression 1-like (LIX1L) protein promotes cholestatic liver injury by regulating bile acid metabolism. J Hepatol 75: 400–413, 2021. doi: 10.1016/j.jhep.2021.02.035. [DOI] [PubMed] [Google Scholar]
- 15. Sosthenes MCK, Diniz DG, Roodselaar J, Abadie-Guedes R, de Siqueira Mendes FCC, Fernandes TN, Bittencourt JC, Diniz CWP, Anthony DC, Guedes RCA. Stereological analysis of early gene expression using Egr-1 immunolabeling after spreading depression in the rat somatosensory cortex. Front Neurosci 13: 1020, 2019. doi: 10.3389/fnins.2019.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu Q, Zhang Q, Gu M, Zhang K, Xia T, Zhang S, Chen W, Yin H, Yao H, Fan Y, Pan S, Xie H, Liu H, Cheng T, Zhang P, Zhang T, You B, You Y. MIR106A-5p upregulation suppresses autophagy and accelerates malignant phenotype in nasopharyngeal carcinoma. Autophagy 17: 1667–1683, 2021. doi: 10.1080/15548627.2020.1781368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding Y, Chen X, Liu C, Ge W, Wang Q, Hao X, Wang M, Chen Y, Zhang Q. Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J Hematol Oncol 14: 19, 2021. doi: 10.1186/s13045-020-01016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banerji R, Saroj SD. Early growth response 1 (EGR1) activation in initial stages of host-pathogen interactions. Mol Biol Rep 48: 2935–2943, 2021. doi: 10.1007/s11033-021-06305-0. [DOI] [PubMed] [Google Scholar]
- 19. Woodson CM, Kehn-Hall K. Examining the role of EGR1 during viral infections. Front Microbiol 13: 1020220, 2022. doi: 10.3389/fmicb.2022.1020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim RW, Varnum BC, Herschman HR. Cloning of tetradecanoyl phorbol ester-induced 'primary response' sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene 1: 263–270, 1987. [PubMed] [Google Scholar]
- 21. Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T. and A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 53: 37–43, 1988. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 22. Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA 85: 4691–4695, 1988. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci USA 85: 7857–7861, 1988. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 28: 2304–2313, 2014. doi: 10.1101/gad.250746.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zandarashvili L, White MA, Esadze A, Iwahara J. Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett 589: 1748–1753, 2015. doi: 10.1016/j.febslet.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang L, Min L, Lin Y, Ping G, Rui W, Ying Z, Xi W, Ting H, Li L, Ke D, Jihong R, Huizhong Z. Downregulation of stathmin expression is mediated directly by Egr1 and associated with p53 activity in lung cancer cell line A549. Cell Signal 22: 166–173, 2010. doi: 10.1016/j.cellsig.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 27. Tur G, Georgieva EI, Gagete A, López-Rodas G, Rodríguez JL, Franco L. Factor binding and chromatin modification in the promoter of murine Egr1 gene upon induction. Cell Mol Life Sci 67: 4065–4077, 2010. doi: 10.1007/s00018-010-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, Goldring MB. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1β. J Biol Chem 278: 17688–17700, 2003. doi: 10.1074/jbc.M301676200. [DOI] [PubMed] [Google Scholar]
- 29. Kasneci A, Kemeny-Suss NM, Komarova SV, Chalifour LE. Egr-1 negatively regulates calsequestrin expression and calcium dynamics in ventricular cells. Cardiovasc Res 81: 695–702, 2009. doi: 10.1093/cvr/cvn357. [DOI] [PubMed] [Google Scholar]
- 30. Wang C, Dostanic S, Servant N, Chalifour LE. Egr-1 negatively regulates expression of the sodium-calcium exchanger-1 in cardiomyocytes in vitro and in vivo. Cardiovasc Res 65: 187–194, 2005. doi: 10.1016/j.cardiores.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 31. Dinkel A, Aicher WK, Haas C, Zipfel PF, Peter HH, Eibel H. Transcription factor Egr-1 activity down-regulates Fas and CD23 expression in B cells. J Immunol 159: 2678–2684, 1997. doi: 10.4049/jimmunol.159.6.2678. [DOI] [PubMed] [Google Scholar]
- 32. Havis E, Duprez D. EGR1 transcription factor is a multifaceted regulator of matrix production in tendons and other connective tissues. Int J Mol Sci 21: 1664, 2020. doi: 10.3390/ijms21051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao XM, Guy GR, Sukhatme VP, Tan YH. Regulation of the Egr-1 gene by tumor necrosis factor and interferons in primary human fibroblasts. J Biol Chem 267: 1345–1349, 1992. doi: 10.1016/S0021-9258(18)48437-2. [DOI] [PubMed] [Google Scholar]
- 34. Cao XM, Koski RA, Gashler A, McKiernan M, Morris CF, Gaffney R, Hay RV, Sukhatme VP. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol Cell Biol 10: 1931–1939, 1990. doi: 10.1128/mcb.10.5.1931-1939.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu J, Zhang SS, Saito K, Williams S, Arimura Y, Ma Y, Ke Y, Baron V, Mercola D, Feng GS, Adamson E, Mustelin T. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J 28: 21–33, 2009. doi: 10.1038/emboj.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA 92: 6873–6877, 1995. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol 16: 3545–3553, 1996. doi: 10.1128/MCB.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ehrengruber MU, Muhlebach SG, Söhrman S, Leutenegger CM, Lester HA, Davidson N. Modulation of early growth response (EGR) transcription factor-dependent gene expression by using recombinant adenovirus. Gene 258: 63–69, 2000. doi: 10.1016/s0378-1119(00)00445-5. [DOI] [PubMed] [Google Scholar]
- 39. Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem 268: 16949–16957, 1993. doi: 10.1016/S0021-9258(19)85286-9. [DOI] [PubMed] [Google Scholar]
- 40. Schwachtgen JL, Campbell CJ, Braddock M. Full promoter sequence of human early growth response factor-1 (Egr-1): demonstration of a fifth functional serum response element. DNA Seq 10: 429–432, 2000. doi: 10.3109/10425170009015615. [DOI] [PubMed] [Google Scholar]
- 41. Sakamoto KM, Bardeleben C, Yates KE, Raines MA, Golde DW, Gasson JC. 5' upstream sequence and genomic structure of the human primary response gene, EGR-1/TIS8. Oncogene 6: 867–871, 1991. [PubMed] [Google Scholar]
- 42. Sassone-Corsi P. The cyclic AMP pathway. Cold Spring Harb Perspect Biol 4: a011148, 2012. doi: 10.1101/cshperspect.a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu L, Zhou J, Zhang G, Huang W, Pei L, Lv F, Zhang Y, Zhang W, Wang H. cAMP/PKA/EGR1 signaling mediates the molecular mechanism of ethanol-induced inhibition of placental 11β-HSD2 expression. Toxicol Appl Pharmacol 352: 77–86, 2018. [Erratum in Toxicol Appl Pharmacol 436: 115866, 2022]. doi: 10.1016/j.taap.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 44. Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev 81: 807–869, 2001. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 45. Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev 92: 689–737, 2012. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 46. Gururajan M, Chui R, Karuppannan AK, Ke J, Jennings CD, Bondada S. c-Jun N-terminal kinase (JNK) is required for survival and proliferation of B-lymphoma cells. Blood 106: 1382–1391, 2005. doi: 10.1182/blood-2004-10-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan C, Chen J, Ding Y, Zhou Z, Li B, Deng C, Yuan D, Zhang Q, Wang X. The crucial role of PPARγ-Egr-1-pro-inflammatory mediators axis in IgG immune complex-induced acute lung injury. Front Immunol 12: 634889, 2021. doi: 10.3389/fimmu.2021.634889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong SH, Kim HJ, Ryu HJ, Ryu WI, Park YH, Bae HC, Jang YS, Son SW. ZnO nanoparticles induce TNF-α expression via ROS-ERK-Egr-1 pathway in human keratinocytes. J Dermatol Sci 72: 263–273, 2013. doi: 10.1016/j.jdermsci.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 49. Xu F, Cao J, Luo M, Che L, Li W, Ying S, Chen Z, Shen H. Early growth response gene 1 is essential for urban particulate matter-induced inflammation and mucus hyperproduction in airway epithelium. Toxicol Lett 294: 145–155, 2018. doi: 10.1016/j.toxlet.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 50. Nose K, Ohba M. Functional activation of the egr-1 (early growth response-1) gene by hydrogen peroxide. Biochem J 316: 381–383, 1996. doi: 10.1042/bj3160381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khachigian LM, Anderson KR, Halnon NJ, Gimbrone MA Jr, Resnick N, Collins T. Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress-response element in the PDGF A-chain promoter. Arterioscler Thromb Vasc Biol 17: 2280–2286, 1997. doi: 10.1161/01.atv.17.10.2280. [DOI] [PubMed] [Google Scholar]
- 52. Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol 193: 287–292, 2002. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 53. Dyson OF, Walker LR, Whitehouse A, Cook PP, Akula SM. Resveratrol inhibits KSHV reactivation by lowering the levels of cellular EGR-1. PLoS One 7: e33364, 2012. doi: 10.1371/journal.pone.0033364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muthusamy S, Shukla S, Amin MR, Cheng M, Orenuga T, Dudeja PK, Malakooti J. PKCδ-dependent activation of ERK1/2 leads to upregulation of the human NHE2 transcriptional activity in intestinal epithelial cell line C2BBe1. Am J Physiol Gastrointest Liver Physiol 302: G317–G325, 2012. doi: 10.1152/ajpgi.00363.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cubero FJ, Nieto N. Arachidonic acid stimulates TNFα production in Kupffer cells via a reactive oxygen species-pERK1/2-Egr1-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 303: G228–G239, 2012. doi: 10.1152/ajpgi.00465.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sakashita A, Nishimura Y, Nishiuma T, Takenaka K, Kobayashi K, Kotani Y, Yokoyama M. Neutrophil elastase inhibitor (sivelestat) attenuates subsequent ventilator-induced lung injury in mice. Eur J Pharmacol 571: 62–71, 2007. doi: 10.1016/j.ejphar.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 57. Ruiz de Sabando A, Wang C, He Y, García-Barros M, Kim J, Shroyer KR, Bannister TD, Yang VW, Bialkowska AB. ML264, a novel small-molecule compound that potently inhibits growth of colorectal cancer. Mol Cancer Ther 15: 72–83, 2016. doi: 10.1158/1535-7163.MCT-15-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zoulikha M, Xiao Q, Boafo GF, Sallam MA, Chen Z, He W. Pulmonary delivery of siRNA against acute lung injury/acute respiratory distress syndrome. Acta Pharm Sin B 12: 600–620, 2022. doi: 10.1016/j.apsb.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mehta HJ, Patel V, Sadikot RT. Curcumin and lung cancer–a review. Target Oncol 9: 295–310, 2014. doi: 10.1007/s11523-014-0321-1. [DOI] [PubMed] [Google Scholar]
- 60. de Miranda LBL, Lima K, Coelho-Silva JL, Traina F, Kobayashi SS, Machado-Neto JA. NT157 exerts antineoplastic activity by targeting JNK and AXL signaling in lung cancer cells. Sci Rep 12: 17092, 2022. doi: 10.1038/s41598-022-21419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bai S, Wen W, Hou X, Wu J, Yi L, Zhi Y, Lv Y, Tan X, Liu L, Wang P, Zhou H, Dong Y. Inhibitory effect of sinomenine on lung cancer cells via negative regulation of α7 nicotinic acetylcholine receptor. J Leukoc Biol 109: 843–852, 2021. doi: 10.1002/JLB.6MA0720-344RRR. [DOI] [PubMed] [Google Scholar]
- 62. Brown KC, Lau JK, Dom AM, Witte TR, Luo H, Crabtree CM, Shah YH, Shiflett BS, Marcelo AJ, Proper NA, Hardman WE, Egleton RD, Chen YC, Mangiarua EI, Dasgupta P. MG624, an α7-nAChR antagonist, inhibits angiogenesis via the Egr-1/FGF2 pathway. Angiogenesis 15: 99–114, 2012. [Erratum in Angiogenesis 15: 331, 2012]. doi: 10.1007/s10456-011-9246-9. [DOI] [PubMed] [Google Scholar]
- 63. Moon Y, Bottone FG Jr, McEntee MF, Eling TE. Suppression of tumor cell invasion by cyclooxygenase inhibitors is mediated by thrombospondin-1 via the early growth response gene Egr-1. Mol Cancer Ther 4: 1551–1558, 2005. doi: 10.1158/1535-7163.MCT-05-0213. [DOI] [PubMed] [Google Scholar]
- 64. Yang W, Kang Y, Zhao Q, Bi L, Jiao L, Gu Y, Lu J, Yao J, Zhou D, Sun J, Zhao X, Xu L. Herbal formula Yangyinjiedu induces lung cancer cell apoptosis via activation of early growth response 1. J Cell Mol Med 23: 6193–6202, 2019. doi: 10.1111/jcmm.14501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hann SS, Tang Q, Zheng F, Zhao S, Chen J, Wang Z. Repression of phosphoinositide-dependent protein kinase 1 expression by ciglitazone via Egr-1 represents a new approach for inhibition of lung cancer cell growth. Mol Cancer 13: 149, 2014. doi: 10.1186/1476-4598-13-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Feng YH, Su YC, Lin SF, Lin PR, Wu CL, Tung CL, Li CF, Shieh GS, Shiau AL. Oct4 upregulates osteopontin via Egr1 and is associated with poor outcome in human lung cancer. BMC Cancer 19: 791, 2019. doi: 10.1186/s12885-019-6014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei C, Li W, He X, Lu KH. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther 25: 739–751, 2017. doi: 10.1016/j.ymthe.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68. Wang W, Xiong Y, Ding X, Wang L, Zhao Y, Fei Y, Zhu Y, Shen X, Tan C, Liang Z. Cathepsin L activated by mutant p53 and Egr-1 promotes ionizing radiation-induced EMT in human NSCLC. J Exp Clin Cancer Res 38: 61, 2019. doi: 10.1186/s13046-019-1054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen S, Xu H, Ye P, Wu C, Ding X, Chen S, Zhang H, Zou Y, Zhao J, Le S, Wu J, Chen S, Xia J. Trametinib alleviates lipopolysaccharide-induced acute lung injury by inhibiting the MEK-ERK-Egr-1 pathway. Int Immunopharmacol 80: 106152, 2020. doi: 10.1016/j.intimp.2019.106152. [DOI] [PubMed] [Google Scholar]
- 70. Hoetzel A, Dolinay T, Vallbracht S, Zhang Y, Kim HP, Ifedigbo E, Alber S, Kaynar AM, Schmidt R, Ryter SW, Choi AM. Carbon monoxide protects against ventilator-induced lung injury via PPAR-γ and inhibition of Egr-1. Am J Respir Crit Care Med 177: 1223–1232, 2008. doi: 10.1164/rccm.200708-1265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ngiam N, Peltekova V, Engelberts D, Otulakowski G, Post M, Kavanagh BP. Early growth response-1 worsens ventilator-induced lung injury by up-regulating prostanoid synthesis. Am J Respir Crit Care Med 181: 947–956, 2010. doi: 10.1164/rccm.200908-1297OC. [DOI] [PubMed] [Google Scholar]
- 72. Shao L, Wu B, Liu C, Chong W. Valproic acid inhibits classical monocyte-derived tissue factor and alleviates hemorrhagic shock-induced acute lung injury in rats. Shock 59: 449–459, 2023. doi: 10.1097/SHK.0000000000002064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harada H, Lama VN, Badri LN, Ohtsuka T, Petrovic-Djergovic D, Liao H, Yoshikawa Y, Iwanaga K, Lau CL, Pinsky DJ. Early growth response gene-1 promotes airway allograft rejection. Am J Physiol Lung Cell Mol Physiol 293: L124–L130, 2007. doi: 10.1152/ajplung.00285.2006. [DOI] [PubMed] [Google Scholar]
- 74. Yoshida O, Yamane M, Yamamoto S, Okazaki M, Toyooka S, Oto T, Sano Y, Miyoshi S. Impact of prolonged cold preservation on the graft function and gene expression levels in an experimental lung transplantation model. Surg Today 43: 81–87, 2013. doi: 10.1007/s00595-012-0234-5. [DOI] [PubMed] [Google Scholar]
- 75. Wu H, Lei S, Yuan J, Liu X, Zhang D, Gu X, Zhang L, Xia Z. Ischemic postconditioning downregulates Egr-1 expression and attenuates postischemic pulmonary inflammatory cytokine release and tissue injury in rats. J Surg Res 181: 204–212, 2013. doi: 10.1016/j.jss.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 76. Waki N, Yamane M, Yamamoto S, Okazaki M, Sugimoto S, Matsukawa A, Oto T, Miyoshi S. Egr1: a novel target for ameliorating acute allograft rejection in an experimental lung transplant model. Eur J Cardiothorac Surg 41: 669–675, 2012. doi: 10.1093/ejcts/ezr030. [DOI] [PubMed] [Google Scholar]
- 77. van der Feen DE, Dickinson MG, Bartelds B, Borgdorff MA, Sietsma H, Lévy M, Berger RM. Egr-1 identifies neointimal remodeling and relates to progression in human pulmonary arterial hypertension. J Heart Lung Transplant 35: 481–490, 2016. doi: 10.1016/j.healun.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 78. Chan IH, Tang NL, Leung TF, Huang W, Lam YY, Wong GW, Chan JC, Chan MH, Wong CK, Zhang YP, Lam CW. Association of early growth response-1 gene polymorphisms with total IgE and atopy in asthmatic children. Pediatr Allergy Immunol 20: 142–150, 2009. doi: 10.1111/j.1399-3038.2008.00757.x. [DOI] [PubMed] [Google Scholar]
- 79. Chan IH, Lee DL, Ho OY, Wong EW, Lam YY, Tang NL, Chan MH, Abdullah VJ, Wong CK, Lam CW. High-level expression of early growth response-1 and association of polymorphism with total IgE and atopy in allergic rhinitis adults. Clin Chim Acta 411: 67–71, 2010. doi: 10.1016/j.cca.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 80. Chen CZ, Ou CY, Wang RH, Lee CH, Lin CC, Chang HY, Hsiue TR. Association of Egr-1 and autophagy-related gene polymorphism in men with chronic obstructive pulmonary disease. J Formos Med Assoc 114: 750–755, 2015. doi: 10.1016/j.jfma.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 81. Kulkarni HS, Lee JS, Bastarache JA, Kuebler WM, Downey GP, Albaiceta GM, et al. Update on the features and measurements of experimental acute lung injury in animals: an official American Thoracic Society Workshop Report. Am J Respir Cell Mol Biol 66: e1–e14, 2022. doi: 10.1165/rcmb.2021-0531ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beitler JR, Thompson BT, Baron RM, Bastarache JA, Denlinger LC, Esserman L, Gong MN, LaVange LM, Lewis RJ, Marshall JC, Martin TR, McAuley DF, Meyer NJ, Moss M, Reineck LA, Rubin E, Schmidt EP, Standiford TJ, Ware LB, Wong HR, Aggarwal NR, Calfee CS. Advancing precision medicine for acute respiratory distress syndrome. Lancet Respir Med 10: 107–120, 2022. doi: 10.1016/S2213-2600(21)00157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ngiam N, Post M, Kavanagh BP. Early growth response factor-1 in acute lung injury. Am J Physiol Lung Cell Mol Physiol 293: L1089–L1091, 2007. doi: 10.1152/ajplung.00265.2007. [DOI] [PubMed] [Google Scholar]
- 84. Overgaard CE, Schlingmann B, Dorsainvil White S, Ward C, Fan X, Swarnakar S, Brown LA, Guidot DM, Koval M. The relative balance of GM-CSF and TGF-β1 regulates lung epithelial barrier function. Am J Physiol Lung Cell Mol Physiol 308: L1212–L1223, 2015. doi: 10.1152/ajplung.00042.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Svee K, White J, Vaillant P, Jessurun J, Roongta U, Krumwiede M, Johnson D, Henke C. Acute lung injury fibroblast migration and invasion of a fibrin matrix is mediated by CD44. J Clin Invest 98: 1713–1727, 1996. doi: 10.1172/JCI118970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yao Y, Yuan Y, Lu Z, Ma Y, Xie Y, Wang M, Liu F, Zhu C, Lin C. Effects of Nervilia fordii extract on pulmonary fibrosis through TGF-β/Smad signaling pathway. Front Pharmacol 12: 659627, 2021. doi: 10.3389/fphar.2021.659627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ohbayashi H. Matrix metalloproteinases in lung diseases. Curr Protein Pept Sci 3: 409–421, 2002. doi: 10.2174/1389203023380549. [DOI] [PubMed] [Google Scholar]
- 88. Kellici TF, Pilka ES, Bodkin MJ. Therapeutic potential of targeting plasminogen activator inhibitor-1 in COVID-19. Trends Pharmacol Sci 42: 431–433, 2021. doi: 10.1016/j.tips.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Faller S, Hoetzel A. Carbon monoxide in acute lung injury. Curr Pharm Biotechnol 13: 777–786, 2012. doi: 10.2174/138920112800399185. [DOI] [PubMed] [Google Scholar]
- 90. Akhter MZ, Chandra Joshi J, Balaji Ragunathrao VA, Maienschein-Cline M, Proia RL, Malik AB, Mehta D. Programming to S1PR1(+) endothelial cells promotes restoration of vascular integrity. Circ Res 129: 221–236, 2021. doi: 10.1161/CIRCRESAHA.120.318412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dong WW, Feng Z, Zhang YQ, Ruan ZS, Jiang L. Potential mechanism and key genes involved in mechanical ventilation and lipopolysaccharideinduced acute lung injury. Mol Med Rep 22: 4265–4277, 2020. doi: 10.3892/mmr.2020.11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, Pinsky DJ, Watkins SC, Pilewski JM, Sciurba FC, Peters DG, Hogg JC, Choi AM. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA 101: 14895–14900, 2004. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huang X, Li Y, Guo X, Zhu Z, Kong X, Yu F, Wang Q. Identification of differentially expressed genes and signaling pathways in chronic obstructive pulmonary disease via bioinformatic analysis. FEBS Open Bio 9: 1880–1899, 2019. doi: 10.1002/2211-5463.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Reynolds PR, Cosio MG, Hoidal JR. Cigarette smoke-induced Egr-1 upregulates proinflammatory cytokines in pulmonary epithelial cells. Am J Respir Cell Mol Biol 35: 314–319, 2006. doi: 10.1165/rcmb.2005-0428OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang W, Yan SD, Zhu A, Zou YS, Williams M, Godman GC, Thomashow BM, Ginsburg ME, Stern DM, Yan SF. Expression of Egr-1 in late stage emphysema. Am J Pathol 157: 1311–1320, 2000. doi: 10.1016/S0002-9440(10)64646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ning W, Dong Y, Sun J, Li C, Matthay MA, Feghali-Bostwick CA, Choi AM. Cigarette smoke stimulates matrix metalloproteinase-2 activity via EGR-1 in human lung fibroblasts. Am J Respir Cell Mol Biol 36: 480–490, 2007. doi: 10.1165/rcmb.2006-0106OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wu D, Yuan Y, Lin Z, Lai T, Chen M, Li W, Lv Q, Yuan B, Li D, Wu B. Cigarette smoke extract induces placental growth factor release from human bronchial epithelial cells via ROS/MAPK (ERK-1/2)/Egr-1 axis. Int J Chron Obstruct Pulmon Dis 11: 3031–3042, 2016. doi: 10.2147/COPD.S120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shen N, Gong T, Wang JD, Meng FL, Qiao L, Yang RL, Xue B, Pan FY, Zhou XJ, Chen HQ, Ning W, Li CJ. Cigarette smoke-induced pulmonary inflammatory responses are mediated by EGR-1/GGPPS/MAPK signaling. Am J Pathol 178: 110–118, 2011. doi: 10.1016/j.ajpath.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, Nakahira K, Pilewski JM, Lee JS, Zhang Y, Ryter SW, Choi AM. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One 3: e3316, 2008. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li CJ, Ning W, Matthay MA, Feghali-Bostwick CA, Choi AM. MAPK pathway mediates EGR-1-HSP70-dependent cigarette smoke-induced chemokine production. Am J Physiol Lung Cell Mol Physiol 292: L1297–L1303, 2007. doi: 10.1152/ajplung.00194.2006. [DOI] [PubMed] [Google Scholar]
- 101. Wang SB, Zhang C, Xu XC, Xu F, Zhou JS, Wu YP, Cao C, Li W, Shen HH, Cao JF, Chen ZH. Early growth response factor 1 is essential for cigarette smoke-induced MUC5AC expression in human bronchial epithelial cells. Biochem Biophys Res Commun 490: 147–154, 2017. doi: 10.1016/j.bbrc.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 102. Hattori N, Nakagawa T, Yoneda M, Hayashida H, Nakagawa K, Yamamoto K, Htun MW, Shibata Y, Koji T, Ito T. Compounds in cigarette smoke induce EGR1 expression via the AHR, resulting in apoptosis and COPD. J Biochem 172: 365–376, 2022. doi: 10.1093/jb/mvac077. [DOI] [PubMed] [Google Scholar]
- 103. Reynolds PR, Kasteler SD, Cosio MG, Sturrock A, Huecksteadt T, Hoidal JR. RAGE: developmental expression and positive feedback regulation by Egr-1 during cigarette smoke exposure in pulmonary epithelial cells. Am J Physiol Lung Cell Mol Physiol 294: L1094–L1101, 2008. doi: 10.1152/ajplung.00318.2007. [DOI] [PubMed] [Google Scholar]
- 104. Polk WW. FH535 potentiation of cigarette smoke condensate cytotoxicity is associated with changes in β-catenin and EGR-1 signaling. Int J Toxicol 31: 380–389, 2012. doi: 10.1177/1091581812447956. [DOI] [PubMed] [Google Scholar]
- 105. Hammad H, Lambrecht BN. The basic immunology of asthma. Cell 184: 1469–1485, 2021. [Erratum in Cell 184: 2521-2522, 2021]. doi: 10.1016/j.cell.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 106. Hamed O, Joshi R, Michi AN, Kooi C, Giembycz MA. β 2-adrenoceptor agonists promote extracellular signal-regulated kinase 1/2 dephosphorylation in human airway epithelial cells by canonical, cAMP-driven signaling independently of β-arrestin 2. Mol Pharmacol 100: 388–405, 2021. [Erratum in Mol Pharmacol 100: 598, 2021]. doi: 10.1124/molpharm.121.000294. [DOI] [PubMed] [Google Scholar]
- 107. Wang KCW, Le Cras TD, Larcombe AN, Zosky GR, Elliot JG, James AL, Noble PB. Independent and combined effects of airway remodelling and allergy on airway responsiveness. Clin Sci (Lond) 132: 327–338, 2018. doi: 10.1042/CS20171386. [DOI] [PubMed] [Google Scholar]
- 108. Kramer EL, Hardie WD, Mushaben EM, Acciani TH, Pastura PA, Korfhagen TR, Hershey GK, Whitsett JA, Le Cras TD. Rapamycin decreases airway remodeling and hyperreactivity in a transgenic model of noninflammatory lung disease. J Appl Physiol (1985) 111: 1760–1767, 2011. doi: 10.1152/japplphysiol.00737.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kramer EL, Mushaben EM, Pastura PA, Acciani TH, Deutsch GH, Khurana Hershey GK, Korfhagen TR, Hardie WD, Whitsett JA, Le Cras TD. Early growth response-1 suppresses epidermal growth factor receptor-mediated airway hyperresponsiveness and lung remodeling in mice. Am J Respir Cell Mol Biol 41: 415–425, 2009. doi: 10.1165/rcmb.2008-0470OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Torres A, Cilloniz C, Niederman MS, Menéndez R, Chalmers JD, Wunderink RG, van der Poll T. Pneumonia. Nat Rev Dis Primers 7: 25, 2021. doi: 10.1038/s41572-021-00259-0. [DOI] [PubMed] [Google Scholar]
- 111. Pang Z, Raudonis R, McCormick C, Cheng Z. Early growth response 1 deficiency protects the host against pseudomonas aeruginosa lung infection. Infect Immun 88, 2019. doi: 10.1128/IAI.00678-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pang Z, Xu Y, Zhu Q. Early growth response 1 suppresses macrophage phagocytosis by inhibiting NRF2 activation through upregulation of autophagy during Pseudomonas aeruginosa infection. Front Cell Infect Microbiol 11: 773665, 2021. doi: 10.3389/fcimb.2021.773665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang J, Cheng W, Wang Z, Xin L, Zhang W. ATF3 inhibits the inflammation induced by Mycoplasma pneumonia in vitro and in vivo. Pediatr Pulmonol 52: 1163–1170, 2017. doi: 10.1002/ppul.23705. [DOI] [PubMed] [Google Scholar]
- 114. Bea F, Puolakkainen MH, McMillen T, Hudson FN, Mackman N, Kuo CC, Campbell LA, Rosenfeld ME. Chlamydia pneumoniae induces tissue factor expression in mouse macrophages via activation of Egr-1 and the MEK-ERK1/2 pathway. Circ Res 92: 394–401, 2003. doi: 10.1161/01.RES.0000059982.43865.75. [DOI] [PubMed] [Google Scholar]
- 115. Yuan L, Fung TS, He J, Chen RA, Liu DX. Modulation of viral replication, apoptosis and antiviral response by induction and mutual regulation of EGR and AP-1 family genes during coronavirus infection. Emerg Microbes Infect 11: 1717–1729, 2022. doi: 10.1080/22221751.2022.2093133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Alexander MR, Brice AM, Jansen van Vuren P, Rootes CL, Tribolet L, Cowled C, Bean AGD, Stewart CR. Ribosome-profiling reveals restricted post transcriptional expression of antiviral cytokines and transcription factors during SARS-CoV-2 infection. Int J Mol Sci 22:3392, 2021. doi: 10.3390/ijms22073392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Jiang Y, Yan Q, Liu CX, Peng CW, Zheng WJ, Zhuang HF, Huang HT, Liu Q, Liao HL, Zhan SF, Liu XH, Huang XF. Insights into potential mechanisms of asthma patients with COVID-19: a study based on the gene expression profiling of bronchoalveolar lavage fluid. Comput Biol Med 146: 105601, 2022. doi: 10.1016/j.compbiomed.2022.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xie TA, He ZJ, Liang C, Dong HN, Zhou J, Fan SJ, Guo XG. An integrative bioinformatics analysis for identifying hub genes associated with infection of lung samples in patients infected with SARS-CoV-2. Eur J Med Res 26: 146, 2021. doi: 10.1186/s40001-021-00609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hoque MN, Sarkar MMH, Khan MA, Hossain MA, Hasan MI, Rahman MH, Habib MA, Akter S, Banu TA, Goswami B, Jahan I, Nafisa T, Molla MMA, Soliman ME, Araf Y, Khan MS, Zheng C, Islam T. Differential gene expression profiling reveals potential biomarkers and pharmacological compounds against SARS-CoV-2: insights from machine learning and bioinformatics approaches. Front Immunol 13: 918692, 2022. doi: 10.3389/fimmu.2022.918692. [DOI] [PMC free article] [PubMed] [Google Scholar]