SUMMARY
Aging results in a decline in neural stem cells (NSCs), neurogenesis, and cognitive function and evidence is emerging to demonstrate disrupted adult neurogenesis in the hippocampus of patients with several neurodegenerative disorders. Here, single-cell RNA-sequencing of the dentate gyrus of young and old mice shows that the mitochondrial protein folding stress is prominent in activated NSCs/neural progenitors (NPCs) among the neurogenic niche, and it increases with aging accompanying dysregulated cell cycle and mitochondrial activity in activated NSCs/NPCs in the dentate gyrus. Increasing mitochondrial protein folding stress results in compromised NSC maintenance and reduced neurogenesis in the dentate gyrus, neural hyperactivity, and impaired cognitive function. Reducing mitochondrial protein folding stress in the dentate gyrus of old mice improves neurogenesis and cognitive function. These results establish the mitochondrial protein folding stress as a driver of NSC aging and suggest approaches to improve aging-associated cognitive decline.
Graphical Abstract
eTOC
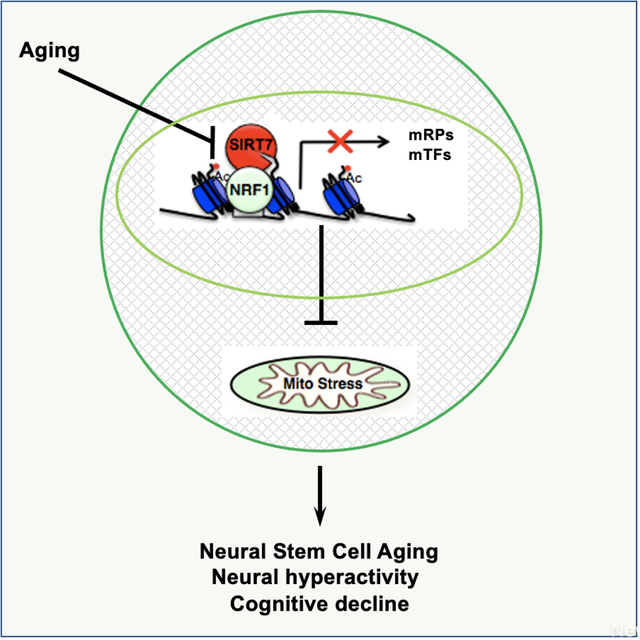
Wang et al. utilize scRNA-seq to uncover elevation of the mitochondrial unfolded protein response in activated neural stem cells with aging. The suppression of mitochondrial protein folding stress by SIRT7 ensures hippocampal neural stem cell maintenance, prevents neural hyperactivity and cognitive decline. This study reveals a mechanism of neural stem cell aging and suggests approaches to improve aging-associated cognitive decline.
INTRODUCTION
One of the most fundamental questions in biology is how we lose the ability to maintain homeostasis and become susceptible to diseases as we age. A hallmark of aging is accumulation of cellular damage1,2. In response to cellular damage, cells have evolved to launch the cellular damage responses to allow cells to return to homeostasis3,4. Failure to do so results in cell death, senescence, and at the organismal level, aging1–8. Addressing cell-specific aging-causing cellular damage is challenging, yet recent advances in single cell technologies are revolutionizing our mechanistic understanding of biological questions at the single cell level and offer an unprecedented opportunity to address this challenge9.
The dentate gyrus of the hippocampus is a brain region involved in learning, memory formation, and spatial coding. Neural stem cells (NSCs) in the subgranular zone of the dentate gyrus generate new granule cells throughout life and this neurogenesis process contributes to learning and memory formation10. Aging results in a decline in NSCs, neurogenesis, and cognitive function due to blood born factors as well as intrinsic insults11–18. Aging brains are also accompanied by neural hyperactivity in the hippocampus, which contributes to aging-associated cognitive decline as well as to the development of neurodegenerative diseases19–29. Importantly, adult hippocampal neurogenesis is disrupted in patients affected by several neurodegenerative disorders30, further motivating the mechanistic understanding of NSC aging that is critical for combating aging-associated neurodegenerative diseases.
In this study, we performed single-cell RNA-sequencing of the dentate gyrus of young and old mice and found that the mitochondrial protein folding stress is prominent in activated NSCs/neural progenitor cells (NPCs) among the neurogenic niche, and it increases with aging accompanying dysregulated cell cycle and mitochondrial activity in activated NSCs/NPCs of the dentate gyrus. Using SIRT7, an NAD+-dependent histone deacetylase that represses the mitochondrial protein folding stress, as a genetic tool combined with two-photon imaging, we show that the mitochondrial protein folding stress is a driver of hippocampal NSC aging, neural hyperactivity, and cognitive decline.
RESULTS
Aging results in increased mitochondrial and cell cycle gene expression in activated NSCs/NPCs of the dentate gyrus
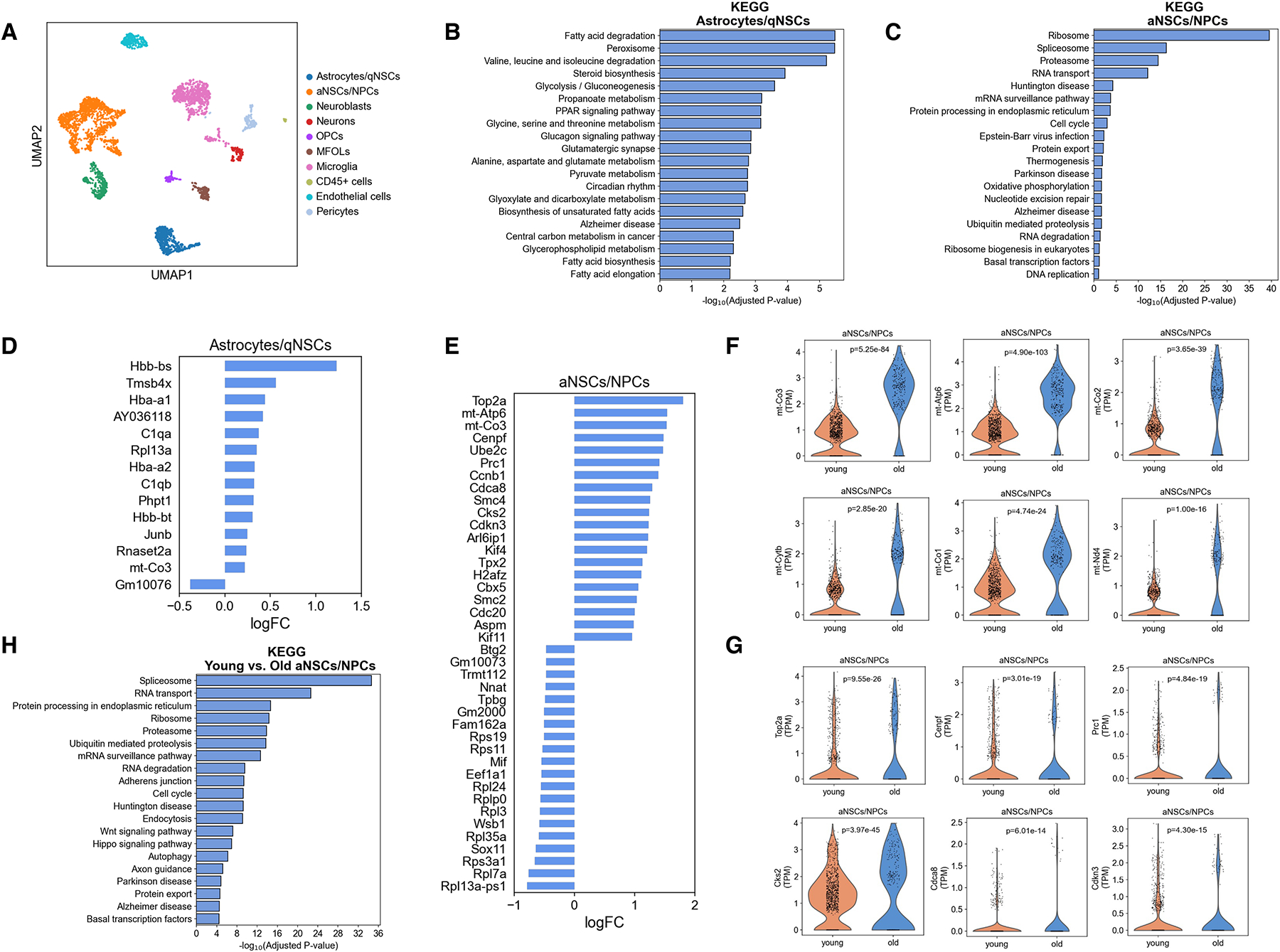
Single-cell RNA-sequencing technology enables the identification of heterogeneous cell populations in complex tissues, including the neurogenic niches12,31–36, and the measurement of cell-to-cell differential gene expression. We performed single-cell RNA-sequencing of the dentate gyrus of young (3 months old) and old (16 months old) mice using the 10x Genomics Chromium platform to uncover aging-related changes (Table S1). The analysis of single-cell transcriptomes with uniform manifold approximation and projection (UMAP) revealed 10 distinct cell types in the dentate gyrus (Figure 1A). These clusters were identified as known cell types in the dentate gyrus based on their marker genes: astrocytes/quiescent NSCs, activated NSCs/NPCs, neuroblasts, neurons, oligodendrocyte progenitor cells (OPCs), myelin-forming oligodendrocytes (MFOLs), endothelial cells, pericytes, microglia, and hematopoietic cells (Figure 1A, S1A, B).
Figure 1.
Aging results in increased mitochondrial and cell cycle gene expression in activated NSCs/NPCs of the dentate gyrus.
A, Single-cell RNA-sequencing of the dentate gyrus of young (3 months old) and old (16 months old) male mice using the 10x Genomics Chromium platform. UMAP clustering of single cell transcriptomes (2463 cells from young and 1430 cells from old) colored by cell type (qNSCs: quiescent neural stem cells, aNSCs: activated neural stem cells, NPCs: neural progenitor cells, OPCs: oligodendrocyte progenitor cells, MFOLs: myelin-forming oligodendrocytes, and CD45+ cells: hematopoietic cells). n=4 mice/age.
B, C, Pathway analysis for the biological functions of cluster-specific genes for astrocytes/quiescent NSCs (B) and activated NSCs/NPCs (C).
D, E, Top 20 upregulated and top 20 downregulated genes in astrocytes/quiescent NSCs (D) and activated NSCs/NPCs (E) with age computed using MAST. Genes were classified as significant with a false-discovery rate less than 0.05. FC: fold change.
F, Violin plots showing the expression of representative mitochondrial genes in activated NSCs/NPCs of the dentate gyrus of young (3 months old) and old (16 months old) mice. Each dot represents the gene expression levels in one cell. n=4 mice/age. Wilcoxon rank-sum test.
G, Violin plots showing the expression of representative cell cycle genes in activated NSCs/NPCs of the dentate gyrus of young (3 months old) and old (16 months old) mice. Each dot represents the gene expression levels in one cell. n=4 mice/age. P values are false discovery rate-corrected, MAST differential expression test.
H, Pathway analysis for the biological functions of differentially expressed genes in activated NSCs/NPCs in the dentate gyrus of young (3 months old) and old (16 months old) mice.
The identity of the cell clusters was further validated by pathway analysis of marker genes to reflect the established biological functions of the corresponding cell type. Consistent with previous reports31,36, astrocytes/quiescent NSCs were enriched for glucose and lipid metabolism genes while activated NSCs/NPCs were enriched for genes related to gene expression, cell cycle, translation, and oxidative phosphorylation (Figure 1B, C). Pseudo-time analysis confirmed the established cellular developmental stages and gene expression changes along the differentiation trajectories (Figure S1C, D)31,36. These results validated the quality of our single-cell RNA-sequencing data.
Aging resulted in few gene expression changes in astrocytes/quiescent NSCs (Figure 1D, Table S2), but significant changes in gene expression in activated NSCs/NPCs (Figure 1E, Table S2). The expression of 5239 genes were significantly changed comparing activated NSCs/NPCs derived from young and old mice (FDR<0.05) (Table S2). Strikingly, cell cycle genes and mitochondrial genes figured prominently among the top upregulated genes in activated NSCs/NPCs with aging (Figure 1E–G, Table S2, 3). Pathway analysis further confirmed that cell cycle genes were significantly altered in activated NSCs/NPCs with aging (Figure 1H). Together, these data suggest that uncontrolled mitochondrial activity and cell cycle may underlie aging-associated decline of activated NSCs/NPCs in the dentate gyrus.
Increased mitochondrial protein folding stress during NSC activation and aging in the dentate gyrus
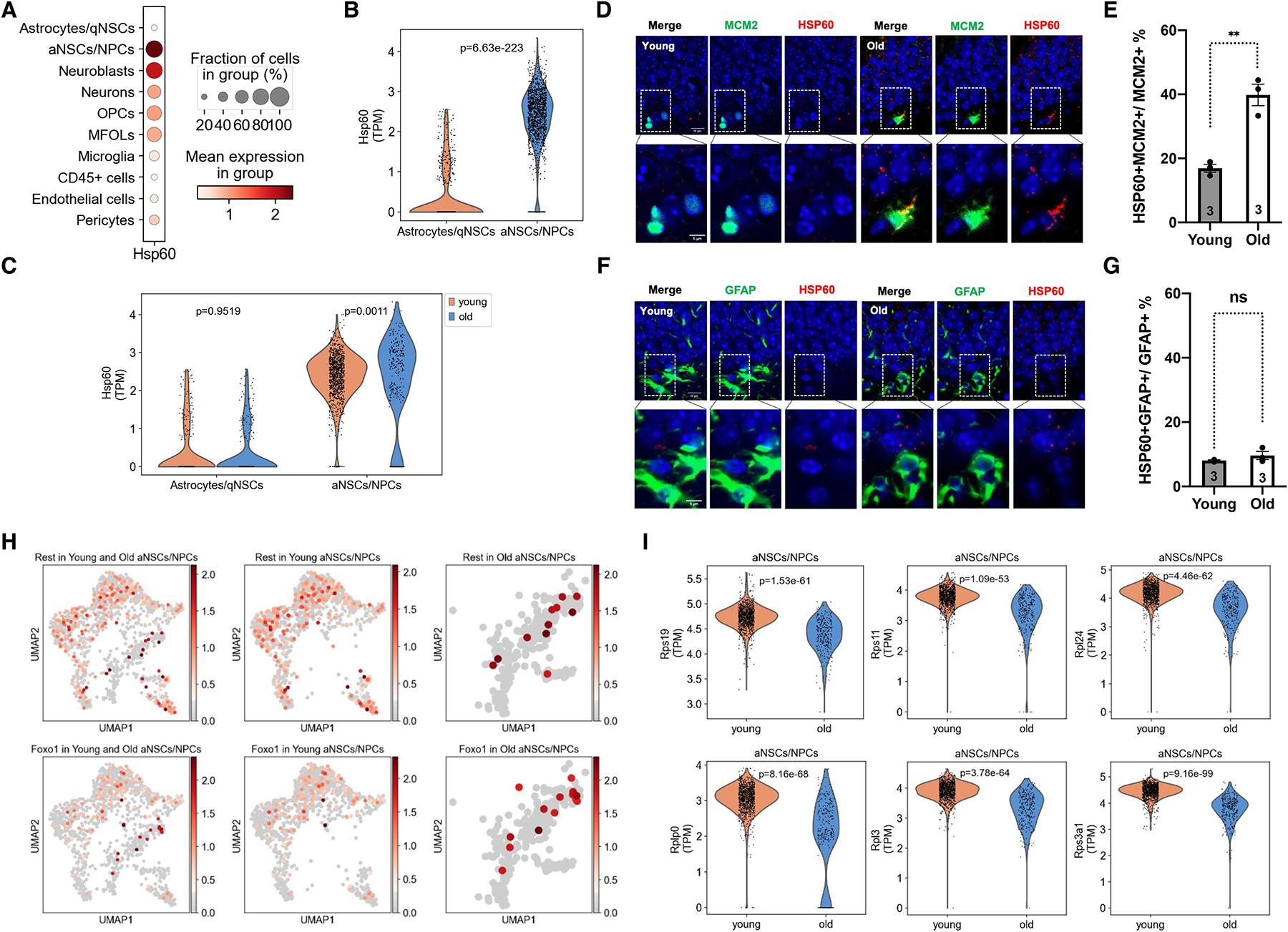
Stem cell transition from quiescence to activation is associated with mitochondrial biogenesis and metabolic reprogramming from glycolysis to mitochondrial metabolism37. Increased mitochondrial biogenesis and activity is associated with the mitochondrial protein folding stress38,39. The analysis of single-cell RNA-sequencing data for the dentate gyrus of young and old mice showed that the mitochondrial chaperone HSP60 was expressed remarkably in activated NSCs/NPCs among all cell populations in the dentate gyrus (Figure 2A). The expression of HSP60 was significantly higher in activated NSCs/NPCs than in astrocytes/quiescent NSCs (Figure 2B), consistent with increased mitochondrial activity upon NSC activation. HSP60 expression was also increased in activated NSCs/NPCs of aged mice compared to young mice (Figure 2C), in keeping with increased expression of cell cycle and mitochondrial genes (Figure 1E–G). HSP60 expression was not increased in astrocytes/quiescent NSCs of the dentate gyrus with aging (Figure 2C, Table S2).
Figure 2.
Increased mitochondrial protein folding stress during NSC activation and aging in the dentate gyrus.
A, Dot plot showing proportion of cells positive for HSP60 and log-normalized expression values of HSP60 in the cell clusters as indicated.
B, C, Violin plots comparing log-normalized expression values of HSP60 in astrocytes/quiescent NSCs and activated NSCs/NPCs of young mice (B) and astrocytes/quiescent NSCs and activated NSCs/NPCs of young and old mice (C). Each dot represents the gene expression levels in one cell. Wilcoxon rank-sum test.
D-G, Immunohistochemistry staining and quantification for HSP60 and MCM2 (D, E) and HSP60 and GFAP (F, G) in the dentate gyrus of young and old mice. Blue: DAPI. Red: HSP60. Green: MCM2 or GFAP. Scale bar: 10 μm (top). 5 μm (bottom). Error bars represent SE. **: p < 0.01. ns: p>0.05. Student’s t test.
H, UMAP plots showing the expression of Rest and Foxo1 in activated NSCs/NPCs of the dentate gyrus of young (3 months old) and old (16 months old) mice. Each dot represents the gene expression levels in one cell. n=4 mice/age.
I, Violin plots showing the differential expression of representative ribosomal proteins in activated NSCs/NPCs of the dentate gyrus of young (3 months old) and old (16 months old) mice. Each dot represents the gene expression levels in one cell. n=4 mice/age. Wilcoxon rank-sum test.
HSP60 expression in the dentate gyrus with aging was further confirmed by immunohistochemistry staining. In 2D images, the frequency of HSP60-positive activated NSCs (MCM2-positive cells) was greater than that of HSP60-positive quiescent NSCs/astrocytes (GFAP-positive cells) (Figure 2D–G, S2, S3). The frequency of HSP60-positive activated NSCs was further increased with aging, but not quiescent NSCs/astrocytes (Figure 2D–G, S2, S3). This pattern of HSP60 expression at the protein level was consistent with its expression at the mRNA level (Figure 2C). To ensure the HSP60 signal and the MCM2 signal were originated from the same cell, we performed close-up imaging and 3D reconstitution of immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of young and old mice. The HSP60 signal in close proximity (<1μm) to the MCM2 signal was increased with aging (Figure S4A–C, Video S1). Furthermore, we isolated NSCs from the dentate gyrus of young and old mice using the neurosphere self-renewal medium (Figure S5A, B). Both immunohistochemistry staining and quantitative real-time PCR showed increased expression of HSP60 in NSCs isolated from the dentate gyrus of old mice (Figure S5C, D, E). Thus, the mitochondrial protein folding stress increases in activated NSCs in the dentate gyrus with aging.
Consistent with increased mitochondrial stress in aged activated NSCs/NPCs, the stress responsive transcription factor REST and its downstream effector Foxo1a were induced to a higher level in aged activated NSCs/NPCs than their young counterparts (Figure 2H)40. The mitochondrial protein folding stress results in attenuated translation, which helps restore proteostasis41. Ribosomal proteins were among the top downregulated genes in activated NSCs/NPCs with aging, and a large number (85) of ribosomal proteins and mitochondrial ribosomal proteins were significantly downregulated (Figure 2I, Table S2, S4), in keeping with increased mitochondrial protein folding stress. Pathway analysis further confirmed ribosomal proteins as significantly changed genes in activated NSCs/NPCs during aging (Figure 1H). Together, these data suggest increased mitochondrial activity and mitochondrial protein folding stress during the hippocampal NSC activation and aging.
Subventricular zone (SVZ) also harbors NSCs that divide to produce neurons in the process of neurogenesis. The analysis of single-cell RNA-sequencing data for the SVZ of young and old mice12 showed that the mitochondrial protein folding stress was also increased during the activation of SVZ NSCs, consistent with the metabolic reprogramming (Figure S6A–C) but was not further increased during aging (Figure S6D). Thus, increased mitochondrial protein folding stress during aging is specific to hippocampal NSCs but not SVZ NSCs.
SIRT7 suppresses the mitochondrial protein folding stress and promotes NSC maintenance and cognition
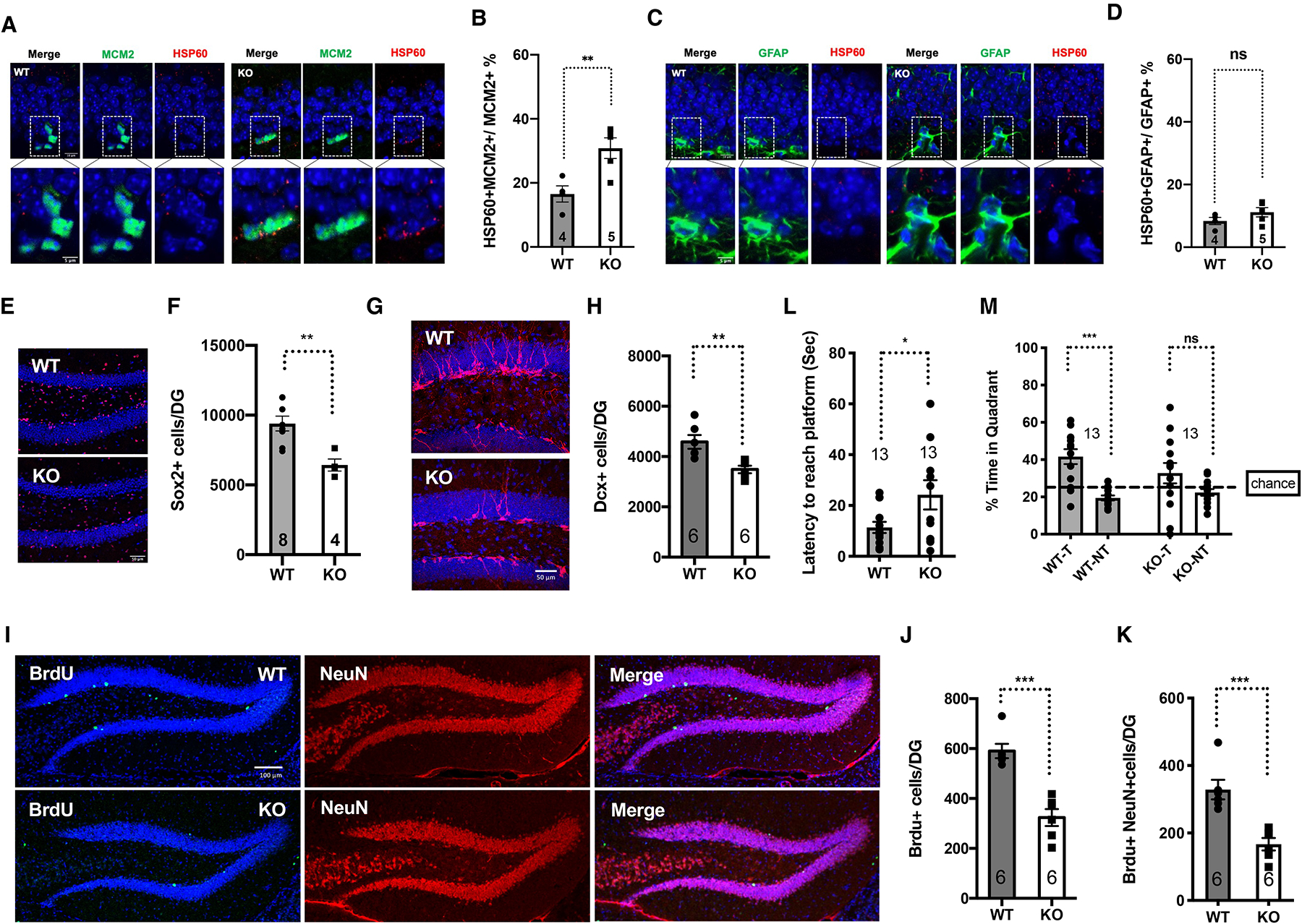
Increased mitochondrial protein folding stress in hippocampal NSCs with aging suggests that the mitochondrial protein folding stress might be a driver of aging-associated decline of NSCs. To test this, we used SIRT7, a histone deacetylase that dampens the mitochondrial protein folding stress, as a genetic tool42. SIRT7 expression was readily detectable in MCM2-positive activated NSCs in the dentate gyrus of young mice, but was diminished in old mice (Figure S7A–C), suggesting that suppressed SIRT7 expression may contribute to increased mitochondrial protein folding stress in activated NSCs with aging. We characterized 3–5-month-old SIRT7−/− mice for the molecular and cellular changes associated with the neurogenic niche in the dentate gyrus. The frequency of HSP60-positive activated NSCs but not quiescent NSCs/astrocytes in the dentate gyrus of SIRT7−/− mice was increased (Figure 3A–D, S8, S9). Close-up imaging and 3D reconstitution of immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus showed that the HSP60 signal in close proximity (<1μm) to the MCM2 signal was increased in SIRT7−/− mice (Figure S10A–C, Video S2), consistent with a role of SIRT7 in suppressing the mitochondrial protein folding stress. The numbers of Sox2-positive cells (Figure 3E, F) and BrdU-positive cells (Figure S10D, E) were both reduced in the dentate gyrus of SIRT7−/− mice. SIRT7−/− mice also exhibited reduced number of newly differentiated Doublecortin (Dcx)-positive neurons in the dentate gyrus (Figure 3G, H). BrdU labeling assay showed reduced number of long-term BrdU-retaining cells in the dentate gyrus of SIRT7−/− mice 4 weeks after BrdU injection (Figure 3I–K). Together, these data suggest that SIRT7 suppresses the mitochondrial protein folding stress and promotes NSC maintenance and neurogenesis in the dentate gyrus.
Figure 3.
SIRT7 suppresses the mitochondrial protein folding stress and promotes NSC maintenance and cognition.
Comparison of 3–5-month-old WT and SIRT7−/− mice.
A-D, Immunohistochemistry staining and quantification of HSP60 and MCM2 (A, B) and HSP60 and GFAP (C, D) in the dentate gyrus. Blue: DAPI. Red: HSP60. Green: MCM2 or GFAP. Scale bar: 10 μm (top). 5 μm (bottom).
E, F, Immunohistochemistry staining (E) and quantification (F) of Sox2-positive cells in the dentate gyrus. Blue: DAPI. Red: Sox2. Scale bar:50 μm.
G, H, Immunohistochemistry staining (G) and quantification (H) of newly differentiated Doublecortin (Dcx)-positive neurons in the dentate gyrus. Blue: DAPI. Red: Dcx. Scale bar:50 μm.
I-K, Immunohistochemistry staining (I) and quantification (J, K) of long-term BrdU-retaining cells in the dentate gyrus. Blue: DAPI. Green: BrdU. Red: NeuN. Scale bar:100 μm.
L, M, Morris Water Maze test. Data shown are latency to reach the platform (L) and percent of time spent in the target and non-target quadrants (M) during probe testing for memory. T: target quadrant. NT: non-target quadrant.
Error bars represent SE. *: p<0.05. **: p < 0.01. ***: p < 0.001. ns: p>0.05. Student’s t test.
Reduced neurogenesis leads to cognitive decline10,18. In a Morris Water Maze test, 3-month-old SIRT7−/− mice showed a clear memory deficit as demonstrated by a longer latency to reach the platform compared to the WT control mice (Figure 3L) during probe testing. Furthermore, while WT mice spent significantly more time in the target quadrant (T) than in the non-target quadrants (NT), SIRT7−/− mice did not demonstrate the preference for the target quadrant (Figure 3M). In contrast, SIRT7−/− mice showed increased total activity in the open field test (Figure S10F, G) and performed competently in the elevated plus maze test (Figure S10H–J). SIRT7−/− mice also exhibited defect in the consolidation of fear memory43. Together, these data suggest that SIRT7 deficiency does not cause defects in exploration and locomotor activity but leads to reduced cognitive function, consistent with compromised NSC maintenance and reduced neurogenesis.
Neural hyperactivity in the hippocampus of aged mice
Neural hyperactivity has been observed in the hippocampus of aged animals, as well as elderly individuals and patients with mild cognitive impairment and Alzheimer’s disease21,23,25,27–29,44. We used ultra-sensitive protein calcium sensor GCaMP6s to image neural activity in vivo45. Mice were subjected to stereotaxic injection in the dorsal dentate gyrus and CA1 with AAV1 expressing GCaMP6s. To provide optical access to the hippocampus for imaging, we locally removed overlying cortical tissue by gentle aspiration and implanted a cranial window (Figure 4A). Two-photon imaging of young (3–5 months old) and old (16–18 months old) wild type (WT) mice showed increased neural activity in both CA1 and the dentate gyrus with age (Figure 4B–F). The percentage of neurons with higher activity was increased in the dentate gyrus and CA1 of old mice compared to the same brain regions of young mice (Figure 4C, D). Wilcoxon rank-sum test showed statistically significant difference in neural activity between the corresponding brain regions of young and old mice (Figure 4E, F).
Figure 4.
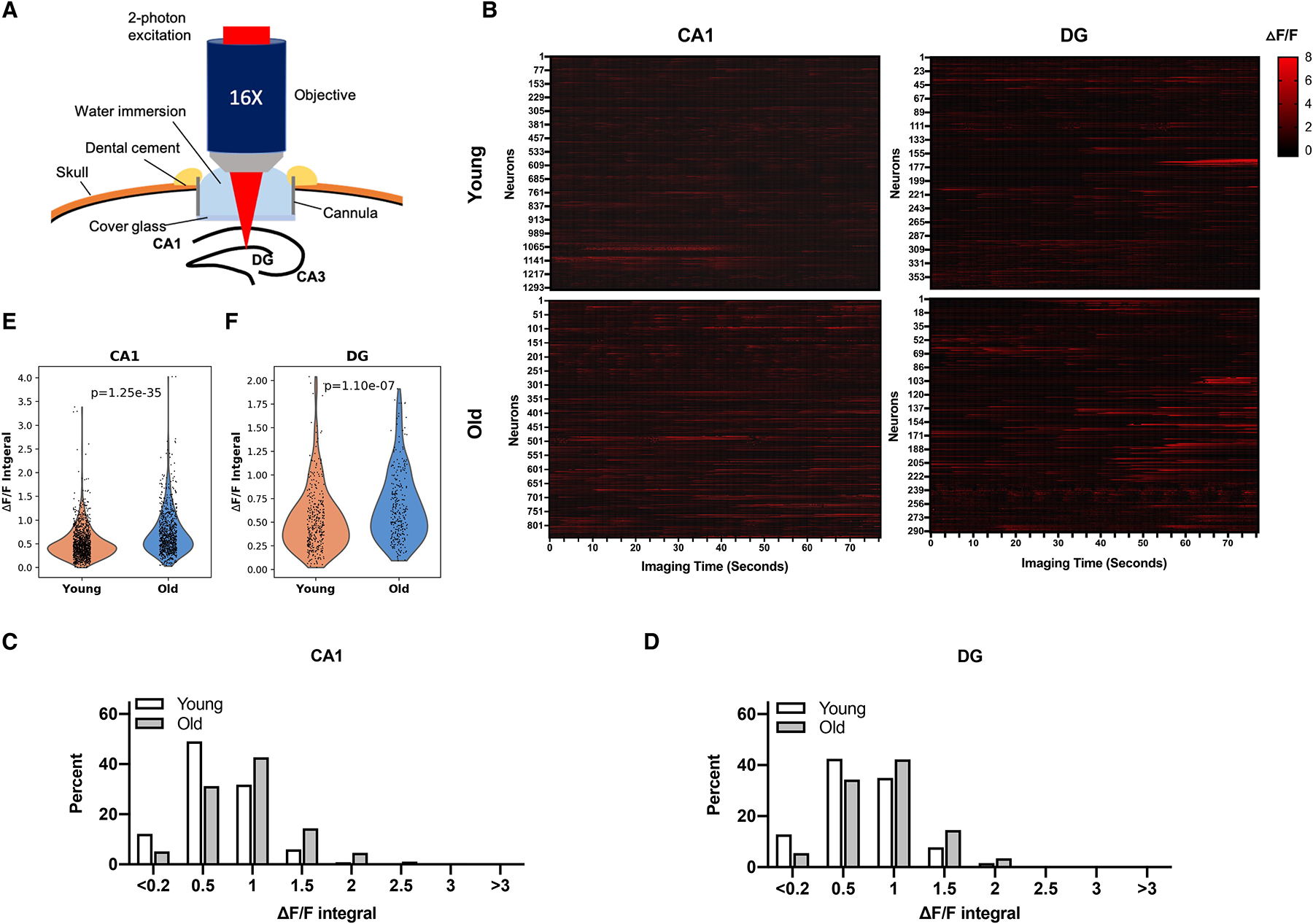
Neural hyperactivity in the hippocampus of aged mice.
A, Schematic illustration for two-photon imaging of the hippocampus.
B-F, Young (3–5 months old) and old (16–18 months old) mice were subjected to stereotaxic injection in the dorsal dentate gyrus and CA1 with AAV1 expressing GCaMP6s and installation of a cranial window, and visualized using two-photon imaging. Data shown are raster plots of ΔF/F from all identified neurons (B), histograms of neural activity (integral of ΔF/F traces) (C, D), and violin plots of neural activity (Wilcoxon rank-sum test) (E, F). CA1, n= 1301 cells of 31 imaging area of six young mice and n=840 cells of 27 imaging area of four old mice; dentate gyrus, n=374 cells of 18 imaging area of four young mice and n=290 cells of 16 imaging area of four old mice. See also Figure S11.
Neural hyperactivity results in compensatory remodeling of inhibitory hippocampal circuits24. Alterations in the expression of neuropeptide Y (NPY) are indicators of imbalances between excitatory and inhibitory neural activities in the hippocampus46. Immunohistochemistry staining showed increased levels of NPY expression in the dentate gyrus of old mice compared to young mice (Figure S11A). Quantitative real time PCR analysis revealed reduced expression of NPY-Y1 receptors, which mediate weak excitatory activity of NPY46, whereas the expression of Y2 receptors, which mediate potent inhibitory activity of NPY46, was unchanged in the dentate gyrus of old mice (Figure S11B). These data provide additional support for neural hyperactivity in the hippocampus of old mice.
SIRT7 suppresses neural hyperactivity
Adult-born neurons inhibit the dentate gyrus network activity by recruiting local interneurons and elimination of adult-born neurons leads to increased neural activity in the dentate gyrus47. Reduced neurogenesis in the dentate gyrus of SIRT7−/− mice suggests that SIRT7 deficiency might result in neural hyperactivity. Two-photon imaging of 4–6-month-old SIRT7−/− mice and their WT controls showed a statistically significant increase in neural activity in both the dentate gyrus and CA1 of SIRT7−/− mice, as evidenced by increased GCaMP6s fluorescence and ΔF/F calcium transient traces of all detected neurons (Figure 5A, B), increased percentage of neurons with hyperactivity (Figure 5C, D), and Wilcoxon rank-sum test (Figure 5E, F) of neural activity between the corresponding brain regions of the two genotypes. The expression of NPY was higher in the dentate gyrus of SIRT7−/− mice than the WT controls (Figure S12A). Together, these data suggest that SIRT7 suppresses neural hyperactivity, consistent with its role in promoting NSC maintenance and neurogenesis.
Figure 5.
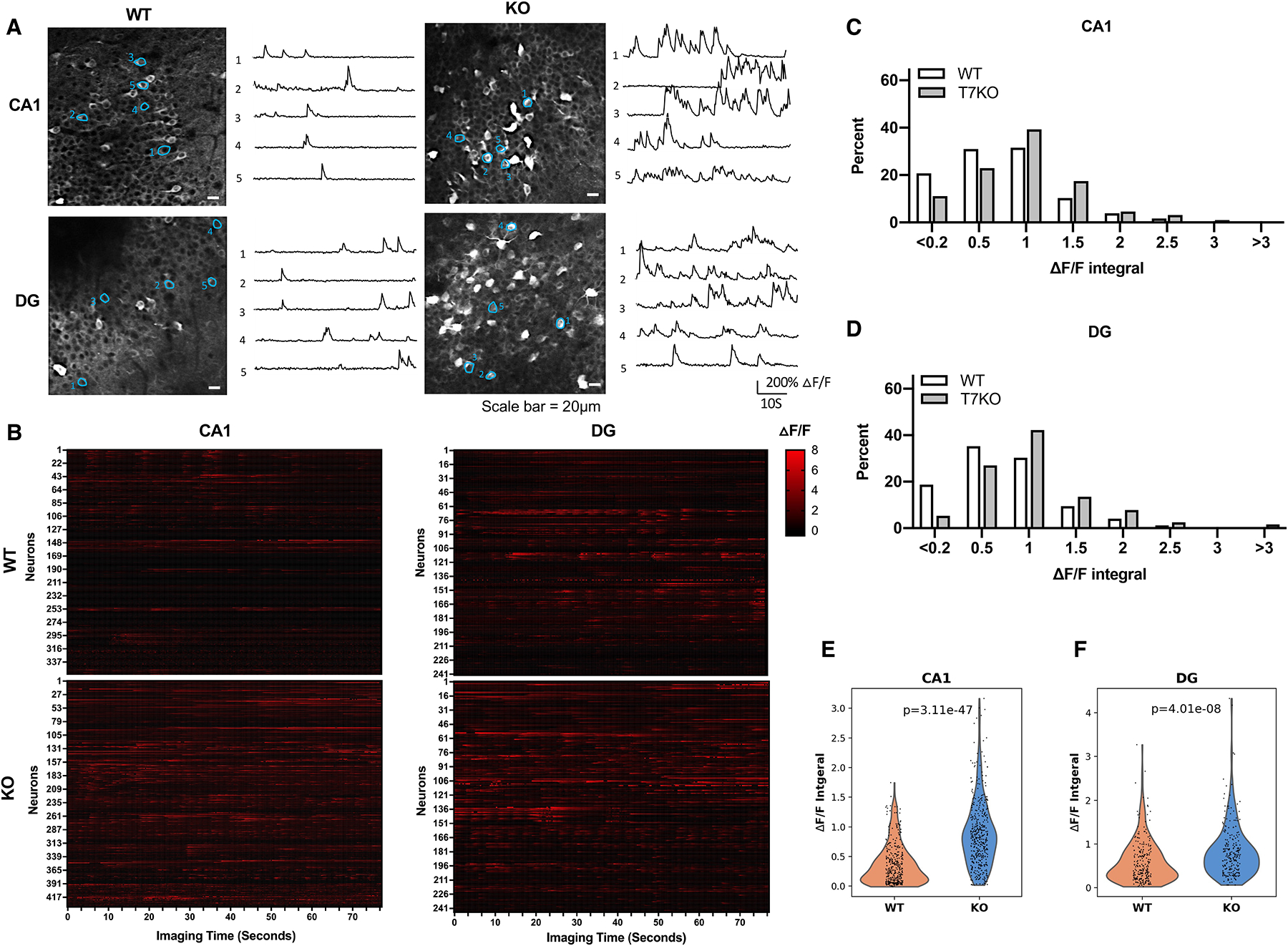
SIRT7 suppresses neural hyperactivity.
A-F, 4–6-month-old WT and SIRT7−/− mice were subjected to stereotaxic injection in the dorsal dentate gyrus and CA1 with AAV1 expressing GCaMP6s and installation of a cranial window, and visualized using two-photon imaging. Data shown are GCaMP6s fluorescence images and ΔF/F calcium transient traces of neurons (scale bar=20 μm) (A), raster plots of ΔF/F from all identified neurons (B), histogram distributions of neural activity (quantified as integral of ΔF/F traces) (C, D), and violin plots of neural activity (Wilcoxon rank-sum test) (E, F) of indicated brain regions. (CA1, n= 356 cells of 19 imaging areas of five WT mice and n=434 cells of 21 imaging areas of four SIRT7−/− mice; dentate gyrus, n=241 cells of 15 imaging areas of four WT mice and n=244 cells of 15 imaging areas of four SIRT7−/− mice). See also Figure S12.
To rule out the possibility that the difference in hippocampal neural activity was due to animal arousal level as reflected by the pupil diameter of the mouse eye, we recorded the pupil diameter while neuronal activity of a head-fixed mouse was imaged under the two-photon microscope (Figure S12B, C). We observed populations of calcium transient either positively or negatively entrained to pupil diameter (Figure S12D, E). Pearson Correlation showed no association between neural activity and pupil diameter in these mice regardless of genotype (Figure S12D, E).
SIRT7 promotes NSC maintenance by suppressing the mitochondrial protein folding stress
SIRT7 promotes NSC maintenance and neurogenesis (Figure 3E–K), correlating with reduced mitochondrial protein folding stress in activated NSCs (Figure 3A–D, S8, S9, S10A–C, Video S2). We next determined whether SIRT7 promoted NSC maintenance and neurogenesis by suppressing the mitochondrial protein folding stress. SIRT7 suppresses the mitochondrial protein folding stress by repressing the activity of the transcription factor NRF142. We infected WT and SIRT7−/− mice with control lentivirus or lentivirus knocking down NRF1, which were delivered to the dentate gyrus via stereotaxic injection, and mice were analyzed 8 weeks after viral injection. NRF1 inactivation reduced the mitochondrial protein folding stress in activated NSCs of SIRT7−/− mice, as evidenced by the immunostaining of HSP60 (Figure 6A–D, S13, S14, S15, Video S3). NRF1 inactivation also increased the number of Sox2-positive cells (Figure 6E, F), long-term BrdU-retaining cells (Figure 6G, H), and newly differentiated Dcx-positive neurons (Figure 6I, J) in the dentate gyrus of SIRT7−/− mice. Together, these data suggest that SIRT7 promotes NSC maintenance and neurogenesis by suppressing the mitochondrial protein folding stress.
Figure 6.
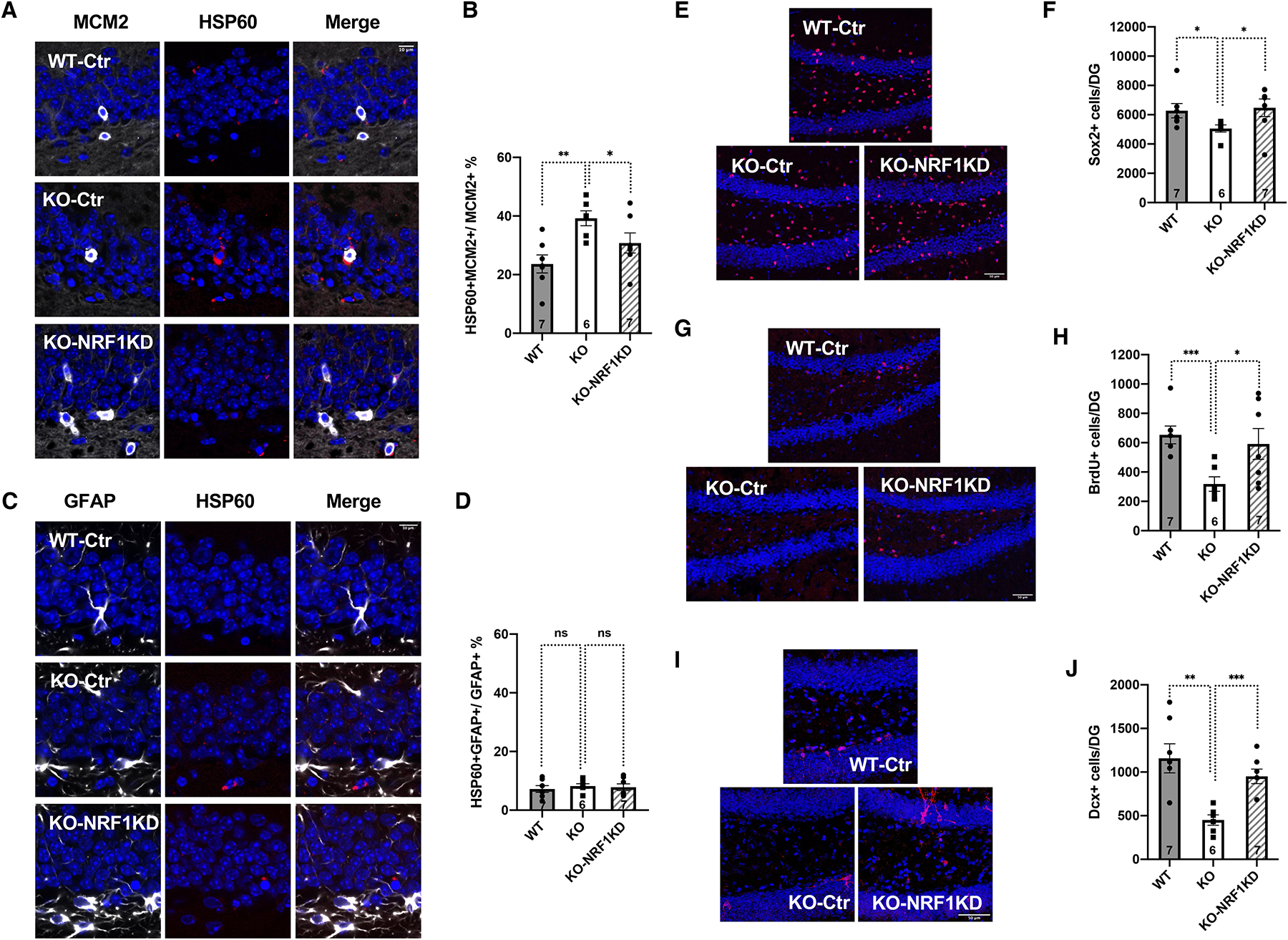
SIRT7 promotes NSC maintenance by suppressing the mitochondrial protein folding stress.
Comparison of 6–7-month-old WT and SIRT7−/− mice that were given stereotaxic injections of control lentivirus or lentivirus knocking down NRF1 into the dentate gyrus.
A-D, Immunohistochemistry staining and quantification of MCM2 and HSP60 (A, B) and GFAP and HSP60 (C, D) in the dentate gyrus. Blue: DAPI. Red: HSP60. White: MCM2 or GFAP. Scale bar: 10 μm.
E, F, Immunohistochemistry staining (E) and quantification (F) of Sox2-positive cells in the dentate gyrus. Blue: DAPI. Red: Sox2. Scale bar: 50 μm.
G, H, Immunohistochemistry staining (G) and quantification (H) of long-term BrdU-retaining cells in the dentate gyrus. Blue: DAPI. Red: BrdU. Scale bar: 50 μm.
I, J, Immunohistochemistry staining (I) and quantification (J) of newly differentiated Dcx-positive neurons in the dentate gyrus. Blue: DAPI. Red: Dcx. Scale bar: 50 μm.
Error bars represent SE. *: p<0.05. **: p < 0.01. ***: p<0.001. ns: p>0.05. Student’s t test. See also Figure S13, S14, S15, Video S3.
SIRT7 overexpression in the dentate gyrus improves neurogenesis and cognitive function of aged mice
Aging is associated with reduced number of Sox2-positive cells, newly differentiated Dcx-positive neurons, and long-term BrdU-retaining cells in the dentate gyrus18. We next determined whether overexpression of SIRT7 and reducing mitochondrial protein folding stress in the dentate gyrus of the aging brain could improve NSC maintenance, neurogenesis, and cognition. We infected 10-month-old mice with control lentivirus or lentivirus overexpressing SIRT7, which were delivered to the dentate gyrus via stereotaxic injection, and mice were analyzed 8 weeks after viral injection (Figure 7A, B). Immunohistochemistry staining showed that SIRT7 overexpression did not change the number of Sox2-positive cells in the dentate gyrus (Figure 7C, D). SIRT7 overexpression increased the number of newly differentiated Dcx-positive neurons (Figure 7E, F) and long-term BrdU-retaining cells in the dentate gyrus 4 weeks after BrdU injection (Figure 7G, H). In a Morris Water Maze test, while control aged mice did not demonstrate the preference for the target quadrant, aged mice overexpressing SIRT7 spent significantly longer time in the target quadrant than in the non-target quadrants, and crossed the platform in the target quadrant significantly more than the non-target quadrants during probe testing (Figure 7I, J), suggesting improved memory. Thus, the mitochondrial protein folding stress drives aging-associated decline of NSCs.
Figure 7.
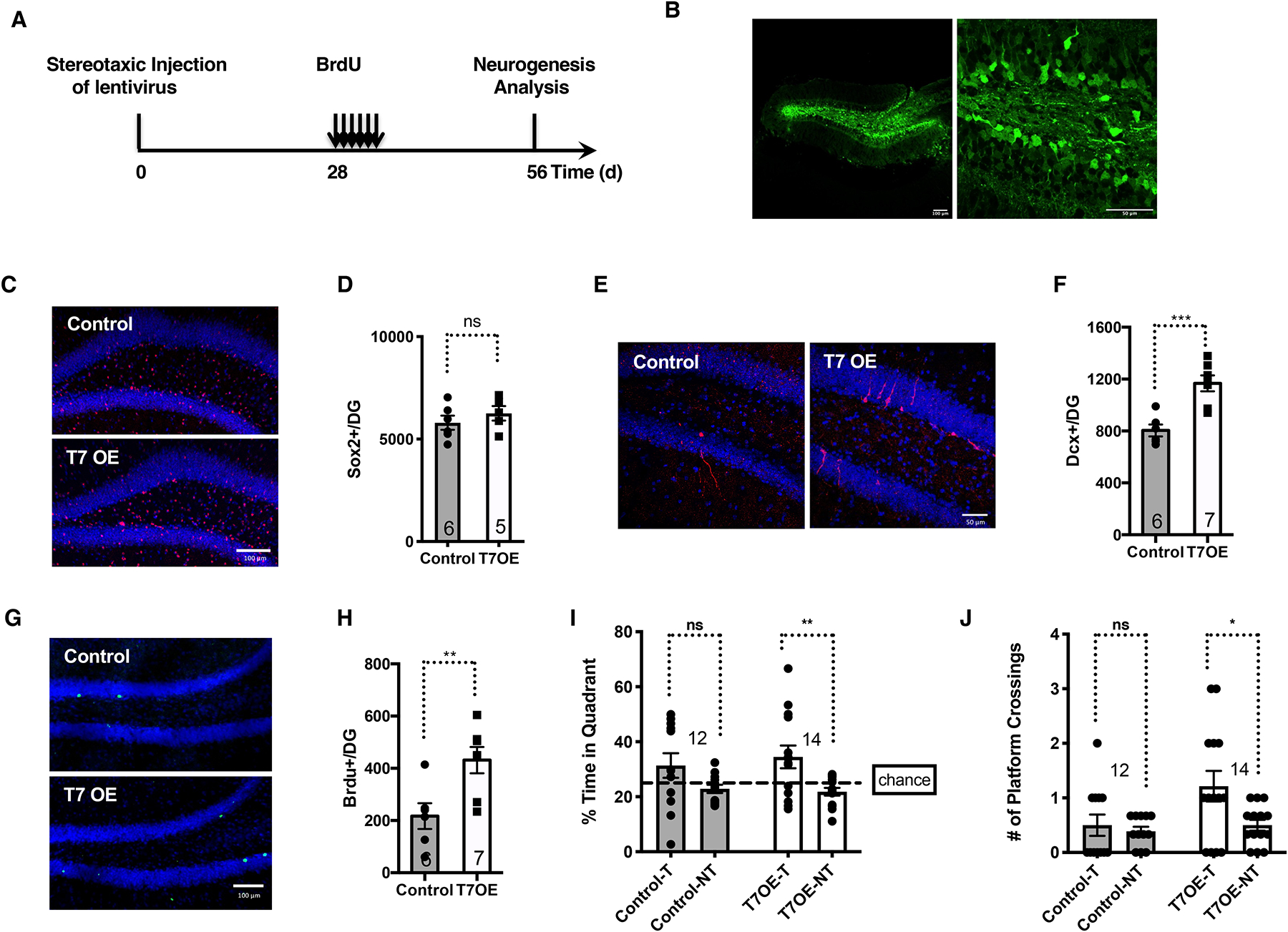
SIRT7 overexpression in the dentate gyrus improves neurogenesis and cognitive function of aged brains.
Comparison of 10-month-old mice that were given stereotaxic injections of control lentivirus or lentivirus expressing SIRT7 into the dentate gyrus.
A, Schematic illustration of the experimental timeline.
B, GFP expression in mice given stereotaxic injections of GFP lentivirus into the dentate gyrus. Scale bar: 100 μm (left) and 50 μm (right).
C, D, Immunohistochemistry staining (C) and quantification (D) of Sox2-positive cells in the dentate gyrus. Blue: DAPI. Red: Sox2. Scale bar:100 μm.
E, F, Immunohistochemistry staining (E) and quantification (F) of newly differentiated Dcx-positive neurons in the dentate gyrus. Blue: DAPI. Red: Dcx. Scale bar:50 μm. G, H, Immunohistochemistry staining (G) and quantification (H) of long-term BrdU-retaining cells in the dentate gyrus. Blue: DAPI. Green: BrdU. Scale bar:100 μm.
I, J, Morris Water Maze test. Data shown are percent of time spent in the target and non-target quadrants (I) and the number of platform crossings in the target and non-target quadrants (J) during probe testing for memory. T: target quadrant. NT: non-target quadrant.
Error bars represent SE. *: p<0.05. **: p < 0.01. ***: p < 0.001. ns: p>0.05. Student’s t test.
DISCUSSION
We have shown that the mitochondrial protein folding stress was increased during hippocampal NSC activation in accord with the metabolic reprogramming (Figure 1B, C, 2A, B). The mitochondrial protein folding stress was further increased in hippocampal activated NSCs/NPCs but not quiescent NSCs/astrocytes of the dentate gyrus during aging (Figure 2C–G, S2, S3, S4, S5, Table S2). The mitochondrial protein folding stress was also increased during SVZ NSC activation, consistent with the shared metabolic reprogramming between NSCs in these two brain regions (Figure S6A–C). However, it was not increased in SVZ NSCs during aging (Figure S6D), which may account for, at least in part, the faster decline of the self-renewal capacity of hippocampal NSCs than SVZ NSCs during aging48–50. Thus, aging-causing cellular damage may be specific to the environment they reside, such as different tissues or even different regions of the same tissue.
Increasing mitochondrial protein folding stress resulted in compromised NSC maintenance and neurogenesis, neural hyperactivity, and cognitive decline (Figure 3, 5, 6). Conversely, reducing mitochondrial protein folding stress in aged animals increased neurogenesis and improved cognition (Figure 7). These findings establish a physiological function of the mitochondrial protein folding stress in regulating NSC aging and neural hyperactivity in the hippocampus, contributing to cognitive decline. Our findings are consistent with the observations that activating the mitochondrial unfolded protein response or repressing neural hyperactivity slows aging and extends lifespan8,29, and suggest that the mitochondrial unfolded protein response slows aging in part by repressing neural hyperactivity.
Interestingly, SIRT7 overexpression in the dentate gyrus of old mice did not change the number of Sox2-positive cells but increased neurogenesis (Figure 7), suggesting that the loss of neural progenitors during aging may be irreversible although their function can be improved. Our finding is consistent with other studies on reversing aged NSCs in the dentate gyrus. For example, administration of plasma from excised mice into aged mice was able to improve neurogenesis in the dentate gyrus but did not change the number of neural progenitors14.
Our study expands the growing physiological functions of SIRT7. In addition to preventing cognitive aging, SIRT7 extends lifespan, prevents fatty liver disease, hematopoietic stem cell aging, hearing loss, exercise intolerance, osteopenia, cardiac dysfunction, and promotes hair growth42,51–56. These findings support SIRT7 as an attractive target for improving healthspan and lifespan, as well as the therapeutic potentials of SIRT7 activators and NAD+ boosters.
LIMITATIONS OF THE STUDY
In the single-cell RNA-sequencing study of the dentate gyrus of young and old mice, quiescent NSCs and astrocytes were combined in one cluster, and activated NSCs and NPCs were combined in one cluster. GFAP staining labels both quiescent NSCs and astrocytes. Sox2 is highly expressed in NSCs/progenitors but not exclusively expressed in NSCs/progenitors. Cognitive defects and neural hyperactivity in SIRT7−/− mice are consistent with the decline in NSCs, although cognition and neural activity can be influenced by factors beyond neurogenesis.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Danica Chen (danicac@berkeley.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The sequencing data reported in this paper has been deposited in NCBI’ s Gene Expression Omnibus and are accessible through GEO: GSE205936.
All raw data used to create all graphs can be found in Data S1.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
SIRT7−/− mice in 129 background have been described previously42,54. Both male and female mice ranging from 3 to 7 months old were used in the study. Mice were bred inhouse. For experiments using young and old mice, both male and female C57BL/6 mice ranging from 3 to 24 months old were obtained from the National Institute on Aging. All mice were housed on a 12:12 hr light:dark cycle at 25°C. Mice were provided with standard chow (5053 - PicoLab® Rodent Diet 20 from LabDiet) and water ad libitum. Onsite veterinarians were overseeing health status checks. All animal procedures were in accordance with the animal care committee at the University of California, Berkeley.
METHOD DETAILS
Immunohistochemistry
Tissue processing and immunohistochemistry was performed on free-floating sections following standard procedure18. Briefly, mice were transcardially perfused with 10 ml of PBS with 10 U/ml of heparin and then with 40 ml of PBS with 4% formaldehyde. Brains were extracted and incubated in PBS with 4% formaldehyde at 4°C overnight. Brains were transferred into PBS with 15% sucrose at 4°C for 6 hr and then transferred into PBS with 30% sucrose at 4°C for a day before cutting. Brains were then sectioned coronally at 40 μm with a cryomicrotome (Leica Camera, Inc.) and stored in cryoprotective medium. For BrdU labelling, brain sections were pre-treated with 2N HCl at 37°C for 30 min before incubation with primary antibody. Primary antibodies were: mouse anti-Dcx (1:750; Abcam), rat anti-BrdU (1:500, Invitrogen), rabbit anti-Sox2 (1:500, Abcam), mouse anti-NeuN (1:500, Millipore), rabbit anti-HSP60 (1:500, CST), rabbit anti-NPY (1:4000, ImmunoStar), mouse anti-MCM2 (1:250, BD Biosciences), chicken anti-GFAP (1:1000, Abcam), rabbit anti-SIRT7 (1:500). After overnight incubation, primary antibody staining was revealed using TSA plus biotin kit (PerkinElmer) or fluorescence conjugated secondary antibodies. To estimate the total number of Dcx- or Sox2- or HSP60-positive cells in the dentate gyrus, immunopositive cells in the granule cell and subgranular cell layer of the dentate gyrus were counted in every sixth coronal hemibrain section through the hippocampus. The average number of positive cells per section was multiplied by the total number of sections and multiplied by 2.
HSP60 was co-stained with MCM2 or GFAP. Both 10x and 40x images were taken. 10x images show the entire dentate gyrus, but do not have the resolution for HSP60. 40x images show clear HSP60 staining. Zoom-ins of 40x images show very clear HSP60 staining. Using 10x images as the references, multiple 40x images were taken to cover the entire dentate gyrus and were used to count manually the number of MCM2+ or GFAP+ cells and the number of HSP60+MCM2+ or HSP60+GFAP+ cells in the dentate gyrus. The criteria for double positive cells is when HSP60 signal is overlapping with MCM2 or GFAP signal and both signals surround the same DAPI signal. The fraction of activated NSCs that are HSP60-positive is determined by the ratio of the number of HSP60+MCM2+ cells to the number of MCM2+ cells. The fraction of quiescent NSCs/astrocytes that are HSP60-positive is determined by the ratio of the number of HSP60+GFAP+ cells to the number of GFAP+ cells.
BrdU administration and quantification of BrdU-positive cells
Mice were given intraperitoneal injection of BrdU (50 mg/kg body weight) daily for 3 days before being euthanized. Alternatively, mice were given intraperitoneal injection of BrdU (50 mg/kg body weight) daily for 6 days and euthanized 28 days later. To estimate the total number of BrdU-positive cells in the dentate gyrus, BrdU staining was performed on every sixth hemibrain section. The average number of positive cells per section was multiplied by the total number of sections and multiplied by 2.
Lentivirus mediated gene transfer via stereotaxic injection
SIRT7 was cloned into the pFUGw lentiviral construct54. NRF1 shRNA was cloned into the pFUGw-H1 lentiviral construct42. Lentivirus was produced as described57, concentrated by centrifugation and resuspended with Hanks’ Balanced Salt Solution (HBSS). Viral titers were determined by HIV-1 p24 antigen ELISA (ZeptoMetrix). Anesthesia was induced using Isoflurane Vaporizer (VETEQUIP) with 3–5% isoflurane in oxygen with a delivery rate of 1 l/min until loss of righting reflex and maintained with 1–2% isoflurane in oxygen with a flow of 1 l/min. Each mouse was injected with 2μl (7 × 107 pg/ml) viral solution via stereotaxic injection into the dentate gyrus (0.5 μl per min) using the following coordinates: a/p, −2.1; m/l, ±1.8; d/v, −2.0. Postoperative analgesic treatments with meloxicam (5 mg/kg body weight) were continued for 2 days after surgery. The mouse status was checked daily in terms of the body weight and general behavior for the first week after surgery. Mice were characterized 8 weeks after lentiviral injections.
Morris Water Maze
Mice were trained on the Morris Water Maze58 with four trials per day over 5 to 7 days. The tank was 122cm in diameter and the platform was hidden 1 cm below the surface of water made opaque with white nontoxic paint. Starting points were changed every trial. Each trial lasted either until the mouse found the platform or for 60 seconds. Mice rested on the platform for 10 seconds after each trial if they could not find the platform. 24 hours and 72 hours after the last training session, the platform was removed for a 60-second probe trial. Mouse swimming was recorded with a digital camera recorder and the videos were analyzed with Ethvision (Noldus Information Technology, Wageningen, The Netherlands).
Open field test
Mice were transferred to and allowed to habituate in the testing room under normal light for 60min before testing. Mice were placed in a clear plastic chamber (50×50 cm) for 15 minutes. Mouse movement was recorded with a digital camera recorder and the videos were analyzed with Ethvision (Noldus Information Technology, Wageningen, The Netherlands).
Elevated plus maze
Mice were transferred to and allowed to habituate in the testing room under normal light for 60min before testing. Mice were allowed to explore an elevated plus maze consisted of open and closed arms for 10 minutes. Entries into closed and open arms were recorded with a digital camera recorder and the videos were analyzed with Ethvision (Noldus Information Technology, Wageningen, The Netherlands).
Single-cell RNA-sequencing of dentate gyrus using 10x Genomics Chromium
Mice were sacrificed, the brains were promptly isolated, and the dentate gyrus was quickly microdissected under a dissection scope. Cells were dissociated using papain neural-tissue dissociation kit (Miltenyi biotechnology), according to the manufacturer’s protocol. After a final wash in HBSS, the cell pellet was resuspended in catching medium: DMEM/F12 (Thermo Fisher) with B27 supplement (Thermo Fisher, no Vitamin A,1:50), N2 supplement (Thermo Fisher, 1:100), 15 mM HEPES buffer, 0.6% glucose, penicillin–streptomycin–glutamine (Life Technologies, 1:100), and insulin–transferrin–selenium (Life Technologies, 1:1,000). The cell suspension was stained with propidium iodide (PI) (biolegend, 1:1000) for 1 minute. PI negative live cells were FACS-sorted on a BD FACS Aria Fusion, using a 100μm nozzle at 13.1 PSI. The sorted cells were then spun down at 300g for 5 min at 4 °C and resuspended in catching medium at a concentration of 300 cells per μl. Cells were loaded onto a 10x Genomics Chromium chip per factory recommendations. Reverse transcription and library preparation were performed using the 10x Genomics Single Cell 3’ reagent kits v3 following the 10x Genomics protocol. The libraries were sequenced using Illumina NovaSeq 6000 28×94 Flow Cell SP.
10x Genomics single-cell RNA-sequencing data pre-processing, UMAP analysis, and identification of cell clusters
RNA reads from sequencing were demultiplexed and aligned to mouse transcriptome (mm10) using the Cell Ranger software (10x Genomics, v.5.0.0). The Scanpy Python package (v.1.6.0) was used for the pre-processing of the single-cell RNA seq data59. Cells with less than 600 unique genes, more than 4500 unique genes, or greater than 15% mitochondrial reads were removed. Genes detected in less than 3 cells were excluded. We included 3893 cells with 2463 cells from young and 1430 cells from old, and 17592 genes for further analysis. The data was normalized such that every cell has 10,000 counts and then log transformed with an offset of 1. The batch correction was done by the bbknn batch-alignment algorithm (v1.3.12)60. We computed the highly variable genes with the top 4,000 genes and the flavor set to ‘cell_ranger’. The highly variable genes were used for principal components analysis. The data were visualized by UMAP (Uniform Manifold Approximation and Projection) projection using Scanpy. Unsupervised clustering was done by the Leiden algorithm (v 0.8.0)61 with a resolution of 0.5. Marker genes for each cluster were calculated by Wilcoxon rank-sum test. The cell identity of each cluster was determined by comparing the marker genes of each cluster with the marker genes identified in the literature12,31.
Differential gene expression analysis, bar plots, violin plots, and dot plots for gene expression in single cells
Adaptive thresholding of the single-cell gene expression data was performed with the MAST R package (v1.12.0), and differential gene expression analysis of young and old cells from each cluster using a hurdle model with the young cells as the reference62. To visualize the expression of genes, log-normalized expressions of genes were extracted from the data after adaptive thresholding and plotted for every cell with a violin plot and an overlying strip plot by the Seaborn Python package (v.0.9.0). The bar plots were generated by Seaborn. The UMAP plots, dot plots, and track plots were generated by Scanpy.
Pathway enrichment analysis and Pseudo-time analysis
The GSEAPY Python package (v.0.10.3) was used for pathway enrichment analysis. The Monocle R package (v.2.14.0)63 was used to reconstruct the differentiation trajectories of the neurogenic lineage and oligodendrocyte lineage. The genes in pseudo-time plots were generated by Monocle with raw counts.
Brain surgery for window installation
All surgical procedures were performed in a stereotaxic apparatus (Kopf instruments) under anesthesia with 1–2% isoflurane mixed in O2 followed by an injection of buprenorphine (0.1 mg/kg) subcutaneously. The mouse head was aligned for both medial-lateral and anterior-posterior directions at Bregma, and the coordinates of implantation center on the right hemisphere (a/p, −2.1; m/l, −1.8) were then marked. A small craniotomy (~3mm diameter) was made with a dental drill. The dura was removed with fine forceps and the cortex was gently aspirated until the external capsule was exposed. The cortical tissue was slowly removed in steps at 50–100 um each time followed by saline irrigation to avoid dehydration of the exposed tissue. Each step was performed until bleeding stopped completely. When the cortex detached from the capsule, the fibers of the cingulum or the corpus callosum mainly extending in the three directions were exposed. The dorsal fibers were carefully peeled away to expose the deepest fibers (alveus of the hippocampus), leaving the inner capsule and the hippocampus undamaged. Then, the tissue was rinsed with saline until the bleeding completely stopped. The virus (pAAV1.Syn.GCaMP6s.WPRE.SV40, titer 2.3 × 1013 GC/ml, Addgene) was injected into DG and CA1 (500nl at a/p, −2.1; m/l, −1.8, d/v −2.0 and −1.4 mm) by stereotaxic injection with a capillary needle. A custom-built cranial window cannula (3-mm diameter, 2-mm height) was designed and printed with a Formlabs 3D printer. The bottom of a window cannula was glued with a 3-mm diameter coverslip (Potomac Photonics) to make a cranial window. A cranial window was implanted using forceps until the glass was in contact with the capsule, and cemented to the skull using C&B Metabond Cement System (Parkell). A titanium head-post (Knick Machining) was attached to the skull with cyanoacrylate glue and Metabond. Postoperative analgesic treatments with meloxicam (5 mg/kg body weight) were continued for 3 days after surgery. The mouse status was checked daily in terms of the body weight and general behavior for the first week after surgery. Four weeks of recovery and habituation to the head fixation were allowed before neuronal activities were imaged under two-photon fluorescence microscopy in awake mice.
Two-photon excitation fluorescence microscopy
Imaging was performed with a homebuilt two-photon fluorescence microscope and custom-written LabVIEW software64. GCaMP6s signal was excited at 940 nm using a femtosecond laser with a built-in dispersion compensation unit (InSight DeepSee, Spectral Physics) and collected by a 16× water-dipping objective (Nikon, 0.8 N.A., 3 mm WD). The excitation power was 13–68 mW.
Image processing and analysis
Imaging data sets were processed with Fiji65 and customized codes written in MATLAB (version R2020b, MathWorks)66. Specifically, images were first registered in Fiji with TurboReg plugin67 for rigid motion correction. Then, non-rigid motion correction was performed using a NoRMcorre function built in the Ezcalcium software68 (https://github.com/porteralab/EZcalcium). After the registration, regions of interest (ROIs) were selected with Ezcalcium ROI detection module, which can manually add and exclude ROIs. The relative fluorescence change (ΔF/F) versus time traces for each ROI were extracted with Ezcalcium software.
Pupil tracking and image analysis
The pupil of mouse left eye was monitored with a digital USB camera (Mako U130B) at 3 frames/sec (1280 × 1024 pixels, 8-bit greyscale images) using a customized video acquisition system implemented in MATLAB. Therefore, the pupil diameter can be recorded while the neuronal activity of a head-fixed mouse was imaged under the 2-photon microscope. To extract the pupil diameter, image frames were centered first and cropped around the pupil using Fiji. Custom MATLAB code was used to extract pupil diameter values from pupil videos. Specifically, the MATLAB function “imfindcircles” from the Image Processing Toolbox was used to locate a circular pupil outline in each image frame. The time traces of each neuron’s ΔF/F were correlated with the mouse’s pupil diameter changed in time. Pearson correlation was used to analyze the correlation coefficient between the neural activity and the pupil diameter.
mRNA analysis
RNA was isolated from tissues using Trizol reagent (Invitrogen). cDNA was generated using the qScript™ cDNA SuperMix (Quanta Biosciences). Gene expression was determined by real time PCR using Eva qPCR SuperMix kit (BioChain Institute) on an ABI StepOnePlus system. All data were normalized to GAPDH expression. Primers used for quantitative real time PCR were:
NPY-Y1-forward: CAGTGAGACCAAGCGAATCAAC
NPY-Y1-reverse: CTGGTGGTTCCAGTCGAACA
NPY-Y2-forward: TGGGCCAGGGCACACTAC
NPY-Y2-reverse: TCACCTGCACCTCGACCA
GAPDH-forward: CCCATCACCATCTTCCAGGAGC
GAPDH-reverse: CCAGTGAGCTTCCCGTTCAGC
Isolation of NSCs using the neurosphere self-renewal medium
NSCs were isolated from the mouse dentate gyrus as described69,70. Briefly, mice were sacrificed by cervical dislocation, and the dentate gyrus was dissected out, minced and dissociated using PDD enzyme mix (Papain 2.5 U/ml, Dispase 1 U/ml, DNaseI 250 U/ml) at 37°C for 15 min and incubated for a further 10 minutes at room temperature. Cells were passed through 40 μm cell strainers to obtain a single cell suspension, followed by pelleting at 100 g for 5 minutes. Cells were further purified through Percoll (Sigma) density gradient centrifugation, washed with DMEM/F12 with 1x B27 (Thermo Fisher Scientific), 1x GlutaMAX (Thermo Fisher Scientific) and 10 units/ml Penicillin/Streptomycin (Gibco), then the cells were resuspended with neural stem cell basal medium (Merck Millipore) with 1x B27, 1x GlutaMAX, 10 units/ml Penicillin/Streptomycin, 20 ng/ml purified mouse EGF (PeproTech), and 20 ng/ml recombinant bovine FGF-2 (PeproTech). Every subsequent 2 days, exchange half of the growth medium with fresh medium to replenish the growth factors. After 2 weeks, neurospheres were formed and used for qPCR or immunofluorescence staining.
For immunofluorescence staining, floating neurospheres were pelleted and dissociated into a single cell suspension by treating with Accutase (Gibco) at 37°C for 5 minutes, followed by trituration. The accutase activity was arrested by neural stem cell basal medium and the cells were plated on poly-D-lysine pre-coated coverslips. Cells grown on coverslips were fixed with 4% PFA in PBS and immunostaining was performed as described (Ahmed et al., 2021). Briefly, cells were washed twice with PBS. Fixed cells were permeabilized in 0.1% Triton X-100 in PBS for 10 minutes, then blocked in 2% bovine serum albumin with 0.2% Tween-20 in PBS for 30 minuets. Primary antibodies were: mouse anti-MCM2 (1:250, BD Biosciences), rabbit anti-HSP60 (1:1000, CST), rabbit anti-Sox2 (1:500, Abcam). After overnight incubation, primary antibody staining was revealed using fluorescence conjugated secondary antibodies. DAPI was used to label nuclei.
For qPCR, RNA was isolated from neurospheres using Trizol reagent (Invitrogen). cDNA was generated using the qScript™ cDNA SuperMix (Quanta Biosciences). Gene expression was determined by real time PCR using Eva qPCR SuperMix kit (BioChain Institute) on an ABI StepOnePlus system. All data were normalized to GAPDH expression. Primers used for quantitative real time PCR were:
HSP60-forward: ACCTGTGACAACCCCTGAAG
HSP60-reverse: TGACACCCTTTCTTCCAACC
GAPDH-forward: CCCATCACCATCTTCCAGGAGC
GAPDH-reverse: CCAGTGAGCTTCCCGTTCAGC
3D reconstruction of confocal images
Sections were observed under a Zeiss LSM 710 Confocal Microscope, using a 63X oilimmersion objective lens. Images were sampled at a resolution of 1024 by 1024 pixels, using a z-step size of 0.6 μm. The region of interest was centered in the image frame and the Z-stack dimensions were set manually by tracking MCM2-labeled activated NSCs. Zeiss image files were imported into Imaris (Bitplane, version 9.9.1). The MCM2 and HSP60 signals were reconstructed as 3D structures using the “create surface” tool. The threshold was selected for each channel and these threshold levels were used for all images. MCM2 structure was identified when it is colocalized with DAPI. HSP60 signals with the distance to the surface of the nucleus (MCM2 signals) less than 1 μm determined using the “shortest distance to surfaces” filter in Imaris were recorded. The fluorescence intensity or volume of HSP60 signals was collected from the statistics tab in Imaris.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mice were randomized to groups and analysis of mice and tissue samples was performed by investigators blinded to the treatment or the genetic background of the animals. No test was used to determine normal distribution. Statistical analysis for each experiment is indicated in the figure legends. Data are presented as means and error bars represent standard errors. In all corresponding figures, * represents p < 0.05. ** represents p < 0.01. *** represents p <0.001. ns represents p > 0.05. Replicate information is indicated in the figures. Wilcoxon rank-sum test was performed using the SciPy Python package (v.1.4.1).
Supplementary Material
Table S1. Normalized counts for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 1.
Table S2. MAST differential gene expression in each cell type for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 1.
Table S3. MAST differential expression of cell cycle genes in activated NSCs/NPCs for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 1.
Table S4. MAST differential expression of ribosomal proteins and mitochondrial ribosomal proteins in activated NSCs/NPCs for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 2.
Video S1. The mitochondrial protein folding stress increases in activated NSCs in the dentate gyrus with age. Related to Figure 2.
3D reconstitution for immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of young and old mice. Red: HSP60. Green: MCM2. Scale bar: 2 μm.
Video S2. SIRT7 suppresses the mitochondrial protein folding stress in activated NSCs in the dentate gyrus. Related to Figure 3.
3D reconstitution for immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of WT and SIRT7−/− mice. Red: HSP60. Green: MCM2. Scale bar: 2 μm.
Video S3. NRF1 knockdown suppresses the mitochondrial protein folding stress in activated NSCs in the dentate gyrus. Related to Figure 6.
3D reconstitution for immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of WT mice, SIRT7−/− mice, and SIRT7−/− mice with NRF1 knockdown in the dentate gyrus. Red: HSP60. White: MCM2. Scale bar: 2 μm.
Data S1. Unprocessed data underlying the display items in the manuscript, related to Figures 2, 3, 4, 5, 6, 7.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BrdU Monoclonal Antibody (BU1/75 (ICR1)) | Invitrogen | MA1–82088, RRID:AB_927214 |
| BrdU antibody [BU1/75 (ICR1) | Abcam | Ab6326, RRID:AB_305426 |
| SOX2 antibody | Abcam | ab97959, RRID:AB_2341193 |
| Doublecortin (Dcx) antibody | Abcam | ab18723, RRID:AB_732011 |
| HSP60 (D6F1) XP® Rabbit monoclonal antibody | Cell Signaling Technology | 12165, RRID:AB_2636980 |
| BM28 (MCM2) | BD Biosciences | 610700, RRID:AB_2141952 |
| GFAP antibody | Abcam | Ab4674, RRID:AB_304558 |
| NeuN Antibody, clone A60 | Millipore | MAB377, RRID:AB_2298772 |
| Neuropeptide Y (NPY) antibody | Immunostar | 22940, RRID:AB_2307354 |
| Sirtuin 7 | This paper | N/A |
| Goat anti-mouse IgG antibody Alexa Fluor 488 | Thermo Fisher Scientific | A10680, RRID:AB_2534062 |
| Goat anti-mouse IgG antibody Alexa Fluor 555 | Thermo Fisher Scientific | A28180, RRID:AB_2536164 |
| Goat anti-Mouse IgG, F(ab’)2 Fragment Antibody, Alexa Fluor® 555 | Thermo Fisher Scientific | A21425, RRID:AB_1500751 |
| Goat anti-rabbit IgG antibody Alexa Fluor® 488 | Thermo Fisher Scientific | A32731, RRID:AB_2633280 |
| Goat anti-rabbit IgG antibody Alexa Fluor® 555 | Thermo Fisher Scientific | A32732, RRID:AB_2633281 |
| Goat anti-rabbit IgG antibody Alexa Fluor® 647 | Thermo Fisher Scientific | A32733, RRID:AB_2633282 |
| Goat anti-Chicken IgY H&L Alexa Fluor® 647 | Abcam | Ab150175 RRID:AB 2732800 |
| Goat anti-rat IgG antibody Alexa Fluor® 488 | Thermo Fisher Scientific | A11006, RRID:AB_2534074 |
| Goat anti-rat IgG (H+L) antibody, Alexa Fluor® 555 | Thermo Fisher Scientific | A21434, RRID:AB_141733 |
| Goat anti-rat IgG antibody HRP | Biolegend | 405405, RRID:AB_315016 |
| Bacterial and Virus Strains | ||
| pAAV.Syn.GCaMP6s.WPRE.SV40 | Addgene | Cat# 100843-AAV1 |
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s Modification of Eagle’s Medium | Gibco | Cat# 11995065 |
| Dulbecco’s Modification of Eagle’s Medium/ Nutrient Mixture F12 | Gibco | Cat# 11320082 |
| Neural Stem Cell Basal Medium | Merck Millipore | Cat# SCM003 |
| Goat serum | Abcam | Cat# ab7481 |
| B27 supplement | Gibco | Cat# 12587010 |
| EGF | PeproTech | Cat# 315–09 |
| FGF2 | PeproTech | Cat# 100–18B |
| N-2 supplement | Gibco | Cat# 17502048 |
| Insulin-transferrin-selenium (ITS-G) (100X) | Gibco | Cat# 41400–045 |
| Penicillin-streptomycin-glutamine (100X) | Gibco | Cat# 10378016 |
| Heparin | Sigma | Cat# H3149 |
| Opti-MEM | Gibco | Cat# 11058021 |
| GlutaMAX supplement | Gibco | Cat# 35050061 |
| DPBS, no calcium, no magnesium | Gibco | Cat# 14190250 |
| Trypsin-EDTA (0.25%) | Gibco | Cat# 25200056 |
| Accutase | Sigma | Cat# 07920 |
| HBSS, calcium, magnesium, no phenol red | Gibco | Cat# 14025092 |
| HBSS, no calcium, no magnesium, no phenol red | Gibco | Cat# 14175095 |
| HEPES | Gibco | Cat# 15630080 |
| Papain | Sigma | Cat# P4762 |
| Dispase I protease | Sigma | Cat# D4818 |
| DNase I | Sigma | Cat# D4263 |
| Percoll | Sigma | Cat# P1644 |
| Lipofectamine 2000 | Invitrogen | Cat# 11668019 |
| Fetal Bovine Serum | Invitrogen | Cat#10437–028 |
| Penicillin Streptomycin Solution (100x) | Invitrogen | Cat# 15140122 |
| TRIzol Reagent | Invitrogen | Cat# 15596026 |
| qScript™ cDNA SuperMix | Quanta Biosciences | Cat# 95048 |
| qPCR SuperMix kit | BioChain Institute | Cat# K5052400 |
| DAPI (4’,6-diamidino-2-phenylindole, dihydrochloride) | Thermo Fisher Scientific | Cat#62247 |
| Propidium Iodide Solution | Biolegend | Cat#421301 |
| BrdU | Sigma | Cat#B5002 |
| C&B Metabond Cement System | Parkell | Cat#S380 |
| Vetbond | 3M | Cat#1469SB |
| Meloxicam | Covertrus | NDC: 11695-6936-1 |
| Buprenorphine Hydrochloride | Par Pharmaceutical, Inc. | NDC: 42023-179-01 |
| Tissue-Tek O.C.T. Compound | Thermo Fisher Scientific | Cat#4585 |
| Formaldehyde | Thermo Fisher Scientific | Cat# F79–500 |
| Sucrose | Thermo Fisher Scientific | Cat# S5-500 |
| Sodium Azide | Sigma | Cat#S2002 |
| Triton X-100 | Sigma | Cat# X100 |
| Tween20 | Sigma | Cat# P7949 |
| Critical Commercial Assays | ||
| HIV Type 1 p24 Antigen ELISA | Zeptometrix | Cat# 0801111 |
| TSA Biotin Kits | PerkinElmer | Cat# NEL700A001KT |
| Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | Cat# 130-093-231 |
| 10× Genomics Single Cell 3’ reagent kits v3 | 10x Genomics | PN-1000075 |
| Deposited Data | ||
| Sequencing data | This paper | GSE205936 |
| Raw data | This paper | Data S1 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Mouse: SIRT7 KO | Shin et al.54 | |
| Mouse: C57BL/6J | National Institute on Aging | |
| Oligonucleotides | ||
| Primers NPY-Y1 Forward: CAGTGAGACCAAGCGAATCAAC Reverse: CTGGTGGTTCCAGTCGAACA |
IDT | N/A |
| Primers NPY-Y2 Forward: TGGGCCAGGGCACACTAC Reverse: TCACCTGCACCTCGACCA |
IDT | N/A |
| Primers HSP60 Forward: ACCTGTGACAACCCCTGAAG Reverse: TGACACCCIIICTTCCAACC |
IDT | N/A |
| Primers GAPDH Forward: CCCATCACCATCTTCCAGGAGC Reverse: CCAGTGAGCTTCCCGTTCAGC |
IDT | N/A |
| Recombinant DNA | ||
| psPAX2 | Addgene | Plasmid: #12260 |
| pMD2.G | Addgene | Plasmid: #12259 |
| pCMV-dR8.2 dvpr | Addgene | Plasmid: #8455 |
| pCMV-VSV-G | Addgene | Plasmid: #8454 |
| pFUGw | Shin et al.54 | |
| pFUGw-H1 | Mohrin et al.42 | |
| pFUGw-H1-NRF1KD | Mohrin et al.42 | |
| pFUGw-SIRT7 | Shin et al.54 | |
| Software and Algorithms | ||
| Etho Vision XT | Noldus | https://www.noldus.com/animal-behavior-research/products/ethovision-xt |
| Fiji | Schindelin et al.65 | https://fiji.sc/ |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| Ezcalcium software | Cantu et al.68 | https://github.com/porteralab/EZcalcium |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| Cell Ranger | 10x Genomics | |
| Scanpy Python package (v.1.6.0) | Wolf et al.59 | https://github.com/scverse/scanpy |
| Bbknn Python package (v1.3.12) | Polański et al.60 | https://github.com/Teichlab/bbknn |
| Leidenalg Python package (v0.8.0) | Traag et al.61 | https://github.com/vtraag/leidenalg |
| MAST R package (v1.12.0) | Finak et al.62 | https://github.com/RGLab/MAST |
| Seaborn Python package (v.0.9.0). | https://seaborn.pydata.org/citing.html | |
| GSEAPY Python package (v.0.10.3) | https://github.com/zgfang/GSEApy/releases | |
| Monocle R package (v.2.14.0) | Trapnell et al.63 | https://github.com/cole-trapnell-lab/monocle3 |
| LabVIEW software | National Instruments | https://www.ni.com/en-us/support/downloads/software-products/download.labview.html#443865 |
| Imaris (v.9.9.1) | Bitplane | https://imaris.oxinst.com |
| Other | ||
| N/A | ||
Highlights.
Mitochondrial protein folding stress is elevated in neural stem cells with aging.
SIRT7 protects neural stem cells by suppressing mitochondrial protein folding stress.
SIRT7 overexpression increases neurogenesis and improves cognition in aged mice.
ACKNOWLEDGEMENTS
We thank Y. Choi and Berkeley Functional Genomics Lab for 10x Genomics libraries. H. Nolla, A. Valeros, and Berkeley FACS Facility for sorting. M. Gill, I. Lo, and Gladstone behavioral core for animal behavioral studies. S. Ruzin, D. Schichnes, and Berkeley Imaging Facility for imaging experiments. L. Xu for neurosphere assays. J. Harris, K. Mosher, and M. Adil for stereotaxic injections. K. Borges and Y. Yang for brain imaging. Supported by NIH R01DK 117481 (D.C.), R01AG063404 (D.C.), R01AG 063389 (D.C.), National Institute of Food and Agriculture (D.C.), Chau Hoi Shuen Foundation (D.C.), NIH U01NS103489 (N.J.), R01NS092918 (D.D.), the ITO Scholarship (R.O., A.M.), the Honjo International Scholarship (R.O.), Dr. and Mrs. James C.Y. Soong Fellowship (W.-C. M.), and Taiwan Government for Study Abroad Scholarship (W.-C. M.), QB3 Frontiers in Medical Research Fellowship (W.-C. M.), JASSO Graduate Scholarship for Degree Seeking Students (A.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes fifteen figures, three videos, four tables, and one data.
REFERENCES
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijg J, and Campisi J (2008). Puzzles, promises and a cure for ageing. Nature 454, 1065–1071. 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa-Mattioli M, and Walter P (2020). The integrated stress response: From mechanism to disease. Science 368. 10.1126/science.aat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou BB, and Elledge SJ (2000). The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439. 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 5.Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, and Bohr VA (2016). Nuclear DNA damage signalling to mitochondria in ageing. Nat Rev Mol Cell Biol 17, 308–321. 10.1038/nrm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaushik S, and Cuervo AM (2015). Proteostasis and aging. Nat Med 21, 1406–1415. 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 7.Korotkov A, Seluanov A, and Gorbunova V (2021). Sirtuin 6: linking longevity with genome and epigenome stability. Trends Cell Biol. 10.1016/j.tcb.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shpilka T, and Haynes CM (2018). The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol 19, 109–120. 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 9.Tanay A, and Regev A (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goncalves JT, Schafer ST, and Gage FH (2016). Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell 167, 897–914. 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Bourdenx M, Martin-Segura A, Scrivo A, Rodriguez-Navarro JA, Kaushik S, Tasset I, Diaz A, Storm NJ, Xin Q, Juste YR, et al. (2021). Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell. 10.1016/j.cell.2021.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dulken BW, Buckley MT, Navarro Negredo P, Saligrama N, Cayrol R, Leeman DS, George BM, Boutet SC, Hebestreit K, Pluvinage JV, et al. (2019). Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210. 10.1038/s41586-019-1362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, et al. (2019). Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci 22, 401–412. 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz AM, Fan X, Bieri G, Smith LK, Sanchez-Diaz CI, Schroer AB, Gontier G, Casaletto KB, Kramer JH, Williams KE, and Villeda SA (2020). Blood factors transfer beneficial effects of exercise on neurogenesis and cognition to the aged brain. Science 369, 167–173. 10.1126/science.aaw2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leon J, Moreno AJ, Garay BI, Chalkley RJ, Burlingame AL, Wang D, and Dubal DB (2017). Peripheral Elevation of a Klotho Fragment Enhances Brain Function and Resilience in Young, Aging, and alpha-Synuclein Transgenic Mice. Cell Rep 20, 1360–1371. 10.1016/j.celrep.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minhas PS, Latif-Hernandez A, McReynolds MR, Durairaj AS, Wang Q, Rubin A, Joshi AU, He JQ, Gauba E, Liu L, et al. (2021). Restoring metabolism of myeloid cells reverses cognitive decline in ageing. Nature 590, 122–128. 10.1038/s41586-020-03160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senatorov VV Jr., Friedman AR, Milikovsky DZ, Ofer J, Saar-Ashkenazy R, Charbash A, Jahan N, Chin G, Mihaly E, Lin JM, et al. (2019). Blood-brain barrier dysfunction in aging induces hyperactivation of TGFbeta signaling and chronic yet reversible neural dysfunction. Sci Transl Med 11. 10.1126/scitranslmed.aaw8283. [DOI] [PubMed] [Google Scholar]
- 18.Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, et al. (2011). The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94. 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakker A, Krauss GL, Albert MS, Speck CL, Jones LR, Stark CE, Yassa MA, Bassett SS, Shelton AL, and Gallagher M (2012). Reduction of hippocampal hyperactivity improves cognition in amnestic mild cognitive impairment. Neuron 74, 467–474. 10.1016/j.neuron.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bero AW, Yan P, Roh JH, Cirrito JR, Stewart FR, Raichle ME, Lee JM, and Holtzman DM (2011). Neuronal activity regulates the regional vulnerability to amyloid-beta deposition. Nat Neurosci 14, 750–756. 10.1038/nn.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busche MA, Eichhoff G, Adelsberger H, Abramowski D, Wiederhold KH, Haass C, Staufenbiel M, Konnerth A, and Garaschuk O (2008). Clusters of hyperactive neurons near amyloid plaques in a mouse model of Alzheimer’s disease. Science 321, 1686–1689. 10.1126/science.1162844. [DOI] [PubMed] [Google Scholar]
- 22.Koh MT, Rosenzweig-Lipson S, and Gallagher M (2013). Selective GABA(A) alpha5 positive allosteric modulators improve cognitive function in aged rats with memory impairment. Neuropharmacology 64, 145–152. 10.1016/j.neuropharm.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller SL, Fenstermacher E, Bates J, Blacker D, Sperling RA, and Dickerson BC (2008). Hippocampal activation in adults with mild cognitive impairment predicts subsequent cognitive decline. J Neurol Neurosurg Psychiatry 79, 630–635. 10.1136/jnnp.2007.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palop JJ, Chin J, Roberson ED, Wang J, Thwin MT, Bien-Ly N, Yoo J, Ho KO, Yu GQ, Kreitzer A, et al. (2007). Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer’s disease. Neuron 55, 697–711. 10.1016/j.neuron.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putcha D, Brickhouse M, O’Keefe K, Sullivan C, Rentz D, Marshall G, Dickerson B, and Sperling R (2011). Hippocampal hyperactivation associated with cortical thinning in Alzheimer’s disease signature regions in non-demented elderly adults. J Neurosci 31, 17680–17688. 10.1523/JNEUROSCI.4740-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, Sanders DW, Cook C, Fu H, Boonen RA, et al. (2016). Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci 19, 1085–1092. 10.1038/nn.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yassa MA, Lacy JW, Stark SM, Albert MS, Gallagher M, and Stark CE (2011). Pattern separation deficits associated with increased hippocampal CA3 and dentate gyrus activity in nondemented older adults. Hippocampus 21, 968–979. 10.1002/hipo.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yassa MA, Stark SM, Bakker A, Albert MS, Gallagher M, and Stark CE (2010). High-resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic Mild Cognitive Impairment. Neuroimage 51, 1242–1252. 10.1016/j.neuroimage.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zullo JM, Drake D, Aron L, O’Hern P, Dhamne SC, Davidsohn N, Mao CA, Klein WH, Rotenberg A, Bennett DA, et al. (2019). Regulation of lifespan by neural excitation and REST. Nature 574, 359–364. 10.1038/s41586-019-1647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terreros-Roncal J, Moreno-Jimenez EP, Flor-Garcia M, Rodriguez-Moreno CB, Trinchero MF, Cafini F, Rabano A, and Llorens-Martin M (2021). Impact of neurodegenerative diseases on human adult hippocampal neurogenesis. Science 374, 1106–1113. 10.1126/science.abl5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artegiani B, Lyubimova A, Muraro M, van Es JH, van Oudenaarden A, and Clevers H (2017). A Single-Cell RNA Sequencing Study Reveals Cellular and Molecular Dynamics of the Hippocampal Neurogenic Niche. Cell Rep 21, 3271–3284. 10.1016/j.celrep.2017.11.050. [DOI] [PubMed] [Google Scholar]
- 32.Dulken BW, Leeman DS, Boutet SC, Hebestreit K, and Brunet A (2017). Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep 18, 777–790. 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochgerner H, Zeisel A, Lonnerberg P, and Linnarsson S (2018). Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci 21, 290–299. 10.1038/s41593-017-0056-2. [DOI] [PubMed] [Google Scholar]
- 34.Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, and Martin-Villalba A (2015). Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 17, 329–340. 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y, Coskun V, Liang A, Yu J, Cheng L, Ge W, Shi Z, Zhang K, Li C, Cui Y, et al. (2015). Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161, 1175–1186. 10.1016/j.cell.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, and Song H (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372. 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavallucci V, Fidaleo M, and Pani G (2016). Neural Stem Cells and Nutrients: Poised Between Quiescence and Exhaustion. Trends Endocrinol Metab 27, 756–769. 10.1016/j.tem.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Haynes CM, and Ron D (2010). The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci 123, 3849–3855. 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 39.Mohrin M, Widjaja A, Liu Y, Luo H, and Chen D (2018). The mitochondrial unfolded protein response is activated upon hematopoietic stem cell exit from quiescence. Aging Cell 17, e12756. 10.1111/acel.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, et al. (2014). REST and stress resistance in ageing and Alzheimer’s disease. Nature 507, 448–454. 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haynes CM, Fiorese CJ, and Lin YF (2013). Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol 23, 311–318. 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, and Chen D (2015). Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377. 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Islam MS, Wei FY, Ohta K, Shigematsu N, Fukuda T, Tomizawa K, Yoshizawa T, and Yamagata K (2018). Sirtuin 7 is involved in the consolidation of fear memory in mice. Biochem Biophys Res Commun 495, 261–266. 10.1016/j.bbrc.2017.10.159. [DOI] [PubMed] [Google Scholar]
- 44.Cheng A, Wang J, Ghena N, Zhao Q, Perone I, King TM, Veech RL, Gorospe M, Wan R, and Mattson MP (2020). SIRT3 Haploinsufficiency Aggravates Loss of GABAergic Interneurons and Neuronal Network Hyperexcitability in an Alzheimer’s Disease Model. J Neurosci 40, 694–709. 10.1523/JNEUROSCI.1446-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vezzani A, Sperk G, and Colmers WF (1999). Neuropeptide Y: emerging evidence for a functional role in seizure modulation. Trends Neurosci 22, 25–30. 10.1016/s0166-2236(98)01284-3. [DOI] [PubMed] [Google Scholar]
- 47.Anacker C, and Hen R (2017). Adult hippocampal neurogenesis and cognitive flexibility - linking memory and mood. Nat Rev Neurosci 18, 335–346. 10.1038/nrn.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahlenius H, Visan V, Kokaia M, Lindvall O, and Kokaia Z (2009). Neural stem and progenitor cells retain their potential for proliferation and differentiation into functional neurons despite lower number in aged brain. J Neurosci 29, 4408–4419. 10.1523/JNEUROSCI.6003-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539. 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker TL, Turnbull GW, Mackay EW, Hannan AJ, and Bartlett PF (2011). The latent stem cell population is retained in the hippocampus of transgenic Huntington’s disease mice but not wild-type mice. PLoS One 6, e18153. 10.1371/journal.pone.0018153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukuda M, Yoshizawa T, Karim MF, Sobuz SU, Korogi W, Kobayasi D, Okanishi H, Tasaki M, Ono K, Sawa T, et al. (2018). SIRT7 has a critical role in bone formation by regulating lysine acylation of SP7/Osterix. Nat Commun 9, 2833. 10.1038/s41467-018-05187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li G, Tang X, Zhang S, Jin M, Wang M, Deng Z, Liu Z, Qian M, Shi W, Wang Z, et al. (2020). SIRT7 activates quiescent hair follicle stem cells to ensure hair growth in mice. EMBO J 39, e104365. 10.15252/embj.2019104365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, et al. (2014). A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab 20, 856–869. 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, et al. (2013). SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep 5, 654–665. 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, and Bober E (2008). Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102, 703–710. 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 56.Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero A, Tischfield JA, and Serrano L (2016). SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J 35, 1488–1503. 10.15252/embj.201593499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo H, Mu WC, Karki R, Chiang HH, Mohrin M, Shin JJ, Ohkubo R, Ito K, Kanneganti TD, and Chen D (2019). Mitochondrial Stress-Initiated Aberrant Activation of the NLRP3 Inflammasome Regulates the Functional Deterioration of Hematopoietic Stem Cell Aging. Cell Rep 26, 945–954 e944. 10.1016/j.celrep.2018.12.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vorhees CV, and Williams MT (2006). Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc 1, 848–858. 10.1038/nprot.2006.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park JE (2020). BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Scientific reports 9, 5233. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji N, Milkie DE, and Betzig E (2010). Adaptive optics via pupil segmentation for highresolution imaging in biological tissues. Nat Methods 7, 141–147. 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 65.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan JL, Rivera JA, Sun W, Peterson J, Haeberle H, Rubin S, and Ji N (2020). High-speed volumetric two-photon fluorescence imaging of neurovascular dynamics. Nat Commun 11, 6020. 10.1038/s41467-020-19851-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thevenaz P, Ruttimann UE, and Unser M (1998). A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing 7, 27–41. 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 68.Cantu DA, Wang B, Gongwer MW, He CX, Goel A, Suresh A, Kourdougli N, Arroyo ED, Zeiger W, and Portera-Cailliau C (2020). EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front Neural Circuits 14, 25. 10.3389/fncir.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed A, Isaksen TJ, and Yamashita T (2021). Protocol for mouse adult neural stem cell isolation and culture. STAR Protoc 2, 100522. 10.1016/j.xpro.2021.100522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker TL, and Kempermann G (2014). One mouse, two cultures: isolation and culture of adult neural stem cells from the two neurogenic zones of individual mice. J Vis Exp, e51225. 10.3791/51225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Normalized counts for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 1.
Table S2. MAST differential gene expression in each cell type for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 1.
Table S3. MAST differential expression of cell cycle genes in activated NSCs/NPCs for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 1.
Table S4. MAST differential expression of ribosomal proteins and mitochondrial ribosomal proteins in activated NSCs/NPCs for the 10x Genomics Chromium single-cell RNA-sequencing data from the dentate gyrus of young and old mice. Related to Figure 2.
Video S1. The mitochondrial protein folding stress increases in activated NSCs in the dentate gyrus with age. Related to Figure 2.
3D reconstitution for immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of young and old mice. Red: HSP60. Green: MCM2. Scale bar: 2 μm.
Video S2. SIRT7 suppresses the mitochondrial protein folding stress in activated NSCs in the dentate gyrus. Related to Figure 3.
3D reconstitution for immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of WT and SIRT7−/− mice. Red: HSP60. Green: MCM2. Scale bar: 2 μm.
Video S3. NRF1 knockdown suppresses the mitochondrial protein folding stress in activated NSCs in the dentate gyrus. Related to Figure 6.
3D reconstitution for immunohistochemistry staining of HSP60 and MCM2 in the dentate gyrus of WT mice, SIRT7−/− mice, and SIRT7−/− mice with NRF1 knockdown in the dentate gyrus. Red: HSP60. White: MCM2. Scale bar: 2 μm.
Data S1. Unprocessed data underlying the display items in the manuscript, related to Figures 2, 3, 4, 5, 6, 7.
Data Availability Statement
The sequencing data reported in this paper has been deposited in NCBI’ s Gene Expression Omnibus and are accessible through GEO: GSE205936.
All raw data used to create all graphs can be found in Data S1.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


