Summary
Primate-specific genes (PSGs) tend to be expressed in the brain and testis. This phenomenon is consistent with brain evolution in primates but is seemingly contradictory to the similarity of spermatogenesis among mammals. Here, using whole-exome sequencing, we identified deleterious variants of X-linked SSX1 in six unrelated men with asthenoteratozoospermia. SSX1 is a PSG expressed predominantly in the testis, and the SSX family evolutionarily expanded independently in rodents and primates. As the mouse model could not be used for studying SSX1, we used a non-human primate model and tree shrews, which are phylogenetically similar to primates, to knock down (KD) Ssx1 expression in the testes. Consistent with the phenotype observed in humans, both Ssx1-KD models exhibited a reduced sperm motility and abnormal sperm morphology. Further, RNA sequencing indicated that Ssx1 deficiency influenced multiple biological processes during spermatogenesis. Collectively, our experimental observations in humans and cynomolgus monkey and tree shrew models highlight the crucial role of SSX1 in spermatogenesis. Notably, three of the five couples who underwent intra-cytoplasmic sperm injection treatment achieved a successful pregnancy. This study provides important guidance for genetic counseling and clinical diagnosis and, significantly, describes the approaches for elucidating the functions of testis-enriched PSGs in spermatogenesis.
Keywords: cynomolgus monkey, male infertility, primate, sperm, SSX1, tree shrew
The importance of primate-specific genes (PSGs) in spermatogenesis is largely unknown. This study identified variants in an X-linked PSG, SSX1, associated with male infertility. By using cynomolgus monkey and tree shrew models, this study provides a powerful experimental system for elucidating the functions of testis-enriched PSGs in spermatogenesis.
Introduction
Infertility has become a growing medical problem, and according to large population surveys, it affects at least 180 million people worldwide.1 Idiopathic sperm abnormalities, which are direct causes of infertility in males, account for approximately 30% of individuals with male infertility.2 Asthenoteratozoospermia, a form of sperm abnormality, has been defined as a disorder with a significant genetic contribution.3 Previous strategies for exploring genetic factors related to asthenoteratozoospermia have mainly relied on the identification of genetic defects commonly shared by multiple affected individuals.4,5 However, human evidence alone based on a common genetic defect among individuals is not enough for proving a pathogenic genetic factor. The use of appropriate model organisms may provide further reliable functional evidence for the genotype-phenotype correlation and facilitate studies on the molecular mechanisms underlying male infertility.6
Mice have been widely used as a model organism in studies on genetic disorders.7,8,9 However, for studies on human mutated genes that are not evolutionarily conserved between mice and humans, a mouse model is not applicable because of the lack of one-to-one mouse orthologs for genetic manipulation and subsequent phenotypic analysis. A recent study on the human genome identified more than 800 primate-specific genes (PSGs), more than 30% of which are predominantly expressed in the testis.10 A similar pattern was also clearly observed with ampliconic genes on the human X chromosome; 69% of these genes have no orthologs in mice, and the majority of them are expressed predominantly or specifically in testicular germ cells.11,12 Therefore, new model organisms are needed to study the potential spermatogenic functions of testis-enriched PSGs and their genetic contributions to male infertility.
As a non-human primate model, cynomolgus monkeys share high similarities with humans in terms of both genetic and physiological characteristics and have been used for modeling human neurodegenerative and cognitive diseases as well as complex behaviors.13 However, the generation of gene-edited offspring and subsequent reproductive phenotyping in adult monkeys take approximately 4–5 years because of long breeding cycles. Instead, in vivo genetic manipulation with adeno-associated virus (AAV) and short hairpin RNA (shRNA) can facilitate short-term observations on genetically related phenotypes. Furthermore, tree shrews are close relatives to primates because of their high degree of similarities in both molecular and physiological aspects.14,15 Moreover, recent studies successively revealed the reference genomes for tree shrews.14,16,17,18 For PSGs, the orthologs in tree shrews have more similarities than these identified in mice. Therefore, cynomolgus monkeys and tree shrews may be used as promising model organisms for genetic manipulation in the testis and studying the functions of PSGs in spermatogenesis.
Here, we identified deleterious variants of X-linked PSG SSX1 (MIM: 312820) as a factor for asthenoteratozoospermia. Furthermore, we achieved Ssx1 testicular knockdown (Ssx1-KD) models by injecting AAV9-Ssx1-shRNA vectors into the seminiferous tubules in the testes of adult male cynomolgus monkeys and tree shrews. Notably, both Ssx1-KD models mimicked the asthenoteratozoospermia phenotypes observed in humans. Interestingly, our bioinformatics analysis revealed that over 1,000 human testis-enriched genes do not have one-to-one orthologs in mice or that their murine orthologs are not testis-enriched, highlighting the need to develop animal models that are phylogenetically closer to human. This study provides a powerful system for the genetic manipulation of germ cells in the testes of cynomolgus monkey and tree shrew models. The described strategy could be used for elucidating the functions of hundreds of testis-enriched PSGs in spermatogenesis.
Material and methods
Cohort description
In total, a cohort of 536 Chinese infertile men affected by asthenoteratozoospermia were enrolled from the First Affiliated Hospital of Anhui Medical University, the Reproductive and Genetic Hospital of CITIC-Xiangya (Changsha), and the First Affiliated Hospital of Nanjing Medical University in China. All the recruited subjects presented with primary infertility accompanied by a variety of abnormal sperm morphologies. Clinical investigation revealed that all the subjects in this study displayed normal male external genitalia, bilateral testicular sizes, hormone levels, and secondary sexual characteristics. The chromosomal karyotypes of all the individuals were also normal (46; XY), and no large-scale deletions were observed in the Y chromosome. This study was approved by the institutional review boards of all the participating institutes. Signed informed consent was obtained from all the subjects who participated in the study.
Whole-exome sequencing (WES)
DNA samples for WES were prepared and sequenced according to a previously described protocol.19 Briefly, genomic DNA was isolated from the peripheral blood samples of human subjects via a DNeasy Blood and Tissue Kit (Qiagen, 51106). Then, we used 1 μg of genomic DNA to enrich the human exome by using the AIExome Enrichment Kit V2 (iGeneTech, China), and we conducted next-generation sequencing with the NovaSeq 6000 platform (Illumina, San Diego, CA). The obtained raw data were mapped to the human genome reference assembly (GRCh37/hg19) with Burrows-Wheeler Aligner (BWA) software, and PCR duplicates were marked and removed via Picard software.20 ANNOVAR software was used for functional annotation with information from OMIM, Gene Ontology, KEGG Pathway, SIFT, PolyPhen-2, MutationTaster, 1000 Genomes Project, and gnomAD.21,22,23,24,25 According the incidence rates of asthenoteratozoospermia in human populations, genetic variants with allele frequencies ≥ 0.01 in the human population genome datasets (e.g., the gnomAD browser and 1000 Genomes Project) were filtered out. Nonsense, frameshift, and essential splice-site variants were preferred. Missense variants that were predicted to be deleterious by bioinformatic tools were also included for further evaluation. Sanger sequencing was performed for variant verification with the primers listed in Table S1.
Semen parameter analysis
Semen analyses were carried out in source laboratories during routine biological examinations of the individuals according to the WHO guidelines.26 Semen samples from the infertile men were collected by masturbation after 2–7 days of sexual abstinence, and the samples were evaluated after liquefaction for 30 min at 37°C. We assessed sperm morphology with hematoxylin and eosin (H&E) staining and scanning electron microscopy (SEM), and we examined at least 200 spermatozoa to evaluate the rates of abnormal sperm morphologies for each subject.
For semen analyses for cynomolgus monkeys, semen samples were acquired by penile electroejaculation before AAV9-Ssx1-shRNA or AAV9-NC-shRNA vectors injection and at 45, 60, and 75 days after injection. For morphology and motility analyses of tree shrew sperm, spermatozoa were extracted from the cauda epididymides through dissection of adult male tree shrews, diluted in 1 mL of HEPES-TL (Cat. # IVL01-10201007, Caisson Labs), and incubated for 15 min at 37°C. Sperm morphologies of cynomolgus monkeys and tree shrews were also analyzed by H&E staining and SEM, and sperm motility was assessed by a CASA system.
Establishment of cynomolgus monkey and tree shrew models
Production of AAV vectors
A previous study indicated that AAV9, one of 12 serotypes (AAV1–AAV12), can penetrate not only the blood-testis barrier (BTB) but also the basement membrane of the seminiferous tubules.27 To KD the expression of Ssx1 in the testes of cynomolgus monkeys and tree shrews, AAV9 vectors containing shRNA were provided by Hanbio Biotechnology (Shanghai, China), and we used these to target the specific sequences of Ssx1. Briefly, Ssx1-shRNAs and control-shRNA were designed, synthesized, and cloned into the pHBAAV-U6-MCS-CMV-EGFP vectors. Then, we cotransfected the vectors with Ssx1 overexpression plasmids into HEK293T cells by using Lipofectamine 2000 reagent for 48 h. We performed subsequent real time qPCR analysis to validate the KD efficacy of the shRNAs (Figure S1). We used the plasmids of the most efficient shRNAs (Table S2) to package AAV9 virus (Hanbio) for the in vivo KD of Ssx1. In addition, to investigate the possible targeting of the selected Ssx1-shRNAs for other Ssx family members, we also cotransfected selected Ssx1-shRNAs with overexpression plasmids of other Ssx family members into HEK293T cells, and we also detected the KD efficacy by real-time qPCR analysis (Figure S2).
Injection of AAV9-Ssx1-shRNA or AAV9-NC-shRNA vectors into the testes
To KD Ssx1 in cynomolgus monkeys, four adult male cynomolgus monkeys (Macaca fascicularis) were provided by the State Key Laboratory of Primate Biomedical Research (Kunming, China). One cynomolgus monkey was designated as the NC (negative control) group, and the other three cynomolgus monkeys were designated as the Ssx1-KD group. The cynomolgus monkey testes were examined under an ultrasound (LOGIQ P5/A5, GE Healthcare, USA), and the testicular reticulum showed a linear echo dense structure. Then, a needle bevel was punctured into the rete of testicular tissue under the guide of ultrasound and 1 mL of CM-AAV9-NC-shRNA solution or CM-AAV9-Ssx1-shRNA solution (a dose of 1.4 × 1012 vector genome/mL) was injected from the rete testis space and scattered to the seminiferous tubules. Sperm samples were collected from male cynomolgus monkeys by penile electroejaculation28 before injection and at 45, 60, and 75 days after injection. Testicular tissues were obtained by puncture for further functional analyses. All the experimental procedures were approved by the Institutional Animal Care and Use Committee of Kunming University of Science and Technology (authorization code: LPBR201701001) and were performed in accordance with the Guide for Care and Use of Laboratory Animals.29
For the KD of Ssx1 in tree shrews, a total of 15 wild-type adult (6 months or older) male Chinese tree shrews (Tupaia belangeri chinensis) were provided by the Kunming Primate Research Center in Kunming Institute of Zoology, Chinese Academy of Sciences. To KD Ssx1 in the testes of tree shrews, each adult tree shrew was injected with 80 μL of TR-AAV9-NC-shRNA solution (nontargeting control) or TR-AAV9-Ssx1-shRNA solution (a dose of 1.5 × 1012 vector genome/mL) into the seminiferous tubules of bilateral testes through the rete testis. For each solution, approximately 0.4% Trypan blue tracer was added to monitor the injection. Then, tree shrews were sacrificed on the 60th day post injection, and testis tissues and epididymal effluents were collected for further phenotypic analyses. All the experimental and animal care procedures were performed according to the protocols approved by the Institutional Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences.
Electron microscopy evaluation
Sperm samples obtained from men and male cynomolgus monkeys and tree shrews were fixed with 2.5% glutaraldehyde for 24 h at 4°C before electron microscopy evaluation. For transmission electron microscopy (TEM), sperm cells were washed and postfixed with 1% osmium tetroxide in 0.1 mol/L PB for 1–1.5 h at 4°C. Then, dehydration was performed with graded ethanol (50%, 70%, 90%, and 100%) and 100% acetone solutions, followed by infiltration with 1:1 acetone and SPI-Chem resin (containing dodecenyl succinic anhydride, N-methylacetamide, SPI-Pon 812, and DMP-30) overnight at 37°C. After infiltration and embedding in Epon 812, the specimens were sliced with an ultramicrotome, stained with uranyl acetate and lead citrate, and then observed and photographed via TEM (TECNAI-10, Philips) with an accelerating voltage of 80 kV.
For the SEM assay, after being deposited on poly-L-lysine-coated coverslips, sperm specimens were fixed in 2.5% glutaraldehyde, washed with 0.1 mol/L phosphate buffer, and postfixed in osmic acid. Then, the samples were progressively dehydrated with an ethanol and isoamyl acetate gradient and dried with a CO2 critical-point drier (Eiko HCP-2, Hitachi). Next, the specimens were mounted on aluminum stubs, sputter-coated with an ionic sprayer meter (Eiko E-1020, Hitachi), and analyzed via SEM (Stereoscan 260) under an accelerating voltage of 20 kV.
Real-time qPCR and immunoblotting analyses
Total RNA and protein were extracted from human spermatozoa or testicular tissues of cynomolgus monkeys and tree shrews via the Allprep DNA/RNA/Protein Mini Kit (Qiagen) according to the manufacturer’s instructions. For real-time qPCR, 1 μg of total RNA was converted into cDNA with HiScript II Q RT Super-Mix for qPCR (Vazyme). Then, the obtained cDNAs were individually diluted by 5-fold to be used as templates for the subsequent real-time fluorescence qPCR, which was performed with AceQ qPCR SYBR Green Master Mix (Vazyme) on a CFX Connect Real-Time PCR Detection System. GAPDH and β-actin were used as the internal controls. The primer sequences used for real-time qPCR are presented in Tables S3 and S4.
For immunoblotting analysis, the protein samples were denatured at 95°C for 10 min, separated with 10% SDS-polyacrylamide gels, and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Then, the membranes were blocked for 1 h with 5% nonfat milk at room temperature and incubated with primary antibodies (SSX1, PA5-61070, Invitrogen, 1:1,000; CCDC39, HPA035364, Sigma, 1:1,000; CEP44, A8317, ABclonal, 1:1,000; DPY19L2, bs-8291R, Bioss, 1:1,000; KIFAP3, A4518, ABclonal, 1:1,000; NR4A2, A22011, ABclonal, 1:1,000; SMURF2, A10592, ABclonal, 1:1,000; TAF5, A7221, ABclonal, 1:1,000; UPF2, A13411, ABclonal, 1:1,000; HRP-conjugated beta actin, HRP-60008, Proteintech, 1:2,000) overnight at 4°C, respectively. After being washed with TBST and incubated with HRP-conjugated secondary antibody (M21002, Abmart, 1:4,000) for 1 h at room temperature, the PVDF membranes were visualized with Chemistar High-sig ECL Western Blotting Substrate (Tanon) by Tanon 5200.
Immunofluorescence (IF) analysis
IF analyses were performed on spermatozoa from human individuals and testicular tissues from tree shrews. For IF analysis of sperm cells, semen samples (fixed in 4% paraformaldehyde at least 24 h) were deposited on slides that were precoated with 0.1% poly-L-lysine (Thermo Fisher Scientific) before being washed two times with PBS and blocked in 10% donkey serum for 1 h at room temperature. Then, the slides were subsequently incubated overnight at 4°C with the following primary antibodies: rabbit polyclonal anti-SSX1 (PA5-61070, Invitrogen, 1:200) and mouse monoclonal anti-α-tubulin (T9026, Sigma, 1:500). For IF analysis of testis tissues, freshly isolated testes were fixed in 4% paraformaldehyde (in PBS) and progressively embedded in optimal cutting temperature compound for frozen sectioning. Then, the cryosections were washed with PBS and blocked in 10% donkey serum containing 3% Triton X-100 before incubation with the following primary antibodies: rabbit polyclonal anti-SSX1 (PA5-61070, Invitrogen, 1:200) and anti-THY1 (LS-B3139, LSBio, 1:200). After the incubation of primary antibodies, both the sperm cell slides and testis cryosections were washed with PBS supplemented with 0.1% (v/v) Tween 20 and incubated with highly cross-adsorbed secondary antibodies Alexa Fluor 488 AffiniPure Donkey Anti-Mouse IgG (34106ES60, Yeasen, 1:1,000) and Cy3-conjugated AffiniPure Goat Anti-Rabbit IgG (111-165-003, Jackson, 1:4,000) for 1 h at room temperature. Images were acquired with a confocal microscope (Zeiss LSM 880).
RNA sequencing (RNA-seq)
For RNA-seq analysis, total RNA was extracted from the testicular tissues of tree shrews via the Allprep DNA/RNA/Protein Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA integrity was evaluated with 2100 Bioanalyzer (Agilent Technologies), and the RNA samples with an RNA integrity number ≥ 7 were used for library construction and subsequent sequencing analysis on the Illumina sequencing platform (NovaSeq 6000) by Oebiotech (Shanghai, China). Raw data (raw reads) were processed with Trimmomatic,30 and reads containing poly-N and low-quality reads were removed. Then, the obtained clean reads were mapped to the tree shrew reference genome (TS_3.0, http://www.treeshrewdb.org/) with HISAT2.31 For transcript-level quantification, the fragments per kilobase per million (FPKM)31 and read count values of each protein-coding transcript were calculated by Bowtie232 and eXpress.33 We identified differentially expressed genes (DEGs) by using the DESeq234 functions to estimate size factors and nbinom test. A fold change of >2 or <0.5 and q < 0.05 were set as the thresholds to identify significant differential expression. We performed hierarchical cluster analysis of DEGs to explore transcript expression patterns. Next, the DEGs were subjected to Gene Ontology enrichment and KEGG35 pathway enrichment analyses by R based on the hypergeometric distribution. Networks were constructed by Cytoscape software (version 3.6.0).
Identification of testis-enriched genes in human and mouse
Published digital gene expression matrices (DGEs) for 27 different tissues from 95 human individuals were obtained from ArrayExpress (E-MTAB-1733). The DGEs of 70 mouse samples containing 13 different tissues were also downloaded from ArrayExpress (E-GEOD-43721, E-GEOD-74747, E-MTAB-2801, E-MTAB-4644, and E-MTAB-3718). Genes specifically or predominantly expressed in the testes of the two species were identified with the R package DESeq236 (FDR < 0.05; >2-fold higher normalized read counts in the testis compared with all other tissues). To define the orthologous genes that probably share conserved functions between human and mouse testes, we downloaded human-mouse one-to-one orthologs of acquired testis-enriched genes from the Ensembl database. For the identified orthologous pairs, only genes with specific or predominant expression patterns in the testis of both humans and mice were considered to have potentially similar testicular functions between the two species. Correspondingly, genes significantly enriched in the testis of one species but without a one-to-one ortholog or a similar testis-enriched expression pattern in the other species were considered to be genes specifically or predominantly expressed in the testis of only that species.
Results
Identification of deleterious variants of primate-specific SSX1 in infertile men with asthenoteratozoospermia
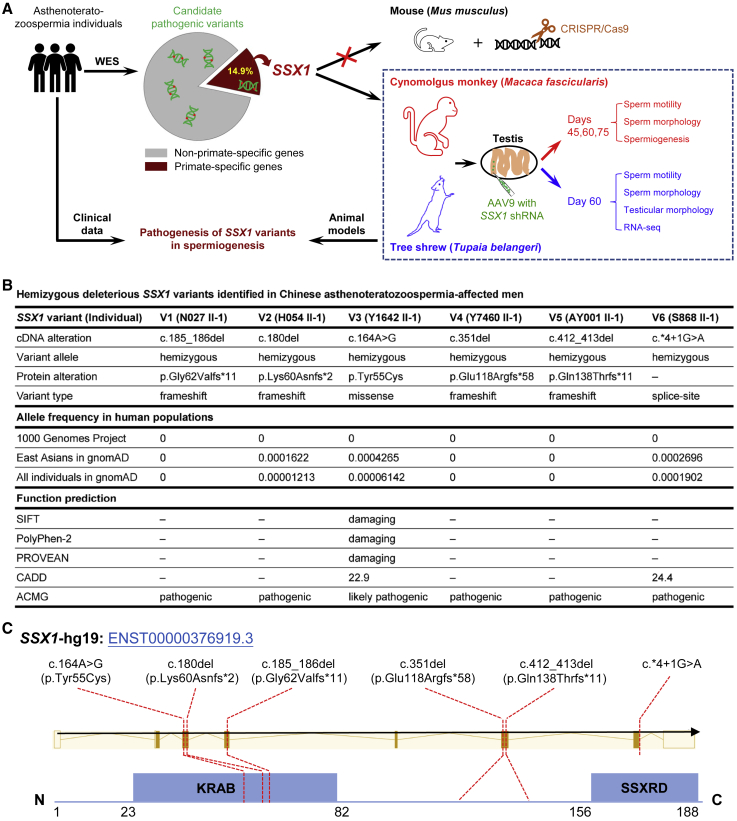
To identify pathogenic genetic variants in asthenoteratozoospermia, we performed high-throughput WES on a cohort of 536 subjects affected by asthenoteratozoospermia (Figure 1A). After stringent bioinformatics analyses,37 we identified six unrelated infertile men harboring hemizygous deleterious variants in SSX1, a PSG that has not been previously associated with any aspects of reproductive physiology in OMIM; these individuals accounted for 1.1% of our cohort. As summarized in Figure 1B, four individuals were found with hemizygous frameshift variants (N027 II-1: c.185_186del [p. Gly62Valfs∗11]; H054 II-1: c.180del [p. Lys60Asnfs∗2]; Y7460 II-1: c.351del [p. Glu118Argfs∗58]; AY001 II-1: c.412_413del [p. Gln138Thrfs∗11]) in SSX1 (GenBank: NM_005635.3). All these variants introduce premature stop codons and are consequently expected to induce nonsense-mediated mRNA decay, which may affect protein synthesis. A hemizygous missense variant of SSX1 (c.164A>G [p. Tyr55Cys]) was identified in the proband of family Y1642, and this variant was predicted by the PolyPhen-2, SIFT, CADD, and PROVEAN tools to be deleterious. Furthermore, we also identified a splice-site variant (c.∗4+1G>A) of SSX1 in subject S868 II-1. According to predictions from the Berkeley Drosophila Genome Project or Splice AI database and real-time qPCR analysis (Figure S3), the c.∗4+1G>A variant abrogates the consensus donor site, leading to alternative splicing and significantly reduced expression of SSX1. All these six variants are absent or extremely rare in the human population genome datasets and predicted as pathogenic or likely pathogenic according to the guidelines from American College of Medical Genetics and Genomics. We performed Sanger sequencing to identify the existence of these variants, and the results are illustrated in Figure S4.
Figure 1.
Identification and functional validation of hemizygous variants in X-linked SSX1 in men with asthenoteratozoospermia
(A) Schematic illustration of the experimental design.
(B) Deleterious hemizygous SSX1 variants identified in six unrelated infertile men affected by asthenoteratozoospermia. The NCBI reference sequence number of SSX1 is GenBank: NM_005635.3. Variants with CADD values greater than 4 were considered to be deleterious. ‒, not applicable.
(C) Locations of the identified SSX1 variants in relation to critical functional domains of SSX1. KRAB, Kruppel-associated box; SSXRD, SSX repression domain.
SSX1 (SSX family member 1; also known as cancer/testis antigen family 5, member 1) is located on the human chromosome X. SSX1 is specifically expressed in the testis and is mainly located in germ cells from spermatogonia to spermatocytes according to the Human Protein Atlas. In addition, SSX1 contains a Kruppel-associated box (KRAB) domain at the N terminus and an SSX repression domain (SSXRD) at the C terminus (Figure 1C), both of which are important for the function of SSX1 as a transcriptional repressor.38 Notably, the SSX1 variants identified in this study were predicted to affect the stability and function of SSX1. Further functional analyses by real-time qPCR and IF also indicated dramatic reduction in expressions of SSX1 mRNA and protein in the spermatozoa of men harboring hemizygous SSX1 variants (Figure S5). Therefore, all these findings suggested that SSX1 is a PSG involved in human spermatogenesis.
SSX1 deficiency caused severe asthenoteratozoospermia phenotypes
Sperm analysis was carried out in source laboratories during routine biological examinations of the individuals according to the World Health Organization (WHO, 2010) guidelines.26 All the men harboring hemizygous deleterious SSX1 variants were shown to have severe-to-complete asthenoteratozoospermia phenotypes (Table 1). Sperm motility and progressive motility were dramatically decreased in these individuals with SSX1 variants (Table 1). The morphology of the sperm cells was assessed by H&E staining and SEM. A variety of abnormal sperm morphologies were observed, including absent, short, coiled flagella and irregular caliber (Figure 2A). The rates of short and coiled flagella in the spermatozoa from men harboring hemizygous SSX1 variants were significantly higher than those from normal reference values (Table 1).
Table 1.
Semen characteristics and sperm flagellar morphology in asthenoteratozoospermia-affected men harboring hemizygous SSX1 variants
| Individual | N027 II-1 | H054 II-1 | Y1642 II-1 | Y7460 II-1 | AY001 II-1 | S868 II-1 | Reference limits |
|---|---|---|---|---|---|---|---|
| Semen parameter | |||||||
| Semen volume (mL) | 4.0 | 3.4 | 2.5 | 3.1 | 2.2 | 4.8 | 1.5a |
| Sperm concentration (106/mL) | 9.5 | 17.7 | 11.6 | 4.9 | 65.0 | 39.8 | 15.0a |
| Motility (%) | 0.0 | 33.8 | 0.0 | 24.0 | 2.0 | 27.2 | 40.0a |
| Progressive motility (%) | 0.0 | 18.2 | 0.0 | 16.0 | 0.5 | 15.4 | 32.0a |
| Sperm flagellar morphology | |||||||
| Absent flagella (%) | 17.5 | – | 9.5 | 9.3 | 2.7 | 2.0 | 5.0b |
| Short flagella (%) | 41.3 | – | 27.5 | 13.5 | 39.8 | 18.0 | 1.0b |
| Coiled flagella (%) | 32.3 | – | 32.3 | 35.3 | 36.3 | 19.8 | 17.0b |
| Angulation (%) | 1.5 | – | 5.8 | 10.8 | 3.3 | 3.3 | 13.0b |
| Irregular caliber (%) | 4.0 | – | 9.3 | 22.0 | 7.0 | 6.3 | 2.0b |
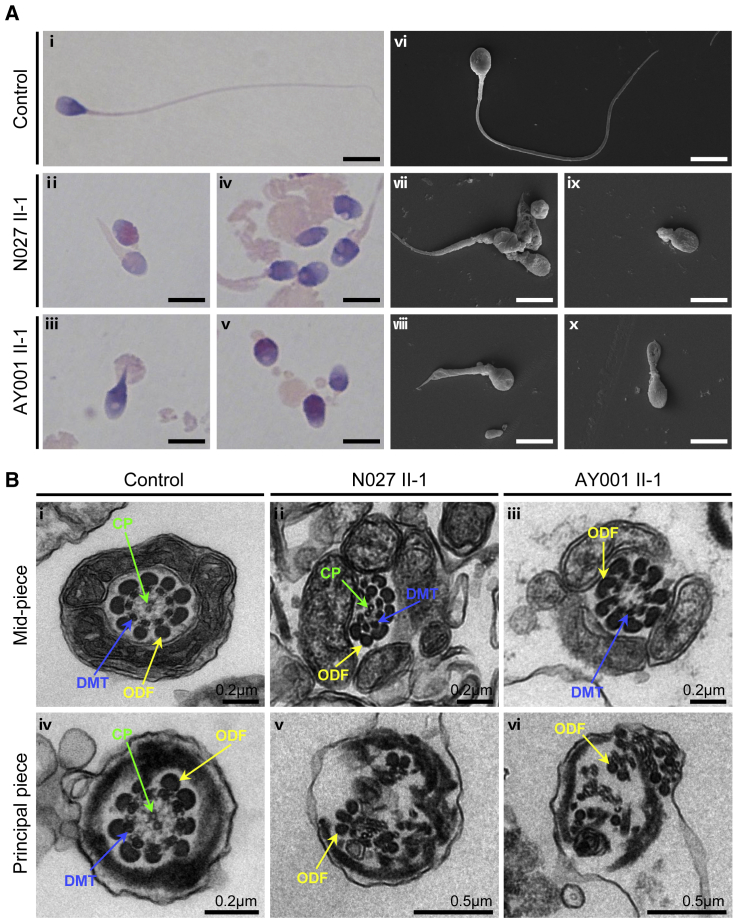
Figure 2.
Morphology and ultrastructure analyses of sperm from men harboring hemizygous SSX1 variants
(A) Morphology analyses of spermatozoa by H&E staining (i–v) and SEM (vi–x). Normal morphology was observed in the spermatozoon from a healthy control male (i and vi). Most spermatozoa from SSX1-deficient men had flagella that were absent (v and ix), short (iv, vii, and viii), coiled (iii and x), or of irregular caliber (iv and v). Scale bars: 5 μm.
(B) TEM analysis of the spermatozoa from a control male individual and men harboring hemizygous SSX1 variants. In the cross-sections of the spermatozoa from a fertile control subject, the typical “9 + 2” microtubule structures were clearly observed (i and iv). Cross-sections of the spermatozoa from men harboring hemizygous SSX1 variants displayed various ultrastructural abnormalities in sperm flagella, including misarranged ODFs (v and vi) and missing DMTs and/or the CP (ii, iii, v, and vi). Abbreviations: CP, central pair of microtubules (green arrows); DMT, peripheral microtubule doublet (blue arrows); ODF, outer dense fiber (yellow arrows).
We further conducted transmission electron microscopy (TEM) to investigate the ultrastructure of the sperm cells from individuals with SSX1 variants. In comparison to the typical “9 + 2” axoneme microtubule structure in the sperm flagella from an unaffected individual, the spermatozoa from men harboring hemizygous SSX1 variants displayed various ultrastructural defects, including the absence or misarrangement of outer dense fibers or microtubules at the midpiece and principal piece of sperm flagella (Figure 2B).
Asthenoteratozoospermia phenotypes were mimicked by KD of Ssx1 expression in the testes of cynomolgus monkeys and tree shrews
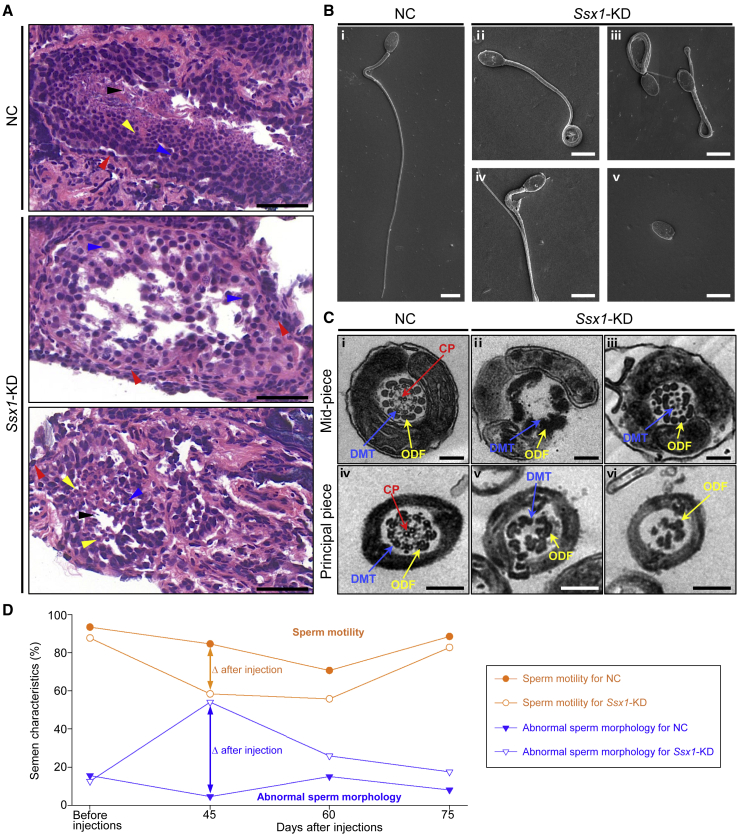
To explore ideal animal models for investigating SSX1, phylogenetic analysis was performed on the basis of protein sequences of SSX family members from mice, tree shrews, cynomolgus monkeys, and humans. As shown in Figure S6, human SSX1 has closer evolutionary distances with cynomolgus monkey SSX1 and tree shrew SSX1 than those of mouse SSX family members. To further investigate the role of primate-specific SSX1 in spermatogenesis, we developed an in vivo KD system based on the microinjection of Ssx1-shRNA-containing, enhanced green fluorescent protein (EGFP)-expressing AAV9 vectors into the seminiferous tubules of the testes of adult male cynomolgus monkeys through the rete testis under ultrasound guidance. Semen samples and testicular biopsy samples were collected before injection and 45, 60, and 75 days after injection. To confirm the KD efficacy of monkey Ssx1 in vivo, we performed a real-time qPCR assay by using testicular biopsy samples, and we observed a dramatically reduced abundance of Ssx1 mRNA in the Ssx1-KD group (Figure S7). H&E staining of testicular sections from the Ssx1-KD male cynomolgus monkeys indicated abnormal spermatogenesis, as shown by abnormal arrangements of germ cells (Figure 3A). Sperm morphology was also analyzed with SEM. Similar to what was observed in men harboring hemizygous SSX1 variants, a higher rate of absent, short, or coiled flagella was observed in the sperm cells from the Ssx1-KD group than that in the NC group (Figure 3B). In addition, TEM observations on spermatozoa from the Ssx1-KD male cynomolgus monkeys also revealed a lack of microtubules or a disorganization of outer dense fibers (Figure 3C). Sperm motility was significantly reduced in the Ssx1-KD group (Figure 3D). Notably, the reduced sperm motility and abnormal sperm morphology were obvious at 45 days after injection but almost recovered at 75 days after injection (Figure 3D).
Figure 3.
Cynomolgus monkey model revealed defective spermatogenesis and abnormal sperm morphologies in the Ssx1-KD testes
(A) H&E staining of testicular tissue sections obtained from male cynomolgus monkeys. Compared to the regular arrangement of various germ cells in the testes from the NC group, spermatogenic defects, including loss or misarranged germ cells, were observed in the testes from Ssx1-KD male cynomolgus monkeys; spermatogonia (red arrowheads), spermatocytes (blue arrowheads), round spermatids (yellow arrowheads), and spermatozoa (black arrowheads). Scale bars: 50 μm.
(B) Abnormal morphologies were also observed in the sperm from Ssx1-KD male cynomolgus monkeys, including coiled (ii, iii), bending (iv), and absent (v) flagella. Scale bars: 5 μm.
(C) Compared to the typical “9 + 2” microtubule structures in the sperm flagella of the NC group (i and iv), Ssx1-KD in the testes also led to various ultrastructural abnormalities in the sperm flagella, including the misarranged or missing ODFs (iii, v, and vi) and the loss of DMTs or CP (ii, iii, v, and vi). Scale bars: 200 nm. Abbreviations: CP, central pair of microtubules (red arrows); DMT, peripheral microtubule doublet (blue arrows); ODF, outer dense fiber (yellow arrows).
(D) Ssx1-KD in the testes of male cynomolgus monkeys led to temporary abnormalities in sperm motility and morphology after injections. The values for the NC group represent three repeated statistical analyses of one cynomolgus monkey, and the values for Ssx1-KD group represent the mean data from three male cynomolgus monkeys. For each cynomolgus monkey, at least 200 spermatozoa were examined to evaluate the rates of morphologically abnormal spermatozoa.
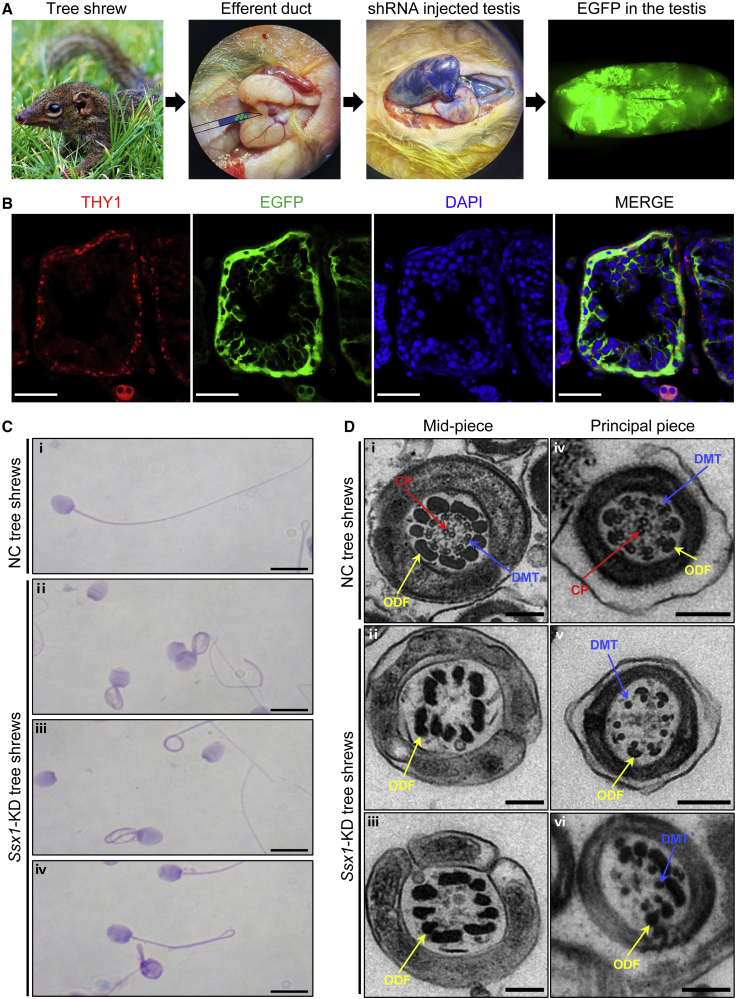
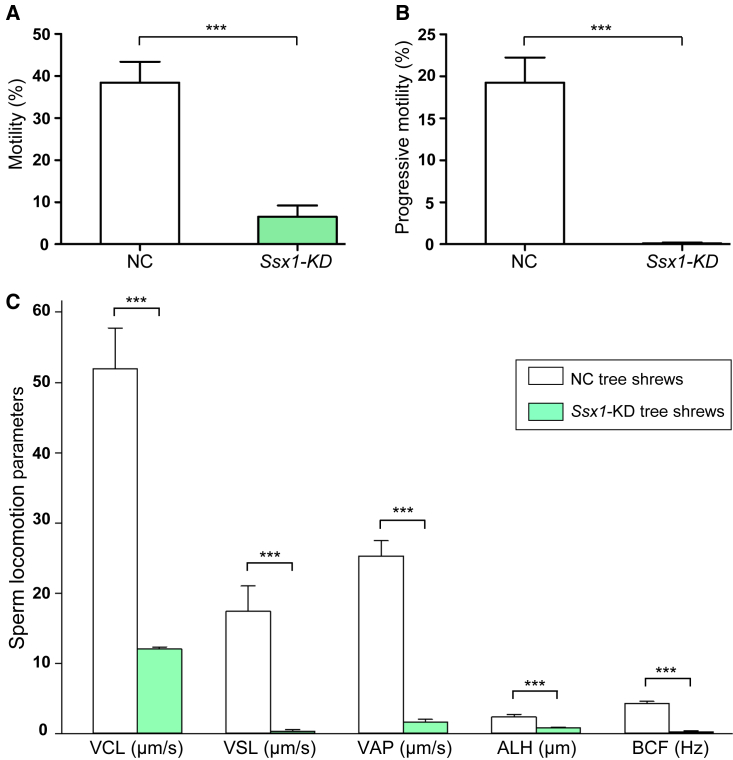
Tree shrews, close relatives of primates, have the advantageous features of small adult body size, short reproductive cycle and life span, and low cost of maintenance40; therefore, tree shrews have been used as a viable alternative animal model to primates in biomedical research.14,41 Ssx1 is also predominantly expressed in the testis of tree shrew according to the Tree shrew database.17 To further confirm the asthenoteratozoospermia phenotypes observed in Ssx1-KD male cynomolgus monkeys, we also constructed an in vivo Ssx1-KD model in tree shrews. Ssx1-shRNA-containing and EGFP-expressing AAV9 vectors were injected into seminiferous tubules via the rete testis. Semen samples and testis tissues were collected at 60 days (after a complete spermatogenic cycle) after injection. As shown in Figure 4A, green seminiferous tubules were clearly visible on the 60th day after injection with the AAV9 vectors. IF staining of testicular tissue sections with an antibody against THY1, which is a marker of spermatogonia, indicated that the AAV9 vectors reached and infected the spermatogonia (Figure 4B). Real-time qPCR assay suggested that the abundance of Ssx1 mRNA was significantly reduced in the testes of the Ssx1-KD group (Figure S8A). Consistently, immunoblotting and immunostaining assays also revealed a dramatically reduced accumulation of SSX1 in the testes of the Ssx1-KD group (Figures S8B and S8C). Testis weight was significantly reduced in the Ssx1-KD group when compared to the NC group (Figure S9). H&E staining of testis sections from the Ssx1-KD male tree shrews revealed abnormal spermatogenesis, including impaired spermatogenesis or the loss of germ cells from seminiferous tubules (Figure S10). H&E staining also revealed various abnormal sperm morphologies, including absent, coiled, or angulated flagella (Figure 4C). Furthermore, TEM observations on the sperm ultrastructure of the Ssx1-KD male tree shrews revealed the absence of peripheral or central microtubules at the midpiece and principal piece of sperm flagella (Figure 4D). Semen characteristics were investigated by a computer-assisted sperm analysis (CASA) system, and significant decreases in sperm motility and progressive motility were observed in the Ssx1-KD group (Figures 5A and 5B). Regarding sperm locomotion parameters, the curvilinear velocity (VCL), straight-line velocity (VSL), average-path velocity (VAP), amplitude of lateral head displacement (ALH), and beat cross frequency (BCF) were significantly lower in the Ssx1-KD group than those in the NC group (Figure 5C).
Figure 4.
Injections of AAV9-Ssx1-shRNA vectors into the testes of male tree shrews and sperm morphological abnormalities in the Ssx1-KD group
(A) AAV9-Ssx1-shRNA vectors or AAV9-NC-shRNA vectors were injected into the seminiferous tubules of tree shrew testes through the rete testis, and Trypan blue was used as a tracer. The signal of enhanced green fluorescent protein (EGFP) expressed by AAV9-Ssx1-shRNA vectors also indicated effective infection of the AAV9-Ssx1-shRNA vectors into the seminiferous tubules.
(B) IF staining of THY1 (a marker of spermatogonia) in testicular tissue sections after injections of AAV9-Ssx1-shRNA vectors or AAV9-NC-shRNA vectors. The colocalization of THY1 and EGFP indicated that the injected AAV9 penetrated the BTB and reached the spermatogonia. DNA was counterstained with DAPI (4′,6-diamidino-2-phenylindole) as a marker of cell nuclei. Scale bars: 50 μm.
(C) H&E staining of spermatozoa from NC and Ssx1-KD male tree shrews. Abnormal sperm morphologies, including absent, short, coiled, or bending flagella, were detected in sperm samples from Ssx1-KD male tree shrews. Scale bars: 20 μm.
(D) TEM analysis of spermatozoa from NC and Ssx1-KD male tree shrews. Consistent with those observed in SSX1-deficient men and Ssx1-KD male cynomolgus monkeys, the ultrastructures of the spermatozoa from Ssx1-KD male tree shrews also displayed misarrangements or loss of DMTs or CP (ii, iii, v, vi). Scale bars: 200 nm. Abbreviations: CP, central pair of microtubules (red arrows); DMT, peripheral microtubule doublet (blue arrows); ODF, outer dense fiber (yellow arrows).
Figure 5.
Semen characteristics of Ssx1-KD male tree shrews
(A–C) Semen characteristics assessed by a CASA system revealed significant reductions in sperm motility (A) and progressive motility (B) in Ssx1-KD male tree shrews when compared with those from NC male tree shrews. The curvilinear velocity (VCL), straight-line velocity (VSL), average-path velocity (VAP), amplitude of lateral head displacement (ALH), and beat cross frequency (BCF) were also dramatically decreased in the Ssx1-KD group (C). For each group, at least five tree shrews were analyzed. The error bars represent the SEM. ∗∗∗p < 0.001.
Abnormal expressions of multiple spermatogenesis-associated factors in the Ssx1-KD model
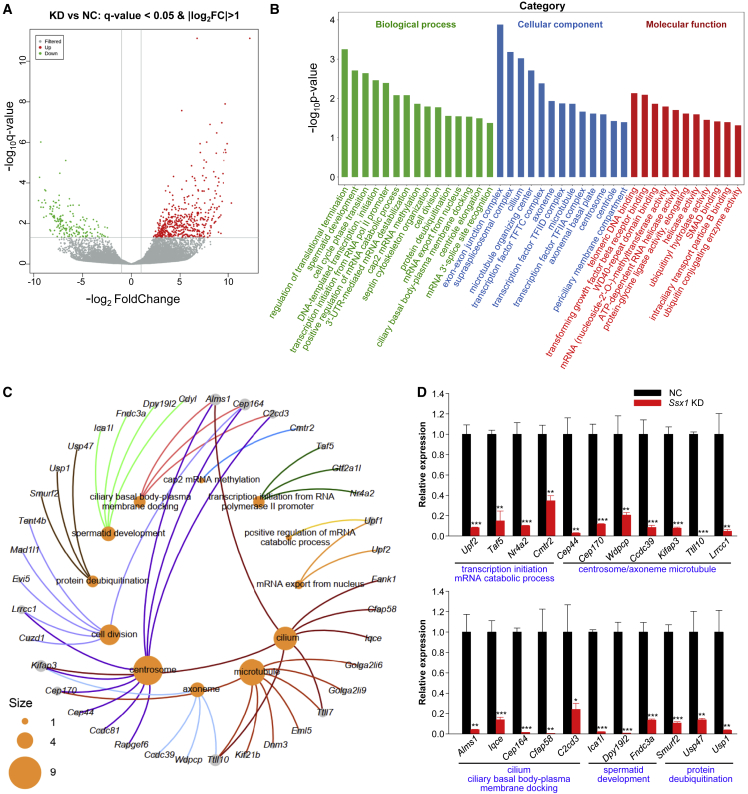
High-throughput RNA-seq was performed with the testicular tissues from Ssx1-KD and NC male tree shrews. A total of 658 genes (484 up-regulated and 174 down-regulated) were differentially expressed (fold-change > 2 or < 0.5 and q < 0.05) in the Ssx1-KD testes compared to the NC testes (Figure 6A). Gene Ontology analysis indicated that these transcriptionally dysregulated genes were significantly enriched in multiple biological processes or components associated with cytogenesis or transcription regulation, the details are shown in Figure 6B. Notably, the downregulated genes included various genes required for spermatogenesis and ciliogenesis, including eight genes involved in “microtubule” (Cep170, Dnm3, Eml5, Golga2li6, Golga2li9, Kif21b, Ttll7, and Ttll10), four genes involved in “spermatid development” (Cdyl, Dpy19l2, Fndc3a, and Ica1l), seven genes involved in “cilium” (Alms1, Cfap58, Fank1, Iqce, Kifap3, Ttll7, and Ttll10), nine genes involved in “centrosome” (Alms1, C2cd3, Ccdc81, Cep44, Cep164, Cep170, Kifap3, Lrrcc1, and Rapgef6), six genes involved in “cell division” (Cep164, Cuzd1, Evi5, Lrrcc1, Mad1l1, and Tent4b), four genes involved in “axoneme” (Ccdc39, Kifap3, Ttll10, and Wdpcp), three genes involved in “ciliary basal body-plasma membrane docking” (Alms1, C2cd3, and Cep164), three genes involved in “protein deubiquitination” (Smurf2, Usp1, and Usp47), and six genes involved in mRNA catabolic process, mRNA export, or transcription initiation (Cmtr2, Gtf2a1L, Nr4a2, Taf5, Upf1, and Upf2) (Figure 6C). We performed real-time qPCR to further verify the significantly decreased expression of these genes (Figure 6D). In addition, Western blotting assay also confirmed the decreased protein levels of multiple selected candidates (Figure S11). Taken together, these findings indicate that Ssx1 deficiency may influence multiple biological processes involved in spermatogenesis.
Figure 6.
Ssx1-KD in the testis alters the expression pattern of genes involved in spermatogenesis
(A) Volcano plot showing the DEGs between NC and Ssx1-KD testes of male tree shrews. A 2-fold difference in expression and q = 0.05 were used as the cutoff.
(B) Gene Ontology (GO) analysis of transcriptionally dysregulated genes.
(C) Network diagram of gene-GO terms revealed that the downregulated genes included various genes required for spermatogenesis or ciliogenesis. The size of the circles represents the number of genes enriched in a particular GO term.
(D) Real-time qPCR validation of a portion of downregulated genes in the testes of Ssx1-KD male tree shrews. The data are presented as the mean ± SE of three independent experiments. Two-tailed Student’s paired or unpaired t tests were used as appropriate (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Damaged male fertility resulting from SSX1 deficiency may be rescued by intra-cytoplasmic sperm injection (ICSI) treatment
ICSI treatment has been suggested as a common clinical therapeutic strategy to circumvent the infertility associated with asthenoteratozoospermia. In this study, five of the six men harboring hemizygous SSX1 variants received assisted reproductive therapy via ICSI. As shown in Table 2, three (H054 II-1, Y7460 II-1, and AY001 II-1) of the five couples achieved a successful pregnancy. For the other two couples (N027 II-1 and S868 II-1), 8-cell embryos or blastocysts were developed and used for embryo transfer while no good ICSI outcome was achieved, which could be partially attributed to older age (e.g., couple S868 II-1), poor quality of the oocytes from the female partners, or other reasons. Overall, ICSI treatment is favorable for the SSX1-associated asthenoteratozoospermia.
Table 2.
Clinical outcomes of ICSI cycles with spermatozoa from men harboring hemizygous SSX1 variants
| Individual | N027 II-1 | H054 II-1 | Y1642 II-1 | Y7460 II-1 | AY001 II-1 | S868 II-1 |
|---|---|---|---|---|---|---|
| Male age (years) | 29 | 26 | 22 | 32 | 31 | 31 |
| Female age (years) | 25 | 28 | 21 | 32 | 29 | 35 |
| Number of ICSI cycle | 1 | 1 | – | 1 | 1 | 1 |
| Number of oocytes injected | 5 | 15 | – | 14 | 28 | 15 |
| Number (and rate) of fertilized oocytes | 5 (100.0%) | 14 (93.0%) | – | 12 (86.0%) | 27 (96.4%) | 14 (93.3%) |
| Number (and rate) of cleavage embryos | 5 (100.0%) | 14 (100.0%) | – | 12 (100.0%) | 26 (96.3%) | 14 (100.0%) |
| Number (and rate) of 8-cells | 4 (80.0%) | 11 (78.6%) | – | 10 (83.3%) | 20 (74.1%) | 12 (85.7%) |
| Number (and rate) of blastocysts | – | 8 (57.1%) | – | 10 (83.3%) | 12 (44.4%) | 9 (64.3%) |
| Number of transfer cycle(s) | 1 | 1 | – | 1 | 1 | 2 |
| Number of embryo(s) transferred per cycle | 2 | 2 | – | 2 | 1 | 1 |
| Implantation rate | 0.0% | 100.0% | – | 50.0% | 100.0% | 0.0% |
| Clinical pregnancy rate | 0.0% | 100.0% | – | 100.0% | 100.0% | 0.0% |
| Miscarriage rate | – | 0.0% | – | 0.0% | 0.0% | – |
‒, not applicable.
Discussion
As new genes that contribute to phenotypic evolution, PSGs, especially X-linked PSGs, are often predominantly or specifically transcribed in the testis.42,43 Among the newly identified PSGs in the human genome, more than 30% of genes are testis enriched.10 Consistently, PSG variants account for 14.9% of all the candidate pathogenic variants identified in the infertile individuals of this study (Figure 1A). Furthermore, we conducted differential expression analysis by using transcriptional data from the Expression Atlas database and revealed 1,477 human testis-enriched genes that lack one-to-one orthologs in mice or whose murine orthologs do not exhibit a significantly testis-enriched expression pattern; these human genes account for 49.7% of all human genes that are specifically or predominantly expressed in the testis (Figure S12). All these findings identified high rates of PSG expressions in the testis, but limited studies have revealed their functions in spermatogenesis.
In the present work, our genetic analyses using WES identified six unrelated infertile men with asthenoteratozoospermia who carried hemizygous variants in SSX1; this gene is PSG preferentially expressed in the testis. Notably, all these SSX1 variants are either rare or absent from human population genome datasets and are predicted to be deleterious variants according to multiple bioinformatic tools. Further analyses indicated that the expression of SSX1 was significantly reduced in the spermatozoa from men harboring hemizygous SSX1 variants. Therefore, all these findings indicate that the phenotypes associated with asthenoteratozoospermia in these individuals are likely to be explained by the hemizygous deleterious variants in primate-specific SSX1.
For studies about PSGs, mouse model is not applicable. To explore the effect of SSX1 deficiency on spermatogenesis, we constructed two Ssx1-KD models by injecting AAV9-Ssx1-shRNA vectors into the seminiferous tubules of the testes of adult male cynomolgus monkeys and tree shrews. Colocalization of green fluorescent protein expressed by AAV9 vectors and THY1 (a marker of spermatogonia) indicated that the AAV9-Ssx1-shRNA vectors penetrated the BTB, reaching and infecting the spermatogonia. Further functional analyses revealed significantly reduced expression of Ssx1 in the testes of Ssx1-KD male cynomolgus monkeys and tree shrews, indicating the successful establishment of the in vivo Ssx1-KD models. Notably, both Ssx1-KD models displayed reduced sperm motility and abnormal sperm morphology, which was consistent with the clinical presentation of men harboring hemizygous deleterious variants in SSX1, further confirming the important role of SSX1 in normal spermatogenesis. In particular, the Ssx1-KD male cynomolgus monkeys displayed obviously reduced sperm motility and abnormal sperm morphologies at 45 days after injection, but this phenotypic abnormality was almost recovered at 75 days after injection. This indicated the flexibility of the AAV-mediated KD system and suggested the possibility of using synthetic biological sequences for highly specific male contraceptives.
As described above, the asthenoteratozoospermia phenotypes associated with SSX1 deficiency were reproduced in Ssx1-KD male cynomolgus monkeys and tree shrews, indicating that AAV9 combined with shRNA-mediated gene expression KD could be a tool for in vivo genetic manipulation of male reproductive systems (especially for the manipulation of genes that are not present in mice or that are evolutionarily differentiated in function). As two model organisms that are close relatives to humans, cynomolgus monkeys and tree shrews had different advantages for research on reproductive diseases via the AAV-shRNA-mediated gene KD system. The cynomolgus monkey model can be used to mimic the steps of semen collection in human, which was convenient for semen parameter analyses at different time points after the gene KD. Moreover, short-term in vivo genetic intervention in adult monkeys significantly shortened the period required for observations on reproductive phenotypes when compared to the previous strategy for germline gene editing; in the latter case, at least 4–5 years are needed for acquiring gene edited offspring and the subsequent appearance of reproductive phenotyping in adults. Furthermore, the efficacy to generate gene-edited monkeys is extremely low so far. Thus, our findings indicated that the monkey model and the AAV-shRNA-mediated gene KD system are suitable for phenotypic analysis and mechanistic studies on human reproductive disorders of genetic origin. For tree shrews, their small adult body size and short reproductive cycle make them easy to collect sperm samples or testis tissues for phenotypic and mechanistic studies. Because of their close proximity to primates, tree shrews with AAV-shRNA-mediated gene expression KD are also ideal models for exploring the molecular mechanisms underlying diseases caused by abnormal gene expressions in human. Therefore, our study, which was based on AAV-shRNA-mediated gene expression KD in two model organisms, provides two experimental systems for research with different purposes.
To gain insights into the molecular mechanisms underlying SSX1-associated asthenoteratozoospermia, we performed high-throughput RNA-seq by using the testicular tissues from Ssx1-KD and NC male tree shrews. Notably, DEGs were enriched in multiple biological processes involved in spermatogenesis, including protein deubiquitination, transcription initiation, and mRNA catabolic processes. For example, C2cd3, which encodes a positive regulator of centriole elongation,44 was found to be expressed at significantly decreased levels in the testes of Ssx1-KD male tree shrews. Furthermore, we also detected a lower abundance of Fndc3a, which encodes a protein required for adhesion between spermatids and Sertoli cells,45 in the Ssx1-KD group than in the NC group. In addition, significantly reduced expressions of two RNA surveillance proteins,46 UPF1 and UPF2, were also observed in the Ssx1-KD group. A previous study reported that the disruption of Upf2 during the early stages of spermatogenesis resulted in the disappearance of nearly all spermatogenic cells through the loss of nonsense-mediated mRNA decay.47 Therefore, all these findings may partially explain the abnormal spermatogenesis observed in the Ssx1-KD group.
ICSI, which is a kind of assisted reproduction technique, has been an effective method for treating male infertility caused by asthenoteratozoospermia because it overcomes the fertilization failure caused by decreased sperm vitality or abnormal sperm morphologies. Previous studies and our recent studies have also revealed good prognosis of ICSI treatment for a series of asthenoteratozoospermia-related genes, including DNAH1, DNAH8, CFAP47, and TTC29.19,37,48,49 In this study, five of the six men harboring hemizygous SSX1 variants received ICSI treatment with their own sperm, and three of the five couples achieved a successful pregnancy. For the other two couples (N027 II-1 and S868 II-1), 8-cell embryos or blastocysts were also developed. Therefore, ICSI treatment is promising and can also be recommended for the individuals with SSX1-associated asthenoteratozoospermia.
In summary, our study identified primate-specific SSX1 as a genetic factor for human male infertility with asthenoteratozoospermia. ICSI treatment is favorable for the SSX1-associated asthenoteratozoospermia. In addition to the PSG SSX1, a high proportion of genes whose expression is testis-enriched but are not evolutionarily conserved between humans and mice were also identified. Specifically, this study established the AAV9-shRNA-mediated gene expression KD system based on the cynomolgus monkey and tree shrew models, which provide a promising strategy for in vivo spermatogenic studies of essential factors that cannot be achieved via the murine models. Overall, combining clinical studies, human genetics, in vivo genetic manipulation, and high-resolution transcriptome analysis, this study provides an approach that might help to further understand the genetic causes of male infertility and develop contraception or personalized therapies for reproductive defects.
Acknowledgments
We would like to thank the families for participating and supporting this study and the Center of Cryo-electron Microscopy at Zhejiang University for technical support. This study was supported by the National Key Research and Development Program of China (2021YFC2701400, 2018YFA0801400, 2018YFC1004901, and 2021YFC2700901), the National Natural Science Foundation of China (32288101, 32100480, 81971447, 82171608, 82101961, 81971441, and 82171607), the Scientific Research (TP202002) from Anhui Medical University, the China Postdoctoral Science Foundation (2020TQ0072), Yunnan Major Scientific and Technological Project (202202AG050018), the Chinese Academy of Sciences (Light of West China Program xbzg-zdsys-201909), and the Key Grant of Prevention and Treatment of Birth Defect from Hunan Province (2019SK1012).
Declaration of interests
The authors declare no competing interests.
Published: February 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.01.016.
Contributor Information
Yunxia Cao, Email: caoyunxia6@126.com.
Feng Zhang, Email: zhangfeng@fudan.edu.cn.
Web resources
1000 Genomes Project, http://www.internationalgenome.org
Berkeley Drosophila Genome Project, https://www.fruitfly.org/
Ensembl, http://www.ensembl.org/index.html
Expression Atlas, https://www.ebi.ac.uk/gxa/home
Human Protein Atlas, https://www.proteinatlas.org
National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/
NNSPLICE 0.9, http://www.fruitfly.org/seq_tools/splice.html
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Tree shrew Database, http://www.treeshrewdb.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental information
References
- 1.Inhorn M.C., Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update. 2015;21:411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 2.Cavallini G. Male idiopathic oligoasthenoteratozoospermia. Asian J. Androl. 2006;8:143–157. doi: 10.1111/j.1745-7262.2006.00123.x. [DOI] [PubMed] [Google Scholar]
- 3.Touré A., Martinez G., Kherraf Z.E., Cazin C., Beurois J., Arnoult C., Ray P.F., Coutton C. The genetic architecture of morphological abnormalities of the sperm tail. Hum. Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 4.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S., et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang S., Wang X., Li W., Yang X., Li Z., Liu W., Li C., Zhu Z., Wang L., Wang J., et al. Biallelic Mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kherraf Z.E., Conne B., Amiri-Yekta A., Kent M.C., Coutton C., Escoffier J., Nef S., Arnoult C., Ray P.F. Creation of knock out and knock in mice by CRISPR/Cas9 to validate candidate genes for human male infertility, interest, difficulties and feasibility. Mol. Cell. Endocrinol. 2018;468:70–80. doi: 10.1016/j.mce.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Young S.A.M., Aitken R.J., Ikawa M. Advantages of using the CRISPR/Cas9 system of genome editing to investigate male reproductive mechanisms using mouse models. Asian J. Androl. 2015;17:623–627. doi: 10.4103/1008-682X.153851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiyozumi D., Noda T., Yamaguchi R., Tobita T., Matsumura T., Shimada K., Kodani M., Kohda T., Fujihara Y., Ozawa M., et al. NELL2-mediated lumicrine signaling through OVCH2 is required for male fertility. Science. 2020;368:1132–1135. doi: 10.1126/science.aay5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan S., Liu Y., Peng H., Tang C., Hennig G.W., Wang Z., Wang L., Yu T., Klukovich R., Zhang Y., et al. Motile cilia of the male reproductive system require miR-34/miR-449 for development and function to generate luminal turbulence. Proc. Natl. Acad. Sci. USA. 2019;116:3584–3593. doi: 10.1073/pnas.1817018116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao Y., Chen C., Shen H., He B.Z., Yu D., Jiang S., Zhao S., Gao Z., Zhu Z., Chen X., et al. GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Res. 2019;29:682–696. doi: 10.1101/gr.238733.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller J.L., Skaletsky H., Brown L.G., Zaghlul S., Rock S., Graves T., Auger K., Warren W.C., Wilson R.K., Page D.C. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vockel M., Riera-Escamilla A., Tüttelmann F., Krausz C. The X chromosome and male infertility. Hum. Genet. 2021;140:203–215. doi: 10.1007/s00439-019-02101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Niu Y., Ji W. Transgenic nonhuman primate models for human diseases: approaches and contributing factors. J. Genet. Genom. 2012;39:247–251. doi: 10.1016/j.jgg.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Fan Y., Huang Z.Y., Cao C.C., Chen C.S., Chen Y.X., Fan D.D., He J., Hou H.L., Hu L., Hu X.T., et al. Genome of the Chinese tree shrew. Nat. Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 15.Rockland K.S., Lund J.S. Widespread periodic intrinsic connections in the tree shrew visual cortex. Science. 1982;215:1532–1534. doi: 10.1126/science.7063863. [DOI] [PubMed] [Google Scholar]
- 16.Fan Y., Ye M.S., Zhang J.Y., Xu L., Yu D.D., Gu T.L., Yao Y.L., Chen J.Q., Lv L.B., Zheng P., et al. Chromosomal level assembly and population sequencing of the Chinese tree shrew genome. Zool. Res. 2019;40:506–521. doi: 10.24272/j.issn.2095-8137.2019.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan Y., Yu D., Yao Y.G. Tree shrew database (TreeshrewDB): a genomic knowledge base for the Chinese tree shrew. Sci. Rep. 2014;4:7145. doi: 10.1038/srep07145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye M.S., Zhang J.Y., Yu D.D., Xu M., Xu L., Lv L.B., Zhu Q.Y., Fan Y., Yao Y.G. Comprehensive annotation of the Chinese tree shrew genome by large-scale RNA sequencing and long-read isoform sequencing. Zool. Res. 2021;42:692–709. doi: 10.24272/j.issn.2095-8137.2021.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C., Tu C., Wang L., Wu H., Houston B.J., Mastrorosa F.K., Zhang W., Shen Y., Wang J., Tian S., et al. Deleterious variants in X-linked CFAP47 induce asthenoteratozoospermia and primary male infertility. Am. J. Hum. Genet. 2021;108:309–323. doi: 10.1016/j.ajhg.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 24.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 26.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W.G., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe S., Kanatsu-Shinohara M., Ogonuki N., Matoba S., Ogura A., Shinohara T. In vivo genetic manipulation of spermatogonial stem cells and their microenvironment by adeno-associated viruses. Stem Cell Rep. 2018;10:1551–1564. doi: 10.1016/j.stemcr.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould K.G., Mann D.R. Comparison of electrostimulation methods for semen recovery in the rhesus monkey (Macaca mulatta) J. Med. Primatol. 1988;17:95–103. [PubMed] [Google Scholar]
- 29.National Research Council (U.S.). Committee for the Update of the Guide for the Care and Use of Laboratory Animals., Institute for Laboratory Animal Research (U.S.), and National Academies Press (U.S.). (2011). Guide for the Care and Use of Laboratory Animals. In. (Washington, D.C., National Academies Press)xxv, 220 p.
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., Robinson M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 35.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C., He X., Liu W., Yang S., Wang L., Li W., Wu H., Tang S., Ni X., Wang J., et al. Bi-allelic mutations in TTC29 cause male subfertility with asthenoteratospermia in humans and mice. Am. J. Hum. Genet. 2019;105:1168–1181. doi: 10.1016/j.ajhg.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auger J., Jouannet P., Eustache F. Another look at human sperm morphology. Hum. Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
- 40.Li C.H., Yan L.Z., Ban W.Z., Tu Q., Wu Y., Wang L., Bi R., Ji S., Ma Y.H., Nie W.H., et al. Long-term propagation of tree shrew spermatogonial stem cells in culture and successful generation of transgenic offspring. Cell Res. 2017;27:241–252. doi: 10.1038/cr.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J., Yang E.B., Su J.J., Li Y., Chow P. The tree shrews: adjuncts and alternatives to primates as models for biomedical research. J. Med. Primatol. 2003;32:123–130. doi: 10.1034/j.1600-0684.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y.E., Vibranovski M.D., Landback P., Marais G.A.B., Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinckenbosch N., Dupanloup I., Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thauvin-Robinet C., Lee J.S., Lopez E., Herranz-Pérez V., Shida T., Franco B., Jego L., Ye F., Pasquier L., Loget P., et al. The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat. Genet. 2014;46:905–911. doi: 10.1038/ng.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obholz K.L., Akopyan A., Waymire K.G., MacGregor G.R. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev. Biol. 2006;298:498–513. doi: 10.1016/j.ydbio.2006.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao S., Amorim R., Niu M., Temzi A., Mouland A.J. The RNA surveillance proteins UPF1, UPF2 and SMG6 affect HIV-1 reactivation at a post-transcriptional level. Retrovirology. 2018;15:42. doi: 10.1186/s12977-018-0425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald C.C., Grozdanov P.N. Nonsense in the testis: multiple roles for nonsense-mediated decay revealed in male reproduction. Biol. Reprod. 2017;96:939–947. doi: 10.1093/biolre/iox033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wambergue C., Zouari R., Fourati Ben Mustapha S., Martinez G., Devillard F., Hennebicq S., Satre V., Brouillet S., Halouani L., Marrakchi O., et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016;31:1164–1172. doi: 10.1093/humrep/dew083. [DOI] [PubMed] [Google Scholar]
- 49.Liu C., Miyata H., Gao Y., Sha Y., Tang S., Xu Z., Whitfield M., Patrat C., Wu H., Dulioust E., et al. Bi-allelic DNAH8 variants lead to multiple morphological abnormalities of the sperm flagella and primary male infertility. Am. J. Hum. Genet. 2020;107:330–341. doi: 10.1016/j.ajhg.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.