Test Submission
How to register a clinical test in the GTR
OMB NO: 0925-0651
EXPIRATION DATE: 01/31/2025
Instructions on this page assume you have logged into the GTR submission site, have registered your laboratory, and are now ready to submit information about a clinical genetic test.
On the home page for your GTR submissions, under 'Tests in this lab', click on 'Submit tests'. To submit a test using the submission wizard, go to the section Submit a single test. The default selection is for a clinical genetic test. Click 'Add a new test'.
You will be presented with a sequence of pages in which to submit data. Each page has a distinct tab, as seen in the image below.

At the bottom of each page is the button Save & Continue. If your entries on that page validate, you will be taken to the next tab. If there is ever a problem with how you completed a value, you will remain on the same tab. Information will be provided to indicate what content needs to be corrected, and the type of error. If at any time you are unsure as to what information to provide in a field you can hover over the help icon (![]() ) to display help information, or review the help page for the tab you are on.
) to display help information, or review the help page for the tab you are on.
Your progress through the submission process is marked as follows. If you have not 'touched' a tab yet, text and tab color remain grey. The tab you are currently processing is shown enlarged. If you have touched a tab (blue text), you can go back to it at any time to update information (Remember click on Save & Continue before leaving the page.). So in the example above, the submitter is currently on Ordering.
The table below summarizes the content included under each tab. The names in the left column are linked to the details about how to submit the data elements in that section.
| Basics | Type of test, names licenses, etc. |
| Ordering | How to order |
| Indication | Condition(s) for which the test is appropriate |
| Methodology | Describe the methods used and what is measured (the test targets) |
| Interpretation | Describe how test results are interpreted |
| Performance | Where the test is performed, clinical validity, clinical utility, and more |
| Review & Submit | Review your data and submit |
How to submit information in the Basics section of the submission form for clinical tests
This page is organized according to the sections you see on the Basics tab. You can click on the name of the section below to navigate to that section of this document.
Please note that most sections in the form provide tool tips, accessible by this icon (![]() ), that provide hints specific to that area.
), that provide hints specific to that area.
Test Information
This test is for: Clinical (minimal). Laboratories in the United States are required to have a current CLIA license in order to provide clinical tests.
If you mistakenly initiate a submission for a research test using the clinical test forms, please discontinue and return to the homepage by clicking the 'GTR Submission' link at the top left of your browser. To submit research tests, see the documentation here.
Laboratory test name (minimal). Enter the name of the test as you want it to be publicly displayed in the GTR. This name will be searchable by users. Examples include: OtoChipTM, CHD7 gene sequencing in CHARGE syndrome.
Short test name (optional). The short test name is the shortened name or mneumonic used by the lab to identify the test. This may be a test name used in conjunction with a test order code for quick test menus, electronic medical records or HL7 systems and programs. Examples include: CHD7Full.
Manufacturer's test name, if any (optional). Enter the manufacturer's name of the commerical test or kit used if in the submitted assay. If the test is an entirely laboratory developed test, there should not be a manufacturer's test name.
Search terms, if any (optional). Enter any search terms you wish to be associated with your submitted test. You do not need to enter keywords such as the disease/condition, gene, variant or test name. These will all automatically be included in a search. Relevant terms may include keywords such as archived or previously used test names. Limit to one search term per entry box. Use the 'add another search term' button to add additional entries.
Purpose of the test (minimal). Select the appropriate test purpose(s) or indication(s) from the choices provided. Multiple selections are allowed.
Diagnosis - Identification or confirmation of disease
Drug response - Evaluation of sequence variation influencing an individual’s reaction to specific medications, as in a pharmacogenetic test. Includes assessment of risk for adverse drug reactions.
Monitoring - Periodic or continuous evaluation of a disease or condition over time, including a patient’s response to medical treatment.
Mutation confirmation - Re-evaluation of a genetic test result to assess the validity of the initial result. For example, research test results or results from another laboratory.
Pre-implantation genetic diagnosis - Genetic testing performed on a small number of cells from a human embryo prior to uterine implantation as part of assisted reproduction procedures.
Pre-symptomatic - Genetic analysis of an asymptomatic or unaffected individual who is at risk of a specific genetic disorder. Applicable to unaffected individuals with a family history of a known (or apparent) high-penetrance single-gene disorder or a pathogenic variant identified in a blood relative. Examples: a family history of Huntington disease; a known pathogenic
Risk assessment - Evaluation of the likelihood of developing a specific condition based on genetic risk. Includes tests that evaluate moderate- to low-penetrance genetic factors (possibly identified through genome-wide association studies), which may be used in combination with other risk factors (e.g., age, gender, environmental factors). Example: testing for pathogenic CFH and ARMS2 variants to predict risk of developing age-related macular degeneration. Also includes carrier testing in affected families. Example: CFTR testing in a male whose pregnant partner is a carrier of a pathogenic CFTR variant.
Screening - Evaluation of a target population to identify a subgroup affected by a genetic condition or that have the potential to transmit the trait to their offspring. Includes newborn screening, ethnicity-based screening and pre-conceptual genetic testing.
Prognostic - Information from the test can be used to determine or predict the aggressiveness of the disease or overall outcome of the disease at the time of initial diagnosis and prior to initiation of treatment. Applicable to prognostic biomarkers used in oncology.
Predictive - Information from the test can be used to determine or predict the potential risk of eventually developing a disease or a disorder. Applicable to predictive biomarkers used in oncology, used to predict treatment response.
Recurrence - Used to detect disease recurrence in a patient who has already been diagnosed and treated for cancer.
Therapeutic management - Information can be used to determine therapeutic decision making.
Target population for this test (recommended). Describe which segment(s) of the population should be tested for the indication and why. Example: Individuals with clinical features consistent with Noonan syndrome.
Citations for target population (recommended). Enter references supporting statements provided in 'Target population for this test'.
Test development (recommended). Select a single option for how the test was developed. Intended to help delineate the differences in certification requirements between the test types. Do not include reflex tests here. Options include:
FDA-reviewed (has FDA test name) - The test, test kit or reagents either have FDA approval or are currently in FDA review.
Manufactured (research use only; not FDA-reviewed) - The test or primary parts of the test are purchased from a commercial entity and have not gone through FDA review.
Modified FDA (has FDA-reviewed entry, but with lab modifications/field changes) - The test, test kit or reagents used in the test have undergone FDA review and are approved, however the submitting laboratory has augmented some part of the test or testing protocol.
Test developed by laboratory (no manufacturer test name) - This test is developed, produced and performed by the submitting laboratory. Also known as LDT.
FDA Review Information
Has there been FDA review of the test? (optional). Select the option (Y/N) corresponding to if the test or any portion of the test including reagents has been reviewed or is pending review by the FDA.
Item reviewed (optional). This field is shown only when 'yes' is selected from the 'Has there been FDA review of this test?'. From the options indicated, select the portion of the test that has been reviewed or is pending review by the FDA. Choices include:
ASR - Analyte-specific reagents
Assay(s)
IVDMIA - In Vitro Diagnostic Multivariate Assay(s)
Instrument(s)
Test kit(s)
Other - Please specify
FDA category designation (optional). Options depend on the selection (Yes or No) for the question 'Has there been FDA review of this test?'.
When the selection is 'No', options include:
FDA exercises enforcement discretion
Not applicable
When the selection is 'Yes', options include:
IUO - Investigational Use Only - The performance characteristics of this product have not been established.
IVD - In Vitro Device
RUO - Research Use Only - Not for use in diagnostic procedures.
FDA regulatory status (optional). This field is shown only when 'Yes' is selected from the 'Has there been FDA review of this test?'. From the options provided, select the status of the item reviewed. Choices include:
FDA cleared/approved
FDA exempt
Pending
Not submitted
Other - Please specify.
FDA application # (optional). This field is shown only when 'yes' is selected from the 'Has there been FDA review of this test?'. Enter the FDA application number associated with the item reviewed. A link to the FDA website will be automatically provided to display documents for the application # provided in this section. Users of the GTR website will be allowed to access this link.
Upload FDA approval document (optional). This field is shown only when 'yes' is selected from the 'Has there been FDA review of this test?'. Upload the final FDA approval summary document. A link to the FDA website will be automatically provided to display documents for the application # provided in this section. Users of the GTR website will be allowed to access this link.
New York State CLEP (NYS CLEP) test approval #
Status (minimal if test holds NYS certification, otherwise optional). If the test has been reviewed by 'NYS CLEP', enter the status of the NYS CLEP approval for the test. Options include:
Approved
Pending
Grandfathered
Exempt
Test approval # (minimal if test holds NYS certification, otherwise optional). Enter the test approval number provided by NYS CLEP for the approved test.
Save your work
Don't forget to save your work by clicking the Save & Continue button at the bottom of the page! If you need to interrupt or delay data entry, you can return at a later time and you will be taken to the last tab you saved.
If any required fields or data inconsistencies are detected on this page, an error message will display after you click Save & Continue indicating what the issue is. The field(s) that need attention will be outlined in red. Please correct your data and click "Save & Continue" again.
How to submit information in the Ordering section of the submission form
This page is organized according to the sections you see on the Ordering tab. You may click on the name of the section below to navigate to that section of this document.
Please note that most sections in the form provide tool tips, accessible by this icon (![]() ), that provide hints specific to that area.
), that provide hints specific to that area.
Ordering information
Each test is a specific, orderable test from a particular laboratory, and each receives a unique GTR accession number. The same or similar test performed by different laboratories is assigned a different accession number. Thus, a laboratory is free to define an orderable test exactly as they represent it in their test catalog.
Test order code (recommended). Enter the test order code, commonly found on the laboratory's requisition form, test menu or test catalog. This is the code health care providers (for example) would use to order the test through the laboratory.
URL of the lab website with information about this test (recommended). Enter the URL for test-specific information on your laboratory's website. URLs should be entered with the appropriate prefix, i.e., http:// or https://.
URL of the lab website with information about how to order this test (recommended). Enter the URL for information on how to order the test. URLs should be entered with the appropriate prefix, i.e., http:// or https://.
How to order (recommended). Enter a description of the test ordering procedure, such as required forms, specimen handling/shipping instructions, etc. Requirements and recommendations about informed consent and genetic counseling can be discussed here. If applicable, a statement similar to the following can be used: “The laboratory recommends that pre- and post-test genetic counseling be offered and informed consent be obtained for this test but does not require documentation to accept and process the sample.”
URL to lab website with information about codes related to this test (recommended). Enter the URL for information on corresponding codes related to the test, including CPT, ICD-9, ICD-10 and HCPCS codes. URLs should be entered with the appropriate prefix, i.e., http:// or https://.
LOINC code(s) (optional). The LOINC (Logical Observation Identifiers Names and Codes) database provides a set of universal names and ID codes for identifying laboratory and clinical test results. Enter the LOINC code. If applicable, the LOINC code can be found by using the 'Search' link, which will take you to the official LOINC website. The LOINC code must be entered manually, even if the submitter searches for the code using the link provided.
Who can order this test? (optional). Identify who can order the test based on the laboratory's policy on how the test can be ordered. Multiple selections can be made.
Choose from the following options:
Genetic counselor
Health care provider
In-state patients
Licensed dentist
Licensed physician
Nurse practitioner
Out-of-state patients
Physician assistant
Public health mandate
Registered nurse
To see the complete, current list of choices for this field, click here.
Ordering requirements
Information in this section conveys whether or not a sample may be accepted and processed based on requirements for informed consent and pre- and post-testing genetic counseling. If specific laboratory forms relating to these types of requirements must accompany the sample, instructions and URLs to relevant forms can be given in the ‘How to order’ text box above. Submitters who want to convey that genetic counseling is recommended can place a statement to that effect in the ‘How to order’ text box.
Informed consent (recommended). Identify if the laboratory requires informed consent prior to performing the test. Select from the following choices: Decline to answer, required, not required, as required by state law. If not required but recommended, you can comment in the 'How to order' text box above.
Pre-test genetic counseling (recommended). Identify if the laboratory requires genetic counseling prior to performing the test. Select from the following choices: Decline to answer, required, not required. If not required but recommended, you can comment in the 'How to order' text box above.
Post-test genetic counseling (recommended). Identify if the laboratory requires genetic counseling after performing the test in order to release test results. Select from the following choices: Decline to answer, required, not required. If not required but recommended, you can comment in the 'How to order' text box above.
Test-specific laboratory services (optional). If applicable, select the laboratory services that are offered in conjunction with the specific test. Multiple selections are allowed. Please select each test service only once (you can provide multiple order codes for one service) in the order code field. This field is different from 'Laboratory services' which allows submitters to select general services the laboratory provides.
Choose from the following options:
Clinical testing/Confirmation of Mutations Identified Previously
Confirmation of Research Findings
Custom Balanced Chromosome Rearrangement Studies
Custom Deletion/Duplication Testing
Custom Sequence Analysis
Data Storage and Backup
Genetic Counseling
Identity Testing
Marker Chromosome Identification
Preimplantation Genetic Diagnosis (PGD)
Result Interpretation
Specimen Source Identification
Uniparental Disomy (UPD) Testing
X-Chromosome Inactivation Studies
Order code (optional). If applicable, enter the order code for the test-specific service, commonly found on the laboratory's requisition form, test menu or test catalog. This is the code health care providers (for example) would use to order the service through the laboratory.
Comment (optional). Enter further information to assist health care providers in ordering the test-specific services. An example includes, 'Pre-test and post-test genetic counseling is available for this test'.
Test-specific additional laboratory services (optional). If applicable, select the additional laboratory services that are offered in conjunction with the specific test. Multiple selections are allowed. Please select each test service only once (you can provide multiple order codes for one service) in the order code field. This field is different from 'Laboratory services' which allows submitters to select general services the laboratory provides. Options include: Custom prenatal testing and Custom mutation-specific/Carrier testing.
Order code (optional). If applicable, enter the order code for the test-specific additional service, commonly found on the laboratory's requisition form, test menu or test catalog. This is the code health care providers (for example) would use to order the service through the laboratory.
Comment (optional). Enter further information to assist health care providers in ordering the additional test-specific services. An example includes, 'Once a mutation is identified, prenatal testing is available'.
Specimen source (recommended). Select the specimen type(s) allowed for the test. Multiple selections allowed. Choices include: Amniocytes, Amniotic fluid, Bone marrow, Buccal swab, Cell culture, Chorionic villi, Cord blood, Cystic hygroma fluid, Dried blood spot (DBS) card, Fetal blood, Fibroblasts, Fresh tissue, Frozen tissue, Paraffin block, Peripheral (whole) blood, Product of conception (POC), Saliva, Serum, Skin, Sputum, Urine, White blood cell prep, Other: please specify.
Specimen source URL (recommended). Enter the URL to the laboratory's website that describes the specimen requirements and handling for the test. URLs should be entered with the appropriate prefix, i.e., http:// or https://.
Testing strategy (recommended). Describe the suggested sequence of ordering tests, discuss reflex testing, testing algorithms and related issues. This field is for recommendations on how to order the test in sequence of relevance to the patient (target population) being tested. This field should not include discussion of methodologies or test procedural protocols. If describing reflex tests, each test component should be delineated. If a test is ordered, additional tests may be performed as necessary under certain circumstances based on initial results and that should be described in this field.
Citations for testing strategy (recommended). Enter references and/or URLs as appropriate to further describe the testing strategy/algorithm. If entering an URL, please specify the appropriate prefix, http:// or https://.
Test-specific contact information
Contact person (recommended). From the personnel listed, select the test-specific contact for the test. This individual's name and contact information will be displayed on the test page for health care providers to view. If no test-specific contact is provided, the laboratory's general contact information will display.
Note: If you do not see the staff member's name you want to select in the personnel list, you will need to add the staff member in the laboratory section. If the staff member is entered on the laboratory personnel list, but does not display in the test-specific contact list, check the staff member's privacy settings by reviewing the selection 'Should this person's information be displayed on the GTR public site?'. If 'No' is selected, the staff member will not display in the test-specific contact list, nor on the public website. If changes to personnel are needed, please save your work on the test and then proceed to editing your laboratory information in order to add new or edit existing staff members. Once submitting the laboratory changes, the newly entered staff information should display in the test contact person field.
Deleting a staff member from the personnel list in the laboratory section will remove that staff member from any tests they are a test-specific contact for. If a staff member has 'No' selected for 'Should this person's information be displayed on the GTR public site?', they will not display as test-specific contacts, even if 'No' is selected after indicating they are a test-specific contact.
Contact policy (recommended). Select the policy of the laboratory regarding who (patients or health care providers) and when (pre-test/post-test/anytime) can contact the lab for this test. Options include:
-
Laboratory can only accept contact from health care providers. Patients/families are encouraged to discuss genetic testing options with their health care provider.
-
Post-test email/phone consultation regarding genetic test results and interpretation is provided to patients/families.
-
Pre-test email/phone consultation regarding genetic test results and interpretation is provided to patients/families.
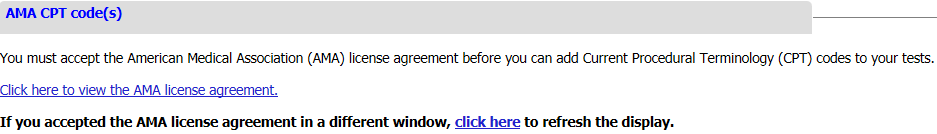
AMA CPT Code(s)
AMA CPT MoPath code(s) (optional). The set of molecular pathology Current Procedural Terminology (CPT®) codes - 'MoPath' codes - from the American Medical Association (AMA) are available for selection into the test record. The MoPath CPT codes were developed to represent nucleic acid analyses to detect germline or somatic variation or for HLA Class I and Class II typing, all of which are in scope for GTR. Tier 1, Tier 2, and 'Unlisted' codes are available in GTR to submitters who accept the AMA license agreement. The so-called 'stacking codes' have been deleted from the AMA CPT codebook.
NOTE: NLM does not have a license to represent other CPT codes, only the MoPath (Molecular Pathology subset) codes. MoPath codes have the format '81XXX'. Please do not submit other CPT codes via spreadsheet or validation errors will result.
In order to view code descriptors and submit AMA CPT codes, the AMA license agreement must be accepted (once per submitter). If you have not yet accepted the agreement, you will see a link 'Click here to view the AMA license agreement', click on the link to review and accept the agreement as shown below. (If your laboratory has a group set up for GTR submissions, other submitters in your group will need to accept the AMA license agreement in order to edit CPT codes.)
Once you have accepted the AMA license agreement, the link "Add AMA CPT Code" will be enabled for you to submit the codes that are applicable to the test.
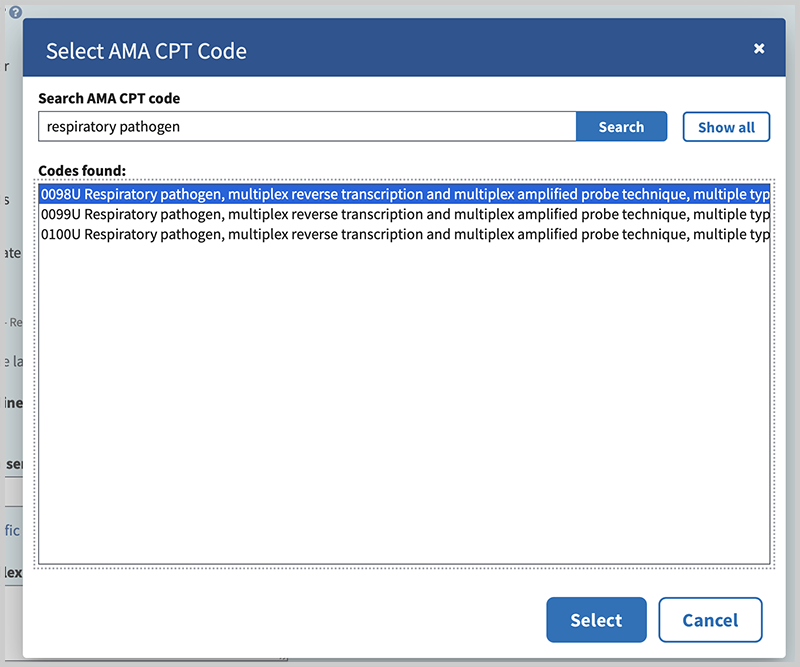
To find and select codes, you can search for any text in the descriptor such as code number, gene symbol or gene name, condition name, common names of variants, or test methods. Use the Enter key or click the Search button. To see all available AMA CPT MoPath code choices, click 'Show all'.
Highlight the appropriate code from the codes found by your search and click "See more". This will display the full text summary description of the specific code. Determine whether the code correctly represents your test; if so then click the "Select" button.
You can enter as many codes as are applicable to you test by clicking the link "Add AMA CPT code". The codes you select will display on the 'How to Order' tab of your test and will be hyperlinked to the AMA descriptor for that code.
Save your work
Don't forget to save your work by clicking the Save & Continue button at the bottom of the page! If you need to interrupt or delay data entry, you can return at a later time and you will be taken to the last tab you saved.
If any required fields or data inconsistencies are detected on this page, an error message will display after you click Save & Continue indicating what the issue is. The field(s) that need attention will be outlined in red. Please correct your data and click "Save & Continue" again.
How to submit information in the Indication section of the submission form for clinical tests
Strategy
To make your test more discoverable, we strongly recommend that you describe any and all of the conditions your test evaluates. GTR users search for tests most frequently by searching for conditions/phenotypes. By enumerating these thoroughly in your submission, you will maximize the chances that your test will appear in a query result.
Organization of this page
This page has two views. The first view (Indication summary status) allows you to specify one or more indications for the test. The second view (Condition/Phenotype Information) enables you to provide additional information about each condition you specify.
Most sections in the form provide tool tips, accessible by this icon (![]() ), that provide hints specific to that area.
), that provide hints specific to that area.
Indication summary status
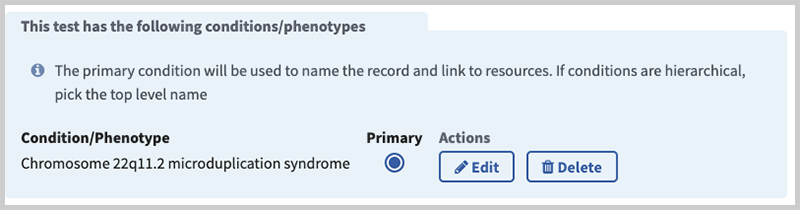
This view contains a table listing of all of Conditions/Phenotypes, a radio button for the condition set as primary, and links to delete or edit the entry.
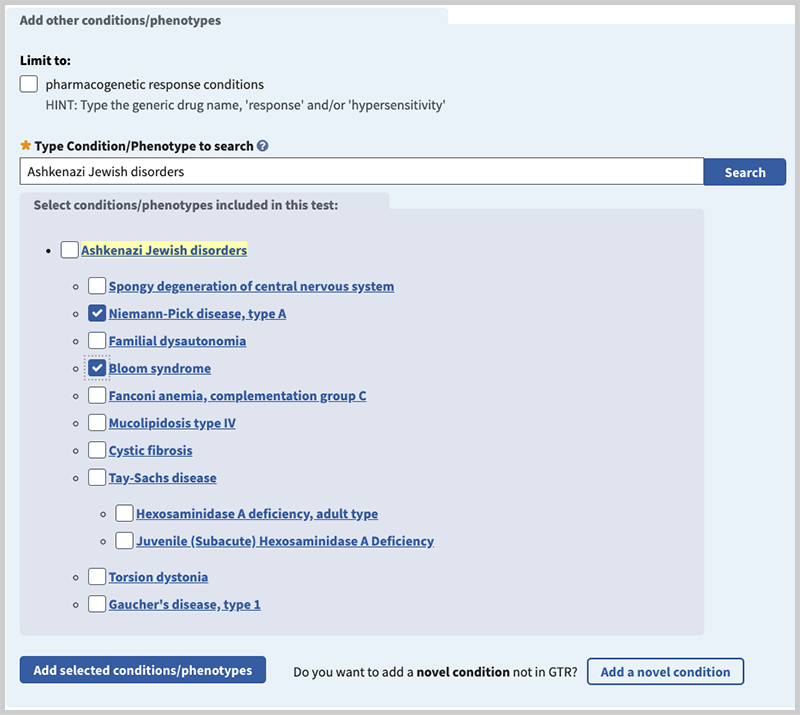
The "Type Condition/Phenotype to search" box has an autocomplete dictionary to help find the condition or phenotype name. Select from the autocomplete dictionary whenever possible. Then select the box next to the condition(s) you want to enter for your test. Select "Add selected conditions/phenotypes".
If the condition or phenotype you want to enter is not on the list, use the "Add a novel condition" button.
Once you select a condition or phenotype from the autocomplete dictionary, you may be presented with a hierarchy of terms you can use to select multiple related conditions to add to your test.
Note: If the test has more than one indication, you must designate one as primary. The primary indication enables NCBI to provide a summary of the condition/phenotype and to populate the discovery panel (right-hand column) next to your test with context-specific information (Reviews, Clinical Resources, etc.).
In the next tab (Methodology), each test indication associated with a target is clearly displayed in the GTR test page and captured into ClinVar. You are encouraged to submit a distinct assertion between the indication and test target.
Choose 'Continue' once all of the indications for your test have been added.
Condition/Phenotype Information
Condition/Phenotype name (minimal). Name of the disease, phenotype, drug response, etc. for which the test is offered. We strongly advise that you select from the list of available choices, if possible, to enable NCBI to provide information links for your test. First, use the autocomplete dictionary to select a suitable term. If the autocomplete dictionary does not have the term you seek, select the link, 'Search GTR for this condition/phenotype' to access all of the terms currently available in GTR (this action launches a new browser). Then either precisely type the term in the submission browser or copy (using 'Ctrl C') and paste ('Ctrl V') the term into the field. If you cannot find the term you seek, you may add a new condition, but NCBI will not be able to provide associated data. NCBI will assign and index a new concept and assuming normal processing, your data will appear on the web within a few days. If you experience any difficulty finding a suitable term, complete the last field in this section 'Private comment about the condition/phenotype to GTR staff'. Note that you have the option of selecting term available in GTR but to display a different term in association with your test.
TIP: Learn how to use MedGen to select condition and phenotype terms.
Indication type (optional). Categories are autopopulated based on selection: blood group, disease, finding, named protein variant, or pharmacologic response.
Condition/Phenotype name to be used for display in the GTR test page, if different from above (optional). Provide a preferred name for the condition for which the test is offered (if desired) to display for your test. For example, if the condition name in the autocomplete dictionary is 'Hb SS disease' and you would like the condition to be titled 'Sickle cell disease', you can enter 'Sickle cell disease' here.
Suggest new synonyms (optional). Once a condition/phenotype has been chosen from the autocomplete dictionary in the 'Condition/phenotype name' field, then synonyms for that condition/phenotype will be automatically provided. Enter any additional synonyms for the condition/phenotype that are not shown, if applicable.
Acronym to be used for display in the GTR test page, if different from above (optional). Once a condition/phenotype has been chosen from the autocomplete dictionary in the 'Condition/phenotype name' field, then the preferred acronym for that condition/phenotype will be automatically provided. Provide a preferred acronym for the condition/phenotype for which the test is offered (if desired) to display for your test.
Suggest new acronyms (optional). Once a condition/phenotype has been chosen from the autocomplete dictionary in the 'Condition/phenotype name' field, then acronyms for that condition/phenotype will be automatically provided. Enter any additional acronyms for the condition/phenotype that are not shown, if applicable.
Mode of inheritance (optional). Select the mode of inheritance used in your analyses for the condition/phenotype.
Disease mechanism (optional). Select the disease mechanism for the condition/phenotype in relation to the test target (measurement). This is the identification of the reasoning or biological process that causes the disease. Options include: loss of function, gain of function, other: please specify.
Prevalence (optional). Enter the prevalence, or the most current estimated number of cases of the disease in the population.
URL for prevalence (optional). If 'prevalence' is provided, either an URL or a citation is recommended to support the prevalence claim. URLs should be entered with the appropriate prefix, i.e., http:// or https://.
Citations for prevalence (optional). If 'prevalence' is provided, either an URL or a citation is recommended to support the prevalence claim.
Private comment about the condition/phenotype to GTR staff (optional). See 'Condition/phenotype name' above. Enter information for GTR staff regarding the addition of a new condtion/phenotype name if you were not able to find the desired name using the autocomplete dictionary.
Save your work
Don't forget to save your work by clicking the Save & Continue button at the bottom of the page! If you need to interrupt or delay data entry, you can return at a later time and you will be taken to the last tab you saved.
If any required fields or data inconsistencies are detected on this page, an error message will display after you click Save & Continue indicating what the issue is. The field(s) that need attention will be outlined in red. Please correct your data and click "Save & Continue" again.
How to submit information in the Methodology section of the submission form
Strategy
To make your test more discoverable, we strongly recommend being as specific as possible in specifying what your test measures (Test targets - genes, chromosomes, proteins and analytes). For example, rather than providing free text about all the alleles or all the analytes being tested, submit each separately in the test targets section. Then use the Test comment section to describe how the combination of the specific targets is evaluated. GTR users will then be able to find your test by searching on specific genes, variants, analytes, etc. and NCBI resources such as ClinVar and MedGen will make more effective linkages to your submitted information.
Organization of this page
This page is organized according to the sections you see on the Methodology tab. You may click on the name of the section below to navigate to that section of this document.
Please note that most sections in the form provide tool tips, accessible by this icon (![]() ), that provide hints specific to that area.
), that provide hints specific to that area.
Test Method(s)
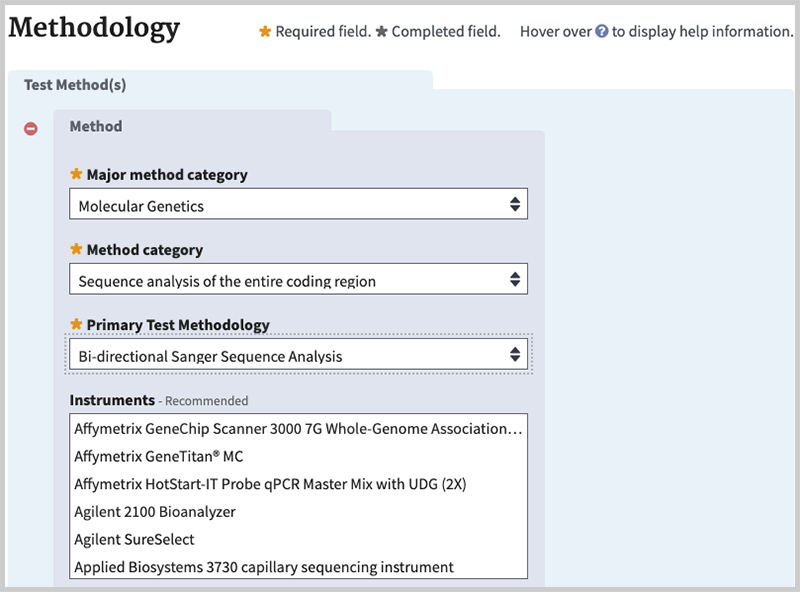
This section organizes specific fields into blocks to capture the hierarchies among methodologies and to enable the description of complex tests. Major method category, Method category, Primary Test Methodology and Instruments are grouped and numbered. You may 'add another method' if more than one group is desired or you may remove a section. Information in the Test Methods section will display in the Methodology tab of the test record.
Major method category (minimal). Choose a higher level method category from the list: Biochemical, Cytogenetic, or Molecular.
Method category (minimal). Select the general category to which the method belongs.
Choices within Biochemical:
Analyte
Enzyme assay
Immunohistochemistry
Protein analysis
Protein expression
Choices within Cytogenetics:
FISH-metaphase
FISH-interphase
Chromosome breakage studies
Sister chromatid exchange
Chromosome Rearrangement
Multicolor FISH (M-FISH)/Spectral Karyotyping™ (SKY™)
Choices within Molecular:
Deletion/duplication analysis
Detection of homozygosity
Linkage analysis
Methylation analysis
Microsatellite instability testing (MSI)
Mutation scanning of select exons
Mutation scanning of the entire coding region
Sequence analysis of select exons
Sequence analysis of the entire coding region
Targeted mutation analysis
Uniparental disomy study (UPD)
Primary Test Methodology (minimal). Provide name of the test method used in the assay e.g., RT-PCR with gel analysis, Metabolite levels, or specify a method not listed.
Instruments (recommended). Name the instrument used for a specific methodology e.g., Roche LightCycler 480, IlluminaHiSeq™2000 system, or specify an instrument not listed. Field is optional for research tests.
Platforms (optional). Select the name of a platform containing pre-defined test targets to provide methodology details and help GTR users find your test. If you use a platform not on the list, choose ‘Other’ and then provide the name. A platform name typically corresponds to a unique catalogue number or a set of catalogue numbers which are bundled with other consumables. The format is usually manufacturer name, product name, and general specifications. Field is optional for research tests.
Test procedure or protocol (optional). Summarize the methodology and describe the specific steps for each method of the assay.
Citations for test procedure or protocol (optional). Provide citations or URL for the protocol.
Confirmation of test results (recommended). Describe whether and how test results are confirmed (beyond initial validation of the assay). For example, 'Positive results are confirmed on a new DNA preparation using repeat sequence analysis'.
NOTE about research tests: Field is optional. State whether or not participants have an opportunity to learn about the results of the research test. If so, describe how participants may find out the test results, whether confirmation may be done in a clinical (e.g. CLIA-certified) lab, and whether or not there may be associated costs.
Test comment(s) (optional). Provide textual description of the test methods or any other information about your test not described elsewhere that may be useful to clinicians ordering your test, e.g. 'Bi-directional sequencing of exons 1-5 with concurrent analysis of NP_000000.0:p.Glu234Gly'.
NOTE about research tests: Use the Test comment(s) field to describe whether or not there a cost to participate in the study and if so, provide details.
NOTE we strongly encourage you to describe the test target(s) your test interrogates in the "Test Targets" section. For the example above, you would follow these steps:
- select 'gene' from the drop down menu for 'target is identified by',
- start typing the gene symbol and select the correct gene from the autocomplete dictionary,
- click the radio button for 'Associated reference sequences and exons',
- start typing the RefSeq accession and select the correct Refseq from the autocomplete dictionary
- enter the exons
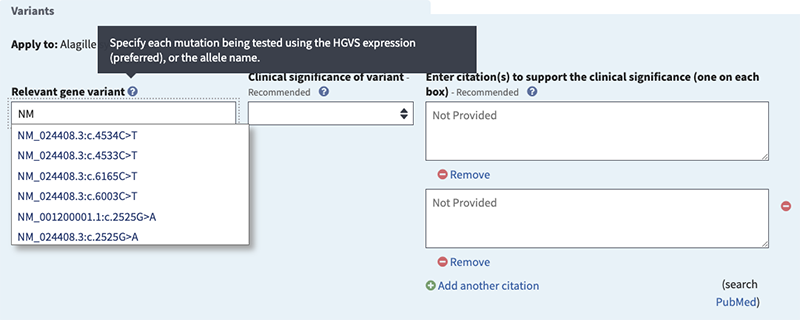
- click the radio button for Variants
- start typing the HGVS expression for a tested variation, beginning with the RefSeq accession (ex. NM or NP),
- select one of the suggestions from the autocomplete dictionary or enter NM_000208.2:c.3079C>T
- click on 'Save target' to save your data
By being specific, rather than writing free text, you make it possible for us to link your test to other resources at NCBI, such as variaton-related reports and ClinVar. This will ensure your test is more readily discoverable by all users of NCBI databases.
Test Targets
Section to describe the analytes, chromosomal or mitochondrial regions, genes and variations, or proteins that are measured in the test. Use the 'Target is associated with' section to match each target with at least one condition. The test must have at least one explicit condition-target relationship.
Note the warning about the need to add at least one test target for every test, and to have at least one explicit condition-target relationship. Click ‘Add Test Target’ to open that section of the Methodology page.
Use the Test target section to enter targets (genes must be entered one at a time). Decide which specific target you are going to enter. For each target, you can provide:
- the type (germline, somatic or both) in ‘Target is’ section
- the associated condition(s) that this target evaluates in ‘Target is associated with’ section
- category (analyte, chromosomal region/mitochondrion, gene, protein) in ‘Target is identified by’ section
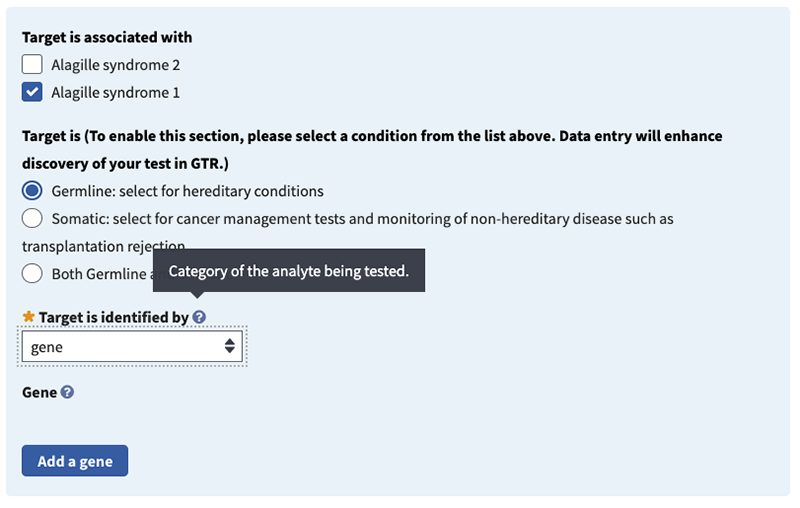
As an example, below are the steps to enter JAG1 for Alagille syndrome 1 and NOTCH2 for Alagille syndrome 2.
Target is (recommended). Select a radio button to indicate the type of target: germline, somatic, or both. Please note that germline is selected by default.
Target is associated with (recommended). Check one or more boxes for the indication(s) that are evaluated by the test target. In this case, Alagille syndrome 1 is associated with the JAG1 gene and Alagille syndrome 2 is associated with the NOTCH2 gene. For this example, check the Alagille syndrome 1 box.
Target is identified by (minimal). Choose 'analyte', 'chromosomal region/mitochondrion', 'gene' or 'protein'. Then specify target(s) tested in field that appears below it (which is dependent on the selection made). In this example, for ‘Target is identified by’ choose ‘gene’.
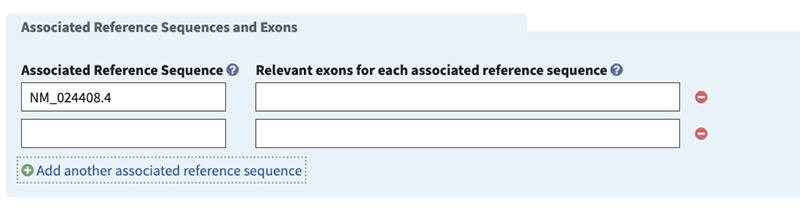
Gene: please specify (minimal if 'gene' is selected in 'Target is identified by' field). Begin typing in the box to activate autocomplete dictionary and select the desired term. Then select radio button for 'Associated Reference Sequences and Exons', 'Variants', or 'None'. The first two radio buttons activate fields to provide additional details. In this example, specify the gene in the pop-up window by typing JAG1 and selecting your choice.
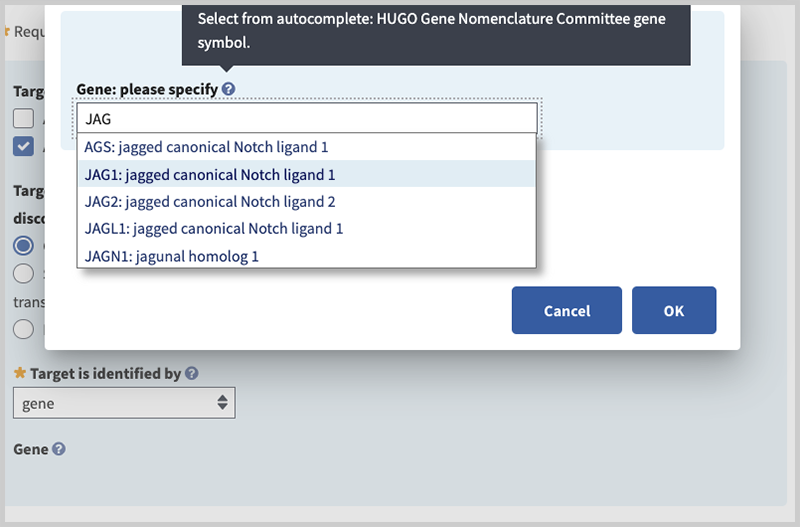
Associated Reference Sequence (minimal if 'Associated Reference Sequence and Exons' selected, otherwise optional). Activate autocomplete dictionary by typing NG or NM or NC to specify the accession and version (e.g. NM_0000001.1) of the sequence used to define the test or test region. Note, NC is applicable only to genes on the mitochondrion.
NOTE: If gene name is provided, the chromosomal location will be automatically provided and does not need to be entered.
Relevant exons for each associated reference sequence (minimal if 'Associated Reference Sequence and Exons' selected, otherwise optional). We encourage the following conventions for reporting the exons being tested:
| What is tested | How to report |
|---|---|
| If a continuous range is tested |
Provide the exon numbers as a range, e.g. 2-5. If the range includes all exons, add (All), e.g. 1-5 (All) |
| If a subset is tested | Enumerate the exons tested. Ranges are allowed, e.g. 3,5,9-12 |
Clinical significance of variant (recommended if 'Variants' selected). Options are:
association
confers sensitivity
drug response
no known pathogenicity
not provided
other
pathogenic
probably not pathogenic
probably pathogenic
risk factor
variant of unknown significance
Citations to support the clinical significance (recommended). Provide citations or URL.
Then ‘Save target’. Your have now completed a target entry (the JAG1 gene) for Alagille syndrome 1.
Complete your work by clicking ‘Add Test Target’ again in order to specify the NOTCH2 gene for Alagille syndrome 2. Specify whether the target is germline, somatic or both. In this example, uncheck Alagille syndrome 1 and check Alagille syndrome 2 to prepare for entering the gene NOTCH2. For ‘Target is identified by’ choose ‘gene’, then specify in the pop-up window by typing NOTCH2 and selecting your choice. Save Target.
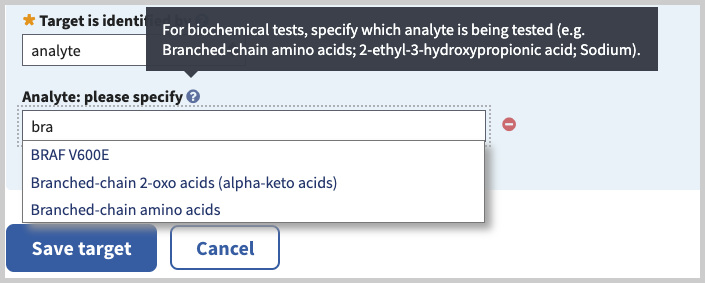
Analyte: please specify (minimal if 'analyte' is selected in 'Target is identified by' field). Begin typing in box to activate autocomplete dictionary, then select desired term. For example, type 'br' and select the analyte 'Branched-chain amino acids' or the enzyme 'Branched-chain ketoacid dehydrogenase'. Use 'Add another analyte' link to add multiple analytes.
Chromosomal region/mitochondrion: please specify (optional; minimal if gene name or analyte not provided). Identify chromosome or chromosome location as precisely as possible. If individual chromosomes are evaluated, enter each one as a distinct target. Use the term 'Complete genome' for comprehensive chromosomal or genomic assessments such as karyotyping, cytogenomic arrays, whole genome sequencing or chromosome breakage analysis.
When identifying a chromosome, begin typing in box 'Chromosome...' to activate autocomplete dictionary and select the desired term from the list that displays. If the desired chromosome is not listed, enter 'Chromosome [number]' in the text box and click the Save button at the bottom of the page.
When identifying a chromosomal region or band, begin typing to activate autocomplete dictionary and select the desired term from the list that displays. For example, enter chromosomal region '11p15' or range '15q11-q13'. If the desired chromosomal region is not listed, enter it in the text box. Click the Save button at the bottom of the page.
GTR uses the same formatting conventions for chromosomal band as HGNC, OMIM and NCBI's Gene. When identifying a cytogenetic band, please represent it in the p-q order and do not repeat the chromosome number. Examples: 22q11.3-q11.4 or 5p15.33-p.15.2.
When your test target is the complete mitochondrial genome, begin typing in box 'mitochondrion'. If the condition is known to result from variants in the mitochondrion, then the choice 'mitochondrion' will appear in the list that displays. (If the condition is not stored in GTR's database as being mitochondrial in origin, 'mitochondrion' will not display so you should enter 'mitochondrion' explicitly.)
NOTE: If you are testing specific mitochondrial genes, please select 'gene' in the Target is identified by section and select the genes by entering the beginning part of the gene symbol, namely MT-.
If gene name is provided, the chromosomal location will be automatically provided and does not need to be entered.
Protein: please specify (minimal if 'protein' is selected in 'Target is identified by' field). Begin typing in box to activate autocomplete dictionary and select the desired term. Add another protein as needed.
Save your work
Don't forget to save your work by clicking the Save & Continue button at the bottom of the page! If you need to interrupt or delay data entry, you can return at a later time and you will be taken to the last tab you saved.
If any required fields or data inconsistencies are detected on this page, an error message will display after you click Save & Continue indicating what the issue is. The field(s) that need attention will be outlined in red. Please correct your data and click "Save & Continue" again.
How to submit clinical test information in the Interpretation section of the submission form
This page is organized according to the sections you see on the Interpretation tab. You may click on the name of the section below to navigate to that section of this document.
Please note that most sections in the form provide tool tips, accessible by this icon (![]() ), that provide hints specific to that area.
), that provide hints specific to that area.
Upload Sample Reports
Sample negative, Sample positive, and Sample VUS reports will display in the Interpretation tab of the test record. GTR staff will review your report for actual or potential identifiers before it becomes public so there may be a delay from the time you upload your report and when you can see it in the public site.
Do not include date of birth, family numbers, sample numbers, medical record numbers, patient name, etc. Format for birth date such as XX/XX/2005 is acceptable but 9/9/1999 will not be accepted as this potentially a real birth date. Most fabricated names are potentially real as well, so please do not include these in your sample reports.
If your laboratory reports typically contain extensive information about variants of uncertain significance, please limit or eliminate these results from the sample report. Submission of more than a few dozen individual level genotypes (e.g., SNPs) can uniquely identify a person with high confidence.
Sample negative report (recommended). Upload a sample report with negative results. Please make sure that all identifiable information (both actual and potential) is absent from the report.
Sample positive report (recommended). Upload a sample report with positive results. Please make sure that all identifiable information (both actual and potential) is absent from the report.
Sample VUS report (optional). Upload a sample report with VUS results. Please make sure that all identifiable information (both actual and potential) is absent from the report.
Variants of Unknown Significance (VUS): Policy and Interpretation
This information will display in the Interpretation tab of the test record.
What is the protocol for interpreting a variation as a VUS? (recommended). Type, or copy and paste from a source you have available, a description of how your lab interprets a variation as a VUS.
What software is used to interpret novel variations? (optional). Type, or copy and paste from a source you have available, the software your lab uses to interpret novel variations. You can include lab-developed software.
What is the laboratory's policy on reporting novel variations? (recommended). Type, or copy and paste from a source you have available, a description of how your lab reports novel variations.
Are family members with defined clinical status recruited to assess significance of VUS without charge? (recommended). Select whether your lab offers testing for a variant of unknown significance to family members free of charge; for example, to affected individuals and/or presumed obligate carriers to help determine clinical significance of a VUS.
Comments about recruiting family members to assess significance of VUS without charge? (recommended). Write an explanation or give information about who would be eligible to testing free of charge and how the lab does it. For example, is the testing offered to affected individuals and/or presumed obligate carriers to help determine clinical significance of a VUS.
Will the lab re-contact the ordering physician if variant interpretation changes? (recommended). Select whether the lab contacts the ordering physician after testing and initial report if a variant interpretation changes.
Comments about the laboratory procedure to re-contact the ordering physician (recommended). Type a description or explanation about how the lab monitors and addresses reinterpretation of genetic tests results over time, after issuing the report. For example, is there a time limit, do you systematically re-evaluate prior interpretations and generate new reports, or does the person ordering the test need to periodically recontact the laboratory to inquire about changes in test interpretation?
Research performed after clinical testing is complete (recommended). Type an explanation or information about any research testing being performed using the submitted sample. For example, research testing is allowed under IRB approved protocols or the patient would have to sign an informed consent for the research protocol. If the protocol is registered in clinicaltials.gov, you can enter that information here. Please note that Quality Control is not considered as research for this question.
Save your work
Don't forget to save your work by clicking the Save & Continue button at the bottom of the page! If you need to interrupt or delay data entry, you can return at a later time and you will be taken to the last tab you saved.
If any required fields or data inconsistencies are detected on this page, an error message will display after you click Save & Continue indicating what the issue is. The field(s) that need attention will be outlined in red. Please correct your data and click "Save & Continue" again.
How to submit information in the Performance characteristics section of the submission form
This page is organized according to the sections you see on the Performance characteristics tab. You may click on the name of the section below to navigate to that section of this document.
Please note that most sections in the form provide tool tips, accessible by this icon (![]() ), that provide hints specific to that area.
), that provide hints specific to that area.
Availability
Information in the Availability section will display in the Performance Characteristics tab of the test record.
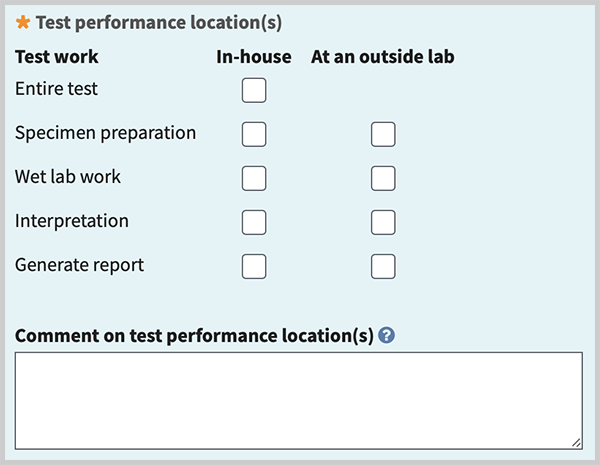
Test performance location(s) (minimal). Describe where test work is performed. You can register tests for which your laboratory performs at least part of the test: specimen preparation, wet lab work, interpretation, and report generation. 'In-house' means within the lab/facility covered by the same CLIA certification number. 'Outside lab' means a different lab/facility than that covered by your CLIA certification, even if both labs have the same parent organization. Tests which are performed entirely at an outside lab/facility (send-out tests) should not be registered in GTR. Send-out tests are expected to be registered by the lab performing them.
Check box(es) to indicate the locations where components of the test work are performed. If 'In-house' is selected for entire test, no other check marks are needed. For other test work choices, you may check one or both for 'In-house' and 'At an outside lab'. If 'In-house' is not selected for 'Entire test', then check marks are needed for each of the remaining components and at least one type of test work must be performed in-house.
Comment on test performance location(s) (minimal if any test work is performed at an outside lab). Briefly describe methods and location for components of test work done at an outside lab.
Analytical validity
How accurately and reliably the test measures the component of interest. This information will display in the Overview and Indication tabs of the test record.
Analytical validity (minimal). Describe the accuracy and reliability of the test for measuring the component of interest. Provide quantitative information as typically available from a validation study required by CAP. Scope includes analytical sensitivity (analytical detection rate) and the number of specimens used to calculate it; analytical specificity (analytical false positive rate); accuracy (how closely the results match those from independent sources known to be the true results); and precision (how closely repeated results match each other).
General statements about methodologies that do not contain specific quantitative information about the test do not satisfy the specifications for this field. Nor do statements such as ‘N/A’, ‘not applicable’, ‘Yes’ or ‘unknown’. Example of an accepted entry: 'The OtoChip is greater than 99% sensitive for detecting substitution variants in the sequence analyzed. In addition, this assay is 95% sensitive for detecting small insertions or deletions (InDels) (100% for 1-2 bp indels, 92.3% for 3-5 bp indels and 87.5% for >10 bp indels)'.
Citations to support analytical validity (recommended). Provide citation(s) to support analytical validity statement. Use search PubMed link to find citation details. Add multiple citations as desired. If no publications are available, provide non-proprietary internal lab data in analytical validity statement, if available.
Assay limitation(s) (recommended). Describe any factors that affect the value of the test for its intended use. Includes limits of detection and validation of test for only specific subpopulations or particular uses.
Citations to support assay limitation (recommended). Provide citations to support assay limitation statement(s). Use search PubMed link to find citation details. Add multiple citations as desired.
Quality Assurance
Describe methods for quality management and quality control. Applicable to preanalytic, analytic, and post-analytic (reporting) phases. Information in the Quality Assurance section will display in the Performance Characteristics tab of the test record.
Is proficiency testing performed for this test? (recommended). Select Yes or No to indicate whether the test is subject to periodic internal or external evaluation of the accuracy of test results.
Proficiency testing method (recommended; minimal if Yes is selected for proficiency testing). Specify which type of proficiency testing is performed. Choices include Alternative Assessment, Formal Proficiency Testing (PT) program, Intra-Laboratory, Inter-Laboratory, and Other (specify). Proficiency testing is a determination of laboratory testing performance by means of interlaboratory comparisons whereby a group of laboratories receive multiple specimens for analysis and/or identification and the program compares the results among laboratories and/or with an assigned value. Alternative assessment is the determination of laboratory testing performance by means other than PT such as split-sample testing or testing by a different method.
Provider for proficiency testing (recommended). Specify the agency or society that administers the proficiency testing program for the test. If Other than listed, please specify. To see the complete, current list of choices for this field, click here.
Major CAP category (recommended). If College of American Pathologists (CAP) is chosen as Provider for proficiency testing, select Major CAP category from list. Add multiple as needed by selecting 'add another CAP test'. If category is not available, specify under 'Category for my test is not listed here'.
CAP category (recommended). If Major CAP category is selected, specify relevant sub-category. Add multiple as needed by selecting 'add another CAP test'.
Description of proficiency testing method (recommended). Explain proficiency testing (PT) method and provide PT scores and/or results, the PT reportable range, the PT interval and the number of specimens tested.
Citations to support the above statement (recommended). Provide citations to support proficiency testing method statement. Use search PubMed link to find citation details. Add multiple citations as desired.
Description of internal test validation method (recommended). Explain how the laboratory validates the test.
Citations to support the above statement (recommended). Provide citations to support internal test validation method statement. Use search PubMed link to find citation details. Add multiple citations as desired.
Clinical Validity
How consistently and accurately the test detects or predicts the intermediate or final outcomes of interest. This information will display in the Overview and Indication tabs of the test record.
Statement of clinical validity (recommended). Describe clinical sensitivity and specificity, positive and negative predictive values, the population(s) assessed, and the number of specimens used to calculate clinical validity. Clinical sensitivity is the proportion of positive test results among patients with the defined clinical presentation. Clinical specificity is the proportion of negative test results among patients without the defined clinical presentation. Positive predictive value is the chance of having the marker among those that test positive. Negative predictive value is the chance of not having the marker among those that test negative.
Citations to support the above statement (recommended). Provide citations to support clinical validity statement. Use search PubMed link to find citation details. Add multiple citations as desired.
Clinical Utility
How likely the test is to significantly improve patient outcomes. This information will display in the Overview and Indication tabs of the test record.
Category of clinical utility (recommended). Select a category from the list and support it with a URL and/or citations. You can enter multiple category sets of information by clicking the "add another clinical utility" link below the box.
Choose from the following options:
Avoidance of invasive testing
Establish or confirm diagnosis
Guidance for management
Guidance for selecting a drug therapy and/or dose
Lifestyle planning
Predictive risk information for patient and/or family members
Reproductive decision-making
Other (specify)
Sufficient research has not been conducted to establish utility of the test
To see the complete, current list of choices for this field, click here.
URL to explain the clinical utility (minimal for each category of clinical utility selected; at least one URL or citation). Cite recommendations or practice guidelines for the test that have been issued by authoritative groups. Some practice guidelines may be available via a URL but not a citation. URL to the laboratory web page with information on the clinical utility of the test may be used.
Citations to support the clinical utility (minimal for each category of clinical utility selected; at least one URL or citation). Cite recommendations or practice guidelines for the test that have been issued by authoritative groups (e.g., U.S. Preventive Services Task Force, professional societies such as the American College of Medical Genetics and Genomics). Use search PubMed link to find citation details. Add multiple citations as desired.
Save your work
Don't forget to save your work by clicking the Save & Continue button at the bottom of the page! If you need to interrupt or delay data entry, you can return at a later time and you will be taken to the last tab you saved.
If any required fields or data inconsistencies are detected on this page, an error message will display after you click Save & Continue indicating what the issue is. The field(s) that need attention will be outlined in red. Please correct your data and click "Save & Continue" again.
How to review information in the Review & Submit section of the submission form
When you reach the Review & Submit tab of your test submission, your data are saved but not yet submitted to the GTR database. Here you can review the data content of your submission. To edit your test information, you must click on the tab name(s) containing the relevant field(s). For example, to edit clinical validity field, you would click on the Performance characteristics tab, scroll down the page to the field, make the changes and click Save and continue at the bottom of the page. Once you are satisfied with your submission, click on Submit at the top or the bottom of the page. Clicking the Submit sends your test record to GTR processing. If you elected to receive notification of updates to submission status, you will receive an email in your inbox entitled Acknowledgement of your submission to GTR with information about your submission: whether your submission was processed or if problems were encountered. After you click the Submit button, you are returned to your GTR submissions homepage.
If you need to delay submission, you can return to your test record at a later time and then submit it. If you need to return to your GTR submission homepage you can click the link "Home" in the GTR Submission header. Your test is not yet submitted to the GTR database! However, your data is saved so you can come back at a later time to submit it.